Abstract
Many conceptual advances in biology have been achieved by experimental studies using planar two-dimensional cell culture systems. Recent adaptations of molecular techniques to three-dimensional model systems are bridging the gap in our understanding of biological events in vitro and in vivo in the study of disease progression. Recently, in vitro studies using Förster resonance energy transfer (FRET) have shown that the prototypical RhoGTPases Cdc42, Rac and RhoA are temporally and spatially synchronized during cell migration, with initial RhoA activity inducing protrusion prior to activation of Rac. This simultaneous FRET approach illustrates the tight control and dynamic regulation of RhoGTPase activity necessary for coordinated cell migration in vitro. Here, we discuss our recent work using FLIM-FRET analysis in a three-dimensional setting to reveal another layer of regulation in which RhoA activity is governed by the extracellular microenvironment. We demonstrate that RhoA is spatially regulated into discrete fractions of activity at the leading edge and rear of cells during invasion in vivo or within three-dimensional matrices. Significantly, this spatial regulation of RhoA was absent in two-dimensional in vitro settings. This distinct sub-cellular regulation of RhoA at the poles of invading cells in three-dimensions sets a precedent that other RhoGTPases or signaling proteins may also be differentially regulated in a con-text-dependent manner during key biological processes such as invasion.
Key words: intravital imaging, RhoGTPases, metastasis, invasion, FLIM, FRET, biosensor
Introduction
In the past 15 years, the application of Förster resonance energy transfer (FRET) for the in vitro study of molecular dynamics has considerably improved our understanding of spatially restricted and temporally regulated protein-protein interactions underlying the etiology of many diseases. FRET is a non-radiative process involving the transfer of energy from an excited donor fluorophore to a second acceptor fluorophore that lie within close proximity (∼10 nm) of each other. Using fluorescently tagged proteins, FRET has allowed the sub-cellular analysis of ‘when and where’ protein interactions may occur in live cells.
A frequent method used to measure FRET during protein-protein interactions is referred to as ratiometric FRET, where the ratio of both fluorescently tagged protein intensities is analyzed in a pixel by pixel manner over the course of the experiment. In this way, the activation and interaction of a number of key signaling proteins have been assessed at sub-cellular resolution in vitro.1 A key caveat to this technique is the requirement that similar and constant levels of each fluorophore must be present throughout the course of the experiment.2 To circumvent this, many experiments use single ‘intramolecular biosensor’ constructs in which the protein of interest is fused, via a linker, to an effector-domain. Upon activation of the biosensor, conformational changes in the sensor lead to the induction or loss of FRET between fluorophores which can be measured within the cell. In this way the activation of numerous signal transduction events have been tracked in live cells in vitro.1 More recently, changes in the fluorescence lifetime of the donor fluorophore induced upon FRET have been measured using time and frequency domain approaches.3 This method is insensitive to artifacts caused by differences or fluctuation in fluorophore levels and can therefore provide a more robust readout of FRET activity within the given specimen.
In vivo FRET Analysis
The employment of fluorescent proteins for imaging and tracking cell behavior in vivo has been applied to many multi-cellular model organisms including Drosophila, Zebrafish and Xenopus embryos. These models are better suited to intravital imaging than mammalian systems due to their transparent nature. However, the use of FRET or FLIM for in vivo or three-dimensional applications has only recently begun to emerge. For example, the chemokine-directed motility of germ cells during zebrafish embryonic development has been investigated using FRET.4 Here, the authors demonstrate the sub-cellular activation of Rac1 and RhoA at the front of migrating cells, where Rac1 functions to induce actin-rich structures while RhoA drives retrograde actin flow. Collectively, the formation and retrograde movement of these actin-rich structures facilitates traction forces between the germ cells and the surrounding tissue, via E-cadherin, allowing directed cell motility necessary for embryonic development.4 This work has implications regarding the synchronized and coordinated role of RhoGTPases in other fundamental three-dimensional processes involving both single and collective cell movement.
