ABSTRACT
Antibiotics are a cost-effective tool for improving feed efficiency and preventing disease in agricultural animals, but the full scope of their collateral effects is not understood. Antibiotics have been shown to mediate gene transfer by inducing prophages in certain bacterial strains; therefore, one collateral effect could be prophage induction in the gut microbiome at large. Here we used metagenomics to evaluate the effect of two antibiotics in feed (carbadox and ASP250 [chlortetracycline, sulfamethazine, and penicillin]) on swine intestinal phage metagenomes (viromes). We also monitored the bacterial communities using 16S rRNA gene sequencing. ASP250, but not carbadox, caused significant population shifts in both the phage and bacterial communities. Antibiotic resistance genes, such as multidrug resistance efflux pumps, were identified in the viromes, but in-feed antibiotics caused no significant changes in their abundance. The abundance of phage integrase-encoding genes was significantly increased in the viromes of medicated swine over that in the viromes of nonmedicated swine, demonstrating the induction of prophages with antibiotic treatment. Phage-bacterium population dynamics were also examined. We observed a decrease in the relative abundance of Streptococcus bacteria (prey) when Streptococcus phages (predators) were abundant, supporting the “kill-the-winner” ecological model of population dynamics in the swine fecal microbiome. The data show that gut ecosystem dynamics are influenced by phages and that prophage induction is a collateral effect of in-feed antibiotics.
IMPORTANCE
This study advances our knowledge of the collateral effects of in-feed antibiotics at a time in which the widespread use of “growth-promoting” antibiotics in agriculture is under scrutiny. Using comparative metagenomics, we show that prophages are induced by in-feed antibiotics in swine fecal microbiomes and that antibiotic resistance genes were detected in most viromes. This suggests that in-feed antibiotics are contributing to phage-mediated gene transfer, potentially of antibiotic resistance genes, in the swine gut. Additionally, the so-called “kill-the-winner” model of phage-bacterium population dynamics has been shown in aquatic ecosystems but met with conflicting evidence in gut ecosystems. The data support the idea that swine fecal Streptococcus bacteria and their phages follow the kill-the-winner model. Understanding the role of phages in gut microbial ecology is an essential component of the antibiotic resistance problem and of developing potential mitigation strategies.
Introduction
The Infectious Diseases Society of America is among the organizations that have recommended that the U.S. government limit the use of antibiotics in agriculture (1, 2). The European Union has banned the use of all agricultural antibiotics that are used for growth promotion (3, 4). However, various factors, notably the cost-effectiveness of antibiotic use for performance benefits in modern, conventional agricultural practices (5), have forestalled similar measures in the United States. Partly driving the ongoing debate on the prudence of the widespread use of antibiotics to improve feed efficiency in agricultural animals (6, 7) is a growing body of research on the collateral effects of these antibiotics, ranging from an increased abundance of antibiotic resistance genes in pigs fed antibiotics (T. P. Looft, T. Johnson, H. K. Allen, D. O. Bayles, D. P. Alt, R. D. Stedtfeld, W.-J. Sul, T. M. Stedtfeld, B. Chai, S. A. Hashsham, J. M. Tiedje, and T. B. Stanton, submitted for publication) to the modulation of bacterial gene expression by subinhibitory concentrations of antibiotics (8). Another collateral effect could be the induction of gene transfer among bacteria.
Horizontal gene transfer is a mechanism by which bacteria exchange genetic material and is known to occur among gut bacteria (9). One type of horizontal gene transfer is mediated by phages. Some antibiotics are known to affect prophage-mediated gene transfer in certain bacteria in vitro. The virus-like gene transfer agent VSH-1 is induced by carbadox in the swine pathogen Brachyspira hyodysenteriae and transfers antibiotic resistance genes (10, 11). Also, beta-lactam antibiotics and fluoroquinolones induce prophages in Staphylococcus aureus, some of which package pathogenicity islands and therefore transfer virulence traits (12, 13). However, the in vivo effects of antibiotics on phages in the gut are unknown.
Phage diversity and function have been studied in water, soil, and animal-associated environments, but only a small fraction of phages have been characterized (14). Metagenomic analyses enable the study of phages without isolating them (15). The phage metagenome, or virome, is the sequenced assemblage of the total phages of a microbial community. Recently, phage metagenomic analyses have launched studies comparing phage ecology between environments. In such studies, phage diversity and functions can be elucidated despite a limited understanding of specific phages in a given community, such as the demonstrated increase in phage diversity in the lungs of cystic fibrosis patients (16).
Our goal was to examine the fecal viromes over time in swine that were fed the common antibiotics carbadox and ASP250. The viromes were compared to those of nonmedicated swine and to the corresponding bacterial communities. The data show that in-feed antibiotics induce prophages in the swine intestine and cause significant shifts in both phage and bacterial community structures. Additionally, analysis of the relative abundance of Firmicutes bacteria and phages, specifically the Streptococcus spp., unexpectedly revealed that the predator-prey population dynamics model called “kill-the-winner” might apply to the swine microbiome.
RESULTS
Diverse phages in swine feces.
