Abstract
Changes in gene expression, coupled with biochemical, physiological, and behavioral alterations, play a critical role in adaptation to environmental stress. Our goal was to explore ways natural populations may have adapted to local, polluted environments. We took advantage of natural populations of Fundulus heteroclitus, one of the few studied fish species in North America that has established resistant populations in highly contaminated urban estuaries. We analyzed morphology, physiology, and gene expression of developing F. heteroclitus embryos during late organogenesis (stage 31); these embryos were from both resistant and sensitive populations and were raised in a common, unpolluted environment. While cardiac heart rates show significant differences between embryos of parents from clean and heavily contaminated Superfund sites, time-to-stage, embryo morphology, and gene expression profile analyses do not differ significantly between untreated embryos from resistant and sensitive populations. Further evaluation that includes tissue-specific approaches in gene expression analysis and larger sample sizes may be necessary to highlight important phenotypes associated with mechanisms of sensitivity and resistance among natural F. heteroclitus embryo populations. Alternatively, population differences may be masked by developmental canalization, and biologically important differences between sensitive and resistant embryos may only manifest with exposure (e.g., be dependent on gene by environment interactions).
Keywords: Fundulus heteroclitus, microarrays, developmental stages, embryo gene expression
Introduction
Chemical contamination of ecosystems and chronic exposure to pollution can have significant negative effects on biodiversity, including population and subpopulation declines, increased mortality and reproductive failures, and overall decreases in variability (Bickham et al. 2000, Wirgin and Waldman 2004). Exposure to pollution may show effects at the molecular level, but also be in part responsible for changes at higher levels, such as tissues, organism health, reproduction, population demographics, and population genetics (Weis and Weis 1989, Smith and Weis 1997, Weis et al. 1999, Bickham et al. 2000, Fox 2001). Individual responses to pollution exposure vary, and while some individuals are sensitive to stress, others survive and reproduce, therefore establishing resistant populations.
Fundulus heteroclitus is a small minnow, which inhabits salt marshes along the east coast of the United States, and is one of the few studied species in North America living in the highly polluted urban estuaries that has shown resistance to pollutants, among both adults and embryos (Prince and Cooper 1995, Elskus et al. 1999, Nacci et al. 1999, Meyer et al. 2003). Due to its coastal habitat, limited home range (Lotrich 1975), large population sizes (Sweeney et al. 1998), hardiness in the laboratory, and extensive background on its physiology (Burnett et al. 2007) and embryology (Oppenheimer 1937, Armstrong and Child 1965, Selman and Wallace 1983, Hsiao et al. 1994), F. heteroclitus is an ecologically relevant and genetically diverse model often used to study pollution effects and genotype-environment interactions within and among populations. While the effects of chemical contamination on this species’ population genetics has been studied in some populations (Mulvey et al. 2003, Roark et al. 2005, McMillan et al. 2006, Williams and Oleksiak 2008) relatively little is known about functionally important variation in embryonic gene expression underlying resistance mechanisms.
Resistance to the lethal effects of pollution has been reported in F. heteroclitus embryos from two highly contaminated US Superfund sites: Elizabeth River, VA and Newark Bay, NJ. The Elizabeth River site is contaminated with creosote, a mixture of polycyclic aromatic hydrocarbons (PAHs) (Vogelbein et al. 1990, Ownby et al. 2002). The lower Hudson River estuary in Newark Bay is mostly contaminated with PAHs, polychlorinated biphenyls (PCBs), pesticides, and metals (Long et al. 1995, Wolfe et al. 1996). F. heteroclitus from these two sites are resistant to the complex pollutant mixtures in their environment and provide an excellent model to identify phenotypes involved in adaptation to pollution. By comparing and contrasting phenotypes of embryos from polluted populations with those of embryos from nearby, flanking reference populations, we can identify altered phenotypes most likely due to pollution.
Most F. heteroclitus embryo studies present evidence for resistance to pollution by comparing embryo responses to contaminated sediments or representative chemicals in the laboratory (Nacci et al. 1999, Meyer et al. 2002, Ownby et al. 2002, Meyer et al. 2003). Our goal was to identify differences in survival, time-to-stage, cardiac physiology, morphology, and gene expression among non-exposed embryos of parents from clean and heavily polluted sites. We compared laboratory reared, common-gardened individual F. heteroclitus embryos of parents from three reference sites (Manteo, NC, King’s Creek, VA, and Succotash, RI) and two polluted Superfund sites (Elizabeth River, VA and Newark Bay, NJ) during a late organogenesis stage to investigate selected phenotypes and common conserved patterns of gene expression among embryos associated with clean and polluted environments.
Materials and Methods
Fish Maintenance and embryo culturing
Adult F. heteroclitus were captured at three reference sites (Manteo, NC; Kings Creek, VA, and Succotash, RI) and two polluted Superfund sites (Elizabeth River, VA, and Newark Bay, NJ) by minnow traps in April 2007 and transported under controlled temperature and aeration condition to the NCSU Aquatic Laboratory. FST values between polluted and reference sites based on AFLP (0.039, and 0.018) are approximately one-half of those found for microsatellites (0.068, and 0.043), for Newark Bay and Elizabeth River, respectively (Adams et al. 2006, Williams and Oleksiak 2008). Fish were acclimated to common conditions of 20° C and 15 ppt salinity in 40 gallon flow-though re-circulating aquatic system tanks for four months prior to embryo culturing. Since F. heteroclitus spawn on a lunar cycle, F. heteroclitus would have spawned up to four times during this four months common gardening period. Effluent from the tanks was passed through an activated charcoal filter system and 20% water was changed weekly. Tanks were maintained and fish were fed (brine shrimp flake, blood meal flake, and Spirulina flake - FOD, Aquatic Biosystems) daily and checked for health. Fish were maintained under a pseudo-summer cycle (8 h dark / 16 h light).
Eggs were stripped from five mature females and fertilized by sperm collected from five individual males within each population, resulting in five family sets, each with multiple embryo offspring. Fertilized embryos were maintained in Petri dishes half submerged in 15 ppt filtered seawater in a 25°C environmental chamber under light during the first two stages of development (2-cell stage). Embryos that successfully reached the 2-cell stage were incubated under a 16 hour light: 8 hour dark photoperiod at 25°C in the environmental chamber (818 Low Temperature Illuminated Incubator, Precision Scientific, USA).
Site exposures
Semi-permeable membrane devices (SPMDs) were deployed in aluminum cages for 28 days in early April 2006 at Newark Bay, NJ, Magotha Bay, VA, Elizabeth River, VA, and Manteo, NC (Hofelt and Shea 1997, Luellen and Shea 2002). At each site, six SPMDs were placed in the marsh during the incoming low tide approximately 20 m apart. Upon retrieval, the SPMDs were wrapped in combusted aluminum foil, placed in a plastic bag, and maintained on ice until frozen. SPMDs were transported to the laboratory, where they were maintained frozen at −20°C until time of analysis. SPMDs were analyzed using established methods (Luellen and Shea 2002).
Embryo survival, developmental delays, heart rate and morphology
Fertilization success and embryo progress was monitored twice daily by examining representative stages during pre-determined time periods (Armstrong and Child 1965); internal data) using a dissecting stereo microscope (Nikon SME1500, Japan). Time to stage, normal versus abnormal development, and mortality also were recorded. Unfertilized eggs and malformed and/or dead embryos were removed from the population, and times and stages of arrest and abnormal development were recorded accordingly. Survival rates were measured within a family of each population and as overall survival rates between populations. Embryos that successfully hatched and survived for two weeks as free-swimming larvae were considered to be surviving embryos.
