ABSTRACT
The assembly of the cell division machinery at midcell is a critical step of cytokinesis. Many rod-shaped bacteria position septa using nucleoid occlusion, which prevents division over the chromosome, and the Min system, which prevents division near the poles. Here we examined the in vivo assembly of the Bacillus subtilis MinCD targeting proteins DivIVA, a peripheral membrane protein that preferentially localizes to negatively curved membranes and resembles eukaryotic tropomyosins, and MinJ, which recruits MinCD to DivIVA. We used structured illumination microscopy to demonstrate that both DivIVA and MinJ localize as double rings that flank the septum and first appear early in septal biosynthesis. The subsequent recruitment of MinCD to these double rings would separate the Min proteins from their target, FtsZ, spatially regulating Min activity and allowing continued cell division. Curvature-based localization would also provide temporal regulation, since DivIVA and the Min proteins would localize to midcell after the onset of division. We use time-lapse microscopy and fluorescence recovery after photobleaching to demonstrate that DivIVA rings are highly stable and are constructed from newly synthesized DivIVA molecules. After cell division, DivIVA rings appear to collapse into patches at the rounded cell poles of separated cells, with little or no incorporation of newly synthesized subunits. Thus, changes in cell architecture mediate both the initial recruitment of DivIVA to sites of cell division and the subsequent collapse of these rings into patches (or rings of smaller diameter), while curvature-based localization of DivIVA spatially and temporally regulates Min activity.
IMPORTANCE
The Min systems of Escherichia coli and Bacillus subtilis both inhibit FtsZ assembly, but one key difference between these two species is that whereas the E. coli Min proteins localize to the poles, the B. subtilis proteins localize to nascent division sites by interaction with DivIVA and MinJ. It is unclear how MinC activity at midcell is regulated to prevent it from interfering with FtsZ engaged in medial cell division. We used superresolution microscopy to demonstrate that DivIVA and MinJ, which localize MinCD, assemble double rings that flank active division sites and septa. This curvature-based localization mechanism holds MinCD away from the FtsZ ring at midcell, and we propose that this spatial organization is the primary mechanism by which MinC activity is regulated to allow division at midcell. Curvature-based localization also conveys temporal regulation, since it ensures that MinC localizes after the onset of division.
Introduction
The assembly of proteins into supramolecular structures is a culminating event of many cellular processes. Understanding how these structures are built requires determining how proteins localize to the right place within the cell at the right time and how they subsequently interact with one another to form complex assemblies (1–3). Such structures may also assume specific residences within the cell, requiring proper localization of assembly-initiating proteins at the very onset of construction (4, 5).
Here we examined the localization and assembly of DivIVA, a protein that forms a critical positional landmark in the Bacillus subtilis cell, recruiting the Min proteins, which spatially regulate cell division, and the RacA protein, which anchors the chromosome to the cell poles at the onset of sporulation (6–9). Constriction of bacterial membranes during cytokinesis is achieved by a tubulin homolog called FtsZ, which polymerizes in a GTP-dependent manner and assembles into a ring at midcell that recruits other division proteins and that also provides force for membrane invagination (10–12). In rod-shaped bacteria, proper placement of a single FtsZ ring (and therefore the cell division septum) at midcell is mediated by two independent mechanisms: nucleoid occlusion, which prevents septum formation over chromosome-containing regions within the cell (13, 14), and a two-protein system which inhibits FtsZ ring assembly, MinC and MinD (15), wherein MinC is thought to actively inhibit FtsZ ring formation and MinD tethers MinC to the plasma membrane, presumably in order to increase the local concentration of MinC (16, 17). Of course, the efficacy of the MinCD system must be absolutely dependent on a mechanism that spatially regulates MinCD activity. In E. coli this is achieved by the MinE protein, which promotes the pole-to-pole oscillation of the MinCD proteins, thereby ensuring that their concentration remains lowest at midcell (10, 11). In Bacillus subtilis, topological specificity is provided by the DivIVA peripheral membrane protein (9, 18), which preferentially assembles on concave (negatively curved) membranes (9, 19) and mediates the subcellular localization of MinCD (20) via MinJ (21, 22).