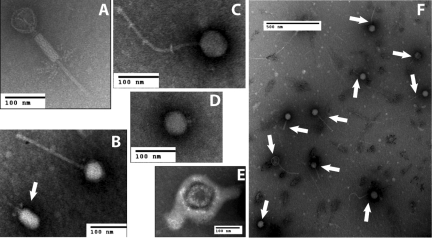
Phages in each sample were visualized by electron microscopy, revealing members of the Siphoviridae, Myoviridae, and Podoviridae phage families based on morphology (Fig. 1A to 1D). Several enveloped viruses were also seen (Fig. 1E). Frequently, many phages could be visualized in a field of view (Fig. 1F). No bacterial cells were seen by electron microscopy. Lack of bacterial cell contamination of phage preparations was further confirmed by the inability to amplify the 16S rRNA gene from virome samples.
FIG 1 .
Electron micrographs of virions isolated from swine feces. (A to D) Representative phages of the Myoviridae (A), Siphoviridae (B [no arrow] and C), and Podoviridae (B [arrow] and D) families. (E) An enveloped virus as seen in numerous fecal samples of young pigs. (F) Ten Siphoviridae (arrows) were visualized in a single field.
ASP250 causes shifts in phage membership.
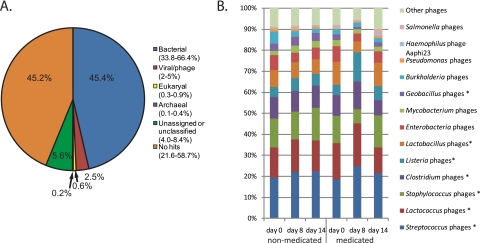
Viromes were analyzed by MG-RAST (17) and GAAS (18) to classify the sequences based on their putative origins. Out of nearly 1 million phage metagenomic sequence reads, 44% of the sequences (average across viromes) had no hits in the databases according to MG-RAST (Fig. 2A). Few virome sequences were solely assignable to phage or virus origins, while considerably more virome sequences were attributed to bacterial origins (Fig. 2A). GAAS was used to infer phage taxonomy based on the best match according to BLASTx. Only phages that occurred at ≥0.1% abundance in at least one virome were analyzed. Approximately 90% of assignable phages were attributed to phages of canonical gut bacteria, and the vast majority of assignments were to bacteria of the Firmicutes phylum (Fig. 2B; see also Fig. S1 in the supplemental material). ASP250 but not carbadox caused considerable shifts in the phage community compared to control and pre-ASP250-fed animals (R = 0.72, P < 0.10).
FIG 2 .
Community structure based on taxonomic inference of phages from swine feces. (A) Phage metagenomic sequence origins. “No hits” is the percentage of reads with no similar sequences in the database, and “unassigned or unclassified” is the percentage of reads with a database hit that has no associated taxonomic information. The values on the graph are the medians, and the values in parentheses are the ranges. (B) Genus-level phylogenetic origins of the phage-derived sequences from the ASP250 experiment. Phage taxa occurring at roughly <0.5% abundance were grouped as “Other phages.” Asterisks denote those of the Firmicutes phylum.
Phage community shifts are paralleled in the bacterial community.
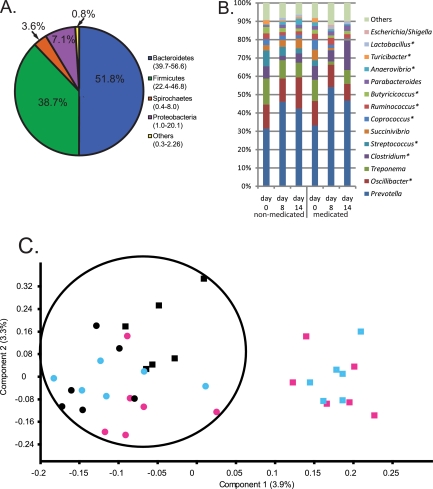
We compared phage diversity to bacterial diversity by amplifying and analyzing 16S rRNA gene sequences from the swine fecal samples. Sequencing the V1-to-V3 region of the 16S rRNA gene of fecal DNAs yielded 1,077,133 sequences from 86 bar-coded samples. All samples were dominated by four phyla: Bacteroidetes, Firmicutes, Spirochaetes, and Proteobacteria (Fig. 3A), and the identified genera are typical for a mammalian gut environment (Fig. 3B). Specific genera detected at lower levels (P < 0.01) in ASP250-treated animals than in corresponding nonmedicated animals (day 0 and nonmedicated control animals) were Coprococcus, Succinivibrio, Streptococcus, Treponema, and Turicibacter spp. (Fig. 3B; see also Table S1 in the supplemental material). Additionally, Escherichia sp. increased with in-feed ASP250, although the overall abundance of this genus remained relatively low (Fig. 3B).
FIG 3 .
Community structure based on taxonomic inference of bacteria (16S rRNA sequences) from swine feces. (A) Phylum-level assignments of assignable 16S rRNA gene sequences from swine feces, averaged across all 86 individual samples. The values on the graph are the medians among treatment groups, and the values in parentheses are the ranges. (B) Average percent abundance of genus-level assignments of 16S rRNA gene sequences from the feces of six swine fed ASP250 and the corresponding nonmedicated animals. Values were normalized to the total number of assignments within a sample. Taxa occurring at roughly <0.3% abundance were grouped as “Others.” Asterisks denote those of the Firmicutes phylum. See Table S1 in the supplemental material for the values of the means and the standard errors. (C) Principal component analysis of OTU-based bacterial 16S rRNA gene sequence abundances in individual pig samples (P < 0.01, R = 0.43). The percent variance accounted for by each component is in parentheses. An ellipse is drawn around the data sets of pigs that did not receive ASP250. Black, day 0 (just prior to treatment); blue, day 8; pink, day 14. Circles, nonmedicated pigs; squares, medicated pigs.
