ABSTRACT
Membrane proteins are involved in numerous essential cell processes, including transport, gene regulation, motility, and metabolism. To function properly, they must be inserted into the membrane and folded correctly. YidC, an essential protein in Escherichia coli with homologues in other bacteria, Archaea, mitochondria, and chloroplasts, functions by incompletely understood mechanisms in the insertion and folding of certain membrane proteins. Using a genome-scale approach, we identified 69 E. coli membrane proteins that, in the absence of YidC, exhibited aberrant localization by microscopy. Further examination of a subset revealed biochemical defects in membrane insertion in the absence of YidC, indicating their dependence on YidC for proper membrane insertion or folding. Membrane proteins possessing an unfavorable distribution of positively charged residues were significantly more likely to depend on YidC for membrane insertion. Correcting the charge distribution of a charge-unbalanced YidC-dependent membrane protein abrogated its requirement for YidC, while perturbing the charge distribution of a charge-balanced YidC-independent membrane protein rendered it YidC dependent, demonstrating that charge distribution can be a necessary and sufficient determinant of YidC dependence. These findings provide insights into a mechanism by which YidC promotes proper membrane protein biogenesis and suggest a critical function of YidC in all organisms and organelles that express it.
IMPORTANCE
Biological membranes are fundamental components of cells, providing barriers that enclose the cell and separate compartments. Proteins inserted into biological membranes serve critical functions in molecular transport, molecular partitioning, and other essential cell processes. The mechanisms involved in the insertion of proteins into membranes, however, are incompletely understood. The YidC protein is critical for the insertion of a subset of proteins into membranes across an evolutionarily wide group of organisms. Here we identify a large group of proteins that depend on YidC for membrane insertion in Escherichia coli, and we identify unfavorable distribution of charge as an important determinant of YidC dependence for proper membrane insertion.
Introduction
How proteins are properly inserted into and folded within membranes is a fundamental and incompletely understood question of cell biology. Escherichia coli genes encode approximately 900 cytoplasmic membrane proteins, constituting ~20% of the proteins produced by the cell (1). Most are inserted into the membrane via a canonical signal recognition particle (SRP)-dependent, Sec translocon-dependent insertion pathway (reviewed in reference 2). Nascent membrane proteins are recognized and bound by SRP, targeted to the membrane via the membrane receptor FtsY, and transferred to the SecYEG translocon for cotranslational insertion, whereby exterior hydrophilic domains are translocated to the periplasm, hydrophobic transmembrane segments partitioned into the lipid bilayer, and interior hydrophilic domains synthesized in the cytoplasm.
Certain cytoplasmic membrane proteins require the membrane protein YidC for proper insertion (2–4). Some YidC substrates are inserted into the membrane via the Sec translocon, while others utilize a YidC-dependent but Sec-independent pathway (5–8). YidC is highly conserved, with homologues in mitochondria and chloroplasts, most bacterial species, and some members of the domain Archaea (9, 10), and is essential for cell viability (7). On the basis of cross-linking to model membrane proteins, YidC has been proposed to act as a chaperone (11), mediating the partitioning of nascent transmembrane segments from the Sec translocon, with which it is physically associated (12), into the lipid bilayer, as well as the proper bundling of transmembrane segments (13). Some substrates strictly require YidC for membrane insertion (14, 15), while others require it for efficient insertion (16, 17) or proper folding (18).
Nearly one-third of E. coli membrane proteins that have been quantified in proteomic studies display significantly altered abundance in the absence of YidC (19, 20), suggesting its involvement in the insertion of a substantial subset of E. coli membrane proteins; however, dependence on YidC for membrane insertion per se has been demonstrated for only 14 proteins in E. coli. While studies employing model membrane proteins have begun to define the role of YidC in membrane protein biogenesis, how it functions and which traits of substrate proteins contribute to YidC dependence remain poorly understood. To address these issues, we identified and characterized a large group of YidC-dependent membrane proteins in E. coli. We show that membrane proteins possessing an unfavorable distribution of positively charged residues are significantly more likely to require YidC for proper insertion and that charge distribution per se can be a necessary and sufficient determinant of YidC dependence.
RESULTS
Screen to identify YidC-dependent cytoplasmic membrane proteins in E. coli.
Bacterial cells synthesizing green fluorescent protein (GFP)-tagged membrane proteins typically exhibit a fluorescent signal that is circumferential around the cell periphery, reflecting uniform distribution of the protein within the membrane. The signal intensity is proportional to the abundance of the protein in the membrane. Based on the principle that a perturbation of proper membrane insertion can alter the abundance, subcellular localization, or distribution of this signal, we designed a screen to identify E. coli cytoplasmic membrane proteins that are dependent on YidC for proper membrane insertion.
We screened a subset of a genome-scale plasmid library consisting of each open reading frame of E. coli strain W3110 cloned with an N-terminal His6 tag and a C-terminal GFP tag and expressed under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible PT5-lac promoter (the ASKA Library [21]). The subcellular localization of each protein in this library had been categorized, with 492 proteins showing a circumferential distribution consistent with localization to the membrane. Using the hidden Markov model topology algorithm Phobius (22), E. coli W3110 is predicted to encode 936 integral cytoplasmic membrane proteins, consistent with published estimates for other E. coli strains (1). Of these 936 proteins, 428 (46%) were included in the membrane-localized subset of the ASKA Library (see Data Set S1 in the supplemental material). Sixty-four additional proteins observed to be membrane-localized proteins but not predicted to be integral membrane proteins are likely to be peripheral membrane proteins and were excluded from our screen. Since GFP folds rapidly in the cytoplasm and cannot be secreted by Sec in its folded state, we restricted our analysis to membrane proteins that were either predicted (22) or have been shown experimentally (23) to have C termini in the cytoplasm. Of the 428 membrane-localized integral cytoplasmic membrane proteins, the 415 that met these criteria constituted the input pool for our screen.
We examined the subcellular localization of each GFP-tagged membrane protein in the absence versus presence of YidC, using a strain in which yidC expression was controlled by the arabinose-inducible ParaBAD promoter (see Fig. S1A in the supplemental material). Synthesis of each GFP-tagged membrane protein was induced with IPTG after 3 h of growth either in the absence of arabinose, at which time no detectable YidC was present, or in the presence of arabinose (Fig. S1B and S1C). Most of the 415 proteins screened exhibited similar fluorescence localization patterns under the two conditions (e.g., YaiZ in Fig. 1B), suggesting that insertion of most membrane proteins was not significantly perturbed in the absence of YidC.