To determine developmental delays to stages 31 and 35 within a population, five embryos from five families were monitored in individual 20-ml scintillation vials. Identification of each stage was determined by scoring embryos at predetermined time-periods for both stages 31 (140 hours post-fertilization) and 35 (212 hours post-fertilization, Armstrong and Childs, 1965; internal unpublished data) using a dissecting stereo microscope (Nikon SME1500, Japan) at 70–80X magnification. Multiple images of developing embryos were taken at different phases of each developmental stage. Images were captured with the Micropublisher 5.0 RTV Camera (QImaging) and catalogued, stored, and analyzed using QCapture Pro imaging software.
The same embryos used to determine time to stage were used to determine heart rates during early organogenesis (stage 31) and pre-hatching (stage 35). A beating heart is formed, with both chambers completely differentiated and in full view, by stage 31, and the heart rate can be accurately measured from that stage on. Embryo vials were labeled to assure that the heart rate was measured from the same embryo at both stages. Individual embryos were placed on a depression slide under the dissecting stereo microscope for one minute prior to taking heart rate measurements so that the stressed embryo could re-establish resting heart beat (most F. heteroclitus embryos temporarily arrest their heart beat due to a sudden change of environment, such as transfer from the petri dish to a well-lit slide surface). The heart rate of each embryo was measured by counting the number of heartbeats for 30 seconds (preliminary results showed no change in the average heart beat when counts were taken at either 30 second or 1 minute intervals).
Embryo Morphology
During development, pictures of three randomly selected embryos from each population were taken and catalogued at each stage, and any embryos showing morphological abnormalities were photographed and catalogued. At stages 31 and 35, three embryos from each family within a population were randomly selected and scored for morphological abnormalities. Only embryos that developed successfully to stage 35 were considered for analysis.
Embryos were scored for normal versus abnormal development. Deformities include various degrees of incompletely differentiated heart chambers, pericardial edema, cranio-facial alterations, loss of pigmentation, tail shortening, and hemorrhaging. Since heart deformities were found to be the most sensitive and reliable endpoint scored in our previous embryo exposure studies, they were more heavily weighted in deformity scorings. Deformities were ranked using a scale from 1 to 5, 1 representing no deformities, 2-mild, 3-moderate, 4-severe, and 5-extreme deformities, respectively.
These experiments were performed according to an approved Institutional Animal Care and Use Committee at North Carolina State University.
Microarrays
Amplified cDNA sequences for approximately 7,000 genes from F. heteroclitus cDNA libraries were spotted onto epoxide slides (Corning) using an inkjet printer (Aj100, ArrayJet, Scotland). Libraries were made from all 40 stages of F. heteroclitus development, immediately post-hatch whole larvae, and adult tissues. Each slide contained four spatially separated arrays of ~7,000 spots (genes) including controls. This is an expanded array of previous F. heteroclitus arrays. These arrays use cDNA probes that have an average length of 1.5 Kb and have a technical variation of less than 5% of the mean (CV < 0.05) (Oleksiak et al. 2002, Oleksiak et al. 2005, Whitehead and Crawford 2006, Fisher and Oleksiak 2007, Oleksiak 2008, Scott et al. 2009a, Scott et al. 2009b). This has allowed us to statistically distinguish less that 1.3 fold differences in expression. All spotted genes were sequenced and represent unique contigs. Thus, even if multiple sequences were annotated identically, they were treated as different genes. Multiple sequences with the same annotation do not contig together because: 1) they really are the same gene, but the sequences do not overlap, 2) they represent duplicate genes with different chromosomal locations, or 3) they share a high similarity (and hence are named based on this similarity) but are not the same gene. We erred on the side of caution and treated every gene-spot as unique.
Embryo RNA Isolation, Amplification, and Labeling
Once the normally developing embryos reached stage 31, they were photographed using a Micropublisher 5.0 RTV Camera (QImaging) fitted on the stereo microscope. Embryos were then immediately placed in pre-chilled 1.5 ml microcentrifuge tubes and snap-frozen at −80°C for later RNA analyses.
One frozen embryo from each of four families within a population was used for RNA isolation, labeling, and microarray hybridization. Embryo RNA was extracted using a Trizol buffer (Invitrogen, Carlsbad, CA, USA) and quantified with a spectrophotometer. On average, we obtained 12.3 +/− 2.8 ug of total RNA from each frozen embryo. RNA quality was assessed by gel electrophoresis, and was prepared for hybridization by one round of amplification (aRNA) using Ambion’s Amino Allyl MessageAmp aRNA Kit to form copy template RNA by T7 amplification. Amino-allyl UTP was incorporated into targets during T7 transcription, and resulting amino-allyl aRNA was coupled to Cy3 and Cy5 dyes (GE Healthcare, Piscataway, NJ, USA).
Labeled aRNA samples (2 pmol dye/ul) were hybridized to slides in 10 ul of hybridization buffer (50% formamide buffer, 5× SSPE, 1% sodium dodecyl sulfate, 0.2 mg/ml bovine serum albumin, 1 mg/ml denatured salmon sperm DNA (Sigma), and 1 mg/ml RNAse free poly(A) RNA (Sigma) for 44 hours at 42° C. Slides were prepared for hybridization by blocking in 5% ethanolamine, 100 mM Tris pH 7.8, and 0.1% SDS added just before use for 30 minutes at room temperature, washed for one hour in 4× SSC, 0.1% SDS at 50° C, and then boiled for 2 minutes in distilled water to denature the cDNAs. Resulting 16 bit Tiff Images were quantified using ImaGene® (Biodiscovery, Inc.) spotfinding software. Controls and any gene that did not have at least one individual with a signal greater than the average signal from all herring sperm control spots (non-specific hybridization signal) plus one standard deviation were removed prior to statistical analyses. In total, 6,659 genes were analyzed.