Until recently, a model suggested that in B. subtilis MinCD are largely sequestered at the extreme ends of the cell by DivIVA, creating a gradient of MinCD such that the concentration is highest at the cell poles (23, 24). In this model, FtsZ ring formation is permissive only at the midcell region, where the MinCD concentration is lowest (and which is also nucleoid free). However, these original articles indicated that DivIVA and MinCD localized to midcell as well as the cell poles, making it unclear how Min activity at midcell was regulated to allow cell division. Two recent observations exacerbated the complications with the simple view that the Min concentration is highest at the cell poles. First, when expressed from its native promoter and viewed by time-lapse microscopy, MinC did not localize to the poles of B. subtilis but rather localized transiently to active cell division sites (25). Second, a protein that mediates the interaction between DivIVA and MinC (called MinJ [21, 22]) also localized preferentially to active division sites (26). Thus, the subcellular localization of FtsZ-ring assembly inhibitors was actually highest where the previous model predicted that it should be largely absent. This indicated that Min activity at midcell must be regulated either temporally, for example by being recruited only after the FtsZ ring is resistant to its activity, or spatially, for example by being held away from the FtsZ ring at midcell until after the completion of cell division.
Here we provide evidence that DivIVA arrives at nascent division sites at the very onset of cell division and forms very stable ring-like structures at midcell, requiring divisome function (membrane constriction) but not simply FtsZ assembly. Using structured illumination microscopy, we observed that DivIVA and MinJ localize as two adjacent rings on either side of active division sites. This localization would ensure that Min activity is both temporally and spatially regulated. We also have demonstrated that unlike many other components of the division machinery, including MinC and FtsZ, DivIVA rings are stable and remain localized at mature septa long after cytokinesis is complete. As chains of B. subtilis cells separated into individual cells and flat septa were transformed into hemispherical poles, DivIVA rings progressively collapsed into patches, responding to the change in the cell’s shape. We propose a model in which changes in cellular architecture dictate the ultrastructure of DivIVA and thereby regulate Min activity.
RESULTS
DivIVA forms rings at septa and patches at poles.
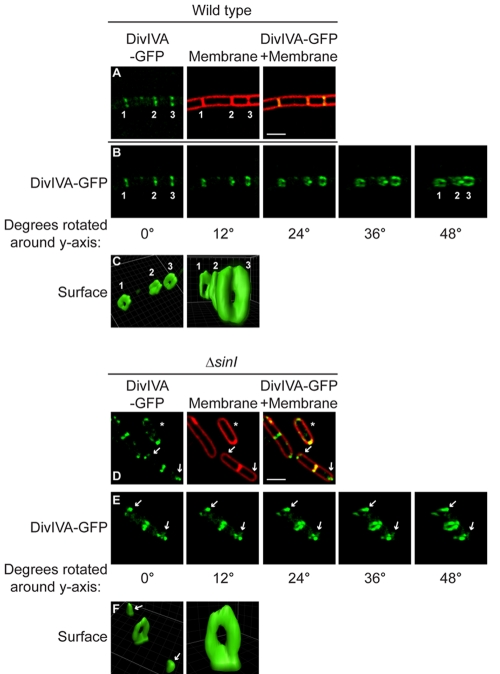
The subcellular localization of DivIVA-green fluorescent protein (GFP) has been previously examined by epifluorescence microscopy (27–29). Although DivIVA-GFP does not fully complement the divIVA null mutant, its localization is similar to that of untagged DivIVA as analyzed by immunofluorescence microscopy (30), and it allows studies of localization and dynamics in living cells. These prior observations revealed that DivIVA localized to septa and, to a lesser extent, to the hemispherical poles of B. subtilis (19). Does the protein assemble into similar structures at the two sites? In order to resolve the supramolecular structure of assembled DivIVA in vivo, we examined the subcellular localization of DivIVA-GFP, produced under control of its native promoter, in B. subtilis by reconstructing Z stacks collected by deconvolution fluorescence microscopy. In otherwise wild-type cells, DivIVA-GFP localized to regularly spaced mature division septa (Fig. 1A, septa 1 and 2) and to sites of ongoing cell division (or incomplete septa) at midcell (Fig. 1A, septum 3; membrane staining revealed that constriction of the septum had not completed). Rotation of the reconstructed images along the y axis revealed that DivIVA-GFP localized at septa formed ring-shaped structures whose outer diameter was roughly equal to the inner diameter of the cell (Fig. 1B and C) (28).
FIG 1 .
