Abstract
The mating efficiency of 50 Aspergillus fumigatus isolates from both clinical and environmental sources was analyzed. Forty isolates completed the sexual cycle in 4 weeks with variable levels of fertility designated high, medium, or low. Two opposite-mating-type strains exhibiting the highest fertility, AFB62 (MAT1-1), isolated from a case of invasive aspergillosis, and AFIR928 (MAT1-2), isolated from the environment, were chosen as the supermater pair. Single cleistothecia obtained from a cross of the two strains harbored a minimum of 1 × 104 ascospores. The viability of ascospores increased with the age of the fruiting body, 17% at 4 weeks and reaching 95% at 20 weeks. AFB62 and AFIR928 were equally virulent in two different murine models, despite differences in their sources. High recombination frequencies were observed when the closely linked genes alb1 (AFUA_2G17600) and abr2 (AFUA_2G17530) were used as genetic markers. Comparative genome hybridization analyses revealed that only 86 genes (ca. 0.86% of the genome) are significantly diverged between AFB62 and AFIR928. The high fertility in a relatively short period, combined with a high degree of virulence and a high recombination frequency, demonstrates that the mating pair AFB62 and AFIR928 provides an excellent tool for genetic studies of A. fumigatus.
Importance Aspergillus fumigatus is a heterothallic fungal pathogen that causes life-threatening infections in immunocompromised hosts. Although heterothallism facilitates genetic study via recombinational analysis, previous work showed that a 6-month incubation period is required for the completion of sexual reproduction in this species. Such a long incubation period impedes progress in genetic research. To discover a highly fertile (supermater) pair that can complete the sexual cycle in a considerably shorter period, we screened 50 strains collected from various geographic regions for mating efficiency. We identified a highly virulent pair of supermaters that can be an invaluable tool for genetic study.
Importance
Aspergillus fumigatus is a heterothallic fungal pathogen that causes life-threatening infections in immunocompromised hosts. Although heterothallism facilitates genetic study via recombinational analysis, previous work showed that a 6-month incubation period is required for the completion of sexual reproduction in this species. Such a long incubation period impedes progress in genetic research. To discover a highly fertile (supermater) pair that can complete the sexual cycle in a considerably shorter period, we screened 50 strains collected from various geographic regions for mating efficiency. We identified a highly virulent pair of supermaters that can be an invaluable tool for genetic study.
Introduction
Aspergillus fumigatus is the primary cause of invasive aspergillosis in immunocompromised patients worldwide (1–3). The species had been known to reproduce only asexually until the recent discovery of its heterothallic pattern of sexual reproduction (4). Although the heterothallic nature of the fungus was evident when the MAT1-1 (MAT-1) and MAT1-2 (MAT-2) idiomorph sequences were identified in the A. fumigatus genome (5, 6), discovery of its sexual state required further experimental work. A. fumigatus fails to produce a sexual state under conventional environmental conditions conducive to sexual reproduction in most other teleomorphic Aspergillus species (7). However, in 2009, O’Gorman et al. reported success in inducing sexual reproduction between strains of opposite mating types isolated from air samples in Dublin, Ireland, and described the teleomorph as Neosartorya fumigata (4). This was consistent with phylogenetic evidence grouping A. fumigatus with species such as Neosartorya fischeri, which has an anamorph indistinguishable from A. fumigatus (8, 9). It was surprising to find that N. fumigata requires a very particular set of environmental conditions for sexual reproduction. Unlike other species, such as N. fischeri, Neosartorya fennelliae, Neosartorya udagawae, or Neosartorya nishimurae, A. fumigatus produced the sexual state only on oatmeal agar among many types of media tested (4, 10). Furthermore, mating culture plates had to be sealed with Parafilm and incubated at 30°C in the dark for 6 months before sexual reproduction was completed (4). Given the requirement of such unusually specific environmental conditions, it is not surprising that attempts to discover the sexual state of A. fumigatus had failed for decades (10).
The discovery of heterothallism in A. fumigatus evoked much excitement, since it could provide an invaluable tool for genetic analysis of this important human pathogen. However, a major hurdle impeding routine genetic studies has been the apparently low rate of fertility coupled with the unusually long period of incubation required to form viable meiotic spores. There is, therefore, an urgent need for a pair of sexually compatible virulent strains that are highly fertile and can complete the sexual cycle in a much shorter time.
In this paper, we describe the mating competency of 50 strains originating from five geographic regions (the United States, Hong Kong, India, England, and Ireland) and the discovery of highly fertile strains that can complete the sexual cycle in only 4 weeks. Three classes of fertility, high, medium, and low, were identified among the 50 strains. In this paper, MAT-1 and MAT-2 are used in place of MAT1-1 and MAT1-2, respectively. A pair of virulent MAT-1 and MAT-2 strains that produced abundant cleistothecia and viable recombinant ascospores in this time period were selected and designated “supermaters.”
RESULTS
Frequency of MAT-1 and MAT-2 mating types.
The mating types of 7 of the 50 strains of A. fumigatus used in this study have been previously identified (4, 5). The mating types of the remaining 43 strains (41 from various locations in the United States, 1 from India, and 1 from Hong Kong) were determined by PCR amplification of the MAT-1 and MAT-2 genes from their genomes. The MAT-1 and MAT-2 strains were in a 1:1 ratio, and each group contained 22 clinical and three environmental isolates (Tables 1 and 2). This ratio of MAT-1 to MAT-2 strains agreed with previous studies (4, 6).
TABLE 1 .
Results of crosses between MAT-1 strains and supermater strain AF928 (MAT-2)
| Fertility typea and isolate |
M13b | Source | Origin |
|---|---|---|---|
| HF | |||
| AFB62 | T2 | Clinical | United States |
| AFN2 | T2 | Clinical | United States |
| AFN9 | T2 | Clinical | United States |
| AFN16 | T1 | Clinical | United States |
| AFN20 | T1 | Clinical | United States |
| AFS2 | T1 | Clinical | United States |
| AFS4 | T2 | Clinical | United States |
| AFS9 | T1 | Clinical | United States |
| B-5233 | T1 | Clinical | United States |
| MF | |||
| AFN8 | T1 | Clinical | United States |
| AFN22 | T2 | Clinical | United States |
| AFN26 | T1 | Clinical | United States |
| AFN27 | T1 | Clinical | United States |
| AFS3 | T1 | Clinical | United States |
| AFS6 | T2 | Clinical | United States |
| AFS10 | T1 | Clinical | United States |
| AFIRB2 | T1 | Environmental | Ireland |
| AFIR974 | T1 | Environmental | Ireland |
| AFIR931 | T1 | Environmental | Ireland |
| LF | |||
| AFB65 | T2 | Clinical | United States |
| AFN10 | T1 | Clinical | United States |
| AFN11 | T1 | Clinical | United States |
| AFN15 | T2 | Clinical | United States |
| AFN17 | T1 | Clinical | United States |
| AFN29 | T2 | Clinical | United States |
Fertility of the crosses was assigned based on the number of cleistothecia per junction zone. HF, >100 cleistothecia with ascospores; MF, <100 cleistothecia with ascospores; LF, 0 to 20 cleistothecia without ascospores.
