Abstract
According to the “generic view” of protein aggregation, the ability to self-assemble into stable and highly organized structures such as amyloid fibrils is not an unusual feature exhibited by a small group of peptides and proteins with special sequence or structural properties, but rather a property shared by most proteins. At the same time, through a wide variety of techniques, many of which were originally devised for applications in other disciplines, it has also been established that the maintenance of proteins in a soluble state is a fundamental aspect of protein homeostasis. Taken together, these advances offer a unified framework for understanding the molecular basis of protein aggregation and for the rational development of therapeutic strategies based on the biological and chemical regulation of protein solubility.
Proteins are present at concentrations at which they are only marginally soluble. Stress or aging can lead to widespread aggregation.
Virtually every complex biochemical process taking place in living cells depends on the ability of the molecules involved to self-assemble into functional structures (Dobson 2003; Robinson et al. 2007; Russel et al. 2009), and a sophisticated quality control system is responsible for regulating the reactions leading to this organization within the cellular environment (Dobson 2003; Balch et al. 2008; Hartl and Hayer-Hartl 2009; Powers et al. 2009; Vendruscolo and Dobson 2009). Proteins are the molecules that are essential for enabling, regulating, and controlling almost all the tasks necessary to maintain such a balance. To function, the majority of our proteins need to fold into specific three-dimensional structures following their biosynthesis in the ribosome (Hartl and Hayer-Hartl 2002). The wide variety of highly specific structures that results from protein folding, and which serve to bring key functional groups into close proximity, has enabled living systems to develop an astonishing diversity and selectivity in their underlying chemical processes by using a common set of just 20 basic molecular components, the amino acids (Dobson 2003). Given the central importance of protein folding, it is not surprising that the failure of proteins to fold correctly, or to remain correctly folded, is at the origin of a wide variety of pathological conditions, including late-onset diabetes, cystic fibrosis, and Alzheimer’s and Parkinson’s diseases (Dobson 2003; Chiti and Dobson 2006; Haass and Selkoe 2007). In many of these disorders proteins self-assemble in an aberrant manner into large molecular aggregates, notably amyloid fibrils (Chiti and Dobson 2006; Ramirez-Alvarado et al. 2010).
PROTEIN FOLDING AND MISFOLDING
In the late 1990s, experimental data started to become available, suggesting that the amyloid state is not simply associated with a few disease-related proteins, but it is a “generic” state accessible in principle to essentially all proteins (Guijarro et al. 1998; Dobson 1999). In addition, because the types of structural interactions within the amyloid state and the native state are similar, their thermodynamic stabilities are likely to be comparable under many circumstances (Dobson 2003; Knowles et al. 2007a). On this assumption, there will be a competition between these two states that results in normal or aberrant biological behavior depending on whether the native or the aggregated state is populated under given circumstances. More generally, the maintenance of the correct balance in the populations of different states of proteins is of great significance, as even marginal alterations in such populations can result in disease in the long term (Dobson 2003; Balch et al. 2008; Geiler-Samerotte et al. 2011). Indeed, it has been recently shown that the limit to the “safe” concentration of proteins in living systems is reached when the stability of the alternative aggregated state becomes comparable to that of the native state (Tartaglia et al. 2007; Powers et al. 2009). It is therefore of great importance to complement the well-established characterization of the structure, folding, and stability of native states with a similar analysis of the structure, assembly, and stability of other states—ranging from unfolded and partially folded species, including natively unfolded states, to aggregated species such as amyloid fibrils (Fig. 1). One particularly effective approach has been to incorporate data from nuclear magnetic resonance (NMR) spectroscopy and other experiments in molecular dynamics simulations (Vendruscolo et al. 2001; Lindorff-Larsen et al. 2005; Vendruscolo and Dobson 2005; Gianni et al. 2010).
Figure 1.
Protein behavior in the cell. Protein homeostasis refers to the ability of cells to generate and regulate the levels of their constituent proteins in terms of conformations, interactions, concentrations, and cellular localization. Failure to fold, or to remain correctly folded, can result in misfolding and aggregation, which are processes associated with a wide range of human disorders, including Alzheimer’s and Parkinson’s diseases (Dobson 2003).