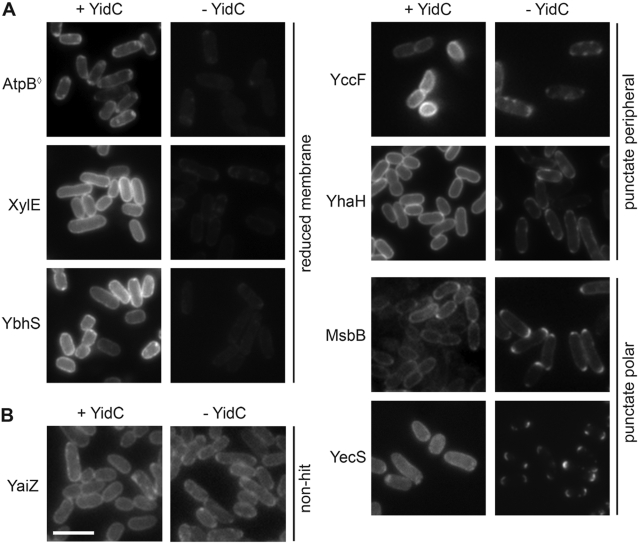
FIG 1 .
YidC-dependent localization of a subset of GFP-tagged membrane proteins. (a) Seven examples of membrane proteins that showed altered GFP signals in the absence of YidC (− YidC). The AtpB protein previously shown to be dependent on YidC for membrane insertion (◊) is shown. (b) Membrane protein that showed comparable circumferential GFP signals in the presence (+) and absence (−) of YidC (YaiZ). Images are representative. Bar, 5 µm.
Sixty-nine membrane proteins (16.6%) showed distinct fluorescence localization patterns in the presence versus absence of YidC (see Table S1 in the supplemental material). Most (40 of 69) showed a >50% decrease in circumferential membrane fluorescence in the absence of YidC with no increase in diffuse cytoplasmic fluorescence (e.g., AtpB, XylE, and YbhS in Fig. 1A), most likely reflecting an overall decrease in protein abundance. A pleiotropic effect of YidC depletion on gene expression was unlikely, since most membrane proteins showed no appreciable decrease in fluorescence, and a panel of cytoplasmic membrane and soluble cytoplasmic proteins showed no change in abundance (see Fig. S2 in the supplemental material). Since membrane proteins that are not inserted into the membrane or that fold improperly are often subject to degradation, the observed reduction in circumferential fluorescence was most consistent with protein degradation consequent to altered membrane insertion or folding. In certain cases, mislocalization may have been due to the effects of YidC depletion on other proteins required for the stability or proper localization of the protein, and reduced fluorescence may have been due to aggregation, since this can lead to fluorescence quenching (24).
Two other patterns that were observed in the absence of YidC were bright punctate fluorescent foci around the periphery of the cell (17 of 69 hits, e.g., YccF and YhaH) or at the cell poles (12 of 69 hits, e.g., MsbB and YecS; Fig. 1A). The presence of punctate foci indicated that the GFP-tagged proteins were unevenly distributed in the membrane and were accumulating at discrete locations, consistent with protein aggregation and strongly suggestive of defects in membrane insertion. Their position at the cell periphery suggested that the proteins either had been partially inserted into the membrane or remained otherwise membrane associated. Misfolded proteins have an increased propensity to form aggregates, particularly when hydrophobic regions of a protein become solvent exposed, as would occur if a hydrophobic transmembrane segment failed to insert into the lipid bilayer.
Among the membrane proteins that were mislocalized in the absence of YidC were two proteins previously shown to be YidC dependent for membrane insertion, AtpB (F0c) (Fig. 1A) and ProW (7, 25). Two additional previously described YidC substrates, MalF and MtlA, were included in the input to the screen but not identified as hits; MalF requires YidC for stability and MalFGK2 complex assembly, but not for insertion or topogenesis (16), and MtlA interacts with YidC during membrane insertion in vitro (13), but whether it depends on YidC for insertion has not been examined; these observations likely account for MalF and MtlA not being identified as hits in our screen. The identification of previously described YidC-dependent membrane proteins as hits in our screen indicated the sensitivity of our assay. Membrane proteins that, in the absence of YidC, fail to insert properly yet remain membrane associated and evenly distributed might be missed by our screen.
Membrane proteins mislocalized in the absence of YidC show biochemical defects in membrane insertion.
To test whether membrane proteins that displayed altered fluorescence localization in the absence of YidC were indeed dependent on YidC for proper membrane insertion, we assayed membrane insertion biochemically, focusing on those screen hits that maintained sufficient abundance in the absence of YidC for this analysis (see Fig. S3A to S3C in the supplemental material). For certain of these hits, as well as for a representative nonhit, we generated strains in which the GFP-tagged protein was expressed from a single copy on the chromosome (Fig. S3C and S1D), allowing us to examine membrane insertion at lower levels of expression. To enable induction of protein synthesis after depletion of YidC, the IPTG-inducible promoter from the parent plasmid was retained in the chromosomally integrated constructs. In all examined cases, the GFP fluorescence localization phenotypes for the single-copy integrants were the same as those observed in the initial microscopy screen (Fig. S1E and S1F). Thus, episomal expression did not contribute significantly to the YidC dependence of these proteins.
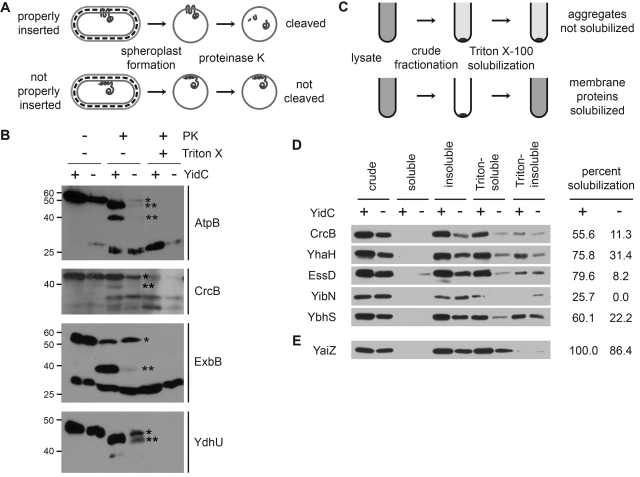
In spheroplasted cells, which lack an outer membrane and cell wall, periplasmic segments of integral membrane proteins are exposed and can be cleaved by an exogenously added protease to which they are sensitive (Fig. 2A). Defects in membrane insertion can result in either protection from proteolysis or altered cleavage. We examined whether, in the absence of YidC, the periplasmic segments of hit proteins had altered accessibility to the protease proteinase K. Because the GFP tag on each fusion was retained in the cytoplasm, it was protected from proteolysis, and C-terminal cleavage products were detected as truncated GFP fusions.
FIG 2 .