Molecular strain typing was done by PCR fingerprinting using microsatellite-specific primer M13. Two groups, T1 and T2, were identified.
TABLE 2 .
Results of crosses between MAT-2 strains and supermater strain AFB62 (MAT-1)
| Fertility typea and isolate |
M13b | Source | Origin |
|---|---|---|---|
| HF | |||
| AFIR928 | T2 | Environmental | Ireland |
| AFB54 | T1 | Clinical | United States |
| AFB61 | T2 | Clinical | United States |
| AFB63 | T1 | Clinical | United States |
| AFN13 | T2 | Clinical | United States |
| AFN18 | T2 | Clinical | United States |
| AFN21 | T1 | Clinical | United States |
| AFS8 | T1 | Clinical | United States |
| MF | |||
| AF293 | T2 | Clinical | England |
| AFB42 | T1 | Clinical | United States |
| AFB43 | T1 | Clinical | India |
| AFB56 | T1 | Clinical | Hong Kong |
| AFB57 | T1 | Clinical | United States |
| AFB58 | T1 | Clinical | United States |
| AFB59 | T1 | Clinical | United States |
| AFB60 | T1 | Clinical | United States |
| AFB64 | T2 | Clinical | United States |
| AFB68 | T1 | Clinical | United States |
| AFN12 | T2 | Clinical | United States |
| AFN28 | T2 | Clinical | United States |
| AFS5 | T1 | Clinical | United States |
| LF | |||
| AFIR964 | T1 | Environmental | Ireland |
| AFN6 | T2 | Clinical | United States |
| AFS1 | T1 | Clinical | United States |
| AFIRB3 | T1 | Environmental | Ireland |
Fertility of the crosses was assigned based on the number of cleistothecia per junction zone. HF, >100 cleistothecia with ascospores; MF, <100 cleistothecia with ascospores; LF, 0 to 20 cleistothecia without ascospores.
Molecular strain typing was done by PCR fingerprinting using microsatellite-specific primer M13. Two groups, T1 and T2, were identified.
Fertility.
We first selected AFIR928, an environmental isolate from Ireland, as a MAT-2 tester strain to evaluate the fertility of 25 MAT-1 isolates. This was based on a previous report that the AFIR928 strain produced abundant cleistothecia upon crossing with MAT-1 Irish strains (4). The initial criterion for evaluating the fertility of each strain was the duration required for completion of the sexual cycle, as defined by the formation of cleistothecia containing mature ascospores. No signs of cleistothecial formation were observed in the first 2 weeks of incubation in any crosses between AFIR928 and the MAT-1 strains. At the third week, however, cleistothecia became visible in some of the crosses. At 4 weeks, the junction zones between AFIR928 and MAT-1 strains displayed various degrees of cleistothecial formation. Fertility was then further categorized according to the numbers of cleistothecia produced. In 9 (36%) crosses, the junction zone (approximately 4.5 cm) was densely populated with cleistothecia harboring mostly free ascospores. A representative example of such a junction zone is shown in Fig. 1A and D. Because of the extremely high number of cleistothecia (>100 per junction zone), these strains were designated the high-fertility (HF) type (Table 1). Significantly lower numbers of cleistothecia (<100 per junction zone) with ascospores were observed in 10 (40%) other crosses, and these strains were designated the medium-fertility (MF) type (Fig. 1B and E; Table 1). The remaining 6 (24%) crosses were considered to be of the low-fertility (LF) type. LF strains produced limited numbers of cleistothecia (0 to 20 per junction zone) with no apparent ascospores in 4 weeks (Fig. 1C and F; Table 1), and their ability to form cleistothecia was often unreproducible. The distribution of the three fertility types (HF, MF, and LF) among the 25 MAT-1 strains assayed did not significantly deviate from equal frequency (P= 0.6, χ2 test). Since over 50% of the MAT-1 strains completed the sexual cycle in 4 weeks, this incubation period was chosen as the endpoint for scoring the fertility of the MAT-2 strains. Among the 25 MAT-1 strains crossed with AFIR928, strain AFB62 was the most fertile (Fig. 1A and D) and thus AFB62 was chosen as the MAT-1 tester strain to score the fertility of the MAT-2 strains. Of the 25 MAT-2 strains (including AFIR928), 8 (32%) were scored as the HF type, 13 (52%) were scored as the MF type, and 4 (16%) were scored as the LF type (Table 2). Therefore, the distribution of the three fertility types among MAT-2 strains deviated from equal frequency (P = <0.001, χ2 test), with MF being the major type. The MF type therefore represents the largest group (46%) among the 50 strains, followed by the HF (34%) and LF (20%) types.
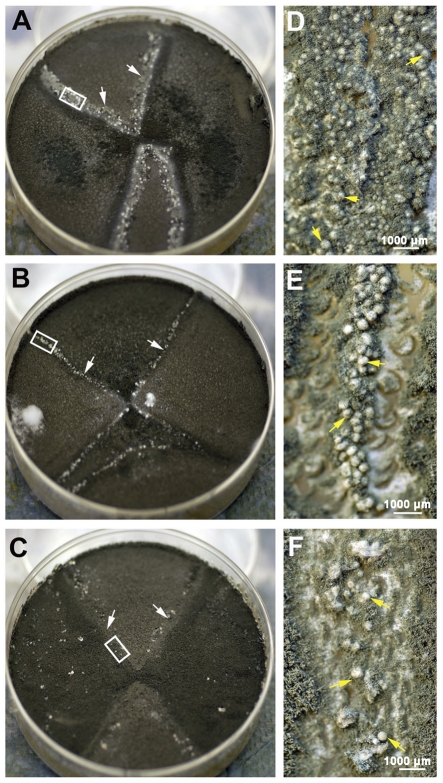
FIG 1 .
Fertility of A. fumigatus. Representative examples of the three levels of fertility (HF, MF, and LF) observed in 50 isolates at 4-week mating are shown in panels A to C. (A) Mating between two isolates of the HF type, AFB62 (MAT-1) and AFIR928 (MAT-2) (supermater pair). The junction zone (white arrows) is heavily populated with fruiting bodies (D, yellow arrows). (B) Mating between HF isolate AFB62 and an isolate of the MF type, AFB60. A significantly lower number of cleistothecia were produced (E, yellow arrows). (C) Mating between HF isolate AFIR928 and AFN15, an isolate of the LF type. Few cleistothecia were found scattered along the junction zone (F, yellow arrows). Panels D, E, and F show higher magnifications of the white insets in panels A, B, and C, respectively.
