Abstract
The postsynaptic side of the synapse is specialized to receive the neurotransmitter signal released from the presynaptic terminal and transduce it into electrical and biochemical changes in the postsynaptic cell. The cardinal functional components of the postsynaptic specialization of excitatory and inhibitory synapses are the ionotropic receptors (ligand-gated channels) for glutamate and γ-aminobutyric acid (GABA), respectively. These receptor channels are concentrated at the postsynaptic membrane and embedded in a dense and rich protein network comprised of anchoring and scaffolding molecules, signaling enzymes, cytoskeletal components, as well as other membrane proteins. Excitatory and inhibitory postsynaptic specializations are quite different in molecular organization. The postsynaptic density of excitatory synapses is especially complex and dynamic in composition and regulation; it contains hundreds of different proteins, many of which are required for cognitive function and implicated in psychiatric illness.
Inhibitory and excitatory synapses differ with regard to their neurotransmitter receptors and morphology, but share some organizing principles (e.g., adhesion-facilitated differentiation and scaffold-based assembly of proteins).
Excitatory synapses on principal neurons of mammalian brain occur mainly on tiny protrusions called dendritic spines (Bourne and Harris 2008). In contrast, inhibitory synapses are formed on the shaft of dendrites, or on cell bodies and axon initial segments. The postsynaptic side of excitatory synapses differs from inhibitory synapses not only in their content of neurotransmitter receptors but also in their morphology and molecular composition and organization. In part because of their greater abundance and distinctive structure, much more is known about the postsynaptic organization of central excitatory (glutamatergic) synapses.
THE POSTSYNAPTIC DENSITY OF EXCITATORY SYNAPSES
Excitatory synapses are characterized by a morphological and functional specialization of the postsynaptic membrane called the postsynaptic density (PSD), which is usually located at the tip of the dendritic spine. The PSD contains the glutamate receptors that are activated by the glutamate neurotransmitter released from the presynaptic terminal, as well as a host of associated signaling and structural molecules. A set of abundant scaffold proteins holds together the PSD by binding to the glutamate receptors, other postsynaptic receptors and adhesion molecules, cytoplasmic signaling enzymes, and cytoskeletal elements. The generally accepted roles for the PSD are to mediate the apposition of pre- and postsynaptic membranes, to cluster postsynaptic receptors, and to couple the activation of postsynaptic receptors to biochemical signaling events in the postsynaptic neuron.
The PSD was first observed in electron micrographs as a fuzzy electron-dense thickening of the postsynaptic membrane that is apposed to the presynaptic active zone (hence the term “asymmetric” synapses) (Gray 1959; Siekevitz 1985). The PSD is essentially a proteinaceous organelle attached to the postsynaptic plasma membrane and held by cytoplasmic actin filaments (Siekevitz 1985; Kennedy 2000; Sheng and Kim 2002; Funke et al. 2004). PSDs typically have a disclike shape, although larger PSDs are often irregular or perforated. They are highly heterogeneous in size: diameter ∼200–800 nm (mean 300–400 nm) and thickness ∼30–60 nm (Carlin et al. 1980). The size of the PSD correlates with the size of the dendritic spine in which they are located and with the abundance of postsynaptic glutamate receptors (Kasai et al. 2003), consistent with the idea that bigger synapses are stronger synapses. The molecular mass of the average PSD has been calculated to be ∼1 gigadalton (Chen et al. 2005), which would correspond to ∼10,000 copies of a 100 kDa protein.
COMPONENTS OF THE PSD
In pioneering experiments of the 1970s, PSDs were purified by detergent treatment of synaptosomes isolated by density gradient centrifugation (Davis and Bloom 1973; Cotman et al. 1974; Blomberg et al. 1977; Cohen et al. 1977). Early analysis of PSD structure and composition by electron microscopy (EM) and gel electrophoresis led Siekevitz to hypothesize that changes in the concentration and conformation of PSD proteins may lead to long-term changes in synapses and neuronal circuits (Siekevitz 1985).
In the 1990s, peptide sequencing of PSD components separated by gel electrophoresis enabled the identification of a variety of abundant PSD proteins, including PSD-95 and calcium calmodulin-dependent kinase II (CaMKII) (Cho et al. 1992; Walsh and Kuruc 1992; Kennedy 2000). Antisera raised against purified synapses also led to cloning of many PSD proteins (Kistner et al. 1993; Langnaese et al. 1996; Garner et al. 2000). Another fruitful approach to find PSD proteins was yeast two-hybrid screens for interacting proteins using known postsynaptic proteins as “bait.” For instance, yeast two-hybrid screens with the cytoplasmic tails of N-methyl-d-aspartate (NMDA) receptors and K+ channels revealed that these membrane proteins bind directly to the PSD-95 family of proteins (Kim et al. 1995; Kornau et al. 1995; Niethammer et al. 1996). Similarly, PSD-95 was found to bind to multiple other PSD proteins, including SynGAP and GKAP; the latter in turn interacts with Shank (Funke et al. 2004; Kim and Sheng 2004).
The development of mass spectrometry (MS) greatly accelerated the identification of PSD components in purified PSDs (Walikonis et al. 2000; Sheng and Hoogenraad 2007; Bayes and Grant 2009). For instance, Peng et al. identified ∼400 PSD proteins and categorized them into 13 functional groups, of which actin-cytoskeletal, kinase signaling and GTPase signaling pathways were highly represented (Fig. 1A) (Peng et al. 2004). Many of these proteins were unexpected and their functional significance in the PSD remains unknown. The total number of putative PSD proteins identified in various studies ranges from a few hundred to approximately 2000 (Jordan et al. 2004; Li et al. 2004; Peng et al. 2004; Yoshimura et al. 2004; Trinidad et al. 2008; Bayes et al. 2011). A common set of PSD proteins (∼460 proteins) has been deduced based on overlapping results from multiple proteomic studies (Collins et al. 2006). However, such protein lists likely contain some false positives arising from impurity of the PSD preparations (e.g., contamination by mitochondria and other organelles). Conversely, MS surveys may have missed true PSD proteins that are rare in abundance or loosely associated with the PSD. In addition to purification by differential centrifugation, PSDs (or subcomplexes thereof) have also been isolated by antibody affinity purification. MS analysis of protein complexes associated with PSD-95 and NMDA receptors have led to the identification of 288 and 77 proteins, respectively, which are largely overlapping and likely to include a core set of proteins of the PSD (Husi et al. 2000; Dosemeci et al. 2007).
Figure 1.
Categorization of proteins of the PSD of excitatory synapses. (A) The percentage of PSD proteins (as identified by mass spectrometry of purified PSD fractions) in various functional classes. Proteins with miscellaneous functions comprise ∼15% (not shown). (B) Copy number of selected proteins in an average size PSD of the forebrain.
In an important advance, quantitative MS and imaging methods enabled neuroscientists to determine the relative molar abundance of key PSD components (Fig. 1B) (Peng et al. 2004; Sugiyama et al. 2005; Cheng et al. 2006). CaMKIIα and CaMKIIβ are by far the most abundant proteins, with ∼4800 and ∼800 copies in an average size PSD, respectively. Other highly abundant molecules are scaffolds of the PSD-95 family (∼400 copies, of which 300 are PSD-95) and SynGAP (a Ras GTPase-activating protein [GAP] that binds to PSD-95; ∼360 copies). The number of PSD-95 family proteins is much greater than the number of NMDA and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor complexes measured in the PSD (∼15–20 of each). This molar surplus of the PSD-95 family proteins more than their glutamate receptor binding partners reflects the fact that PSD-95 family proteins have many additional interactors in synapses, including multiple signaling molecules and adhesion molecules (Kim and Sheng 2004).
MS is also useful for discovery of post-translational modifications (such as phosphorylation) of PSD proteins (Yoshimura et al. 2002; Jaffe et al. 2004; Collins et al. 2005; Trinidad et al. 2006; Kang et al. 2008; Trinidad et al. 2008). For instance, activation of NMDA receptors led to the phosphorylation of >100 PSD proteins, and activation of different postsynaptic receptors (i.e., NMDA vs. metabotropic glutamate receptors) induces differential phosphorylation in overlapping PSD proteins (Coba et al. 2009). Such proteomic approaches will be valuable to understand the molecular mechanisms of synaptic plasticity and of neurological diseases affecting synaptic function.
It should be noted that PSD protein composition varies considerably between different brain regions and probably between different cell types (Zhang et al. 1999; Cheng et al. 2006; Vullhorst et al. 2009). For instance, CaMKIIα is selectively present at the PSDs of glutamatergic neurons, whereas ErbB4 and citron are mainly present at postsynaptic sites on GABAergic interneurons in the hippocampus. This suggests that mechanisms of excitatory postsynaptic signaling differ between neuronal cell types.
MEMBRANE PROTEINS OF THE PSD
Only a small fraction of the protein components of the PSD have been extensively studied. Among the better understood groups are the membrane proteins, which include the cardinal functional proteins of the excitatory synapse—the glutamate receptors (AMPA, NMDA, and metabotropic glutamate receptors)—as well as other ion channels and adhesion molecules (see Smart and Paoletti 2011; Missler et al. 2011).