Membrane proteins mislocalized in the absence of YidC are YidC dependent for membrane insertion. (A and B) Proteinase K accessibility assay of spheroplasts depleted of YidC or not depleted of YidC (see Materials and Methods). (A) Experimental approach. (B) Screen hits that displayed reduced protease accessibility following YidC depletion. The spheroplasts were treated with proteinase K (PK) (+) in the protease accessibility assay. Cells pretreated with Triton X-100 (+) to promote lysis served as a control for proteolysis. In the absence of YidC (−), increased abundance of full-length protein (*) and decreased abundance of major cleavage product(s) (**) was observed. The positions of molecular mass standards (in kilodaltons) are shown to the left of the gels. (C to E) Differential fractionation of screen hits into Triton X-100-soluble and -insoluble membrane fractions following synthesis in cells depleted of YidC or not depleted of YidC. (C) Experimental approach. (D) Screen hits that displayed reduced Triton X-100 solubility when synthesized in the absence of YidC. (E) A screen nonhit that displayed comparable Triton X-100 solubilities when synthesized in the presence and absence of YidC. Western blots using antibody to GFP are shown. The cell fractions are as follows: crude, whole-cell proteins; soluble, soluble cytoplasmic and periplasmic proteins; insoluble, membranes and other insoluble proteins; Triton-soluble, Triton X-100-soluble fraction of insoluble proteins; Triton-insoluble, Triton X-100-insoluble fraction of insoluble proteins. The loads were proportional and normalized to the optical density at 600 nm (OD600) of culture. Percent solubilization, the ratio of Triton-soluble band to crude fraction band as determined by band densitometry. Images are representative.
Of the 19 GFP-tagged membrane proteins that we examined, four—AtpB, CrcB, ExbB, and YdhU—demonstrated cleavage by proteinase K when synthesized in the presence of YidC that was distinct from that observed when synthesized in the absence of YidC or following cell lysis. In the absence of YidC, proteolysis of each of these four proteins was dramatically reduced [Fig. 2B, increase in the abundance of the full-length protein (asterisk) relative to the major cleavage product(s) (two asterisks)], indicating that they were protected from proteolysis and suggesting that they were either not inserted into the membrane or were inserted with an altered, protease-resistant conformation. Even in the presence of YidC, some CrcB and some ExbB were inaccessible to proteinase K cleavage, suggesting either that the proteins were incompletely inserted under our synthesis conditions or, in the case of CrcB, that two conformations exist (see below). Nevertheless, altered protease accessibility in the absence of YidC indicated that these proteins are dependent on YidC for proper membrane insertion.
Membrane proteins that exhibit punctate localization in the absence of YidC form insoluble protein aggregates.
Many GFP-tagged membrane proteins that were mislocalized in the absence of YidC exhibited punctate fluorescence, suggestive of protein aggregation. As an indicator of protein aggregation, we examined solubility in the detergent Triton X-100, in which cytoplasmic membrane proteins can generally be solubilized but outer membrane proteins, protein aggregates, and inclusion bodies cannot (26). After separating membranes and other insoluble material from soluble cytoplasmic and periplasmic contents by crude fractionation, we selectively solubilized nonaggregated cytoplasmic membrane proteins from the total insoluble fraction with Triton X-100 (Fig. 2C). Each of the proteins examined was solubilized in the presence of YidC but showed greatly reduced solubilization in the absence of YidC (Fig. 2D). Where both assays could be performed, these results consistently correlated with those of protease accessibility experiments (Fig. 2B). The nonhit YaiZ showed similar solubility under the two conditions (Fig. 2E).
Together, the results of our biochemical analyses demonstrate that mislocalization of GFP-tagged membrane proteins as observed by microscopy was strongly indicative of membrane insertion defects. Our screen hits thus represent a newly identified group of 67 E. coli cytoplasmic membrane proteins (in addition to AtpB and ProW) that are dependent on YidC for proper membrane insertion.
Multiple proteins essential for cell growth are YidC dependent.
YidC is essential for cell growth and viability (7). The Sec-independent membrane insertion pathway, substrates of which include subunit Foc of the F1Fo ATPase (8), has been implicated in mediating this essentiality (27). In addition, several membrane proteins that we identified as YidC dependent are known or predicted to be essential—CorA, CydD, FtsX, Lnt, RodA, YbhN, and YgaP (28–33). Thus, the essentiality of YidC may result from the loss of function of multiple essential cytoplasmic membrane proteins that require it for proper membrane insertion.
Membrane proteins containing unbalanced transmembrane segments are more likely to require YidC for proper membrane insertion.
We searched for determinants of YidC dependence by examining commonalities among the 69 proteins identified as hits in our screen. There was no significant enrichment for gene ontological function and process categories, or for protein families, groups, or domains, as determined using the Swiss-Prot database, nor was there significant enrichment in the number of transmembrane segments, the sizes of predicted periplasmic and cytoplasmic protein segments, or overall topology (N in versus N out), as determined using Phobius (22), in the hydrophobicity of predicted transmembrane segments, as determined using Kyte-Doolittle and JTT2 hydropathy scales (1, 34), or in the distribution of negatively charged residues.
A characteristic that was significantly enriched among YidC-dependent membrane proteins was an atypical distribution of positively charged residues. Positively charged residues serve as determinants of membrane protein topology (35, 36), with a strong topological preference for lysine and arginine residues to reside in the cytoplasm (the “positive inside rule”). A transmembrane segment for which the number of positively charged residues in the adjacent periplasmic segment is greater than the number of positively charged residues in the adjacent cytoplasmic segment is referred to as “unbalanced.” Such a topology is unfavorable and is predicted to occur at low frequency.
Within our screen input pool, 34 of 415 proteins (8.2%) were predicted by Phobius to contain unbalanced transmembrane segments (see Data Set S1 and Fig. S4 in the supplemental material). Two-thirds of these (67.6% [23 of 34]) were YidC dependent (Fig. S5). Unbalanced membrane proteins constituted 33% of our screen hits (23 of 69) (Table S1) and were >5-fold more likely to be YidC dependent than balanced membrane proteins (12.1% [46 of 381]) (P = 0.0001). Of these 23 proteins, all but 2 were also predicted to have an unbalanced topology when examined by a second algorithm, OCTOPUS (data not shown) (37). These findings strongly suggest that unbalanced charge distribution can be an important determinant of YidC dependence.
Rendering an unbalanced YidC-dependent membrane protein balanced promotes YidC-independent membrane insertion.