We next investigated whether polymorphism(s) in the MAT loci could cause differences in fertility between HF and LF strains. Sequences of the MAT locus from LF strains of each mating type were compared to those of the highly fertile tester strains, AFIR928 and AFB62. No polymorphism was found in the MAT-1 locus between the LF strains and the AFB62 strain. In the MAT-2 locus, however, three of the four LF strains contained an insertion of three nucleotides (GCC) not present in the AFIR928 strain (data not shown). The insertion is located between the two divergently transcribed open reading frames (ORFs) (AFUA_3G06160 and AFUA_3G06170) within the MAT-2 locus. Whether this insertion affects the transcription of the two genes is currently being investigated.
We then carried out molecular strain typing by PCR fingerprinting using M13 microsatellite-specific primers (11, 12) to determine if there was a possible correlation between mating competency and the strain type within each mating type, as this could provide a valuable diagnostic tool. DNA fingerprinting of the 50 strains revealed low genetic diversity, i.e., only two molecular types, T1 and T2, differing in only one band (see Figure S1 in the supplemental material). Thirty-two strains (64%) were of the T1 type, and 18 (36%) were of the T2 type. In MAT-1 strains, the T1 and T2 types were distributed equally among the HF and LF isolates, whereas the T1 type was predominant among the MF strains (Table 1). In MAT-2 strains, T1 was the predominant molecular type among the isolates of MF, as well as LF, while the two molecular types were equally distributed among the HF strains (Table 2).
Supermaters.
AFB62 (MAT-1) and AFIR928 (MAT-2) were designated the supermater pair based on their ability to produce abundant cleistothecia containing viable ascospores in 4 weeks and reproducible mating efficiency. In fact, the number of cleistothecia produced at the junction of the two mating partners was so high as to prevent accurate counting of the fruiting bodies (Fig. 1A and D). However, it was possible to assess the number of ascospores per fruiting body by isolating single cleistothecia and scoring the number of ascospores. Although this number varied greatly among cleistothecia within the same cross, the minimum number obtained was 1 × 104 per fruiting body (see Table S1 in the supplemental material). Figure 2A shows a cloud of ascospores released from a cleistothecium, and the ascospores released are shown at a higher magnification in the right half of Fig. 2B. The interwoven hyphae of the cleistothecial wall and contaminating conidia from the mated strains are shown on the left side of Fig. 2B. The two equatorial crests and the rigid surface ornamentation of ascospores previously described for N. fumigata (4) are shown in Fig. 2C. Ascospores were approximately twice as large and were not as hydrophobic as conidia. Thus, the detergent Tween 20 was not required to minimize clustering, as in the case of conidia. Occasionally, intact asci containing ascospores were observed in the midst of released ascospores (Fig. 2D). Strains AFIR928 and AFB62 were deposited in the Agricultural Research Service Culture Collection USDA, Peoria, IL, under accession numbers NRRL 62427 and NRRL 62428, respectively.
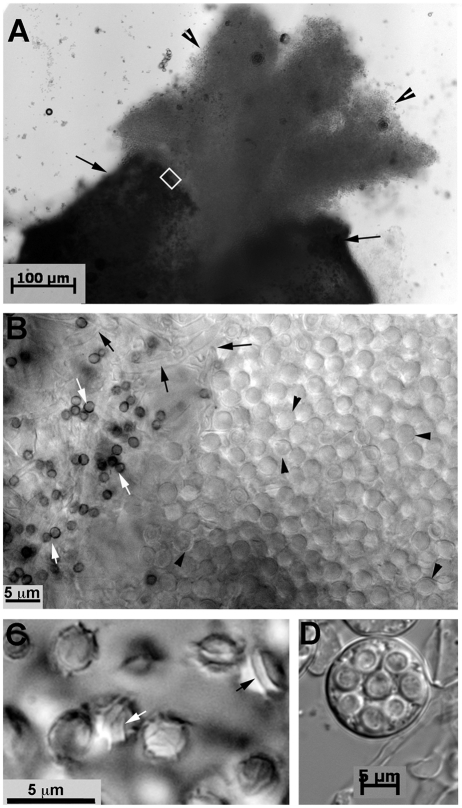
FIG 2 .
Micrographs of the progeny from the AFB62 × AFIR928 cross. (A) A single cleistothecium (black arrows) and a cloud of ascospores (arrowheads) released from the fruiting body. (B) Higher magnification of the inset in panel A. The abundant number of ascospores (black arrowheads) is shown on the right side of the field. Contaminating asexual spores from the parent strains (white arrows) and the interwoven hyphae from a cleistothecial wall (black arrows) are shown on the left side. (C) Ascospores showing two equatorial crests (black arrow) and a surface ornamented with ridges (white arrow). (D) An ascus containing eight ascospores.
Viability and germination of ascospores.
Since a 100% pure ascospore preparation is difficult to obtain, heat treatment at 70°C for 30 min was carried out to eliminate contaminating conidia. After treatment at 70°C, A. fumigatus ascospores are still able to germinate (4) while conidia are killed at 65°C (13). Control assays showed that heat treatment at 70°C for 30 min inactivated 100% of the conidia without affecting ascospore viability (see Text S1a in the supplemental material). These findings indicated that treatment neither deterred nor enhanced the viability of ascospores. Since heat treatment at 70°C for 30 min did not affect ascospore viability but killed conidia very effectively, the remaining assays were therefore carried out under these conditions. Ascospore viability assays showed that the population of viable ascospores from crosses with the supermater pair increased proportionally with the age of the mated cultures (Fig. 3A), 17% by 4 weeks, 25% by 6 weeks, 40% by 8 weeks, and 95% by 20 weeks.
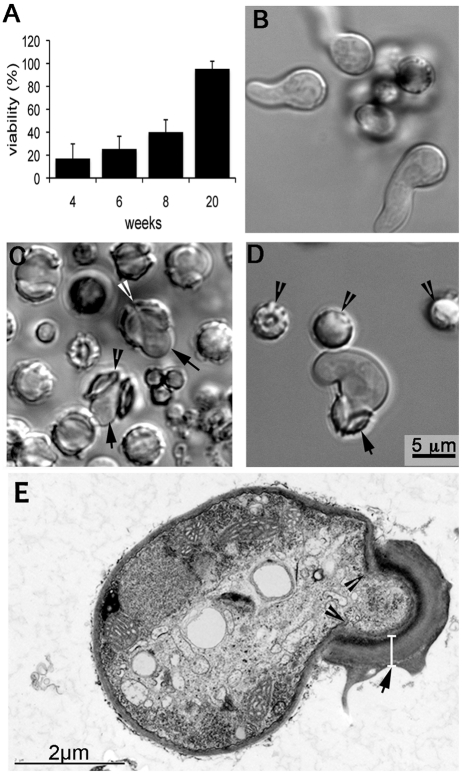
FIG 3 .