To visualize changes in bacterial diversity, a principal component analysis of operational taxonomic units (OTUs) was performed. The bacterial community shifted in animals medicated with ASP250 compared to that in nonmedicated animals (both ASP250 on day 0 and all corresponding control animals; P < 0.01, R = 0.43; Fig. 3C), mirroring the results shown for the phage communities. The shift was driven in part by total bacterial OTU diversity decreases in the day 14 ASP250-treated samples compared to the control animals (see Table S2 in the supplemental material). No significant changes in bacterial diversity were detected with in-feed carbadox treatment (Table S2).
Kill-the-winner population dynamics are displayed in the swine fecal microbiome.
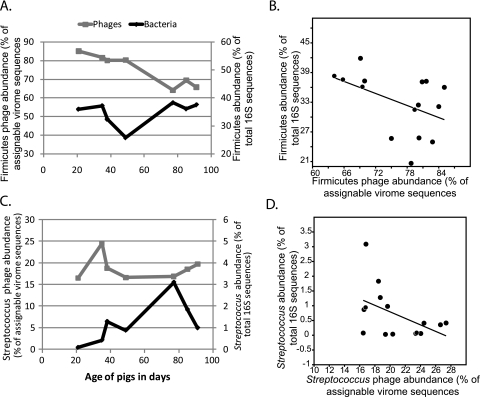
Predator-prey relationships between swine fecal phages and bacteria were examined. Firmicutes bacteria and their associated phages were analyzed because they made up a large proportion of the assignable virome data (Fig. 2B). Even at the phylum level, the dynamic relationship between phages and bacteria was apparent in the nonmedicated swine viromes over time (Fig. 4A). A regression analysis of all viromes revealed that when the Firmicutes phages were present in relatively high abundance, the abundance of Firmicutes bacteria tended to be lower (r2 = 0.21, P < 0.1; Fig. 4B). Of the Firmicutes, the Streptococcus genus was analyzed because its members were relatively abundant in both the phage and bacterial data sets. As with the Firmicutes, Streptococcus bacteria decreased as Streptococcus phages increased (r2 = 0.23, P < 0.1; Fig. 4C and 4D). Only three other genera were present in both the phage and bacterial relative abundance data, and they all showed the same trend to various degrees (Escherichia, r2 = 0.45; Lactobacillus, r2 = 0.13; Clostridium, r2 = 0.03). These data suggest that the kill-the-winner model of population dynamics applies to swine gut microbial communities.
FIG 4 .
Population dynamics of bacteria and phages in swine fecal microbiomes. (A and C) Abundances of Firmicutes (A) and Streptococcus (C) bacteria and phages in the nonmedicated swine are plotted against time. (B and D) Regression analyses of the abundances of Firmicutes (B) and Streptococcus (D) phages against the respective bacterial abundances in all treatments and time points (r2 = 0.21 and 0.23, respectively; P < 0.1 for both). In all figures, the bacterial abundances are pooled data from six animals.
Functional analysis of swine viromes reveals fitness genes.
Putative functions of coding sequences were collated by CAMERA (19) and analyzed. Clusters of orthologous groups (COGs) revealed an emphasis on DNA replication and transcription in the viromes (see Fig. S3 in the supplemental material). Multivariate analyses, in addition to statistical analyses of the COG assignments using ShotgunFunctionalizeR (20), revealed no significant patterns of COGs based on time or treatment.
We hypothesized that antibiotic resistance would be one type of bacterial fitness trait encoded by phages; therefore, we searched for antibiotic resistance functions in all viromes. Sequences encoding resistance to antibiotics and toxins were annotated by MG-RAST (see Table S3 in the supplemental material) (17), and further details were acquired by comparing the viromes to the antibiotic resistance gene database (ARDB [21]) (Table 1). According to the ARDB, most viromes harbored few antibiotic resistance genes (107 genes out of 1,036,084 total reads [0.01%]), and two viromes had no detectable resistance genes. Eight resistance genes occurred more than twice across all viromes, and most of these encoded efflux pumps (Table 1). Normalized resistance gene frequencies were analyzed in multivariate analyses but showed no discernable pattern based on time or treatment.
TABLE 1 .
Antibiotic resistance genes detected more than twice across all viromes, as annotated by the antibiotic resistance gene database (21)
| Mechanism of resistance |
Resistance gene |
Resistance conferred by gene product |
No. of hits to virome reads |
|---|---|---|---|
| Efflux pumps | |||
| ABC transporter system | bcrA | Bacitracin | 8 |
| RND family transporter | macB | Macrolides | 22 |
| MFS family transporter | mef(A) | Macrolides | 4 |
| Target evasion | |||
| Drug-insensitive dihydrofolate reductase | dfrAa | Trimethoprim | 7 |
| Ribosomal protection protein | tet(W) | Tetracycline | 10 |
| Modification of peptidylglycan biosynthetic pathway | vanb | Vancomycin | 26 |
| Enzymatic deactivation | |||
| NADPH-dependent oxidoreductase | tet37 | Tetracycline | 3 |
The following DfrA-encoding genes were detected: dfrA20, dfrA22, dfrA24, and dfrA26.
Portions (in parentheses) of the following vancomycin resistance pathways were detected in various viromes: VanA (vanHA, vanRA, vanYA); VanB (vanB, vanHB, vanRB, vanYB); VanC (vanRC); VanD (vanHD); VanE (vanRE, vanSE, vanTE); VanG (vanG, vanRG, vanTG, vanUG).
