Abstract
Bacteria belonging to the Vibrionaceae family are widespread in the marine environment. Today, 128 species of vibrios are known. Several of them are infamous for their pathogenicity or symbiotic relationships. Despite their ability to interact with eukaryotes, the vibrios are greatly underexplored for their ability to produce bioactive secondary metabolites and studies have been limited to only a few species. Most of the compounds isolated from vibrios so far are non-ribosomal peptides or hybrids thereof, with examples of N-containing compounds produced independent of nonribosomal peptide synthetases (NRPS). Though covering a limited chemical space, vibrios produce compounds with attractive biological activities, including antibacterial, anticancer, and antivirulence activities. This review highlights some of the most interesting structures from this group of bacteria. Many compounds found in vibrios have also been isolated from other distantly related bacteria. This cosmopolitan occurrence of metabolites indicates a high incidence of horizontal gene transfer, which raises interesting questions concerning the ecological function of some of these molecules. This account underlines the pending potential for exploring new bacterial sources of bioactive compounds and the challenges related to their investigation.
Keywords: Vibrio, marine bacteria, bioactive, antibiotics, siderophores
1. Introduction
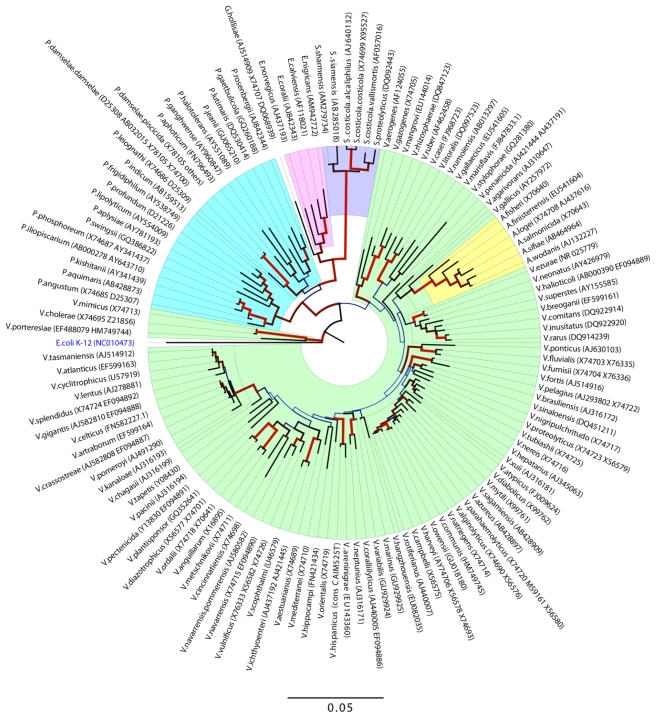
Marine Vibrionaceae are Gram-negative, rod-shaped γ-proteobacteria that are usually motile and possess a chemoheterotrophic metabolism [1]. Members of this family are widespread in the marine environment, including estuaries, coastal waters, and sediments [1]. At this writing, the family includes seven genera (Figure 1): Allivibrio (6 species), Enterovibrio (4 species), Salinivibrio (6 species), Catenococcus (1 species), Grimontia (1 species), Vibrio (89 species), and Photobacterium (21 species) [1,2].
Figure 1.
1.1. Occurrence and Ecological Significance
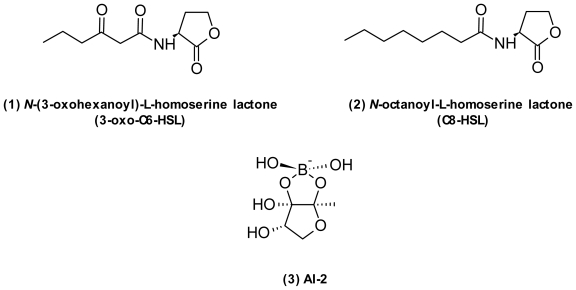
Vibrios are particularly abundant on the surface of marine macroorganisms such as corals, fish, seagrass, sponges, and zooplankton, where they form commensal, symbiotic, or pathogenic associations [1]. Excellent books and reviews have been published on the taxonomy [8], ecology [1,9,10], and pathogenesis of vibrios [11–14]. Several species are well studied and serve as model systems for understanding symbioses [15,16], interspecies signaling [17], and pathogen persistence [11,18]. A particularly famed vibrio is Vibrio fischeri, known for its light organ symbiosis with the Hawaiian squid Euprymna scolopes [15]. V. fischeri colonizes the squid light organ and provides bioluminescence for the squid to use as countershading in order to evade predators. In return, the bacteria gain a protected nutrient environment. The discovery of N-acylhomoserine lactones (Figure 2) as quorum sensing signals was first made in V. fischeri and the LuxI/R system of V. fischeri is the paradigm of Gram-negative QS systems even though it is not found in all vibrios [18]. In V. fischeri, there are three distinct QS signals; 3-oxo-C6-HSL (1), C8-HSL (2), and AI-2 (3). These are used to control a regulatory cascade leading to induction of luminescence. Vibrios also use QS to control traits such as virulence and biofilm formation [17].
Figure 2.
Structures common quorum sensing molecules from Vibrio sp.
The Vibrionaceae include species that are opportunistic pathogens of humans and marine animals. V. vulnificus, V. parahaemolyticus, and V. cholerae are serious human pathogens. V. cholerae probably has the greatest impact on human health, causing the acute diarrheal disease cholera that can result in epidemics [11]. It is a very persistent bacterium that can survive on a variety of vectors, including zooplankton [19] and cyanobacteria [18]. V. parahaemolyticus [20] and V. vulnificus [12] are food-borne pathogens associated with the ingestion of raw seafood. V. anguillarum, V. salmonicida, and V. vulnificus are important fish pathogens and are widespread in aquaculture settings, where conditions seem to enhance their virulence [1].
The ability to form biofilms is widespread among vibrios and plays a significant role in the pathogenicity of V. cholera [21], V. parahaemolyticus, and V. vulnificus [22], as well as in the symbiotic colonization by V. fischeri [23,24]. Key proteins include pili, lectins, exopolysaccharides, and components involved in the formation of flagella [22]. Though vibrios share a high number of regulatory systems of biofilm formation, there are differences that could reflect different niche specificity or ecological roles [25]. For example, it appears that vibrios produce species-specific exopolysaccharides, the major component of bacterial biofilms, and often have the potential to produce more than one type [22].
High densities of Vibrio and Photobacterium on the surface of zooplankton [26] have in part been ascribed to the ability of vibrios to utilize chitin, an N-acetyl d-glucosamine polymer in zooplankton exoskeletons, as carbon and nitrogen source [27]. The presence of chitinases and chitinase encoding genes has been confirmed for several members of the family [27,28]. Chitin controls several genetic and physiological characteristics of vibrios [19] including antagonistic activity [29]. Also, vibrios are able to degrade other complex carbohydrates such as fucoidan and laminarin found in algal species [29,30]. Thus, this superior nutrient utilization may be one of the reasons for the ubiquitous presence of vibrios in the marine environment [31].
1.2. Genomic Diversity and Phylogeny
In contrast to most γ-proteobacteria, vibrios possess two circular chromosomes [32,33]. Essential functions and housekeeping genes are usually located on the large chromosome ChrI, which is rather constant in size (~3 Mb), while the smaller ChrII is flexible in size, ranging between 0.8–2.4 Mb [9]. ChrII contains accessory genes related to transcriptional regulation, for example, pathogenicity and antimicrobial resistance [9,34]. Genes encoding chitin metabolism and quorum sensing are split between the two chromosomes [9]. The ability of vibrios to vary the copy numbers of the two chromosomes is suspected to be involved in the adaptation to varying environmental conditions [34]. Horizontal gene transfer is involved in the genetic flexibility of vibrios, including transduction by phages, plasmid conjugation [35], and so-called “super-integrons” [36,37]. In addition, Meibom et al. (2005) [38] showed that vibrios become naturally competent when grown in the presence of chitin, allowing uptake of free DNA from the environment. Chitin-induced competence has been demonstrated in V. cholera [38], V. vulnificus [39], and V. fischeri [40].
