SUMMARY
We present the draft 273 Mb genome of the migratory monarch butterfly (Danaus plexippus) and a set of 16, 866 protein-coding genes. Orthology properties suggest that the Lepidoptera are the fastest evolving insect order yet examined. Compared to the silkmoth Bombyx mori, the monarch genome shares prominent similarity in orthology content, microsynteny, and protein family sizes. The monarch genome reveals: a vertebrate-like opsin whose existence in insects is widespread; a full repertoire of molecular components for the monarch circadian clockwork; all members of the juvenile hormone biosynthetic pathway whose regulation shows unexpected sexual dimorphism; additional molecular signatures of oriented flight behavior; microRNAs that are differentially expressed between summer and migratory butterflies; monarch-specific expansions of chemoreceptors potentially important for long-distance migration; and a variant of the sodium/potassium pump that underlies a valuable chemical defense mechanism. The monarch genome enhances our ability to better understand the genetic and molecular basis of long-distance migration.
INTRODUCTION
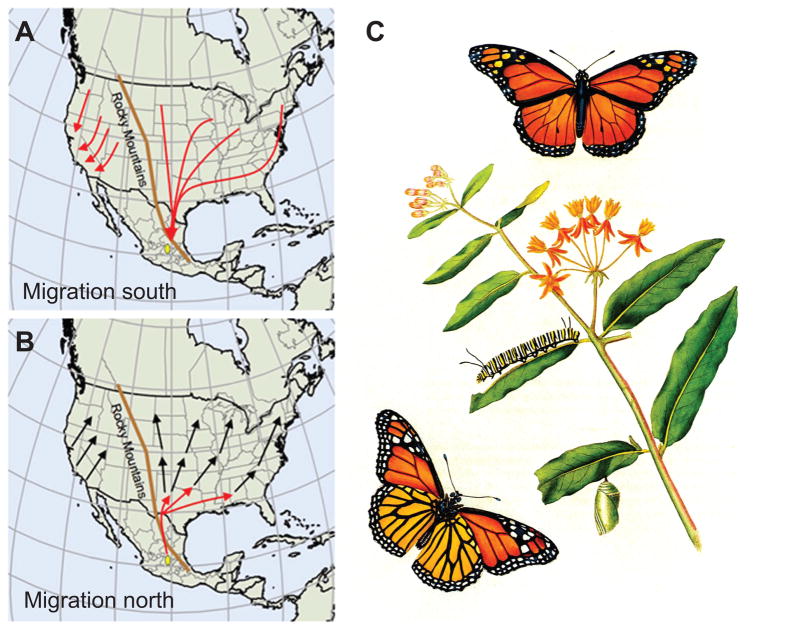
Each fall, millions of eastern North American monarch butterflies undergo a vertebrate-like, long-distance migration, traveling up to 4000 km to reach their overwintering grounds in central Mexico (Brower, 1995; Reppert et al., 2010; Urquhart and Urquhart, 1978) (Fig. 1A). Migratory monarchs are in reproductive diapause. Migrants also have a striking increase in longevity, increased abdominal fat stores and cold tolerance, and an overpowering drive to fly south. Diapause persists at the overwintering sites until spring when the migrants reproduce and then fly northward to oviposit fertile eggs on newly emerged milkweed plants in the southern United States (Fig. 1B). Monarchs are milkweed specialists (Fig. 1C), and their evolved chemical defense mechanism has led to the monarch’s widely known involvement in a mimicry complex with the viceroy butterfly (Limenitis archippus) (Ritland and Brower, 1991).
Figure 1. Natural history of the monarch butterfly.
A. Migration south. The eastern North American monarch butterfly undergoes a long-distance fall migration to a restricted site in central Mexico (yellow oval). The population of monarchs west of the Rocky Mountains undergo a truncated fall migration. Red arrows, flight paths. From Reppert et al., 2010.
B. Journey north. Eastern migrants remain at the overwintering areas until spring, when the same butterflies reproduce and migrate northward to lay fertilized eggs on newly emerged milkweed in the southern United States (red arrows). Successive generations of spring and summer monarchs re-populate the home range (black arrows). From Reppert et al., 2010.
C. Life cycle. Complete metamorphosis from egg to larva (5 instars) to pupa (chrysalis) to adult. The male butterfly (upper right) has visible black spots on their hind wings that are missing in females (lower left, underwing view). The larvae feed on milkweed (plants of the genus Asclepias). Photograph of lithograph, John Abbot, 1797.
A major compass system monarchs use for directional information for the migration is a time-compensated sun compass (Froy et al., 2003; Mouritsen and Frost, 2002; Perez et al., 1997). Sun compass components involve the eye’s sensing of skylight cues for direction and the brain integration of skylight-stimulated neural responses in the central complex, the presumed site of the sun compass (Heinze and Reppert, 2011). Sun compass output in brain is time compensated by the circadian clock that allows flight direction to be constantly adjusted to maintain a southerly flight direction.
The other lepidopteran genome that is publically available for comparison with the monarch is that of the commercial silkmoth Bombyx mori (Consortium, 2008). Because moths are usually olfactory-centric and butterflies vision-centric, due in part to their respective nocturnal and diurnal behaviors, comparison of the genes involved in these sensory modalities may be informative.
Here we present the draft 273 Mb genome of the migratory monarch butterfly, including its assembly, a set of 16, 866 protein-coding genes, and evolutionary analyses. We focus our gene annotation on gene families likely involved in major aspects of the seasonal migration. The biological interpretation of the monarch genome advances our understanding of the genes and regulatory elements important for the remarkable fall migration.
RESULTS AND DISCUSSION
Genome Assembly and Gene Content
We used a whole-genome shotgun approach with next-generation sequencing platforms to generate the draft genome of the monarch butterfly (Table 1 and supplemental Table S1). The combined assembly of 14.7 Gb pairs of Illumina reads (equal to 53.3X coverage of the whole genome) and 6.2 Gb Roche 454 reads (22.3X) resulted in 273 megabases (Mb) of genomic sequence (combined total coverage of 74.7X) (Table S1A). This was termed the v1 assembly and was used for all subsequent analyses (Table S1B). Assessment of the completeness and quality of the assembly v1 is described in the Experimental Methods (Table 1).
Table 1.
Features of assembled genome and gene set
| Monarch (v1) | Bombyx | |
|---|---|---|
| Genome size (Mb) | 273 | 432 |
| Number of Chromosomes | 29–30+ | 28 |
| Quality control (covered by assembly) | ||
| % ESTs | 96 | 98† |
| % CEGMA genes | 98.5 | 98.7 |
| % CRP genes | 100 | 100 |
| Genomic features | ||
| % Allele frequency | 0.55 | ND† |
| % Repeat | 13.1 | 43.6 |
| % G+C | 31.6 | 37.7 |
| CpG [O/E] | 1.13 | 1.14 |
| % Coding | 7.51 | 4.14 |
| % Intron | 22.8 | 16.3 |
| Number of miRNA | 116* | ND† |
| Number of tRNA | 431 | 441† |
| Gene repertoire | ||
| Number of protein-coding genes | 16,866 | 14,623 |
| With InterPro domains | 10,999 | 9,892 |
| With GO terms | 11,210 | 10,148 |
| Universal orthologues lost | 50 | 194 |
| Species-specific genes | 2,511 | 1,598 |
From Brown et al., 2004.
Only adult miRNAs were identified.
Defined independently or not determined in the latest version of Bombyx genome (Consortium. 2008). See also Table S1A, S1B, S1C, S1D, S1E, S1F, and S1G for details. CEGMA genes, core eukaryotic genes (Parra et al., 2007); CRP genes, cytoplasmic ribosomal protein genes; CpG[O/E], ratio of observed-to-expected CpG; GO, gene ontology.
See also Figure S1 and Table S1A-J.
In comparison with the 432 Mb Bombyx genome (Consortium, 2008), the monarch genome had much less repeat content (13.1% of the whole-genome versus 43.6% in Bombyx, Table S1E) and GC content (31.6 % in the monarch versus 37.7 % in Bombyx, Table S1F) (Table 1). Like Bombyx and Tribolium, but unlike Apis mellifera, GC content distribution was uniform but showed a bias of occurrence in coding regions (Supplemental Fig. S1A & B). In addition, the distribution plots of CpG ratios (observed/expected CpG dinucleotide density) of the monarch, Bombyx, and Tribolium genomes clustered together, leaving Drosophila and A. mellifera with two different patterns (Fig. S1C & D). These features are consistent with the fact that the monarch, Bombyx and Tribolium each encode two types of DNA methyltransferases (Dnmt1 and Dnmt2), while Drosophila only has Dnmt2 and A. mellifera has Dnmt1-3 (Fig. S1D). The monarch may thus have a Bombyx-like epigenetic system, with a predicted low methylation level (Xiang et al., 2010).