In-feed antibiotics induce prophages in the swine gut.
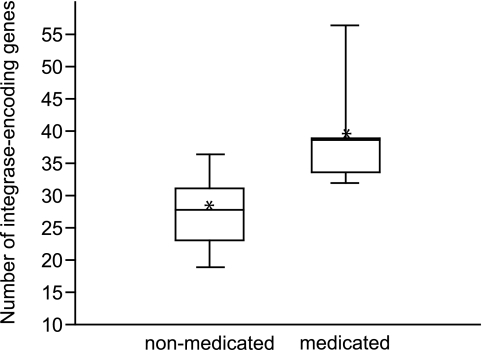
A potential indicator of the effect of antibiotics on prophages could be genes encoding phage integrases (S. Casjens, personal communication). Genes annotated as encoding an integrase were enumerated by MG-RAST and normalized by the total number of reads per virome. To gain statistical power, both carbadox- and ASP250-treated swine viromes were grouped as “medicated.” The viromes of medicated pigs (n = 5) harbored more integrase genes than did the viromes of nonmedicated pigs (n = 10, P < 0.01) (Fig. 5). This indicates that in-feed antibiotics induced prophages from gut bacteria, with a surge in the abundance of integrase-encoding genes in the ASP250-treated swine virome at day 8.
FIG 5 .
Box plot of integrase-encoding gene abundance in nonmedicated (n = 10) and medicated (n = 5) swine viromes (P < 0.01). Asterisks denote the means. The number of integrase-encoding genes was normalized by the total number of reads per virome.
DISCUSSION
This is the first report of the effect of antibiotics on total phage diversity in a microbial community. The results show that a collateral effect of antibiotic treatment is increased abundance of phage integrase-encoding genes, reflecting the induction of prophages from gut bacteria. Integrases are an appropriate marker for prophage induction because they are required for temperate phages transitioning from lysis to lysogeny (22). Integrases are also associated with pathogenicity islands, which are often mobilized by prophages (23). A greater abundance of integrases with antibiotic treatment, therefore, indicates that antibiotics are inducing phage-mediated bacterial lysis in the gut. Integrase abundance increased regardless of the type of in-feed antibiotic; further research is required to determine the specificity of perturbations that result in increased integrase abundance.
Additional consequences of in-feed antibiotic-mediated phage induction could be increased abundance of bacterial fitness or virulence genes, such as those encoding antibiotic resistance. Various homologues of antibiotic resistance genes were detected in the swine viromes at a frequency corresponding to approximately 1/50 of the frequency of antibiotic resistance genes in an Escherichia coli genome. Particularly in the context of no detection of 16S rRNA genes by PCR in the viromes, the apparent low number of resistance-encoding virome reads seems surprisingly frequent. Resistance genes were identified slightly more frequently in human fecal viromes (0.1% [24]). With selective pressure, any phage-transferred resistance genes could accelerate the evolution of resistance in the gut microbiome. Despite the potential relevance of the transduction of resistance genes in an antibiotic-containing environment, the swine viromes provided no evidence of a treatment effect. Increased sequencing depth may be required to detect differences among the viromes, such as the effect of generalized transduction on a virome. Additionally, transcriptomic analyses would demonstrate which phage genes have altered expression as a result of antibiotic treatment, revealing those genes important for fitness in an antibiotic environment.
Phages have been shown to play an important role in ecosystem dynamics (25), and one dynamic is the relationship between phages and bacteria. A widely investigated model for this relationship is called kill-the-winner (26). This model predicts that an increase in a given bacterial host population (winning prey) results in an increase in its phages (predators) and subsequent predation of the winner. Kill-the-winner population dynamics have been supported in marine ecosystems (27), but it is unclear if the model holds true for gut ecosystems. The extended sampling (seven viromes over time) of nonmedicated animals’ viromes presented a ripe data set for investigating phage-bacterial population dynamics. Overall, the nonmedicated swine viromes showed taxonomic and functional stability over time, as seen in aquatic microbiomes (27). Despite this apparent stability, examination of the relative abundances of Streptococcus phages compared to Streptococcus bacterial abundances over time revealed a dynamic process resembling kill-the-winner. Indeed, the swine viromes suggest that the kill-the-winner process might be detectable at the phylum level. This is consistent with other work that has shown kill-the-winner dynamics at the strain level in aquatic microbiomes (27) and horse feces (28). However, a fecal phage metagenomic study from pairs of twins and their mothers revealed little intravirome change across three sampling dates, and the authors refuted the model (29). Two major differences between the present study and the twin study are that we isolated phages from fresh (not frozen) feces and that we did not employ a DNA amplification step prior to sequencing. These protocol improvements were designed to reduce bias in analyses of phage diversity (30), enabling us to view population dynamics even in complex ecosystems such as the swine microbiome. The results tentatively support kill-the-winner dynamics in swine microbiomes, but more research is required to resolve the applicability of the kill-the-winner model across mammalian gut ecosystems.