Postsynaptic cell adhesion molecules within and surrounding the PSD bind to specific proteins in the presynaptic membrane to mediate the synaptic contact and alignment. During the initial contact of axons and dendrites, the interaction of pre- and postsynaptic cell adhesion molecules may determine the specificity of axodendritic contacts and drive synapse formation and maturation (Craig and Kang 2007; Sudhof 2008; Shen and Scheiffele 2010). The best-known example of heterophilic and transsynaptic adhesion is that mediated by presynaptic neurexins and postsynaptic neuroligins. The cytoplasmic tails of neuroligins bind to PSD scaffold proteins including PSD-95 (Irie et al. 1997), whereas presynaptic neurexins bind to scaffolds such as CASK (Hata et al. 1996); such sets of protein–protein interactions provide a transsynaptic link between the PSD and the presynaptic active zone.
Given the diversity of synapse types in the nervous system, it is not surprising that there are multiple additional sets of adhesion molecules in the PSD that can induce synaptogenesis (see below). Perhaps different combinations of these transsynaptic adhesion molecules regulate synaptic specificity and affinity.
The classic adhesion molecule N-cadherin is also present in the PSD (∼30 copies in an average size PSD) and critical for synapse integrity (Tai et al. 2008). In addition to binding in homophilic fashion to presynaptic N-cadherin, N-cadherin can also bind to AMPA receptors (Saglietti et al. 2007) and to β-catenin, a protein of the PSD that links N-cadherin to actin filaments. N-cadherin is dynamically involved in transsynaptic signaling and structural/functional synaptic plasticity (Tai et al. 2008).
SCAFFOLD PROTEINS OF THE PSD
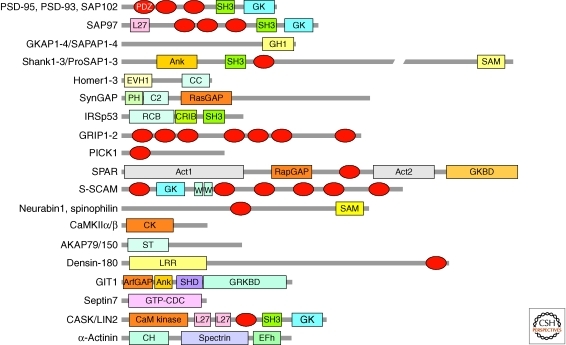
The PSD contains a large number of scaffolding proteins including PSD-95 (also known as SAP90), GKAP (or SAPAP), Shank (or ProSAP), and Homer (or Vesl). All these proteins are represented by multiple family members and splice variants. Synaptic scaffolding proteins are usually abundant in the PSD and contain multiple domains for protein–protein interaction (Fig. 2). A frequent feature of PSD scaffold proteins is the PDZ domain—a ∼90 amino acid-long module that usually interacts with a peptide motif located at the very carboxyl terminus of binding partners (Funke et al. 2004; Kim and Sheng 2004; Feng and Zhang 2009). Three PDZ domains are found in PSD-95 (a founding member of the PDZ protein family), allowing it to interact with many other proteins of the PSD (Kim and Sheng 2004).
Figure 2.
Scaffold proteins of the PSD and their domains. Major scaffold proteins and cytoskeleton-associated proteins of the PSD of excitatory synapses, scaled approximately by size. Act, actin-binding; Ank, ankyrin repeats; ArfGAP, GTPase-activating protein for Arf small GTPase; CC, coiled coil; CH, calponin homology; CK, calmodulin-dependent kinase (CaMK)-like domain; CRIB, Cdc42/Rac interactive-binding; EFh, EF-hand; EVH1, Ena/VASP homology 1; GH1, GKAP homology 1; GK, guanylate kinaselike domain; GKBD, GKAP-binding domain; GRKBD, GRK2-binding domain; GTP-CDC, GTP-cell division protein; L27, domain initially found in LIN-2 and LIN-7; LRR, leucine-rich repeat; PDZ domains are shown as red ellipses. PH, pleckstrin homology; RapGAP, GTPase-activating protein for Rap small GTPases; RasGAP, GTPase-activating protein for Ras small GTPases; RCB, Rac binding; SAM, sterile alpha motif; SH3, Src homology 3; SHD, Spa2 homology domain; Spectrin, spectrin repeat; ST, subcellular targeting domain; WW, domain with two conserved Trp (W) residues.
PSD-95
PSD-95 is the best studied of PSD scaffolds since its identification in the early 1990s (Cho et al. 1992; Kistner et al. 1993). A widely accepted function of PSD-95 and its family members (PSD-93/chapsyn-110, SAP97, and SAP102) is to bind to and tether or stabilize various membrane proteins and signaling molecules in the PSD (Fig. 3) (Kim and Sheng 2004). It is therefore centrally involved in multiple aspects of synaptic function (Kim and Sheng 2004; Fitzjohn et al. 2006; Nicoll et al. 2006). Via its first two PDZ domains, PSD-95 binds directly to the carboxy-terminal tails of GluN2 (also known as NR2) subunits of NMDA receptors (Kornau et al. 1995; Niethammer et al. 1996). In this way, PSD-95 appears to stabilize NMDA receptors at the cell surface (Roche et al. 2001; Prybylowski et al. 2005). Phosphorylation of GluN2 subunits affects their interaction with PSD-95 and regulates GluN2 subunit composition at synapses (Chen and Roche 2007; Sanz-Clemente et al. 2010). PSD-95 clusters GluN2 subunits in heterologous cells (Kim et al. 1996); however, it remains uncertain whether PSD-95 plays a role in synaptic localization of NMDA receptors in vivo.
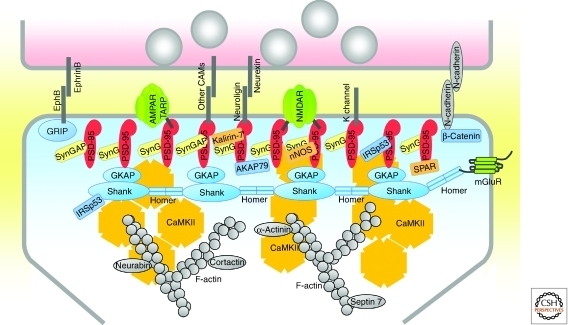
Figure 3.
Molecular organization of the PSD of excitatory synapses. Schematic diagram of the major proteins of the PSD, with protein interactions indicated by direct contacts or overlaps between the proteins. The relative numbers of the proteins shown correlate roughly with their relative abundance in PSDs of forebrain. Copy numbers of most adhesion molecules and ion channels are unknown (indicated by dark gray lines). Each CaMKII shape represents a dodecamer.
Perhaps more important than receptor clustering is the ability of PSD-95 to assemble an NMDA receptor-associated protein complex, thereby facilitating the functional coupling of NMDA receptors with downstream signaling molecules in the PSD, such as nitric oxide synthase (Aarts et al. 2002), Kalirin-7, a Rac guanine nucleotide exchange factor (GEF) that regulates dendritic spine morphology (Penzes et al. 2008), and SynGAP, a GTPase activating protein (GAP) for Ras (Chen et al. 1998; Kim et al. 1998, 2005). Conversely, PSD-95 can also recruit signaling proteins (e.g., AKAP79/150 and calcineurin [also known as protein phosphatase 2B]) that modulate the function of glutamate receptors (Bhattacharyya et al. 2009). It is possible that different members of the PSD-95 family bind to different overlapping sets of proteins in synapses.
By binding to the carboxyl terminus of transmembrane TARPs (accessory subunits of AMPA receptors), PSD-95 recruits AMPA receptors to synapses and can thereby determine synaptic strength (Chen et al. 2000; Schnell et al. 2002; Nicoll et al. 2006; Bats et al. 2007). Overexpression of PSD-95 increases AMPA receptor-mediated excitatory synaptic transmission (El-Husseini et al. 2000), whereas RNAi knockdown of PSD-95 decreases it (Elias et al. 2006; Futai et al. 2007), with relatively little effect on NMDA receptor-mediated transmission. Consistent with the control of synaptic strength by PSD-95, stronger synapses are correlated with larger PSDs and a higher content of PSD-95 and AMPA receptors. The crucial role of PSD-95 in organization of the PSD is shown by the PSD disintegration that occurs on RNAi knockdown of PSD-95 (Chen et al. 2011).
PSD-95-dependent increase in synaptic transmission appears to share common mechanisms with long-term potentiation (LTP), a form of activity-dependent increase in synaptic strength. For instance, overexpression of PSD-95 in hippocampal slices increases AMPA receptor-mediated synaptic transmission that occludes LTP (Stein et al. 2003; Ehrlich and Malinow 2004; Beique et al. 2006). However, it seems unlikely that an increase in postsynaptic PSD-95 accounts for the early phase of LTP (E-LTP), because PSD-95 does not accumulate in synapses during E-LTP (Steiner et al. 2008).