We estimated 16,866 protein-coding genes (Table 1) by combining both homology-based and ab initio methods (Table S1G), along with ~228X coverage of the monarch transcriptome. The gene model accounted for the full complement of conserved cytoplasmic ribosomal proteins genes (Marygold et al., 2007), with only one pseudogene and two incomplete predictions (Table S1H), and matched 89.1% of 5,415 manually annotated exons (Table S1G). Nearly 85% of the predicted genes detected homology in the public databases (Table S1I). Moreover, more than 93% of the monarch genes were supported by our transcriptome sequence. These attributes show that the gene models were predicted with accuracy and completeness (Table 1).
Lepidopteran Orthology and Evolution
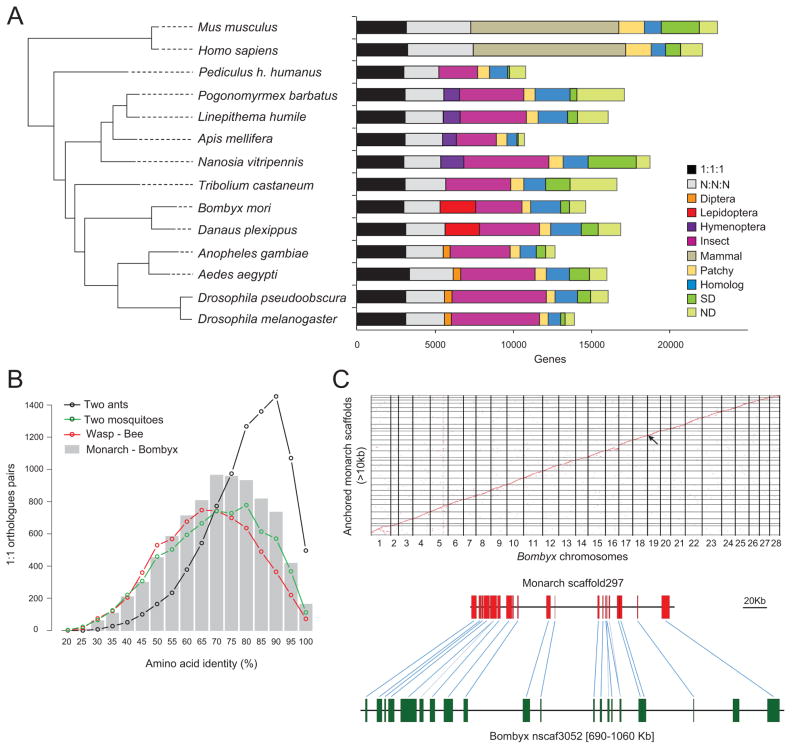
To understand the lepidopteran proteomes of the monarch and Bombyx in the context of other insect species, we compared the reported gene sets of twelve insects and two mammalian species (Fig. 2A). Orthology was then assigned according to the predicted evolutionary relationships. The monarch gene set contained 3,138 (18.6%) single-copy genes and 2,514 (14.9%) many-to-many universal genes, compared to 20.4% and 15.9%, respectively, for Bombyx. We found significant coverage bias of the transcriptome sequence for the monarch universal orthologs (Table S1J), indicating that they constitute a core set of proteins with conserved functions. Transcriptome coverage also showed a higher distribution of many-to-many universal orthologs than single-copy genes, indicating that the universal orthologs with duplication contributed greatly to basic biological processes compared to the contribution from duplication of recently evolved insect genes.
Figure 2. Lepidopteran orthology and evolution.
A. Orthology assignment of 12 insect and two mammal genomes. Bars are subdivided to represent different types of orthology relationships: 1:1:1 indicates universal single-copy genes, but absence and/or duplication in a single genome is tolerated; N:N:N indicates other universal genes, but absence in a single genome or two genomes within the different orders is tolerated; Diptera indicates dipteran-specific genes, presence in at least one mosquito and one fly genome; Lepidoptera indicates lepidopteran-specific genes, presence in both the monarch and Bombyx genomes; Hymenoptera indicates hymenopteran-specific genes, presence in at least one bee or wasp genome, and one ant genome; Insect indicates all other insect-specific orthologs; Mammal indicated mammalian-specific orthologs; Patchy indicates orthologs that are present in at least one insect and one mammal genome; Homolog indicates partial homology detected with E<10−5 but no orthology grouped; SD, species-specific duplicated genes; ND, species-specific genes. The phylogeny on the left was calculated using maximum likelihood analyses of a concatenated alignment of 1,642 single-copy proteins from the 1:1:1 subgroup. The tree was rooted using mammals as outgroup. Bootstrap values based on 1,000 replicates are equal to 1,000 for each node.
B. The distribution of pairwise amino acid identity. Histogram shows the distribution of sequence identity of 8,221 1:1 orthologs between the monarch and Bombvx (diverged ~ 65 million years ago; (Grimaldi and Engel, 2005)). To highlight the similar level of molecular divergence, 8,897 orthologs between two ants (Linepithema humile and Pogonomyrmex barbatus, which diverged ~100–150 million years ago; (Moreau et al., 2006)), 6,875 orthologs between two mosquitoes (Anopheles gambiae and Aedes aegypti, which diverged ~150 million years ago; (Krzywinski et al., 2006)), and 6,520 orthologs between bee and wasp (Apis mellifera and Nasonia vitripennis, which diverged ~ 180 million years ago; (Werren et al., 2010)) were plotted in black, green, and red, respectively.
C. Microsynteny between monarch and Bombyx genomes. Alignment of monarch scaffolds and silkworm chromosomes is shown by pairwise dot plots based on gene homology. 1,802 monarch scaffolds (>10 kb) were anchored to the corresponding position based on the consensus order of gene homology. The arrow denotes the position of a scaffold between the monarch and Bombyx that showed particularly strong microsynteny, which is magnified below. See also Figure S2.
To address lepidopteran-specific evolution, we identified 1,962 lepidopteran-specific orthologs, which was about twice the number of hymenopteran-specific orthologs and five times the number of dipteran-specific orthologs. In addition, the lepidopteran lineage lacked 223 orthologs that exist widely in other insects and in mammals. In comparison, there were 167 and 103 orthologs missing in the Diptera and Hymenoptera, respectively, suggesting that the Lepidoptera are more derived than the other insect orders.
The Lepidoptera have rapidly evolved. The monarch and Bombyx shared 70.8% average amino acid identity between 8,221 1:1 orthologs (Fig. 2B), comparable to two mosquitoes (69.4% for 6,875 Anopheles gambiae/Aedes aegypti orthologs) or wasp-bee identity (67.2% for 6,520 A. mellifera/Nasonia vitripennis orthologs), but significantly lower than such comparison between two ants (82.2% for 8,897 orthologs between Linepithema humile and Pogonomyrmex barbatus). Because all these three genome pairs diverged at least 100 million years ago (Krzywinski et al., 2006; Moreau et al., 2006; Werren et al., 2010) and the monarch radiated from Bombyx ~65 million years ago (Grimaldi and Engel, 2005), the Lepidoptera appear to be the fastest evolving insect order sequenced to date.
The monarch and Bombyx genomes exhibited a surprisingly high degree of microsynteny (Fig. 2C). Approximately 80% of the monarch genes have identifiable Bombyx homologs, while less than 5% of the co-existing orthologs are duplicated in the monarch or Bombyx. According to the consensus gene order shared between the monarch and Bombyx, we successfully mapped 1,802 >10kb monarch scaffolds spanning 142.7Mb of the genome to the corresponding Bombyx scaffolds. We found strong co-linearity in most of the putative chromosomes except for the sex chromosome Z (Chr. 1 in Fig. 2C). A total of 8,290 monarch genes (75% of 11,017 genes located in mapped scaffolds with Bombyx homology) were found in microsynteny blocks, versus 75% for A. gambiae and A. aegypti (Zdobnov and Bork, 2008), and 63% for A. mellifera and N. vitripennis single-copy orthologs (Werren et al., 2010). Although we cannot exclude large-scale chromosomal rearrangements because of the lack of a monarch linkage map, the existent extensive microsynteny reveals that most regions of conserved gene neighborhood were retained after divergence.
Comparison of protein family sizes also showed prominent similarities between the monarch and Bombyx, from the global view of proteome domain content (Fig. S2). Only 17 InterPro (IPR)-defined families had significant size differences between the two Lepidoptera (Fig. S2A), most of which were related to proteinase activity (Fig. S2B). Interesting IPR expansions included insect pheromone-binding proteins in the monarch and insulin-like peptides in Bombyx (Fig. S2B). The major contribution to the lepidopteran phenotypic changes was also apparent by comparing the IPR family size of the Lepidoptera with Drosophila or Tribolium (Fig. S2C). Overall, most IPR families that exhibited variation have general functions involved in transcriptional regulation, protein interactions, and cell-cell communication.
We also compared the evolutionary rate between the monarch and Bombyx based on amino acid substitutions, using Drosophila as a common outgroup. The results showed that the monarch shares similar sequence identity with Bombyx in 1:1 orthologs (50.4% versus 50.7%). This analysis suggests that the ~ 5,000 year of human domestication of the silkworm (Xiang, 1995) has not had a strong influence on the overall evolutionary rate of non-selected traits in Bombyx.
