Abstract
Small RNA regulators (sRNAs) have been identified in a wide range of bacteria and found to play critical regulatory roles in many processes. The major families of sRNAs include true antisense RNAs, synthesized from the strand complementary to the mRNA they regulate, sRNAs that also act by pairing but have limited complementarity with their targets, and sRNAs that regulate proteins by binding to and affecting protein activity. The sRNAs with limited complementarity are akin to eukaryotic microRNAs in their ability to modulate the activity and stability of multiple mRNAs. In many bacterial species, the RNA chaperone Hfq is required to promote pairing between these sRNAs and their target mRNAs. Understanding the evolution of regulatory sRNAs remains a challenge; sRNA genes show evidence of duplication and horizontal transfer but also could be evolved from tRNAs, mRNAs or random transcription.
Small RNA regulators control many bacterial processes. They include true antisense RNAs and sRNAs that show limited complementarity to their mRNA targets and are more akin to eukaryotic microRNAs.
1. INTRODUCTION
In the third edition of The RNA World book, we described how many small RNA regulators (sRNAs), ranging in length from approximately 50 to 500 nucleotides, were found in bacteria, and summarized what was then known about the functions of the sRNAs. As has been seen for regulatory RNAs in eukaryotes, many bacterial sRNAs act by base pairing, having either extensive or more limited complementarity with their target mRNAs. However, others modulate the activity of proteins by mimicking secondary structures of other nucleic acids. The pace of sRNA discovery in a wide range of bacteria has continued to accelerate and the functions of increasing numbers of sRNAs are being elucidated. Here we cite reviews that discuss older work and emphasize what has been found in the past 5 yr. We will focus on the sRNAs encoded on bacterial chromosomes, particularly in the model organisms Escherichia coli, Salmonella enterica and Staphylococcus aureus, but will point out parallels to phage and plasmid-encoded sRNAs. We also will focus on RNA regulators that act as independent transcripts as opposed to riboswitches, which are part of the mRNA they regulate and are described in (Breaker, Riboswitches and the RNA World). CRISPR RNAs, which are central to a defense against foreign DNA in many bacteria and archaea, are yet another class of sRNAs that will not be covered here because they are the topic of (Jone, Brouns and van der Oost, RNA in Defense: CRISPRS protect prokaryotes against alien nucleic acids).
2. HOW MANY sRNAs ARE THERE?
The initial screens for sRNA genes in bacteria relied primarily on computational searches of intergenic regions for conserved sequences or orphan promoter and terminator sequences (reviewed in Livny and Waldor 2007). However, with the advances in whole genome expression profiling using tiling arrays or deep sequencing, approaches that rely on direct detection are superseding the computational approaches (reviewed in Sharma and Vogel 2009). Nevertheless, despite the many searches that have now been conducted, the exact number of sRNAs present is still not known for any bacterium. A number of 80–100 is generally cited for E. coli (compared with around 4300 proteins); whereas numbers two or three times higher have been reported for other bacteria. Unless a large class of sRNAs has been missed (for instance, sRNAs derived from mRNAs), it seems likely that bacteria will have on the order of a few hundred rather than thousands of sRNAs.
Part of the challenge in establishing the true number of regulatory sRNAs is that many short transcripts have only been detected by one approach and have not been shown to have functions. For example, there are reports of tens or hundreds of antisense RNAs encoded opposite annotated protein-coding genes, based on detection by microarray analysis or deep sequencing (Georg et al. 2009; Guell et al. 2009; Toledo-Arana et al. 2009; Sharma et al. 2010). Chromatin immunoprecipitation experiments similarly suggest that E. coli RNA polymerase is transcribing the strands opposite a number of protein-coding genes (Peters et al. 2009). However, few of these signals—some of which potentially could be caused by cross hybridization or spurious cDNA synthesis by reverse transcriptase—have been verified in independent experiments, particularly in experiments with control strains in which the putative gene or promoter has been deleted. In addition, only a limited number of antisense sRNAs have been assigned functions. On the other hand, some sRNAs with true regulatory functions might still be missed because they are only expressed under very specific conditions, their structures make them recalcitrant to the predominant methods of detection, or they are processed from mRNAs and hard to distinguish from 5′ or 3′ UTRs.
With increased sRNA detection and characterization in a wide range of bacteria, we will undoubtedly be able to obtain more definitive answers to questions about the number of sRNAs across bacterial species and whether, as we would predict, all bacteria have sRNA regulators.
3. TRUE ANTISENSE sRNAs
As mentioned earlier, and as has also been noted for eukaryotic organisms, large numbers of antisense RNAs, also referred to as cis-encoded RNAs, are being reported to be transcribed opposite annotated genes and thus share extensive complementarity with the corresponding transcripts (Georg et al. 2009; Guell et al. 2009; Toledo-Arana et al. 2009; Sharma et al. 2010). However, clear physiological roles have only been established for a small number of antisense sRNAs. Nevertheless, these examples serve as models for what a cis-encoded antisense sRNA is likely to do.
Thus far, the most prevalent role for antisense sRNAs in bacteria has been the repression of genes that encode potentially toxic proteins (reviewed in Gerdes and Wagner 2007; Fozo et al. 2008). This was one of the first functions described for the plasmid-encoded sRNAs, where antisense sRNAs control the expression of small hydrophobic proteins, such as the 50-amino-acid Hok protein encoded on the E. coli R1, R100, and F plasmids and the 33 amino acid Fst protein encoded on the Enterococcus faecalis plasmid pAD1. Bacterial chromosomes have also been found to encode homologs of these proteins and their corresponding antisense sRNAs. In addition, an increasing number of other small hydrophobic protein-antisense sRNA pairs are being discovered in a variety of organisms, often tandemly duplicated in the same region of the chromosome (Fozo et al. 2010; Sharma et al. 2010). The physiological roles of these gene pairs are not yet known, but in all cases that have been examined, the antisense sRNA represses synthesis of the corresponding protein, which is toxic at high levels. The mechanism by which repression occurs has only been characterized for a limited number of pairs and generally appears to involve a block in translation. However, the mechanism may vary between toxin-antitoxin pairs because the antisense sRNA can overlap the 5′ end of the mRNA, the entire mRNA, or the 3′ end of the mRNA. In many cases both the toxin mRNA and antisense sRNA are predicted to have extensive secondary structures (reviewed in Gerdes and Wagner 2007; Weaver 2007) raising questions about the mechanism of base pairing. The antisense sRNAs appear to be critical for keeping the basal level expression of the toxins low, and where it has been examined, the levels of the antisense sRNA have been reported to be in excess of the mRNA. Thus, another open question is under what conditions antisense regulation is overcome to allow toxin synthesis.
Another role of characterized antisense sRNAs is the directed cleavage of the mRNA encoded on the opposite strand. The first such sRNA to be characterized was the OOP RNA of the bacteriophage λ, where a 77-nucleotide sRNA encoded opposite the cII-O mRNA of λ represses cII expression by directing cleavage of this transcript in a mechanism that is partially dependent on RNase III (Krinke and Wulff 1987; Krinke and Wulff 1990). The E. coli GadY RNA encoded opposite the gadX-gadW mRNA may similarly be directing cleavage of the mRNA, in this case leading to increased levels of the gadX transcript (Opdyke et al. 2004; Tramonti et al. 2008). In contrast, the antisense IsrR RNA of Synechocystis negatively impacts the levels of the oppositely encoded isiA mRNA (Duhring et al. 2006). IsrR synthesis is repressed by iron stress, allowing isiA accumulation and translation under these conditions. In addition to blocking translation and directing mRNA processing, one can imagine that simply the act of transcribing the antisense RNA might impact transcription of the mRNA encoded opposite.
4. BASE PAIRING sRNAs WITH LIMITED COMPLEMENTARITY
The most well studied bacterial sRNA regulators are those that act by base pairing but only have limited complementarity with their target RNAs because they are encoded at a different genomic location. They have sometimes been referred to as trans-encoded sRNAs, to distinguish them from the antisense, cis-encoded sRNAs. Almost all of the sRNAs that fall into this class are expressed under specific growth conditions ranging from limiting iron, oxidative stress, and anaerobic conditions to cell envelope stress, low magnesium, or high glucose-6-phosphate concentrations and glucose starvation. These observations have led to the expectation that base-pairing sRNAs with limited complementarity with their targets will be found to be associated with almost every global response in bacteria. In the past 5 yr, there has been significant progress in characterizing the mechanisms of action and physiological roles of this class of sRNA.