The high genomic diversity of vibrios can be directly translated into high phenotypic variability [41]. This makes it difficult to obtain meaningful groupings of vibrios at genus and species level based on isolated phenotypical markers [1]. Also, the 16S rRNA gene is highly conserved, and present in serveral alleles, among the Vibrionaceae and not well suited for identification to the species level [41]. Attempts to improve the taxonomy include sequencing and comparison of various housekeeping genes, including recA, rpoA, toxR, which hold greater sequence variability than 16S [41,42]. Taxonomy of vibrios by genetic markers has been supplemented by chemical analyses, including fatty acid methyl ester (FAME) profiling, and more recently by whole-cell MALDI-TOF MS [43,44], and LC-UV/MS chemical profiling [29,45]. Chemotyping was found to be especially useful at sub-species level, identifying differences in antibiotic production [29]. Whole-cell MALDI-TOF MS was able to distinguish closely related species like V. parahaemolyticus and V. alginolyticus or V. cholerae and V. mimicus scouting potential biomarkers within a 4000–14,000 Da mass range [43]. Closely related species (V. coralliilyticus and V. neptunius) could be distinguished based on their secondary metabolite production (Figure 3) [45]. Despite intra-species differences [29], the chemical profiles corroborated the phylogenetic relationship and clearly showed that production of secondary metabolites in vibrios is more than a strain specific trait.
Figure 3.
LC-MS profiles of a V. coralliilyticus (A) and V. neptunius (B), showing significant differences in secondary metabolite production. Andrimid (RT 10.02) was only found in V. coralliilyticus strains. Figure modified from Wietz et al. (2010) [45].
2. Natural Product Production by Members of the Vibrionaceae Family
Considering their widespread presence in the marine environment and their genomic flexibility, vibrios are largely underexplored for their proclivity to produce secondary metabolites. So far, a total of 93 compounds have been isolated from Vibrionaceae. The majority of these compounds have been isolated from only three species; V. parahaemolyticus, V. anguillarum, and V. vulnificus, which is likely a reflection of their importance as pathogens. In the following, all metabolites reported from Vibrionaceae (Table 1) will be presented and interesting compounds highlighted in an attempt to give an overview of the chemical diversity and assess the biosynthetic potential of this group of bacteria.
Table 1.
Bioactive compounds produced by marine Vibrionaceae. Excluded from the list are sugars, fatty acids, and small peptides commonly found in marine culturable bacteria. Excluded are also compounds from AntiBase 2010 whose presence could not be confirmed in any reference referring to Vibrionaceae.
| Bioactivities | Name | Compound class | Source | Other activities | Ref. |
|---|---|---|---|---|---|
| Antibacterial | Andrimid (4) | Pyrrolidinedione | V. coralliilyticus | [45,46] | |
| Aqabamycin A (6) | Nitro maleimide | Vibrio sp. | Anticancer | [47,48] | |
| Aqabamycin B (7) | |||||
| Aqabamycin C (8) | |||||
| Aqabamycin D (9) | |||||
| Aqabamycin E (10) | Maleimide oxime | ||||
| Aqabamycin E’ (11) | |||||
| Aqabamycin F (12) | |||||
| Aqabamycin G (13) | Nitro maleimide | ||||
| B-4607-C | Phenazine | Vibrio sp. | [49] | ||
| Cycloprodigiosin (15) | Prodiginine | V. gazogenes | [50] | ||
| 3,5-Dibromo-2-(3′,5′-dibromo-2′-methoxyphenoxy)-phenol | Diphenyl ether | Vibrio sp. | Antifungal | [51,52] | |
| 2,2-Di-(3-indolyl)-3-indolone | Indole | V. parahaemolyticus | [53,54] | ||
| Griseoluteic acid | Phenazine | Vibrio sp. | [49] | ||
| Holomycin (5) | Pyrrothine | P. halotolerans | [45] | ||
| Indazole-3-carbaldehyde | Indazole | Vibrio sp. | Anticancer | [47] | |
| Magnesidin A (16) | Tetramic acid Mg2+ salt | V. gazogenes | Antialgal | [55] | |
| Moiramide B | Pyrrolidinedione | Vibrio sp. | [56] | ||
| Ngercheumicin A (19) | Depsipeptide | Photobacterium sp. | [57] | ||
| Ngercheumicin B (20) | |||||
| Ngercheumicin C (21) | |||||
| Ngercheumicin D (22) | |||||
| Ngercheumicin E (23) | |||||
| Pelagiomicin C | Phenazine | Vibrio sp. | Anticancer | [49,58,59] | |
| Prodigiosin (14) | Prodiginine |
V. psychroerythrus V. gazogenes V. ruber |
Antiprotozoan antifungal anticancer | [60–62] | |
| Turbomycin | Indole | Vibrio sp. (V. parahaemolyticus) | Antifungal | [54] | |
| Unnarmicin A (17) | Depsipeptide | Photobacterium sp. | Antifungal | [63] | |
| Unnarmicin C (18) | |||||
| Vibrindole A | Indole | V. parahaemolyticus | Antifungal | [53] | |
| Siderophore | Anguibactin (24) | Catechol hydroxamate | V. anguillarum | Anticancer | [64,65] |
| Aerobactin | Hydroxamate | Vibrio sp. | [66] | ||
| Amphibactin B | Hydroxamate (amphiphilic) | Vibrio sp. | [67] | ||
| Amphibactin C | |||||
| Amphibactin D | |||||
| Amphibactin E | |||||
| Amphibactin F | |||||
| Amphibactin G | |||||
| Amphibactin H | |||||
| Amphibactin I | |||||
| Bis-[3-(2,3-dihydroxybenzoylamino)-propyl]-amin | Catechol | V. fluvialis | [68] | ||
| Bisucaberin (29) | Hydroxamate | V. salmonicida | Anticancer | [69,70] | |
| Divanchrobactin | Catechol | Vibrio sp. | [65] | ||
| Fluvibactin (28) | Catechol Hydroxyphenyloxazolone | V. fluvialis | [66] | ||
| Trivanchrobactin | Catechol | Vibrio sp. | [65] | ||
| Vanchrobactin (25) | Catechol | V. anguillarum | [71] | ||
| Vibriobactin (27) | Catechol Hydroxyphenyloxazolone | V. cholerae | [72] | ||
| Vibrioferrin | Carboxylate | V. parahaemolyticus | [73] | ||
| Vulnibactin (26) | Catechol Hydroxyphenyloxazolone | V. vulnificus | [74] | ||
| Vulnibactin 2 | Vulnibactin precursor | ||||
| Vulnibactin 3 | |||||
| Anticancer | Kahalalide F (30) | Depsipeptide | V. mediterranei (V. shilonii) | Antibacterial antimalarial antifungal | [75] |
| Kahalalide H | [76] | ||||
| Kahalalide J | |||||
| Quorum sensing interference | AI-2 (3) | Furanosyl borate diester | Vibrio | QS | [76] |
| N-hexanoyl-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [77] | |
| N-(3-hydroxybutanoyl)-l-homoserine lactone | Homoserine lactone | V. harveyi | QS | [78] | |
| N-(3-hydroxyhexanoyl)-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [79] | |
| [1-(2′-methylpropoxy)-2-hydroxy-2-methylpropoxy] butane (41) | P. angustum (V. angustum) | QS | [80] | ||
| N-(3-oxodecanoyl)-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [81] | |
| N-(3-oxohexanoyl)-l-homoserine lactone (1) | Homoserine lactone |
V. fischeri V. cholerae V. harveyi V. anguillarum |
QS | [17,82] | |
| N-octanoyl-l-homoserine lactone (2) | Homoserine lactone | V. fischeri | QS | [77] | |
| Solonamide A (32) | Depsipeptide | P. halotolerans | QSI Gram pos | [83] | |
| Solonamide B (33) | |||||
| Na channel blocker | Anhydro-tetrodotoxin | Vibrio sp. | [84,85] | ||
| 4-epi-tetrodotoxin | Vibrio sp. | [84,85] | |||
| Tetrodonic acid | Vibrio sp. | [85,86] | |||
| Tetrodotoxin (31) |
V. harveyi V. alginolyticus V. fischeri |
[84,85,87] | |||
| Riboflavin synthase inhibitor | 7-hydroxy-6-methyl-8-(1-d-ribityl)lumazine | Pteridine | P. phosphoreum | [88] | |
| Photolumazine A | |||||
| Photolumazine B | |||||
| Photolumazine C | |||||
| Misc. | Arundine | Indole | V. parahaemolyticus | [54] | |
| Benzoic acid | Aromatic | Vibrio sp. | [47] | ||
| 3,3-Bis-(3-indolyl)butan-2-one | Indole | V. parahaemolyticus | [54] | ||
| 3,3′-Bisindolylmethane | |||||
| 1,4-dithiane | Vibrio sp. | [47] | |||
| 3-hydroxybenzoic acid | Aromatic | Vibrio sp. | [47] | ||
| 4-hydroxycinnamic acid | |||||
| p-Hydroxyphenyl-acetamide | Aromatic | V. parahaemolyticus | [54] | ||
| Indole-3-carboxaldehyde | Indole | V. parahaemolyticus | [53] | ||
| Indole-3-acetic acid | Indole | Vibrio sp. | [89] | ||
| 6-methyl-8-d-ribityl-2,4,7-trioxopteridine | Pteridine | P. phosphoreum | [90] | ||
| 3-nitro-4-hydroxy-benzaldehyde | Nitro aromatic | Vibrio sp. | [47] | ||
| 3-nitro-4-hydroxycinnamic acid | |||||
| 3-nitro-1H-indazole | |||||
| Pharacine (43) | Terephthalic ester | V. parahaemolyticus | [54] | ||
| Phenylacetic acid | Aromatic | Vibrio sp. | [47] | ||
| Phenyl-2-bis-indolylmethane | Indol | ||||
| Photopterin A | Pteridine | P. phosphoreum | [90] | ||
| 8-d-ribityl-2,4,7-trioxopteridine | |||||
| Trisindoline | Indole | V. parahaemolyticus | [54] | ||
| 1,1,3-Tris-(3-indolyl)butane | |||||
| 1,1′,1″-Trisindolyl-methane (42) |
2.1. Compounds with Antibacterial Activity
Some marine vibrios produce antibacterial compounds [91–93] that are believed to contribute to their abundance in surface-associated communities [94]. Long and Azam (2001) [92] studied anta-gonistic interactions among pelagic bacteria and found that vibrios produced broad-range antibacterial compounds. Similar capabilities have been observed for coral-associated vibrios [95]. Yet, none of these compounds were isolated and structurally characterized. The relatively widespread production of antibiotics in marine vibrios [45] indicates that antagonistic activity is of ecological importance [29].