Sensory Input to the Sun Compass
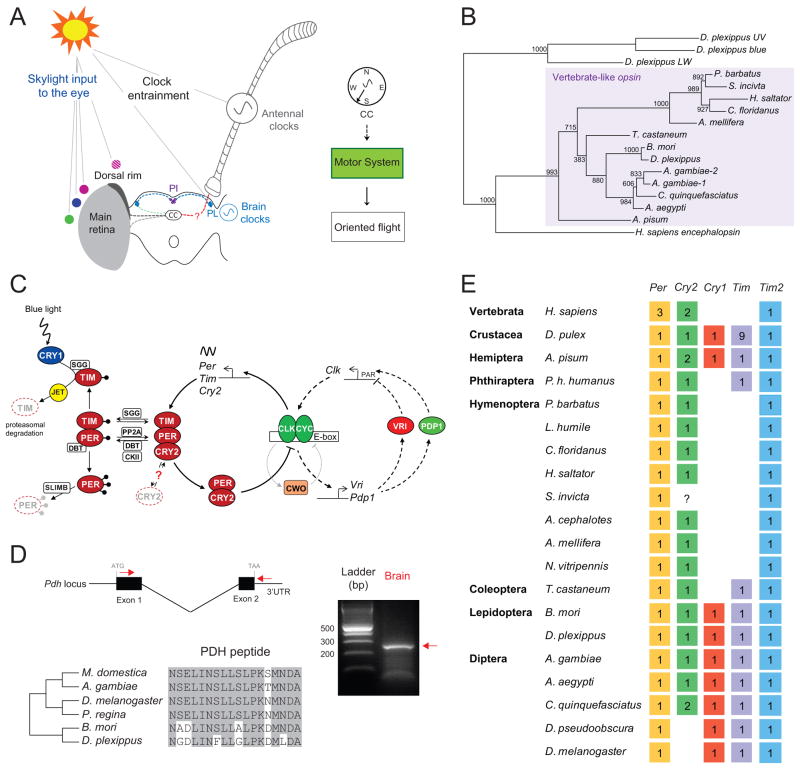
We began our manual annotation by focusing on genes involved in the formation and function of visual input into the sun compass system (Fig. 3A). In migrating monarchs, the horizontal position of the sun (solar azimuth) and the derived polarized skylight pattern provide directional cues for the sun compass (Heinze and Reppert, 2011). The solar azimuth is likely sensed by the main retina, while polarized light is sensed by the specialized dorsal rim area, a small region of the compound eye anatomically specialized for sensing the angle of plane-polarized skylight (Labhart and Meyer, 1999; Reppert et al., 2004) (Fig. 3A).
Figure 3. Sun compass components focusing on the circadian clock.
A. Model delineating the components used for sun-compass navigation. The compass mechanism involves the monarch eye sensing of skylight cues, including color gradient or the sun itself (violet, blue and green circles) and the polarization pattern of ultraviolet (UV) light (violet circle crosshatched), and the brain integration of skylight-cue stimulated neural response in the central complex (CC; grey dashed lines). In addition, time-compensation is provided by circadian clocks located in the antenna. The integrated time-compensated sun compass information is relayed to the motor system to induce oriented flight. The brain circadian clocks are located in the pars lateralis (PL) and communicate with the pars intercerebralis (PI). The PL may also communicate with the central complex. Modified from Reppert et al., 2010.
B. Maximum-likelihood phylogenetic tree of insect vertebrate-like opsins (pteropsins). The tree was rooted using the monarch UV, blue and long-wavelength opsins.
C. Schematic of the proposed clockwork mechanism in the monarch butterfly, including the core transcriptional/translational feedback loop (thick arrows) and the modulatory feedback loop (dashed arrows), both incorporating monarch orthologs of all described Drosophila clock genes (Dubruille and Emery, 2008). CLOCK (CLK) and CYCLE (CYC) heterodimers drive the transcription of period (per), timeless (tim), and type-2 cryptochrome (cry2), which upon translation form complexes and 24 hours later cycle back into the nucleus where CRY2 inhibits CLK:CYC-mediated transcription. Light entrainment is mediated by type-1 cryptochrome (CRY1), which promotes TIM degradation. Casein kinase II (CKII), doubletime (DBT) and the protein phosphatase 2A (PP2A) are involved in the posttranslational modifications of PER and TIM, and supernumerary limbs (SLIMB) and jetlag (JET) signal their degradation. The gene(s) involved in CRY2 degradation are unknown (red question mark). The modulatory feedback loop regulates the expression of clock through VRILLE (VRI) and PDP1. Monarch vri has five consensus CACGTG E-box elements in its promoter and pdp1 has five in its first intron, through which CLK and CYC could drive their transcription; each transcription factor also contains PAR DNA binding domains that could modulate CLK transcription by binding to PAR-like binding sites present in the monarch clk promoter. Clockworkorange (CWO) modulates the amplitude of the clock.
D. Pdh expression in monarch brain. Top, The primers used for RT-PCR amplification are shown (red arrows) on a schematic of the pdh locus identified from the genome assembly. (Right) Agarose gel showing the pdh amplicon migrating at ~250bp (red arrow). Bottom, Alignment of monarch PDH peptide sequence with those previously described in insects.
E. Insights into the evolution of the arthropod circadian clock. The presence/absence of the core clock components period (Per), type-2 cryptochrome (Cry2), type-1 cryptochrome (Cry1), timeless (Tim) and timeout (Tim2) has been assessed in all the published arthropod genomes, including 17 insect species. The presence of a given gene is represented by a colored box and the numbers represent the number of copies found in the genome. The question mark represents an absence that may be due to a degree of incompleteness in the genome given the presence of this gene in all others ant genomes. See also Figure S3, Table S2, Table S3, Table S4, Table S5 and Table S7.
In the monarch butterfly genome, we identified orthologs of most genes involved in eye development in Drosophila (Supplemental Table S2A) with some notable differences. For development of the main retina, only two genes were not detected in either the monarch or Bombyx. The lens crystalline protein Drosocrystallin, which is restricted to the Diptera, was predictably missing from the two Lepidoptera. The other missing gene was phyllopod that is involved in photoreceptor cell fate commitment. Of the 53 genes examined, seven are duplicated in Drosophila and none in either the monarch or Bombyx, supporting less genetic complexity in the Lepidoptera eye.
Many of the genes necessary for the formation of the dorsal rim area in Drosophila are present in the monarch butterfly genome, including two counterparts of homothorax (both also present in Bombyx), a transcription factor that is both necessary and sufficient for the formation of the fly dorsal rim (Table S2A) (Wernet et al., 2003). However, members of the spalt-related and the irC gene families that are involved in Drosophila dorsal rim formation were not found in either the monarch or Bombyx. The one duplication and two gene contractions in the two Lepidoptera genomes suggest a modified pattern of dorsal rim development from that in Drosophila.
In contrast to eye development, there were interesting differences in the genes involved in phototransduction between the monarch and Bombyx (Table S2B). The majority of the genes involved in phototransduction in Drosophila were present in the monarch butterfly genome (Table S2B). However, there were duplications of six genes in the monarch only. Most paralogs exhibited lower expression than their counterpart (Table S2B), suggesting that these duplications are relatively recent. Because these duplications were not found in either Bombyx or Drosophila, the monarch-specific expansions may be involved in the phototransduction mechanisms for sensing skylight cues.
In addition to the monarch butterfly genes encoding each of the three major opsin subfamilies (ultraviolet, blue, and long-wavelength) previously identified (Sauman et al. 2005), we found a monarch gene encoding a vertebrate-like opsin, called pteropsin in A. mellifera (Velarde et al., 2005) (Fig. 3B; Table S2B). Further examination of other insect genomes revealed its presence in Bombyx, mosquitoes, Tribolium and several Hymenoptera (but not all) beyond A. mellifera (Fig. 3B), suggesting that this putative light-detecting system is widespread in insects. Interestingly, a sea urchin ortholog was recently shown to play a role in photosensitive larval swimming vertical migration (Ooka et al., 2010).
Central Processing by the Sun Compass
The central neuronal processing of skylight cues in the monarch occurs in the central complex, the sun compass structure in central brain (Heinze and Reppert, 2011) (Fig. 3A). With some exceptions, most of the proteins encoding Drosophila genes in which mutations lead to altered structure of the central complex and/or locomotion defects were present in the monarch genome (Table S3). There were no lepidopteran homologs of tay bridge, whose loss causes defects in the protocerebral bridge, and polyhomeotic, a complex locus encoding two transcription factors that are part of Polycomb group involved in segment identity. There were lepidopteran expansions in fused lobes, which encodes a hexosaminidase involved in N-glycan processing, and SNF4, which encodes an AMP-activated protein kinase gamma subunit. Many of the Drosophila genes whose mutations cause central complex defects have broad developmental defects. Nonetheless, the identified set of monarch orthologs and paralogs provides a starting point for more extensive analyses of the sun compass complex network and its development.
Several peptides (including tachykinins, allostatins, pyrokinin, and neuropeptide F) and neurotransmitters (e.g., serotonin and GABA) have been identified by immunocytochemistry in the central complex of locusts (Homberg, 2002), grasshoppers (Herbert et al., 2010) and Drosophila (Kahsai and Winther, 2011) that are likely to be important for neural function and circuitry. We annotated monarch genes that encode orthologs of the vast majority of neuropeptides, polypeptides (Table S4A), and the enzymes involved in biogenic amine synthesis (Table S4B) that are collectively used for neural signaling. There was good agreement between these neural signaling molecules and their corresponding G protein-coupled receptors (Table S4C & D). Specific antibodies can now be developed to map the molecular substrates for central complex neural signaling.