The evidence is stronger that PSD-95 is functionally involved in long-term depression (LTD). LTD is impaired by RNAi knockdown or genetic disruption of PSD-95 in mice, and it can be enhanced by PSD-95 overexpression (Migaud et al. 1998; Beique and Andrade 2003; Stein et al. 2003; Ehrlich et al. 2007; Carlisle et al. 2008; Xu et al. 2008). Activity-dependent dephosphorylation of PSD-95 (leading to destabilization of PSD-95 in the PSD), and PSD-95-mediated interactions with key signaling proteins (such as AKAP79/150 and protein phosphatases), may underlie the importance of PSD-95 in LTD (Kim et al. 2007; Xu et al. 2008; Bhattacharyya et al. 2009; Han et al. 2009).
Another function of PSD-95 is to organize synaptic adhesion by binding to the carboxy-terminal tails of many of the postsynaptic adhesion molecules including neuroligins, NGLs (netrin-G ligands), SALMs (synaptic adhesionlike molecules), and LRRTMs (leucine-rich repeat transmembrane neuronal) (Irie et al. 1997; Craig and Kang 2007; Sudhof 2008; Brose 2009; Shen and Scheiffele 2010; see also Missler et al. 2011). By interacting simultaneously with adhesion molecules, glutamate receptors and signaling proteins, PSD-95 family proteins can play a central role in the morphological and functional maturation of synapses at sites of axon–dendrite contact (Han and Kim 2008).
GKAP, Shank, and Homer
The GKAP family of scaffolding proteins (GKAP/SAPAP1-4) interacts with the carboxy-terminal GK-like domain of PSD-95 family proteins (Kim et al. 1997; Takeuchi et al. 1997). The carboxyl terminus of GKAP, in turn, interacts with the PDZ domain of Shank, which binds to Homer. These three stoichiometrically abundant proteins form an axis of interacting scaffolds in the deeper part of the PSD, linked to PSD-95 that is attached to the postsynaptic membrane (Fig. 3).
The Shank family of scaffolds (Shank1–3) (Sheng and Kim 2000; Boeckers et al. 2002) are large proteins (∼200 kDa) with multiple protein interaction domains that undergo differential splicing (Fig. 2). Functionally, Shank promotes dendritic spine growth and synaptic transmission in collaboration with Homer (Sala et al. 2001; Hung et al. 2008).
The Homer family of scaffolding proteins (Homer1–3) are scaffolding proteins that associate with Shank and group I metabotropic glutamate receptors (mGluR1 and mGluR5), thereby linking Shank with mGluRs (Naisbitt et al. 1999; Tu et al. 1999). This set of protein interactions could facilitate a functional interaction between PSD-95-associated NMDA receptors and mGluRs. Homer also interacts with dynamin-3 to link the PSD with the endocytic zone, a specialized region for endocytosis located outside the PSD and within the dendritic spine (Lu et al. 2007; Newpher and Ehlers 2008).
Other PSD Scaffolds
Other scaffolds of the PSD include IRSp53 (insulin substrate protein of 53 kDa), which directly interacts with PSD-95 and Shank, and acts as a downstream effector of Rac1 for the regulation of actin polymerization in dendritic spines (Soltau et al. 2004; Choi et al. 2005). AKAP79/150 interacts with PSD-95 and functions as an adaptor bringing AKAP-associated enzymes such as protein kinase A and calcineurin to the PSD for regulation of glutamate receptors and synaptic function (Tavalin et al. 2002; Bhattacharyya et al. 2009). AMPA receptor binding scaffolds GRIP and PICK1 are present but not selectively enriched in the PSD, suggesting they have roles primarily in AMPA receptor trafficking (Shepherd and Huganir 2007).
SIGNALING PROTEINS OF THE PSD
The PSD contains a diversity of cytoplasmic signaling molecules such as kinases (e.g., CaMKIIα and nonreceptor tyrosine kinases) and phosphatases (e.g., serine/threonine protein phosphatase 1 and protein tyrosine phosphatases) (Sheng and Hoogenraad 2007; Bayes and Grant 2009). CaMKIIα binds a variety of PSD proteins including NMDA receptors; the latter interaction not only promotes synaptic localization of CaMKIIα but also “locks” CaMKII in an active conformation (Shen and Meyer 1999; Bayer et al. 2001). CaMKIIα—which is typically activated by calcium influx through NMDA receptors—stimulates synaptic delivery of AMPA receptors and is critical for LTP (Hayashi et al. 2000). CaMKIIα also acts as a scaffold to bind and recruit proteasomes to activated dendritic spines, and is important in this way for activity-induced turnover of ubiquitinated proteins in spines (Bingol et al. 2010). This scaffolding function of CaMKIIα provides an explanation for the extremely high abundance of CaMKIIα at the synapse. CaMKIIβ also performs a structural role by binding to F-actin and stabilizing dendritic spine structure (Okamoto et al. 2007).
Also highly represented in the PSD are small GTPases (Ras, Rap, Rac, Rho, Ral, and Arf) and numerous GEFs and GAPs that regulate them (Newpher and Ehlers 2008; Penzes et al. 2008; Saneyoshi et al. 2008). Synaptic small GTPases regulate multiple aspects of synaptic structure and function; for instance, Ras and Rap regulate AMPA receptor trafficking during LTP and LTD, respectively (Zhu et al. 2002), whereas Rac and Rho control F-actin polymerization and the structure and dynamics of dendritic spines (Newey et al. 2005; Tada and Sheng 2006). Related GEFs and GAPs also affect synaptic structure and function; Kalirin-7, βPIX, and Tiam1 are GEFs that positively regulate Rac1 activity and promote dendritic spine morphogenesis (Penzes et al. 2008). The RasGAP SynGAP dampens Ras signaling and in this way influences mitogen-associated protein (MAP) kinase signaling and synaptic plasticity (Chen et al. 1998; Kim et al. 1998, 2005). The Rap-specific GAP, SPAR, regulates actin cytoskeleton and dendritic spine morphology and is the target for phosphorylation-induced degradation by pololike kinase 2 (Plk2) (Pak et al. 2001; Pak and Sheng 2003).
THREE-DIMENSIONAL STRUCTURE OF THE PSD
Advanced EM studies have revealed details of the three-dimensional (3D) structure of the PSD (Petersen et al. 2003). The synaptic cleft side of the PSD contains granular particles (5–15 nm in diameter, likely membrane protein complexes), as well as membrane patches (50–100 nm in diameter) that might represent lipid rafts. The cytoplasmic face of the PSD is convoluted and contains irregular protrusions extending into the cytoplasm, which are probably formed largely from CaMKIIα.
Specific proteins are differentially positioned in the perpendicular axis of the PSD, revealing a laminar organization of the PSD (Fig. 3). The PDZ domains of PSD-95 are located close to the postsynaptic membrane surface, consistent with PSD-95 interactions with many membrane proteins. In contrast, GKAP, Shank, and CaMKIIα proteins are localized nearer the cytoplasmic side of the PSD (∼24–26 nm from the membrane) (Valtschanoff and Weinberg 2001; Petersen et al. 2003; Rostaing et al. 2006). This difference has been confirmed using super-resolution fluorescence imaging techniques (Dani et al. 2010).
In the horizontal plane of the PSD, NMDA receptors appear on average more centrally located than AMPA receptors (Kharazia and Weinberg 1997; Racca et al. 2000; Tarusawa et al. 2009). This differential distribution is consistent with the notion that NMDA receptors are relatively stably incorporated into the PSD, whereas AMPA receptors are undergoing more dynamic exchange with extrasynaptic receptors (Cognet et al. 2006; Triller and Choquet 2008). PSD-95 is more evenly distributed throughout the horizontal plane of the PSD (DeGiorgis et al. 2006).
The 3D structures of individual PSD proteins can assist our understanding of PSD organization. Purified recombinant PSD-95 and its relative SAP97 exist in a C-shaped or extended conformations (Nakagawa et al. 2004). Shank3, which is known to self-multimerize (Naisbitt et al. 1999), can form large sheetlike structures in which individual helical fibers are stacked side by side (Baron et al. 2006). Zn2+ markedly increases the assembly of the sheet structures. Zn2+ is often released together with glutamate from presynaptic terminals and enters into postsynaptic sites through NMDA/AMPA glutamate receptors or calcium channels (Frederickson et al. 2005), raising the possibility that glutamatergic transmission may regulate PSD structure through Zn2+ (Grabrucker et al. 2011). Homer forms tetramers in which two parallel dimers interact in a tail-to-tail manner. Interestingly, Homer added to Shank in a 1:1 ratio leads to formation of meshlike structures visible under EM, which may contribute to scaffolding of the PSD (Hayashi et al. 2009).