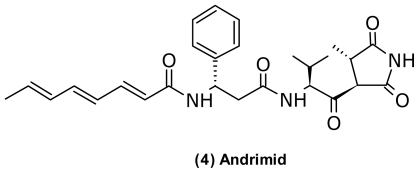
Probably the best studied antibiotic produced by vibrios is the hybrid NRPS-PKS peptide antibiotic andrimid (4) (Figure 4) [46]. The compound interferes with fatty acid biosynthesis [96] and is effective against a wide range of bacteria [97]. Structure-activity studies by Pohlmann et al. (2005) [96] revealed that the pyrrolidinedione head was essential for activity, while variations in the fatty acid tail were more tolerable. This suggested that these two structural moieties play different roles in target binding.
Figure 4.
Structure of andrimid isolated from Vibrio coralliilyticus.
Andrimid is a cosmopolitan antibiotic found in distantly related bacteria, including a symbiotic Enterobacter sp. from the planthopper Nilaparvata lugens [98], Pseudomonas flourescens [99], Pantoea agglomerans [100], and Vibrio sp. [45,46]. The andrimid biosynthetic gene cluster was conserved in two different producers [101]. Interestingly, the cluster encodes resistance genes [102] as well as specific transposases that could be responsible for the diverse occurrence of this antibiotic [100,101]. From Vibrio species, the compound was first isolated by Oclarit et al. in 1994 [46], and Long et al. (2005) [93] identified andrimid as the compound responsible for the growth inhibition of V. cholerae by an unidentified Vibrio strain. Production of andrimid was for the first time linked to a specific vibrio species by Wietz et al. (2010) [45] that isolated the compound from the culture broth of a V. coralliillyticus strain S2052. Within V. coralliilyticus, the production of andrimid is a marker of different chemotypes [29]. Two V. coralliilyticus strains S2052 and S4053 from two distant geographical locations produced andrimid [45], while the type strain and a close relative did not [29]. Interestingly, V. coralliilyticus S2052 focused its production of secondary metabolites to the production of andrimid when grown on chitin and also increased the yield of the antibiotic [29]. The bacterium was capable of producing andrimid in a live chitin model system with Artemia [29]. This indicated that andrimid potentially contributes to different niche-specificities of V. coralliilyticus.
Another example of cosmopolitan antibiotics from Vibrionaceae is the highly potent pyrrothine antibiotic, holomycin (5) (Figure 5) isolated from a strain closely related to Photobacterium halotolerans [45]. Prior to this isolation, holomycin had only been isolated from actinomycetes, including Streptomyces clavuligerus [103], S. griseus [104], and a marine Streptomyces sp. [105].
Figure 5.

Structure of holomycin isolated from Photobacterium halotolerans.
The NRPS biosynthetic cluster encoding holomycin in S. clavuligerus was recently identified by Li and Walsh (2010) [106], and this allows for the comparison of the holomycin clusters in other producers, including Photobacterium. Holomycin has a broad spectrum of antibacterial activity against pathogenic bacteria such as Staphylococcus aureus, S. pneumoniae, S. epidermis, Enterococcus faecalis, and Escherichia coli [107]. The mode-of-action in E. coli includes inhibition of RNA chain elongation, but holomycin is suspected to act as prodrug rather than a direct inhibitor of the RNA polymerase [107]. Holomycin is also strongly inhibitory against several marine strains from the Roseobacter-clade, Pseudoalteromonas, and Vibrio, including pathogens such as V. harveyi, V. vulnificus, and V. parahaemolyticus [45], altogether suggesting that holomycin plays a role in antagonism in the marine environment.
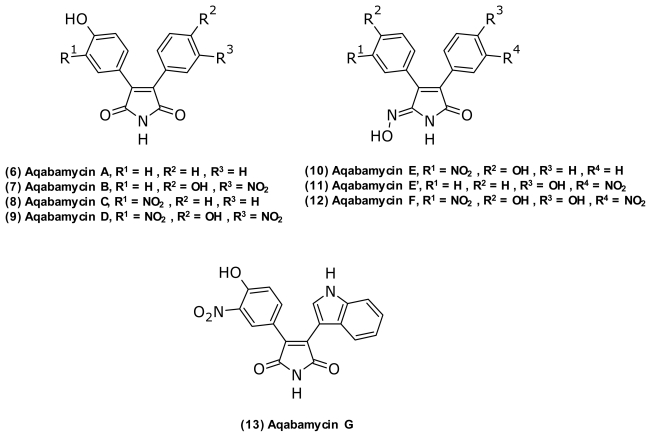
Yao and Al-Zereini recently (2010) [47,48] isolated a series of nitrosubstrituted maleimides called aqabamycins (6–13) (Figure 6) from a coral-associated Vibrio sp. The analogues had varying antibacterial activity against Gram-positive bacteria, including Micrococcus luteus, Bacillus subtilis, and B. brevis as well as cytotoxic activity [48]. The aqabamycins represent a unique structural group both due to their high degree of nitrosubstitution which is rare in nature [108] and the maleimide monoxime present in aqabamycin E/E′ (10–11) and F (12).
Figure 6.
Structures of aqabamycin A–G isolated from coral-associated Vibrio sp.
The red pigment and antibiotic prodigiosin (14) (Figure 7) has been isolated from V. psychroerythreus [60], V. gazogenes (originally termed Beneckea gazogenes but later revised) [61], and V. ruber [62]. Additional producers of this compound include Alteromonas rubra/Pseudoalteromonas rubra [109,110], Hahella chejuensis [111], and different Serratia [112], and Streptomyces species [113,114]. Prodigiosin and its cyclized analogue (15) [50,115] have a broad range of biological activities, including antimicrobial, antimalarial, immunosuppressive, and anticancer [116–118]. Prodiginines have clinical potential in anticancer therapy [118], and prodigiosin is currently in preclinical trials (Aida Pharmaceuticals) for pancreatic cancer [116]. The clinical potential as antibiotics is, however, limited due to a low therapeutic window and considerable toxic effects [119]. Starič et al. (2010) [120] recently demonstrated that the production of prodiginines in a Vibrio sp. isolated from estuaries conferred competitiveness against a Bacillus sp. from the same sample, suggesting that prodigiosin might act as a antibiotic in the natural environment. Interestingly, the prodigiosin producing V. gazogenes also produced the unique magnesium containing antibiotic, magnesidin (16) (Figure 7) [55,121–123].
Figure 7.
Structures of prodigiosins and magnesidin.
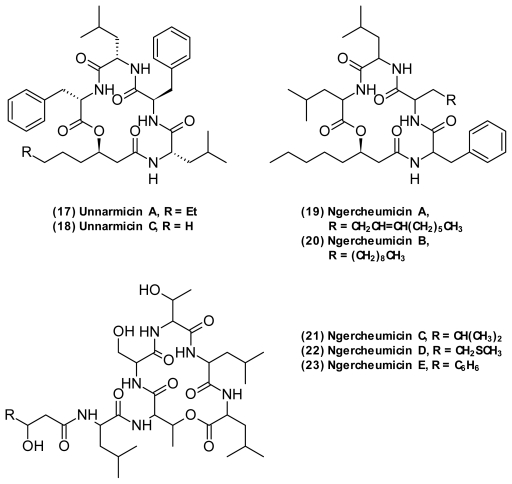
Shizuri and co-workers isolated two distinct groups of depsipeptides (Figure 8), the unnarmicins [63] and ngercheumicins [57] from a Photobacterium sp. with potent, but narrow-spectrum antibacterial effect against strains of Pseudovibrio. The unnarmicin A (17) and C (18) consist of four amino acids (l-Phe, l-Leu, d-Phe, l-Leu) and a 3-hydroxyoctanoic and 3-hydroxyhexanoic fatty acid, respectively. The ngercheumicins A–E have a depsipeptide macrocycle and either a fatty acid (19–20) or peptide tail (21–23). They have been patented for treating infections caused by Pseudovibrio denitrificans, though no literature describes pathogenic traits of this bacterium [124].