Ultrastructure of DivIVA rings at division septa and DivIVA patches at hemispherical poles. (A) Localization of DivIVA-GFP (green) in wild-type B. subtilis growing as a chain of cells that have completed cytokinesis but not cell separation (strain KR515, left). The center panel shows a membrane visualized with the fluorescent dye FM4-64 (red). The right panel shows overlay of fluorescence from DivIVA-GFP and FM4-64; regions of fluorescence overlap are yellow. (B and E) Rotation of the images in panels A and D, respectively, around the y axis (degrees rotated are indicated below each panel). Two older septa are labeled “1” and “2,” and a nascent division septum is labeled “3.” (C and F) DivIVA-GFP fluorescence in panels B and E, respectively, represented as a three-dimensional surface. (D) Localization of DivIVA-GFP in ΔsinI cells that grow as individual cells during exponential phase (strain KR528). At center and right, membrane stain and overlay are shown. Arrows indicate DivIVA patches at the poles. A cell that had not elaborated a septum is marked with an asterisk. Scale bar, 2 µm. Strains are described in Materials and Methods.
Does DivIVA-GFP at the extreme poles also form similar structures? Since wild-type cells, which grow as unseparated chains during exponential phase, rarely elaborate hemisphere-shaped poles, we examined DivIVA-GFP localization in a mutant strain (ΔsinI) that grows as separated cells during vegetative growth (31) in order to increase the chances of visualizing hemispherical cell poles. In dividing ΔsinI cells, DivIVA-GFP localized preferentially to septa and to a lesser extent to the hemispherical cell poles (Fig. 1D) (19). In cells that were not dividing, DivIVA-GFP was concentrated at the hemispherical cell poles (Fig. 1D, asterisk). In actively dividing individual cells, DivIVA-GFP formed rings at partial septa, similar to wild-type cells (Fig. 1E and F). However, at cell poles, DivIVA-GFP formed patches rather than rings (Fig. 1E and F, arrows). Cell-to-cell comparisons revealed that these patches were not uniform and varied widely in size and intensity. Thus, DivIVA forms two distinct ultrastructures: rings at septa (here called “A rings”) and patches at hemispherical poles (here called “A patches”). The formation of one structure or the other appears to depend on the local architecture of the cell.
DivIVA localizes to midcell at the onset of cell division.
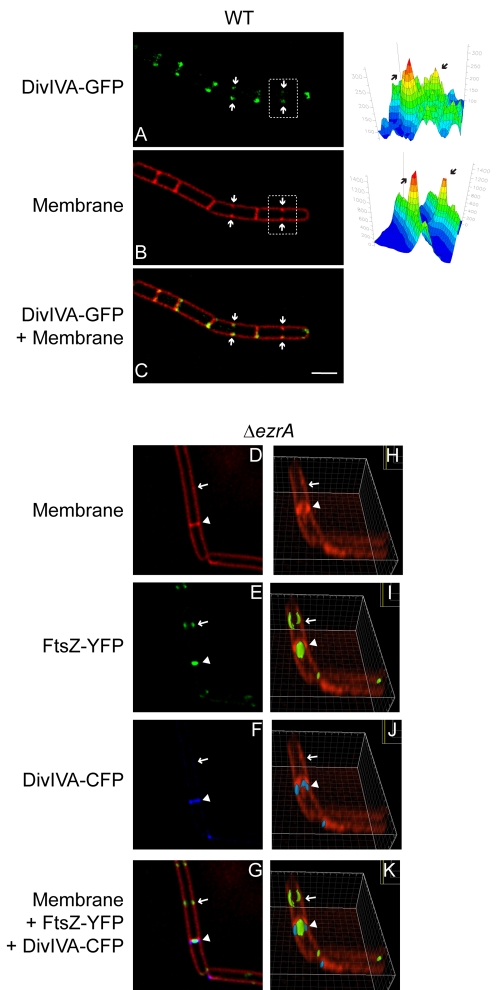
Recognition of negative membrane curvature drives the localization of DivIVA, resulting in the preferential accumulation of the protein at sharply curved septa (9, 19). Does localization, however, require full elaboration of a mature septum, or can DivIVA begin to localize to future division sites at the very onset of membrane constriction? In order to test this, we examined the localization of DivIVA-GFP in cells during septal biogenesis. By deconvolving the fluorescence micrographs, slight increases in membrane staining by the fluorescent membrane stain FM4-64, evidence of membrane invagination, were made readily apparent. Figure 2A to C show a chain of B. subtilis cells as they began to divide (arrows indicate division sites). Quantifying the fluorescence intensity of the membrane stain (Fig. 2B, right) revealed increased fluorescence at each midcell position, consistent with invagination of the membrane at the onset of cell division. DivIVA-GFP colocalized with each of these membrane invagination sites, suggesting that the protein localizes to active septa at the very onset of cell division.
FIG 2 .