Circadian Rhythms
Circadian clocks and their output pathways play an essential role in migratory processes (Fig. 3A). Circadian clocks located in the antennae provide time compensation for the sun compass system (Merlin et al., 2009). In addition, brain clocks located in the pars lateralis of central brain are likely involved in initiating the migratory generation by sensing decreasing daylength in the fall (Goehring and Oberhauser, 2002; Reppert et al., 2010).
In Drosophila and mammals, the clock mechanism is comprised of a core negative transcriptional feedback loop, which drives self-sustaining rhythms of essential clock components, and a modulatory, interlocking second feedback loop (Allada and Chung, 2010; Reppert and Weaver, 2002). The monarch genome contained the components of both loops (Fig. 3C; Table S5). The monarch core feedback loop possesses all the critical clock genes found in Drosophila: clock (clk), cycle (cyc), period (per), timeless (tim), type-1 cryptochrome (designated cry1) but differs in that it also possesses a type-2 vertebrate-like cry (cry2) previously shown to encode the main transcriptional repressor in the monarch clock (Zhu et al., 2008b); a function fulfilled by per in Drosophila (Allada and Chung, 2010), which does not possess cry2. We also identified genes encoding orthologs of all of the major proteins involved in posttranslational modifications of the core clock proteins (PER and TIM) (Fig. 3C). We further identified the major components of a Drosophila-like, secondary clock feedback loop in the monarch. This included genes encoding orthologs of VRILLE and PDP1, the major regulators of CLK transcription in Drosophila (Cyran et al., 2003), along with the appropriate cis- and trans-regulatory elements (Fig. 3C).
A special focus of our manual annotation was the identification of genes encoding pigment-dispersing hormone (PDH), a circadian output signal in Drosophila brain essential for clock circuitry and driving locomotor activity rhythms (Helfrich-Forster et al., 2000; Shafer and Taghert, 2009), and its G protein-coupled receptor. Although PDH-like immunostaining has been detected in a many other insects, including silkmoths (Zavodska et al., 2003), previous immunocytochemical studies have failed to identify PDH staining in the monarch brain (Sauman and Reppert, unpublished) likely due to the divergence in the monarch PDH sequence (Fig. 3D). Although the pdh transcript that encodes the prepropeptide was not present in our transcriptome, we verified that it is expressed in the monarch brain (Fig. 3D). Mapping monarch PDH expression and clock circuitry is now feasible.
The discovery of type-2 vertebrate-like CRYs in insects, derived from the discovery of CRY2 in monarchs ( Zhu et al., 2005), altered our view of how circadian clocks of non-drosophilid insects work (Yuan et al., 2007). To further our understanding of animal clock evolution, we reinvestigated the existence of type-1 and type-2 CRYs in all arthropods in which a draft genome has been published. Virtually all possess a type-2 CRY (the fire ant, Solenopsis invicta, genome did not reveal any cry genes), except all Drosophila species, which only possess the light-sensitive type-1 CRY (Fig. 3E; Fig. S3). This supports the existence of both CRY types at the base of arthropod evolution. In addition, type-1 CRY and TIM appear to have been lost prior to the radiation of the hymenopterans, suggesting that the Hymenoptera have evolved different mechanism(s) for photic entrainment. Perhaps the TIMELESS paralog, TIMEOUT, which has some influence on the light input pathway in Drosophila is the key (Benna et al., 2010), as it is expressed in all available insect genomes (Fig. 3E).
Juvenile Hormone Regulatory System
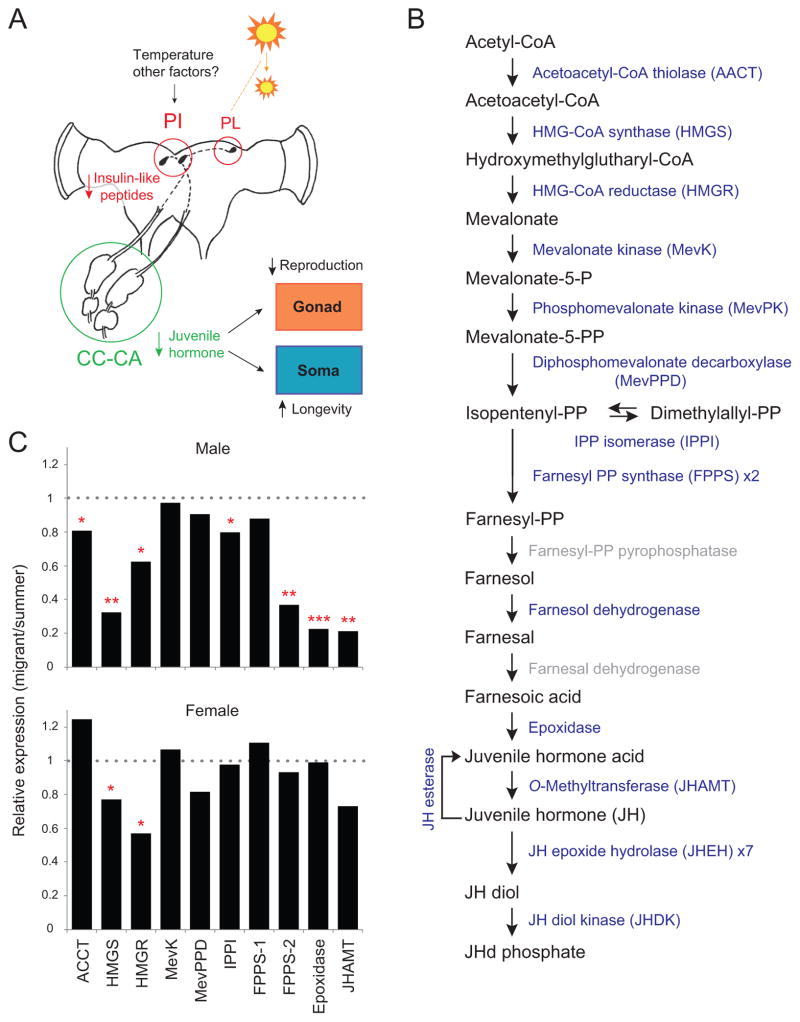
Endocrine regulation is crucial in migratory butterflies to coordinate the multiple physiological processes required for a successful long-distance migration, including reproductive arrest, an increase in life span and a metabolic change increasing fat stores used for flight. These traits are induced in the migratory monarch by a likely downregulation of the insulin signaling pathway and demonstrated juvenile hormone (JH) deficiency (Herman, 1975; Herman and Tatar, 2001), as documented in flies (Flatt et al., 2005). In response to environmental factors (i.e., temperature and photoperiod), insulin signaling could be reduced through a decrease in the production and/or secretion of insulin-like peptides and/or in the expression of their associated receptors, which would reduce JH biosynthesis in the corpora cardiaca-corpora allata complex leading to both reproductive quiescence and aging (Fig. 4A) (Reppert et al., 2010). We thus focused on annotating genes involved in this endocrine regulation and comparing their expression profiles between summer and migrant butterflies, based on the previous microarray data of corresponding ESTs (GSE14041 of GEOdatabase).
Figure 4. Juvenile Hormone regulatory pathway.
A. Proposed endocrine regulation of reproductive arrest and longevity in migratory monarch butterflies. Decreasing daylength (decrease in sun size) is sensed by circadian clocks in the pars lateralis (PL). This information is relayed to the pars intercerebralis (PI) in which the production and/or secretion of insulin-like peptides is decreased resulting in a decrease juvenile hormone (JH) biosynthesis in the corpora cardiac-corpora allata (CC-CA) complex. JH deficiency affects target tissues, leading to reproductive quiescence and increased longevity.
B. JH biosynthetic and degradation pathways (Belles et al., 2005). Enzymes in the monarch genome are in blue. Gray denotes enzymes proposed to catalyze Farnesyl-PP to farnesoic acid, but their actual existence has not yet been verified.
C. Sexually dimorphic pattern of JH biosynthesis. Bars indicate relative gene expression levels of enzymes, as annotated in B, in migrants relative to summer counterparts (data calculated from GSE14041 of GEOdatabase); n=5 per sex per enzyme). Significance was estimated by two-sample t-tests. *, p<0.05; **, p<0.01; ***, p<0.001.
See also Table S6.