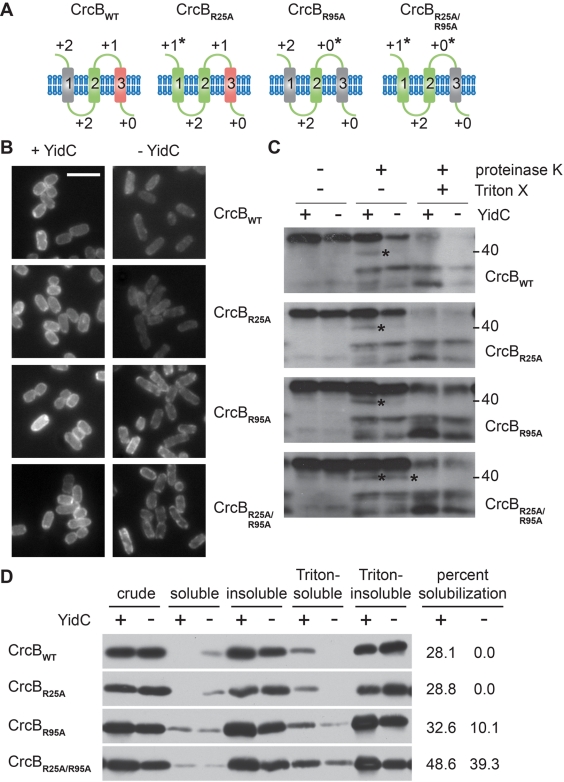
To test whether YidC dependence can be determined by unbalanced charge distribution per se, we altered the charge distribution of the YidC-dependent unbalance membrane protein CrcB. CrcB is predicted by Phobius to be an N-out, C-terminus-in (C-in) three-span polytopic membrane protein possessing a charge-neutral first transmembrane segment, a balanced second transmembrane segment, and an unbalanced third transmembrane segment (Fig. 3A). Other algorithms suggest that the CrcB signal peptide is not cleaved, resulting in an N-in four-span membrane protein. Importantly, whether the signal peptide is cleaved has no effect on the charge balance of wild-type CrcB (CrcBWT) or the charge-altered variants described below. Substituting alanines for positively charged periplasmic residues, we created variants of CrcB that were successively more balanced (Fig. 3A); each construct was integrated onto the chromosome.
FIG 3 .
The distribution of positively charged residues determines the dependence of CrcB on YidC for membrane insertion. (A) Topological illustrations of the cytoplasmic membrane protein CrcB (22) and variants with altered charge balances. Balanced transmembrane segments (green), charge-neutral transmembrane segments (grey), and charge-unbalanced transmembrane segments (red) are shown. Charge balance altered by mutagenesis is indicated by an asterisk. (B) Subcellular localization of GFP-tagged CrcB (CrcB-GFP) variants in the presence or absence of YidC. Bar, 5 µm. (C and D) Protease susceptibility (C) or differential fractionation into Triton X-100-soluble and -insoluble membrane fractions (D) of CrcB-GFP variants following synthesis in the presence or absence of YidC (see Fig. 2 legend and Materials and Methods). The position of a proteolytic product detected only in the absence of YidC for CrcBWT but in increasing amounts in the absence of YidC for the balanced variants is indicated by an asterisk. Images are representative; all images in each panel are from the same Western blot or same microscopy experiment.
The localization of balanced variants of CrcB was less affected by YidC depletion than that of CrcBWT. CrcB with the R95A substitution (CrcBR95A) and CrcB with the R25A and R95A substitutions (CrcBR25A/R95A), in which the third transmembrane segment is no longer unbalanced, showed substantial membrane localization in the absence of YidC, whereas CrcBWT and CrcBR25A, which each retain an unbalanced third transmembrane segment, were highly mislocalized (Fig. 3B). In addition, while CrcBWT exhibited loss of proteinase K sensitivity in the absence of YidC (Fig. 3C, proteolytic product of CrcBWT detected in the presence, but not absence, of YidC [asterisk]), indicating a membrane insertion defect, the balanced CrcBR25A/R95A variant exhibited equivalent proteolysis in the presence and absence of YidC (Fig. 3C). Partially balanced variants CrcBR25A and CrcBR95A exhibited an intermediate phenotype (Fig. 3C). Similarly, while CrcBWT showed no detectable solubilization in Triton X-100 in the absence of YidC, the balanced CrcBR25A/R95A variant showed near-equivalent solubilization in the presence and absence of YidC, with the partially balanced CrcBR95A variant exhibiting an intermediate phenotype (Fig. 3D). As shown above (Fig. 2B), even in the presence of YidC, some protein is inaccessible to proteinase K cleavage, suggesting that either two conformations exist or that the protein is incompletely inserted under our synthesis conditions. Nevertheless, whereas the unbalanced variants of CrcB were YidC dependent, the balanced variants no longer required YidC for proper membrane insertion, indicating that the distribution of positively charged residues in CrcB, and likely in other membrane proteins, serves as a strong determinant of YidC dependence.
The CrcBWT and CrcB variants used in these experiments all contained a C-terminal GFP fusion. When the GFP tag was absent, CrcBWT exhibited dual topology, consistent with recent observations (38), as a C-terminal LacZ fusion formed blue colonies on agar containing 5-bromo-4-chloro-3-indolyl-galactopyranoside (BCIG or X-Gal), and a C-terminal PhoA fusion formed blue colonies on agar that contained 5-bromo-4-chloro-3-indolyl phosphate (XP) (see Fig. S6 in the supplemental material). It is likely that under our experimental conditions the GFP-tagged CrcB constructs exhibited a C-in topology, as indicated by fluorescence signal from the GFP; however, the GFP tag does not prevent the remainder of the proteins from assuming dual topologies. In any case, the dual topology of CrcB appeared to be independent of YidC dependence and the presence of unbalanced transmembrane segments, as the partially balanced and balanced variants all displayed dual topology (Fig. S6).
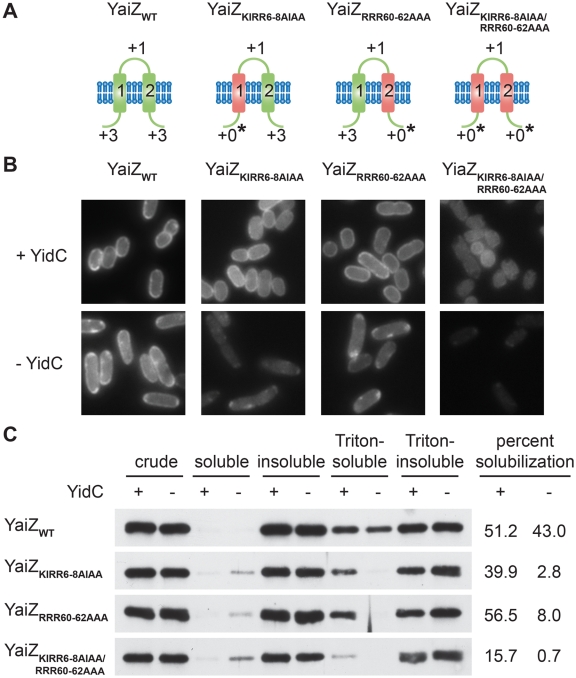
Rendering a balanced cytoplasmic membrane protein unbalanced is sufficient to promote dependence on YidC.
To determine whether an unbalanced distribution of positively charged residues can also be sufficient to render a membrane protein YidC dependent, we altered the charge distribution of YaiZ, a charge-balanced cytoplasmic membrane protein that did not require YidC for membrane insertion. Substituting alanines for positively charged residues in the cytoplasm, we created variants of YaiZ that were unbalanced in one or both transmembrane segments (Fig. 4A). Each construct was integrated into the chromosome. In contrast to the balanced wild-type protein, all unbalanced variants of YaiZ showed a dramatic reduction in membrane localization (Fig. 4B) and Triton X-100 solubility (Fig. 4C) in the absence of YidC, consistent with YidC-dependent defects in membrane insertion. Thus, the introduction of unbalancing mutations appeared to be sufficient to promote a dependence on YidC for membrane insertion. The YaiZ variant possessing two unbalanced transmembrane segments, and to a lesser extent the N-terminal unbalanced variant, also showed solubility defects in the presence of YidC (Fig. 4C), suggesting that YidC was limited in its capacity to promote stable membrane insertion of these more severely unbalanced variants.