4.1. Mechanisms
These base-pairing sRNAs are reminiscent of eukaryotic microRNAs (miRNAs) and small interfering RNAs (siRNAs) in their ability to modulate mRNA stability and translation. However, they differ in the details of how they are generated and where they pair with their target mRNAs. The bacterial base pairing sRNAs are generally transcribed as single entities around 100 nucleotides in length. Unlike miRNAs and siRNAs, they usually are not processed, although for a few sRNAs, cleavage to a shorter form does occur. In addition, bacterial sRNAs frequently base pair with the 5′ end, rather than with the 3′ end, of target mRNAs. In most cases, the bacterial sRNAs also appear to act stoichiometrically, being degraded along with the mRNA after pairing.
Outcomes of productive base pairing
Pairing between sRNA and mRNA generally involves at least a seed region of 6–8 contiguous base pairs, although significantly longer pairing regions have been predicted in some cases (but rarely tested). Within the sRNA, it is common for the region involved in base pairing, often with multiple targets, to be highly conserved. For instance, the Salmonella and E. coli GcvB RNAs have a conserved G/U rich, single-stranded region, which is required for base pairing with C/A-rich sequences in the 5′ untranslated regions of the target mRNAs (Sharma et al. 2007).
Base pairing between sRNAs and mRNAs can have a number of regulatory outcomes. Many of the sRNAs base pair at the ribosome binding site, thus blocking translation by preventing ribosome binding. However, systematic shifting of the region involved in Salmonella RybB RNA pairing with the ompN mRNA showed that RybB RNA can block translation even when the region of pairing is as far as the fifth codon into the open reading frame (ORF) (Bouvier et al. 2008). Translation can also be blocked when the region of pairing is 50 or more nucleotides upstream of the ribosome binding site (Sharma et al. 2007). In most cases where ribosome binding is blocked, an associated decrease in the stability of the mRNA is also observed, possibly as an indirect result of blocking ribosome entry. There are, however, target mRNAs in which base pairing occurs downstream of the Shine-Dalgarno sequence such that ribosome binding would not be blocked. For example, base pairing between the Salmonella MicC and the ompD mRNA within codons 23–26 accelerates RNase E-dependent decay of the mRNA without blocking ribosome binding (Pfeiffer et al. 2009). In another example, base pairing between E. coli RyhB and the iscRSUA mRNA in the iscR-iscS intergenic region leads to degradation of the downstream region of the polycistronic mRNA (Desnoyers et al. 2009).
In addition to repressing translation, sRNAs can also activate translation, in many cases by preventing or overcoming the formation of an inhibitory secondary structure (Morfeldt et al. 1995; Majdalani et al. 2005; Prévost et al. 2007). Binding of the sRNA leads to remodeling of the mRNA structure, uncovering the ribosome binding site and allowing translation. In some of these cases, binding of the sRNA can be quite distant from the start of translation, making it more difficult to identify targets computationally. In addition, because many targets are found by changes in mRNA abundance in microarrays, targets for positive regulation, which may only show a modest increase in mRNA stability, may be missed. Nonetheless, a growing number of sRNAs have been found to have positive effects on some targets while negatively regulating others (Morfeldt et al. 1995; Lease et al. 1998; Huntzinger et al. 2005; Prévost et al. 2007).
The interactions among and competition between sRNAs, ribosomes, ribonucleases, and the RNA chaperone Hfq and the target mRNAs, as well as the structures of both RNAs are all likely to influence how effective regulation is going to be, but are not well understood. It is interesting to note that effective SgrS repression of the ptsG mRNA requires either membrane localization of the target mRNA or a reduction in translation, suggesting that associated changes in mRNA structure or the slower binding of ribosomes is needed for SgrS accessibility to the region of pairing (Kawamoto et al. 2005). It is also intriguing that a surprising number of mRNAs are the targets of multiple sRNAs; perhaps only a subset of mRNAs have features (such as an Hfq binding site) that allow them to be regulated by base pairing sRNAs.
Role of Hfq
Although complex protein machineries are required for miRNA and siRNA processing and to effect the functions of these eukaryotic regulators, the majority of the sRNAs that act via limited base pairing in enteric bacteria have only been found to require the RNA chaperone Hfq, a close relative of the Sm/Lsm family of proteins involved in splicing and mRNA decay (reviewed in Brennan and Link 2007). Hfq binds both sRNAs and mRNAs, and, in vitro, stimulates their pairing. However, a number of questions remain about how this doughnut-shaped hexamer facilitates interactions between RNAs.
When and where is Hfq bound? The sites of Hfq binding on both sRNAs and target mRNAs have only been mapped for a limited number of transcripts, but thus far all are AU-rich single stranded regions. One binding study of the rpoS mRNA leader showed that Hfq binds to two sites, one near the region of base pairing with the DsrA RNA as well as another site significantly further upstream (Soper and Woodson 2008). The high affinity upstream Hfq binding site was found to significantly increase the rate of DsrA binding to rpoS. This has led to the model that once base-pairing between DsrA and rpoS mRNA is established, the second binding site stabilizes the base-paired complex by titrating Hfq away from the sRNA. Several mutational studies indicate that both the proximal and distal face of the Hfq ring bind to RNA (reviewed in Brennan and Link 2007). This view is now supported by structural studies. A previous structure of S. aureus Hfq bound to AU5G shows the RNA bound to the proximal face encircling the pore (Schumacher et al. 2002), whereas the RNA is bound to the distal face in a recent structure of E. coli Hfq bound to A15 (Link et al. 2009).
How does Hfq act to facilitate base pairing? Hfq binding has been shown to affect RNA secondary structure and has also been proposed to increase the local concentration of mRNA and sRNA by binding both transcripts (reviewed in Brennan and Link 2007). It is not yet known which mechanism facilitates sRNA-mRNA pairing or whether both are used together or independently. Hfq has also been reported to interact with ribosomes and with the RNase E endonuclease responsible for degrading the regulated mRNAs; to what extent these interactions are critical for Hfq action are not clear. It seems likely that the coming years will lead to a much better understanding of the mechanism of Hfq-stimulated pairing and regulation.
Hfq-independent base pairing
Unlike in enteric bacteria, several of the base pairing sRNAs characterized in Gram-positive bacteria such as S. aureus and Bacillus subtilis do not require Hfq for function, even when Hfq is present in the organism (Silvaggi et al. 2005; Boisset et al. 2007; Heidrich et al. 2007). However, Hfq does bind sRNAs in Listeria monocytogenes and hfq mutants do affect gene expression in both B. subtilis and L. monocytogenes, suggesting a possible role in RNA folding or pairing (Christiansen et al. 2004; Christiansen et al. 2006; Heidrich et al. 2006; Nielsen et al. 2009). It is not yet completely clear why Hfq is required in some cases of limited base pairing and not others. Possibly, more extended pairing and a higher proportion of G:C base pairs obviate the need for Hfq for a subset of sRNAs. Alternatively, another RNA chaperone may be playing a parallel role in these cases, as has been proposed for three small basic proteins in B. subtilis (Gaballa et al. 2008). This may well be the case in other bacterial species in which sRNAs, but not Hfq, have been found.
4.2. Physiological Roles
The anticipation that base pairing sRNAs would have a variety of physiological roles has been met; both the roles in previously known responses have expanded and new roles have been found.
Repression of outer membrane protein synthesis
In the third edition of The RNA World, we noted that the expression of several membrane proteins in E. coli is controlled by sRNAs. The list of sRNAs that modulate the expression of membrane proteins, particularly outer membrane β-barrel proteins, many of which function as porins, has continued to expand (Table 1). In E. coli and Salmonella, all the major outer membrane porins have been found to be down-regulated by one or more sRNAs (reviewed in Guillier et al. 2006; Vogel and Papenfort 2006), and new studies of sRNAs continue to uncover additional examples of regulation of outer membrane porins (Johansen et al. 2008; Papenfort et al. 2008; De Lay and Gottesman 2009; Figueroa-Bossi et al. 2009; Overgaard et al. 2009). Why outer membrane proteins are such predominant targets of sRNA regulation is still not completely clear, especially because many porin proteins are thought to be stable. However, given the abundance of examples, there must be a regulatory advantage to this mode of regulation.