Figure 8.
Cyclodepsipeptides isolated from Photobacterium sp.
2.2. Siderophores
Many vibrios produce siderophores as a strategy to sequester iron in the marine environment, where the iron level is extremely low [125,126]. This is necessary to maintain important enzymatic processes (with iron as cofactor) and a prerequisite for pathogenicity for many vibrios. It should be mentioned that siderophores also may have antibacterial activity but are dealt with in a separate section due to their specific metabolic function.
A great structural diversity has been observed among the siderophores produced by Vibrio species (Figure 9). V. anguillarum has at least two different siderophore-mediated systems, namely anguibactin (24) [64,127] and vanchrobactin (25) [128]. The non-ribosomal peptide anguibactin represents a unique structural class of siderophores with both a catechol and hydroxamate ligand and a thiazole core [64]. The biosynthetic genes encoding this compound are found on a 65-kb virulence plasmid in some V. anguillarum strains. Knock-out of genes involved in anguibactin production attenuated virulence, confirming that anguibactin is a prerequisite for successful host-invasion of this bacterium [129]. In contrast, the catechol vanchrobactin is chromosome-encoded, and interestingly, the coding genes are silenced in anguibactin producing strains [71,130]. Recently, dimeric and trimeric versions of vanchrobactin were isolated from an unidentified Vibrio by Sandy et al. (2010) [65]. Also, they found anguibactin to possess cytotoxic activities against P388 murine leukemia cells [65].
Figure 9.
Siderophores isolated from Vibrio sp.
Vibriobactin (27) [72], vulnibactin (26) [74], and fluvibactin (28) (Figure 9) [68] are unique siderophores produced by V. cholerae, V. vulnificus, and V. fluvialis, respectively. They are all catechol hydroxyphenyloxalone siderophores that share a rare norspermidine backbone, giving them a propeller-like structure. In vibriobactin and vulnibactin, two of the hydroxybenzoyl moieties are linked to the backbone through an l-threonine, forming an oxazoline ring. Fluvibactin only has one oxazoline ring, with one hydroxybenzoyl directly linked to the norspermidine terminal. Vibriobactin and vulnibactin differ only in the number of hydroxylations, and this high structural similarity enables cross-utilization of these two siderophores [74].
Bisucaberin (29) (Figure 9) [69] is a symmetric cyclic dihydroxamate produced by the fish pathogen V. salmonicida [70]. Unlike most other vibrio siderophores [129], bisucaberin is produced through an NRPS-independent route [131] where alternating dicarboxylic acids and diamine or amino alcohols are assembled through amide or ester bonds [132]. Bisucaberin was found to be useful in combinatorial anticancer therapy by sensitizing tumor cells to macrophage-mediated cytolysis [69,133].
Siderophores are used as therapeutic deferration agents to treat iron overload in chronically transfused thalassemia patients. A stereochemically modified version of fluvibactin efficiently removed iron without increasing microbial growth [134]. It has been suggested that siderophores can be used for the development of a new class of “trojan horse” antibiotics [135]. Siderophore-antibiotic conjugates exploit the iron transport system of the pathogen to penetrate the bacterial outer membrane, increasing the antibacterial activity of the antibiotic [136]. Recently, Bergeron et al. (2009) [137] made a conjugate linking antibodies to vulnibactin as a strategy towards a vaccine against V. vulnificus.
2.3. Compounds with Other Activities
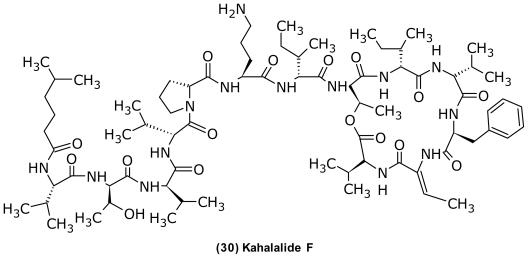
Another interesting group of compounds produced by a member of the Vibrionaceae is the kahalalides. These cyclic depsipeptides were originally isolated from the herbivorous mollusc Elysia refescens and its diet, the green algae Bryopsis sp. In particular, kahalalide F (30) (Figure 10) has an attractive spectrum of activities against AIDS-related opportunistic infections and against cancer cell lines [138]. Kahalalide F is currently undergoing Phase II clinical trials (PharmaMar) for the treatment of prostate, lung, and liver cancer [138] and in patients with severe psoriasis (PharmaMar/Marinomed) [139]. Interestingly, Hill and Hamann (2005) [75] found kahalalide F as well as two analogues to be produced by a V. mediterranei/shilonii. The finding of a microbial origin for this compound allows for the large-scale industrial fermentation of this compound rather than arduous organic synthesis.
Figure 10.
Structure of kahalalide F isolated from Vibrio mediterranei/shilonii.
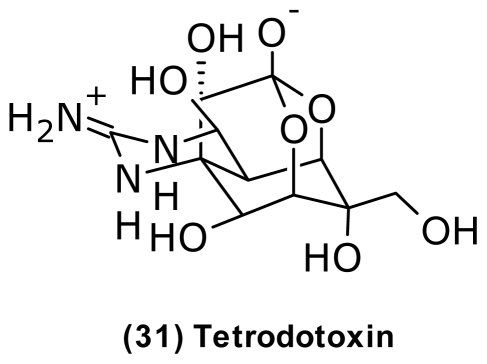
Several vibrios produce the potent neurotoxin tetrodotoxin (TTX) (31) (Figure 11), also known as the pufferfish poison [85]. The true origin of TTX has been the subject of much debate [140], nonetheless V. harveyi and V. alginolyticus isolated from different species of pufferfish produced the toxin as well as several analogues [84]. Also, V. fischeri isolated from the intestines of the xanthid crab, Atergatis floridus produced TTX [87]. Vibrios dominated the intestinal microbiota of the pufferfish, Fugu vermicularis vermicularis [84], suggesting that the toxification is caused by TTX-producing bacteria accumulated through the food web [85]. The role of these compounds to vibrio itself is still unclear, though it has been suggested to play a role in regulating sodium transport [85].
Figure 11.
Structure of tetrodotoxin isolated from Vibrio harveyi and Vibrio alginolyticus.
Vibrios produce compounds that interfere with the quorum sensing system of Gram-positive bacteria. From a strain related to P. halotolerans two novel depsipeptides (Figure 12), solonamides A and B (32–33) that interfere with QS regulated virulence genes in S. aureus were isolated [83]. In particular, solonamide B dramatically reduced expression of both hla encoding α-hemolysin and RNAIII, while increasing expression of spa encoding Protein A. This suggested that the depsipeptides interfere with agr, the global virulence regulator in S. aureus. High structural similarity of the solonamides to the natural autoinducers of the agr system suggested that they might be competitive inhibitors. Interestingly, the solonamides had a pronounced effect on virulence gene expression in S. aureus strain USA300, which is the predominant community-acquired MRSA (CA-MRSA) strain in the USA [141]. The solonamides strongly resemble the unnarmicins (17–18) found in an unidentified Photobacterium sp. (Section 2.3.1) [63]. Thus, it is possible that the unnarmicins also possess QSI activity.
Figure 12.
Structures of solonamides isolated from Photobacterium halotolerans related strain.
Several small molecules isolated from vibrios induce Gram-negative quorum sensing systems. That includes various diketopiperazines (DKP) (Figure 13). For example, cyclo(l-Pro, l-Leu) (34), cyclo(l-Pro, l-Val) (35), and cyclo(l-Pro, l-Tyr) (36), DKPs commonly isolated from vibrios [53], modulated LuxR-type protein activity though at higher concentrations than AHLs [142]. It is speculated that these dipeptides represent a new class of naturally occurring QS signals potentially involved in interspecies signaling, as DKPs are found in most culturable marine bacteria [143]. However, some DKPs are likely to be artifacts generated from media components during work-up procedures [144]. De Nys et al. (2001) [80] isolated [1-(2′-methylpropoxy)-2-hydroxy-2-methylpropoxy]-butane (37) (Figure 13) from P. angustum (V. angustum) S14 with the ability to mediate expression in two AHL-regulated systems, inducing bioluminescence in V. harveyi and the AHL reporter system in Agrobacterium tumefaciens.