FIG 4 .
The distribution of positively charged residues is sufficient to determine the dependence of YaiZ on YidC for membrane insertion. (A) Topological illustrations of the cytoplasmic membrane protein YaiZ (22) and variants with altered charge balances (see Fig. 3 legend). (B) Subcellular localization of YaiZ-GFP variants in the presence or absence of YidC. (C) Differential fractionation of YaiZ-GFP variants into Triton X-100-soluble and -insoluble membrane fractions following synthesis in the presence or absence of YidC (see Fig. 2 legend and Materials and Methods). Images are representative.
For YaiZ and its variants, protease accessibility was uninformative due to the absence of cleavage products distinct from those observed following cell lysis. As determined by C-terminal LacZ and PhoA fusions, YaiZWT and the partially unbalanced variant YaiZKIRR6-8AIAA maintained C-in topology, whereas the unbalanced variant YaiZKIRR6-8AIAA/RRR60-62AAA and to a lesser extent the partially unbalanced variant YaiZRRR60-62AAA displayed dual topology (see Fig. S6 in the supplemental material), suggesting that the RRR60-62AAA mutation contributed to both YidC dependence and the disruption of normal YaiZ topology.
DISCUSSION
YidC is required for the membrane insertion of certain cytoplasmic membrane proteins in E. coli and other bacteria. Our results indicate that while YidC is dispensable for a majority of cytoplasmic membrane proteins in E. coli, it is required to facilitate proper insertion of a sizeable subset. We identified 69 cytoplasmic membrane proteins (16.6% of 415 examined) whose membrane insertion and/or folding is severely impaired in the absence of YidC.
Consistent with a substantial percentage of membrane proteins inserting in a YidC-dependent manner, in recent studies utilizing isotope labeling (19) or two-dimensional (2D) blue native/SDS-PAGE (20) and mass spectroscopy, sizeable subsets of examinable cytoplasmic membrane proteins (38/120 [19] and 20/44 [20], respectively) have exhibited significantly reduced abundance in the absence versus presence of YidC (19). In addition, analysis of a subset of membrane proteins of less than 50 amino acids indicates that many are dependent on YidC for membrane insertion (39). Our approach permitted systematic analysis of a substantially larger fraction of the E. coli membrane proteome and, importantly, more direct examination of YidC dependence for membrane insertion per se, through the combination and cross-validation of microscopic visualization of protein distribution, detergent solubilization and protease accessibility analyses. There was partial but incomplete overlap in the proteins shown by Price et al. (19) and Wickström et al. (20) to have altered abundance in the absence of YidC, as well as between these sets and the proteins identified as YidC dependent in our study (see Data Set S1 in the supplemental material). These differences likely result from differences in approach and highlight the importance and difficulty of defining YidC substrates.
Unlike some YidC substrates that have been characterized, most of the membrane proteins we identified as being YidC dependent are likely inserted via the canonical SRP/Sec-dependent pathway. Most are polytopic membrane proteins possessing three or more transmembrane segments (59 of 69 hits; see Data Set S1 in the supplemental material). Among the 10 that possess one or two transmembrane segments, half are large (>300 residues in length), and one possesses a long periplasmic segment (>50 residues). These traits are inconsistent with insertion via the Sec-independent pathway, whose substrates are small (<150 residues) and either single-span or bitopic membrane proteins with short periplasmic loops (<20 residues) (5, 7, 15). All proteins we analyzed carried an N-terminal His tag; although we did not directly examine whether this affected N-terminal translocation across the cytoplasmic membrane, neither proteins with predicted N-terminus-in (N-in) nor N-terminus-out (N-out) topologies were enriched in either our screen input pool or hits, suggesting that the His tag was not a significant factor in YidC dependence.
Membrane proteins that were predicted to contain charge-unbalanced transmembrane segments were significantly more likely to depend on YidC for proper membrane insertion (67.6% of unbalanced versus 12.1% of balanced membrane proteins) and constituted 33% (23 of 69) of the YidC-dependent membrane proteins that we identified overall. Correcting the charge distribution across the unbalanced transmembrane segment of CrcB abrogated dependency on YidC for proper membrane insertion, indicating that an unbalanced distribution of positively charged residues was sufficient to explain its YidC dependence. Moreover, introduction of unbalancing mutations into the balanced membrane protein YaiZ appeared to render the protein YidC dependent. Thus, an unbalanced distribution of positively charged residues can act as a necessary and sufficient determinant of YidC dependence.
Positively charged residues act as strong determinants of membrane protein topology (the “positive inside rule”). Membrane proteins that cannot be inserted into the membrane in an orientation such that all predicted transmembrane segments remain both in the membrane and charge-balanced have been referred to as “topologically frustrated” (40, 41). Most of the unbalanced transmembrane proteins we identified as YidC dependent fall into this category. While not directly examined here, in the absence of YidC, some unbalanced membrane proteins may adopt an inverted topology, where it is more favorable. While the positive inside rule has been recognized for over two decades, there have been few insights into the mechanism that underlies the rule or into the mechanism by which certain proteins are able to possess a topology that defies it.