The vast majority of the genes involved in the insulin signaling pathway described in Drosophila was represented in the monarch genome (Table S6A). We identified seven insulin-like peptides (ILPs), a number matching that in Drosophila but lower than the 20 genes present in Bombyx, which, as mentioned previously, represents an expansion for this gene family. Monarch ILP-1 was expressed in the transcriptome at a ~10-fold higher level than any of the other six (Table S6A). In addition, ILP-1 was the only one found in the brain EST library, and its levels were generally decreased in migrants compared to summer butterflies. This suggests that ILP-1 is the main peptide involved in the monarch insulin signaling pathway and likely the one ultimately regulating JH biosynthesis. Genes encoding downstream target molecules such as the transcription factor forkhead have been identified in the monarch (Table S6A). As reported in flies (Flatt et al., 2005), a de-repression of forkhead by a decrease in insulin-like peptides in the migratory monarch would result in increased longevity by inducing JH deficiency (Fig. 4A). We annotated from the monarch 71 of the 81 genes that are involved in longevity in Drosophila (Table S6B). An ortholog of one of the longevity genes in flies, rosy (which encodes xanthine dehydrogenase), has been shown previously to be upregulated in migrant brains (Zhu et al., 2009); rosy loss-of-fuction mutant flies have decreased life span (Geiger-Thornsberry and Mackay, 2004). The potential involvement of other JH-regulated genes in migrant longevity can now be evaluated more extensively.
We also identified and annotated genes involved in the biosynthesis of JH. The entire repertoire of known enzymes involved in the JH biosynthetic pathway proposed in insects (Belles et al., 2005) is also represented in the monarch genome (Fig. 4B; Table S6C). Transcriptional profiles between summer and migratory monarchs of both sexes with confirmed reproductive status revealed an unexpected sexually dimorphic pattern of JH biosynthesis regulation (Fig. 4C); male migrants exhibit an overall down-regulation of biosynthesis, while female migrants appear to use instead an increased turnover (involving JH esterase and/or the epoxide hydrolases; Fig. 4B) and/or a putative regulation by JH-binding proteins (Table S6D) to maintain low JH levels. Even though JH has been previously shown to play a role in the control of sexual dimorphism in fly locomotor activity (Belgacem and Martin, 2002), our findings represent novel evidence for a sexual dimorphism in the molecular pathway of JH regulation itself, which could be common in insects.
Sun Compass Orientation Genes
Besides JH-regulated genes, what are the genes that show seasonal changes in expression that define the migratory state? To address this question in the monarch, microarray analysis of unique cDNA sequences in a brain EST library was recently performed (Zhu et al., 2009). By treating migrants with a JH analog it was possible to isolate genes involved in sun compass-oriented flight (not affected by JH status) from those involved in other, JH-dependent aspects of the migration, like reproductive function and longevity. Using this approach, 40 cDNAs were identified whose differential expression in the brain correlated with sun compass-oriented flight behavior in individual migrants, independent of JH activity (Zhu et al., 2009). At the time of publication, only 25 of them could be annotated.
With the monarch genome, we have successfully annotated all 40 cDNAs (Table S7); two ESTs were found to be parts of the same gene leaving 39 orientation genes. The 14 previously unannotated orientation genes included genes upregulated in migrants that encode two transcription factors, bric a brac-like protein and methoprene-tolerant protein 1, a cGMP subunit, and a monarch-specific protein of unknown function. Down-regulated orientation genes in migrants include a beta-arrestin and a monarch-specific protein of unknown function. The most exciting outcome of this complete annotation is two differentially expressed monarch-specific proteins of unknown function that may be unique to the sun compass orientation mechanism.
Small Non-Coding RNAs and Monarch Migration
In concert with protein coding genes, regulatory elements in the genome could be responsible for the initiation and/or maintenance of the migratory state. RNA interference (RNAi) is a double-stranded RNA-mediated process that ultimately regulates the expression of target mRNAs by gene silencing (Meister and Tuschl, 2004). The primary gene silencing-regulators are microRNAs (miRNAs), small interfering RNAs, or piwi-interacting RNAs. Of 31 RNAi pathway-related genes found in Drosophila and/or C. elegans, 21 were annotated in the monarch and 18 in Bombyx (Table S8). Our manual annotation suggests that the Lepidoptera may possess the machinery for effective systemic RNAi-mediated gene silencing, in spite of variable success reported between and within lepidopteran species (Terenius et al., 2011).
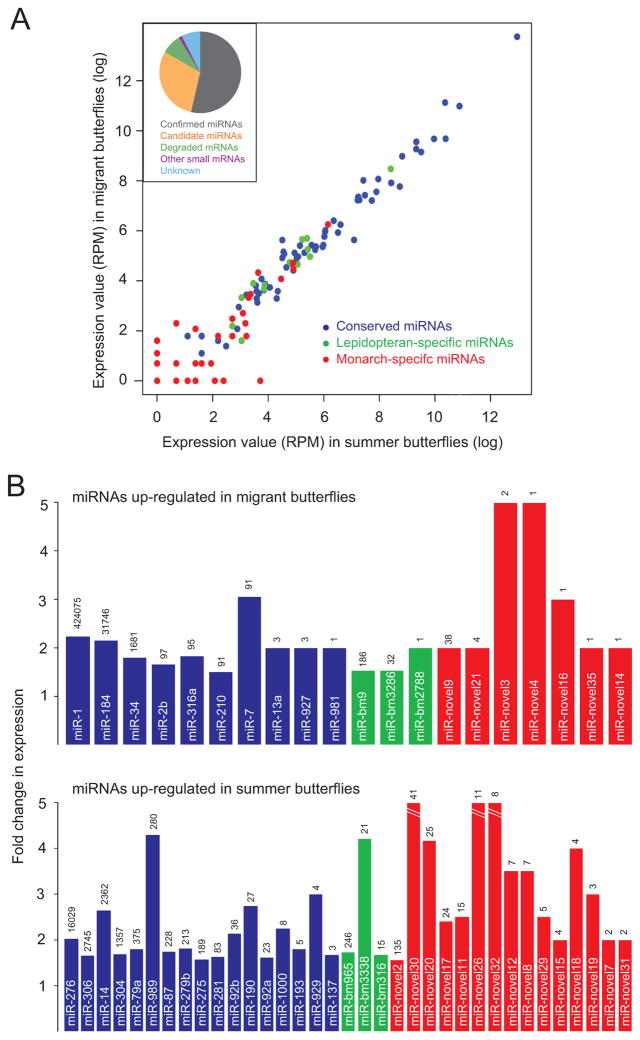
To investigate the potential for gene regulation by endogenous non-coding RNAs in the migratory process, we used Illumina sequencing and computational methods to characterize these regulators in summer and migratory butterflies. miRNAs accounted for the vast majority of the reads from the whole-body small RNA libraries (Fig. 5A, inset). We identified 116 miRNAs from monarch, including 66 conserved, 15 lepidopteran-specific, and 35 novel miRNAs (Fig. 4A). A total of 55 miRNAs had ≥ 1.5-fold differences in mean expression levels between summer and migratory monarchs, with 35 up-regulated in summer butterflies and 20 up-regulated in migrants (Fig. 5B). The conserved miRNAs were the most highly expressed in the monarch (Fig. 5), similar to other invertebrates (Ruby et al., 2006; Ruby et al., 2007; Wei et al., 2009).
Fig. 5. miRNAs in summer and migratory monarchs.
A. Expression of monarch miRNAs. The relationship between the expression values in reads per million (RPM) of summer butterflies and migrants plotted on a natural logarithmic (log) scale. Each of the 116 identified miRNAs is represented as a colored dot (blue, conserved; green, lepidopteran specific; red, monarch specific). Inset is a pie chart showing the classification of non-coding small RNA sequencing reads from the merged summer and migratory samples.
B. miRNAs expressed differentially between a pool of 10 summer butterflies and a pool of 10 migratory monarchs. Top, bars show the mean miRNA levels that were up-regulated ≥ 1.5-fold in migrants compared to summer butterflies. The normalized expression value (in RPM) of migrants is displayed above each bar. Bottom, mean miRNA levels up-regulated in summer butterflies compared to migrants.
See also Table S8.
Of the 27 conserved miRNAs whose mean levels were expressed differentially between summer and migratory monarchs (Fig. 5B), three were of interest because of their reported functions in other species. miR-1, a muscle-specific miRNA that is upregulated in gregarious locusts (Wei et al., 2009), showed the highest abundance of all the monarch miRNAs, and its mean value was up-regulated by 2.2-fold in migrants; miR-1 may be involved in the prolonged muscle-dependent flight required of the monarch during their long-distance migration. In Drosophila, miR-7 stabilizes regulatory processes against temperature perturbations (Li et al., 2009); it showed the largest up-regulation of mean values of the conserved group in migrants, and it may help with the increased cold tolerance manifested by migrants and necessary for their existence at the overwintering grounds atop the transvolcanic mountains in central Mexico. We also found that miR-14, a regulator of fat metabolism in Drosophila (Xu et al., 2003), showed relatively high expression and its mean value was down-regulated in migrants. Because migrant butterflies have increased lipid stores that can be used as fuel during the migration, the metabolic effects of miR-14 could be important for the migration.
Similar to the low coverage of non-universal genes (Table S1J), most novel miRNAs showed weak expression (Fig. 5A), indicating that they evolved recently and/or that their distribution is restricted. Because these novel miRNAs were confidently predicted by three independent measures, it is possible that some have functions unique to the migratory state of monarchs and require further study.