Experimental Design for Microarrays
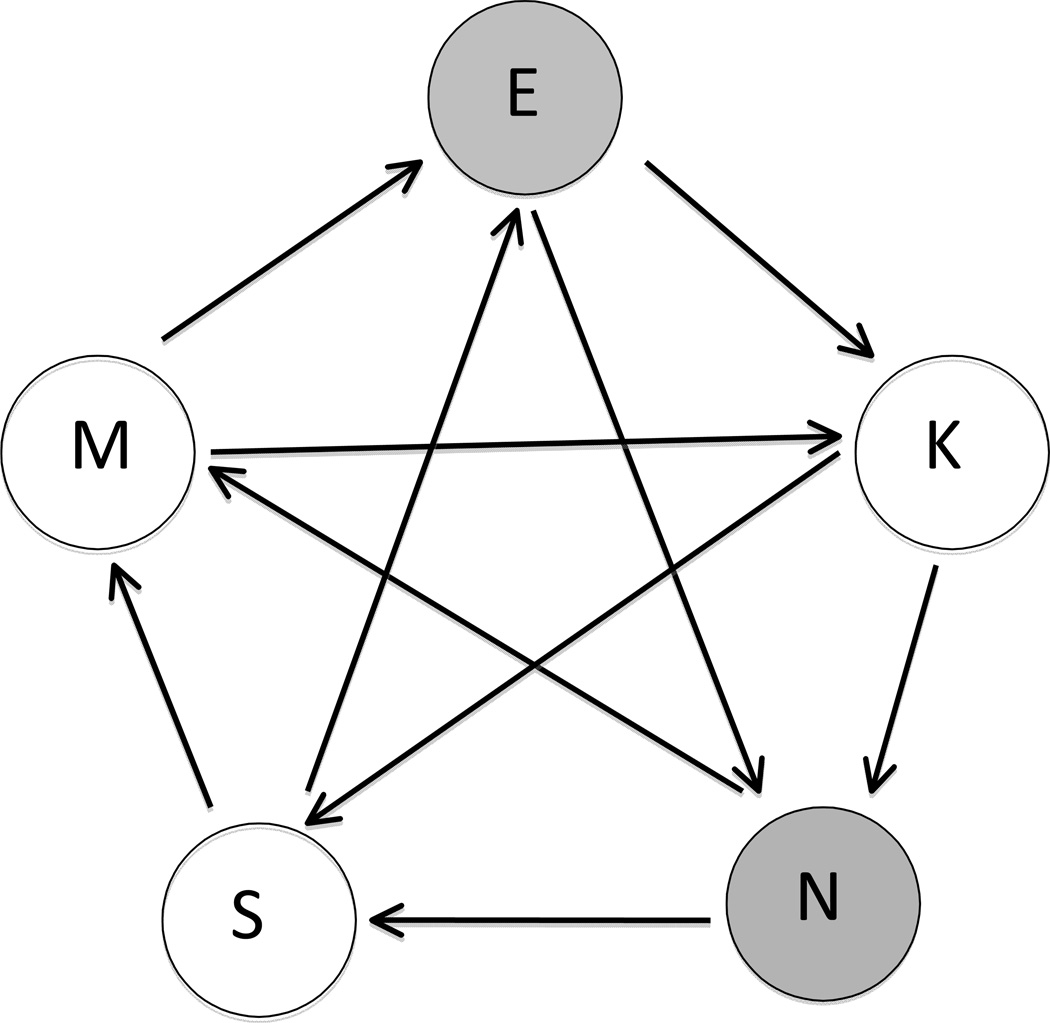
A loop design (Figure 1) was used for the microarray hybridizations where each sample is hybridized to 2 arrays using both Cy3 and Cy5 labeled fluorophores (Kerr and Churchill 2001a, Kerr and Churchill 2001b). The loop consisted of Cy3 and Cy5 labeled embryo aRNAs from 4 embryos (biological samples), one from each family within a population. In total, 20 embryos were hybridized to 10 microarrays. Each array had a different combination of biological samples, so that the most direct comparisons of geographically related populations, and reference versus polluted populations are hybridized to the same array. The loop formed was M→ E → K→ N →S→ M→ K→ S→ E→ N → M (Figure 1), where each arrow represents a separate hybridization (array) with the biological sample at the base of the arrow labeled with Cy3 and the biological sample at the head of the arrow labeled with Cy5. M represents the Manteo, NC population, E represents the Elizabeth River, VA population, K represents the King’s Creek, VA population, N represents the Newark Bay, NJ population, and S represents the Sandwich, MA population.
Figure 1. Loop Design for Embryo gene expression analysis.
Loop consists of 4 biological replicates (embryo aRNA), each representing a family within a population, totaling 20 biological replicates (4 embryos × 5 populations). Two embryo RNAs are labeled with Cy3 and two with Cy5 within each population. Each arrow represents an array, and the direction of the arrow is from Cy3 to Cy5, so that two embryo RNAs from different populations labeled with different dyes are hybridized to the same array. M – Manteo, NC; E – Elizabeth River, VA; K – King’s Creek, VA; N – Newark Bay, NJ, S – Succotash, RI. Clear circles represent reference populations; gray circles represent polluted populations.
Statistical Analyses
Survival, Heart Rate, Time to Stage, and Morphology
Differences in the survival, heat rate, time to stage, and morphology among five embryo populations were analyzed with Prism Statistical Software (GraphPad Prism®, Version 5.0) using one-way Analysis of Variance (1-way ANOVA, p < 0.05); a pairwise t-test was used to test the differences of means between embryo populations while Dunnett’s one-tailed t-test was used to evaluate differences between reference embryos and “polluted” embryos, respectively. Bartlett’s test of equal variance was used to compare variation within populations.
Microarrays
Log2 measures of gene expression were normalized using a linear mixed model in SAS (JMP v3.2 with a microarray platform beta-version in SAS v9.1.3) to remove the effects of dye (fixed effect) and array (random effect) following a joint regional and spatial Lowess transformation in MAANOVA Version 0.98.8 for R to account for both intensity and spatial bias (Wu et al. 2003).
The model was of the form yij = μ + Ai + Dj + (AxD)ij + εij, where, yij is the signal from the ith array with dye j, μ is the sample mean, Ai and Dj are the overall variation in arrays and dyes (Cy3 and Cy5), (AxD)ij is the array × dye interaction and εij is the stochastic error (Jin et al. 2001, Wolfinger et al. 2001).
Residuals from the above model were used for gene-by-gene analyses of population effect during stage 31, using population and dye as fixed effects, and array as a random effect. The model was rijn = μ + Ai + Dj + Pn + εijn where Pn is the nth population. Residuals also were used for gene-by-gene analyses of treatment effect during stage 31, using treatment and dye as fixed effects, and array as a random effect. The model was rijk = μ + Ai + Dj + Tk + εijk where Tk is the kth treatment (polluted or reference). This model was run using all five populations (embryos from the two polluted populations versus those from the three reference populations) and the southern and northern populations alone (Elizabeth River embryos versus Manteo, NC and Magotha, VA embryos and Newark Bay embryos versus Succotash, RI ones).
For all mixed model analyses, we used a nominal p-value cut-off for significant genes of p < 0.01. Using this p-value reveals more genes that may be differentially expressed but risks identifying genes that may be false positives.
Results
Site Exposures
Exposure to 48 PAHs at the different sites were measured using a modified semipermeable membrane device (SPMD) (Hofelt and Shea 1997, Luellen and Shea 2002). These devices serve as passive accumulators of the dissolved and/or bioavailable fraction of organic chemicals and allow estimation of time-weighted average exposure of PAHs at each site. Accumulation in SPMDs correlates well with accumulation in caged mussels and is not susceptible to biological variations found in living organisms such as respiration rate and lipid mass which affect bioaccumulation (Hofelt and Shea 1997).
The Elizabeth River site was highly polluted with a variety of PAHs, including both low and high molecular weight compounds (Table 1). The total PAH concentration was highest in the Elizabeth Riversite (3599.20 ng/L), followed by the Newark Bay site (110.72 ng / L) (Table 1). Two reference sites, Magotha Bay, Va (3.64 ng / L) and Manteo, NC (17.89 ng / L), had relatively low total PAH concentrations, compared to the two polluted sites (Table 1). PAHs were not measured at the Succotash, RI site, a previously characterized reference site (McMillan et al. 2006).
Table 1.
Estimated PAH concentrations in water (ng/L).
