Abstract
Small Rho GTPases regulate a diverse range of cellular behavior within a cell. Their ability to function as molecular switches in response to a bound nucleotide state allows them to regulate multiple dynamic processes, including cytoskeleton organization and cellular adhesion. Because the activation of downstream Rho GTPase signaling pathways relies on conserved structural features of target effector proteins (i.e., CRIB domain), these pathways are particularly vulnerable to microbial pathogenic attack. Here, we discuss new findings for how the bacterial virulence factor EspG from EHEC O157:H7 exploits a CRIB-independent activation mechanism of the Rho GTPase effector PAK. We also compare this mechanism to that of EHEC EspFU, a bacterial virulence factor that directly activates N-WASP. While both virulence factors break the inhibitory interaction between the autoinhibitory and activity-bearing domains of PAK or WASP, the underlying mechanics are very distinct from endogenous Cdc42/Rac GTPase regulation. The ability of bacterial proteins to identify novel regulatory principles of host signaling enzymes highlights the multi-level nature of protein activation, and makes them effective tools to study mammalian Rho GTPase signaling pathways.
Key words: EPEC, EHEC, EspG, EspFU, PAK, type III effector, WASP, N-WASP, toxin
The capacity of a eukaryotic cell to survive and function is directly tied to its ability to rapidly adapt cellular processes in response to both external and internal stimuli. One class of proteins in particular, the Rho family GTPases, plays a critical role in signal transduction. Through interaction with downstream effectors, Rho GTPases regulate the assembly and branching of actin filaments at the membrane interface, creating a dynamic structure that is responsible for cell to cell adhesion in tissues, cytokinesis, cell morphology, phagocytosis and others.1 In addition, this family of proteins is also capable of modulating the activity of transcriptional regulators both directly and indirectly, exhibiting control over cell cycle progression and survival.2,3 Their significance is especially highlighted by many diseases linked to their absence or dysregulation.4,5
Small GTPases exist in either active GTP-bound or inactive GDP-bound forms, acting as biochemical switches and turning specific signaling pathways on or off, respectively. The current question in cell biology is how GTPases interact with different downstream target proteins to induce a wide range of cellular responses, while at the same time maintaining signaling specificity. The details of these interactions and, in particular, how the balance between the conservation of activation mechanism and the specificity for downstream substrate is achieved, are essential for our understanding of complex signaling events.
An analysis of known downstream GTPase effectors revealed a common structural feature known as the Cdc42/Rac Interactive Binding (CRIB) domain for a number of proteins, which include kinases, actin-binding scaffolds and adaptor proteins.6 Whereas these CRIBdomain containing proteins are otherwise distinct both structurally and functionally, they are related in that activation is dependent on direct interaction of CRIB with active Cdc42 or Rac GTPases. A simplified model view for effector protein inhibition vs. activation can thus be presented by defining the protein through two core domains—an inhibitory CRIB-containing GTPase binding domain (GBD) and an activity-bearing domain (AD).
Significant insight into the intramolecular changes happening during activation of target proteins by GTPases came from structural and thermodynamic studies of WASP and PAK1.7,8 The findings show that in an inactive state, GBD is folded onto AD, maintaining AD in a passive conformation and preventing the access of the substrate to the catalytic site. Following binding of Cdc42/Rac to CRIB, GBD is dissociated from AD, removing the constraint and relieving the inhibition.9,10
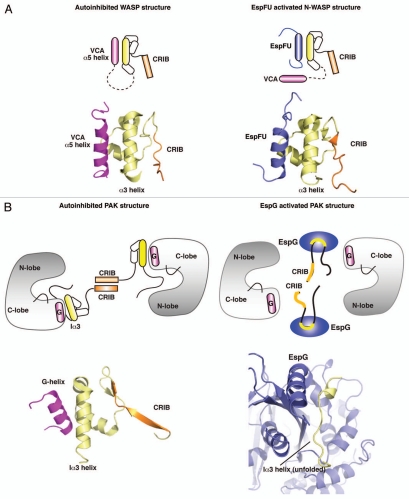
In free WASP, autoinhibitory domain folds onto the hydrophobic α5-helix of the activity bearing VCA domain, preventing its recruitment of Arp2/3 actin nucleation complex (Fig. 1A). Binding of Cdc42 to CRIB disrupts the GBD/α5-helix interface by destabilizing GBD and making it unfold, which exposes VCA residues for interaction with Arp2/3.11 Interestingly, studies of WASP have shown that isolated GBD and VCA domains are largely unfolded in solution, thus suggesting a potential role for the inhibitory interaction in stabilizing the protein.7,9
Figure 1.