Ascospore viability and germination. (A) Ascospores from 4-, 6-, 8-, and 20-week-old crosses were plated on MEA after heat treatment at 70°C for 30 min. Viability was assessed by the number of colonies grown after 48 h at 37°C. (B) Germinating asexual spores. (C) Germinating ascospores with germlings. Ascospores isolated from 20-week-old crosses were heat treated and incubated at 37°C for 8 to 10 h to allow germination. Germlings (arrows) emerge from the opening between the crests on the ascospores (arrowhead). (D) Growth of an ascospore germling into a short hyphal filament. The wall of the ascospore remains attached to the growing germling (arrow). Arrowheads point to nongerminated ascospores. (E) TEM of a germinating ascospore. The ascospore wall (arrow) is composed of two layers, an electron-dense inner wall covered by a thick outer wall. The cell wall of the emerged germling is contiguous with the innermost wall of the ascospores (arrowheads). The magnification in panels B and C is the same as in panel D.
We also monitored morphogenesis of ascospore germination following incubation of heat-treated ascospores in RPMI─25 mM HEPES for 8 to 10 h at 37°C. Bright-field microscopy revealed that ascospores germinated in a manner distinct from that of conidia, in which swelling is followed by protrusion of a hyphal filament (Fig. 3B). In contrast, ascospores do not appear to swell before germination but germination is initiated by the opening of the cell between the equatorial crests (Fig. 3C; see Movie S1 in the supplemental material). The opened walls of germinating ascospores resemble two halves of a cracked nutshell. In some ascospores, the two shells completely separate during the emergence of the germling (Fig. 3C), while in others, the two halves of the shell are still attached at one end (Fig. 3C and D). The emerged germlings grow into hyphae (Fig. 3D), and the shells of the spore wall often remain attached even after extensive hyphal elongation (data not shown). Figure 3E shows an ultrathin section of a germinating ascospore observed by transmission electron microscopy (TEM). The cell wall of the emerged germling is contiguous with the innermost wall of the ascospores (Fig. 3E). The ascospore wall is composed of two layers, one electron-dense inner layer covered by an approximately 0.5-µm-thick outer layer (excluding the surface ornaments). The width of germlings that emerge from ascospores is mostly 1.5 to 2 times that of those germinated from conidia (Fig. 3B to D). This pattern of ascospore germination was consistent regardless of whether they were isolated from 4- or 20-week-old crosses.
Virulence.
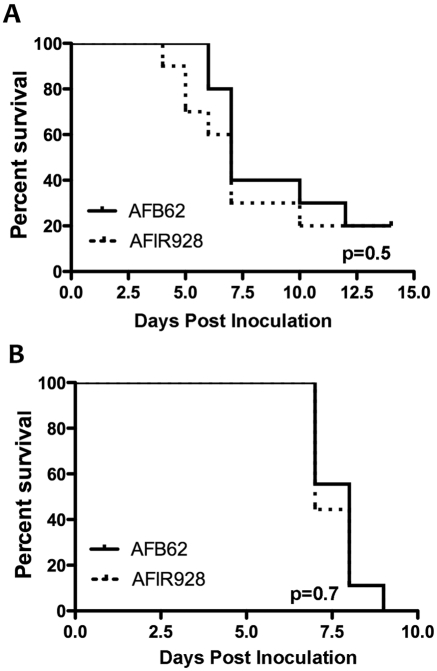
Because strain AFB62 (MAT-1) had originated from a fatal case of invasive aspergillosis while strain AFIR928 (MAT-2) was isolated from the environment, the two strains were compared for virulence using two different mouse models that represent two major risk groups for invasive aspergillosis: BALB/c mice immunosuppressed with hydrocortisone and p47phox knockout chronic granulomatous disease (CGD) mice. CGD mice are susceptible to Aspergillosis due to NADPH oxidase deficiency (14). The survival curves of both models suggested that these two isolates are equally virulent (Fig. 4A and B) and they are as virulent as strain B-5233, which was used as a reference strain for our previous studies (15–17). Thus, the supermater pair AFB62 and AFIR928 make a valuable tool for genetic studies on the pathobiology of A. fumigatus.
FIG 4 .
Virulence of the supermater pair AFB62 and AFIR928. (A) BALB/c mice treated with hydrocortisone and inoculated with 6 × 105 conidia/mouse. (B) p47phox knockout mice inoculated with 3 × 104 conidia/mouse. Animals in both models received conidia via pharyngeal aspiration.
Recombinational analysis.
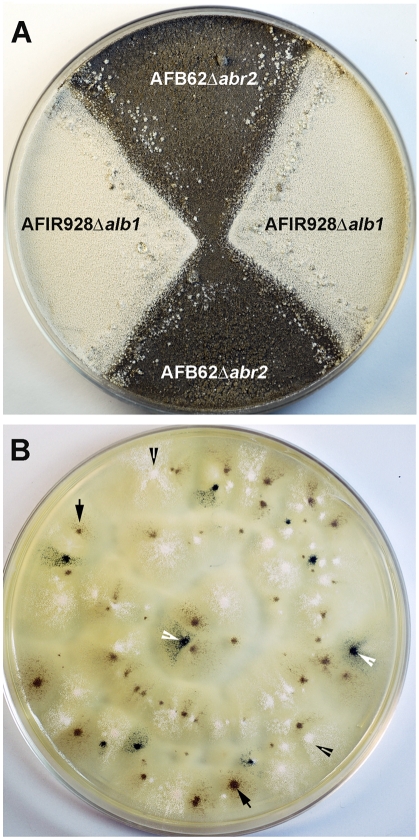
To confirm whether the supermater pair is also suitable for recombinational analysis, we created deletion mutant forms of the alb1 (pksP) gene, AFUA_2G17600, in MAT-2 strain AFIR928, and the abr2 gene, AFUA_2G17530, in MAT-1 strain AFB62. Both genes are components of a 6-gene cluster within a 17-kb region of chromosome 2. The genes in this cluster encode proteins involved in the biosynthesis of 1,8-dihydroxynaphthalene (DHN)-melanin, which gives rise to the bluish-green conidial color of A. fumigatus. The alb1 gene encodes a polyketide synthase, which catalyzes the first step in the biosynthesis of DHN-melanin. Deletion of this gene results in white conidia (18). The abr2 gene encodes an oxidase that is hypostatic to alb1, which is involved in the later stages of DHN-melanin synthesis. Deletion of abr2 results in a brown conidial color (18). The two genes are transcribed in the same direction, with 8.3 kb separating the two ORFs. Since the phenotypes of the two mutants are clearly distinguishable and the physical distance between the two genes is known, the frequency of recombination between these closely linked genes can be accurately determined. Three independent alb1Δ clones of the MAT-2 strain (AFIR928 alb1Δ-1, -2, and -3) were crossed with the MAT-1 abr2Δ mutant strain (AFB62abr2Δ) for recombinational analysis (Fig. 5A). The progeny are expected in three different phenotypes if crossing over occurred between the two genes: first, recombinant wild-type progeny with green conidia (alb1 abr2); second, those with white conidia (alb1Δ or alb1Δ abr2Δ); and third, clones with brown conidia (abr2Δ) (Fig. 5B). Thus, the number of white progeny is expected to be equal to the sum of the brown and green progeny since the white phenotype can result from segregation, as well as recombination events in which alb1 is epistatic to abr2. The number of green progeny represents the minimum number of recombinants (i.e., not including the white progeny of alb1Δ abr2Δ genotype resulting from crossover between the two genes), given that crossing over between the alb1Δ and abr2Δ loci is required for wild-type conidial color. When the ascospores were harvested at 4 weeks, recombination was confirmed by the presence of progeny producing green conidia. Interestingly, the number of white progeny was lower than the expected sum of green and brown progenies in all three independent crosses. The number of white progeny, however, was equal to or higher than the sum of the brown and green progeny when harvested at 8 weeks (Table 3). The minimal number of recombinants based on green progeny alone was between 5 and 7% at 8 weeks (Table 3). These results suggest that ascospores carrying alb1Δ may require longer than 4 weeks to fully mature or become hardy enough to survive heat treatment since the albino progeny increased approximately 5% from 4- to 8-week-old crosses. Analysis of the MAT-1-to-MAT-2 ratios among the progeny from the 8-week-old mating plates was then carried out. Thirty progenies, 10 of each conidial color, were randomly selected and analyzed by PCR using MAT-specific primers. The proportion of MAT-1 to MAT-2 among the progeny of each color was nearly 1:1 (data not shown), confirming independent assortment of the mating-type alleles located on chromosome 3 and the conidial color genes on chromosome 2.