DivIVA localizes to septa at the onset of membrane constriction. (A) DivIVA-GFP localization in strain KR515. (B) Membrane visualized with FM4-64. (C) Overlay of DivIVA-GFP and FM4-64. Arrows indicate invaginating membranes at nascent cell division sites. Quantification of fluorescence intensity from FM4-64 and DivIVA-GFP for one incomplete septum (white box) is on the right (arrows). (D to G) Localization of DivIVA-CFP and FtsZ-YFP in the ΔezrA strain (PE99). (D) Membrane visualized with FM4-64. (E) FtsZ-YFP. (F) DivIVA-CFP. (G) Overlay of FM4-64, FtsZ-YFP, and DivIVA-CFP. (H to K) Rotation of the images in (D to G), respectively, around the x axis. The arrow indicates location of an assembled but nonfunctional FtsZ ring; the arrowhead indicates an actively constricting FtsZ ring. Scale bar, 2 µm.
Does DivIVA assemble preferentially at division sites by directly recognizing FtsZ, or is its localization directly mediated by constriction of the membrane during cell division? Previous studies reported that DivIVA localized to hemispherical poles of germinating B. subtilis spores, suggesting that FtsZ is not required for DivIVA localization, but the dependence on FtsZ at septa during vegetative growth remained unresolved (28, 32). In order to distinguish between these possibilities, we examined the localization of DivIVA in cells that harbored a deletion in the ezrA gene. In these mutants, FtsZ rings assemble, but approximately half are unable to constrict and form division septa, resulting in slightly elongated cells (33) (elongated cells are not to be confused with chains of B. subtilis; in chains, cytokinesis completes faithfully, but cells simply remain attached by the cell wall). At actively constricting septa (Fig. 2D and H, arrowhead), FtsZ-yellow fluorescent protein (YFP) was detected as a band that was narrower than the width of the cell (Fig. 2E). DivIVA-cyan fluorescent protein (CFP) localized to these septa as well but as a band whose width was similar to the width of the cell (Fig. 2F). Reconstruction of Z stacks revealed that FtsZ-YFP at these septa formed a disk around the long axis of the cell (Fig. 2I), which we interpret as a constricting Z ring, whereas DivIVA-CFP began assembling into an A ring (Fig. 2J) whose diameter remained larger than the constricting Z ring (Fig. 2K). In contrast, DivIVA-CFP rarely localized (4.4%; n = 113) to sites at which Z rings assembled but did not measurably constrict the membrane (Fig. 2H to K, arrow). Since these results did not rule out a mechanism in which DivIVA could be recruited by a divisome component that assembled downstream of EzrA (34), we repeated the experiment in otherwise wild-type cells (strain PE97). Again, DivIVA-CFP colocalized with FtsZ-YFP in cells with evident membrane constriction and rarely localized (2.4%; n = 42) to sites where Z rings had formed but constriction was not yet evident. Taken together, the data are consistent with a model in which DivIVA assembly at septa requires a functional divisome that produces membrane constriction, not simply formation of an FtsZ ring.
Assembly and stability of A rings at septa.
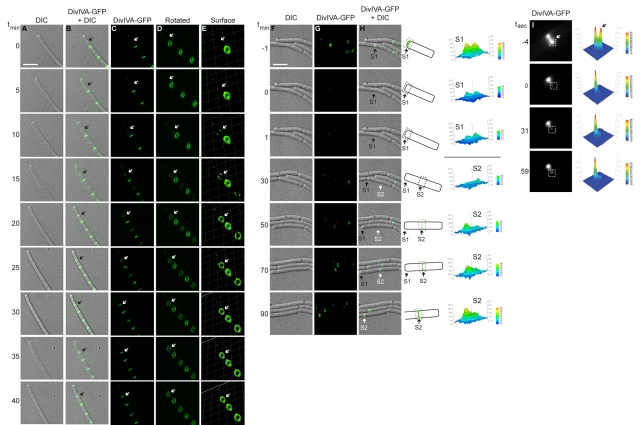
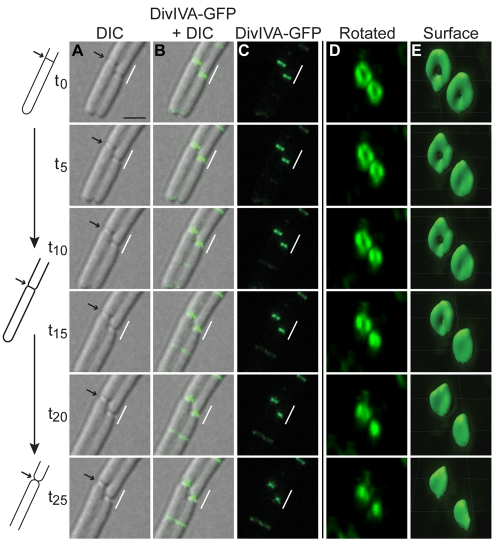
We next monitored localization of DivIVA-GFP at active division septa using time-lapse fluorescence microscopy. To avoid photobleaching, time-lapse experiments required the overexpression of divIVA-gfp from an inducible promoter that resulted in an approximately 75-fold increase in the production of the fusion protein (see Fig. S1 in the supplemental material). Despite the overproduction, the pattern of DivIVA-GFP localization in these cells was similar to the pattern of localization in cells in which DivIVA-GFP was produced at native levels (see Fig. S1). DivIVA-GFP fluorescence was observed, faintly at first, at midcell between existing A rings (or, in one case as