There is an increasing interest in developing an understanding of how proteins fold correctly to generate normal biological function, or misfold to generate disease. To achieve this objective, it is necessary to create highly interdisciplinary research environments that use wide ranges of advanced procedures, both experimental and theoretical (Morimoto 2008; Hartl and Hayer-Hartl 2009; Powers et al. 2009). In our own group we initially used these methods in the fields of biophysics and structural biology (Dobson 2003; Jaroniec et al. 2004; Carulla et al. 2005, 2009; Smith et al. 2006; Knowles et al. 2007a, 2009; Orte et al. 2008); they have now been extended into biochemistry and cell biology (Chiti et al. 1999; Fandrich et al. 2001; de la Paz et al. 2002; Dumoulin et al. 2003; Dedmon et al. 2005; Wright et al. 2005; Baglioni et al. 2006), and the results have led to fundamentally new ideas about the nature of protein structures, how they have evolved to be as they are, and why they are now increasingly associated with emerging diseases in the modern world (Chiti and Dobson 2006). Most recently, these various ideas have been tested in living systems by using model organisms such as Caenorhabditis elegans (Roodveldt et al. 2009) and Drosophila melanogaster (Luheshi et al. 2007, 2010). These developments, as well as many others (Balch et al. 2008; Hartl and Hayer-Hartl 2009; Powers et al. 2009), have made it possible to begin to relate in detail the mechanisms of protein folding and misfolding, uncovered from in vitro studies, with the origins and progression in vivo of the most common and debilitating neurodegenerative diseases. In addition to promoting a deeper understanding of these rapidly proliferating disorders, such an understanding will form a novel and robust basis for new approaches to their prevention and therapy (Dobson 2004).
PROTEIN AGGREGATION AND DISEASE
Strategies based on biophysical approaches are likely to play an increasingly important role in the study of protein homeostasis (Fig. 1). This view has been prompted by the realization, made through initial observations of protein aggregation by us and by others, that the kinetics of this phenomenon are of critical importance but have great complexity (Harper and Lansbury 1997; Kelly 1998; Serio et al. 2000; Dobson 2003; Haass and Selkoe 2007; Knowles et al. 2009). The experience we gained in our studies of protein folding, in which biophysical and theoretical methods have together been crucial to establish a firm understanding of this process (Wolynes et al. 1995; Dobson et al. 1998), suggested that to make advances in the field of protein aggregation, similar strategies were needed. It is now important to bring together methods of biochemistry and cell biology with those of physical chemistry and biophysics, to develop approaches for observing the fundamental molecular events in the process of aggregation, and thus be in a position to carry out a quantitative analysis of the cellular and organismal responses associated with this phenomenon.
A particularly clear example of the great potential of this approach has been the study of the microscopic processes underlying protein aggregation. Here, the analytical solution of the equations describing the time evolution of the fibrillar assemblies identified the three principal events that play a key role in determining this phenomenon—nucleation, elongation, and fragmentation (Knowles et al. 2009). Specific biophysical methods for measuring each of them in a quantitative manner were developed, including the quartz crystal microbalance (QCM) approach for quantifying the elongation process (Knowles et al. 2007b) and atomic force microscopy methods for measuring fragmentation (Smith et al. 2006; Knowles et al. 2007a). These developments are enabling the quantification not only of directly observable macroscopic parameters, such as lag times and growth rates, but also of the microscopic rates characterizing the fundamental molecular processes underlying aggregation. By building further on these achievements, it will now be possible to extend this approach to include processes that are expected to play a central role in vivo.
THE KEY ROLE OF PROTEIN SOLUBILITY IN PROTEIN HOMEOSTASIS
The “generic view” of protein aggregation (Dobson 1999) offers a unified framework for understanding the fundamental inherent molecular basis of protein aggregation and for identifying possible ways to regulate this phenomenon (Fig. 2). This goal can be achieved by linking theory and experiment to develop a comprehensive conceptual basis of misfolding diseases to suggest rational means for their avoidance and therapy. This approach is based on the realization that major advances can be made by exploiting the opportunities offered by bringing together a series of technical and conceptual developments in this area of science:
new ideas about protein aggregation, including the finding that the ability to assemble into stable and highly organized structures (e.g., amyloid fibrils) is not an unusual feature exhibited by a small group of peptides and proteins with special sequence or structural properties, but rather a property likely to be shared by most proteins (Dobson 1999)
the discovery that specific aspects of the behavior of proteins, including their propensities for aggregation (Chiti et al. 2003; Tartaglia et al. 2008) and for interacting with molecular chaperones (Tartaglia et al. 2010), as well as the degree of toxicity associated with the aggregation (Luheshi et al. 2007), can be predicted with a remarkable degree of accuracy from the knowledge of their amino acid sequences
the realization that a wide variety of techniques, originally devised for applications in other disciplines, can be used to probe the nature of protein aggregation and assembly, and of the structures that emerge from these events (Knowles et al. 2007a,b, 2009)
the observation that proteins are present at concentrations at which they are only marginally soluble (Tartaglia et al. 2007) and that stress or aging can lead to widespread aggregation in living organisms (Gidalevitz et al. 2006; Ben-Zvi et al. 2009; Narayanaswamy et al. 2009; David et al. 2010; Geiler-Samerotte et al. 2011; Olzscha et al. 2011)
the recognition that a fundamental understanding of these issues offers unique opportunities for the rational development of therapeutic strategies (Dumoulin et al. 2003; Dobson 2004)
Figure 2.