| Analyte (ng/L) | Newark Bay, NJ |
Magotha, VA |
Elizabeth River, VA |
Manteo, NC |
|---|---|---|---|---|
| Naphthalene | 0.73 | 0.46 | 3.10 | 0.28 |
| 2-Methylnaphthalene | 0.79 | 0.29 | 2.03 | 0.18 |
| 1-Methylnaphthalene | 0.49 | 0.15 | 1.97 | 0.10 |
| Biphenyl | 1.22 | 0.00 | 0.88 | 0.20 |
| 2,6-Dimethylnaphthalene | 0.98 | 0.10 | 1.80 | 0.20 |
| Acenaphthylene | 0.00 | 0.00 | 1.07 | 0.00 |
| Acenaphthene | 6.55 | 0.00 | 67.03 | 0.00 |
| Dibenzofuran | 1.93 | 0.14 | 9.56 | 0.10 |
| 2,3,5-Trimethylnaphthalene | 1.66 | 0.00 | 5.49 | 0.00 |
| C1 - Naphthalenes | 1.60 | 0.55 | 4.23 | 1.02 |
| C2 - Naphthalenes | 6.21 | 0.77 | 13.21 | 1.53 |
| C3 - Naphthalenes | 5.90 | 0.00 | 16.03 | 1.30 |
| C4 - Naphthalenes | 4.06 | 0.00 | 12.15 | 0.00 |
| Fluorene | 2.99 | 0.15 | 18.38 | 0.14 |
| 1-Methylfluorene | 0.94 | 0.00 | 4.80 | 0.00 |
| C1 - Fluorenes | 3.31 | 0.00 | 38.65 | 0.00 |
| C2 - Fluorenes | 0.00 | 0.00 | 87.38 | 0.00 |
| C3 - Fluorenes | 0.00 | 0.00 | 0.00 | 0.00 |
| Dibenzothiophene | 0.74 | 0.00 | 10.03 | 0.00 |
| C1 - Dibenzothiophene | 1.72 | 0.00 | 18.73 | 0.00 |
| C2 - Dibenzothiophene | 2.65 | 0.00 | 23.43 | 0.00 |
| C3 - Dibenzothiophene | 0.00 | 0.00 | 42.39 | 0.00 |
| Phenanthrene | 6.35 | 0.58 | 84.43 | 0.55 |
| Anthracene | 1.26 | 0.00 | 30.55 | 0.00 |
| 1-Methylphenanthrene | 0.81 | 0.00 | 14.02 | 0.00 |
| C1 - Phenanthrenes/Anthracenes | 6.10 | 0.00 | 113.48 | 0.00 |
| C2 - Phenanthrenes/Anthracenes | 7.75 | 0.00 | 128.44 | 0.00 |
| C3 - Phenanthrenes/Anthracenes | 6.80 | 0.00 | 75.17 | 0.00 |
| C4 - Phenanthrenes/Anthracenes | 0.00 | 0.00 | 0.00 | 0.00 |
| Fluoranthene | 12.85 | 0.45 | 1026.30 | 0.42 |
| Pyrene | 8.23 | 0.00 | 279.47 | 0.50 |
| C1 - Fluoranthenes/Pyrenes | 7.60 | 0.00 | 495.35 | 0.00 |
| Retene | 0.00 | 0.00 | 0.00 | 11.34 |
| Benz[a]anthracene | 1.28 | 0.00 | 92.61 | 0.00 |
| Chrysene | 2.01 | 0.00 | 74.46 | 0.00 |
| C1 - Chrysenes | 3.12 | 0.00 | 78.96 | 0.00 |
| C2 - Chrysenes | 0.00 | 0.00 | 22.78 | 0.00 |
| C3 - Chrysenes | 0.00 | 0.00 | 0.00 | 0.00 |
| C4 - Chrysenes | 0.00 | 0.00 | 0.00 | 0.00 |
| Benzo[b]fluoranthene | 0.00 | 0.00 | 194.12 | 0.00 |
| Benzo[k]fluoranthene | 0.00 | 0.00 | 189.80 | 0.00 |
| Benzo[e]pyrene | 2.07 | 0.00 | 105.10 | 0.00 |
| Benzo[a]pyrene | 0.00 | 0.00 | 30.66 | 0.00 |
| Perylene | 0.00 | 0.00 | 7.70 | 0.00 |
| Indeno[1,2,3-cd]pyrene | 0.00 | 0.00 | 130.39 | 0.00 |
| Dibenz[a,h,]anthracene | 0.00 | 0.00 | 0.00 | 0.00 |
| Benzo[g,h,i]perylene | 0.00 | 0.00 | 39.34 | 0.00 |
| Coronene | 0.00 | 0.00 | 3.72 | 0.00 |
| Total PAH | 110.72 | 3.64 | 3599.20 | 17.88 |
Embryo Survival
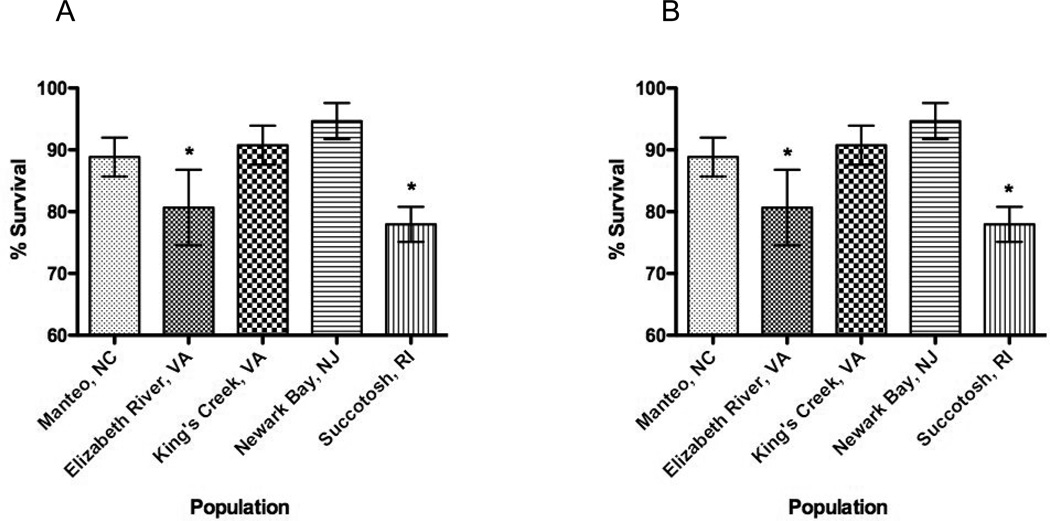
Survival rates were significantly different among populations at both stage 31 and 35 (Figure 2, 1-way ANOVA, p<0.01). For both stages, the highest % survival rate (94.7 ± 6%) was among Newark Bay, NJ embryos, while the lowest % survival (78.6 ± 6%) was among Succotash, RI embryos, which was significantly lower when compared to both King’s Creek, VA and Newark Bay, NJ embryos. Variance within populations did not differ significantly (Bartlett’s test for equal variances, p = 4.42).
Figure 2. Embryo survival among five populations at stages 31 and 35.
A. Stage 31 embryos. B. Stage 35 embryos. 1-way ANOVA (p<0.05) indicates significant differences among embryo population survival rates. Dunnett’s t-test indicates that Elizabeth River, VA and Succotash, RI embryos have significantly lower survival rates when compared to Newark Bay, NJ embryos.