FIG 5 .
Recombination analysis. (A) The AFIR928alb1Δ (MAT-2, white conidia) and AFB62abr2Δ (MAT-1, brown conidia) strains were crossed and incubated for 4 to 8 weeks. (B) A representative plate showing progeny with three conidial colors, white, brown, and green. Ascospores were isolated from 8-week-old crosses, heat treated, and plated on MEA. The colonies producing white (black arrowheads), brown (black arrows), and green (white arrowheads) conidia were counted after 72 h of incubation at 37°C.
TABLE 3 .
Phenotypic analysis of the progeny derived from the mating pair AFB62 abr2Δ and AFIR928 alb1Δ
| Crossa | % of progeny at: |
|||||
|---|---|---|---|---|---|---|
| 4 wk |
8 wk |
|||||
| White | Brown | Green | White | Brown | Green | |
| 1 | 43 | 47 | 10 | 50 | 43 | 7 |
| 2 | 48 | 48 | 4 | 51 | 44 | 5 |
| 3 | 44 | 50 | 6 | 50 | 45 | 5 |
Crosses 1, 2, and 3 refer to mating between AFB62 abr2Δ and AFIR928 alb1Δ-1, -2, and -3, respectively.
CGH.
We compared the genomes of the supermater pair AFB62 and AFIR928 using comparative genome hybridization (CGH) based on the A. fumigatus AF293 microarray (5). Although this approach does not reveal potential divergence between the two supermaters due to the genes which may not be present in the AF293 array, the analysis showed high similarity (99%), with only 86 genes diverged between or absent from the two genomes (see Table S2 in the supplemental material). Among the 86 genes, 46 were present in AFB62 but absent or diverged in AFIR928 and 40 showed the opposite distribution. While these genes were distributed across all chromosomes, 50% of them were concentrated at genomic loci that were found to be variably present among A. fumigatus and other sequenced aspergilli (5, 19, 20). As expected, the CGH results showed that the AFB62 mating locus had diverged from that in AF293 (MAT-2) and AFIR928. Surprisingly, however, was the finding that two genes required for sexual reproduction, steA and vosA (7), also showed divergence between the strains (see Table S2 in the supplemental material). A PCR-based analysis of eight genes predicted to have diverged between AFB62 and AFIR928 found all of these genes to be absent in either of these two genomes (data not shown), thus validating the CGH results. Out of the 86 absent or highly diverged genes, 34 encode proteins of unknown function. The functions of the remaining 52 genes varied greatly and appeared to be mutually exclusive. One noteworthy observation is that 14 of the 46 AFB62-specific genes are located adjacent to each other on chromosome 6. This gene cluster contains at least two genes putatively associated with chitinolytic activity and is variably present among A. fumigatus strains (5).
DISCUSSION
The first report of sexual reproduction in A. fumigatus strains was based on the crossing of Irish environmental strains from a population in which recombination had been detected (4). It was then speculated that the sexual fertility of the species A. fumigatus could be restricted to isolates from particular environments and/or specific geographic locations (4). Subsequently, however, Szewczyk and Krappmann (21) reported the successful crossing of two clinical strains, one from England and the other from Germany, indicating that sexual reproduction in A. fumigatus is restricted neither to environmental strains nor to those of a certain geographic origin. Considering the very specific requirements for completion of the sexual cycle of A. fumigatus, such as particular environmental conditions coupled with a 6-month incubation period, we speculated that there could be a wide range of levels of fertility among global populations. Our findings showed first that environmental and clinical isolates from separate global locations (the United States, India, Hong Kong, England, and Ireland) exhibited sexual fertility, confirming that sexual reproduction is not linked to the origin of the isolates. Second, our findings showed a wide variability in mating efficiency among the 50 strains. In particular, to our surprise, the sexual cycle in 80% of the strains took just 4 weeks to complete. Interestingly, this included crosses between AFIR928 and two MAT-1 environmental Irish strains studied by O’Gorman et al., which had previously taken 6 months to complete the sexual cycle (4). One explanation for this difference could be the compositions of the mating media. Among the environmental conditions conducive to mating (oatmeal agar, incubation at 30°C, oxygen restriction, and darkness), oatmeal agar medium seems to be the most variable factor. We found that different batches of oatmeal obtained from the same manufacturer did not always yield the same degree of fertility in certain crosses. Since the oatmeal used by O’Gorman et al. (4) and that used in the present study were from different sources, the disparity in fertility results obtained with the Irish strains is most likely due to differences in the composition of the oatmeal, as well as methods of grain processing. In fact, oatmeal agar obtained from Difco (BD) was not as effective for cleistothecial formation as the medium prepared in-house using Quaker oats or the original Irish pinhead oats (4; data not shown).