The relative ease of imaging in these model systems compared with mammalian models is partly due to the challenges of high autofluorescence, light scattering and poor tissue penetration found within mammalian tissue. Live animal imaging is also inherently demanding due to sample instability caused by movement of the heart, respiration system and muscle contractions. To overcome problems associated with sample instability, small incisions in the skin surrounding xenograft tumors are made and a ‘skin flap’ is created, allowing images to be acquired at a distance from the body of the mouse.5–7 Using this technique, images can be acquired over a short time period and an average readout of several parameters can be generated using a number of animals. Recently this approach has been used in combination with multiphoton-based FLIM-FRET for deep tissue imaging.8 Here, the investigators generated a series of z-stack sections ranging from 20 to 100 µm within the tumor tissue and provided accompanying lifetime maps representing the interaction between PKCα and the pro-inflammatory chemokine receptor CXCR4.8 This analysis, within a population of tumor cells, provides a representative image of protein-protein interplay governed by local environmental cues from the surface to the interior of the tumor. Similarly, FLIM-FRET has been used at the single cell level within a tumor population to assess chemotherapy-induced resistance to apoptosis.9
Analysis of RhoA Activity in vivo
Recently, we used FLIM-FRET, for the first time, at the sub-cellular level in vivo, to examine RhoA activity during cancer cell invasion,7 using a genetically defined live animal model of pancreatic cancer driven by mutations in Kras and p53.10,11 Since this model replicates human pancreatic tumorigenesis in terms of disease profile and metastatic burden12,13 it serves as an excellent system to examine the role of RhoA which is thought to play a vital role in mutant p53-driven invasion.14–17 Here, we specifically identified at high resolution a small yet important pool of active RhoA at the poles of invading cells, not observed in vitro, that correlates with invasion in vivo.7
Our initial finding that RhoA was spatially regulated during invasion was achieved using organotypic three-dimensional matrices involving co-culture of pancreatic tumor cells with fibrillar collagen I and fibroblasts. Organotypic matrices have previously been utilized to mimic cell-ECM interactions with regards to physical and mechanical forces driven by the elasticity of the fibrillar matrix.18,19 They also provide a source of growth factors and appropriate integrin engagement, resulting in bidirectional signaling between the cells and the surrounding stromal fibroblasts and ECM (see Fig. 1).20,21 This approach allowed the three-dimensional manipulation and fluorescent lifetime imaging of RhoA activity during invasion to be examined in an intermediate-system prior to full in vivo exploration of this biological process. Expansion of our initial findings led to the first use of FLIM-FRET to monitor molecular dynamics of RhoA in live animal tumors upon therapeutic intervention. In particular, dasatinib which shows anti-metastatic activity in this pancreatic model, inhibited RhoA activity at the poles of mutant p53 cells in vivo and this effect was independent of changes in basal activity within the cell body.7,22 Moreover, inhibition of this pool of RhoA activity correlated with invasion. These results demonstrate that RhoA is not only necessary for invasion, but suggest that sub-cellular spatial regulation of RhoA activity, as opposed to its global activity, may govern invasion efficiency in situ.
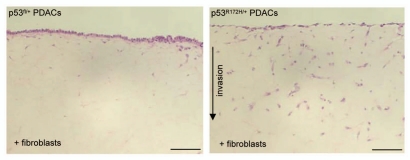
Figure 1.
Mutant p53R172H drives PDAC invasion. H&E stained sections of non-invasive p53fl and invasive p53R172H PDAC cells on organotypic matrix. Scale bar represent 100 µm.
Whether this polarized regulation of RhoA to the front or rear of cells is driving protrusion, retraction or both in vivo is currently unknown. This, however, could be assessed in the future by simultaneously imaging cancer cell invasion in live animals, in real-time using FLIM-FRET. Recently, we have performed real-time FLIM-FRET to examine the spatial and temporal regulation of RhoA activity on three-dimensional cell derived matrix (CDM).18 Similar to our initial observation in live animals, we cannot only demonstrate the spatial, but now the temporal regulation of RhoA during cell movement (Fig. 2). The capacity to quantitatively assess molecular behavior in live animals both temporally and spatially will aid in attributing key cellular modes of movement in vivo with signaling events and should assist the future development of site-specific drug targeting.
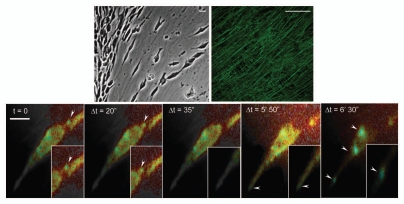
Figure 2.
Real-time FLIM-FRET imaging of RhoA activity during motility on 3D-matrix. Mutant p53R172H PDAC on cell derived matrix (CDM) expressing the Raichu-RhoA reporter with corresponding lifetime map of RhoA activity acquired in real-time. Blue represents the active biosensor and yellow is inactive. Scale bar represents 10 µm.