Further aspects of PSD 3D structure have been revealed by EM tomography (Fig. 4A–C) (Chen et al. 2008a,b). The PSD contains numerous filaments (∼5 nm × ∼20 nm) oriented perpendicularly to the synaptic membrane (Fig. 4D,E); these “vertical filaments” probably correspond to PSD-95 molecules in extended conformation, as evidenced by direct immuno-EM labeling (Chen et al. 2011). A PSD of 400 nm diameter contains ∼400 evenly spaced vertical filaments, which matches the number of PSD-95 family proteins found in an average PSD. EM tomography also detects membrane particles (Fig. 4F,G) that have dimensions, numbers, and distributions consistent with them being AMPA- and/or NMDA-type glutamate receptor complexes (Nakagawa et al. 2005; Chen et al. 2008b). These putative glutamate receptor complexes often associate on the intracellular side with the vertical filaments, and on their extracellular side with putative adhesion molecules (3–4-nm-wide filaments) that span the synaptic cleft (Chen et al. 2008b). Short (∼20 nm) and long (30–35 nm) “horizontal filaments” are found in the deep part of the PSD, often contacting the vertical filaments (Fig. 4H,I). It is tempting to speculate that the horizontal filaments are made up of GKAP and/or Shank and/or Homer scaffolds, which link to each other and PSD-95 (Kim et al. 1997; Naisbitt et al. 1999). Specific knockdown of PSD-95 in cultured neurons leads to a patchy loss of PSD structures, including loss of PSD-95-containing vertical filaments, associated horizontal filaments, and putative AMPA receptor-type but not NMDA receptor-type structures (Chen et al. 2011).
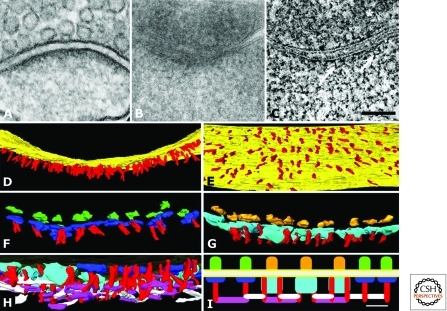
Figure 4.
3D organization of the excitatory PSD revealed by EM tomography. (A) EM of excitatory synapse from neuron in hippocampal culture prepared with conventional fixation and staining. (B) Section (200 nm thick)—typical of those used for tomography—through a dendritic spine of cultured hippocampal neuron prepared by freeze substitution. (C) Virtual sections (1.4 nm thick) derived from a tomographic reconstruction (tomogram) of synapse shown in (B). Fine structural details become apparent within the PSD, including vertical filaments (arrows). Asterisk indicates a synaptic vesicle. Scale bar, 100 nm. (D) Rendering of vertical filaments (colored red) from the tomogram (on-edge view of the PSD, with the postsynaptic membrane in yellow). (E) En-face view of the postsynaptic membrane (yellow), showing the regular distribution of the vertical filaments (red). (F) On-edge view of the PSD at higher magnification in which one type of transmembrane structure has been rendered in green on its extracellular side and in blue on the cytoplasmic side of the postsynaptic membrane. These putative AMPA receptorlike structures are usually contacted by one vertical filament (red). (G) A second type of transmembrane structure (putative NMDA receptors) is shown rendered in gold on its extracellular side and azure on its larger cytoplasmic side. These putative NMDAR-type structures are typically contacted by two vertical filaments (red). (H) Cross section of PSD slightly tilted to reveal its cytoplasmic side. The vertical filaments (red) contact two types of horizontal filament, rendered in purple and white. (I) Schematic diagram showing the distribution and interactions of the two types of transmembrane structures, the vertical filaments and the two types of horizontal filaments. The azure and dark blue structures represent the cytoplasmic aspects of putative NMDA receptor and AMPA receptor complexes, respectively, and the red vertical filaments are likely composed of PSD-95. The vertical filaments connect transmembrane structures to an orthogonal, interlinked scaffold at the core of the PSD. (Figure adapted, with permission, from Chen et al. 2008b and reprinted, with permission, from the Society of Neuroscience © 2008.)
DEVELOPMENTAL CHANGES IN THE PSD
The structure and composition of PSDs change during maturation of synapses (Petralia et al. 2005; Swulius et al. 2010). Expression levels of many PSD proteins (e.g., PSD-95, CaMKIIα, and AMPA receptor subunits) increase during development, reaching their peaks at ∼2–4 wk after birth and correlating with the formation and maturation of synapses in the brain (Sans et al. 2000; Petralia et al. 2005). Synaptic localization of PSD proteins typically increases during postnatal development. Some PSD proteins, however, show reduced expression during postnatal development; e.g., the GluN2B (NR2B) subunit of NMDA receptors and SAP102 (a member of the PSD-95 family) (Monyer et al. 1994; Sheng et al. 1994; Petralia et al. 2005; Zheng et al. 2011). As the brain develops, the GluN2B-SAP102 complex is increasingly replaced with the GluN2A (NR2A)-PSD-95 complex at synapses, which could lead to changes in receptor function and synaptic plasticity (Sans et al. 2000; Yoshii et al. 2003). Likewise, the synaptic localization of AMPA receptors at different developmental stages appears to involve different PSD-95 family proteins; at immature synapses, SAP102 plays a critical role for synaptic AMPA receptor recruitment, whereas PSD-95 and PSD-93 are more important in mature synapses (Elias et al. 2006).
As sensory systems mature and sensory input increases, sensory experience- and activity-dependent changes take place in synapse structure and function, accompanied by protein changes in the PSD. Exposure of dark-reared rats to light causes a rapid increase in synaptic levels of GluN2A (Quinlan et al. 1999). Eye opening in rats promotes recruitment of PSD-95 to synapses and its association with GluN2A, but suppresses its association with GluN2B (Yoshii et al. 2003). Furthermore, LTP-inducing stimuli can rapidly (within seconds) increase the ratio of GluN2A-containing NMDA receptors over GluN2B-containing receptors in neonatal (P2-9), but not in older (P16-21), slices (Bellone and Nicoll 2007). The activity-dependent GluN2B-to-GluN2A switch requires activation of NMDA receptors and mGluR5 metabotropic glutamate receptors (Matta et al. 2011). Thus, NMDA receptor composition and associated signaling complexes can be dynamically regulated in the PSD by activity, especially in immature synapses.
BASAL AND ACTIVITY-DEPENDENT PROTEIN TURNOVER IN THE PSD
Even in mature neurons, the PSD undergoes continual molecular turnover under basal conditions and shows larger changes in response to activity (Inoue and Okabe 2003). The dynamic behavior of PSDs in living neurons in culture and in vivo has been typically visualized by GFP-tagged PSD-95, which localizes specifically in the PSD. This approach revealed that overall PSD structure undergoes continuous remodeling over a timescale of minutes to days (Blanpied et al. 2008; Minerbi et al. 2009). Within the PSD, however, mixing of existing PSD-95 molecules is relatively limited, suggesting that individual PSD-95 molecules, once integrated, maintain stable positions within the PSD (Blanpied et al. 2008).
Proteins of the PSD turn over in large part by continuous exchange with counterparts outside of the PSD. Among the most dynamic are the AMPA-type glutamate receptors, which show rapid lateral diffusion in and out of the postsynaptic membrane (Cognet et al. 2006). Regulated AMPA receptor insertion into, and removal from, the PSD are major mechanisms underlying the strengthening and weakening of synaptic transmission (Shepherd and Huganir 2007; see also Lüscher and Malenka 2011). Although many molecular details have emerged about AMPA receptor trafficking, a remaining question is how the structural organization of the PSD is altered to enable AMPA receptor incorporation and removal during LTP and LTD. It is likely that remodeling of the actin cytoskeleton and regulated proteolysis of PSD components are crucial for synaptic rearrangements underlying plasticity (Cingolani and Goda 2008; Li et al. 2010; Bingol and Sheng 2011).
PSD-95 is dynamically exchanged between neighboring PSDs in cortical neurons in vivo, with median synaptic retention times of PSD-95 within individual spines of 20–100 min depending on age (Gray et al. 2006). Measured by FRAP (fluorescence recovery after photobleaching) or by PAGFP (photoactivatable green fluorescent protein) imaging, PSD-95 is relatively stable within the PSD, compared with other PSD components, such as SAP102, GKAP, Shank, Homer, CaMKIIα, GluA2 (GluR2), and stargazin (Kuriu et al. 2006; Sturgill et al. 2009; Zheng et al. 2010).
PSD composition can also be rapidly modified by mechanisms including protein phosphorylation, palmitoylation, ubiquitination, and proteasome-mediated protein degradation. Phosphorylation of PSD-95 on Ser-295, mediated by JNK1, promotes localization of PSD-95 in the PSD (Kim et al. 2007), whereas PSD-95 phosphorylation on Ser-73 by CaMKIIα mobilizes it (Steiner et al. 2008). Palmitoylation of PSD-95 favors its synaptic localization (El-Husseini Ael et al. 2002). Activity-dependent degradation of synaptic proteins—which is required for LTP and LTD, as well as learning and memory (Colledge et al. 2003; Fonseca et al. 2006; Karpova et al. 2006; Lee et al. 2008)—may be aided by the rapid redistribution of proteasomes to postsynaptic sites following synaptic stimulation (Bingol and Schuman 2006; Bingol et al. 2010).