Schematic illustration of the link between solubility and homeostasis of proteins. One of the major consequences of the generic view of protein aggregation is that protein solubility is expected to play a crucial role in protein homeostasis. By carrying out accurate measurements of the kinetics and thermodynamics of protein aggregation, it will be possible to define the key microscopic processes underlying this phenomenon and to identify those proteins that are particularly prone to aggregate under stress conditions. These advances will open avenues by which the behavior of such proteins could be potentially regulated, both by enhancing the activity of natural cellular defenses and by pharmacological intervention.
In the light of these advances, it is becoming increasingly clear that protein homeostasis is crucially associated with the maintenance of proteins in their soluble state, and that even relatively small impairments in the quality control mechanisms that regulate the concentration of proteins in living systems will eventually lead to disease. If this view is correct, an effective type of therapeutic intervention will involve the biological and chemical regulation of protein solubility.
KINETICS OF PROTEIN AGGREGATION
A major challenge in the study of protein aggregation is to define the microscopic processes underlying this phenomenon. There have been great advances recently in methods for analytically solving the system of coupled differential equations that describe the conversion of normally soluble peptides and proteins into amyloid structures (Knowles et al. 2009). This approach is very accurate for the intermediate stages of protein aggregation, when there is a close interplay between nucleation, elongation, and fragmentation (Fig. 3). The mathematical method used to achieve this result is based on the power of fixed-point analysis to treat systems of differential equations having great complexity, and appears to be of very general applicability. Because this approach allows solutions to be derived to highly nonlinear equations, which form the majority of cases encountered in biology, it is now possible to explore a series of fundamental problems to clarify the microscopic processes underlying protein aggregation in increasingly complex situations, ranging from in vitro to in vivo environments. For instance we have recently proposed a master equation for the early stages of protein aggregation describing the primary nucleation processes and the dynamics of oligomeric and prefibrillar aggregate populations, which are likely to be directly associated with toxic effects (N Cremades, SIA Cohen, AY Chen, et al. in prep.).
Figure 3.
(A) Illustration of the nucleation-elongation-fragmentation model of fibril formation, for which (B) an analytical solution has recently been obtained (Knowles et al. 2009).
This approach is uniquely suited for establishing a clear connection between the microscopic processes that determine the molecular mechanisms of protein aggregation, and the corresponding macroscopic processes, which are those that are directly measurable (Knowles et al. 2009). This theoretical analysis can be combined with a range of biophysical experiments that allow key properties governing protein aggregation to be defined in vitro and in vivo in a highly quantitative manner by using a variety of sophisticated fluorescence methods (van Ham et al. 2010), as well as label-free techniques such as atomic force microscopy (Smith et al. 2006; Knowles et al. 2007b), dynamic light scattering, and QCM measurements (Knowles et al. 2007b).
One of the key challenges in elucidating the molecular mechanisms that underlie protein aggregation is the difficulty in quantifying the rates of the individual steps that are involved in the overall process. Specific biophysical methods for measuring each of them in a quantitative manner were developed, including the QCM approach for quantifying the elongation process (Knowles et al. 2007b), and atomic force microscopy methods for measuring fragmentation (Smith 2006; Knowles et al. 2007a).
THERMODYNAMICS OF PROTEIN AGGREGATION
One of the most powerful implications of the generic view of protein aggregation is that the behavior of proteins, in vitro as well as in vivo, should depend in an essential manner on the nature and relative stabilities of their different conformational states. On this perspective, the suggestion that most peptides and proteins are capable of converting into the amyloid state implies that this state can be thermodynamically stable under a wide range of conditions. If this is true, then the conversion in vivo of proteins from their native states into fibrils could well be limited by kinetic factors, a conclusion that differs from many current views but is consistent with much that is known about misfolding diseases, including the fact that such disorders tend to be age related (Dobson 2003; Haass and Selkoe 2007; Balch et al. 2008; Powers et al. 2009).