Structural principles for the activation of WASP and PAK proteins by bacterial effectors. (A) Left part: cartoon and structural models of autoinhibited WASP, showing the interaction of VCA (magenta) with GBD (yellow). (PDB ID: 1EJ5) Right part: cartoon and structural models showing the binding of EspFu helix (blue) to GBD, leading to the release of VCA and activation of WASP signaling. (PDB ID: 2K42). (B) Left part: cartoon and structural models of autoinhibited PAK1 homodimer, and close up view of the autoinhibitory domain organization (yellow) and G-helix of the Kinase Domain. (PDB ID: 1F3M) Right part: Binding of EspG (blue) to PAK unfolds Ia3-helix (yellow) and leads to the disruption of inhibitory contacts, activating the kinase. (PDB ID: 3PCS).
This general mechanism of activation through unfolding and rearrangement of GBD/AD interface is also applicable to another CRIB-containing protein family, p21-Activated Kinases. PAK consists of an autoinhibitory GBD and a catalytic Kinase Domain (KD). In free form PAK is a homodimer, where GBD domains interact with KD and also with each other through CRIB dimerization domains (Fig. 1B). Binding of Cdc42 disrupts the CRIB interface and induces structural changes to the GBD architecture, partially unfolding it and leading to the release of KD.8,10 Analogous to WASP, the isolated GBD of PAK appears unstable in solution, but stable when folded onto KD in an autoinhibited homodimer. These observations make the unstable nature of GBD one of the characteristic features of CRIB proteins, and implicate CRIB-domains in being able to not only regulate the activation state of the protein, but also control its stability and dynamics.
The nature and the extent of processes regulated by Rho GTPases also makes them prime targets for bacterial virulence factors, allowing pathogens to subvert host machinery in order to facilitate entry, attachment to cells, and to promote bacterial replication and infection.12 An impressive list of secreted bacterial effectors aimed at mimicking or modifying the behavior of host small GTPases illustrates the essential role of this class of proteins and suggests that there are still features that have not yet been characterized.13,14 It is interesting, therefore, to focus at how the mechanisms of action may be different between eukaryotic and bacterial activators. One of the most logical and obvious means for the bacteria to regulate the pathways dependent on Rho GTPase signaling is to activate the GTPase itself, allowing it to then signal through endogenous pathways. Indeed, literature describes multiple examples where type III secreted effectors function as Guanine-nucleotide Exchange Factors (GEFs) for host GTPases, facilitating the exchange of GDP to GTP and activating the enzyme.13,15 This strategy alone, however, cannot explain how bacteria are able to overcome a significant setback of having secreted effectors compete for binding with endogenous regulatory molecules. During infection, a limited number of virulence factors are secreted into host cytoplasm, which means their specificity and efficiency in modifying host pathways needs to be extremely high. Mimicking or inducing the activity of global regulators, such as Rho GTPases, may result in the activation of multiple downstream pathways, which would sequester the effector function into non-productive pathways, reducing critical efficiency. Instead, secreted virulence factors can perform focused pathway activation through their specificity toward only a particular class of signaling enzymes. And because this strategy relies on the endogenous activation mechanisms to propagate the signal, it ensures the preservation of signal integrity throughout the pathway.
We have recently discovered a novel mechanism of PAK activation by the bacterial effector EspG.16 Notably, this mechanism can be compared directly to PAK activation by Cdc42/Rac, as well as the newly discovered mechanism of N-WASP activation by EHEC EspFu.10,17 The structures of EspG and EspFu, in complex with their CRIB-containing host targets PAK and N-WASP, respectively, highlight how bacterial proteins use unique recognition and binding of GBD to dissociate AD and activate target proteins.16,17 Surprisingly, they do so by interacting with binding sites completely distinct from those targeted by endogenous Cdc42/Rac. For example, EspFu hijacks the ability of WASP family of proteins to activate actin nucleation factor Arp2/3 and induces the formation of an actin pedestal at the site of bacterial attachment.18,19 Unlike Cdc42, EspFu does not destabilize GBD through binding to the CRIB-domain, but instead acts as a competitive substrate for GBD by mimicking the hydrophobic helix of VCA. The amphipathic helix of EspFu binds in a similar orientation to the same binding site as the α5-helix of VCA, displacing VCA and relieving inhibitory association (Fig. 1A).17,20 At the same time, EspFu acts as a scaffold and maintains the stability of host protein by providing a substrate for WASP GBD to fold onto.
In our recent study we have identified EspG as an activator of class I p21-family kinases and showed that it binds to the Ia3-helix of the autoinhibitory domain.16 All class I PAKs share the CRIB domain and dissociate into active monomers upon binding of Cdc42/Rac. Dissociation of inactive homodimer is necessary for the removal of the autoinhibitory loop (AI) that is linked to autoinhibitory domain and runs through the catalytic site of PAK KD.8 The KD is fully active on its own, but can be inhibited by titrating in GBD, again showing the importance of interaction between inhibitory and active domains in modulating protein activity. As in case of EspFu and WASP, EspG does not overlap with the Cdc42 binding site of PAK GBD, but instead recognizes and unwinds the Iα3-helix, which is directly connected to the AI loop (Fig. 1B). Our crystal structure provides a mechanistic insight into how this interaction affects the orientation of AI loop, exposing the catalytic residues of the active site, while at the same time disrupting the inhibitory contacts between GBD and KD. In other words, both bacterial virulence factors exploit the basic principles of a unique endogenous mechanism and activate their downstream host targets through separation of inhibitory domain from activity-bearing domain, but define new regulatory sites through which this activation is achieved.