Figure 13.
Structures of common diketopiperazines from Vibrio sp. and [1-(2′-methylpropoxy)-2-hydroxy-2-methylpropoxy]-butane.
2.4. Compounds with Unknown Activities
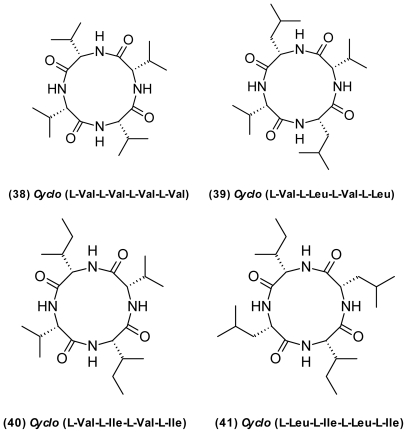
Vibrios also produce numerous compounds for which no biological activity has been reported so far. That includes small-molecule by-products, for example some nitro-substituted compounds such as 3-nitroindazole and 3-nitro-4-hydroxycinnamic acid [47]. From P. halotolerans S2753, we isolated a series of cyclic tetrapeptides (Figure 14); cyclo(l-Val-l-Val-l-Val-l-Val) (38), cyclo(l-Val-l-Leu-lVal- l-Leu) (39), cyclo(l-Val-l-Ile-l-Val-l-Ile) (40), and cyclo(l-Leu-l-Ile-l-Leu-l-Ile) (41) (Kjaerullf and Mansson, unpublished data). These types of peptides are often found in marine culturable bacteria [145–147], suggesting that they are storage compounds accumulated during growth under excess nutrients.
Figure 14.
Structures of cyclotetrapeptides isolated from Photobacterium.
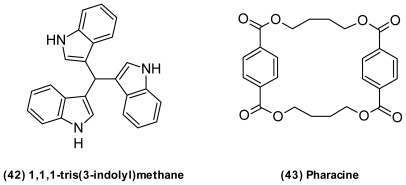
Many of compounds isolated from vibrios are suspected to be artifacts generated from media components during work-up procedures [54,144,148]. These include several bis- and trisindole derivatives from a V. parahaemolyticus strain, Bio249 [54]. An example is 1,1,1-tris (3-indolyl) methane (42) (Figure 15) that could easily be formed by simple condensation of indole-3-carbaldehyde and indole, both having been isolated from V. parahaemolyticus [54]. From the same V. parahaemolyticus strain, the cyclic terephthalic acid ester, pharacine (43) [148] was isolated [54]. This was suspected to be an artifact from plastic material contaminants; however, fermentation results were reproducible with no contact with plastic. Until biosynthetic studies have been performed, the true origin of these molecules remains uncertain.
Figure 15.
Structures of 1,1,1-tris (3-indolyl) methane and pharacine, examples of potential artefacts from work-up of Vibrio extracts.
3. Conclusion
The versatility and widespread occurrence of vibrios can be ascribed to different characteristics such as their superior nutrient utilization, their excellent biofilm formation, and their genetic construction. High genomic flexibility in Vibrionaceae makes this group of bacteria very apt to resist various environmental changes, for example through the acquisition of biosynthetic genes linked to the production of antibiotics or siderophores. So, rampant horizontal gene transfer occurs in these bacteria. As a reflection, most compounds isolated from vibrios have also been found in other types of bacteria, in many cases from distantly related taxa [45]. Sometimes the mobile genetic elements are even incorporated in the biosynthetic cluster itself, making it even more prone to gene-exchange. The antibiotic andrimid is an example of a compound encoded by such a “nomadic gene cluster” [149].
Production of secondary metabolites in vibrios has been linked to antagonism, intraspecies communication, and pathogenicity. The compounds produced by vibrios are mainly non-ribosomal peptides or hybrids hereof, with examples of N-containing compounds produced by NRPS-independent pathways. Despite this narrow structural span compared to metabolites produced by other marine bacteria, vibrios produce compounds with a broad range of interesting biological activities. For example the solonamides, cyclic depsipeptides from P. halotolerans were found to attenuate virulence in a CA-MRSA strain [83] and the cyclic depsipeptide kahalalide F from V. medierranei [75] that is undergoing Phase II clinical trials for the treatment of prostate, lung, and liver cancer [138].
Many vibrios have multiple lifestyles, including a planktonic (free swimming), sessile (attached to zooplankton or other surfaces), and a pathogenic form [1]. As production of secondary metabolites often confers a selective advantage to the producing organism [150], the diverse lifestyles of these bacteria are reflected in their metabolic capabilities. There are intraspecies variations in the compounds produced, with different chemotypes potentially reflecting niche adaptation. For example, antagonistic strains of V. coralliilyticus were found to produce andrimid in high yields, while pathogenic related strains did not have the ability to produce the antibiotic [29].
The cosmopolitan occurrence of several vibrio metabolites raises the question whether there are unique Vibrionaceae metabolites. Of the 227 vibrio genomes sequenced so far [2,151], only a fraction has been fully assembled [33], mainly pathogenic V. cholerae strains [9], and none have been functionally annotated with regard to the presence of biosynthetic clusters. Thus, it is still uncertain whether these bacteria represent a novel “hotspot” of secondary metabolites. For the future, it will be of utmost interest to extend full-genome sequencing to other vibrios and investigate the prevalence of biosynthetic genes linked to secondary metabolism. Also, this will make it possible to compare homology of biosynthetic genes between diverse producers of cosmopolitan antibiotics. Overall, this will allow insight into the ecological roles of these bacteria and the environmental and physiological parameters governing production of their secondary metabolites.
Acknowledgments
We thank Matthias Wietz for critical reading of the manuscript and Bruno Gòmez-Gil from the Association of Vibrio Biologist for making Figure 1. Funding from the Programme Committee for Food, Health and Welfare under the Danish Strategic Research Council is acknowledged. The present work was carried out as part of the Galathea 3 expedition under the auspices of the Danish Expedition foundation. This is Galathea 3 contribution P83.
References and Notes
- 1.Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell R, Swings J, Thompson FL. Association of Vibrio Biologists (AViB) [accessed on 16 August 2011]. Available online: http://www.vibriobiology.net/
- 3.Evolutionary relationship of Vibrionaceae species. The evolutionary history was inferred using the Neighbor-Joining method [4]. The optimal tree with the sum of branch length = 1.64144427 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the brances [5]. The tree is drawn to scale, with branch lengths in the same units as those if the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method [6] and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.46). The analysis involved 127 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1596 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [7]. Dendogram was made with the Figtree v. 1.3.1 program (http://tree.bio.ed.ac.uk/) where line color and width are related to the bootstrap support.