The challenge now is to gather quantitative evidence to probe this overall concept, and to establish how proteins have evolved (or rather coevolved with their cellular environments) to remain soluble on the timescales required for the successful functioning of living organisms (Tartaglia et al. 2007). At high concentrations, proteins will be thermodynamically unstable in their native states and prone to undergo rapid aggregation. Therefore, there will be strong evolutionary pressure on their amino acid sequence to maintain their solubility. In contrast, at low concentrations, proteins will be highly soluble as their native states are thermodynamically very stable. There is consequently no evolutionary pressure acting against a natural drift, resulting, e.g., from random mutations, toward less soluble sequences. This drift, however, must stop when proteins reach a regime in which their native states are still kinetically, even though not thermodynamically stable. Indeed, when a regime of kinetic instability is reached, protein solubility is no longer guaranteed, and the functioning of a biological system is compromised.
An essential requirement to establish the principle of metastability of native states against aggregation (Tartaglia et al. 2007; Olzscha et al. 2011) is the ability to quantify the relative stabilities of the native, unfolded, oligomeric, and fibrillar states. Experimental measurements of these stabilities can be obtained by using denaturant-induced disaggregation experiments (O’Nuallain et al. 2005). Our initial studies have already indicated that it is possible to exploit this strategy successfully to probe the thermodynamic characteristics of a wide variety of proteins (Baldwin et al. 2011). These experiments establish a close connection between earlier biophysical studies that allowed the nature of protein folding to be elucidated (Wolynes et al. 1995; Dobson et al. 1998), and the phenomenon of amyloid growth, whose general mechanistic principles are still emerging.
These investigations have been greatly stimulated by our observation of a close relationship between the messenger RNA (mRNA) expression levels and the aggregation rates of corresponding series of human proteins (Tartaglia et al. 2007), which further suggests that the concentration of proteins in the cell should be closely related to the critical concentration under the corresponding conditions. We have referred to this relationship as the “life on the edge” hypothesis, because according to this view, proteins are just soluble enough to remain in their active states at the concentrations required for their optimal function (Tartaglia et al. 2007).
THE METASTABILITY OF THE HUMAN PROTEOME
A key implication of the life on the edge hypothesis (Tartaglia et al. 2007) is that the human proteome might include proteins that are highly metastable, as their solubility is significantly lower than would be expected on the basis of their cellular concentration. This view is receiving support from a series of recent studies that have shown how protein aggregation is a widespread phenomenon, involving, in particular, metastable proteins (Gidalevitz et al. 2006; Narayanaswamy et al. 2009; David et al. 2010; Geiler-Samerotte et al. 2011; Olzscha et al. 2011).
These results provide new insight into the observation that our biochemical pathways are highly susceptible to the formation of misfolded or aggregated proteins as a consequence of aging or stress conditions (Haass and Selkoe 2007; Ben-Zvi et al. 2009). Because misfolding and its consequences put the cellular quality control systems under further pressure, these initial findings suggest that misfolding diseases can result from a “misfolding cascade,” giving rise to a variety of unregulated events that can lead to biochemical dysfunction and cell death. This view provides a novel perspective of neurodegenerative conditions, according to which events cause the accumulation of the peptide or protein aggregates that initiate the disease (e.g., amyloid β and tau in the case of Alzheimer’s disease) activate a set of downstream cellular signaling and metabolic events that ultimately damage or destroy nerve cells (Hardy and Selkoe 2002).
BIOLOGICAL REGULATION OF PROTEIN AGGREGATION
A series of quantitative techniques has been recently developed to study the structures of the states involved in the interactions between misfolded proteins and chaperones and other protective mechanisms, as well as to probe their effects on the aggregation process (Dumoulin et al. 2003; Knowles et al. 2007b; Balch et al. 2008; Hartl and Hayer-Hartl 2009; Powers et al. 2009; Roodveldt et al. 2009). One of the key difficulties in studying the effects of cellular components on the aggregation of proteins is the interfering effects that these species can exercise on the assays that are used to monitor protein aggregation itself. In particular, chaperones can compete for the binding of dye labels and molecular crowding can influence the affinity of fluorescent labels to amyloid structures (White et al. 2010; Buell et al. 2011). We have recently proposed the application of a variety of biophysical techniques to overcome these difficulties. Using an alternative readout to the prevalent optical signals used in traditional aggregation studies, namely, direct measurements of the changes in mass of an ensemble of aggregates by means of a QCM, we have been able to develop aggregation assays that function reliably even in a complex environment (Knowles et al. 2007b). The success of this concept has been shown in early experiments, in which we showed that the inhibition of filament growth by the heat shock protein αB-crystallin occurs through interactions with the aggregated species and primarily with their monomeric precursors as had commonly been assumed (Knowles et al. 2007b). In addition, by using this biosensor-based approach, we have been able to probe directly the effect that molecular crowding has on protein assembly (White et al. 2010). In combination with atomic force microscopy studies, which allow individual aggregates to be resolved prior, during, and after growth, and enable a correlation to be established between kinetic signals and the changes in the morphology of the aggregates that are formed, these biosensor-based approaches provide a robust platform for understanding the effects of biological modulators of protein aggregation (Fig. 4).