PSD composition also changes on a timescale of hours to days during synaptic scaling, which is a homeostatic adjustment of synaptic strength in response to long-term alterations in activity (Turrigiano 2008; see also Turrigiano 2011). In addition to compensatory changes in synaptic AMPA receptor content (Turrigiano et al. 1998), chronic elevation of synaptic activity leads to large-scale changes in PSD composition, due in part to ubiquitination and proteasome-mediated degradation of GKAP and Shank scaffolds (Ehlers 2003; Hung et al. 2010). PSD-95, which docks AMPA receptors at excitatory synapses via transmembrane AMPA receptor regulatory protein (TARP), and whose synaptic content changes by neuronal activity, is required for homeostatic synaptic scaling (Sun and Turrigiano 2011).
BRAIN DISEASE AND PSD PROTEINS
Given the functional relationship of PSD proteins to synapse development, structure, and function, it is not surprising that mutations in many PSD proteins are associated with human neurologic and psychiatric disease (Bayes et al. 2011). Autism spectrum disorders have been linked to mutations in genes encoding Shank2 and Shank3, PSD-93, DLGAP2/SAPAP2, and SynGAP1, as well as synaptic adhesion molecules neuroligin 3, neuroligin 4, and neurexin 1 (Durand et al. 2007; Sudhof 2008; Berkel et al. 2010; Pinto et al. 2010). Mutations in SAPAP3 are associated with obsessive compulsive disorder (Welch et al. 2007; Zuchner et al. 2009). It is likely that other neuropsychiatric illnesses will be found to be related to mutation or dysfunction of PSD components. Reduction of PSD protein levels in brain is a hallmark of Alzheimer’s disease, presumably reflecting the profound synapse loss in this neurodegenerative disorder (Haass and Selkoe 2007; see also Zoghbi and Bear 2011 and Sabatini et al. 2011).
POSTSYNAPTIC ORGANIZATION OF INHIBITORY SYNAPSES
Inhibitory synapses are mainly formed on the shaft of dendrites or around the neuronal cell body, and by EM, they show only a slight electron-dense thickening associated with the postsynaptic membrane and hence were described as symmetric (type II) synapses (Gray 1959). This presumably reflects the fact that the inhibitory postsynaptic specialization is much less elaborate than the PSD of excitatory synapses.
The cardinal neurotransmitter receptors of central inhibitory synapses are the GABAA receptors and glycine receptors. These receptors interact directly with gephyrin, a well-known postsynaptic scaffold protein of inhibitory synapses (Prior et al. 1992; Luscher and Keller 2004; Craig and Kang 2007; Fritschy et al. 2008; Jacob et al. 2008). Gephyrin is critical for glycine receptor clustering at inhibitory synapses, but appears less important for GABAA receptor clustering (Kneussel et al. 2001; Levi et al. 2004). Conversely, however, GABAA receptors can recruit gephyrin to synapses (Essrich et al. 1998; Levi et al. 2004). Thus GABAA receptors clustered at early inhibitory synaptic sites by mechanisms independent of gephyrin may be able to recruit gephyrin and other gephyrin-associated proteins to drive inhibitory postsynaptic differentiation (Fig. 5).
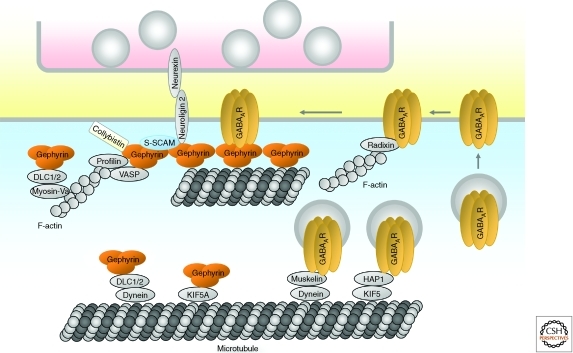
Figure 5.
Postsynaptic organization of inhibitory GABAergic synapses. Diagram of major proteins at a GABAergic synapse, with protein interactions indicated by direct contacts or overlaps between the shapes. Note that distinct cytoplasmic domains of neuroligin 2 bind to both gephyrin and S-SCAM. Glycinergic synapses likely show a similar molecular organization including the interaction of glycine receptors with gephyrin. See text for details.
Reminiscent of the major scaffolds in excitatory PSDs, gephyrin can self-multimerize and form a hexagonal lattice, and thus may function as a postsynaptic scaffold through which GABAA/glycine receptors functionally interact with gephyrin-associated proteins. Similar to PSD-95, gephyrin clustering is regulated by phosphorylation. GSK3β phosphorylation of Ser-270 impairs gephyrin clustering and inhibitory transmission (Tyagarajan et al. 2011). Lithium chloride, an inhibitor of GSK3β, enhances gephyrin clustering in cultured neurons, suggesting that the mood-stabilizing effect of lithium may partly involve this mechanism.
About a dozen binding partners of gephyrin have been identified (Fritschy et al. 2008). Although the functions of these protein interactions are largely unknown, some binding partners such as actin-associated proteins profilin and Mena/VASP are thought to link gephyrin to actin filaments (Giesemann et al. 2003). In addition, gephyrin-associated dynein light chains have been implicated in motor-dependent transport of the gephyrin-receptor complex along microtubules/actin filaments and translocation of gephyrin from the cytoplasmic to a submembraneous compartment (Fig. 5) (Fuhrmann et al. 2002).
As at excitatory synapses, synaptic adhesion molecules may guide the differentiation and maturation of inhibitory synapses by coupling axodendritic adhesion events with the recruitment of specific membrane and signaling proteins (Craig and Kang 2007; Siddiqui and Craig 2011). A specific isoform of neuroligin (neuroligin 2) is specifically localized at inhibitory postsynaptic sites, and overexpression of neuroligin 2 promotes formation and function of GABAergic synapses in cultured neurons (Graf et al. 2004; Varoqueaux et al. 2004; Chih et al. 2005; Levinson et al. 2005; Chubykin et al. 2007). Neuroligin 2 interacts on the extracellular side with presynaptic neurexins on GABAergic axons; via intracellular interactions it recruits postsynaptic gephyrin and gephyrin-bound collybistin, a GEF for Cdc42 (Graf et al. 2004; Varoqueaux et al. 2004; Chih et al. 2005; Craig and Kang 2007; Poulopoulos et al. 2009). This is thought to enhance collybistin activation, membrane tethering of the gephyrin–collybistin complex, and recruitment of other gephyrin-associated proteins (Fig. 5) (Poulopoulos et al. 2009). Neuroligin 2 was also found to interact with S-SCAM, a PDZ domain protein present at inhibitory and excitatory synapses (Sumita et al. 2007). Neuroligin 2 deletion in mice does not eliminate inhibitory synapses, although it leads to impairments in inhibitory synaptic transmission (Varoqueaux et al. 2006; Gibson et al. 2009; Hoon et al. 2009; Poulopoulos et al. 2009), suggesting that neuroligin 2 is more important for the functional maturation of inhibitory synapses than for their formation.
In addition to neuroligin 2, neuroligin 3 and neuroligin 4 are found at inhibitory synapses. Neuroligin 3 is detected at both excitatory and inhibitory synapses (Budreck and Scheiffele 2007). The impaired inhibitory transmission induced by triple knockdown of neuroligins 1–3 is rescued by neuroligin 3 expression, although to a lesser extent than with neuroligin 2, suggesting that neuroligin 3 contributes to inhibitory transmission (Shipman et al. 2011). Neuroligin 4 is primarily localized at glycinergic synapses in various brain regions (Hoon et al. 2011). Neuroligin 4 forms a complex with gephyrin and collybistin in vivo, and functionally modulates glycinergic transmission, as shown by the slowed glycinergic currents in neuroligin 4-deficient mice (Hoon et al. 2011). Lastly, independent of neuroligins, neurexin has been found to interact with GABAA receptors and suppress GABAergic transmission (Zhang et al. 2010).
Regulation of GABAA receptor trafficking may determine inhibitory synaptic strength and hence neuronal excitability (Luscher et al. 2011). Newly synthesized GABAA receptors translocate to the plasma membrane through the secretory pathway, aided by specific proteins such as PLIC, GODZ, and BIG2, which regulate protein degradation, protein palmitoylation, and trans-Golgi exit of GABAA receptors, respectively. GABAA receptors are initially inserted into the extrasynaptic plasma membrane, and laterally diffuse into synaptic sites (Bogdanov et al. 2006). Extrasynaptic GABAA receptors can be stabilized at the surface by interaction with radixin, an actin filament-associated protein (Fig. 5) (Loebrich et al. 2006). More than 20% of extrasynaptic GABAA receptors are internalized within 15 min via phosphorylation-dependent interaction with endocytic proteins. Internalized GABAA receptors are recycled back to the plasma membrane or trafficked to the lysosomal pathway for degradation, depending on their interaction with huntingtin-associated protein 1 (HAP1). HAP1 functions as an adaptor protein linking GABAA receptors to the KIF5 kinesin motor for microtubule-dependent and anterograde trafficking of GABAA receptors to the plasma membrane (Twelvetrees et al. 2010). Conversely, muskelin, a multidomain protein, promotes retrograde trafficking of GABAA receptors by linking GABAA receptors to the actin-dependent motor myosin VI and the microtubule-dependent motor dynein for receptor endocytosis and degradation (Heisler et al. 2011).