- 4.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Jukes TH, Cantor CR. In: Mammalian Protein Metabolism. Munro HN, editor. Academic Press; New York, NY, USA: 1969. pp. 21–132. [Google Scholar]
- 7.Tamura K, Peterson D, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson CC, Vicente ACP, Souza RC, Vasconcelos ATR, Vesth T, Alves N, Ussery DW, Iida T, Thompson FL. Genomic taxonomy of vibrios. BMC Evol. Biol. 2009;9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimes DJ, Johnson CN, Dillon KS, Flowers AR, Noriea NF, Berutti T. What genomic sequence information has revealed about vibrio ecology in the ocean—A review. Microb. Ecol. 2009;58:447–460. doi: 10.1007/s00248-009-9578-9. [DOI] [PubMed] [Google Scholar]
- 10.Colwell R. Vibrios in the Environment. John Wiley & Sons, Inc; Hoboken, NJ, USA: 1984. [Google Scholar]
- 11.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linkous DA, Oliver JD. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 1999;174:207–214. doi: 10.1111/j.1574-6968.1999.tb13570.x. [DOI] [PubMed] [Google Scholar]
- 13.Blake PA, Weaver RE, Hollis DG. Diseases of humans (other than cholera) caused by Vibrios. Annu. Rev.Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 14.Thompson FL. The Biology of Vibrios. ASM Press; Washington, DC, USA: 2006. [Google Scholar]
- 15.Ruby EG. Lessons from a cooperative, bacterial-animal association: The Vibrio fischeri Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 16.Nyholm SV, McFall-Ngai MJ. The Winnowing: Establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 17.Milton DL. Quorum sensing in vibrios: Complexity for diversification. Int. J. Med. Microbiol. 2006;296:61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 18.Islam MS, Mahmuda S, Morshed MG, Bakht HBM, Khan MNH, Sack RB, Sack DA. Role of Cyanobacteria in the Persistence of Vibrio cholerae O139 in saline microcosms. Can. J. Microbiol. 2004;50:127–131. doi: 10.1139/w03-114. [DOI] [PubMed] [Google Scholar]
- 19.Pruzzo C, Vezzulli L, Colwell RR. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 2008;10:1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 20.Su YC, Liu CC. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007;24:549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Faruque SM, Biswas K, Udden SMN, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. Transmissibility of cholera: In vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yildiz FH, Visick KL. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator rscs controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayles KW. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 26.Heidelberg JF, Heidelberg KB, Colwell RR. Bacteria of the gamma-subclass proteobacteria associated with zooplankton in chesapeake bay. Appl. Environ. Microbiol. 2002;68:5498–5507. doi: 10.1128/AEM.68.11.5498-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt DE, Gevers D, Vahora NM, Polz MF. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 2008;74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svitil AL, Chadhain SMN, Moore JA, Kirchman DL. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wietz M, Mansson M, Gram L. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ Microbiol Rep. 2011 doi: 10.1111/j.1758-2229.2011.00259.x. 10.11117j.1758-2229.2011.00259.x. [DOI] [PubMed] [Google Scholar]
- 30.Goecke F, Labes A, Wiese J, Imhoff JF. chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 2010;409:267–299. [Google Scholar]
- 31.Riemann L, Azam F. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 2002;68:5554–5562. doi: 10.1128/AEM.68.11.5554-5562.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. J. Bacteriol. 2005;187:752–757. doi: 10.1128/JB.187.2.752-757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirkup BC, Chang LA, Chang S, Gevers D, Polz MF. Vibrio chromosomes share common history. BMC Microbiol. 2010;10:137. doi: 10.1186/1471-2180-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagomori K, Iida T, Honda T. comparison of genome structures of vibrios, bacteria possessing two chromosomes. J. Bacteriol. 2002;184:4351–4358. doi: 10.1128/JB.184.16.4351-4358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazen TH, Pan L, Gu JD, Sobecky PA. The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol. Ecol. 2010;74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 36.Rowe-Magnus DA, Guerout AM, Mazel D. Super-integrons. Res. Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 37.Mazel D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 38.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 39.Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. User friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 2009;75:4936–4949. doi: 10.1128/AEM.02564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollack-Berti A, Wollenberg MS, Ruby EG. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. 2010;12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, Munn CB, Swings J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascual J, Macian MC, Arahal DR, Garay E, Pujalte MJ. Multilocus sequence analysis of the central clade of the genus vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 2010;60:154–165. doi: 10.1099/ijs.0.010702-0. [DOI] [PubMed] [Google Scholar]
- 43.Dieckmann R, Strauch E, Alter T. Rapid identification and characterization of vibrio species using whole-cell MALDI-TOF mass spectrometry. J. Appl. Microbiol. 2010;109:199–211. doi: 10.1111/j.1365-2672.2009.04647.x. [DOI] [PubMed] [Google Scholar]
- 44.Hazen TH, Martinez RJ, Chen YF, Lafon PC, Garrett NM, Parsons MB, Bopp CA, Sullards MC, Sobecky PA. Rapid identification of Vibrio parahaemolyticus by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2009;75:6745–6756. doi: 10.1128/AEM.01171-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs. 2010;8:2946–2960. doi: 10.3390/md8122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oclarit JM, Okada H, Ohta S, Kaminura K, Yamaoka Y, Iizuka T, Miyashiro S, Ikegami S. Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios. 1994;78:7–16. [PubMed] [Google Scholar]
- 47.Yao CBFF, Al Zereini W, Fotso S, Anke H, Laatsch H. Aqabamycins A–G: Novel nitro maleimides from a marine Vibrio species: II. Structure elucidation. J. Antibiot. 2010;63:303–308. doi: 10.1038/ja.2010.35. [DOI] [PubMed] [Google Scholar]
- 48.Al-Zereini W, Yao CBFF, Laatsch H, Anke H. Aqabamycins A–G: Novel nitro maleimides from a marine Vibrio species: I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2010;63:297–301. doi: 10.1038/ja.2010.34. [DOI] [PubMed] [Google Scholar]
- 49.Sato A. Annual Report Sankyo Research Laboratories. Daiichi Sankyo; Tokyo, Japan: 1995. [Google Scholar]
- 50.Gerber NN. Cycloprodigiosin from Beneckea Gazogenes. Tetrahedron Lett. 1983;24:2797–2798. [Google Scholar]
- 51.Elyakov GB, Kuznetsova T, Mikhailov VV, Maltsev II, Voinov VG, Fedoreyev SA. Brominated diphenyl ethers from a marine bacterium associated with the sponge Dysidea sp. Experientia. 1991;47:632–633. [Google Scholar]
- 52.Sionov E, Roth D, Sandovsky-Losica H, Kashman Y, Rudi A, Chill L, Berdicevsky I, Segal E. Antifungal effect and possible mode of activity of a compound from the marine sponge Dysidea herbacea. J. Infect. 2005;50:453–460. doi: 10.1016/j.jinf.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 53.Bell R, Carmeli S, Sar N. Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod. 1994;57:1587–1590. doi: 10.1021/np50113a022. [DOI] [PubMed] [Google Scholar]
- 54.Veluri R, Oka I, Wagner-Döbler I, Laatsch H. New indole alkaloids from the north sea bacterium Vibrio parahaemolyticus Bio249. J. Nat. Prod. 2003;66:1520–1523. doi: 10.1021/np030288g. [DOI] [PubMed] [Google Scholar]
- 55.Imamura N, Adachi K, Sano H. Magnesidin A, a component of marine antibiotic magnesidin, produced by Vibrio gazogenes Atcc29988. J. Antibiot. 1994;47:257–261. doi: 10.7164/antibiotics.47.257. [DOI] [PubMed] [Google Scholar]
- 56.Laatsch H. AntiBase 2010. Wiley-VCH; Weinheim, Germany: 2010. [Google Scholar]
- 57.Adachi K, Kawabata Y, Kasai H, Katsuta M, Shizuri Y. Novel ngercheumicin or its salt useful for treating infection caused by Pseudovibrio denitrificans. JP2007230911-A. Patent. 2007 Sep;
- 58.Imamura N, Nishijima M, Takadera T, Adachi K, Sakai M, Sano H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J. Antibiot. 1997;50:8–12. doi: 10.7164/antibiotics.50.8. [DOI] [PubMed] [Google Scholar]
- 59.Singh MP, Menendez AT, Petersen PJ, Ding WD, Maiese WM, Greenstein M. Biological and mechanistic activities of phenazine antibiotics produced by culture LL-14I352. J. Antibiot. 1997;50:785–787. doi: 10.7164/antibiotics.50.785. [DOI] [PubMed] [Google Scholar]
- 60.Daoust JY, Gerber NN. Isolation and purification of prodigiosin from Vibrio psychroerythrus. J. Bacteriol. 1974;118:756–757. doi: 10.1128/jb.118.2.756-757.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harwood CS. Beneckea gazogenes sp. nov., a red, facultatively anaerobic, marine bacterium. Curr. Microbiol. 1978;1:233–238. [Google Scholar]
- 62.Shieh WY, Chen YW, Chaw SM, Chiu HH. Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol. 2003;53:479–484. doi: 10.1099/ijs.0.02307-0. [DOI] [PubMed] [Google Scholar]
- 63.Oku N, Kawabata K, Adachi K, Katsuta A, Shizuri Y. Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J. Antibiot. 2008;61:11–17. doi: 10.1038/ja.2008.103. [DOI] [PubMed] [Google Scholar]
- 64.Jalal MAF, Hossain MB, Vanderhelm D, Sandersloehr J, Actis LA, Crosa JH. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J. Am. Chem. Soc. 1989;111:292–296. [Google Scholar]
- 65.Sandy M, Han A, Blunt J, Munro M, Haygood M, Butler A. Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. J. Nat. Prod. 2010;73:1038–1043. doi: 10.1021/np900750g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haygood MG, Holt PD, Butler A. Aerobactin production by a planktonic marine Vibrio sp. Limnol. Oceanogr. 1993;38:1091–1097. [Google Scholar]
- 67.Martinez JS, Carter-Franklin JN, Mann EL, Martin JD, Haygood MG, Butler A. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc. Natl. Acad. Sci. USA. 2003;100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto S, Okujo N, Fujita Y, Saito M, Yoshida T, Shinoda S. Structures of two polyamine containing catecholate siderophores from Vibrio fluvialis. J. Biochem. 1993;113:538–544. doi: 10.1093/oxfordjournals.jbchem.a124079. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi A, Nakamura H, Kameyama T, Kurasawa S, Naganawa H, Okami Y, Takeuchi T, Umezawa H. Bisucaberin, a new siderophore, sensitizing tumor-cells to macrophage-mediated cytolysis. 2. Physicochemical properties and structure determination. J. Antibiot. 1987;40:1671–1676. doi: 10.7164/antibiotics.40.1671. [DOI] [PubMed] [Google Scholar]
- 70.Winkelmann G, Schmid DG, Nicholson G, Jung G, Colquhoun DJ. Bisucaberin—A dihydroxamate siderophore isolated from Vibrio salmonicida, an important pathogen of farmed Atlantic salmon (Salmo salar) Biometals. 2002;15:153–160. doi: 10.1023/a:1015206419613. [DOI] [PubMed] [Google Scholar]
- 71.Soengas RG, Anta C, Espada A, Paz V, Ares IR, Balado M, Rodriguez J, Lemos ML, Jimenez C. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett. 2006;47:7113–7116. [Google Scholar]
- 72.Griffiths GL, Sigel SP, Payne SM, Neilands JB. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 73.Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S. structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem. 1994;115:868–874. doi: 10.1093/oxfordjournals.jbchem.a124432. [DOI] [PubMed] [Google Scholar]
- 74.Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7:109–116. doi: 10.1007/BF00140480. [DOI] [PubMed] [Google Scholar]
- 75.Hill RT, Hamann MT, Enticknap JJ, Rao KV. PCT/US2004/036201. [accessed on 16 August 2011];Kahalalide-Producing Bacteria. 2005 May; Available online: http://www.wipo.int/patentscope/search/en/WO2005042720.