shown, between an existing ring and a cell pole; Fig. 3A to E, arrow) and became brighter over time. Reconstruction of Z stacks (Fig. 3D and E) revealed that DivIVA-GFP fluorescence in a newly forming A ring was nonuniform, consistent with nucleation of DivIVA on a point along the circumference of a new septum (Fig. 3E, t15), as previously reported for MinC (25). DivIVA-GFP then continued assembling circumferentially along the new septum and completed formation of an intact circle in approximately 15 min (Fig. 3E, t10 to t20). Over the next 20 min, DivIVA-GFP fluorescence increased until it achieved maximal intensity compared to that of previously formed A rings at adjacent septa.
FIG 3 .
Assembly of stable A rings at septa. (A to E) Time-lapse micrographs of DivIVA-GFP localization in strain KR541. (A) Differential interference contrast (DIC). (B) Overlay, DivIVA-GFP and DIC. (C) DivIVA-GFP. (D) Images in panel C rotated to view A rings. (E) DivIVA-GFP fluorescence intensity in panel D, represented as a three-dimensional surface. Time (minutes) is shown at left. The arrow indicates the site of a nascent A ring assembling. (F to H) DIC, DivIVA-GFP, and overlay in which fluorescence recovery was monitored after photobleaching a field of cells (strain KR528). Time (minutes) before and after photobleaching is shown at left. “S1” is a preexisting septum, and “S2” is a septum formed after photobleaching. A schematic representation of DivIVA-GFP localization in panel H at either S1 or S2 is shown to the right, along with quantification of fluorescence intensities of S1 and S2 before and after photobleaching. Scale bar, 5 µm. (I) Fluorescence recovery was monitored after photobleaching half of an A ring (white box). Time (seconds) before or after photobleaching is indicated at left, and quantification of fluorescence intensity is at right.
To test if new A rings are constructed from newly synthesized DivIVA molecules or by recycling DivIVA molecules that had been incorporated elsewhere, we used fluorescence recovery after photobleaching (FRAP) to assess the ability of bleached A rings to exchange subunits during growth. Fluorescence intensity of existing A rings was diminished after photobleaching (Fig. 3F to H, t-1 to t0; see septum “S1,” indicated with an arrow), with little recovery seen throughout the 90-min experiment, suggesting that previously assembled A rings failed to incorporate newly synthesized DivIVA-GFP molecules. However, a new A ring was formed in this cell (composed of newly synthesized DivIVA-GFP molecules) 50 min after photobleaching (Fig. 3F to H, t50; septum “S2”) and achieved maximal fluorescence intensity 40 min later. The final fluorescence intensity of septum S2 after recovery of fluorescence was comparable to that of septum S1 before photobleaching. This suggests that new A rings are constructed almost entirely of newly synthesized DivIVA molecules and that assembled A rings are stable.
Many proteins at the division septa, such as the FtsZ ring, form dynamic structures in which monomers may be rapidly exchanged within seconds (35). We used FRAP to test the stability of existing A rings, bleaching an area corresponding to just half of a single A ring, and measured recovery of fluorescence at that spot over time (Fig. 3I). After 59 s, the photobleached area had not appreciably recovered fluorescence, suggesting that exogenous DivIVA-GFP was not significantly incorporated into a mature A ring and also that the A ring did not rapidly rotate around the long axis of the cell. In contrast, FtsZ rings show fluorescence recovery with a half-time of 8 s (36). Thus, unlike previously described cytoskeletal structures that assemble at division septa, we conclude that A rings are stable, long-lived structures that are constructed largely of newly synthesized DivIVA molecules.
Assembly of adjacent A rings at division septa.