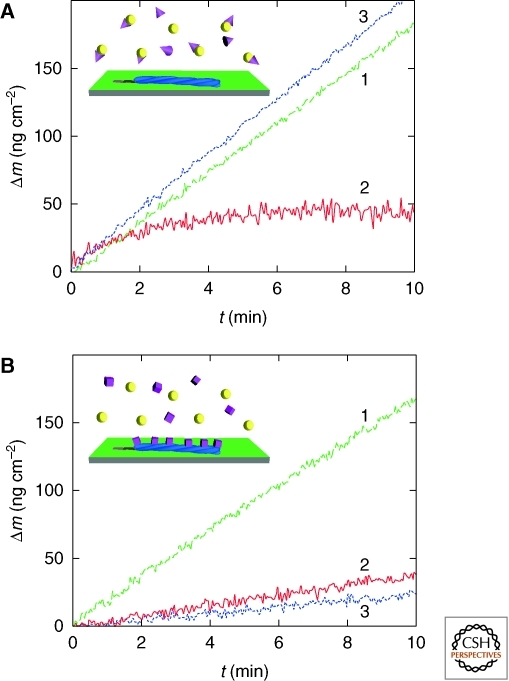
Figure 4.
Biosensor assays allow the mechanisms of amyloid growth inhibition to be investigated (White et al. 2009). (A) Growth inhibition by a chemical chaperone (trimethylamine oxide [TMAO]). Insulin fibrils are first grown in the presence of soluble protein (1), then in the presence of both protein and chaperone (2), and finally, in the presence of protein alone (3). In the absence of chaperone, the growth resumes to the initial level, indicating no permanent interactions between the fibrils and the chaperone. (B) The growth inhibition assay is repeated for αB-crystallin. The growth rate (3) after exposure to chaperone (2) is significantly lower than that measured for pristine fibrils (1), revealing a strong interaction between the fibrils and αB-crystallin as the basis of the growth inhibition (Knowles et al. 2007b).
These quantitative approaches have opened up the possibility of probing more generally the effects of specific cellular components on the factors that modulate protein aggregation. Biosensor-based assays have the potential to dissect the aggregation pathway and elucidate the steps with which given components interfere. Following the development of these methods and their shown success in quantifying protein aggregation in our earlier studies, it has become possible to use them to obtain information on the interactions of a range of cellular components, including clusterin, heat shock proteins, ATP-dependent chaperone systems, with aggregates formed from various polypeptide chains, including short peptides, protein fragments, protein domains, and full-length proteins. With this approach it will become possible to elucidate the general physical principles that govern biological regulation of protein aggregation and understand how nature has developed mechanisms able to counter protein aggregation in a highly effective way.
Furthermore, it is becoming crucial to acquire a better understanding of the nature of protein stability and solubility. One of the expectations inherent in the life on the edge hypothesis (Tartaglia et al. 2007) is that the presence of chaperones should enhance the effective solubility of proteins in the cellular environment (Gidalevitz et al. 2006; Olzscha et al. 2011). In this sense, chaperones act to protect the cell from the pathological effects of misfolded proteins and aberrant aggregates, which are particularly abundant in the context of specific pathways (Dobson 2003; Hartl and Hayer-Hartl 2009; Powers et al. 2009). The presence of a general relationship between protein solubility and abundance (Fig. 5) provides further insight by understanding how the specific cellular conditions in which proteins carry out their functions can modulate this relationship.
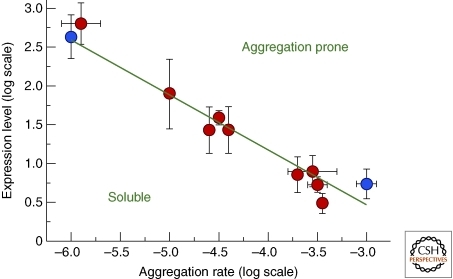
Figure 5.
Correlation between in vivo mRNA expression levels and in vitro protein aggregation rates for a set of 11 human proteins, either related (red) or unrelated (blue) to disease (Tartaglia et al. 2007).