Table 1.
Physiological roles of Hfq-binding RNAs in E. coli and S. typhimurium
| RNA name | Length | Regulation | Physiological Response* | References |
|---|---|---|---|---|
| MicF | 93 | Induced by high osmolarity; Activated by OmpR, SoxS, MarA | Repression of porin synthesis (ompF) | (Andersen and Delihas 1990; Coyer et al. 1990) |
| MicC | 109 | Increased at low temperature; Repressed by OmpR | Repression of porin synthesis (ompC, ompD) | (Chen et al. 2004) |
| MicA | 72 | Cell envelope stress; σE-regulated |
Repression of porin synthesis and PhoPQ (ompA, ompX, phoP) | (Rasmussen et al. 2005; Udekwu et al. 2005; Cornaert et al. 2010) |
| RybB | 80 | Cell envelope stress; σE-regulated |
Repression of porin synthesis (ompC, ompW) | (Johansen et al. 2006; Papenfort et al. 2006; Thompson et al. 2007) |
| RseX | 91 | Not reported | Repression of porin synthesis (ompC, ompA) | (Douchin et al. 2006) |
| MicM/ChiX | 85 | Repressed by chitosugars; chbBCARFG decoy mRNA stimulates degradation | Repression of chitoporin synthesis (chiP) (also dpiBA) | (Figueroa-Bossi et al. 2009; Overgaard et al. 2009) |
| CyaR | 87 | Induced by low glucose; Activated by CRP (σE-regulated) | Repression of porin synthesis and group behavior (ompX, luxS, nadE) | (Johansen et al. 2008; Papenfort et al. 2008; De Lay and Gottesman 2009) |
| OmrA | 88 | Induced by high osmolarity; Activated by OmpR | Repression of outer membrane protein synthesis (cirA, fecA, fepA, ompT, gntP, ompR) | (Guillier and Gottesman 2006; Guillier and Gottesman 2008) |
| OmrB | 82 | Induced by high osmolarity; Activated by OmpR | Repression of outer membrane protein synthesis (cirA, fecA, fepA, ompT, gntP, ompR) | (Guillier and Gottesman 2006; Guillier and Gottesman 2008) |
| GcvB | 205 | Induced by high glycine levels; Activated by GcvB | Repression of peptide transport (oppA, dppA, gltI, livK, livJ, argT, cycA, sstT) | (Urbanowski et al. 2000; Sharma et al. 2007; Pulvermacher et al. 2009a; Pulvermacher et al. 2009b) |
| RydC | 64 | Not reported | Repression of putative ABC transporter (yejABEF) | (Antal et al. 2005) |
| MgrR | 99 | Induced by low magnesium; Activated by PhoP | Repression of LPS modification gene (eptB, ygdQ) | (Moon and Gottesman 2009) |
| SgrS | 227 | Induced by glucose-phosphate or analogs; Activated by dedicated SgrR Bifunctional RNA | Protection against glucose-phosphate stress (ptsG) | (Vanderpool and Gottesman 2004; Wadler and Vanderpool 2007) |
| GlmZ | 207 | Repressed by high nitrogen; GlmY decoy sRNA blocks degradation | Induction of GlcN-6-P synthase (discoordinate regulation of glmUS operon) | (Kalamorz et al. 2007; Reichenbach et al. 2008; Urban and Vogel 2008) |
| Spot42 | 109 | Repressed by low glucose; Repressed by CRP | Repression of galactokinase blocks degradation (discoordinate regulation of galETKM operon) | (Møller et al. 2002) |
| GadY | 105, 90, 59 | Induced in stationary phase; σS-regulated | Activation of acid response (gadX) | (Opdyke et al. 2004; Tramonti et al. 2008) |
| RyhB | 90 | Induced by limiting iron; Repressed by Fur | Iron-sparing (sodB, sdhC, frdA, activates shiA) | (Massé and Gottesman 2002; Massé et al. 2005; Prévost et al. 2007) |
| FnrS | 113 | Induced under anaerobic conditions; Activated by FNR, ArcA and CRP | Repression of unneeded enzymes (sodB, maeA, gpmA, folE, folX) | (Durand and Storz 2010; Boysen et al. 2010) |
| OxyS | 109 | Induced by oxidative stress; Activated by OxyR | Repression of unneeded activities (fhlA, yobF-cspC, ybaY wrbA, rpoS) | (Altuvia et al. 1997; Altuvia et al. 1998) |
| ArcZ | 120, 50 | Induced under aerobic conditions; Repressed by ArcA | Activation of σS; repression of alternative activities (rpoS, sdaCB, tpx) | (Papenfort et al. 2009; Mandin and Gottesman 2010) |
| DsrA | 87 | Increased at low temperature | Activation of σS; repression of hns (rpoS, hns) | (Sledjeski and Gottesman 1995; Sledjeski et al. 1996; Majdalani et al. 1998; Lease and Belfort 2000) |
| RprA | 105 | Induced by cell surface stress; Activated by RcsB | Activation of σS (rpoS) | (Majdalani et al. 2001; Majdalani et al. 2002) |
| DicF | 53 | Not reported (cryptic prophage gene) | Inhibition of cell division (ftsZ) | (Bouché and Bouché 1989; Faubladier et al. 1990; Tetart and Bouché 1992) |
* Some, but not all, published targets of these sRNAs are listed in parentheses.
The synthesis of the porin-regulating sRNAs is induced by various growth conditions and regulators (Table 1). For example the micA and rybB genes are transcribed by σE, a sigma factor that regulates genes involved in both the trafficking of outer membrane proteins through the periplasm and quality control when the periplasm accumulates unfolded proteins (Johansen et al. 2006; Papenfort et al. 2006; Thompson et al. 2007; Udekwu and Wagner 2007). These findings have led to the satisfying model that disruptions to the integrity of the outer membrane lead to the induction of sRNAs that are capable of down-regulating the expression of porins, thereby easing the strain on the trafficking machinery and the membrane. The regulatory circuits for some of the other sRNAs in this group, such as CyaR, whose levels increase on glucose starvation, have less obvious connections to membrane stress, but nevertheless may help to down-regulate the σE response by keeping synthesis of outer membrane porins low.
Remodeling metabolism
Another theme that is becoming more and more prevalent among the targets of base pairing sRNAs is metabolic remodeling upon environmental shifts. One of the clearest examples of this comes from the Fur-regulated RyhB RNA in E. coli, whose levels increase in response to low iron and which represses the synthesis of many nonessential iron-containing enzymes such as aconitase B and succinate dehydrogenase. This allows the limited iron in the cell to be used by the critical enzymes (reviewed in Massé et al. 2007). Other sRNAs have also been found to impact what metabolic enzymes are synthesized. For example, the CRP-regulated Spot42 RNA, whose levels are repressed during glucose starvation, is responsible for the discordant regulation of genes in the galactose operon (Møller et al. 2002). Similarly, the FNR-regulated FnrS RNA, whose levels are induced by low oxygen, represses the expression of enzymes such as lactate dehydrogenase that are not needed during anaerobic growth (Boysen et al. 2010; Durand and Storz 2010).