Developmental Delays
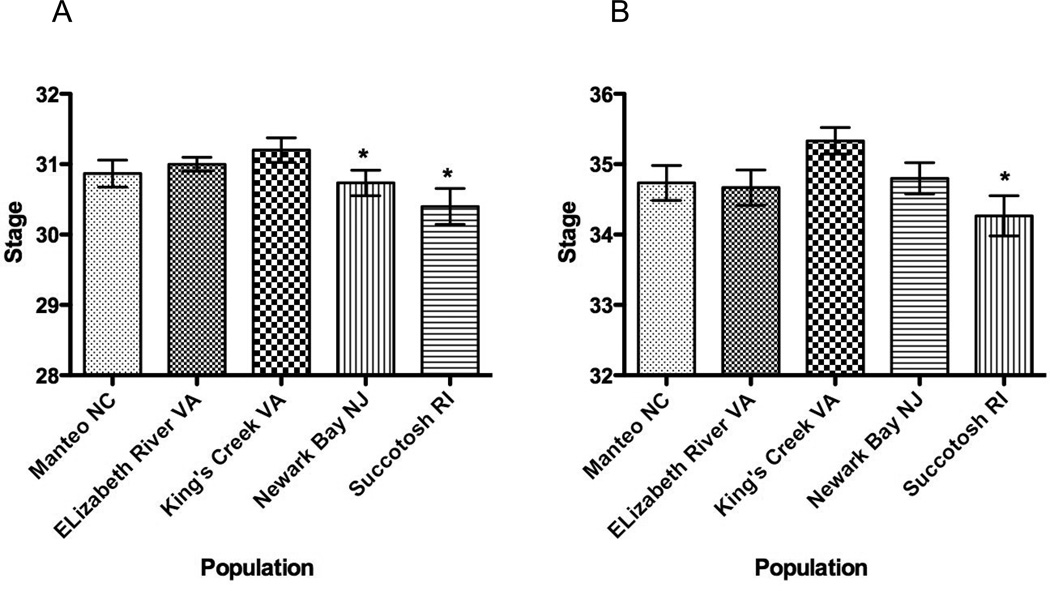
Although most of the embryos reached stages 31 and 35 within an expected time period, time-to-stage was significantly different among populations (Figure 3). King’s Creek, VA embryos reached stage 31 first while both northern embryo populations (Newark Bay, NJ; Succotash, RI) developed significantly more slowly (1-way ANOVA, p < 0.05) when compared to the King’s Creek embryo population. The variance within populations differed significantly (p < 0.05) with the most variance noted in the northern most population (Succotash, RI, 30.40 ± 0.99). Time-to-stage also was significantly different among populations (1-way ANOVA, p < 0.05) at stage 35; variance did not differ significantly (Bartlett’s test for equal variance, p = 2.560). The most variance was noted among King’s Creek embryos; on average, King’s Creek embryos reached stage 35 the soonest (35.33±0.72), while Succotash embryos developed most slowly (34.27±1.10) among the five populations. Pairwise comparisons between populations revealed significant differences in the onset of developmental stage before hatching between King’s Creek, and Succotash, RI embryos (Bonferroni’s Multiple Comparisons test, p < 0.05; t = 3.13).
Figure 3. Developmental delays among embryos from five populations.
A. Stage 31 embryos. 1-way ANOVA (p<0.05) indicates significant differences among populations. Dunnett’s t-test indicates that Newark Bay, NJ and Succotash, RI embryos are significantly delayed when compared to King’s Creek, VA embryos. B. Stage 35 embryos. 1-way ANOVA (p<0.05) indicates significant differences among populations. Pairwise comparisons between populations revealed significant differences in the onset of developmental stage before hatching between King’s Creek, and Succotash, RI embryos (Bonferroni’s Multiple Comparisons test, p < 0.05; t = 3.13).
Embryo Heart Rate
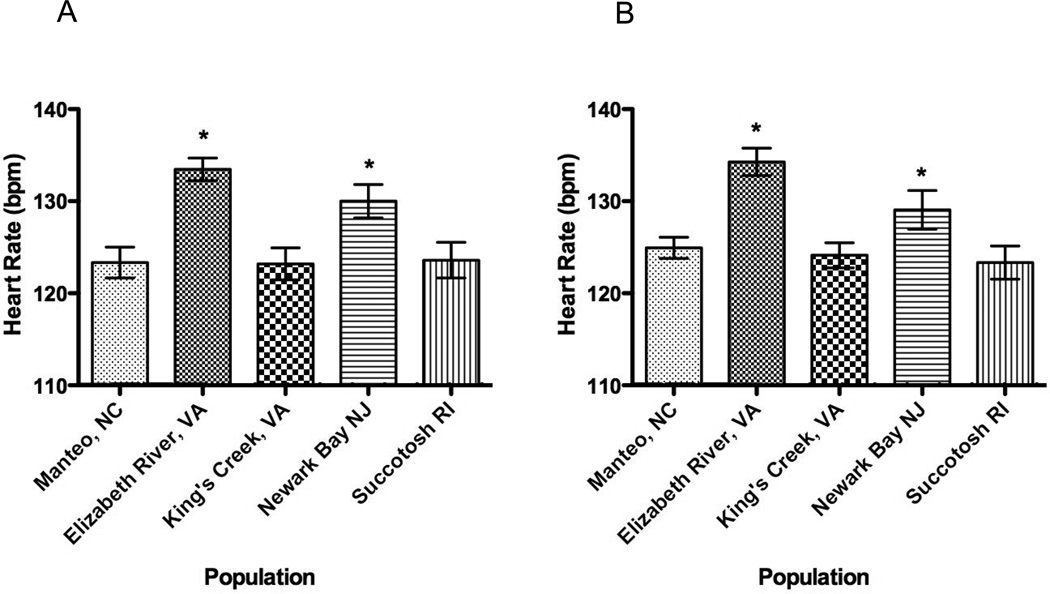
Heart rates at stage 31 also were significantly different among embryo populations (Figure 4, 1-way ANOVA, p < 0.01) while variance within populations was not (Bartlett’s test, p = 0.55). Interestingly, both embryo populations of parents caught at the polluted sites showed significantly higher heart rates (Elizabeth River, VA 133.5 ± 4.7 bpm; Newark Bay, NJ 130 ± 7.0 bmp), when compared to three reference embryo populations. Heart Rates differed significantly among the five embryo populations at stage 35 (1-way ANOVA, p < 0.05), while variance did not. The highest heart rates were noted among Elizabeth River embryos (134.3 ± 5.75), while the lowest heart rates were among Succotash, RI embryos (123.3±7.00). Bonferroni’s multiple comparisons tests between populations revealed that Elizabeth River embryos have significantly higher heart rates then Manteo, NC (p < 0.01; t = 4.084), King’s Creek, NC (p < 0.001; t = 4.43), and Succotash, RI embryos p < 0.001; t = 4.78).
Figure 4. Heart rate (beats/min) differences during late organogenesis among five embryo populations.
A. Stage 31 embryos. Two embryo populations from polluted sites (Elizabeth River, VA and Newark Bay, NJ) have significantly faster (p<0.05) heart rates when compared to embryos from clean reference sites. B. Stage 35 embryos. Heart rates differed significantly among 5 embryo populations at stage 35 (1-way ANOVA, p < 0.05). Bonferroni’s Multiple Comparisons Tests comparisons test between populations revealed that Elizabeth River embryos have significantly higher HRs then Manteo, NC (p < 0.01; t = 4.084), King’s Creek, NC (p < 0.001; t = 4.43), and Succotash, RI embryos p < 0.001; t = 4.78).