Isolates of both mating types of A. fumigatus could be divided into three different levels of fertility, HF, MF, and LF. While the HF type was not clearly associated with a specific mating type, the MF type was more prominent in MAT-2 populations than in the MAT-1 population. The LF type was slightly more prominent in the MAT-1 population. In an attempt to identify genomic differences that might explain differences in mating efficiency, the mating locus of all 10 LF strains was sequenced and compared to the MAT-1 and MAT-2 sequences of AFB62 and AFIR928, respectively. While no polymorphism was identified between the MAT-1 locus of the LF strains and AFB62, a GCC insertion was found at the MAT-2 locus in three out of four strains that failed to produce ascospores within 4 weeks. Since the GCC insertion is located between the HMG domain of the MAT1-2 gene (AFUA_3g06170), which is essential for mating (21), and a second mating gene (AFUA_3g06160) present within the MAT-2 locus, the insertion might affect the expression of both genes. We are currently investigating the relationship between the GCC insertion and the LF phenotype in the MAT-2 strains. Although strains AFB62 and AFIR928 were chosen as the principal supermater pair, recent screening of a larger worldwide collection of isolates (C. M. O’Gorman, J. A. Sugui, S. S. Swilaiman, K. J. Kwon-Chung, and P. S. Dyer, unpublished data) has revealed other MAT-1 and MAT-2 isolates with high fertility and the ability to cross with isolates of A. fumigatus from diverse global locations. Although fertility may vary, a remarkable variation in phenotype/genotype among worldwide isolates is not expected, since A. fumigatus is known to be a species with a global distribution without any correlation between genotype and geographic location or clinical history (22, 23). Detailed characterization of these strains is underway in order to determine their suitability as the alternate supermaters for A. fumigatus genetic studies.
In order for the supermater pair to be useful for routine genetic analysis, a standardized optimum method for mating is needed. Since the duration required to isolate meiotic spores was significantly reduced, the next important step was to devise a way to obtain pure ascospore populations free of contaminating parental conidia present on the cleistothecial surface. Our findings showed that incubation of an ascospore suspension containing contaminating conidia at 70°C for 30 min killed 100% of the conidia without affecting the viability of the ascospores. The thick and dense ascospore wall shown by TEM suggests that it may serve to protect spores from environmental stress, including high temperature. For instance, the ascospores would likely be able to tolerate the elevated temperatures within compost piles that would inactivate the conidia (24). It is also possible that this wall becomes more protective as the ascospore ages, which tracks with the gradual temperature increase in a self-heating compost pile over time. This possibly explains why the viability of ascospores increased as mated cultures were incubated for more than 4 weeks. In fact, an association between ascospore age and resistance to heat has been reported in N. fischeri, a close relative of A. fumigatus (25). Other factors reported to affect the heat resistance of ascospores are pH and the composition of the medium used during incubation at high temperatures (26, 27). We tested the effect of a 10% sucrose solution as the incubation medium, as well as lowering the pH of the medium from 7 to 3 during heat treatment, and found no difference in ascospore viability (data not shown). Ascospores with thick, melanized walls, such as those produced by Neurospora crassa, require heat activation before they can effectively germinate (28). Our findings demonstrated that high temperature was not required to activate the germination of A. fumigatus ascospores from 4-week-old crosses since ascospores were able to germinate and produce colonies without heat treatment. Our findings corroborate those of O’Gorman et al., which showed that the ascospores from 6-month-old crosses germinated with or without heating at 70°C for 90 min (4). It is possible, though, that ascospores become dormant with age and heat treatment may be required to affect germination.
Another factor that appears to be associated with ascospore viability is the protein encoded by the gene alb1 (AFUA_2G17600). Alb1p catalyzes the first step of DHN-melanin biosynthesis, which is responsible for the bluish-green conidial pigment of A. fumigatus. Mutants of alb1 produce white conidia (18). Recombination analyses of ascospores harvested at 4 weeks showed a slightly lower-than-expected number of albino progeny than that of brown and green progeny combined. At 8 weeks, however, such a discrepancy was not observed. It is therefore plausible that the product(s) of this protein or of the pathway helps to confer on developing propagules resistance against environmental stress such as high temperature.
The ability to obtain viable progeny within 4 to 8 weeks is highly significant for facilitating classical genetic analysis. The previous study of O’Gorman et al. demonstrated recombination of DNA fingerprint and mating-type markers in a Mendelian fashion, attributed to independent assortment (4). In the present study, we have provided evidence of recombination arising from genetic crossover between closely linked alb1 and abr2 loci. The frequency of recombination between the two loci presented in this study is the first report for A. fumigatus demonstrating the relationship between physical distance in the genome and genetic distance assessed by the number of recombinants. Genetic distances between pairs of linked markers are usually established by crossover frequencies reflected in the recombinant population (29). We assessed recombination between two loci 8.3 kb apart in the AFIR928 alb1Δ × AFB62 abr2Δ cross and found a minimum of 5% of the progeny to be recombinant. Since this is the first recombinational study of A. fumigatus, it is not known if recombinational hot spots exist between the two conidial color markers. Further recombinational analysis would provide additional information as to the relationship between physical distance and genetic map distance in the species.
Finally, genetic variability between AFB62 and AFIR928 was evaluated using CGH. When two A. fumigatus strains, AF293 and A1163, were compared by CGH, at least 90% of the sequences overlapped (19, 20). Thus, it is not surprising that the supermater pair showed close to 99% identity based on the AF293 gene probes. Although there might be many other differences that cannot be ascertained by CGH, the low genetic diversity between the supermater pair corroborates previous findings that the genomes of A. fumigatus are highly conserved (19, 20). Of the 86 genes highly diverged between or absent from the supermater pair, 15 were previously described as unique to AF293 (MAT-2) but not to A1163 (19). Therefore, it is likely that a more comprehensive CGH analysis using a larger number of MAT-1 and MAT-2 strains would reveal that the genes identified as exclusive to one or the other strain of the supermater pair are not necessarily mating type dependent.
In conclusion, although the viability of the ascospores of A. fumigatus collected at 4 weeks is low, the abundance of cleistothecia and the number of ascospores per cleistothecium (≥1 × 104) are high enough to perform recombinational analysis. However, in order to avoid possible bias in genetic data, such an analysis can be delayed until mated cultures are 8 weeks old, if necessary. Furthermore, it is reassuring to know that both of the supermater strains proposed in this study are equally virulent and as virulent as B-5233, which was extensively used for previous virulence studies in our laboratory (15–17). The high virulence, high fertility, and genetic amenability of the supermater pair AFB62 and AFIR928 render them an invaluable tool for future genetic studies on the pathobiology of A. fumigatus.
MATERIALS AND METHODS
Strains and mating.