CHEMICAL REGULATION OF PROTEIN AGGREGATION
A powerful idea that has recently emerged is that fundamental physicochemical properties of peptides and proteins are important in determining the toxicity of aberrant assemblies by studying in vivo models (Bucciantini et al. 2002; Dobson 2004; Luheshi et al. 2007, 2010). In this context, we anticipate that processes that activate particular pathways, such as oxidative phosphorylation or proteasomal degradation, without affecting the supply of proteins into those pathways, will not be fully effective by themselves in treating misfolding diseases. Our view is that early interventions may be targeted and specific, but that later strategies must restore health to the proteome as a whole. Recent studies performed by Jeffrey Kelly, Richard Morimoto, and coworkers have indeed provided initial evidence that favoring the return to normal homeostasis may represent a powerful strategy in combating neurodegenerative diseases (Balch et al. 2008; Morimoto 2008; Powers et al. 2009).
The palette of biophysical techniques that we have developed and applied to the phenomenon of protein aggregation allows a uniquely detailed understanding to be established of the chemical factors that regulate this process in vivo and in vitro. A key requirement to restore biological function is the solubilization or elimination of aggregates that have formed in an aberrant way. Quantitative assays of both of these processes are therefore of paramount importance in this context. We have already shown that biosensor approaches allow the effect on protein aggregation of small molecules to be defined in a highly quantitative way (Knowles et al. 2007b). This methodology should also be highly suited for probing the disaggregation of aberrant assemblies under the action of a variety of components, and hence forms an ideal platform for studying the mechanism of action of antiaggregation compounds, such as β breakers (Soto et al. 1998).
More generally, a central question in devising strategies against protein aggregation is about the relative effectiveness of intervention at the different specific stages involved in the overall process. Studies of the inhibition of aggregation in bulk solution assays, even though they frequently show evidence for the inhibitory action of various compounds, do not in general allow precise information to be obtained on the specific molecular events on the aggregation pathway that have been affected, leading to difficulties for a rational design of strategies to prevent loss of protein solubility. A combination of innovative biophysical approaches, including biosensing and atomic force microscopy techniques, is becoming available to dissect the effects of antiaggregation compounds at the level of individual microscopic rates and processes. Biosensing approaches allow for the growth and potential degradation of aggregates to be probed in the absence of nucleation events, whereas atomic force microscopy provides a connection to structural and morphological changes that accompany aggregation or disaggregation under the effect of chemical agents favoring solubilization.
Finally, the information obtained from these core biophysical and theoretical investigations can be used in the context of treatments for misfolding diseases based on targeting amyloidogenic proteins or on improving the capacity of the cell to protect unfolded proteins against a range of aggregation pathways. The fundamental concepts emerging from biophysical studies can be applied to in vivo model systems including C. elegans. By using this type of technique, it will become possible to enhance protein homeostasis by antibodies (Dumoulin et al. 2003; Luheshi et al. 2010) or by small molecule inhibitors of aggregation (Dobson 2004; Lendel et al. 2009).
A UNIFIED VIEW OF PROTEIN MISFOLDING AND ITS ASSOCIATED DISORDERS
The studies that we have discussed are creating a general framework for understanding protein misfolding disorders in terms of the properties of the molecules involved in them. This approach is inspired by one of the major consequences of the generic view of amyloid aggregation (Dobson 1999)—that the study of the factors that influence the stability of the various states of proteins in the cell is capable not only of providing an understanding of the molecular basis of misfolding disorders, but also of suggesting general strategies for their treatment. This approach is based on the advances that have been recently made in developing a range of novel biophysical, theoretical, and computational techniques.
The approach that we have presented, instead of being targeted to the specific processes involved in a particular disease, is aimed at establishing a unified theory of protein misfolding disorders. This theory is inspired by the general association of misfolding diseases with the problem of protein solubility and the metastability of the human proteome. It is also based on the premise that comparison of the molecular events associated with related diseases will enable us to understand the key features that define the pathogenesis at the molecular level. These advances are clarifying how misfolding diseases are caused by the failure of the natural cellular protective mechanisms to provide sufficient defenses against the naturally limited solubility of our proteins.