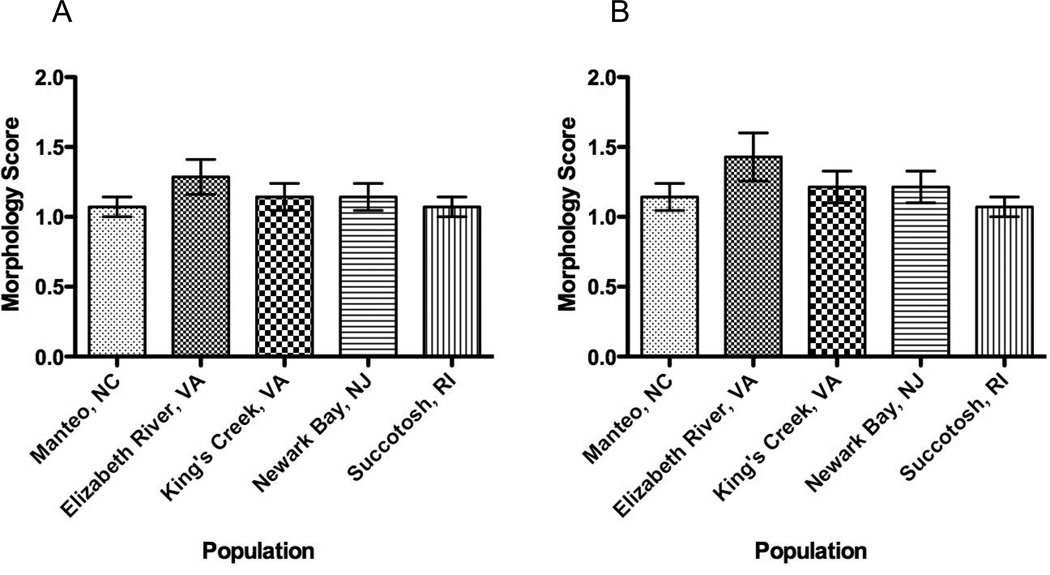
Morphology
Slight, but not statistically significant in vivo morphological differences were noted in 26% of embryos from Elizabeth River, while > 90% of embryos from the remaining populations developed normally (N=15, Figure 5). In each case, only the mild abnormalities, including cranio-facial imperfections, lack of pigmentation in the head regions, and near-complete heart chambers at stage 31 were noted among all embryo populations. None of the noted abnormalities were scored higher than 2 (mild abnormality). Variation within a population did not differ when compared among populations (Bartlett’s test, p = 0.22). At stage 35, embryos exhibited no significant differences in morphology score (1-way ANOVA, p < 0.01), but the variances were significantly different (Bartlett’s test for equal variances, p = 0.034). 26.6 % of the Elizabeth River embryos were mildly deformed (score 2), while 6.7% (1/15) of these embryos showed signs of moderate heart and cranio-facial abnormalities (score 3). Greater than 86% of embryos developed normally in other populations, with greater than 93% of the embryos developing normally in the Succotash, RI population.
Figure 5. Embryo morphology among five populations during late organogenesis (stage 31).
A. Stage 31 embryos. B. Stage 35 embryos. Most embryos developed normally, and there were no significant differences in morphology (1-way ANOVA, p<0.05) among the three clean and two polluted embryo populations. Criteria used when scoring embryo morphology: 1 = normal; 2 = mildly-deformed; 3 = moderately-deformed; 4 = severely deformed; 5 = extremely deformed.
Gene Expression
Among five embryo populations during late organodifferentiation (stage 31), only 77 genes (1.2% of 6,659 genes) had a significant population effect (p < 0.01). Expression of 5.7 % (380) of genes was significantly different between any two pairs of these embryo populations (p<0.01). No genes were significantly differently expressed at an adjusted pFDR (p < 0.0001). Differences in heart rates among embryo populations did not correlate significantly with any of the significant genes.
Pairwise comparison results of variation in gene expression between embryo populations are summarized in Table 1. The highest numbers of significantly differently expressed genes occur between Manteo, NC and Succotash, RI embryos (164 genes, 2.5%). Surprisingly, our results show similar differences between Manteo, NC embryos and nearby sites, Elizabeth River, VA (145 genes, 2.2%), and King’s Creek, VA (142 genes, 2.1%). Less than 1.1 % of genes were found to be significantly different in other population comparisons. We found no significant differences in gene expression between embryo populations when applying pFDR adjustment (p < 0.00001).
Expression levels of 54 genes (0.8%) were significantly different (p < 0.01) when comparing embryos from the southern polluted site, Elizabeth River, VA, to embryos from two nearby reference sites, Manteo, NC, and King’s Creek, VA. Because these two reference sites flank the polluted site, the genetic distance between the two clean reference populations is greater than the genetic distance between the polluted population and either reference population. Thus, divergence in a polluted population compared to both paired reference populations suggests that pollution might be causative. Forty-two genes (0.6%) were significantly different when comparing the northern polluted site, Newark Bay, NJ, to its reference site, Succotash, RI. Because we do not have two flanking reference sites in this comparison, differences might be due pollution (or another difference between the sites) or simply due to genetic drift. We found no significant genes at pFDR < 0.00001 in either analysis.
To determine shared responses between the polluted sites, both polluted sites were compared to all three, reference sites. Again, the greater genetic distance among the reference sites as compared to the polluted sites should obviate difference simply due to genetic drift. Expression of 37 genes (0.6%) was significantly different when comparing the two polluted sites to the three reference sites. We found no significant genes at pFDR < 0.00001.
Discussion
Chemical contamination of the environment, which includes long-term chronic exposure and short-term, acute exposures to natural populations, could be one of the important factors contributing to changes in a population’s genetic diversity (Dieter 1993, Herbert and Luiker 1996, Nacci et al. 1999, Bickham et al. 2000). Pollution exposure can lead to transgenerational effects and reduce population variability via somatic and heritable mutations and non-genetic modes of toxicity (Bickham and Smolen 1994, Cronin and Bickham 1998). The traits favoring individuals that successfully cope with the environmental stress are inherited over time, and the exposed populations eventually consist of individuals contributing to subsequent generations whose phenotypes favor adaptations to current conditions. However, it is often difficult to prove natural selection among natural populations (Endler 1986). We hypothesized that F. heteroclitus embryos of parents chronically exposed to pollution may differ from embryo populations of parents from clean sites, and we searched for significant differences in selected in vivo morphological, physiological, and gene expression phenotypes. We analyzed late organogenesis stages (stages 31 and 35) of F. heteroclitus embryos whose parents were collected at three clean and two highly contaminated urban Superfund sites.
Only heart rate was consistently and significantly different in the two polluted populations (Figure 4); in both the Elizabeth River and Newark Bay embryos, heart rates were significantly higher than in the reference embryos at stages 31 and 35. Survival rates were statistically lower for two embryo populations, one polluted and one reference at stages 31 and 35 (Figure 2). However, the highest survival rate was noted among embryos from the other highly polluted Superfund site, Newark Bay. Thus, polluted populations do not show a consistent difference in survival.
Time to stage was significantly longer for the two northern populations, Newark Bay, NJ and Succotash, RI, compared to the southern populations at stage 31 (Figure 3A). Average temperature differences between Southern and Northern F. heteroclitus sites (1° C change / degree latitude; (Powers et al. 1993)) affect developmental rates and metabolism (DiMichele and Powers 1982), yet our results contradict the counterbalance latitude effect of water temperature that is associated with shorter hatching time among northern F. heteroclitus populations (Massachusetts and Delaware, 10.5–12.5 days) than southern populations (Georgia, 14–15 days) (DiMichele and Westerman 1997). At stage 35, only Succotash, RI embryos had a significantly longer time to stage compared to all other populations (Figure 3B). Finally, there were no significant differences in in vivo morphology among any of the five populations (Figure 5). Thus, for the physiological phenotypes that we measured, only heart rate was correlated with pollution exposure.
Only 77 genes (1.2% of 6,659 genes) had a significant population effect (p < 0.01) among the five embryo populations, regardless of pollutant status, and most differences in gene expression among embryo populations are due to the Manteo embryos. From 1.1 to 2.4% of genes differ in expression between the Manteo embryos and embryos from other populations (p < 0.01, Table 2). Theses differences are unlikely to be physiologically induced because the embryos were common-gardened in the laboratory during their development. This is supported by our time-to-stage, morphology, and survival data suggesting no significant differences between Manteo and other embryo populations. Another possibility for the observed differences in gene expression patterns between embryos from the Manteo population and those from the other populations is the overall health of the field-collected adults coupled with acclimatization to laboratory conditions. If Manteo embryos are excluded from the analysis, less than 1% of genes are different when comparing expression of any two populations (Table 2).
Table 2.