All of the A. fumigatus strains used in this study (Tables 1 and 2) were maintained on Aspergillus minimal medium (15). The AFN, AFB, AFS, and AFIR strains were obtained from the United States National Institutes of Health, B. L. Wickes (the University of Texas Health Science Center at San Antonio, San Antonio, TX), J. S. Klutts (University of Iowa Carver College of Medicine, Iowa City, IA), and P. S. Dyer (University of Nottingham, Nottingham, United Kingdom), respectively. Oatmeal agar was prepared using Quaker oatmeal (Quaker Oats, Chicago, IL) according to the method described by the CBS-KNAW Fungal Biodiversity Centre (http://www.cbs.knaw.nl/collections/Biolomics.aspx?Table=Growth+media).
Matings were performed as follows. Conidia from strains of opposite mating types were inoculated pairwise (two crosses per plate) on oatmeal agar medium, sealed with Parafilm, and incubated at 30°C in the dark (4). Each cross was repeated 2 to 4 times. Cleistothecia were made visible by “hoovering” the junction zones with a vacuum line to remove hyphae and conidia obscuring the fruiting bodies. Malt extract agar (MEA; Sigma, St. Louis, MO) was used to analyze ascospore viability, as well as conidial color in recombination studies.
Determination of mating type.
The mating type of each strain was identified by PCR amplification of mating-type-specific genes from genomic DNA (see Text S1b in the supplemental material). For the MAT-1 type, primers specific for the sequence of the alpha mating-type gene AFUB_04290 were used (forward primer CTGGAGGAGCTTCTGCAGTAC and reverse primer GGAGTACGCCTTCGCGAG). The MAT-2 type was identified with primers specific for the sequence of the MAT-2 gene AFUA_3G06170 (forward primer CTCTTGTGGCAGGATGCTCT and reverse primer TTGCTGGTAGAGGGCAGTCT).
M13 PCR fingerprinting.
Molecular typing was carried out as described previously (12), with minor modification (11), using the microsatellite-specific primer M13 (5′ GAGGGTGGCGGTTCT 3′). PCR fingerprint profiles were visualized and compared on 1.4% agarose gels stained with ethidium bromide.
Sequencing of MAT loci.
The MAT loci of strains AFIR928, AFIR964, AFIRB3, AFS1, AFN6, AFB62, AFB65, AFN10, AFN11, AFN15, AFN17, and AFN29 were sequenced with primers MATa (CGCGCCGGTATGGTACA) and MATb (ATGTCAATTGTAGAACCATGAAAGAC), located in the conserved regions flanking the mating-type locus. The sequences of amplicons were compared using the software SeqMan Pro from DNASTAR, Inc.
Analysis of viability and germination of ascospores.
To estimate the number of ascospores per fruiting body, single cleistothecia from a 4-week-old mated culture of AFB62 × AFIR928 were isolated with a needle, transferred to a vial with 20 to 50 µl of sterile distilled water, and crushed with a sterile loop to release the ascospores, which were then diluted and counted using a hemocytometer. The ascospore suspensions prepared by this method were invariably contaminated with conidia. However, due to the size and morphological differences between the two types of spores, the number of ascospores could be assessed accurately. To determine the viability of ascospores, the spore suspension was heat treated at 70°C for 30 min to inactivate contaminating conidia. The effectiveness of heat treatment for elimination of conidia without affecting ascospore viability was determined in control experiments (see Text S1a in the supplemental material). Viability of the progeny from AFB62 × AFIR928 was analyzed using 4-, 6-, 8-, and 20-week-old mated cultures. Cleistothecia were harvested at the designated times, suspended in water, and ruptured to release ascospores. The number of ascospores was assessed before adjusting the concentration to 1 × 103 ascospores/ml. Aliquots of 100 µl were heat treated, vortexed briefly, plated on MEA plates in quadruplicate, and incubated for 48 h at 37°C. Ascospore viability was determined by counting the colonies on the plates. Three crosses per time point were used to assay the viability of ascospores from 4-, 6-, and 8-week-old plates. In the case of ascospores from 20-week-old plates, two crosses were assayed. Morphogenesis of ascospore germination was studied using RPMI─25 mM HEPES at 37°C in 5% CO2, conditions which are conducive to A. fumigatus spore germination (11). Ascospores were harvested from 4- and 20-week-old crosses, heat treated, suspended in RPMI─25 mM HEPES, and inoculated in an 8-well chamber slide. Samples were visualized by bright-field microscopy after 8 to 10 h of incubation.
TEM.
Heat-treated ascospores were incubated in RPMI─25 mM HEPES at 37°C in 5% CO2 for 8 to 10 h to allow germination. Ascospores were then fixed with 4% glutaraldehyde for 3 h at 4°C, double-fixed in phosphate-buffered-saline-buffered glutaraldehyde (2.5%) and osmium tetroxide (0.5%), dehydrated, and embedded in Spurr’s epoxy resin. Ultrathin (90-nm) sections were double stained with uranyl acetate and lead citrate and viewed in a Philips CM10 transmission electron microscope.
Virulence studies.
The animal experiments were carried out with the approval and oversight of the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases, United States National Institutes of Health, Bethesda MD. The virulence of strains AFB62 (MAT-1) and AFIR928 (MAT-2) was assessed in two murine models, BALB/c mice (National Cancer Institute Division of Cancer Treatment) treated with hydrocortisone acetate and p47phox knockout (B6.129S2-Ncf1tm1Shl N14) mice (30) (Taconic Farms, Inc.). For the hydrocortisone treatment, 6-week-old BALB/c mice were injected subcutaneously with 100 µl of 20 mg/ml hydrocortisone acetate (Sigma, St. Louis, MO) on days −4, −2, 0, and +2 of fungal inoculation. Mice in both models were inoculated with freshly harvested conidia via pharyngeal aspiration (31). BALB/c and p47phox knockout mice received 6 × 105 and 3 × 104 conidia/mouse, respectively. Survival was monitored for 20 days, and the results were analyzed by log-rank test.
Recombinational analysis.
The AFB62 abr2Δ (MAT-1) strain was crossed with three independent alb1 deletion strains derived from MAT-2 supermater AFIR928, AFIR928 alb1Δ-1, -2, and -3, and the progeny was analyzed for recombination frequency (see Text S1c in the supplemental material for the deletion strains). Analysis of the progeny for conidial color and mating type was done with ascospores isolated from 4- and 8-week-old crosses. Heat-treated ascospores were plated on MEA and incubated at 37°C for 72 h. This prolonged incubation time, from 48 to 72 h, ensured full development of conidial pigment, which allowed accurate color scoring. Conidial color was scored in 300 to 1,000 progenies from each cross. Ten single colonies of each conidial color were randomly selected, and their mating types were determined by PCR as described above.
CGH.