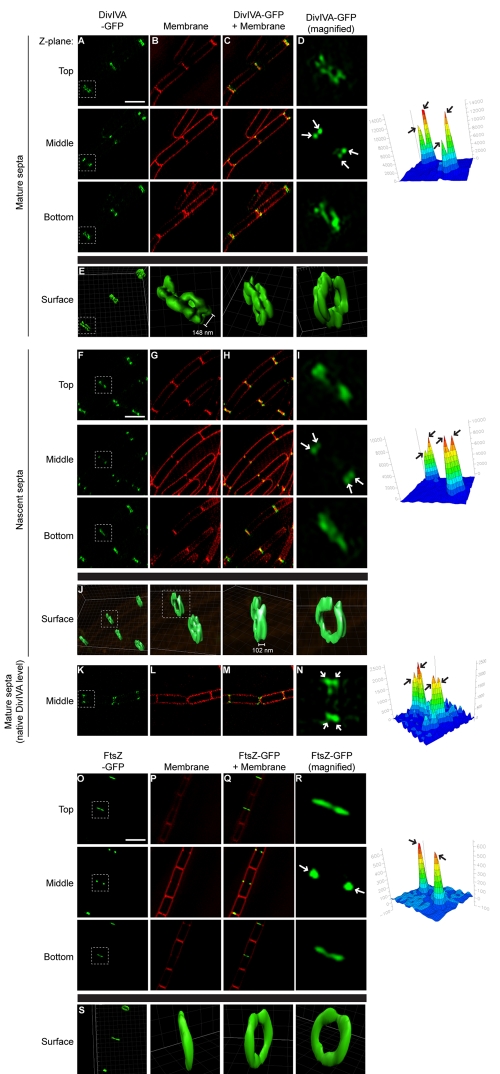
The hypothesis that DivIVA localizes by recognizing negative membrane curvature makes a very specific prediction that A rings should actually assemble on either side of a division septum, since negative curvature is produced on both sides of a forming septum. Indeed, Harry and Lewis suggested that DivIVA could occasionally be seen as “closely juxtaposed” bands at midcell by immunofluorescence microscopy of fixed cells (28). However, since the width of the division septum (~80 nm [37]) is smaller than the resolution limit of light microscopy, we and others were unable to distinguish two adjacent A rings at division septa in live cells using this technique (24). We therefore examined the localization of DivIVA-GFP in live cells using a superresolution technique called three-dimensional structured illumination microscopy (3D-SIM), wherein a known pattern is overlaid onto a sample and a subdiffraction image is then computationally restored from the interference pattern (38). In wild-type cells, when top and bottom Z planes were viewed, DivIVA-GFP formed two adjacent bands at mature division septa (Fig. 4A to D, “Top” and “Bottom”; the highlighted septum in 4A is magnified in 4D). At an intermediate Z plane, DivIVA-GFP appeared as four foci, corresponding to the four regions of negative membrane curvature on either side of the division septum where it meets the lateral surface of the cell (Fig. 4A to D, “Middle”). Reconstruction of the Z stacks revealed two A rings on either side of the mature division septum (Fig. 4E) separated by a space of 148 nm ± 20 nm (n = 24). At incomplete septa, adjacent A rings were also present, although at this stage they were separated by only 102 nm ± 14 nm (n = 8) (Fig. 4F to J).
FIG 4 .
Adjacent A rings at division septa. DivIVA-GFP localization (strain KR541, produced under control of an IPTG-inducible promoter) at mature (A to E) or nascent (F to J) division septa, DivIVA-GFP localization (strain KR515, produced at native levels) at mature division septa (K to N), and FtsZ-GFP localization (strain AD3007) (O to S) viewed by 3D-SIM at three Z planes (indicated as “Top,” “Middle,” or “Bottom”). (B, G, L, and P) Membranes in panels A, F, K, and O, respectively, visualized using FM4-64. (C, H, M, and Q) Overlay of membrane stain and GFP fluorescence. (D, I, N, and R) Magnification of one septum (white box) in panels A, F, K, and O, respectively. (E, J, and S) GFP fluorescence for selected septa in panels A, F, and O, represented as a three-dimensional surface. Separation between A rings indicated is the average distance between regions of peak fluorescence (n = 24 for doublet A rings at mature septa, ±20 nm; n = 8 for doublet A rings at incomplete septa, ±14 nm). GFP fluorescence intensity for the selected septa in panels D, I, N, and R at the intermediate Z plane was quantified and is shown on the right to highlight the number of foci (arrows). Scale bars, 2 µm.