Interestingly, in our initial in vitro studies we also found that fluorescence lifetime imaging could accurately detect subtle changes in RhoA activity within a large tumor population, as small subpopulations of mutant p53R172H+/− cells proximal to a wound edge showed significant RhoA activation while cells at the rear of a wound demonstrated a low basal activity.7 In line with this, our preliminary in vivo findings using quantum dots to identify blood vessels show that in non-invasive p53fl/+cells there is a low basal level of RhoA activity independent of their location to the tumor blood supply (Fig. 3A–D). Invasive mutant p53R172H/+ PDAC cells however have significantly higher RhoA activity when in close proximity to the host vasculature compared with cells at distal sites (Fig. 3E). The enhanced activation of RhoA in invasive mutant p53R172H/+ cells proximal to the host vasculature (Fig. 3) and at the wound edge,7 suggests a gradient of activity may exist within the local environment of tumors.
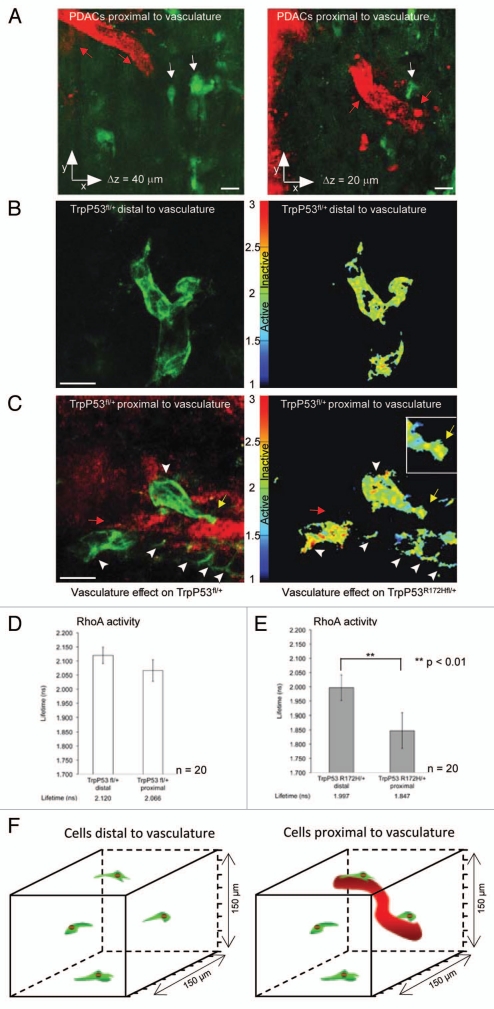
Figure 3.
Spatial regulation of RhoA activity within tumor population. (A) Representative image of cells (green) proximal to host vasculature (red). (B) p53fl PDAC cells distal to host vasculature expressing the Raichu-RhoA reporter (green) with corresponding in vivo lifetime map of RhoA activity, respectively. (C) p53fl PDAC cells expressing the Raichu-RhoA reporter (green) proximal to host vasculature (red) with corresponding in vivo lifetime map of RhoA activity, respectively. (D and E) Quantification of lifetime measurements of RhoA activity within the tumor environment. Lifetime pseudo-colors; blue represents active biosensor and yellow represents inactive biosensor. Figure partially reproduced from original Cancer Res article in reference 7. Scale bar, 30 µm. Columns, mean; bars, SE. **p < 0.01.
In this regard, it is feasible that a similar scenario could exist regarding drug targeting within the tumor environment, whereby response to therapeutic intervention may be affected by a similar spatial gradient. The investigation of drug response, at the single cell level, using FLIM-FRET could therefore be used to assess the efficiency of cancer drug delivery within different zones of the tumor. Akin to the vasculature governing drug delivery and targeting efficiency, the density and topography of the ECM that surrounds tumor cells has also recently been shown to contribute to the perfusion and access of drugs to the tumor tissue.23 Consistent with this, we find that not all cells in vivo are inactive upon dasatinib treatment.7 As tumor cells which are refractory to initial drug treatment may recolonise or progress to form micro-metastases, it would be beneficial to identify and isolate regions of poor drug delivery and response within various solid tumor environments using FLIM-FRET. We are currently using this technique to investigate aspects of the tumor environment that may contribute to poor drug targeting such as the density of ECM component surrounding the cells, their proximity to host vasculature or their orientation with regards to tumor cell cortex or border. This work could assist in the design, scheduling and stratification of future combination therapy to sensitize the tumor for efficient drug delivery.
Applications of FLIM or FRET for Drug Discovery
Using FLIM-FRET alone or multiplexed with additional functional reporters, the groups of French and Bastiaens have adapted Time domain or frequency domain based-FLIM, respectively, to allow for fast FLIM in a 96-well high-throughput format suitable for screening drug response or molecular profiling, such as the in situ identification of tyrosine phosphorylation networks in response to EGFR signaling.24,25 Adaptation of this type of high throughput FLIM-FRET imaging for three-dimensional settings may provide further context-dependent detail regarding drug response not feasible in vitro. In line with this, French and colleagues have also extended the application of lifetime imaging from cell based disease models to compare the intrinsic lifetime of matrix proteins within spatially distinct areas of human tumor samples, such as the cortex vs. interior of the tumor.26 This use of FLIM to document changes in the surrounding stromal environment has been recently utilized in human tissue microarrays (TMA) studies.8 Using FLIM in vivo to assess the interactions of molecules that are known to initiate and/or accelerate disease progression and that confer poor patient prognosis will rapidly enhance its development for the clinical setting.