Inhibitory synapses are fundamentally different from excitatory synapses with regard to the nature of neurotransmitter receptors and associated proteins. However, many of the organizing principles appear to be shared between excitatory and inhibitory postsynaptic specializations, including the involvement of adhesion-facilitated differentiation, scaffold-based assembly of receptors and signaling proteins, and dynamic and activity-regulated trafficking of neurotransmitter receptors in and out of synapses.
CONCLUSIONS
Although the protein components of the excitatory PSD are now largely revealed, the molecular organization of the postsynaptic specialization of excitatory synapses is still not well understood in terms of 3D structure and functional significance. Inhibitory synapses lag even further behind excitatory synapses in these respects. How the composition, geometry, and function of postsynaptic specializations are altered during activity-dependent plasticity—and by neurological and psychiatric disease—remain important questions. Further studies are needed to understand how diverse signaling mechanisms within neurons (e.g., neurotrophin, apoptosis, autophagy, and transcription factor signaling pathways) impinge on the structure, composition, and function of excitatory and inhibitory postsynaptic specializations. Aberrant development and regulation of the PSD owing to genetic defects in key proteins, which can lead to imbalance between excitation and inhibition in neuronal circuits, are emerging as common etiological factors in autism, intellectual disability, and mental illness. Thus understanding the functional organization and the diversity of PSDs is of utmost importance for revealing the neurobiological basis of human brain disorders.
Footnotes
Editors: Morgan Sheng, Bernardo Sabatini, and Thomas Südhof
Additional Perspectives on The Synapse available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aarts M, Liu Y, Liu L, Besshoh S, Arundine M, Gurd JW, Wang YT, Salter MW, Tymianski M 2002. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science 298: 846–850 [DOI] [PubMed] [Google Scholar]
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU 2006. An architectural framework that may lie at the core of the postsynaptic density. Science 311: 531–535 [DOI] [PubMed] [Google Scholar]
- Bats C, Groc L, Choquet D 2007. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 53: 719–734 [DOI] [PubMed] [Google Scholar]
- Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H 2001. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature 411: 801–805 [DOI] [PubMed] [Google Scholar]
- Bayes A, Grant SG 2009. Neuroproteomics: Understanding the molecular organization and complexity of the brain. Nat Rev Neurosci 10: 635–646 [DOI] [PubMed] [Google Scholar]
- Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG 2011. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 14: 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beique JC, Andrade R 2003. PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J Physiol 546: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beique JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL 2006. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci 103: 19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellone C, Nicoll RA 2007. Rapid bidirectional switching of synaptic NMDA receptors. Neuron 55: 779–785 [DOI] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. 2010. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 42: 489–491 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Biou V, Xu W, Schluter O, Malenka RC 2009. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci 12: 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Schuman EM 2006. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Bingol B, Sheng M 2011. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 69: 22–32 [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M 2010. Autophosphorylated CaMKIIα acts as a scaffold to recruit proteasomes to dendritic spines. Cell 140: 567–578 [DOI] [PubMed] [Google Scholar]
- Blanpied TA, Kerr JM, Ehlers MD 2008. Structural plasticity with preserved topology in the postsynaptic protein network. Proc Natl Acad Sci 105: 12587–12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg F, Cohen RS, Siekevitz P 1977. The structure of postsynaptic densities isolated from dog cerebral cortex. II. Characterization and arrangement of some of the major proteins within the structure. J Cell Biol 74: 204–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED 2002. ProSAP/Shank proteins—A family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 81: 903–910 [DOI] [PubMed] [Google Scholar]
- Bogdanov Y, Michels G, Armstrong-Gold C, Haydon PG, Lindstrom J, Pangalos M, Moss SJ 2006. Synaptic GABAA receptors are directly recruited from their extrasynaptic counterparts. EMBO J 25: 4381–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, Harris KM 2008. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci 31: 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N 2009. Synaptogenic proteins and synaptic organizers: Many hands make light work. Neuron 61: 650–652 [DOI] [PubMed] [Google Scholar]
- Budreck EC, Scheiffele P 2007. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci 26: 1738–1748 [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P 1980. Isolation and characterization of postsynaptic densities from various brain regions: Enrichment of different types of postsynaptic densities. J Cell Biol 86: 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Fink AE, Grant SG, O’Dell TJ 2008. Opposing effects of PSD-93 and PSD-95 on long-term potentiation and spike timing-dependent plasticity. J Physiol 586: 5885–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BS, Roche KW 2007. Regulation of NMDA receptors by phosphorylation. Neuropharmacology 53: 362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB 1998. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20: 895–904 [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA 2000. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408: 936–943 [DOI] [PubMed] [Google Scholar]
- Chen X, Vinade L, Leapman RD, Petersen JD, Nakagawa T, Phillips TM, Sheng M, Reese TS 2005. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci 102: 11551–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winters C, Azzam R, Li X, Galbraith JA, Leapman RD, Reese TS 2008a. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci 105: 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winters CA, Reese TS 2008b. Life inside a thin section: Tomography. J Neurosci 28: 9321–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Nelson CD, Li X, Winters CA, Azzam R, Sousa AA, Leapman RD, Gainer H, Sheng M, Reese TS 2011. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci 31: 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, et al. 2006. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics 5: 1158–1170 [DOI] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P 2005. Control of excitatory and inhibitory synapse formation by neuroligins. Science 307: 1324–1328 [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB 1992. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs–large tumor suppressor protein. Neuron 9: 929–942 [DOI] [PubMed] [Google Scholar]
- Choi J, Ko J, Racz B, Burette A, Lee JR, Kim S, Na M, Lee HW, Kim K, Weinberg RJ, et al. 2005. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci 25: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Sudhof TC 2007. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron 54: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y 2008. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 9: 344–356 [DOI] [PubMed] [Google Scholar]
- Coba MP, Pocklington AJ, Collins MO, Kopanitsa MV, Uren RT, Swamy S, Croning MD, Choudhary JS, Grant SG 2009. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal 2: ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognet L, Groc L, Lounis B, Choquet D 2006. Multiple routes for glutamate receptor trafficking: Surface diffusion and membrane traffic cooperate to bring receptors to synapses. Sci STKE 2006: e13. [DOI] [PubMed] [Google Scholar]
- Cohen RS, Blomberg F, Berzins K, Siekevitz P 1977. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J Cell Biol 74: 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, Choudhary JS, Grant SG 2005. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem 280: 5972–5982 [DOI] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG 2006. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem 97: 16–23 [DOI] [PubMed] [Google Scholar]
- Cotman CW, Banker G, Churchill L, Taylor D 1974. Isolation of postsynaptic densities from rat brain. J Cell Biol 63: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Kang Y 2007. Neurexin–neuroligin signaling in synapse development. Curr Opin Neurobiol 17: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X 2010. Superresolution imaging of chemical synapses in the brain. Neuron 68: 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GA, Bloom FE 1973. Isolation of synaptic junctional complexes from rat brain. Brain Res 62: 135–153 [DOI] [PubMed] [Google Scholar]
- DeGiorgis JA, Galbraith JA, Dosemeci A, Chen X, Reese TS 2006. Distribution of the scaffolding proteins PSD-95, PSD-93, and SAP97 in isolated PSDs. Brain Cell Biol 35: 239–250 [DOI] [PubMed] [Google Scholar]
- Dosemeci A, Makusky AJ, Jankowska-Stephens E, Yang X, Slotta DJ, Markey SP 2007. Composition of the synaptic PSD-95 complex. Mol Cell Proteomics 6: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. 2007. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 39: 25–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6: 231–242 [DOI] [PubMed] [Google Scholar]
- Ehrlich I, Malinow R 2004. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci 24: 916–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Klein M, Rumpel S, Malinow R 2007. PSD-95 is required for activity-driven synapse stabilization. Proc Natl Acad Sci 104: 4176–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS 2000. PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364–1368 [PubMed] [Google Scholar]
- El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS 2002. Synaptic strength regulated by palmitate cycling on PSD-95. Cell 108: 849–863 [DOI] [PubMed] [Google Scholar]
- Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA 2006. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron 52: 307–320 [DOI] [PubMed] [Google Scholar]
- Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B 1998. Postsynaptic clustering of major GABAA receptor subtypes requires the γ2 subunit and gephyrin. Nat Neurosci 1: 563–571 [DOI] [PubMed] [Google Scholar]
- Feng W, Zhang M 2009. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci 10: 87–99 [DOI] [PubMed] [Google Scholar]
- Fitzjohn SM, Doherty AJ, Collingridge GL 2006. Promiscuous interactions between AMPA-Rs and MAGUKs. Neuron 52: 222–224 [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV 2006. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52: 239–245 [DOI] [PubMed] [Google Scholar]
- Frederickson CJ, Koh JY, Bush AI 2005. The neurobiology of zinc in health and disease. Nat Rev Neurosci 6: 449–462 [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Harvey RJ, Schwarz G 2008. Gephyrin: Where do we stand, where do we go? Trends Neurosci 31: 257–264 [DOI] [PubMed] [Google Scholar]