Numbers and percentages of significantly differently expressed genes between pair wise embryo population comparisons (p < 0.01).
| Population | Manteo, NC |
Elizabeth River, VA |
King’s Creek, VA |
Newark Bay, NJ |
Succotash, RI |
|---|---|---|---|---|---|
| Manteo, NC | * | 145 | 142 | 73 | 164 |
| Elizabeth River, VA | 2.1% | * | 30 | 61 | 39 |
| King’s Creek, VA | 2.1% | 0.4% | * | 44 | 33 |
| Newark Bay, NJ | 1.1% | 0.9% | 0.7% | * | 42 |
| Succotash, RI | 2.4% | 0.5% | 0.5% | 0.6% | * |
When both polluted site embryos are compared to all reference site embryos, only 0.6% of the genes differ significantly in expression. When the polluted Elizabeth River, VA embryos are compared to both flanking reference sites (King’s Creek, VA and Manteo, NC), 0.8% of the genes differ significantly in expression; when the polluted Newark Bay, NJ embryos are compared to one nearby reference site (Succotash, RI), 0.6% of the genes differ significantly in expression. These percentages are less than the 1% expected by chance and suggest that parent exposure history does not have a significant effect on developmental gene expression in unexposed embryos at stage 31.
One can reduce the number of queried genes by applying any number of stringent quality filters on the data (Hackstadt and Hess 2009) and thus reduce the stringency of the multiple test correction. However, doing this will have no effect on the p-values calculated from the general linear model used in the data analysis. For all of our comparisons of polluted and reference populations, the p-values are quite large (the smallest p-value is 0.0003). Thus, even if we analyzed only 100 of the ~7,000 genes, no gene would be significantly differently expressed with a correction for multiple testing.
The percentage of significantly differently expressed genes is surprisingly low, considering the large variation in gene expression reported within and among adult F. heteroclitus populations (Oleksiak et al. 2002, Oleksiak et al. 2005, Whitehead and Crawford 2006, Fisher and Oleksiak 2007, Oleksiak 2008), including populations with no history of pollutant exposure (Oleksiak et al. 2002, Oleksiak et al. 2005, Whitehead and Crawford 2006, Fisher and Oleksiak 2007, Oleksiak 2008). These studies with adult F. heteroclitus used discrete tissues, and the lack of observed differences among embryo populations could be due to the analysis of whole embryos rather than a discrete tissue, especially since we did measure significantly higher heart rates in embryos from parents collected from polluted environments. However, except for heart rate, other in vivo morphological measurements showed no consistent differences among polluted versus reference embryo populations. Thus, in a common garden environment, developmental gene expression may be well conserved and supersede population differences.
It is possible that an increase in the number of biological samples and hence greater statistical power could help discern changes among unexposed embryos of parents from polluted sites. We only analyzed four, unrelated embryos from each population. However, previous studies on adults using only one more individual (five versus four F. heteroclitus from these same populations) have found up to 40% of genes that differ due to treatment (population) (Fisher and Oleksiak 2007, Oleksiak 2008). Additionally, other studies using only four F. heteroclitus embryos have found up to 25% of genes that differ due to treatment (unpublished). Finally, the most interesting comparison of polluted versus reference populations is in fact a comparison of eight polluted embryos versus twelve reference individuals. Thus, while a greater number of biological replicates would make our study more powerful, the lack of observed differences is unlikely to be due solely to too small of a sample size of outbred individuals.
The stage we examined also could explain the lack of differences we found between polluted and reference populations. We only measured gene expression at one of the forty developmental stages of F. heteroclitus. Critical differences in gene expression between polluted and reference embryos may occur at an earlier or later stage. For instance, gene expression changes that cause increased heart rates in the embryos from polluted populations might occur when the heart first starts to beat at stage 25.
Other F. heteroclitus embryo studies implicate cardiac abnormalities as one of the hallmarks of embryotoxicity associated with the chemicals found at both Elizabeth River and Newark Bay Superfund sites (Prince and Cooper 1995, Ownby et al. 2002, Meyer et al. 2003). Notably, these studies all examined embryos that were treated with polluted sediments. Our studies used embryos from polluted parents, cultured in a clean environment, i.e. untreated embryos. Thus, a likely explanation for the lack of differences in gene expression between polluted and reference embryos is that most of the differences are induced by pollutant exposure, and without this exposure, there are few differences in developmental gene expression among embryos.
Conclusion
Natural F. heteroclitus populations, including both embryos and adults, provide an elegant model to explore the ways individuals and populations respond to challenges of their ever changing environment. With the exception of heart rates, our findings lead us to reject the hypothesis that the unexposed F. heteroclitus embryo populations differ based on their parental exposure to pollution, at least during stage 31 of development. However, these results open possibilities for further inquiry: because the analysis of the chosen endpoints may not reveal subtle, important changes in gene expression contributing the resistance phenotypes, a more targeted approach that includes a tissue-specific analysis of gene expression may expose important phenotypes among F. heteroclitus embryo populations associated with the mechanisms of sensitivity and resistance to pollution. Additionally, in a common garden environment canalization of developmental gene expression may mask population differences, and gene by environment interactions may be necessary to clarify differences among embryos from polluted and reference populations. Thus, without pollutant exposure, we found few differences in developmental gene expression among embryos from polluted and reference populations.
Acknowledgements
Funding for this work was received from NIH 5 RO1 ES011588, 2P42 ES010356, and 2 P42 ES007381. The authors thank Douglas L. Crawford for help with production of the arrays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributors:
Goran Bozinovic preformed fish collection and embryo culturing, mortality, heart rate and morphology analysis, mRNA isolation, array hybridization, and assisted in cDNA library construction and gene expression data analysis. Marjorie Oleksiak preformed fish collection, cDNA library construction and microarry printing, RNA-array hybridizations, and gene expression experimental design and data analysis. The article was co-written by the authors. Both authors have approved the final article.