Modulating the synthesis of key transcription factors
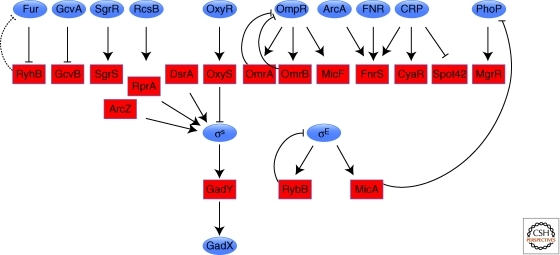
The rpoS gene encoding the stationary phase sigma factor σS in E. coli was one of the first targets of base pairing sRNA targets to be characterized, being positively regulated by DsrA and RprA and negatively regulated by OxyS (reviewed in Majdalani et al. 2005). Yet another sRNA, denoted ArcZ, has recently been found to increase the levels of σS (Mandin and Gottesman, submitted). DsrA and RprA are induced in response to low temperature and cell surface stress, respectively, whereas OxyS is induced by oxidative stress and ArcZ is repressed under anaerobic conditions. Thus, the sRNAs can finely tune σS synthesis in response to a range of environmental signals (Fig. 1). Another transcription factor whose synthesis is under the control of multiple sRNAs is the LuxR regulator of quorum sensing in Vibrio harveyi and its homolog HapR in V. cholerae. In this case the Qrr RNAs (five in V. harveyi and four in V. cholerae), which repress translation, are all homologous, but again the redundancy allows for nuanced regulation (Lenz et al. 2004; Tu and Bassler 2007; Svenningsen et al. 2008; Tu et al. 2008; Svenningsen et al. 2009). In S. aureus, RNA III has a significant impact on virulence by repressing the synthesis of Rot, a repressor of exotoxin genes (Boisset et al. 2007). sRNAs also directly negatively regulate a number of two-component systems; OmrA and OmrB repress ompR expression as part of a negative feed-back loop (Guillier and Gottesman 2008) and the σE- regulated MicA RNA connects one regulatory input (outer membrane stress) to another (the PhoPQ regulon) (Cornaert et al. 2010) (Fig. 1). We predict that the interconnections between sRNAs and transcription regulators will only increase as more sRNA targets are identified.
Figure 1.
Regulatory circuits for base pairing sRNAs. sRNAs are shown as red boxes, and the transcription regulators known to regulate their synthesis as blue ovals. Both positive and negative regulators are shown. Feedback regulation can be direct (as for OmrA and OmrB regulation of OmpR) or indirect (as for RyhB regulation of Fur).
4.3. Variations
With the characterization of an increasing number of base pairing sRNAs, more and more regulatory variations are being discovered; here three are described in more detail.
Dual functions as mRNA and sRNA:SgrS and RNAIII
First, not all base pairing sRNAs are “noncoding.” The E. coli SgrS RNA was initially identified as an Hfq binding sRNA whose expression was induced by the presence of sugar phosphates and which base paired with and blocked translation of ptsG mRNA, encoding a sugar phosphate transporter (reviewed in Vanderpool 2007). However, it was noted that the sRNA is unusually long for a base pairing sRNA and further inspection of the sequence revealed that SgrS also encodes a 43 amino acid protein denoted SgrT (Wadler and Vanderpool 2007). Because expression of just the small protein has some of the same effects as expression of the noncoding region of SgrS involved in base pairing (such as decreased glucose uptake), it was hypothesized that SgrT reinforces the regulation brought about by the sRNA; SgrS inhibits PtsG synthesis by base pairing with the ptsG mRNA whereas SgrT inhibits the activity of the pre-existing transporter. Interestingly, a phylogenetic analysis of SgrS from a variety of related bacteria showed that the most conserved feature of the sRNA is the region of base pairing whereas the region encoding the small protein is much more variable (Horler and Vanderpool 2009).
The S. aureus RNA III is one of the longest base pairing sRNAs and also encodes a small protein, in this case the exotoxin hemolysin δ (reviewed in Romby and Charpentier 2009). Here again, the base pairing function and mRNA function of the sRNA are complementary, with pairing indirectly leading to increased synthesis of exotoxins by repressing expression of Rot (Boisset et al. 2007). These two examples open up the likelihood that RNAs now considered to be only mRNAs might in fact be bifunctional, also having a role as regulatory RNAs. This possibility significantly complicates the search strategies for regulatory RNAs, because we do not currently know how to distinguish a bifunctional RNA from an mRNA.
Base-pairing riboswitch: SreA, SreB
Bifunctional RNAs exist in other forms as well. As described in (Breaker, Riboswitches and the RNA World), riboswitches are leader sequences that control expression of the downstream genes through metabolite (ranging from flavin mononucleotide to molybdenum cofactor) induced changes in secondary structures. The changes in mRNA structure can alter ribosome binding or transcription termination. The short transcripts generated by riboswitch promoted transcription termination have been detected in a number of sRNA screens, raising the possibility that they also could act in trans (Vogel et al. 2003; Kawano et al. 2005a). Recently S-adenosylmethionine riboswitches of Listeria monocytogenes, denoted SreA and SreB, have been reported to base pair with the mRNA encoding the PrfA virulence regulator, leading to an approximately twofold reduction in the levels of this regulator (Loh et al. 2009). This example raises the possibility that other riboswitches have additional functions.
Regulating sRNA levels by mimics: chbBC and GlmY
Although the synthesis of many base-pairing sRNAs is regulated by specific transcription factors, this is not the only means by which the levels of the sRNAs can be controlled. The E. coli and S. enterica sRNA, alternatively called ChiX and MicM, was shown to strongly repress expression of the mRNA encoding the ChiP chitoporin (also denoted YbfM) (Rasmussen et al. 2009). Surprisingly, genetic screens for regulators of ChiX did not result in the identification of transcription factors. Instead an intergenic region in the chbBCARFG (chitobiose use) operon was found to modulate ChiX levels (Figueroa-Bossi et al. 2009; Overgaard et al. 2009). Further characterization of this region revealed that ChiX can base pair with the mRNA in the chbBC intergenic region and that this base pairing triggers decay of ChiX. Thus ChiX levels are regulated by an “mRNA target mimic” that promotes degradation of the sRNA. In a slightly different twist, the levels of the GlmZ RNA, which base pairs with the glmUS mRNA to promote translation of the glmS-encoded GlcN-6-P synthase, are modulated by the GlmY “sRNA mimic” (Kalamorz et al. 2007; Urban and Vogel 2008). In this case, high levels of the GlmY RNA appear to titrate a factor responsible for GlmZ degradation (Reichenbach et al. 2008; Urban and Vogel 2008). In both examples, the transcription of the mimics, chbBCARFG and glmY, is controlled by environmental signals sensed by transcription regulators (Figueroa-Bossi et al. 2009; Reichenbach et al. 2009).
Regulation of sRNAs solely by target mimics may be rare, but if the sRNAs are generally used stoichiometrically (Massé et al. 2003), all mRNA targets will act as competitors with one another, unless the sRNA is in excess. Thus, it seems quite possible that hierarchies of mRNA regulation exist, and are not fully appreciated under the usual laboratory test conditions of sRNA overproduction.
5. sRNAs THAT MODIFY PROTEIN ACTIVITY
In addition to acting by base pairing, sRNAs can interact with proteins, modifying their activities by mimicking and thus competing with RNA or DNA targets. In bacteria, two families of regulatory sRNAs that act by mimicking other nucleic acids have been characterized most extensively and will be discussed here. The first, exemplified by the E. coli 6S RNA, mimics a DNA promoter open complex and interacts with RNA polymerase. The second, the sRNAs that regulate the CsrA/RsmA family of translation regulatory proteins by competing with mRNA targets, will be referred to here as the CsrB family of sRNAs.
5.1. 6S sRNA
Although the E. coli 6S RNA was first identified as a distinct RNA species in 1967, the function of this sRNA was not understood until more than 30 years later, when 6S was found to bind tightly to the RNA polymerase holoenzyme containing σ70 but not the related σS holoenzyme (reviewed in Wassarman 2007). Binding of 6S inhibits RNA polymerase activity in vitro. In vivo, it preferentially inhibits a subset of promoters (those with weak -35 regions) (Cavanagh et al. 2008). The 6S structure, determined both by probing experiments and by comparing the 6S RNA in a variety of bacteria (see the following), is a double-stranded RNA hairpin with a critical bubble that mimics the DNA in an open complex promoter, binding at the active site of RNA polymerase. Evidence that this sRNA can mimic DNA is provided by the finding that, both in vitro and in vivo, RNA polymerase can synthesize a short transcript encoded by the 6S RNA, starting within the bubble (Wassarman and Saecker 2006; Gildehaus et al. 2007; Sharma et al. 2010). Interaction of the 6S RNA with RNA polymerase requires region 4.2 of σ70 (Cavanagh et al. 2008). The same region of σ70 is involved in binding to the -35 region of promoter DNA, although the amino acids required for the interactions with the promoter DNA and 6S RNA are somewhat distinct (Klocko and Wassarman 2009).