Analysis of the in-feed ASP250 viromes suggests that there is an antibiotic effect on the relative abundance of fecal phages. The only component of ASP250 known to have an effect on phage lysis is penicillin. Subinhibitory concentrations of penicillin were shown to weaken Streptococcus spp. such that even so-called phage-resistant strains in mixed cultures were susceptible to phage lysis by exogenous phages (31). This could account for the significant decrease in Streptococcus spp. with ASP250, although it provides no evidence for the concomitant increase in Streptococcus phages. A related phenomenon is called phage-antibiotic synergy (PAS) and has been demonstrated with diverse phages of E. coli in the presence of subinhibitory concentrations of various cephalosporin-type beta-lactam antibiotics (32). The result of PAS is phage induction, and it is independent of an SOS response and dependent on a filamentation phenotype resulting from certain antibiotic treatments (32). Taken together, these data suggest that penicillin is the active component of ASP250 that is affecting the phage population, perhaps by numerous and complex mechanisms in the bacterial milieu.
Analysis of the structure of the bacterial communities shows that a small, important fraction of the data is driving the shift in diversity with ASP250 treatment. The decrease in the lactic acid bacterium (LAB) Streptococcus is particularly intriguing and agrees with the reported decrease in the abundance of LABs with certain oral antibiotic treatments (33–35). This decrease is often accompanied by an increase in Proteobacteria, specifically in Escherichia populations as shown here and elsewhere (33; Looft et al., submitted). In addition to the immediate effects on the microbiota, oral antibiotic treatment was shown to decrease the immune response in mice, even in distant locations such as the lungs (34, 35). The interaction of LABs with the gut mucosa is thought to be immunomodulatory (36), so perhaps there is a connection between the abundance of LABs and immune function. Interestingly, a recent study evaluating in-feed fumaric and formic acids showed a trend towards increased abundance of coliforms and decreased lactobacilli in plate counts (37), mirroring the effect of ASP250 on the microbiota. Fumaric acid has been demonstrated to improve weight gain despite no changes in available energy in the gut (38). Furthermore, in a study of irritable bowel syndrome, subjects with a higher body mass index than that of normal subjects had fewer lactobacilli (a type of LAB) (39). Considering that one mechanism of antibiotic-mediated growth promotion could be suppressed immune response due to decreased bacterial load (40), a decrease in immunomodulatory LABs might also decrease the energy spent on immunity and allow for increased feed efficiency.
The fecal bacterial diversity in the current study supports what has been shown previously: the swine gut is dominated by Firmicutes (~30%), Bacteroidetes (~50%), and Proteobacteria (~10%). However, the proportion of assignable phage sequences does not mirror this distribution, with nearly 80% of reads called from phages of bacteria of the Firmicutes phylum. Their inflation in our data set could result from the overrepresentation of phages of Firmicutes in public databases compared to those of Bacteroidetes, perhaps because of increased research interest due to their potential biotechnological applications (41). Additionally, phage sequences are simply lacking in the databases compared to bacterial sequences, limiting the pool of potential homologues for the swine viromes.
A relatively large proportion of assignable virome sequences were of bacterial origin despite no detectable bacterial contamination of the viromes. Phages harbor more bacterial genes than previously appreciated (42), making it reasonable that the assignable sequences of the swine viromes have 46% bacterial genes even in the absence of bacterial contamination. Also, generalized transducing phages package host bacterial DNA, contributing an unknown proportion to the counted bacterial genes.
No statistically significant effect of carbadox was detected on swine fecal phages or bacteria. Different sampling intervals and minor protocol improvements between the carbadox and ASP250 experiments (see Materials and Methods) may have affected the results. Our results suggest that 1 week following the commencement of in-feed antibiotics is an appropriate time to detect changes in the fecal microbiome.
This study provides evidence that a collateral effect of some in-feed antibiotics, such as ASP250, is the induction of prophages. Additionally, antibiotic resistance genes were detected in the phage metagenomes. Further work is required to determine the implications of prophage induction on the transfer of antibiotic resistance genes. Surprisingly, the data also support the kill-the-winner model for phage-bacterium population dynamics. Taken together, the data underscore the importance of phages in complex microbial communities.
MATERIALS AND METHODS
Swine.
Three rooms that would house pigs were decontaminated prior to the beginning of the study. All animals were managed in accordance with National Animal Disease Center Animal Care and Use Committee guidelines. Three pregnant sows farrowed on site, and piglets were weaned after 14 days. Weaned pigs were divided into three groups with approximately equal representation of littermates and gender, with two groups of six pigs housed in their own clean rooms and the remaining pigs housed in a third room (see Fig. S2 in the supplemental material). All pigs were fed the same diet (TechStart 17-25; Kent Feeds, Muscatine, IA) for 1 week after weaning and until the start of their respective study, at which point six control pigs continued to receive TechStart while three groups of six experimental pigs received one of the following in-feed supplements: subtherapeutic carbadox, 10 g/ton; therapeutic carbadox, 50 g/ton; or ASP250 (chlortetracycline, 100 g/ton; sulfamethazine, 100 g/ton; penicillin, 50 g/ton). For a 4.5-kg pig, this equaled the following concentrations of antibiotic per gram of pig per day: 8.2 µg therapeutic carbadox; 1.6 µg subtherapeutic carbadox; and 16.4 µg chlortetracycline, 16.4 µg sulfamethazine, and 8.2 µg penicillin (ASP250). Note that these concentrations decreased as the pigs increased in size. Freshly voided fecal samples were collected from control and medicated animals just before treatment (day 0) and 3, 8, 14, or 28 days after continued treatment (Fig. S2). Feces were transported on ice from the barn to the lab and were immediately processed for phage isolation and stored at −20°C for bulk DNA extraction.
Phage isolation.