Chemoreception
Chemoreception is likely critical for a successful fall migration (Reppert et al., 2010). The detection of chemical cues is mediated by multigene families of olfactory receptors (Ors), ionotropic receptors (IRs), and gustatory receptors (Grs) (Fig. 6). The molecular underpinnings of lepidopteran chemoreception have been extensively studied in moths (Wanner and Robertson, 2010), while it has received little attention in butterflies.
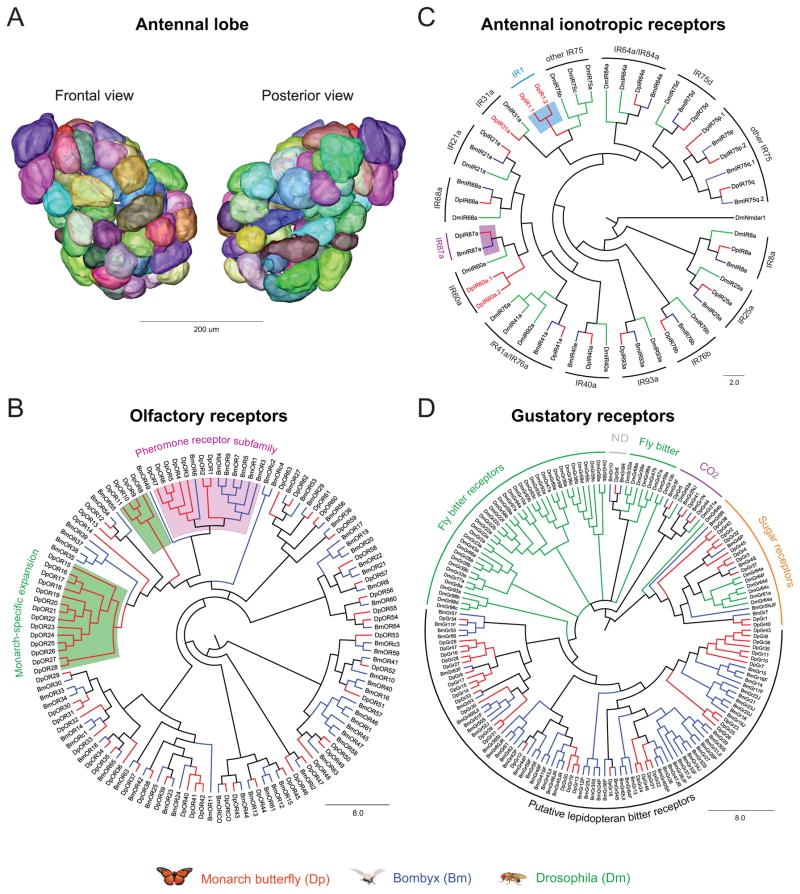
Figure 6. Insights into chemosensory function in the monarch butterfly.
A. Three-dimentional reconstruction of right antennal lobe (AL) of a female migrant monarch. Each glomerulus is highlighted with a unique color without physiological significance.
B. Unrooted tree of candidate monarch (Dp, red lines) and Bombyx mori (Bm, blue lines) olfactory receptors (Ors). Green boxes, monarch-specific expansions. Purple box, pheromone receptor candidates. BmOR2 was renamed as BmORCO. ORl, OR-like; ORc, OR candidate.
C. Phylogenetic relationship of the monarch antennal ionotropic receptor (IR) candidates with Bombyx and Drosophila antennal IRs. Red lines, monarch; blue, Bombyx; green, Drosophila. The monarch IR names in red represent genes present in the monarch genome but not in Bombyx.
D. Phylogenetic relationship of the monarch gustatory receptor (Gr) candidates with Bombyx and Drosophila Grs. Red, monarch; blue, Bombyx; green, Drosophila.
We identified and manually annotated a repertoire of 64 Or candidates (~60% are full-length), which constitutes the first Or repertoire in a butterfly (Fig. 6B). The number of Ors identified is comparable to the number of Ors found in Bombyx (66; Tanaka et al., 2009) and in close agreement with the number of glomeruli in the antennal lobe of the migratory female monarch (Fig. 6A). Indeed, 68 and 69 glomeruli were counted in each lobe (right one) of two respective specimens (Fig. 6A). This supported the 1:1 relationship from the axonal projections of the neurons expressing a given Or to a single glomerulus (Gao et al., 2000). The identified monarch ortholog of the highly conserved Drosophila DmOr83b was designated DpORCO, based on a unified nomenclature for this co-receptor (Vosshall and Hansson, 2011). Phylogenetic analysis including Bombyx (Bm)Ors revealed two monarch-specific subfamily expansions (Fig. 6B; green boxes) that may be used for species-specific recognition behaviors. For example, these Ors could be involved in the recognition of their overwintering sites, nectaring sources for feeding or milkweed host plants for oviposition.
Interestingly, we also identified seven Or genes that form monarch-specific expansions clustering with the moth pheromone receptor subfamily. Unlike Bombyx which relies on pheromones for sexual communication, butterflies use multisensory modalities, including vision and olfaction. However, the use of pheromone cues during monarch courtship is unclear (Pliske, 1975). We thus hypothesize that these Ors may be involved in social behavior, such as in the roosting behavior that migratory monarchs manifest at night during their migration south and at the overwintering sites (Reppert et al., 2010).
The chemosensory ionotropic receptor (IR) family may also be involved in monarch chemosensory behaviors. We identified and manually annotated 19 antennal IRs that appear to be functional genes (no pseudogenes were detected), compared to 14 in Bombyx (Croset et al., 2010; Olivier et al., 2011) (Fig. 6C). Phylogenetic analysis revealed that 16 monarch IRs are putative orthologs of conserved antennal IRs (Croset et al., 2010). Two of the three other monarch antennal IR candidates, DpIR1.1 and DpIR1.2, cluster together in a lineage previously thought to be unique to noctuids (Olivier et al., 2011), which now appears instead to be lepidopteran specific (Fig. 6C; blue box). Another IR candidate, DpIR87a, might define with its BmIR87a ortholog another subtype of lepidopteran-specific antennal IR, as proposed previously (Olivier et al., 2011) (Fig. 6C; purple box).
Gustatory receptors (Grs) mediate contact chemoreception that is used by insects for feeding behavior, host plant selection and oviposition. Bombyx and the monarch present similarities in that their larvae feed exclusively on mulberry and milkweed leaves, respectively. We annotated 47 monarch Gr candidates (Fig. 6D), compared to 65 BmGrs (Wanner and Robertson, 2008). Phylogenetic analysis revealed that 14 putative DpGrs are from the three conserved lineages in insects: the DmGr43a protein subfamily of unknown function, the carbon dioxide receptors and the sugar receptors subfamilies functionally characterized in flies (Dahanukar et al., 2007; Jones et al., 2007). Despite a lower number of Grs identified, the monarch possesses twice as many sugar receptors as found in Bombyx, which is consistent with its ecology as flower nectaring butterfly.
Remarkably, the 33 remaining monarch Gr candidates cluster in our phylogenetic analysis with the 55 BmGrs that form a monophyletic subfamily distinct from those of other insects. This subfamily has been proposed to be putative silkmoth bitter receptors for secondary plant compounds (Wanner and Robertson, 2008) (Fig. 6D). Our annotation extends this discovery to butterflies and therefore supports the hypothesis of a specialization in deterrent bitter compounds detection basal to the lepidopteran lineage. Bombyx and monarch putative bitter Grs exhibit species-specific small expansions that could reflect different specificity in host plant recognition (mulberry vs. milkweed).
The Molecular Basis of the Monarch’s Chemical Defense Mechanism
Predators take a heavy toll on the yearly migration, and the monarch has evolved a special chemical defense mechanism to limit the loss. The P type Na+/K+-ATPase is an essential enzyme that maintains the proper balance of ions on opposite sides of the cell that is critical for normal cellular function (Skou, 1998). As a milkweed specialist, monarch larvae are exposed to cardiac glycosides that are sequestered in adults, making the butterfly bitter and toxic. Although these cardenolide glycosides block the sodium/potassium pump and cause death (Prassas and Diamandis, 2008), the monarch enzyme is completely resistant to inhibition by the cardiac glycoside ouabain (Holzinger et al., 1992). The molecular basis for this was originally proposed to be a point mutation in the α subunit changing Asn-193 to His (numbering based on the full-length monarch protein; Fig. S4), which is a critical residue for ouabain binding. In fact, mutating Asn to His at the homologous position in Drosophila converts the fly enzyme to the monarch version highly resistant to ouabain binding (Holzinger and Wink, 1996). In addition, there was no mutation at this residue in the DNA of the α subunit of other Danaus species whose larvae also feed on milkweed (Mebs et al., 2000). It is possible that the α subunit variant allows for the higher sequestration of cardenolides that are found in the monarch, compared to the lower sequestration in the non-migrating Queen butterfly (D. gilippus) (Cohen, 1985).