Unlike most other E. coli regulatory sRNAs, 6S is processed from a longer mRNA, although the function of the downstream gene (ygfA) is unknown. The RNA accumulates in stationary phase and acts as one of the multiple regulatory inputs for down-regulating vegetative genes and allowing preferential expression of stationary phase genes (transcribed by RNA polymerase containing σS). Recovery from 6S inhibition appears to be caused by the release of 6S from RNA polymerase on transcription from the inhibited complex; it may be that this is the purpose of this transcription. Alternatively or in addition, the short 6S templated RNAs, termed pRNAs, may have independent functions. In vitro, transcription from 6S requires high levels of nucleotides and therefore may only occur during the transition from stationary phase into exponential growth when nucleotide levels rise. Once released, 6S becomes sensitive to degradation (Wassarman and Saecker 2006; Wassarman 2007; Wurm et al. 2010).
6S RNAs are found fairly broadly in other bacterial species, including Gram-positive as well as Gram-negative bacteria (Barrick et al. 2005; Trotochaud and Wassarman 2005; Shi et al. 2009). However, primary sequence conservation is very limited. Thus computational searches for 6S homologs have depended on both secondary structure conservation and the linkage to the downstream ygfA ORF, found in some but not all organisms. Because 6S RNA is generally 180–200 nucleotides, not overlapping in size with tRNAs, it has also been possible to find 6S as an abundant and stable RNA (as it was first found in E. coli) or by its ability to bind to RNA polymerase (Watanabe et al. 1997; Trotochaud and Wassarman 2005; Wilkomm et al. 2005). Some organisms have multiple 6S RNAs (Barrick et al. 2005). For example, two 6S-like RNAs are present in B. subtilis, both able to bind the vegetative RNA polymerase (Trotochaud and Wassarman 2005), whereas Clostridium has three 6S-like RNAs (Barrick et al. 2005). The purpose of multiple RNAs is not yet known, but it is certainly possible that 6S-like RNAs in other organisms will bind different forms of RNA polymerase, opening up the possibility of a broader role for 6S-like RNAs.
5.2. CsrB Family of RNAs
The CsrB family of sRNAs counteracts the activities of the small CsrA protein and its homologs (such as RsmA) by titrating them away from their mRNA binding sites. In E. coli, CsrA was first identified as a critical posttranscriptional regulator of the switch between gluconeogenesis and glycolytic growth, inhibiting glycogen synthesis (reviewed in Babitzke et al. 2009). Most of the studied regulation by CsrA is negative; the protein inhibits translation, usually by binding near and thus blocking the binding site. There are reported cases of positive regulation that appear to be direct because they occur in a coupled transcription/translation system (Wei et al. 2001), but the mechanism for positive regulation has not been reported. Binding of CsrA may block other negative translational regulators from binding or may remodel the mRNA. The CsrA protein has a preference for binding GGA in the loop of an RNA hairpin (Dubey et al. 2005; Gutierrez et al. 2005; Heeb et al. 2005; Schubert et al. 2007), and target mRNAs frequently contain multiple binding motifs.
Although synthesis of the family of CsrA proteins may change with environmental conditions (Cui et al. 2005), the major regulation appears to be via inhibition of CsrA activity by the CsrB family of RNAs. These RNAs contain as many as 18 CsrA binding motifs. When they are present at high levels, CsrA binds to them and therefore is not available for interacting with the target mRNAs. Thus, mutations in csrB and csrC, the genes for the two E. coli sRNAs in this family, lead to stringent repression of CsrA targets, and overexpression of CsrB (the more abundant of the two sRNAs) mimics a csrA mutation, leading to increased glycogen synthesis (reviewed in Babitzke and Romeo 2007).
Synthesis of the CsrB and CsrC RNAs is dependent on the two-component system BarA/UvrY, and this regulatory cascade is conserved in many bacteria, although the signals feeding into this cascade are not well defined. Additionally, the stability of CsrB and CsrC is regulated. The turnover of the sRNAs is generally fairly rapid (half-life of 2 minutes for CsrB, around 4 min for CsrC in E. coli), but these sRNAs are fully stabilized in cells mutant for either RNase E, the essential endonuclease involved in degradation of many mRNAs, or CsrD, a protein with homology to, but lacking the activity of, proteins associated with synthesis and degradation of the signaling molecule cyclic diGMP (Suzuki et al. 2009). The effects of CsrD in destabilizing these RNAs appear to be fairly specific, raising a number of questions about how this occurs, as well as the question of whether or not this is a regulatory point and if so, what regulates CsrD (Suzuki et al. 2009).
CsrA-like proteins are fairly widespread, found in both Gram-negative and Gram-positive bacteria, sometimes in more than one copy, although the majority of studies have been with the Gram-negative proteins (White et al. 1996). The CsrB/Rsm RNAs have been found in Enterobacteria, Pseudomonads, and Legionella (Kulkarni et al. 2006; Babitzke and Romeo 2007; Sahr et al. 2009); possibly they are more widespread but have diverged sufficiently to make identification of the equivalent RNAs difficult in more distantly related species. The network of CsrA-like proteins, the BarA/UvrY family of two-component regulators, and downstream sRNA regulators seem to play central roles in the selection of bacterial life-styles between swarming, free-swimming, and biofilm formation, as well as modulating virulence, although the precise roles vary between bacterial species. Much remains to be understood about this network, including how environmental signals impinge on it to up-regulate or down-regulate the sRNAs by transcription or RNA turnover.
5.3. More sRNAs that Regulate Proteins?
As additional sRNA regulators are characterized, it seems likely that more will be found to interact with and regulate proteins, as has been found with the 6S and CsrB RNAs. Certainly there are a large number of known RNA-binding proteins involved in translation, RNA splicing, and trafficking, and generally affecting mRNA fates; many more are likely to exist. It is easy to imagine that sRNAs will be found that regulate these proteins. Other sRNAs may well mimic DNA, as 6S does, and therefore may act as regulators of a variety of DNA binding proteins. In principle, regulatory sRNAs could also fold into a form that can interact with proteins in other ways, such as acting as a scaffold or tether to bring proteins together.
6. sRNAs WITH INTRINSIC ACTIVITIES
Bacteria also have a small number of sRNAs with known activities that do not involve pairing or the regulation of proteins. The ribozyme RNase P has been well characterized (reviewed in Gopalan and Altman 2006; Altman 2007). A detailed description of 4.5S, the RNA component of bacterial signal recognition particle can also be found elsewhere (reviewed in Egea et al. 2005). Here we will discuss tmRNA (also denoted 10Sa and SsrA RNA), which is widespread within bacteria, is also found in some eukaryotic organelles, and carries out both quality control and regulatory roles.
tmRNA was named for its hybrid characteristics; it has both a tRNA-like domain that is charged by alanine synthetase and an mRNA segment encoding a short ORF that is used as an extension for cotranslational tagging of proteins (reviewed in Moore and Sauer 2007). The mechanism by which tmRNA is recruited to a ribosome when translation stalls and/or the mRNA end lacks a translation termination codon is well studied. On recognizing a stalled ribosome, tmRNA acts first like a tRNA, adding an alanine to the stalled polypeptide chain, and then like a mRNA by encoding a short amino acid tag (10 amino acids in E. coli). Because the short ORF ends in a termination codon, it allows release of the polypeptide and re-use of the stalled ribosome. The amino acid tag on the released polypeptide is efficiently recognized by a number of bacterial ATP-dependent proteases, particularly ClpXP. This leads to rapid degradation of the tagged protein and therefore rescue from protein fragments that might otherwise accumulate (reviewed in Moore and Sauer 2007; Keiler 2008). One of the tmRNA roles is reducing stress associated with ribosome stalling/sequestration. This is reflected in the induction of the heat-shock response in cells mutant for tmRNA and in an increased need for tmRNA in response to antibiotics or genetic changes that lead to more stalled ribosomes (reviewed in Keiler 2008). In Neisseria and Helicobacter, tmRNA is essential (Huang et al. 2000; Thibonnier et al. 2008).