Footnotes
Editors: Richard I. Morimoto, Dennis Selkoe, and Jeff Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Baglioni S, Casamenti F, Bucciantini M, Luheshi LM, Taddei N, Chiti F, Dobson CM, Stefani M 2006. Prefibrillar amyloid aggregates could be generic toxins in higher organisms. J Neurosci 26: 8160–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW 2008. Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Knowles TPJ, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas S, Waudby CA, Mossuto MF, Meehan S, Gras SL 2011. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc (in press) [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci 106: 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo JS, Taddei N, Ramponi G, Dobson CM, Stefani M 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416: 507–511 [DOI] [PubMed] [Google Scholar]
- Buell AK, Dobson CM, Knowles TPJ, Welland ME 2010. Interactions between amyloidophilic dyes and their relevance to studies of amyloid inhibitors. Biophys J 99: 3492–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla N, Caddy GL, Hall DR, Zurdo J, Gairi M, Feliz M, Giralt E, Robinson CV, Dobson CM 2005. Molecular recycling within amyloid fibrils. Nature 436: 554–558 [DOI] [PubMed] [Google Scholar]
- Carulla N, Zhou M, Arimon M, Gairi M, Giralt E, Robinson CV, Dobson CM 2009. Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation. Proc Natl Acad Sci 106: 7828–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: 333–366 [DOI] [PubMed] [Google Scholar]
- Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM 1999. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci 96: 3590–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM 2003. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424: 805–808 [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8: e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedmon MM, Christodoulou J, Wilson MR, Dobson CM 2005. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J Biol Chem 280: 14733–14740 [DOI] [PubMed] [Google Scholar]
- de la Paz ML, Goldie K, Zurdo J, Lacroix E, Dobson CM, Hoenger A, Serrano L 2002. De novo designed peptide-based amyloid fibrils. Proc Natl Acad Sci 99: 16052–16057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM 1999. Protein misfolding, evolution and disease. Trends Biochem Sci 24: 329–332 [DOI] [PubMed] [Google Scholar]
- Dobson CM 2003. Protein folding and misfolding. Nature 426: 884–890 [DOI] [PubMed] [Google Scholar]
- Dobson CM 2004. In the footsteps of alchemists. Science 304: 1259–1260 [DOI] [PubMed] [Google Scholar]
- Dobson CM, Sali A, Karplus M 1998. Protein folding: A perspective from theory and experiment. Angew Chem Intl Ed 37: 868–893 [DOI] [PubMed] [Google Scholar]
- Dumoulin M, Last AM, Desmyter A, Decanniere K, Canet D, Larsson GR, Spencer A, Archer DB, Sasse J, Muyldermans S, et al. 2003. A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme. Nature 424: 783–788 [DOI] [PubMed] [Google Scholar]
- Fandrich M, Fletcher MA, Dobson CM 2001. Amyloid fibrils from muscle myoglobin— Even an ordinary globular protein can assume a rogue guise if conditions are right. Nature 410: 165–166 [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SA, Hartl DL, Drummond DA 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci 108: 680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S, Ivarsson Y, De Simone A, Travaglini-Allocatelli C, Brunori M, Vendruscolo M 2010. Structural characterization of a misfolded intermediate populated during the folding process of a PDZ domain. Nature Struct Mol Biol 17: 1431–1437 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM 1998. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci 95: 4224–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ 2007. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nature Rev Mol Cell Biol 8: 101–112 [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ 2002. Medicine—The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 297: 353–356 [DOI] [PubMed] [Google Scholar]
- Harper JD, Lansbury PT 1997. Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem 66: 385–407 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2002. Protein folding—Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2009. Converging concepts of protein folding in vitro and in vivo. Nature Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG 2004. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci 101: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JW 1998. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol 8: 101–106 [DOI] [PubMed] [Google Scholar]
- Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME 2007a. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 318: 1900–1903 [DOI] [PubMed] [Google Scholar]
- Knowles TPJ, Shu WM, Devlin GL, Meehan S, Auer S, Dobson CM, Welland ME 2007b. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc Natl Acad Sci 104: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TPJ, Waudby CA, Devlin GL, Cohen SIA, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM 2009. An analytical solution to the kinetics of breakable filament assembly. Science 326: 1533–1537 [DOI] [PubMed] [Google Scholar]