Purified genomic DNA was labeled and hybridized as previously described (5). Labeling reactions and hybridizations were conducted using protocols as described in the J. Craig Venter Institute microarray standard operating procedures available at http://pfgrc.jcvi.org. All hybridizations with the two biological replicates were repeated in dye swap sets. Hybridized slides were scanned and analyzed to obtain relative hybridization profiles. Normalized data were averaged over replications, and genes with differential hybridizations at the 95% confidence level were determined using intensity-dependent Z scores (with Z = 1.96). The resulting data were analyzed to identify genes variably present in any of the probed genomes. Any gene with hybridization signals with a >1.4-fold difference (−0.75 ≤ log2 ≤ 0.75) between two different strains was considered either absent or highly diverged (20). A PCR-based approach was carried out to validate the CGH results. Eight diverged genes, 4 from each strain, were chosen for this assay. These genes are listed in Table S2 in the supplemental material. PCR amplification of the genes from genomic DNA was carried out with primers specific for each gene.
SUPPLEMENTAL MATERIAL
Supplemental methods. Text S1, DOCX file, 0.8 MB.
Germination of ascospores. Cleistothecia from cross between AFB62 and AFIR928 were harvested and crushed to release ascospores. Ascospores were treated at 70°C for 30 min to kill contaminating conidia and then incubated at 37°C and 5% CO2 for 8-10h. Microscopic observation was carried out with a Zeiss Axiovert microscope and Axiovision (version 4.0) software.
M13 molecular strain typing. Molecular strain typing by PCR fingerprinting using microsatellite-specific primer M13. Two groups, T1 and T2, were identified. The white arrowhead points to the single band that differentiates the two groups. Download Figure S1, DOCX file, 0.1 MB.
Number of ascospores per cleistothecium from a 4-week-old cross between AFB62 and AFIR928. Eight cleistothecia were analyzed. Average values ± the standard errors of the means are shown. Table S1, DOCX file, 12.7 MB.
The 86 genes absent from or highly diverged between A. fumigatus strains AFB62 and AFIR928 as identified by CGH analysis.
ACKNOWLEDGMENTS
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, United States National Institutes of Health (Bethesda, MD), and the Wellcome Trust (London, United Kingdom).
We thank J. S. Klutts, University of Iowa Carver College of Medicine, for providing A. fumigatus isolates.
Footnotes
Citation Sugui JA, et al. 2011. Identification and characterization of an Aspergillus fumigatus “supermater” pair. mBio 2(6):e00234-11. doi:10.1128/mBio.00234-11.
REFERENCES
- 1. Kontoyiannis DP, Bodey GP. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161–172 [DOI] [PubMed] [Google Scholar]
- 2. Lin SJ, Schranz J, Teutsch SM. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358–366 [DOI] [PubMed] [Google Scholar]
- 3. Kwon-Chung KJ, Bennett JE. 1992. Medical mycology, p. 201–247 Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 4. O’Gorman CM, Fuller HT, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474 [DOI] [PubMed] [Google Scholar]
- 5. Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 6. Paoletti M, et al. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242–1248 [DOI] [PubMed] [Google Scholar]
- 7. Dyer PS, O’Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 8. Raper KB, Fennell DI. 1965. The genus Aspergillus. William and Wilkins Co., Baltimore, MD. [Google Scholar]
- 9. Samson RA, Hong S, Peterson SW, Frisvad JC, Varga J. 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud. Mycol. 59:147–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon-Chung KJ, Sugui JA. 2009. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends Microbiol. 17:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J, et al. 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 14:755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer W, et al. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA─a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790–1799 [DOI] [PubMed] [Google Scholar]
- 13. Chai LY, et al. 2010. Aspergillus fumigatus conidial melanin modulates host cytokine response. Immunobiology 215:915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Romani L, et al. 2008. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature 451:211–215 [DOI] [PubMed] [Google Scholar]
- 15. Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sugui JA, et al. 2007. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 6:1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sugui JA, et al. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rokas A, et al. 2007. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud. Mycol. 59:11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fedorova ND, et al. 2009. Using aCGH to study intraspecific genetic variability in two pathogenic molds, Aspergillus fumigatus and Aspergillus flavus. Med. Mycol. 47(Suppl. 1):S34–S41 [DOI] [PubMed] [Google Scholar]
- 21. Szewczyk E, Krappmann S. 2010. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot. Cell 9:774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pringle A, et al. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899 [PubMed] [Google Scholar]
- 23. Rydholm C, Szakacs G, Lutzoni F. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hogland W, Bramryd T, Persson I. 1996. Physical, biological and chemical effects of unsorted fractions of industrial solid waste in waste fuel storage. Waste Manag. Res. 14:197–210 [Google Scholar]
- 25. Conner DE, Beuchat LR. 1987. Efficacy of media for promoting ascospore formation by Neosartorya fischeri, and the influence of age and culture temperature on heat resistance of ascospores. Food Microbiol. 4:229–238 [Google Scholar]
- 26. Kavanagh J, Larchet N, Stuart M. 1963. Occurrence of a heat-resistant species of Aspergillus in canned strawberries. Nature 198:1322 [Google Scholar]
- 27. Kikoku Y, Tagashira N, Gabriel AA, Nakano H. 2009. Heat activation of Neosartorya and Talaromyces ascospores and enhancement by organic acids. Biocontrol Sci. 14:87–95 [DOI] [PubMed] [Google Scholar]
- 28. Shear CL, Dodge B. 1927. Life histories and heterothallism of the red breadmold fungi of the Monilia sitophila group. J. Agric. Res. 34:1019–1042 [Google Scholar]
- 29. Mortimer RK, Schild D. 1981. Genetic mapping in Saccharomyces cerevisiae , In p. 11 Strathern JN, Jones EW, Broach JR, The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor [Google Scholar]
- 30. Jackson SH, Gallin JI, Holland SM. 1995. The p47phox mouse knock-out model of chronic granulomatous disease. J. Exp. Med. 182:751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rao GV, et al. 2003. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J. Toxicol. Environ. Health A 66:1441–1452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Text S1, DOCX file, 0.8 MB.
Germination of ascospores. Cleistothecia from cross between AFB62 and AFIR928 were harvested and crushed to release ascospores. Ascospores were treated at 70°C for 30 min to kill contaminating conidia and then incubated at 37°C and 5% CO2 for 8-10h. Microscopic observation was carried out with a Zeiss Axiovert microscope and Axiovision (version 4.0) software.
M13 molecular strain typing. Molecular strain typing by PCR fingerprinting using microsatellite-specific primer M13. Two groups, T1 and T2, were identified. The white arrowhead points to the single band that differentiates the two groups. Download Figure S1, DOCX file, 0.1 MB.
Number of ascospores per cleistothecium from a 4-week-old cross between AFB62 and AFIR928. Eight cleistothecia were analyzed. Average values ± the standard errors of the means are shown. Table S1, DOCX file, 12.7 MB.
The 86 genes absent from or highly diverged between A. fumigatus strains AFB62 and AFIR928 as identified by CGH analysis.