Full reconstruction of A rings by this technique required the overexpression of divIVA-gfp from an inducible promoter to avoid photobleaching. To ensure that elaboration of double A rings was not due to overproduction, we examined cells producing DivIVA-GFP under control of the native promoter at a single medial focal plane (Fig. 4K to N). As expected, DivIVA-GFP formed four foci at division septa, consistent with the formation of adjacent A rings. As a control, we also examined the localization of FtsZ-GFP by 3D-SIM. In contrast to DivIVA-GFP, FtsZ-GFP localized as a single band at active division sites in both top and bottom focal planes and as two foci in the medial focal plane (Fig. 4O to S). We conclude that DivIVA assembles two adjacent A rings on either side of mature septa and active division sites and that it is recruited at the very onset of membrane constriction.
A rings prevent aberrant cell division adjacent to new septa.
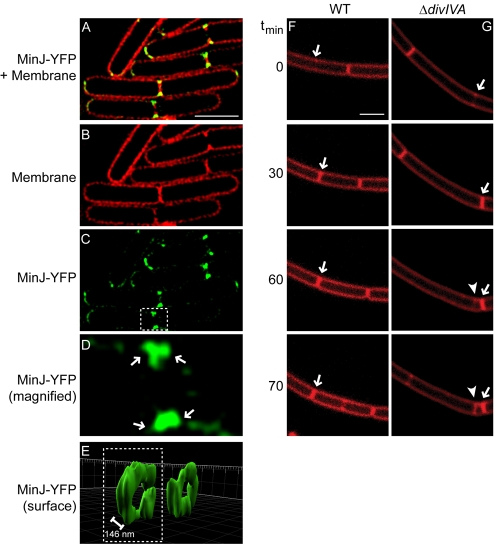
DivIVA binds and recruits the cell division inhibitors MinCD via a recently discovered protein called MinJ (21, 22). Does MinJ also form double rings at division septa? To test this, we examined the localization of MinJ-YFP, produced at native levels, by 3D-SIM (Fig. 5A to E). Like DivIVA, MinJ localized primarily to division septa and, at an intermediate Z plane, appeared as four foci on either side of division septa. Reconstruction of the Z stacks revealed that, similar to DivIVA, MinJ-YFP formed adjacent rings separated by 146 nm ± 16 nm (n = 8), which flanked mature division septa. At newly forming septa that had not finished constriction, doublet MinJ rings were also present but were separated by only 101 nm ± 16 nm (n = 9). Gregory et al. have proposed that the role of MinCD in B. subtilis is to prevent the formation of aberrant septa immediately adjacent and subsequent to recent cell division events (25). Do A rings serve as a platform to recruit MinJ (and by extension MinCD) to prevent incorrect cell division next to recently formed septa? To test this, we examined the frequency of aberrant septum formation in cells that harbored a deletion of divIVA. Whereas in wild-type cells we observed aberrant membrane constriction adjacent to only 2% of recently elaborated septa (n = 246 septa), in ΔdivIVA cells inappropriately positioned septa were elaborated adjacent to 49% (n = 1,150) of active division sites (Fig. 5F and G). Taking these findings together, we conclude that the A rings that flank active division sites and septa provide ring-shaped scaffolds that recruit the Min proteins that prevent cell division from occurring immediately adjacent to recently completed septa.
FIG 5 .
A rings mediate fidelity of cell division by recruiting MinJ to both sides of active division sites. (A) Overlay of FM4-64 and MinJ-YFP fluorescence, viewed by 3D-SIM at an intermediate Z plane (strain DS3609). (B) Membrane stain. (C) MinJ-YFP. (D) Magnification of one septum (white box) in panel C. (E) YFP fluorescence for selected septa in panel C, represented as a three-dimensional surface. Separation between MinJ rings indicated is the average distance between regions of peak fluorescence, ±16 nm (n = 8). Time-lapse images of dividing wild-type (WT) (F) (strain PY79) or ΔdivIVA (G) (strain KR546) cells were visualized using FM4-64. Time (minutes) after the initiation of the experiment is indicated on the left. Arrows indicate recently completed septa; arrowhead indicates a mispositioned septum. Scale bars, 2 µm.
Assembly and stability of A patches at poles.