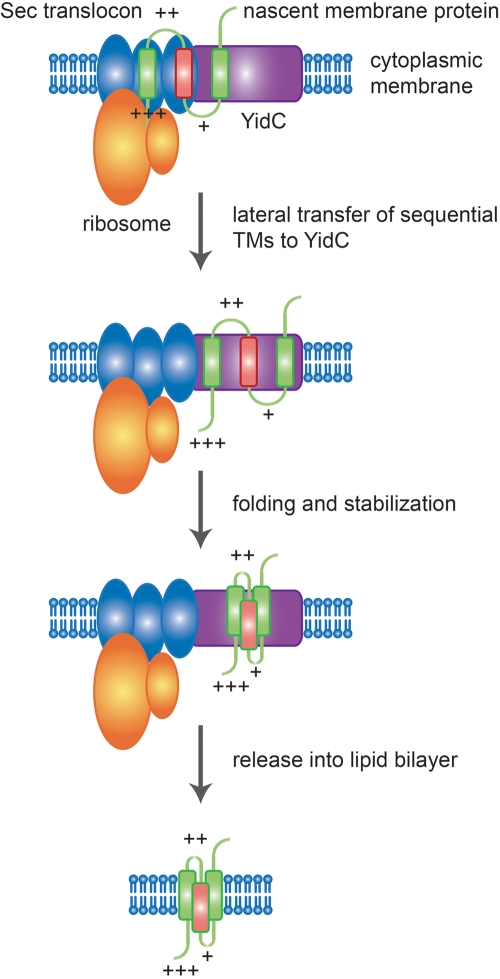
Our data strongly suggest that the mechanism by which many unbalanced transmembrane proteins are properly inserted into and folded in the membrane depends on YidC. We propose a model in which YidC protects the charge-unbalanced segments of these proteins from the influence of electrostatic and/or other topogenic forces that would normally compel more positively charged extramembrane segments to reside in the cytoplasm (Fig. 5), thereby allowing folding events that could not occur if sequential transmembrane segments were inserted directly from the Sec translocon into the lipid bilayer. We postulate that YidC thus serves as a stabilizing environment that lies intermediate between the translocon channel and the lipid bilayer. Prior work lends additional support to this model. YidC is physically associated with the Sec translocon (12, 42), the Sec translocon possesses a lateral gate that has been proposed to serve as an exit site for transmembrane segments (43), and a projection structure of YidC, when docked with that of the Sec translocon, shows the potential positioning of YidC adjacent to this gate (44), which would ideally position YidC to carry out the functions we propose; nascent transmembrane segments of YidC substrates interact first with SecY and then with YidC (45); and YidC can interact with multiple transmembrane segments simultaneously (13) and promote proper folding (18). Although not examined in this paper, it is possible that YidC plays a similar role in the insertion of balanced YidC-dependent membrane proteins, promoting proper topology, folding, and membrane insertion under circumstances where insertion of sequential transmembrane segments directly into the lipid bilayer would not permit proper biogenesis.
FIG 5 .
Model for YidC-dependent insertion of membrane proteins containing unbalanced transmembrane segments. Transmembrane segments (TMs) of nascent cytoplasmic membrane proteins targeted to the Sec translocon are sequentially inserted into the translocon and then partitioned into the lipid bilayer via a lateral gate. YidC, docked at the lateral gate, interacts with transmembrane segments as they exit the translocon, allowing the stabilization of unbalanced transmembrane segments and folding of the protein with correct topology into an intrinsically stable conformation, which is then released into the lipid bilayer. Balanced transmembrane segments (green) and charge-unbalanced transmembrane segments (red) are shown. Positively charged residues within the flanking extramembrane domain are indicated by plus signs.
Since most YidC-dependent membrane proteins that we identified do not or are not predicted to contain unbalanced transmembrane segments, other features must also contribute to YidC dependence. A recent study demonstrated that negatively charged residues within transmembrane segments can act as such a determinant (46). However, neither this feature nor any of the other potential determinant topological features that we examined were enriched among our screen hits. Another feature of cytoplasmic membrane proteins that has been shown to correlate with reduced abundance in the absence of YidC is soluble domains shorter than 100 residues (20); this parameter was not included in our analyses. It is likely that in most cases, YidC dependence is a complex trait or is determined by features that we did not examine or that are less well defined. It should also be noted that some of the screen hits that were classified as balanced membrane proteins may actually be unbalanced, since the empirical bias against unbalanced transmembrane segments is likely to influence the topology predictions of Phobius and other hidden Markov model algorithms (22).
Our identification of a large set of YidC-dependent membrane proteins in E. coli greatly improves our understanding of which and what proportion of cytoplasmic membrane proteins in E. coli require YidC for proper membrane insertion. Given their high degree of conservation and, at least in certain cases, functional complementarity (9, 10), it is likely that YidC homologues in other organisms, mitochondria, and chloroplasts play similar roles and function by similar mechanisms in promoting proper membrane protein biogenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table S2 in the supplemental material.
Genetic methods.
Site-directed mutagenesis was performed by two-step overlap extension PCR. Integration of ASKA library clones into the chromosome was performed using a strain generated to contain homology to the plasmids at the attphi80 phage attachment site. Additional genetic methods are described in the supplemental material.
Fluorescence microscopy screen.
In 96-well plate format, cells were grown to exponential phase, washed, and resuspended in medium containing arabinose (0.2%) or not containing arabinose; growth was continued for 3 h to allow depletion of YidC. Synthesis of GFP-tagged membrane proteins was induced by the addition of IPTG to 100 µM, and growth was continued for an additional 30 min. The cultures were transferred to glass-bottom 96-well plates, and bacteria were brought to the bottom of the wells by gentle centrifugation. Live-cell microscopy and imaging were performed using a Nikon TE300 microscope.
Protease accessibility assay.
Exponential-phase bacteria were grown in the presence or absence of arabinose for 90 to 180 min, so as to maintain or deplete YidC. Synthesis of GFP-tagged membrane proteins was induced by the addition of IPTG to 100 µM, and growth was continued for an additional 30 min. Spheroplasts were generated by cold osmotic shock and treatment with 3 mM EDTA and 40 µg/ml lysozyme, and then were treated with 100 to 200 µg/ml proteinase K for 1 h at 4°C, followed by protease quenching with 2 mM phenylmethylsulfonyl fluoride (PMSF) and trichloroacetic acid (TCA) precipitation of proteins. Additional details are provided in the supplemental material.
Membrane fractionation.
Membranes and insoluble material were isolated from crude lysates by centrifugation. Cytoplasmic membrane proteins were recovered by solubilization in 1% Triton X-100. Additional details are provided in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental information. Download Text S1, PDF file, 0.151 MB.
Summary of screen input pool selection and Phobius topology predictions for GFP-tagged membrane proteins. Download Data Set S1, XLS file, 0.2 MB.
Experimental design of approaches used in this study. (A to C) Fluorescence-based microscopy screen. (A) Genetic organization of the ASKA library clone plasmids and the YidC depletion strain used in the microscopy screen. (B) Western blot demonstrating the absence of YidC in the YidC depletion strain following 3.5 h of growth in the absence of arabinose. Whole-cell lysates of wild-type (WT) and YidC depletion strains. Loading was normalized to OD600. (C) Layout of the glass bottom 96-well plates used for the microscopy screen. The distribution of the signal from the GFP tag on each ASKA library clone was documented following either depletion of YidC or expression of YidC. (D to F) Single-copy expression of genes encoding YidC-dependent membrane proteins. (D) Genetic cross leading to stable chromosomal integration of gfp fusions to genes encoding membrane proteins of interest. (E) Subcellular localization of GFP-tagged screen hit membrane proteins expressed from chromosomal integrants in the presence or absence of YidC. (F) Subcellular localization of GFP-tagged screen nonhit membrane protein expressed from chromosomal integrants in the presence or absence of YidC. Bar, 5 µm. Download Figure S1, JPG file, 1 MB.
YidC depletion does not lead to pleiotropic effects on protein abundance. Relative abundance of GFP-tagged membrane and cytosolic proteins following the depletion of YidC or expression of YidC. Whole-cell lysates were analyzed by Western blotting using antibody to GFP. (A) Abundance of representative GFP-tagged membrane proteins that were not identified as hits in the microscopy screen. (B) Abundance of a sample of GFP-tagged cytosolic proteins. Download Figure S2, JPG file, 0.3 MB.