- Lendel C, Bertoncini CW, Cremades N, Waudby CA, Vendruscolo M, Dobson CM, Schenk D, Christodoulou J, Toth G 2009. On the mechanism of nonspecific inhibitors of protein aggregation: Dissecting the interactions of alpha-synuclein with Congo red and Lacmoid. Biochemistry 48: 8322–8334 [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M 2005. Simultaneous determination of protein structure and dynamics. Nature 433: 128–132 [DOI] [PubMed] [Google Scholar]
- Luheshi LM, Tartaglia GG, Brorsson AC, Pawar AP, Watson IE, Chiti F, Vendruscolo M, Lomas DA, Dobson CM, Crowther DC 2007. Systematic in vivo analysis of the intrinsic determinants of amyloid beta pathogenicity. PLoS Biol 5: 2493–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luheshi LM, Hoyer W, de Barros TP, van Dijk Härd I, Brorsson AC, Macao B, Persson C, Crowther DC, Lomas DA, Ståhl S, et al. 2010. Sequestration of the Abeta peptide prevents toxicity and promotes degradation in vivo. PLoS Biol 8: e1000334 10.1371/journal.pbio.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci 106: 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Worner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78 [DOI] [PubMed] [Google Scholar]
- O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R 2005. Thermodynamics of Aβ(1–40) amyloid fibril elongation. Biochemistry 44: 12709–12718 [DOI] [PubMed] [Google Scholar]
- Orte A, Birkett NR, Clarke RW, Devlin GL, Dobson CM, Klenerman D 2008. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc Natl Acad Sci 105: 14424–14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991 [DOI] [PubMed] [Google Scholar]
- Ramirez-Alvarado M, Kelly JW, Dobson CM (eds). 2010. Protein misfolding diseases. Current and emerging principles and therapies. Wiley, Hoboken, NJ [Google Scholar]
- Robinson CV, Sali A, Baumeister W 2007. The molecular sociology of the cell. Nature 450: 973–982 [DOI] [PubMed] [Google Scholar]
- Roodveldt C, Bertoncini CW, Andersson A, van der Goot AT, Hsu ST, Fernandez-Montesinos R, de Jong J, van Ham TJ, Nollen EA, Pozo D, et al. 2009. Chaperone proteostasis in Parkinson’s disease: Stabilization of the Hsp70/alpha-synuclein complex by Hip. EMBO J 28: 3758–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Phillips J, Schneidman-Duhovny D, Velazquez-Muriel JA, Sali A 2009. The structural dynamics of macromolecular processes. Curr Opin Cell Biol 21: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL 2000. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321 [DOI] [PubMed] [Google Scholar]
- Smith JF, Knowles TPJ, Dobson CM, MacPhee CE, Welland ME 2006. Characterization of the nanoscale properties of individual amyloid fibrils. Proc Natl Acad Sci 103: 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C, Sigurdsson EM, Morelli L, Kumar RA, Castano EM, Frangione B 1998. Beta-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: Implications for Alzheimer’s therapy. Nature Med 4: 822–826 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M 2007. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci 32: 204–206 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pawar A, Campioni S, Chiti F, Vendruscolo M 2008. Prediction of aggregation-prone regions of structured proteins. J Mol Biol 308: 425–436 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Dobson CM, Hartl FU, Vendruscolo M 2010. Physicochemical determinants of chaperone requirements. J Mol Biol 400: 579–588 [DOI] [PubMed] [Google Scholar]
- van Ham TJ, Esposito A, Kumita JR, Hsu STD, Schierle GSK, Kaminsk CF, Dobson CM, Nollen EAA, Bertoncini CW 2010. Towards multiparametric fluorescent imaging of amyloid formation: Studies of a YFP model of alpha-synuclein aggregation. J Mol Biol 395: 627–642 [DOI] [PubMed] [Google Scholar]
- Vendruscolo M, Dobson CM 2005. Towards complete descriptions of the free-energy landscapes of proteins. Phil Trans R Soc A 363: 433–450 [DOI] [PubMed] [Google Scholar]
- Vendruscolo M, Dobson CM 2009. Quantitative approaches to defining normal and aberrant protein homeostasis. Faraday Disc 143: 277–291 [DOI] [PubMed] [Google Scholar]
- Vendruscolo M, Paci E, Dobson CM, Karplus M 2001. Three key residues form a critical contact network in a protein folding transition state. Nature 409: 641–645 [DOI] [PubMed] [Google Scholar]
- White DA, Buell AK, Dobson CM, Welland ME, Knowles TPJ 2009. Biosensor-based label-free assays of amyloid growth. FEBS Lett 583: 2587–2592 [DOI] [PubMed] [Google Scholar]
- White DA, Buell AK, Knowles TPJ, Welland ME, Dobson CM 2010. Protein aggregation in crowded environments. J Am Chem Soc 132: 5170–5175 [DOI] [PubMed] [Google Scholar]
- Wolynes PG, Onuchic JN, Thirumalai D 1995. Navigating the folding routes. Science 267: 1619–1620 [DOI] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM 2005. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438: 878–881 [DOI] [PubMed] [Google Scholar]
- Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR 2007. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J 21: 2312–2322 [DOI] [PubMed] [Google Scholar]