- Fuhrmann JC, Kins S, Rostaing P, El Far O, Kirsch J, Sheng M, Triller A, Betz H, Kneussel M 2002. Gephyrin interacts with Dynein light chains 1 and 2, components of motor protein complexes. J Neurosci 22: 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS 2004. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem 74: 219–245 [DOI] [PubMed] [Google Scholar]
- Futai K, Kim MJ, Hashikawa T, Scheiffele P, Sheng M, Hayashi Y 2007. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat Neurosci 10: 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL 2000. PDZ domains in synapse assembly and signalling. Trends Cell Biol 10: 274–280 [DOI] [PubMed] [Google Scholar]
- Gibson JR, Huber KM, Sudhof TC 2009. Neuroligin-2 deletion selectively decreases inhibitory synaptic transmission originating from fast-spiking but not from somatostatin-positive interneurons. J Neurosci 29: 13883–13897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesemann T, Schwarz G, Nawrotzki R, Berhorster K, Rothkegel M, Schluter K, Schrader N, Schindelin H, Mendel RR, Kirsch J, et al. 2003. Complex formation between the postsynaptic scaffolding protein gephyrin, profilin, and Mena: A possible link to the microfilament system. J Neurosci 23: 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Knight MJ, Proepper C, Bockmann J, Joubert M, Rowan M, Nienhaus GU, Garner CC, Bowie JU, Kreutz MR, et al. 2011. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J 30: 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM 2004. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119: 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EG 1959. Axo-somatic and axo-dendritic synapses of the cerebral cortex: An electron microscope study. J Anat 93: 420–433 [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Weimer RM, Bureau I, Svoboda K 2006. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol 4: e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ 2007. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat Rev Mol Cell Biol 8: 101–112 [DOI] [PubMed] [Google Scholar]
- Han K, Kim E 2008. Synaptic adhesion molecules and PSD-95. Prog Neurobiol 84: 263–283 [DOI] [PubMed] [Google Scholar]
- Han K, Kim MH, Seeburg D, Seo J, Verpelli C, Han S, Chung HS, Ko J, Lee HW, Kim K, et al. 2009. Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD. PLoS Biol 7: e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Butz S, Sudhof TC 1996. CASK: A novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci 16: 2488–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R 2000. Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science 287: 2262–2267 [DOI] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y 2009. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 137: 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler FF, Loebrich S, Pechmann Y, Maier N, Zivkovic AR, Tokito M, Hausrat TJ, Schweizer M, Bahring R, Holzbaur EL, et al. 2011. Muskelin regulates actin filament- and microtubule-based GABAA receptor transport in neurons. Neuron 70: 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M, Bauer G, Fritschy JM, Moser T, Falkenburger BH, Varoqueaux F 2009. Neuroligin 2 controls the maturation of GABAergic synapses and information processing in the retina. J Neurosci 29: 8039–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M, Soykan T, Falkenburger B, Hammer M, Patrizi A, Schmidt KF, Sassoe-Pognetto M, Lowel S, Moser T, Taschenberger H, et al. 2011. Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc Natl Acad Sci 108: 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, et al. 2008. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci 28: 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sung CC, Brito IL, Sheng M 2010. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One 5: e9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG 2000. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 3: 661–669 [DOI] [PubMed] [Google Scholar]
- Inoue A, Okabe S 2003. The dynamic organization of postsynaptic proteins: Translocating molecules regulate synaptic function. Curr Opin Neurobiol 13: 332–340 [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC 1997. Binding of neuroligins to PSD-95. Science 277: 1511–1515 [DOI] [PubMed] [Google Scholar]
- Jacob TC, Moss SJ, Jurd R 2008. GABAA receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci 9: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe H, Vinade L, Dosemeci A 2004. Identification of novel phosphorylation sites on postsynaptic density proteins. Biochem Biophys Res Commun 321: 210–218 [DOI] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA 2004. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics 3: 857–871 [DOI] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456: 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova A, Mikhaylova M, Thomas U, Knopfel T, Behnisch T 2006. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. J Neurosci 26: 4949–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H 2003. Structure-stability-function relationships of dendritic spines. Trends Neurosci 26: 360–368 [DOI] [PubMed] [Google Scholar]
- Kennedy MB 2000. Signal-processing machines at the postsynaptic density. Science 290: 750–754 [DOI] [PubMed] [Google Scholar]
- Kharazia VN, Weinberg RJ 1997. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci Lett 238: 41–44 [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M 2004. PDZ domain proteins of synapses. Nat Rev Neurosci 5: 771–781 [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M 1995. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378: 85–88 [DOI] [PubMed] [Google Scholar]
- Kim E, Cho KO, Rothschild A, Sheng M 1996. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron 17: 103–113 [DOI] [PubMed] [Google Scholar]
- Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M 1997. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol 136: 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau LF, Huganir RL 1998. SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20: 683–691 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M 2005. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron 46: 745–760 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Futai K, Jo J, Hayashi Y, Cho K, Sheng M 2007. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron 56: 488–502 [DOI] [PubMed] [Google Scholar]
- Kistner U, Wenzel BM, Veh RW, Cases-Langhoff C, Garner AM, Appeltauer U, Voss B, Gundelfinger ED, Garner CC 1993. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. J Biol Chem 268: 4580–4583 [PubMed] [Google Scholar]
- Kneussel M, Brandstatter JH, Gasnier B, Feng G, Sanes JR, Betz H 2001. Gephyrin-independent clustering of postsynaptic GABAA receptor subtypes. Mol Cell Neurosci 17: 973–982 [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH 1995. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269: 1737–1740 [DOI] [PubMed] [Google Scholar]
- Kuriu T, Inoue A, Bito H, Sobue K, Okabe S 2006. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J Neurosci 26: 7693–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langnaese K, Seidenbecher C, Wex H, Seidel B, Hartung K, Appeltauer U, Garner A, Voss B, Mueller B, Garner CC, et al. 1996. Protein components of a rat brain synaptic junctional protein preparation. Brain Res Mol Brain Res 42: 118–122 [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Kim H, Kaang BK 2008. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science 319: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Levi S, Logan SM, Tovar KR, Craig AM 2004. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci 24: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson JN, Chery N, Huang K, Wong TP, Gerrow K, Kang R, Prange O, Wang YT, El-Husseini A 2005. Neuroligins mediate excitatory and inhibitory synapse formation: Involvement of PSD-95 and neurexin-1β in neuroligin-induced synaptic specificity. J Biol Chem 280: 17312–17319 [DOI] [PubMed] [Google Scholar]
- Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, et al. 2004. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem 279: 987–1002 [DOI] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, Cho K, Sheng M 2010. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebrich S, Bahring R, Katsuno T, Tsukita S, Kneussel M 2006. Activated radixin is essential for GABAA receptor α5 subunit anchoring at the actin cytoskeleton. EMBO J 25: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Helton TD, Blanpied TA, Racz B, Newpher TM, Weinberg RJ, Ehlers MD 2007. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron 55: 874–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Keller CA 2004. Regulation of GABAA receptor trafficking, channel activity, and functional plasticity of inhibitory synapses. Pharmacol Ther 102: 195–221 [DOI] [PubMed] [Google Scholar]
- *.Lüscher C, Malenka RC 2011. NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, Kilpatrick CL 2011. GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70: 385–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta JA, Ashby MC, Sanz-Clemente A, Roche KW, Isaac JT 2011. mGluR5 and NMDA receptors drive the experience- and activity-dependent NMDA receptor NR2B to NR2A subunit switch. Neuron 70: 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, et al. 1998. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396: 433–439 [DOI] [PubMed] [Google Scholar]
- Minerbi A, Kahana R, Goldfeld L, Kaufman M, Marom S, Ziv NE 2009. Long-term relationships between synaptic tenacity, synaptic remodeling, and network activity. PLoS Biol 7: e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Missler M, Südhof TC, Biederer T 2011. Synaptic cell adhesion. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH 1994. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12: 529–540 [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M 1999. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23: 569–582 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel HA, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M 2004. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron 44: 453–467 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T 2005. Structure and different conformational states of native AMPA receptor complexes. Nature 433: 545–549 [DOI] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek EE, Van Aelst L 2005. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol 64: 58–74 [DOI] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD 2008. Glutamate receptor dynamics in dendritic microdomains. Neuron 58: 472–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS 2006. Auxiliary subunits assist AMPA-type glutamate receptors. Science 311: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Niethammer M, Kim E, Sheng M 1996. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci 16: 2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y 2007. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci 104: 6418–6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Sheng M 2003. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302: 1368–1373 [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M 2001. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31: 289–303 [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M 2004. Semi-quantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 279: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Srivastava DP 2008. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol 18: 405–413 [DOI] [PubMed] [Google Scholar]