Relative abundance in the presence and absence of YidC of those GFP-tagged membrane proteins that were identified as screen hits based on their mislocalization in the absence of YidC. The relative abundance of membrane proteins that carry an N-terminal His tag and a C-terminal GFP tag expressed following either depletion of YidC or expression of YidC is shown. Whole-cell lysates were analyzed by Western blotting. (A to C) Tagged proteins were expressed from episomal constructs (A and B) or from chromosomal integrants (C). Western blot analysis using antibody to GFP (A and C) or His6 (B). Loading was proportional and was normalized to OD600 of culture. All strips in each panel are from the same Western blot. Download Figure S3, JPG file, 2.1 MB.
Topological comparison of predicted unbalanced membrane proteins identified as requiring YidC versus not requiring YidC for membrane insertion. (A to C) Topological illustrations of unbalanced membrane proteins identified in this study as requiring YidC (A) or not requiring YidC (B) for proper membrane insertion or folding, or of RstB, for which YidC dependence could not be determined due to poor GFP signal (C). Balanced transmembrane segments (green), charge-neutral transmembrane segments (grey), and charge-unbalanced transmembrane segments (red) (see text for definitions) are shown. Download Figure S4, JPG file, 2.4 MB.
Localization of predicted unbalanced membrane proteins identified as requiring YidC versus not requiring YidC for membrane insertion. Protein localization following expression in the presence or absence of YidC, as determined by the GFP fluorescence signal from the C-terminal GFP tag on each protein. (A) Unbalanced membrane proteins that showed altered GFP signals in the absence of YidC. (B) Unbalanced membrane proteins that did not show altered GFP signals in the absence of YidC. (C) RstB, unbalanced membrane protein for which GFP signal was poor in both the presence and absence of YidC. For selected membrane proteins, the GFP signal was weak following expression in the absence of YidC; in these cases, the images are also shown after 2-fold (2×) to 5-fold (5×) enhancement of the signal (−YidC enhanced). Images are representative. Download Figure S5, JPG file, 2.2 MB.
Beta-galactosidase and alkaline phosphatase phenotypes of C-terminal LacZ and PhoA fusions to CrcBWT, CrcB variants, YaiZWT, and YaiZ variants. (A and B) Plasmid gene-encoded in-frame C-terminal fusions of LacZ (A) or PhoA (B) to CrcBWT, YaiZWT, and charge-altered variants of CrcB and YaiZ analyzed in this study. These plasmids are carried in strain MC4100 (A), which is ΔlacZ (A), and strain CC118 (B), which is PhoA-minus. LacZ fusions were plated on media containing the beta-galactosidase indicator X-Gal (A), and PhoA fusions were plated on media containing the alkaline phosphatase indicator XP (B). Blue colony color indicates beta-galactosidase activity on X-Gal plate and alkaline phosphatase activity on XP plate; white colony color indicates the absence of beta-galactosidase or alkaline phosphatase activity. Negative controls (A and B), E. coli MC4100 or CC118 carrying plasmid-encoded CrcBWT-GFP or YaiZWT-GFP; positive controls (B), CC118 carrying PhoA fusions to the C terminus of NuoM or FtsQ, each of which is C terminus out. Download Figure S6, JPG file, 1.3 MB.
GFP-tagged membrane proteins exhibiting aberrant localization in the absence of YidC.
Strains and plasmids used in this study.
ACKNOWLEDGMENTS
We thank R. E. Dalbey, M. Mueller, and B. L. Wanner for providing reagents used in this study, and Y. Kurita, A. Kopynec, and T. Wu for technical assistance.
This work was supported by NIH grant AI035817 (to M.B.G.) and by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.N.).
Footnotes
Citation Gray AN, et al. 2011. Unbalanced charge distribution as a determinant for dependence of a subset of Escherichia coli membrane proteins on the membrane insertase YidC. mBio 2(6):e00238-11. doi:10.1128/mBio.00238-11.
REFERENCES
- 1. Boyd D, Schierle C, Beckwith J. 1998. How many membrane proteins are there? Protein Sci. 7:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie K, Dalbey RE. 2008. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6:234–244 [DOI] [PubMed] [Google Scholar]
- 3. Kol S, Nouwen N, Driessen AJ. 2008. Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J. Biol. Chem. 283:31269–31273 [DOI] [PubMed] [Google Scholar]
- 4. Yi L, Dalbey RE. 2005. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (review). Mol. Membr. Biol. 22:101–111 [DOI] [PubMed] [Google Scholar]
- 5. Facey SJ, Neugebauer SA, Krauss S, Kuhn A. 2007. The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365:995–1004 [DOI] [PubMed] [Google Scholar]
- 6. Yi L, Celebi N, Chen M, Dalbey RE. 2004. Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279:39260–39267 [DOI] [PubMed] [Google Scholar]
- 7. Samuelson JC, et al. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637–641 [DOI] [PubMed] [Google Scholar]
- 8. van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. 2004. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yen MR, Harley KT, Tseng YH, Saier MH., Jr 2001. Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol. Lett. 204:223–231 [DOI] [PubMed] [Google Scholar]
- 10. Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuhn A, Stuart R, Henry R, Dalbey RE. 2003. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13:510–516 [DOI] [PubMed] [Google Scholar]
- 12. Scotti PA, et al. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beck K, et al. 2001. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2:709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price CE, Driessen AJ. 2008. YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J. Biol. Chem. 283:26921–26927 [DOI] [PubMed] [Google Scholar]
- 15. van der Laan M, et al. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U. S. A. 100:5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner S, et al. 2008. Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283:17881–17890 [DOI] [PubMed] [Google Scholar]
- 17. Fröderberg L, et al. 2003. Versatility of inner membrane protein biogenesis in Escherichia coli. Mol. Microbiol. 47:1015–1027 [DOI] [PubMed] [Google Scholar]
- 18. Nagamori S, Smirnova IN, Kaback HR. 2004. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 165:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price CE, et al. 2010. Differential effect of YidC depletion on the membrane proteome of Escherichia coli under aerobic and anaerobic growth conditions. Proteomics 10:3235–3247 [DOI] [PubMed] [Google Scholar]
- 20. Wickström D, et al. 2011. Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE. J. Mol. Biol. 409:124–135 [DOI] [PubMed] [Google Scholar]
- 21. Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 22. Käll L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 23. Daley DO, et al. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323 [DOI] [PubMed] [Google Scholar]
- 24. Waldo GS, Standish BM, Berendzen J, Terwilliger TC. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17:691–695 [DOI] [PubMed] [Google Scholar]
- 25. Yi L, et al. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42:10537–10544 [DOI] [PubMed] [Google Scholar]
- 26. Schnaitman CA. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Bloois E, et al. 2005. The Sec-independent function of Escherichia coli YidC is evolutionary-conserved and essential. J. Biol. Chem. 280:12996–13003 [DOI] [PubMed] [Google Scholar]
- 28. Schmidt KL, et al. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robichon C, Vidal-Ingigliardi D, Pugsley AP. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280:974–983 [DOI] [PubMed] [Google Scholar]
- 30. Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78–89 [DOI] [PubMed] [Google Scholar]
- 31. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:20060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hashimoto M, et al. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 55:137–149 [DOI] [PubMed] [Google Scholar]
- 33. Gerdes SY, et al. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 35. Boyd D, Beckwith J. 1989. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 86:9446–9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Heijne G. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5:3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668 [DOI] [PubMed] [Google Scholar]
- 38. Rapp M, Granseth E, Seppälä S, von Heijne G. 2006. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 13:112–116 [DOI] [PubMed] [Google Scholar]
- 39. Fontaine F, Fuchs RT, Storz G. 2011. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 286:32464–32474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gafvelin G, von Heijne G. 1994. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell 77:401–412 [DOI] [PubMed] [Google Scholar]
- 41. von Heijne G. 2006. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7:909–918 [DOI] [PubMed] [Google Scholar]
- 42. Nouwen N, Driessen AJ. 2002. SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44:1397–1405 [DOI] [PubMed] [Google Scholar]
- 43. Van den Berg B, et al. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44 [DOI] [PubMed] [Google Scholar]
- 44. Lotz M, Haase W, Kühlbrandt W, Collinson I. 2008. Projection structure of yidC: a conserved mediator of membrane protein assembly. J. Mol. Biol. 375:901–907 [DOI] [PubMed] [Google Scholar]
- 45. Urbanus ML, et al. 2001. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Price CE, Driessen AJ. 2010. Conserved negative charges in the transmembrane segments of subunit K of the NADH: ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J. Biol. Chem. 285:3575–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information. Download Text S1, PDF file, 0.151 MB.