- 76.Chen X, Schauder S, Potier N, van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 77.Kuo A, Blough NV, Dunlap PV. Multiple N-acyl-l-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 1994;176:7558–7565. doi: 10.1128/jb.176.24.7558-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 79.Milton DL, Chalker VJ, Kirke D, Hardman A, Camara M, Williams P. The luxM homologue vanM from Vibrio anguillanrum directs the synthesis of N-(3-hydroxyhexanoyl) homoserine Lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 2001;183:3537–3547. doi: 10.1128/JB.183.12.3537-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Nys R, Kumar N, Sharara KA, Srinivasan S, Ball G, Kjelleberg S. A new metabolite from the marine bacterium Vibrio angustum S14. J. Nat. Prod. 2001;64:531–532. doi: 10.1021/np000512r. [DOI] [PubMed] [Google Scholar]
- 81.Milton DL, Hardman A, Camara M, Chhabra SR, Bycroft BW, Stewart GSAB, Williams P. Quorum sensing in Vibrio anguillarum: Characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 83.Mansson M, Nielsen A, Kjærulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine Photobacterium. Mar Drugs. 2011 doi: 10.3390/md9122537. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noguchi T, Hwang DF, Arakawa O, Sugita H, Deguchi Y, Shida Y, Hashimoto K. Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu-Vermicularis vermicularis. Mar. Biol. 1987;94:625–630. [Google Scholar]
- 85.Lee MJ, Jeong DY, Kim WS, Kim HD, Kim CH, Park WW, Park YH, Kim KS, Kim HM, Kim DS. A tetrodotoxin-producing Vibrio strain, LM-1, from the puffer fish Fugu vermicularis radiatus. Appl. Environ. Microbiol. 2000;66:1698–1701. doi: 10.1128/aem.66.4.1698-1701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noguchi T, Ali AE, Arakawa O, Miyazawa K, Kanoh S, Shida Y, Nishio S, Hashimoto K. Tetrodonic acid-like substance-a possible precursor of tetrodotoxin. Toxicon. 1991;29:845–855. doi: 10.1016/0041-0101(91)90221-c. [DOI] [PubMed] [Google Scholar]
- 87.Noguchi T, Jeon JK, Arakawa O, Sugita H, Deguchi Y, Shida Y, Hashimoto K. Occurrence of tetrodotoxin and anhydrotetrodotoxin in Vibrio sp isolated from the intestines of a xanthid crab, Atergatis floridus. J. Biochem. 1986;99:311–314. doi: 10.1093/oxfordjournals.jbchem.a135476. [DOI] [PubMed] [Google Scholar]
- 88.Suzuki A, Goto M. Photolumazines, new naturally occurring inhibitors of riboflavin synthetase. Biochim. Biophys. Acta. 1973;313:229–234. doi: 10.1016/0304-4165(73)90205-5. [DOI] [PubMed] [Google Scholar]
- 89.Gutierrez CK, Matsui GY, Lincoln DE, Lovell CR. Production of the phytohormone indole-3-acetic acid by estuarine species of the genus Vibrio. Appl. Environ. Microbiol. 2009;75:2253–2258. doi: 10.1128/AEM.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matsuura S, Odaka M, Sugimoto T, Goto T. The structure of pteridines from Photobacterium phosphorium. Chem. Lett. 1973;2:343–346. [Google Scholar]
- 91.Gram L, Melchiorsen J, Bruhn JB. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 2010;12:439–451. doi: 10.1007/s10126-009-9233-y. [DOI] [PubMed] [Google Scholar]
- 92.Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Long RA, Rowley DC, Zamora E, Liu JY, Bartlett DH, Azam F. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl. Environ. Microbiol. 2005;71:8531–8536. doi: 10.1128/AEM.71.12.8531-8536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 96.Pohlmann J, Lampe T, Shimada M, Nell PG, Pernerstorfer J, Svenstrup N, Brunner NA, Schiffer G, Freiberg C. Pyrrolidinedione derivatives as antibacterial agents with a novel mode of action. Bioorg. Med. Chem. Lett. 2005;15:1189–1192. doi: 10.1016/j.bmcl.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 97.Singh MP, Mroczenski-Wildey MJ, Steinberg DA, Andersen RJ, Maiese WM, Greenstein M. Biological activity and mechanistic studies of andrimid. J. Antibiot. 1997;50:270–273. [PubMed] [Google Scholar]
- 98.Fredenhagen A, Tamura SY, Kenny PTM, Komura H, Naya Y, Nakanishi K, Nishiyama K, Sugiura M, Kita H. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J. Am. Chem. Soc. 1987;109:4409–4411. [Google Scholar]
- 99.Needham J, Kelly MT, Ishige M, Andersen RJ. Andrimid and moiramides A–C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: Structure elucidation and biosynthesis. J. Org. Chem. 1994;59:2058–2063. [Google Scholar]
- 100.Jin M, Fischbach MA, Clardy J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu XY, Fortin PD, Walsh CT. Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic. Proc. Natl. Acad. Sci. USA. 2008;105:13321–13326. doi: 10.1073/pnas.0806873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kenig M, Reading C. Holomycin and an antibiotic (Mm-19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J. Antibiot. 1979;32:549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 104.Ettlinger L, Gaumann E, Hutter R, Kellerschierlein W, Kradolfer F, Neipp L, Prelog V, Zahner H. Stoffwechselprodukte von actinomyceten. 17. Holomycin. Helv. Chim. Acta. 1959;42:563–569. [Google Scholar]
- 105.Hou YH, Li FC, Wang SJ, Qin S, Wang QF. Intergeneric conjugation in holomycin-producing marine Streptomyces sp. strain M095. Microbiol. Res. 2008;163:96–104. doi: 10.1016/j.micres.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 106.Li B, Walsh CT. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. USA. 2010;107:19731–19735. doi: 10.1073/pnas.1014140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oliva B, O’Neill A, Wilson JM, O’Hanlon PJ, Chopra I. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob. Agents Chemother. 2001;45:532–539. doi: 10.1128/AAC.45.2.532-539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Winkler R, Hertweck C. Biosynthesis of nitro compounds. ChemBioChem. 2007;8:973–977. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 109.Vynne NG, Mansson M, Nielsen KF, Gram L. Bioactivity, chemical profiling, and 16S rRNA based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol. 2011 doi: 10.1007/s10126-011-9369-4.. [DOI] [PubMed] [Google Scholar]
- 110.Gerber NN, Gauthier MJ. New prodigiosin-like pigment from Alteromonas rubra. Appl. Environ. Microbiol. 1979;37:1176–1179. doi: 10.1128/aem.37.6.1176-1179.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim D, Lee JS, Park YK, Kim JF, Jeong H, Oh TK, Kim BS, Lee CH. Biosynthesis of antibiotic prodiginines in the marine bacterium Hahella chejuensis KCTC 2396. J. Appl. Microbiol. 2007;102:937–944. doi: 10.1111/j.1365-2672.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- 112.Bennett JW, Bentley R. Seeing red: The story of prodigiosin. Adv. Appl. Microbiol. 2000;47:1–32. doi: 10.1016/s0065-2164(00)47000-0. [DOI] [PubMed] [Google Scholar]
- 113.Tsao SW, Rudd BAM, He XG, Chang CJ, Floss HG. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 1985;38:128–131. doi: 10.7164/antibiotics.38.128. [DOI] [PubMed] [Google Scholar]
- 114.Gerber NN, Lechevalier MP. Prodiginine (prodigiosin-like) pigments from Streptomyces and other aerobic Actinomycetes. Can. J. Microbiol. 1976;22:658–667. doi: 10.1139/m76-097. [DOI] [PubMed] [Google Scholar]
- 115.Laatsch H, Thomson RH. A revised structure for cycloprodigiosin. Tetrahedron Lett. 1983;24:2701–2704. [Google Scholar]