With the monarch genome and transcriptome available, we have now been able to obtain the entire sequence of the coding region of the α subunit (1193 aa) and its genomic structure (Fig. S4). We have established an additional amino acid replacement in the coding sequence, which would confer even greater resistance to ouabain binding than the original Asn193His change; no other amino acid changes exist among the conserved regions of the monarch, Drosophila and sheep proteins. We previously identified a Glu182Val change (Zhu et al., 2008a); amino acid substitutions at both 182 and 193 (Fig. S4) confer a higher degree of resistance to ouabain binding (Price et al., 1990). This Glu182Val change was missed in previous work (Holzinger and Wink, 1996; Mebs et al., 2000) because only genomic DNA was amplified and the splicing of the intron 3′ to position 182 was incorrectly predicted (Fig. S4). We therefore have a full explanation for the difference in what we and others have reported for residue 182. Furthermore, our transcriptome revealed both the Glu182Val and Asn193His changes in all >1,000X transcriptome coverage (Table S9). We also annotated two α subunit paralogs in the monarch genome (Table S9), but neither had a conserved ouabain binding site, and the transcriptome coverage of both was low (≤ 3).
Since a functional Na+/K+-ATPase depends on heterodimerization between α and β subunits, we identified in the monarch all three forms of the β subunit described in Drosophila (nervana 1, 2 and 3). There were four homologs of nervana 2, of which two were highly expressed in the transcriptome (Table S9). Moreover, the most highly expressed β subunit was nervana 3 that has been recently shown in Drosophila to be exclusively expressed in the nervous system, especially sensory neurons (Baumann et al., 2010).
The unique structure of the major α subunit of Na+/K+-ATPase provides a molecular substrate for the ability of the monarch butterfly to sequester toxic cardenolides. This would help protect the monarch against predation during its migration and overwintering period. We also propose that this molecular substrate has allowed the monarch to participate in the well-known mimicry complex with the viceroy butterfly. This mimicry system was first described as Batesian, with the monarch being unpalatable and toxic, and the viceroy, first defined as palatable, exploiting the model species through its shared coloration pattern and display behavior to predators (Brower, 1958). This view was modified with the finding that the monarch and viceroy bodies are equally unpalatable to birds, suggesting a Müllarian mimicry, in which both species are co-mimics (Ritland and Brower, 1991).
Conclusions
We have performed deep sequencing and de novo assembly of the monarch genome to provide the first characterized genome of a butterfly and of a long-distance migratory species. Overall, the attributes of the monarch genome and its proteome provide a treasure trove for furthering our understanding of monarch butterfly migration; a solid background for population genetic analyses between migratory and non-migratory populations; and a basis for future genetic comparison of the genes involved in navigation yet to be discovered in other long-distance migrating species, including vertebrates like migratory birds.
EXPERIMENTAL PROCEDURES
See the Extended Experimental Procedures for detailed protocols.
Genome sequencing
We used wild-caught, migratory female butterflies (the heterogametic sex) for sequencing; laboratory-generated butterfly lines were not available. Although the use of a single butterfly for all sequencing runs would have been optimal, it was precluded by the need of different libraries for the different sequencing runs from different platforms and vendors. Genomic DNA (34–65 μg) was isolated from individual thoraces using standard protocols with RNase treatment. We employed both Illumina technology and Roche 454 sequencing technology (Table S1A). For deep sequence coverage, we used Illumina sequencing. Short insert paired-end (200 bp; SIPES) and long insert mate-pair (3–5 kb; LIPES) libraries were constructed from the DNA of female F-2 (34μg DNA yield). Sequencing runs from three lanes of SIPES and three lanes of LIPES were performed by Eureka Genomics (Hercules, CA, USA). To overcome the probable repetitive regions, we also employed Roche 454 sequencing to obtain longer reads. From female F-9 (63 μg DNA), 12 shotgun fragment runs were performed on the 454 FLX/titanium platform (conducted by Virginia Bioinformatics Institute, Blacksburg, VA, USA), as well as three runs from a 20-kb insert paired-end library. From a third female (F-4; 65 μg DNA), we generated DNA for two Roche sequencing runs from an 8-kb insert paired-end library.
Genome assembly
Initial assemblies were generated by CLC bio’s de novo assembler (Katrinebjerg, Denmark) and Newbler (Roche, Inc.) for Illumina and Roche 454 reads, respectively (Table S1B). We then used the Illumina paired-end reads, step-by-step from 200-bp to 5-kb insert size, to join the initial Illumina contigs into scaffolds by SSPACE 1.0 (Boetzer et al., 2010). Remaining gaps within these scaffolds were iteratively filled with paired-end SIPES reads and the Roche 454 contigs using GapCloser available in SOAPdenovo (Li et al., 2010). The resulting v1 assembly included all scaffolded contigs and had a final scaffold N50 length of 53,032 bp (spanning 272.7 Mb) and contig N50 length of 50,721 bp (spanning 272.2 Mb). A second version of the assembly (v2) provided additional extension of the scaffolds using reads from the Roche 8 kb and 20 kb paired-end libraries, improving the scaffold assembly to a N50 length of 207,025 bp (spanning 277.7 Mb) (Table S1B). In the v2 assembly, 7780 scaffolds contain 3,598 gaps, spanning 5,393,193 bp. We continue to improve the assembly and will update as appropriate.
We evaluated the completeness of coverage of our assembly using homologs of other insects. The monarch assembly covered 457 core eukaryotic genes (CEGMA) (Parra et al., 2007) (TBLASTN, E<10−5) of Drosophila at a level comparable to four other well-organized insect genomes, Bombyx, Tribolium, P. barbatus, and S. invicta (Table S1C) even though those genomes have substantially larger scaffold sizes. Moreover, the fraction of bases in the CEGMA genes present in single scaffolds was also very similar among the five species (Table S1D). We also aligned the entire Drosophila and Tribolium gene sets to the monarch and Bombyx assembled genomes (TBLASTN, E<10−5) (Table S1C). Both Lepidoptera showed very similar levels of coverage (using genblastA v1.0.4) and the percentage of mapped genes located in a single scaffold. Coverage of above alignments was automatically calculated using genblastA v1.0.4 with “-e 1e-5 –a 0.5 -r 1 -c 0.5” option. In addition, all 79 conserved cytoplasmic ribosomal protein genes were completely present in the v1 assembly (Table S1H), with only one gene that was lacking 30 amino acids. We also assessed the completeness and accuracy of our assembly using 9,484 independently sequenced and assembled monarch ESTs (Zhu et al., 2008a). A total of 9,072 ESTs (~96%) could be mapped to the assembly (BLASTN, E<10−50) and none of them mapped to more than one scaffold. In terms of accuracy, we found only 28 exons (0.3% of all mapped ESTs) were located in the opposite orientation with their neighboring exons, as candidates inverted assembly. There were 64,380 single-base mismatches (0.94%) and 5,660 indels (0.08%) found in the 6.85 Mb region mapped. Taking into account the high level of heterozygosity of the monarch genome (0.55%), our assembly exhibits a low level of assembly error. Taken together, the monarch genome assembly appears quite complete and accurate compared to the gene coverage in other genomes.
Genome annotation
A total of 5.4 Gb transcriptome sequence was generated by Illumina RNA-seq (The National Center for Genome Resources, Santa Fe NM USA), representing all stages of monarch development. The official gene set (OGS1.0; Table S1G) was based on a GLEAN consensus model (Elsik et al., 2007), which combined transcriptome, homology, and five ab-initio sets (Table S1G). Automatic orthology was determined using the OrthoMCL pipeline (Li et al., 2003). More than 1,000 genes of biological interest were manually annotated. Genomes and genesets for comparative analysis were listed in Table S1I. To identify and profile miRNAs, Illumina small RNA-seq was performed independently for summer butterflies and migrants samples, each being a pool of 10 animals. We primarily used the miRDeep algorithm (Friedlander et al., 2008) for miRNA prediction. The genome assembly, gene set and annotations have been deposited at GenBank under accession number GDsub16004.