References
- Adams SM, Lindmeier JB, Duvernell DD. Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Mol Ecol. 2006;15:1109–1123. doi: 10.1111/j.1365-294X.2006.02859.x. [DOI] [PubMed] [Google Scholar]
- Armstrong PB, Child JS. Stages of normal development of Fundulus heteroclitus. Biological Bulletin. 1965;128:143–168. [Google Scholar]
- Bickham JW, Sandhu S, Hebert PD, Chikhi L, Athwal R. Effects of chemical contaminants on genetic diversity in natural populations: implications for biomonitoring and ecotoxicology. Mutat Res. 2000;463:33–51. doi: 10.1016/s1383-5742(00)00004-1. [DOI] [PubMed] [Google Scholar]
- Bickham JW, Smolen MJ. Somatic and heritable effects of environmental genotoxins and the emergence of evolutionary toxicology. Environ Health Perspect. 1994;102 Suppl 12:25–28. doi: 10.1289/ehp.94102s1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gomez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, Maclatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the Premier Teleost Model in Environmental Biology: Opportunities for New Insights Using Genomics. Comp Biochem Physiol Part D Genomics Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin MA, Bickham JW. A population genetic analysis of the potential for a crude oil spill to induce heritable mutations and impact natural populations. Ecotoxicology. 1998;7:259–278. [Google Scholar]
- Dieter MP. Identification and quantification of pollutants that have the potential to affect evolutionary processes. Environ Health Perspect. 1993;101:278. doi: 10.1289/ehp.93101278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMichele L, Powers DA. LDH-B genotype-specific hatching times of Fundulus heteroclitus embryos. Nature. 1982;296:563–564. doi: 10.1038/296563a0. [DOI] [PubMed] [Google Scholar]
- DiMichele L, Westerman ME. Geographic variation in development rate between poopulations of the teleost Fundulus hetericlitus. Marine Biology. 1997;128:1–7. [Google Scholar]
- Elskus AA, Monosson E, McElroy AE, Stageman JJ, Woltering DS. Altered CYP1A expression in Fundulus heteroclitus adults and larvae: a sign of pollutant resistance? Aquatic Toxicology. 1999;45:99–113. [Google Scholar]
- Endler JA. Natural selection in the wild. Princeton, NJ: Princeton University Press; 1986. [Google Scholar]
- Fisher MA, Oleksiak MF. Convergence and divergence in gene expression among natural populations exposed to pollution. BMC Genomics. 2007;8:108. doi: 10.1186/1471-2164-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GA. Effects of encocrine disrupting chemicals on wildlife in Canada: Past, Present, and Future. Water Quality Research Journal of Canada. 2001;36:233–251. [Google Scholar]
- Hackstadt AJ, Hess AM. Filtering for increased power for microarray data analysis. BMC Bioinformatics. 2009;10:11. doi: 10.1186/1471-2105-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert P, Luiker M. Genetic effects of contaminant exposure - towards an assessment of impacts on animal populations. Science of the Total Environment. 1996;191:23–58. doi: 10.1016/0048-9697(96)05169-8. [DOI] [PubMed] [Google Scholar]
- Hofelt CS, Shea D. Accumulation of organochlorine pesticides and PCBs by semipermeable membrane devices and Mytilus edulis in New Bedford Harbor. Environmental science and technology. 1997;31:154–159. [Google Scholar]
- Hsiao SM, Greeley MSJ, Wallace RA. Reproductive cycling in female Fundulus heteroclitus. Biological Bulletin. 1994;186:271–284. doi: 10.2307/1542273. [DOI] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Kerr K, Churchill G. Experimental design for gene expression analysis. Biostatistics. 2001a;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Kerr M, Churchill G. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Long ER, Wolfe DA, Scott KJ, Thursby GB, Stern EA, Peven C, Schwartz T. Magnitude and Extent of Sediment Toxicity in the Hudson-Raritan Estuary. 1995 [Google Scholar]
- Lotrich VA. Summer home range and movements of Fundulus heteroclitus (Family: Cyprinonodontidae) in a tidal creek. Ecology. 1975;56:191–198. [Google Scholar]
- Luellen DR, Shea D. Calibration and field verification of semipermeable membrane devices for measuring polycyclic aromatic hydrocarbons in water. Environ. Sci. Technol. 2002;36:1791–1797. doi: 10.1021/es0113504. [DOI] [PubMed] [Google Scholar]
- McMillan AM, Bagley MJ, Jackson SA, Nacci DE. Genetic diversity and structure of an estuarine fish (Fundulus heteroclitus) indigenous to sites associated with a highly contaminated urban harbor. Ecotoxicology. 2006;15:539–548. doi: 10.1007/s10646-006-0090-4. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P450A (CYP1A)in Killifish (Fundulus heteroclitus): Heritability of Altered Expression and Relatioship to Survival in Contaminated Sediments. Toxicological Sciences. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Smith JD, Winstin GW, Di Giulio RT. Antioxidant defenses in killifish (Fundulus heteroclitus) exposed to contaminated sediments and model prooxidants: short-term and heritable responses. Aquatic Toxicology. 2003;65:377–395. doi: 10.1016/j.aquatox.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Mulvey M, Newman MC, Vogelbein WK, Unger MA, Ownby DR. Genetic structure and mtDNA diversity of Fundulus heteroclitus populations from polycyclic aromatic hydrocarbon-contaminated sites. Environ Toxicol Chem. 2003;22:671–677. [PubMed] [Google Scholar]
- Nacci DE, Coiro L, Champlin D, Jayaraman S, Mckinney R, Gleason TR, Munns J, Specker WRJL, Cooper KR. Adaptation of wild populations of the estuarine fish Fundulus heteroclitus to persistant environmental contaminants. Marine Biology. 1999;134:9–17. [Google Scholar]
- Oleksiak MF. Changes in gene expression due to chronic exposure to environmental pollutants. Aquat Toxicol. 2008;90:161–171. doi: 10.1016/j.aquatox.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Roach JL, Crawford DL. Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nature Genetics. 2005;37:67–72. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer JM. The Normal Stages of Fundulus heteroclitus. Anat. Rec. 1937;68:1–15. [Google Scholar]
- Ownby DR, Newman MC, Mulvey M, Vogelbein WK, Unger MA, Arzayus LF. Fish (Fundulus heteroclitus) populations with different exposure histories differ in tollerance of creosote-contaminated sediments. Enironmental Toxicology and Chemistry. 2002;21:1897–1902. [PubMed] [Google Scholar]
- Powers DA, Smith M, Gonzalez-Villasenor I, DiMichele L, Crawford DL, Bernardi G, Lauerman TA, editors. A multidisciplinary approach to the selectionsit/neutralist contraversy using the model teleost, Fundulus heteroclitus. New York, NY: Oxford University Press; 1993. [Google Scholar]
- Prince R, Cooper KR. Comparison of the effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on chemically impacted and nonimpacted subpopulations of Fundulus heteroclitus: II Metabolic Considerations. Environmental Toxicology and Chemistry. 1995;14:589–595. [Google Scholar]
- Roark SA, Nacci D, Coiro L, Champlin D, Guttman SI. Population genetic structure of a nonmigratory estuarine fish (Fundulus heteroclitus) across a strong gradient of polychlorinated biphenyl contamination. Environ Toxicol Chem. 2005;24:717–725. doi: 10.1897/03-687.1. [DOI] [PubMed] [Google Scholar]
- Scott CP, VanWye J, McDonald MD, Crawford DL. Technical analysis of cDNA microarrays. PLoS One. 2009a;4:e4486. doi: 10.1371/journal.pone.0004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CP, Williams DA, Crawford DL. The effect of genetic and environmental variation on metabolic gene expression. Mol Ecol. 2009b;18:2832–2843. doi: 10.1111/j.1365-294X.2009.04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman K, Wallace RA. Oogenesis in Fundulus heteroclitus. III. Vitellogenesis. Journal of Experimental Zoology. 1983;226:441–457. doi: 10.1002/jez.1402260315. [DOI] [PubMed] [Google Scholar]
- Smith GM, Weis SJ. Predator-prey relationships in mumischogs (Fundulus heteroclitus): Effects of living in a polluted environment. Journal of Expreimental Marine Biology and Ecology. 1997;209:75–87. [Google Scholar]
- Sweeney J, Deegan L, Gattitt R. Population size and site fidelity of Fundulus heteroclitus in a macrotidal saltmarsh creek. Biol. Bull. 1998;195:238–239. doi: 10.2307/1542858. [DOI] [PubMed] [Google Scholar]
- Vogelbein WK, Fournie JW, Van Veld PA, Huggett RJ. Hepatic neoplasms in the mummichog Fundulus heteroclitus from a creosote-contaminated site. Cancer Res. 1990;50:5978–5986. [PubMed] [Google Scholar]
- Weis JS, Smith GM, Zhou T. Altered predator/pray behavior in polluted environments: Implications for conservation. Environmental Biology of Fishes. Environmental Biology of Fishes. 1999;55:43–51. [Google Scholar]
- Weis JS, Weis P. Effects of environmental pollutants on early fish development. Reviews in Aquatic Sciences. 1989;1:45–73. [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Oleksiak MF. Signatures of selection in natural populations adapted to chronic pollution. BMC Evol Biol. 2008;8:282. doi: 10.1186/1471-2148-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Resistance to contaminants in North American fish populations. Mutat Res. 2004;552:73–100. doi: 10.1016/j.mrfmmm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wolfe DA, Long ER, Thursby GB. Sediment toxicity in the Hudson-Raritan Estuary: Distribution and correlations with chemical contamination. Estuaries. 1996;19:901–912. [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr K, Cui X, Churchill G. "MAANOVA: a software package for the analysis of spotted cDNA microarray experiments". The Analysis of Gene Expression Data: Methods and Software. 2003 [Google Scholar]