Potency in the hijacking of cellular machinery is one of the hallmarks of bacterial virulence factors. The complexity of eukaryotic signaling networks includes spatial and temporal regulation of activating molecules, co-factors and substrate availability. While multiple checkpoints ensure precise and accurate response, they put pressure on bacteria to overcome endogenous inhibitory mechanisms. Post-translational modification of host regulatory molecules by secreted bacterial effectors usually results in a constitutively active or inhibited enzyme, producing an all-or-nothing effect.14,21 By itself, this would make it extremely difficult for a pathogen to fine tune an entire network of signaling events. On the other hand, however, this makes a good platform for bacterial effectors that can then target specific downstream proteins of the Rho GTPase pathways for activation. The precision of targeting is particularly striking, considering that both WASP and PAK can be activated by Cdc42/Rac through conserved binding to CRIB, whereas EspFu and EspG exclusively activate their respective targets. By reducing “background” host signaling, the effect of virulence factors is thus amplified, allowing pathogens to be remarkably efficient despite limited repertoire of secreted effectors. This supports the concept that multiple bacterial proteins function in concert, with some affecting the global regulatory hubs while others initiating specific pathways, and suggests that more secreted effectors are likely to be discovered that exploit conserved host mechanisms of activation through novel regulatory sites.
Studying the molecular mechanisms of bacterial effectors that activate downstream targets is particularly interesting for our understanding of multi-level regulation of GTPase signaling. Multiple factors can be involved in a signaling pathway within a cell, making it extremely difficult to observe and study under in vitro conditions. Bacterial proteins have evolved the ability to maximize the signaling potential due to limited availability, and bring into focus new ways in which signaling may potentially be controlled in vivo. For example, EspFu contains multiple domains that mimic the amphipathic helix of VCA in tandem, and was found to display much higher actin-nucleation ability than endogenous activator, Cdc42.20 This has lead to the discovery that oligomerized WASP has a higher affinity for Arp2/3, introducing dimerization factors into the signaling equation of WASP activation.20,22 Similarly, understanding of how scaffolding properties of EspG come into play during its activation of PAKs may provide insight into additional levels of kinase regulation. Bacterial virulence factors can function as distinct, but equivalent activators of GTPase targets, in as much as they utilize similar global mechanism of relieving inhibition, but display a stronger activity and precise specificity for their substrate through novel regulatory sites.
Acknowledgements
This work was supported by grants from the Welch foundation (#I-1704) and the National Institute of Health (NIAID; 1RO1AI083359-01) to N.M.A.
Extra View to: Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593.
References
- 1.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/S0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 3.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factorkappaB by Rho, CDC42 and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 4.Nadif Kasri N, Van Aelst L. Rho-linked genes and neurological disorders. Pflugers Arch. 2008;455:787–797. doi: 10.1007/s00424-007-0385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pai SY, Kim C, Williams DA. Rac GTPases in human diseases. Dis Markers. 2010;29:177–187. doi: 10.3233/DMA-2010-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 7.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35010088. [DOI] [PubMed] [Google Scholar]
- 8.Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/S0092-8674(00)00043-X. [DOI] [PubMed] [Google Scholar]
- 9.Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, et al. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 10.Morreale A, Venkatesan M, Mott HR, Owen D, Nietlispach D, Lowe PN, et al. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat Struct Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- 11.Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boquet P. Small GTP binding proteins and bacterial virulence. Microbes Infect. 2000;2:837–843. doi: 10.1016/S1286-4579(00)90369-1. [DOI] [PubMed] [Google Scholar]
- 13.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Visvikis O, Maddugoda MP, Lemichez E. Direct modifications of Rho proteins: deconstructing GTPase regulation. Biol Cell. 2010;102:377–389. doi: 10.1042/BC20090151. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, et al. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol. 2009;16:853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng HC, Skehan BM, Campellone KG, Leong JM, Rosen MK. Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U) Nature. 2008;454:1009–1013. doi: 10.1038/nature07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Garmendia J, Carlier MF, Egile C, Didry D, Frankel G. Characterization of TccP-mediated N-WASP activation during enterohaemorrhagic Escherichia coli infection. Cell Microbiol. 2006;8:1444–1455. doi: 10.1111/j.1462-5822.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 20.Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, Mayer BJ, et al. The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature. 2008;454:1005–1008. doi: 10.1038/nature07170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aktories K, Schmidt G, Just I. Rho GTPases as Targets of Bacterial Protein Toxins. Biol Chem. 2000;381:421–426. doi: 10.1515/BC.2000.054. [DOI] [PubMed] [Google Scholar]
- 22.Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]