However, recent findings in a number of systems suggest that tmRNAs also may play specific regulatory roles. For example, in Yersinia, the lack of tmRNA is accompanied by avirulence and a defect in the transcription of effector proteins (Okan et al. 2006). In Caulobacter crescentus, a bacterium with a complex developmental cycle, tmRNA expression is regulated with the cell cycle and tmRNA mutants have aberrant timing of DNA replication (Keiler and Shapiro 2003). This disruption seems to be because of misregulation of dnaA transcription, encoding a key regulator of DNA replication; it may be that an unidentified transcription factor is inappropriately synthesized in the tmRNA mutants (Cheng and Keiler 2009).
There are several possibilities for how tmRNAs might play specific roles in regulation. It is known that some ORFs contain sequences that promote tmRNA-dependent tagging and therefore degradation (Roche and Sauer 2001; Sunohara et al. 2002). Conceivably, the absence of tagging leads to overproduction of the untagged protein under inappropriate conditions. Another route to tagging and therefore tmRNA-dependent degradation has been studied in the lacI gene encoding the Lac repressor. In this case, the Lac repressor binds near and within the lacI gene, blocking full transcription and therefore translation to the end of the gene; this then leads to tagging and degradation of the partially translated protein (Abo et al. 2000). These regulatory mechanisms do not require changes in the levels of tmRNA action, but rather that tmRNA maintains low levels of specific proteins. If tmRNA is transmitting regulatory information, we would guess that this is at the level of general translational stress (stalled ribosomes), possibly leading to changing efficiencies of tagging of a given protein.
7. EVOLUTIONARY CONSIDERATIONS
One interesting question for bacterial sRNAs is whether they have their origins in the RNA world or are newly evolving regulators. Although one can imagine that base pairing between RNAs was one of the earliest forms of regulation, the question of whether any of the bacterial sRNAs date from the RNA world cannot yet be answered. However, the rapid accumulation of genome sequence information together with the interest in uncovering sRNAs in a wide range of bacteria have provided glimpses into the evolution of these RNAs that may help to discern their history.
The protein-binding regulatory 6S and CsrB RNAs, as well as the functional RNase P RNA, SRP/4.5S, and SsrA/tmRNAs, are more broadly conserved than those that act by base pairing. These RNAs are a bit larger, and, in the case of 6S RNA and tmRNA, there are constraints on structure which have facilitated the tracking of these sRNAs in different species. The ability to find 6S-like RNAs over a wide range of bacteria both supports an early evolutionary origin for this regulatory RNA and points out the difficulties in tracing the evolution of sRNAs. Although the structure of 6S is reasonably well conserved, the sequence is not (only 23% of positions conserved at >80% throughout eubacteria) (Barrick et al. 2005). The linkage of this sRNA to a protein-coding gene in many organisms, more easily identified by homology, allowed the robust development of a structural model, therefore facilitating further searches for RNAs unlinked to this ORF.
7.1. Recent Evolution
For the Hfq-binding sRNAs, evolution of the regulatory RNAs appears to be rapid. As a result, neither sequence similarities nor structural similarities are sufficient to provide a clear picture of their evolution, even between relatively closely related species (such as in Gram-negative γ-Proteobacteria). Nevertheless some general constraints on, or contributions to, their evolution can be noted.
Constraints imposed by the targets
The Hfq-binding sRNAs have to pair with specific mRNA targets, and so the evolution of the sRNA and the targets must be linked. The observation that many, but not all, of these sRNAs contain a highly conserved core region, and that this region is frequently involved in pairing with targets (Sharma et al. 2007), supports the idea that pairing helps constrain evolution. For sRNAs with many targets, it may be that some particular target or targets are particularly critical and therefore may correlate better in terms of pairing with the sRNA. In this context, it is intriguing that a number of mRNAs are the targets of multiple sRNAs. This overlap complicates the possibilities for evolution and suggests that these multiple sRNAs may have a common ancestor.
Conserved regulation
The regulators controlling the expression of the sRNAs are other factors that can be used to trace the evolution of sRNAs. The most broadly studied class of potentially related base-pairing sRNAs are those regulated by iron availability via the Fur repressor. This repressor and the sites to which it binds are strikingly well conserved from Gram-negative to Gram-positive bacteria (reviewed in Carpenter et al. 2009). It is becoming increasingly apparent that one or more sRNA is part of the Fur regulon in many organisms. The E. coli RyhB RNA was the first example of a Fur-regulated RNA found. Other Enterobacterial species have sRNAs clearly related to RyhB, sharing the Fur binding site in the promoter and a conserved core region involved in pairing to many but not all of the same targets (reviewed in Gottesman et al. 2007). Second copies of RyhB-like RNAs are found in some of these species. In organisms such as Pseudomonas, the Fur-regulated sRNAs, called PrrF, have Fur binding sites in the promoter and regulate similar types of genes to those regulated in E. coli, but have no obvious sequence similarity to the other RyhB RNAs (Wilderman et al. 2004). Either these sRNAs evolved independently, certainly a possibility, or they have diverged significantly. More recently, Fur-regulated sRNAs have been found in Neisseria and B. subtilis; again, these sRNAs bear no sequence similarity to RyhB beyond the Fur binding sites (Mellin et al. 2007; Gaballa et al. 2008), also suggesting either independent evolution or rapid divergence.
Gene duplication
Similar to eukaryotic miRNAs, where proliferation of related families has occurred, there is evidence of sRNA gene duplication in bacteria. As mentioned earlier, two Fur-regulated sRNAs with significant overlap in sequence are found in some bacteria. In most Pseudomonads, the two prrF genes are located at two different chromosomal locations. In P. aeruginosa, however, the two highly similar genes are located adjacent to each other. One evolutionary scenario is that the original prrF gene duplicated and then moved to a new site. In an ancestor of P. aeruginosa, one of these copies was lost or did not survive the move, and the original gene then duplicated more recently (Gottesman et al. 2007). There are other duplicated sRNAs as well. The highly similar omrA and omrB genes are adjacent and commonly regulated in E. coli. Vibrio species have four to five copies of the Qrr genes, at separate chromosomal locations, with varying degrees of divergence (Lenz et al. 2004). Thus duplication of genes has played a role in the distribution of the several of the sRNAs. Having more than one copy of an sRNA gene can have regulatory implications, allowing for increased induction or differential regulation of the copies.
Conserved gene neighborhood
Given the lack of sequence similarity between similarly regulated sRNAs with related functions, the use of gene context is proving to be another means of investigating the functional conservation of sRNAs. For example, tracking of the Hfq-binding GcvB RNA has been simplified because it is always found divergently transcribed from its transcription regulator, GcvA. GcvB is found throughout γ-Proteobacteria, except for Pseudomonads (Sharma et al. 2007). Spot42, a conserved sRNA found by homology only in Enterobacterial species, occupies an intergenic region between two highly conserved protein-coding genes; in Pseudomonas, this same intergenic neighborhood encodes an sRNA of unknown function, with no sequence similarity to Spot42 (Gottesman et al. 2007). One explanation is that Spot42 and this sRNA are evolutionarily related but have diverged rapidly. Alternatively, this is a location into which sRNA genes are inserted from different (and unknown) sources. The characterization of more mRNA targets of both of these sRNAs may give insights into whether they are indeed related.
Horizontal transfer
Are sRNAs more or less likely than other genes to be part of mobile genetic elements, and might they be subject to horizontal transfer? In E. coli, DicF, an Hfq-binding sRNA, is found within a cryptic prophage, but regulates the host ftsZ gene (Bouché and Bouché 1989). IpeX, an Hfq-independent sRNA that regulates porin genes, was also found to be encoded in a cryptic E. coli prophage (Castillo-Keller et al. 2006). In specific searches for sRNAs in pathogenicity islands in S. enterica, a number of such regulatory RNAs were found, including a second copy of a Fur-regulated RyhB-like sRNA (Padalon-Brauch et al. 2008). Both the RyhB-related sRNA and at least one other pathogenicity island sRNA, InvR, regulate genes encoded in the core genome (Pfeiffer et al. 2007). Pathogenicity island sRNAs have also been discovered for S. aureus (Pichon and Felden 2005) and are likely to be present in other pathogens. The reciprocal case where a host-encoded sRNA regulates the expression of an island-encoded gene has been found as well (Papenfort and Vogel, unpublished). A number of the chromosomally located sRNAs lie adjacent to regions into which phage or transposons have been shown to insert, suggesting that they may either be picked up by these elements when the elements excise, or that they were first brought into the genome with these elements (De Lay and Gottesman 2009).