The following protocol was adapted from previous reports (43–45). Approximately equal amounts of feces from each of the six animals in a treatment group were pooled to roughly 10 g. Feces were pooled because of the low biomass of phages and to avoid biasing the samples by amplifying the DNA in a later step (30). Pooled feces were blended in 50 ml SM buffer (8 mM MgSO4, 100 mM NaCl, 50 mM Tris-Cl, and 0.002% [wt/vol] gelatin) in a Waring blender (Torrington, CT) for 30 s. Fecal slurries were poured through a Nitex mesh (~118-µm pore size; Wildlife Supply, Yulee, FL) into a sterile centrifuge bottle. Samples were centrifuged three times at 4°C, first at 3,000 × g for 10 min and then twice at 10,000 × g for 10 min. The supernatants were carefully transferred to sterile centrifuge bottles between spins or to 50-ml Falcon tubes following the final spin. CsCl was added to the final supernatant to a density of 1.15 g/ml.
Meanwhile, two CsCl gradients were prepared per fecal sample (44, 45). Gradients were ultracentrifuged at 37,946 × g for 2 h at 4°C in a Beckman SW28 rotor. A 20-gauge needle on a 1-ml syringe was used to draw off 1 ml containing concentrated virions from the interface between the 1.35- and 1.5-g/ml layers. To improve DNA yield, the needle was inserted in the middle of the 1.5-g/ml layer and the volume of virions extracted was increased to 5 ml for the six samples taken during the ASP250 experiment. Because of a brown, stringy substance suspended vertically through the gradient, all virions were gently filtered through a 0.45-µm syringe filter into an Ultracel 3K regenerated cellulose concentrator (Millipore, Billerica, MA). Samples were gently centrifuged (<3,000 × g) until the sample volume was ~1 ml.
TEM visualization of phage particles.
An aliquot of virions was washed and concentrated for transmission electron microscopy (TEM) visualization. Briefly, 50 µl of virion-containing CsCl was applied to a Microcon YM-100 filter tube (Millipore) and washed twice with 500 µl SM buffer. Virions were resuspended in 10 µl SM buffer and stored at 4°C until TEM. On the day of TEM, 10 µl of virions was mixed with 10 µl of fresh 2% phosphotungstic acid (pH 7.0) and incubated at room temperature for 3 minutes. A Formvar- and carbon-coated 200-mesh copper grid (Electron Microscopy Sciences, Hatfield, PA) was introduced into this mixture for 1 min. The excess fluid was wicked away with filter paper. Grids were viewed on a Tecnai G2 Biotwin transmission electron microscope (FEI, Hillsboro, OR).
Phage DNA isolation.
Phage DNAs were isolated as described previously (45). Briefly, intact virions were treated with DNase to eliminate free DNA. Following DNase inactivation, virions were lysed with formamide and the DNA was precipitated. DNA was extracted by sequential SDS, cetyltrimethylammonium bromide (CTAB), and chloroform treatments.
Metagenomic sequencing and analysis.
Individual preparations of phage DNA were quantified using Quant-iT PicoGreen (Invitrogen, Carlsbad, CA). Five hundred nanograms of DNA from each preparation was used in the Rapid Library Preparation method (454 Life Sciences, Branford, CT). Libraries were created with Roche multiplex identifier (MID)-labeled adaptors. Individual libraries were pooled into one of two groups in an equimolar fashion. Pooled preparations were used to prepare DNA beads for sequencing using a two-region picotiter plate on a Roche GS-FLX instrument using Titanium chemistry (454 Life Sciences).
The metagenome sequences were processed with the 454 Replicate Filter, extract_replicates.py script, to remove artificially replicated sequences (46). Sequences in a cluster were removed as artificial replicates if the first three bases were identical and there was greater than 90% identity over the length of the shortest sequence in a cluster (see Table S3 in the supplemental material). Dereplicated reads were uploaded to CAMERA (https://portal.camera.calit2.net [19]), and the RAMMCAP pipeline sorted the reads for encoded functions, including separation by clusters of orthologous groups (COGs). Domain taxonomy and integrases were determined by MG-RAST (http://metagenomics.anl.gov/ [17]). The dereplicated virome reads were analyzed with GAAS to obtain estimates of the viral genome sizes as well as confidence intervals of the estimates (18). PAST (47) was used to perform one-way ANOSIM and principal component analyses on the normalized (percentage of the total number of assigned reads per virome) phage relative abundance data from GAAS. ShotgunFunctionalizeR was used to make statistical comparisons of COG assignments among viromes (20). To draw statistical conclusions, the following cutoffs were indicated: P < 0.1, trend; P < 0.01, significance; R = 0 to 0.3, slight correlation; R = 0.3 to 0.5, medium correlation.
16S rRNA gene sequence analysis.