The long time scale and sporadic nature of tumor dissociation and invasion has resulted in a need for methods to facilitate longer, non-invasive, repeated imaging of cell activity in vivo. The use of observation windows27 or dorsal skinfold chambers,28 in which tumors are established externally between glass and the underlying tissue, has partially helped in this respect. By applying controlled anesthesia and observation windows, repeated non-invasive single cell imaging in real-time is possible and has recently been used to examine cancer cell motility and intravasation over time.29 This approach has facilitated in the assessment of therapeutic intervention on many tumor types and has also been utilized to monitor the integrity of surrounding ECM components at different stages of treatment within the same animal.27,30,31 Moreover, the success of imaging injected cells expressing GFP-based reporters in vivo has resulted in a rapid expansion of genetically engineered mice that recapitulate human diseases, being crossed with tissue-specific fluorescent reporter mice.11,32–35 In this setting, the ability to image recombined fluorescent cells within the host tissue and organ specific micro-environment has lead to more faithful imaging of cell fate in vivo. In the future, the ability to combine genetically engineered mouse models expressing FRET biosensors with observation windows may allow for repeated non-invasive, longitudinal studies of specific molecular events that act as dynamic and quantitative in vivo biomarkers of drug response in the context of various genetically defined diseases.
Finally, the capacity to use multiphoton-based FLIM-FRET at depths of up to 150 µm to investigate RhoA activity during invasion in live animals and the added contextual detail this affords, highlight the need to advance FLIM-FRET imaging in the future by adapting FRET biosensors to incorporating the use of red-and far red FRET pairs. In this way, the reduced light scattering, autofluorescence and absorption by tissue found at longer wavelengths should provide a powerful tool for directly observing fundamental events at even greater depth in intact native tissue than can currently be achieved using the fluorescent lifetime of low wavelength biosensors such as the GFP-RFP based Raichu-RhoA probe reported here in reference 7.
Future Perspective
Our findings demonstrate the potential utility of FLIM-FRET in the analysis of dynamic biomarkers during the assessment of various therapeutic drug regimes. Such detailed analysis in vivo enables the detection and precise quantification of subtle changes in protein activity following therapeutic intervention that cannot always be detected in vitro. In this regard, the future application of complex three-dimensional in vitro and in vivo systems, which are amenable to molecular manipulation may improve our understanding of cell behavior in a more physiological and functional setting and therefore partially reduce the current attrition rate of new compounds entering clinical trials due to lack of appropriate initial experimental model systems in which to test drug efficacy.36
Acknowledgements
The authors would like to thank Dr. Haley Bennett for critical reading of the manuscript and Dr. Patrick Caswell for providing images of cell derived matrix. E.J.M., J.P.M., A.V.K., J.P.S., S.A.K., O.J.S., P.T. and K.I.A. were supported by a Cancer Research UK core grant. N.O.C was supported by a CRUK fellowship award.
Extra View to: Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 2011;71:747–757. doi: 10.1158/0008-5472.