It is worth remembering that antisense sRNAs are intimately involved in regulating both plasmid copy number and plasmid conjugation (reviewed in Wagner et al. 2002; Brantl 2007). As in eukaryotes, where RNA interference plays a critical role in limiting the spread of transposable elements, transposons in bacteria are also limited in their activity by antisense RNAs. Thus, the elements for horizontal transfer carry within them regulatory sRNAs that could evolve to have complementarity with targets other than the mobile elements.
7.2. Speculation on Deeper Evolution
The examples described in the previous section provide a glimpse at the recent evolution of the base-pairing sRNAs in bacteria, and suggest that the constraints on their evolution are loose enough so that they change sequence and structure relatively rapidly. Given a rapid rate of change, it is not yet possible to develop hard evidence for original sources of these sRNAs, leaving us free to speculate. There is no reason to assume a single evolutionary pathway for the rather divergent sRNAs already identified in bacteria, so all (or none) of the possibilities may turn out to be correct.
Capture of random transcription for the purpose of regulation
As more deep sequencing and tiling array analysis is performed, a certain level of what might be considered transcriptional “noise,” low-level transcription antisense to genes and within spacer regions, with no assigned function thus far, is apparent. The hypothesis that there is low level transcription that does not lead to productive transcripts is supported by the finding of antisense promoter activity without the detection of stable transcripts (Kawano et al. 2005b). It is relatively easy to imagine that if some low-level promoter activity were sufficient to give advantageous regulation under some condition, both the promoter and the RNA might evolve into a highly expressed regulatory sRNA.
Capture of transcripts of other function: sRNA-tRNA connections
Another possible source of regulatory sRNAs is tRNAs, or the ancestors of tRNAs. These are folded, stable RNAs of about the same size as many of the regulatory RNAs. Several characteristics are suggestive of a relationship between regulatory sRNAs and tRNAs. As for tRNAs, both tmRNA (clearly tRNA-related) and E. coli CyaR, an Hfq-dependent sRNA, act as attachment sites for phage integration (Kirby et al. 1994; De Lay and Gottesman 2009). Because it is not really clear why tRNA genes are used this way, the implications of this are not known (Campbell 1992). In eukaryotic cells, retrotransposons make use of tRNA primers. A number of studies have suggested that specific tRNAs also interact with Hfq, possibly because Hfq modulates their processing (Zhang et al. 2003; Lee and Feig 2008). This ability of tRNAs to bind Hfq could be considered support for a common ancestry. One prediction is that tRNAs (or more specifically tRNA fragments) will be found to have regulatory roles. This prediction is supported by recent studies in eukaryotic cells (Cole et al. 2009; Lee et al. 2009; Thompson and Parker 2009); investigations of functions for tRNA fragments in bacteria have not yet been reported. A second prediction is that genes which appear to be similar to, but not quite like, tRNAs will be shown to be regulatory sRNAs rather than functional tRNAs.
Capture of transcripts of other function: sRNA-mRNA connections
A third possible source for regulatory sRNAs are mRNAs. The sRNAs that also encode proteins and the bifunctional riboswitches recently described in L. monocytogenes (see earlier) provide examples for what precursors for free-standing regulatory sRNAs might be. In E. coli, mRNAs that are targets for Hfq-dependent sRNA regulation all appear to have Hfq-binding sites; it is not difficult to imagine that an Hfq-binding UTR or intergenic region could be both the target of regulation and, under other conditions, the regulator. An example of an mRNA regulator has already been described (see earlier discussion, section on RNA mimics). Separation from the mRNA or loss of the downstream ORF might then lead to what we now detect as a free-standing sRNA. This model might predict that further sequencing will reveal cases where sequences similar to sRNAs are found as UTRs or within mRNAs. Certainly, in the eukaryotic RNA world, the appearance of miRNAs encoded within introns is consistent with the possibility that pieces of evolving mRNAs may become regulatory sRNAs, or vice versa.
8. PERSPECTIVES
Although regulatory RNAs have been a major topic of interest in eukaryotic cells, studies of bacterial sRNAs have been equally exciting. Exactly why the cell regulates a given gene or set of genes via regulatory RNAs, rather than at the level of transcription initiation, is not yet understood, but the availability of multiple sRNAs, identification of their upstream regulators and of their targets, is allowing investigators to begin to test the advantages of sRNA as regulators in mathematical models (Levine and Hwa 2008; Mehta et al. 2008; Mitarai et al. 2009). In many instances, it may not be a question of one type of regulation over another, but that the requirement for many bacteria to respond rapidly to changing environments has led to multiple levels of regulation, among them the very versatile and easily adaptable regulatory sRNAs.
ACKNOWLEDGMENTS
The writing of this review was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank members of our laboratories, C. Vanderpool and J. Vogel for comments on the manuscript, and thank J. Vogel for sharing unpublished results.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Abo T, Inada T, Ogawa K, Aiba H 2000. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J 19: 3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S 2007. A view of RNase P. Mol Biosyst 3: 604–607 [DOI] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G 1997. A small stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 90: 43–53 [DOI] [PubMed] [Google Scholar]
- Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G 1998. The Escherichia coli oxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 17: 6069–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Delihas N 1990. micF RNA binds to the 5′ end of ompF mRNA and to a protein from Escherichia coli. Biochemistry 29: 9249–9256 [DOI] [PubMed] [Google Scholar]
- Antal M, Bordeau V, Douchin V, Felden B 2005. A small bacterial RNA regulates a putative ABC transporter. J Biol Chem 280: 7901–7908 [DOI] [PubMed] [Google Scholar]
- Babitzke P, Baker CS, Romeo T 2009. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol 63: 27–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Romeo T 2007. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10: 156–163 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR 2005. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F, Bouché JP 1989. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol 3: 991–994 [DOI] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32: 827–837 [DOI] [PubMed] [Google Scholar]
- Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P, Overgaard M 2010. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem 285: 10690–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Campbell AM 1992. Chromosomal insertion sites for phages and plasmids. J Bacteriol 174: 7495–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, Merrell DS 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect Immun 77: 2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Keller M, Vuong P, Misra R 2006. Novel mechanism of Escherichia coli porin regulation. J Bacteriol 188: 576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh AT, Klocko AD, Liu X, Wassarman KM 2008. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of σ70. Mol Microbiol 67: 1242–1256 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang A, Blyn LB, Storz G 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol 186: 6689–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Keiler KC 2009. Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. J Bacteriol 191: 4268–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH 2004. The RNA-binding protein Hfq of Listeria monocytogenes: Role in stress tolerance and virulence. J Bacteriol 186: 3355–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M 2010. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 76: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyer J, Andersen J, Forst SA, Inouye M, Delihas N 1990. micF RNA in ompB mutants of Escherichia coli: different pathways regulate micF RNA levels in response to osmolarity and temperature change. J Bacteriol 172: 4143–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chatterjee A, Hasegawa H, Dixit V, Leigh N, Chatterjee AK 2005. ExpR, a LuxR homolog of Erwinia caratovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA-binding protein. J Bacteriol 187: 4792–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S 2009. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prevost K, Massé E 2009. Small RNA-induced differential degradation of the polycistronic mRNA. iscRSUA. EMBO J 28: 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchin V, Bohn C, Bouloc P 2006. Down-regulation by porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J Biol Chem 281: 12253–12259 [DOI] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhring U, Axmann IM, Hess WR, Wilde A 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci 103: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Storz G 2010. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 75: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P 2005. Targeting proteins to membranes: Structure of the signal recognition particle. Curr Opin Struct Biol 15: 213–220 [DOI] [PubMed] [Google Scholar]
- Faubladier M, Cam K, Bouché J-P 1990. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J Mol Biol 212: 461–471 [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Bossi L 2009. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 1981–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Hemm MR, Storz G 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G 2010. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song K-B, Smaldone GT, Helmann JD 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci 105: 11927–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J, Vos B, Scholz I, Mitschke J, Wilde A, Hess WR 2009. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Sys Biol 5: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Wagner EG 2007. RNA antitoxins. Curr Opin Microbiol 10: 117–124 [DOI] [PubMed] [Google Scholar]
- Gildehaus N, Neusser T, Wurm R, Wagner R 2007. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res 35: 1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan V, Altman S 2006. Ribonuclease P: Structure and catalysis. In The RNA world (ed. Gesteland R.F., Cech T.R., Atkins J.F.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ 2007. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp Quant Biol 71: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen W-H, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, et al. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59: 231–247 [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36: 6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G 2006. Modulating the outer membrane with small RNAs. Genes Dev 20: 2338–2348 [DOI] [PubMed] [Google Scholar]
- Gutierrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol 187: 3496–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P 2005. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol 355: 1026–1036 [DOI] [PubMed] [Google Scholar]
- Heidrich N, Moll I, Brantl S 2007. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res 35: 4331–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich N, Chinali A, Gerth U, Brantl S 2006. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol 62: 520–536 [DOI] [PubMed] [Google Scholar]