Supplementary Material
Acknowledgments
We thank Stanley Heinze and Surainder Asokaraj for providing the three-dimensional reconstruction images of the monarch antennal lobe shown in Figure 6A; Alan Ritacco for use of the computing server; and Susan Fuerstenberg and Amy Casselman for assistance. Author contributions: SMR initiated and oversaw the project. JLB designed the sequencing strategy, provided sequence quality control, and generated the v0 assemblies. SZ performed the v1 and v2 assemblies, generated the gene sets, provided quality control and the data for all Figures, except Figure 6A. CM performed the genomic DNA preparations, collected and prepared RNA for the transcriptome library, helped with the annotation of the chemosensory receptors, and properly formatted all the Figures. SZ, CM, JLB and SMR performed data analyses. SZ, CM and SMR wrote the paper. This work was supported by NIH grant GM086794-02S1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Salvaterra PM, Takeyasu K. Developmental changes in beta-subunit composition of Na,K-ATPase in the Drosophila eye. Cell Tissue Res. 2010;340:215–228. doi: 10.1007/s00441-010-0948-x. [DOI] [PubMed] [Google Scholar]
- Belgacem YH, Martin JR. Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc Natl Acad Sci USA. 2002;99:15154–15158. doi: 10.1073/pnas.232244199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles X, Martin D, Piulachs MD. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu Rev Entomol. 2005;50:181–199. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- Benna C, Bonaccorsi S, Wulbeck C, Helfrich-Forster C, Gatti M, Kyriacou CP, Costa R, Sandrelli F. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr Biol. 2010;20:346–352. doi: 10.1016/j.cub.2009.12.048. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2010;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Brower JVZ. Experimental studies of mimicry in some North American butterflies. Part 1. The monarch, Danaus plexipus, and the viceroy, Limenitis archippus archippus. Evolution. 1958;12:32–47. [Google Scholar]
- Brower LP. Understanding and misunderstanding the migration of the monarch butterfly (Nymphalidae) in North America. J Lepidopt Soc. 1995;49:1857–1995. [Google Scholar]
- Brown KS, Jr, Von Shoultz B, Suomalainen E. Chromosome evolution in neotropical Danainae and Ithomiinae (Lepidoptera) Hereditas. 2004;141:216–236. doi: 10.1111/j.1601-5223.2004.01868.x. [DOI] [PubMed] [Google Scholar]
- Cohen JA. Differences and similarities in cardenolide contents of queen and monarch butterflies in florida and their ecological and evolutionary implications. J Chem Ecol. 1985;11:85–103. doi: 10.1007/BF00987608. [DOI] [PubMed] [Google Scholar]
- International Silkworm Genome Corsortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R, Emery P. A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol Neurobiol. 2008;38:129–145. doi: 10.1007/s12035-008-8035-y. [DOI] [PubMed] [Google Scholar]
- Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- Friedlandeer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Disovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Froy O, Gotter AL, Casselman AL, Reppert SM. Illuminating the circadian clock in monarch butterfly migration. Science. 2003;300:1303–1305. doi: 10.1126/science.1084874. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Geiger-Thornsberry GL, Mackay TF. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech Ageing Dev. 2004;125:179–189. doi: 10.1016/j.mad.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Goehring L, Oberhauser KS. Effects of photoperiod, temperature, and host plant age on induction of reproductive diapause and development time in Danaus plexippus. Ecol Entomol. 2002;27:674–685. [Google Scholar]
- Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10:332. doi: 10.1186/1471-2164-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the Insects. C.U. Press; New York, NY: 2005. [Google Scholar]
- Heinze S, Reppert SM. Sun compass integration of skylight cues in migratory monarch butterflies. Neuron. 2011;69:345–358. doi: 10.1016/j.neuron.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert Z, Rauser S, Williams L, Kapan N, Guntner M, Walch A, Boyan G. Developmental expression of neuromodulators in the central complex of the grasshopper Schistocerca gregaria. J Morphol. 2010;271:1509–1526. doi: 10.1002/jmor.10895. [DOI] [PubMed] [Google Scholar]
- Herman WS. Endocrine regulation of posteclosion enlargement of the male and female reproductive glands in Monarch butterflies. Gen Comp Endocrinol. 1975;26:534–540. doi: 10.1016/0016-6480(75)90176-8. [DOI] [PubMed] [Google Scholar]
- Herman WS, Tatar M. Juvenile hormone regulation of longevity in the migratory monarch butterfly. Proc Biol Sci. 2001;268:2509–2514. doi: 10.1098/rspb.2001.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger F, Frick C, Wink M. Molecular basis for the insensitivity of the Monarch (Danaus plexippus) to cardiac glycosides. FEBS Lett. 1992;314:477–480. doi: 10.1016/0014-5793(92)81530-y. [DOI] [PubMed] [Google Scholar]
- Holzinger F, Wink M. Mediation of cardiac glycoside insensitivity in the monarch butterfly (Danaus plexippus): Role of an amino acid substitution in the ouabain binding site of Na+,K+-ATPase. J Chem Ecol. 1996;22:1921–1937. doi: 10.1007/BF02028512. [DOI] [PubMed] [Google Scholar]
- Homberg U. Neurotransmitters and neuropeptides in the brain of the locust. Microsc Res Tech. 2002;56:189–209. doi: 10.1002/jemt.10024. [DOI] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kahsai L, Winther AM. Chemical neuroanatomy of the Drosophila central complex: distribution of multiple neuropeptides in relation to neurotransmitters. J Comp Neurol. 2011;519:290–315. doi: 10.1002/cne.22520. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Labhart T, Meyer EP. Detectors for polarized skylight in insects: a survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc Res Tech. 1999;47:368–379. doi: 10.1002/(SICI)1097-0029(19991215)47:6<368::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebs D, Zehner R, Schneider M. Molecular Studies on the Oubain Binding Site of the Na+, K+-ATPase in Milkweed Butterflies. Chemoecology. 2000;10:201–203. [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Merlin C, Gegear RJ, Reppert SM. Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science. 2009;325:1700–1704. doi: 10.1126/science.1176221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the ants: diversification in the age of angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- Mouritsen H, Frost BJ. Virtual migration in tethered flying monarch butterflies reveals their orientation mechanisms. Proc Natl Acad Sci U S A. 2002;99:10162–10166. doi: 10.1073/pnas.152137299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2011;20:189–199. doi: 10.1111/j.1365-2583.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- Ooka S, Katow T, Yaguchi S, Yaguchi J, Katow H. Spatiotemporal expression pattern of an encephalopsin orthologue of the sea urchin Hemicentrotus pulcherrimus during early development, and its potential role in larval vertical migration. Dev Growth Differ. 2010;52:195–207. doi: 10.1111/j.1440-169X.2009.01154.x. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Perez SM, Taylor OR, Jander R. A sun compass in monarch butterflies. Nature. 1997;387:29. [Google Scholar]
- Pliske TE. Courtship behavior of the monarch butterfly, Danaus plexippus L. Ann. Entomol. Soc. America. 1975;68:143–151. [Google Scholar]
- Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- Price EM, Rice DA, Lingrel JB. Structure-function studies of Na,K-ATPase. Site-directed mutagenesis of the border residues from the H1-H2 extracellular domain of the alpha subunit. J Biol Chem. 1990;265:6638–6641. [PubMed] [Google Scholar]
- Reppert SM, Gegear RJ, Merlin C. Navigational mechanisms of migrating monarch butterflies. Trends Neurosci. 2010;33:399–406. doi: 10.1016/j.tins.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Zhu H, White RH. Polarized light helps monarch butterflies navigate. Curr Biol. 2004;14:155–158. doi: 10.1016/j.cub.2003.12.034. [DOI] [PubMed] [Google Scholar]
- Ritland DB, Brower LP. The viceroy butterfly is not a batesian mimic. Nature. 1991;350:497–498. [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauman I, Briscoe AD, Zhu H, Shi D, Froy O, Stalleicken J, Yuan Q, Casselman A, Reppert SM. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron. 2005;46:457–467. doi: 10.1016/j.neuron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide’s circadian functions. PLoS One. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skou JC. Nobel Lecture. The identification of the sodium pump. Biosci Rep. 1998;18:155–169. doi: 10.1023/a:1020196612909. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, Touhara K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881–890. doi: 10.1016/j.cub.2009.04.035. [DOI] [PubMed] [Google Scholar]
- Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, Albrechtsen M, An C, Aymeric JL, Barthel A, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Urquhart FA, Urquhart NR. Autumnal migration routes of the eastern population of the monarch butterfly (Danaus p. plexippus L.; Danaidae; Lepidoptera) in North America to the overwintering site in the Neovolcanic Plateau of Mexico. Can J Zool. 1978;56:1759–1764. [Google Scholar]
- Velarde RA, Sauer CD, Walden KK, Fahrbach SE, Robertson HM. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem Mol Biol. 2005;35:1367–1377. doi: 10.1016/j.ibmb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Hansson BS. A Unified Nomenclature System for the Insect Olfactory Coreceptor. Chem Senses. 2011;36:497–498. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Robertson HM. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol Biol. 2008;17:621–629. doi: 10.1111/j.1365-2583.2008.00836.x. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Robertson HM. Molecular biology and genetics of the lepidoptera (Taylor and Francis Group) 2010. Lepidopteran chemoreceptors; pp. 153–168. [Google Scholar]
- Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJ, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- Xiang Z. Genetics And Breeding Of The Silkworm. Chinese Agriculture Press; Beijing, PR China: 1995. [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- Zavodska R, Sauman I, Sehnal F. Distribution of PER protein, pigment-dispersing hormone, prothoracicotropic hormone, and eclosion hormone in the cephalic nervous system of insects. J Biol Rhythms. 2003;18:106–122. doi: 10.1177/0748730403251711. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Bork P. Quanti cation of insect genome divergence. TRENDS in Genetics. 2008;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zhu H, Casselman A, Reppert SM. Chasing migration genes: a brain expressed sequence tag resource for summer and migratory monarch butterflies (Danaus plexippus) PLoS One. 2008a;3:e1345. doi: 10.1371/journal.pone.0001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Gegear RJ, Casselman A, Kanginakudru S, Reppert SM. Defining behavioral and molecular differences between summer and migratory monarch butterflies. BMC Biol. 2009;7:14. doi: 10.1186/1741-7007-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, Reppert SM. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008b;6:e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Yuan Q, Briscoe AD, Froy O, Casselman A, Reppert SM. The two CRYs of the butterfly. Curr Biol. 2005;15:R953–954. doi: 10.1016/j.cub.2005.11.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.