16S rRNA gene sequences were amplified from the fecal samples of individual pigs. PCR amplification of the V1-to-V3 region of bacterial 16S rRNA genes was carried out with the conserved primers 8F (5′-AGAGTTTGATCCTGGCTCAG [48]) and 518R (5′-ATTACCGCGGCTGCTGG [49]) with sequence tags (bar codes) and sequencing primers incorporated into each PCR primer (see Table S4 in the supplemental material). PCR mixtures contained 200 µM (each) deoxyribonucleotide triphosphate, 2.0 µM (each) primer, 2.0 U Ampligold Taq polymerase (Applied Biosystems, Foster City, CA), 2.5 mM MgCl2, 50 ng template DNA, Ampligold Taq buffer (Applied Biosystems), and water to 50 µl. PCRs were performed in a PTC-225 thermal cycler (MJ Research, Watertown, MA) with the following protocol: 3 min at 95°C; 21 cycles of 1 min at 95°C, 30 s at 56°C, and 45 s at 72°C; and a final elongation step for 3 min at 72°C. PCR for the 16S rRNA gene was also performed on purified phage metagenomic DNAs, but the number of cycles was increased to 40. PCR products were separated by gel electrophoresis and purified using the MinElute kit (Qiagen Inc., Valencia, CA). They were then sequenced on a 454 Genome Sequencer FLX, using the manufacturer’s protocol for Titanium chemistry (Roche Diagnostics, Branford, CT). Data were processed per manufacturer’s protocols, and AmpliconNoise (50) was used to reduce sequence artifacts produced during PCR and sequencing.
Phylotype analysis.
After binning the samples by bar code, phylogenetic analysis and taxonomic assignments of the 16S rRNA gene sequences were made using the Ribosomal Database Project (RDP) web tools (51). Additional phylotype comparisons and hypothesis testing were performed with the software package mothur (52). OTU abundances (97% similarity) were normalized to the total number of OTUs per sample, and these data were subjected to principal component analyses in mothur (52). PAST (47) was used to plot the data, allowing for the visualization of the relationships between samples. The estimated total diversity of operational taxonomic units was calculated with Catchall (53). Regression analyses were performed in Excel (P < 0.1, trend; P < 0.01, significance).
Nucleotide sequence accession numbers.
All sequences (phage metagenomes and 16S rRNA gene sequences) were deposited in NCBI (BioProject PRJNA72355; Sequence Read Archive accession number SRA045429) and are additionally available through CAMERA and MG-RAST.
SUPPLEMENTAL MATERIAL
GAAS (18) was used to infer phylogeny of the phage-derived sequences from the subtherapeutic (A) and therapeutic (B) carbadox experiments. Phage taxa occurring at roughly <0.5% abundance are grouped as “Other phages.” Asterisks denote those of the Firmicutes phylum. Note that because of the experimental design, nonmedicated day 14 in panel A is the same virome as nonmedicated day 0 in panel B. Download Figure S1, PDF file, 0.4 MB.
Schematic of swine fecal phage metagenomic study. Phages were isolated from fecal samples that were pooled from 6 pigs at each sampling point per treatment. Bacterial 16S rRNA genes were amplified and sequenced from individual pigs at each sampling point. Download Figure S2, PDF file, 0.1 MB.
Clusters of orthologous groups (COGs) identified by CAMERA in assignable virome reads. Download Figure S3, PDF file, 0.5 MB.
Mean and standard error (SE) of the most abundant bacteria in the feces of swine fed ASP250 and their nonmedicated counterparts.
Estimated operational taxonomic unit (OTU) diversity (± standard error) of 16S rRNA gene sequences in the ASP250 experiment, averaged within each sampling date and treatment.
Summary of swine fecal phage metagenomic (virome) data.
Bar codes used in this study for parallel 16S rRNA gene sequencing.
ACKNOWLEDGMENTS
We thank Judi Stasko for electron microscopy; Lea Ann Hobbs, Deb Lebo, and the NADC animal caretakers for technical support; and Tom Casey, Sherwood Casjens, and Dion Antonopoulos for helpful discussions and comments on the manuscript.
This work was supported by the Agricultural Research Service.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
Citation Allen HK, et al. 2011. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2(6):00260-11. doi:10.1128/mBio.00260-11.
REFERENCES
- 1. Infectious Diseases Society of America 14 July 2010. The Infectious Diseases Society of America’s (IDSDA) statement on antibiotic resistance: promoting judicious use of medically important antibiotics in animal agriculture. Testimony before the House Committee on Energy and Commerce Subcommittee on Health. House Committee on Energy and Commerce, Washington, DC: http://democrats.energycommerce.house.gov/documents/20100714/Johnson.Testimony.07.14.2010.pdf [Google Scholar]
- 2. Center for Veterinary Medicine, Food and Drug Administration, US Department of Health and Human Services 2010. The judicious use of medically important antimicrobial drugs in food-producing animals. Draft guidance no. 209. Center for Veterinary Medicine, Food and Drug Administration, US Department of Health and Human Services, Rockville, MD [Google Scholar]
- 3. Aarestrup FM, et al. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. US Government Accountability Office 2011. Antibiotic resistance: agencies have made limited progress addressing antibiotic use in animals. Report no. GAO-11-801. US Government Accountability Office, Washington, DC. [Google Scholar]
- 5. Cromwell GL. 2002. Why and how antibiotics are used in swine production. Anim. Biotechnol. 13:7–27 [DOI] [PubMed] [Google Scholar]
- 6. Levy SB, O’Brien TF, Alliance for the Prudent Use of Antibiotics 2005. Global antimicrobial resistance alerts and implications. Clin. Infect. Dis. 41 (Suppl. 4):S219–S220 [DOI] [PubMed] [Google Scholar]
- 7. Lipsitch M, Singer RS, Levin BR. 2002. Antibiotics in agriculture: when is it time to close the barn door? Proc. Natl. Acad. Sci. U. S. A. 99:5752–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445–453 [DOI] [PubMed] [Google Scholar]
- 9. Salyers AA, Gupta A, Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412–416 [DOI] [PubMed] [Google Scholar]
- 10. Matson EG, Thompson MG, Humphrey SB, Zuerner RL, Stanton TB. 2005. Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J. Bacteriol. 187:5885–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stanton TB, Humphrey SB, Sharma VK, Zuerner RL. 2008. Collateral effects of antibiotics: carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl. Environ. Microbiol. 74:2950–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maiques E, et al. 2006. Beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188:2726–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ubeda C, et al. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56:836–844 [DOI] [PubMed] [Google Scholar]