- Horler RS, Vanderpool CK 2009. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res 37: 5465–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI 2000. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J 19: 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Boisset S, Saveneau C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 24: 824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P 2008. Down-regulation of outer membrane proteins by noncoding RNAs: Unraveling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J Mol Biol 383: 1–9 [DOI] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P 2006. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J Mol Biol 364: 1–8 [DOI] [PubMed] [Google Scholar]
- Kalamorz F, Reichenbach B, Marz W, Rak B, Gorke B 2007. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol 65: 1518–1533 [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H 2005. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev 19: 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Reynolds AA, Miranda-Rios J, Storz G 2005a. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 33: 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Storz G, Rao BS, Rosner JL, Martin RG 2005b. Detection of low-level promoter activity within open reading frame sequences of Escherichia coli. Nucleic Acids Res 33: 6268–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC 2008. Biology of trans-translation. Annu Rev Microbiol 62: 133–151 [DOI] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L 2003. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol 185: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JE, Trempy JE, Gottesman S 1994. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol 176: 2068–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko AD, Wassarman KM 2009. 6S RNA binding to Eσ70 requires a positively charged surface of σ70 region 4.2. Mol Microbiol 73: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke L, Wulff DL 1987. OOP RNA, produced from multicopy plasmids, inhibits λ cII gene expression through an RNase III-dependent mechanism. Genes Dev 1: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Krinke L, Wulff DL 1990. RNase III-dependent hydrolysis of λ cII-O gene mRNA mediated by λ OOP antisense RNA. Genes Dev 4: 2223–2233 [DOI] [PubMed] [Google Scholar]
- Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res 34: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Belfort M 2000. Riboregulation by DsrA RNA: Trans-actions for global economy. Mol Microbiol 38: 667–672 [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick M, Belfort M 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA 95: 12456–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Feig AL 2008. The RNA binding protein Hfq interacts specifically with tRNAs. RNA 14: 514–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Levine E, Hwa T 2008. Small RNAs establish gene expression thresholds. Curr Opin Microbiol 11: 574–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA 106: 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Waldor MK 2007. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol 10: 96–101 [DOI] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139: 770–779 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 46: 813–826 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S 2005. Bacterial small RNA regulators. CRC Crit Rev Biochem 40: 93–113 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S 2001. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol Microbiol 39: 1382–1394 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S 2010. submitted [Google Scholar]
- Massé E, Gottesman S 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Salvail H, Desnoyers G, Arguin M 2007. Small RNAs controlling iron metabolism. Curr Opin Microbiol 10: 140–145 [DOI] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Wingreen NS 2008. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol 4: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA 2007. A novel Fur- and Iron-regulated small RNA, NrrF, is required for indirect Fur-medicated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol 189: 3686–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai N, Benjamin J-AM, Krishna S, Semsey S, Csiszovszki Z, Massé E, Sneppen K 2009. Dynamic features of gene expression control by small regulatory RNAs. Proc Natl Acad Sci USA 106: 10655–10659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Molec Microbiol 74: 1314–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124 [DOI] [PubMed] [Google Scholar]
- Morfeldt E, Taylor D, von Gabain A, Arvidson S 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14: 4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH 2009. Defining a role for Hfq in gram-positive bacteria: Evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res 38: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Bliska JB, Karzai AW 2006. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog 2: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186: 6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P 2009. Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800 [DOI] [PubMed] [Google Scholar]
- Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S 2008. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36: 1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68: 890–906 [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JCD, Vogel J 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62: 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J 2009. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158 [DOI] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA 106: 15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16: 840–846 [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66: 1174–1191 [DOI] [PubMed] [Google Scholar]
- Pichon C, Felden B 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci USA 102: 14249–14254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis Mol Microbiol 64: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV 2009a. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology 155: 106–114 [DOI] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV 2009b. The small RNA GcvB regulates sstT mRNA expression in Escherichia coli. J Bacteriol 191: 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Ericksen M, Gilany K, Udesen C, Franch T, Peterson C, Valentin-Hansen P 2005. Regulation of ompA mRNA stability: The role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol 58: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P 2009. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol 72: 566–577 [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Gopel Y, Gorke B 2009. Dual control by perfectly overlapping σ54- and σ70-promoters adjusts small RNA GlmY expression to different environmental signals. Mol Microbiol 74: 1054–1070 [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res 36: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem 276: 28509–28515 [DOI] [PubMed] [Google Scholar]
- Romby P, Charpentier E 2009. An overviews of RNAs with regulatory functions in gram-positive bacteria. Cell Mol Life Sci 67: 217–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol 72: 741–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Alain FH 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14: 807–813 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J 21: 3546–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Vogel J 2009. Experimental approaches for the discovery and characterization of regulatory small RNAs. Curr Opin Microbiol 12: 536–546 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J 2007. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 21: 2804–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffman S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, DeLong EF 2009. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459: 266–269 [DOI] [PubMed] [Google Scholar]
- Silvaggi JM, Perkins JB, Losick R 2005. Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol 187: 6641–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D, Gottesman S 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci 92: 2003–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15: 3993–4000 [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14: 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T, Abo T, Inada T, Aiba H 2002. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA 8: 1416–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T 2009. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL 2009. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J 28: 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Waters CM, Bassler BL 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev 22: 226–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetart F, Bouché JP 1992. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol 6: 615–620 [DOI] [PubMed] [Google Scholar]
- Thibonnier M, Thiberge J-M, De Reuse H 2008. Trans-translation in Helicobacter pylori: Essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PloS One 3: e3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009. Stressing out over tRNA cleavage. Cell 138: 215–219 [DOI] [PubMed] [Google Scholar]
- Thompson KM, Rhodius VA, Gottesman S 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol 189: 4243–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tensuu T, Vaitkevicius K, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, De Biase D 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol 70: 965–982 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12: 313–319 [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev 21: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL 2008. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol 70: 896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Wagner EG 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res 35: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV 2000. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol 37: 856–868 [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol 10: 146–151 [DOI] [PubMed] [Google Scholar]
- Vogel J, Papenfort K 2006. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9: 605–611 [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang HH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31: 6435–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA 104: 20454–20459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG, Altuvia S, Romby P 2002. Antisense RNAs in bacteria and their genetic elements. Adv Genet 46: 361–398 [DOI] [PubMed] [Google Scholar]
- Wassarman KS 2007. 6S RNA: A regulator of transcription. Mol Microbiol 65: 1425–1431 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Saecker RM 2006. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314: 1601–1613 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sugiura M, Sugita M 1997. A novel small stable RNA, 6Sa RNA, from the cyanobacterium Synechococcus sp. strain PCC3601. FEBS Lett 416: 302–306 [DOI] [PubMed] [Google Scholar]
- Weaver KE 2007. Emerging plasmid-encoded antisense RNA regulated systems. Curr Opin Microbiol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40: 245–256 [DOI] [PubMed] [Google Scholar]
- White D, Hart ME, Romeo T 1996. Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene 182: 221–223 [DOI] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci 101: 9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkomm DK, Minnerup J, Huttenhofer A, Hartmann RK 2005. Experimental RNomics in Aquifex aeolicus: Identification of small non-coding RNAs and the putative 6S RNA homolog. Nucleic Acids Res 33: 1949–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm R, Neusser T, Wagner R 2010. 6S RNA-dependent inhibition of RNA polymerase is released by RNA-dependent synthesis of small de novo products. Biol Chem 391: 187–196 [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]