References
- 1.Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Berney C, Danuser G. FRET or no FRET: a quantitative comparison. Biophys J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakowicz JR, Szmacinski H, Nowaczyk K, Berndt KW, Johnson M. Fluorescence lifetime imaging. Anal Biochem. 1992;202:316–330. doi: 10.1016/0003-2697(92)90112-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kardash E, Reichman-Fried M, Maître JL, Boldajipour B, Papusheva E, Messerschmidt EM, et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol. 2010;12:47–53. doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- 5.Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1:155–167. doi: 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serrels A, Timpson P, Canel M, Schwarz JP, Carragher NO, Frame MC, et al. Real-time study of E-cadherin and membrane dynamics in living animals: implications for disease modeling and drug development. Cancer Res. 2009;69:2714–2719. doi: 10.1158/00085472.CAN-08-4308. [DOI] [PubMed] [Google Scholar]
- 7.Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 2011;71:747–757. doi: 10.1158/00085472.CAN-10-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelleher MT, Fruhwirth G, Patel G, Ofo E, Festy F, Barber PR, et al. The potential of optical proteomic technologies to individualize prognosis and guide rational treatment for cancer patients. Target Oncol. 2009;4:235–252. doi: 10.1007/s11523-009-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keese M, Yagublu V, Schwenke K, Post S, Bastiaens P. Fluorescence lifetime imaging microscopy of chemotherapy-induced apoptosis resistance in a syngenic mouse tumor model. Int J Cancer. 2010;126:10413. doi: 10.1002/ijc.24730. [DOI] [PubMed] [Google Scholar]
- 10.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:46983. doi: 10.1016/j. ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/00085472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 13.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12:5277–5287. doi: 10.1158/10780432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 14.Mizuarai S, Yamanaka K, Kotani H. Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells. Cancer Res. 2006;66:6319–6326. doi: 10.1158/0008-5472.CAN-05-4629. [DOI] [PubMed] [Google Scholar]
- 15.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:132741. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Nalbant P, Chang YC, Birkenfeld J, Chang ZF, Bokoch GM. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol Biol Cell. 2009;20:4070–4082. doi: 10.1091/mbc.E09-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat Struct Mol Biol. 2007;14:215–223. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- 18.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 19.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 20.Edward M, Gillan C, Micha D, Tammi RH. Tumour regulation of fibroblast hyaluronan expression: a mechanism to facilitate tumour growth and invasion. Carcinogenesis. 2005;26:1215–1223. doi: 10.1093/carcin/bgi064. [DOI] [PubMed] [Google Scholar]
- 21.Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. http://dx.doi.org/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton JP, Karim SA, Graham K, Timpson P, Jamieson N, Athineos D, et al. Dasatinib inhibits the development of metastases in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2010;139:292–303. doi: 10.1053/j.gastro.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 23.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grecco HE, Roda-Navarro P, Girod A, Hou J, Frahm T, Truxius DC, et al. In situ analysis of tyrosine phosphorylation networks by FLIM on cell arrays. Nat Methods. 2010;7:467–472. doi: 10.1038/nmeth.1458. [DOI] [PubMed] [Google Scholar]
- 25.Talbot CB, McGinty J, Grant DM, McGhee EJ, Owen DM, Zhang W, et al. High speed unsupervised fluorescence lifetime imaging confocal multiwell plate reader for high content analysis. J Biophotonics. 2008;1:514–521. doi: 10.1002/jbio.200810054. [DOI] [PubMed] [Google Scholar]
- 26.Elson D, Requejo-Isidro J, Munro I, Reavell F, Siegel J, Suhling K, et al. Time-domain fluorescence lifetime imaging applied to biological tissue. Photochem Photobiol Sci. 2004;3:795–801. doi: 10.1039/b316456j. [DOI] [PubMed] [Google Scholar]
- 27.Brown E, McKee T, diTomaso E, Pluen A, Seed B, Boucher Y, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 28.Lehr HA, Leunig M, Menger MD, Nolte D, Messmer K. Dorsal skinfold chamber technique for intravital microscopy in nude mice. Am J Pathol. 1993;143:1055–1062. [PMC free article] [PubMed] [Google Scholar]
- 29.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canel M, Serrels A, Miller D, Timpson P, Serrels B, Frame MC, et al. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res. 2010;70:9413–9422. doi: 10.1158/00085472.CAN-10-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perentes JY, McKee TD, Ley CD, Mathiew H, Dawson M, Padera TP, et al. In vivo imaging of extra-cellular matrix remodeling by tumor-associated fibroblasts. Nat Methods. 2009;6:143–145. doi: 10.1038/nmeth.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed F, Wyckoff J, Lin EY, Wang W, Wang Y, Hennighausen L, et al. GFP expression in the mammary gland for imaging of mammary tumor cells in transgenic mice. Cancer Res. 2002;62:7166–7169. [PubMed] [Google Scholar]
- 33.Hoffman RM. Recent advances on in vivo imaging with fluorescent proteins. Methods Cell Biol. 2008;85:485–495. doi: 10.1016/S0091-679X(08)85021-2. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad I, Morton JP, Singh LB, Radulescu SM, Ridgway RA, Patel S, et al. beta-Catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene. 2011;30:178–189. doi: 10.1038/onc.2010.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole AM, Ridgway RA, Derkits SE, Parry L, Barker N, Clevers H, et al. p21 loss blocks senescence following Apc loss and provokes tumourigenesis in the renal but not the intestinal epithelium. EMBO Mol Med. 2010;2:472–486. doi: 10.1002/emmm.201000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamb A. What's wrong with our cancer models? Nat Rev Drug Discov. 2005;4:161–165. doi: 10.1038/nrd1635. [DOI] [PubMed] [Google Scholar]