- 14. Clokie MR, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage 1:31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willner D, et al. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. 2010. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb. Protoc. 2010 (1):pdb.prot5368 [DOI] [PubMed] [Google Scholar]
- 18. Angly FE, et al. 2009. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 5:e1000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. 2007. CAMERA: a community resource for metagenomics. PLoS Biol. 5:e75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kristiansson E, Hugenholtz P, Dalevi D. 2009. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics 25:2737–2738 [DOI] [PubMed] [Google Scholar]
- 21. Liu B, Pop M. 2009. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 37:D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groth AC, Calos MP. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667–678 [DOI] [PubMed] [Google Scholar]
- 23. Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 24. Minot S, et al. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616-1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohwer F, Prangishvili D, Lindell D. 2009. Roles of viruses in the environment. Environ. Microbiol. 11:2771–2774 [DOI] [PubMed] [Google Scholar]
- 26. Rodriguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828–836 [DOI] [PubMed] [Google Scholar]
- 27. Rodriguez-Brito B, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739–751 [DOI] [PubMed] [Google Scholar]
- 28. Golomidova A, Kulikov E, Isaeva A, Manykin A, Letarov A. 2007. The diversity of coliphages and coliforms in horse feces reveals a complex pattern of ecological interactions. Appl. Environ. Microbiol. 73:5975–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reyes A, et al. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim KH, Bae JW. 2011. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77:7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verhue WM. 1978. Interaction of bacteriophage infection and low penicillin concentrations on the performance of yogurt cultures. Appl. Environ. Microbiol. 35:1145–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Comeau AM, Tétart F, Trojet SN, Prère MF, Krisch HM. 2007. Phage-antibiotic synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2:e799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antonopoulos DA, et al. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hill DA, et al. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ichinohe T, et al. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A. 108:5354–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perdigon G, Alvarez S, Rachid M, Agüero G, Gobbato N. 1995. Immune system stimulation by probiotics. J. Dairy Sci. 78:1597–1606 [DOI] [PubMed] [Google Scholar]
- 37. Franco LD, Fondevila M, Lobera MB, Castrillo C. 2005. Effect of combinations of organic acids in weaned pig diets on microbial species of digestive tract contents and their response on digestibility. J. Anim. Physiol. Anim. Nutr. (Berl.) 89:88–93 [DOI] [PubMed] [Google Scholar]
- 38. Radecki SV, Juhl MR, Miller ER. 1988. Fumaric and citric acids as feed additives in starter pig diets: effect on performance and nutrient balance. J. Anim. Sci. 66:2598–2605 [DOI] [PubMed] [Google Scholar]
- 39. Malinen E, et al. 2010. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 16:4532–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dibner JJ, Richards JD. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634–643 [DOI] [PubMed] [Google Scholar]
- 41. Klumpp J, Lavigne R, Loessner MJ, Ackermann HW. 2010. The SPO1-related bacteriophages. Arch. Virol. 155:1547–1561 [DOI] [PubMed] [Google Scholar]
- 42. Sharon I, et al. 2011. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 5:1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Breitbart M, et al. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367–373 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 1, p 2.47–2.50 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4:470–483 [DOI] [PubMed] [Google Scholar]
- 46. Gomez-Alvarez V, Teal TK, Schmidt TM. 2009. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314–1317 [DOI] [PubMed] [Google Scholar]
- 47. Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9 [Google Scholar]
- 48. Wilmotte A, Van der Auwera G, De Wachter R. 1993. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF”) strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96–100 [DOI] [PubMed] [Google Scholar]
- 49. Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 1991. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinform. 12:38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bunge J. 2011. Estimating the number of species with Catchall. Pac. Symp. Biocomput. 11:121–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GAAS (18) was used to infer phylogeny of the phage-derived sequences from the subtherapeutic (A) and therapeutic (B) carbadox experiments. Phage taxa occurring at roughly <0.5% abundance are grouped as “Other phages.” Asterisks denote those of the Firmicutes phylum. Note that because of the experimental design, nonmedicated day 14 in panel A is the same virome as nonmedicated day 0 in panel B. Download Figure S1, PDF file, 0.4 MB.
Schematic of swine fecal phage metagenomic study. Phages were isolated from fecal samples that were pooled from 6 pigs at each sampling point per treatment. Bacterial 16S rRNA genes were amplified and sequenced from individual pigs at each sampling point. Download Figure S2, PDF file, 0.1 MB.
Clusters of orthologous groups (COGs) identified by CAMERA in assignable virome reads. Download Figure S3, PDF file, 0.5 MB.
Mean and standard error (SE) of the most abundant bacteria in the feces of swine fed ASP250 and their nonmedicated counterparts.
Estimated operational taxonomic unit (OTU) diversity (± standard error) of 16S rRNA gene sequences in the ASP250 experiment, averaged within each sampling date and treatment.
Summary of swine fecal phage metagenomic (virome) data.
Bar codes used in this study for parallel 16S rRNA gene sequencing.