- 116.Pandey R, Chander R, Sainis KB. Prodigiosins as anti cancer agents: Living upto their name. Curr. Pharm. Des. 2009;15:732–741. doi: 10.2174/138161209787582192. [DOI] [PubMed] [Google Scholar]
- 117.Perez-Tomas R, Vinas M. New insights on the antitumoral properties of prodiginines. Curr. Med. Chem. 2010;17:2222–2231. doi: 10.2174/092986710791331103. [DOI] [PubMed] [Google Scholar]
- 118.Williamson NR, Fineran PC, Gristwood T, Chawrai SR, Leeper FJ, Salmond GPC. Anticancer and immunosuppressive properties of bacterial prodiginines. Future Microbiol. 2007;2:605–618. doi: 10.2217/17460913.2.6.605. [DOI] [PubMed] [Google Scholar]
- 119.Furstner A. Chemistry and biology of roseophilin and the prodigiosin alkaloids: A survey of the last 2500 years. Angew. Chem. Int. Ed. 2003;42:3582–3603. doi: 10.1002/anie.200300582. [DOI] [PubMed] [Google Scholar]
- 120.Staric N, Danevcic T, Stopar D. Vibrio sp. DSM 14379 pigment production—a competitive advantage in the environment? Microb. Ecol. 2010;60:592–598. doi: 10.1007/s00248-010-9671-0. [DOI] [PubMed] [Google Scholar]
- 121.Gandhi NM, Nazareth J, Divekar PV, Kohl H, Desouza NJ. Magnesidin, a novel magnesium-containing antibiotic. J. Antibiot. 1973;26:797–798. doi: 10.7164/antibiotics.26.797. [DOI] [PubMed] [Google Scholar]
- 122.Bhat SV, Kohl H, Ganguli BN, Desouza NJ. Magnesidin-related tetramic acids-synthesis and structural requirements for antibacterial activity. Eur. J. Med. Chem. 1977;12:53–57. [Google Scholar]
- 123.Kohl H, Bhat SV, Patell JR, Gandhi NM, Nazareth J, Divekar PV, Souza NJD, Berschei HG, Fehlhabe HW. Structure of magnesidin, a new magnesium-containing antibiotic from Pseudomonas magnesiorubra. Tetrahedron Lett. 1974;12:983–986. [Google Scholar]
- 124.Shieh WY, Lin YT, Jean WD. Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int. J. Syst. Evol. Microbiol. 2004;54:2307–2312. doi: 10.1099/ijs.0.63107-0. [DOI] [PubMed] [Google Scholar]
- 125.De Carvalho CCCR, Fernandes P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs. 2010;8:705–727. doi: 10.3390/md8030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hider RC, Kong XL. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 127.Actis LA, Fish W, Crosa JH, Kellerman K, Ellenberger SR, Hauser FM, Sandersloehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(Pjm1) J. Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lemos ML, Balado M, Osorio CR. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: Evolution and ecology of a fish pathogen. Environ. Microbiol. Rep. 2010;2:19–26. doi: 10.1111/j.1758-2229.2009.00103.x. [DOI] [PubMed] [Google Scholar]
- 129.Di Lorenzo M, Poppelaars S, Stork M, Nagasawa M, Tolmasky ME, Crosa JH. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibio anguillarum. J. Bacteriol. 2004;186:7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Naka H, Lopez CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 2008;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kadi N, Song LJ, Challis GL. Bisucaberin biosynthesis: An adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem Commun. 2008:5119–5121. doi: 10.1039/b813029a. [DOI] [PubMed] [Google Scholar]
- 132.Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 133.Kameyama T, Takahashi A, Kurasawa S, Ishizuka M, Okami Y, Takeuchi T, Umezawa H. Bisucaberin, a new siderophore, sensitizing tumor-cells to macrophage-mediated cytolysis. 1. Taxonomy of the producing organism, isolation and biological properties. J. Antibiot. 1987;40:1664–1670. doi: 10.7164/antibiotics.40.1664. [DOI] [PubMed] [Google Scholar]
- 134.Bergeron RJ, Xin MG, Weimar WR, Smith RE, Wiegand J. Significance of asymmetric sites in choosing siderophores as deferration agents. J. Med. Chem. 2001;44:2469–2478. doi: 10.1021/jm010019s. [DOI] [PubMed] [Google Scholar]
- 135.Miller MJ, Malouin F. Siderophore-Mediated Drug Delivery: The Design, Synthesis, and Study of Siderophore-Antibiotic and Antifungal Conjugates. In: Bergeron RJ, Brittenham GM, editors. The Development of Iron Chelators for Clinical Use. CRC Press; Boca Raton, FL, USA: 1994. pp. 275–306. [Google Scholar]
- 136.Wittmann S, Schnabelrauch M, Scherlitz-Hofmann I, Mollmann U, Ankel-Fuchs D, Heinisch L. New synthetic siderophores and their beta-lactam conjugates based on diamino acids and dipeptides. Bioorg. Med. Chem. 2002;10:1659–1670. doi: 10.1016/s0968-0896(02)00044-5. [DOI] [PubMed] [Google Scholar]
- 137.Bergeron RJ, Bharti N, Singh S, McManis JS, Wiegand J, Green LG. Vibriobactin antibodies: A vaccine strategy. J. Med. Chem. 2009;52:3801–3813. doi: 10.1021/jm900119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hamann MT. Technology Evaluation: Kahalalide F, PharmaMar. Curr. Opin. Mol. Ther. 2004;6:657–665. [PMC free article] [PubMed] [Google Scholar]
- 139.PharmaMar. PharmaMar Licenses Analogs of Kahalalide F to Marinomed for Uses Outside of Oncology/Neurology. [accessed on 19 August 2011]. Available online: http://www.pharmamar.com/pdf/MarinomedMAR_ENG.pdf.
- 140.Matsumura K. Reexamination of tetrodotoxin production by bacteria. Appl. Environ. Microbiol. 1995;61:3468–3470. doi: 10.1128/aem.61.9.3468-3470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother. 2009;64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 142.Campbell J, Lin Q, Geske GD, Blackwell HE. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 2009;4:1051–1059. doi: 10.1021/cb900165y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Unson MD, Faulkner DJ. Cyanobacterial symbiont biosynthesis of chlorinated metabolites from Dysidea herbacea (Porifera) Experientia. 1993;49:349–353. [Google Scholar]
- 144.Laatsch H. In: Frontier in Marine Biotechnology. Proksch P, editor. Horizon Bioscience; Norfolk, UK: 2006. pp. 225–288. [Google Scholar]
- 145.Mitova M, Popov S, De Rosa S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod. 2004;67:1178–1181. doi: 10.1021/np049900+. [DOI] [PubMed] [Google Scholar]
- 146.Rungprorn W, Siwu ERO, Lambert LK, Dechsakulwatana C, Barden MC, Kokpol U, Blanchfield JT, Kita M, Garson MJ. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron. 2008;64:3147–3152. [Google Scholar]
- 147.Shin J, Seo Y, Lee HS, Rho JR, Mo SJ. A new cyclic peptide from a marine-derived bacterium of the genus Nocardiopsis. J. Nat. Prod. 2003;66:883–884. doi: 10.1021/np030075r. [DOI] [PubMed] [Google Scholar]
- 148.Shaaban M, Maskey RP, Wagner-Döbler I, Laatsch H. Pharacine, a natural p-cyclophane and other indole derivatives from Cytophaga sp. strain AM13.1. J. Nat. Prod. 2002;65:1660–1663. doi: 10.1021/np020019a. [DOI] [PubMed] [Google Scholar]
- 149.Fischbach MA. Antibiotics from microbes: Converging to kill. Curr. Opin. Microbiol. 2009;12:520–527. doi: 10.1016/j.mib.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Firn RD, Jones CG. A darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot. 2009;60:719–726. doi: 10.1093/jxb/erp002. [DOI] [PubMed] [Google Scholar]
- 151.Entrez Genome Project Database. National Center of Biotechnology; [accessed on 16 August 2011]. Available online: http://www.ncbi.nlm.nih.gov/sites/genome. [Google Scholar]