- Petersen JD, Chen X, Vinade L, Dosemeci A, Lisman JE, Reese TS 2003. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci 23: 11270–11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Sans N, Wang YX, Wenthold RJ 2005. Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol Cell Neurosci 29: 436–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. 2009. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63: 628–642 [DOI] [PubMed] [Google Scholar]
- Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, et al. 1992. Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron 8: 1161–1170 [DOI] [PubMed] [Google Scholar]
- Prybylowski K, Chang K, Sans N, Kan L, Vicini S, Wenthold RJ 2005. The synaptic localization of NR2B-containing NMDA receptors is controlled by interactions with PDZ proteins and AP-2. Neuron 47: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan EM, Philpot BD, Huganir RL, Bear MF 1999. Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci 2: 352–357 [DOI] [PubMed] [Google Scholar]
- Racca C, Stephenson FA, Streit P, Roberts JD, Somogyi P 2000. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J Neurosci 20: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche KW, Standley S, McCallum J, Dune Ly C, Ehlers MD, Wenthold RJ 2001. Molecular determinants of NMDA receptor internalization. Nat Neurosci 4: 794–802 [DOI] [PubMed] [Google Scholar]
- Rostaing P, Real E, Siksou L, Lechaire JP, Boudier T, Boeckers TM, Gertler F, Gundelfinger ED, Triller A, Marty S 2006. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur J Neurosci 24: 3463–3474 [DOI] [PubMed] [Google Scholar]
- *.Sabatini B, Sheng M, Südhof T 2011. Synapses and Alzheimer's disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. 2007. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54: 461–477 [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M 2001. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115–130 [DOI] [PubMed] [Google Scholar]
- Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR 2008. Activity-dependent synaptogenesis: Regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron 57: 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos Jn, Hell JW, Wenthold RJ 2000. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci 20: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Clemente A, Matta JA, Isaac JT, Roche KW 2010. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron 67: 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA 2002. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci 99: 13902–13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Meyer T 1999. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science 284: 162–166 [DOI] [PubMed] [Google Scholar]
- Shen K, Scheiffele P 2010. Genetics and cell biology of building specific synapse connectivity. Annu Rev Neurosci 33: 473–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC 2007. The postsynaptic architecture of excitatory synapses: A more quantitative view. Annu Rev Biochem 76: 823–847 [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E 2000. The shank family of scaffold proteins. J Cell Sci 113: 1851–1856 [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim MJ 2002. Postsynaptic signaling and plasticity mechanisms. Science 298: 776–780 [DOI] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY 1994. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368: 144–147 [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL 2007. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol 23: 613–643 [DOI] [PubMed] [Google Scholar]
- Shipman SL, Schnell E, Hirai T, Chen BS, Roche KW, Nicoll RA 2011. Functional dependence of neuroligin on a new non-PDZ intracellular domain. Nat Neurosci 14: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Craig AM 2011. Synaptic organizing complexes. Curr Opin Neurobiol 21: 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz P 1985. The postsynaptic density: A possible role in long-lasting effects in the central nervous system. Proc Natl Acad Sci 82: 3494–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Smart TG, Paoletti P 2011. Synaptic neurotransmitter-gated receptors. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a009662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltau M, Berhorster K, Kindler S, Buck F, Richter D, Kreienkamp HJ 2004. Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J Neurochem 90: 659–665 [DOI] [PubMed] [Google Scholar]
- Stein V, House DR, Bredt DS, Nicoll RA 2003. Postsynaptic density-95 mimics and occludes hippocampal long-term potentiation and enhances long-term depression. J Neurosci 23: 5503–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner P, Higley MJ, Xu W, Czervionke BL, Malenka RC, Sabatini BL 2008. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron 60: 788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill JF, Steiner P, Czervionke BL, Sabatini BL 2009. Distinct domains within PSD-95 mediate synaptic incorporation, stabilization, and activity-dependent trafficking. J Neurosci 29: 12845–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455: 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Kawabata I, Sobue K, Okabe S 2005. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat Methods 2: 677–684 [DOI] [PubMed] [Google Scholar]
- Sumita K, Sato Y, Iida J, Kawata A, Hamano M, Hirabayashi S, Ohno K, Peles E, Hata Y 2007. Synaptic scaffolding molecule (S-SCAM) membrane-associated guanylate kinase with inverted organization (MAGI)-2 is associated with cell adhesion molecules at inhibitory synapses in rat hippocampal neurons. J Neurochem 100: 154–166 [DOI] [PubMed] [Google Scholar]
- Sun Q, Turrigiano GG 2011. PSD-95 and PSD-93 play critical but distinct roles in synaptic scaling up and down. J Neurosci 31: 6800–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Kubota Y, Forest A, Waxham MN 2010. Structure and composition of the postsynaptic density during development. J Comp Neurol 518: 4243–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M 2006. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol 16: 95–101 [DOI] [PubMed] [Google Scholar]
- Tai CY, Kim SA, Schuman EM 2008. Cadherins and synaptic plasticity. Curr Opin Cell Biol 20: 567–575 [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y 1997. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem 272: 11943–11951 [DOI] [PubMed] [Google Scholar]
- Tarusawa E, Matsui K, Budisantoso T, Molnar E, Watanabe M, Matsui M, Fukazawa Y, Shigemoto R 2009. Input-specific intrasynaptic arrangements of ionotropic glutamate receptors and their impact on postsynaptic responses. J Neurosci 29: 12896–12908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalin SJ, Colledge M, Hell JW, Langeberg LK, Huganir RL, Scott JD 2002. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J Neurosci 22: 3044–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triller A, Choquet D 2008. New concepts in synaptic biology derived from single-molecule imaging. Neuron 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL 2006. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics 5: 914–922 [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL 2008. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics 7: 684–696 [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, et al. 1999. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron 23: 583–592 [DOI] [PubMed] [Google Scholar]
- Turrigiano GG 2008. The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell 135: 422–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Turrigiano G 2011. Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB 1998. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391: 892–896 [DOI] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, et al. 2010. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron 65: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagarajan SK, Ghosh H, Yevenes GE, Nikonenko I, Ebeling C, Schwerdel C, Sidler C, Zeilhofer HU, Gerrits B, Muller D, et al. 2011. Regulation of GABAergic synapse formation and plasticity by GSK3beta-dependent phosphorylation of gephyrin. Proc Natl Acad Sci 108: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtschanoff JG, Weinberg RJ 2001. Laminar organization of the NMDA receptor complex within the postsynaptic density. J Neurosci 21: 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N 2004. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol 83: 449–456 [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N 2006. Neuroligins determine synapse maturation and function. Neuron 51: 741–754 [DOI] [PubMed] [Google Scholar]
- Vithlani M, Moss SJ 2009. The role of GABAAR phosphorylation in the construction of inhibitory synapses and the efficacy of neuronal inhibition. Biochem Soc Trans 37: 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D, Neddens J, Karavanova I, Tricoire L, Petralia RS, McBain CJ, Buonanno A 2009. Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci 29: 12255–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW Jr, Mercer JA, Kennedy MB 2000. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci 20: 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MJ, Kuruc N 1992. The postsynaptic density: Constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J Neurochem 59: 667–678 [DOI] [PubMed] [Google Scholar]
- Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, Luo J, et al. 2007. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 448: 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Schluter OM, Steiner P, Czervionke BL, Sabatini B, Malenka RC 2008. Molecular dissociation of the role of PSD-95 in regulating synaptic strength and LTD. Neuron 57: 248–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii A, Sheng MH, Constantine-Paton M 2003. Eye opening induces a rapid dendritic localization of PSD-95 in central visual neurons. Proc Natl Acad Sci 100: 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Shinkawa T, Taoka M, Kobayashi K, Isobe T, Yamauchi T 2002. Identification of protein substrates of Ca2+/calmodulin-dependent protein kinase II in the postsynaptic density by protein sequencing and mass spectrometry. Biochem Biophys Res Commun 290: 948–954 [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T 2004. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem 88: 759–768 [DOI] [PubMed] [Google Scholar]
- Zhang W, Vazquez L, Apperson M, Kennedy MB 1999. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci 19: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, Sudhof TC 2010. Neurexins physically and functionally interact with GABAA receptors. Neuron 66: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Petralia RS, Wang YX, Kachar B, Wenthold RJ 2010. SAP102 is a highly mobile MAGUK in spines. J Neurosci 30: 4757–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Seabold GK, Horak M, Petralia RS 2011. MAGUKs, synaptic development, and synaptic plasticity. Neuroscientist 17: 493–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R 2002. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 110: 443–455 [DOI] [PubMed] [Google Scholar]
- *.Zoghbi HY, Bear MF 2011. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a009886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Wendland JR, Ashley-Koch AE, Collins AL, Tran-Viet KN, Quinn K, Timpano KC, Cuccaro ML, Pericak-Vance MA, Steffens DC, et al. 2009. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol Psychiatry 14: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]