Summary of screen input pool selection and Phobius topology predictions for GFP-tagged membrane proteins. Download Data Set S1, XLS file, 0.2 MB.
Experimental design of approaches used in this study. (A to C) Fluorescence-based microscopy screen. (A) Genetic organization of the ASKA library clone plasmids and the YidC depletion strain used in the microscopy screen. (B) Western blot demonstrating the absence of YidC in the YidC depletion strain following 3.5 h of growth in the absence of arabinose. Whole-cell lysates of wild-type (WT) and YidC depletion strains. Loading was normalized to OD600. (C) Layout of the glass bottom 96-well plates used for the microscopy screen. The distribution of the signal from the GFP tag on each ASKA library clone was documented following either depletion of YidC or expression of YidC. (D to F) Single-copy expression of genes encoding YidC-dependent membrane proteins. (D) Genetic cross leading to stable chromosomal integration of gfp fusions to genes encoding membrane proteins of interest. (E) Subcellular localization of GFP-tagged screen hit membrane proteins expressed from chromosomal integrants in the presence or absence of YidC. (F) Subcellular localization of GFP-tagged screen nonhit membrane protein expressed from chromosomal integrants in the presence or absence of YidC. Bar, 5 µm. Download Figure S1, JPG file, 1 MB.
YidC depletion does not lead to pleiotropic effects on protein abundance. Relative abundance of GFP-tagged membrane and cytosolic proteins following the depletion of YidC or expression of YidC. Whole-cell lysates were analyzed by Western blotting using antibody to GFP. (A) Abundance of representative GFP-tagged membrane proteins that were not identified as hits in the microscopy screen. (B) Abundance of a sample of GFP-tagged cytosolic proteins. Download Figure S2, JPG file, 0.3 MB.
Relative abundance in the presence and absence of YidC of those GFP-tagged membrane proteins that were identified as screen hits based on their mislocalization in the absence of YidC. The relative abundance of membrane proteins that carry an N-terminal His tag and a C-terminal GFP tag expressed following either depletion of YidC or expression of YidC is shown. Whole-cell lysates were analyzed by Western blotting. (A to C) Tagged proteins were expressed from episomal constructs (A and B) or from chromosomal integrants (C). Western blot analysis using antibody to GFP (A and C) or His6 (B). Loading was proportional and was normalized to OD600 of culture. All strips in each panel are from the same Western blot. Download Figure S3, JPG file, 2.1 MB.
Topological comparison of predicted unbalanced membrane proteins identified as requiring YidC versus not requiring YidC for membrane insertion. (A to C) Topological illustrations of unbalanced membrane proteins identified in this study as requiring YidC (A) or not requiring YidC (B) for proper membrane insertion or folding, or of RstB, for which YidC dependence could not be determined due to poor GFP signal (C). Balanced transmembrane segments (green), charge-neutral transmembrane segments (grey), and charge-unbalanced transmembrane segments (red) (see text for definitions) are shown. Download Figure S4, JPG file, 2.4 MB.
Localization of predicted unbalanced membrane proteins identified as requiring YidC versus not requiring YidC for membrane insertion. Protein localization following expression in the presence or absence of YidC, as determined by the GFP fluorescence signal from the C-terminal GFP tag on each protein. (A) Unbalanced membrane proteins that showed altered GFP signals in the absence of YidC. (B) Unbalanced membrane proteins that did not show altered GFP signals in the absence of YidC. (C) RstB, unbalanced membrane protein for which GFP signal was poor in both the presence and absence of YidC. For selected membrane proteins, the GFP signal was weak following expression in the absence of YidC; in these cases, the images are also shown after 2-fold (2×) to 5-fold (5×) enhancement of the signal (−YidC enhanced). Images are representative. Download Figure S5, JPG file, 2.2 MB.
Beta-galactosidase and alkaline phosphatase phenotypes of C-terminal LacZ and PhoA fusions to CrcBWT, CrcB variants, YaiZWT, and YaiZ variants. (A and B) Plasmid gene-encoded in-frame C-terminal fusions of LacZ (A) or PhoA (B) to CrcBWT, YaiZWT, and charge-altered variants of CrcB and YaiZ analyzed in this study. These plasmids are carried in strain MC4100 (A), which is ΔlacZ (A), and strain CC118 (B), which is PhoA-minus. LacZ fusions were plated on media containing the beta-galactosidase indicator X-Gal (A), and PhoA fusions were plated on media containing the alkaline phosphatase indicator XP (B). Blue colony color indicates beta-galactosidase activity on X-Gal plate and alkaline phosphatase activity on XP plate; white colony color indicates the absence of beta-galactosidase or alkaline phosphatase activity. Negative controls (A and B), E. coli MC4100 or CC118 carrying plasmid-encoded CrcBWT-GFP or YaiZWT-GFP; positive controls (B), CC118 carrying PhoA fusions to the C terminus of NuoM or FtsQ, each of which is C terminus out. Download Figure S6, JPG file, 1.3 MB.
GFP-tagged membrane proteins exhibiting aberrant localization in the absence of YidC.
Strains and plasmids used in this study.