How does DivIVA form two completely different structures at different locations within a cell (rings at septa versus patches at poles)? In order to investigate the assembly of A patches, we examined the localization of DivIVA-GFP by time-lapse fluorescence microscopy. At the onset of our observations, DivIVA-GFP localized to completed division septa in two adjacent cells, and upon rotation of the image, elaborated A rings were evident (Fig. 6A to E, t0). It is important to note that cytokinesis had already been completed in these cells (as evidenced by the formation of a flat division septum) but that they simply had not separated into individual cells (and therefore had not elaborated a hemispherical pole). Next, the divided cells began to separate (Fig. 6A to E, t5), gradually transforming the local architecture from a flat septum to a hemispherical pole (t5 to t25). Concomitantly, the width of A rings associated with septa in the micrographs decreased (Fig. 6C, compare t0 and t25), suggesting that the structure of the A ring changed over time coincident with the change in the cell’s shape at this site. Reconstruction of Z stacks confirmed this notion (Fig. 6D and E), since A rings steadily decreased in diameter and finally collapsed into a patch at the now mature hemispherical pole. Of course, it is possible that the A patch is still formally a ring, albeit with a central cavity that is too small to be measured by our technique. In any case, in contrast to components of the cell division machinery that decrease in size during cell division to drive the constriction of membranes during cytokinesis, the diameter of A rings remains constant during and after cell division and decreases only in response to changes in the cell’s shape when flat septa become hemispherical poles.
FIG 6 .
Assembly of A patches at hemispherical poles. (A to E) Time-lapse micrographs of DivIVA-GFP localization in strain KR541. (A) DIC image. Arrow indicates a septum whose architecture is represented schematically on the left. (B) Overlay of DivIVA-GFP and DIC. (C) DivIVA-GFP. (D) Indicated region in panel C, magnified and rotated. (E) DivIVA-GFP fluorescence intensity from the deconvolved images in panel D, represented as a three-dimensional surface. Scale bar, 2 µm.
DISCUSSION
The three-dimensional shapes of supramolecular structures are typically predetermined by the amino acid sequences of the proteins that comprise it. Here, we have described a scenario in which the local architecture of the cell instead appears to dictate the supramolecular structure of assembled DivIVA. Using superresolution microscopy, we observed that DivIVA assembled into two adjacent rings that flanked active division sites and complete septa. The cell division protein MinJ, which is recruited by DivIVA, also formed similar double rings that flanked septa. Time-lapse microscopy revealed that as chains of B. subtilis cells separated and as division septa were gradually transformed into hemispherical poles, rings of DivIVA progressively shrank and finally collapsed into a patch. We observed that rings at septa were extremely stable, in contrast to previously described components of the cell division machinery. Taking these findings together, we conclude that the ultrastructure of assembled DivIVA responds to changes in the cell’s shape to coincide with its function.
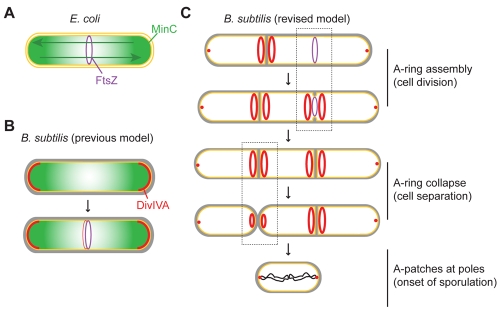
What are the implications of DivIVA localization and assembly for models of cell division in B. subtilis? In E. coli, the cell division inhibitors MinC and MinD oscillate from one pole to the other, creating a time-averaged concentration gradient of MinCD that is highest at the poles and lowest at midcell, where cell division is permissible (Fig. 7A) (39–42). In B. subtilis, MinC does not display a similar oscillatory behavior. Instead, it was initially suggested that MinC stably localized to the extreme poles of the cell and formed a static concentration gradient, wherein the MinC concentration was lowest at midcell, resulting in a role for MinC that was analogous to that in E. coli (Fig. 7B) (23, 26). However, recent data have indicated that B. subtilis MinC localization is dynamic, that it transiently localizes to sites of ongoing cell division rather than to cell poles, and that the primary role of MinCD is to prevent aberrant Z-ring assembly immediately adjacent to newly completed septa (25). The observation that MinC localizes to midcell suggests that its activity must be regulated, since it fails to inhibit FtsZ activity at midcell. The curvature-based DivIVA localization pattern we report here could both spatially and temporally regulate Min activity. Specifically, we propose that DivIVA, which recruits MinCD via MinJ (20–22), interprets a local increase in negative membrane curvature as evidence for the initiation of cell division, thereby localizing only after cell division has commenced (Fig. 7C) and temporally regulating Min activity. Curvature-based localization also spatially regulates Min activity, since the A rings that flank the invaginating septum hold the Min proteins away from FtsZ. We propose that this previously unanticipated spatial regulation is central to the ability of the B. subtilis Min proteins to localize to midcell without inhibiting cell division.
FIG 7 .
Model for the architecture-driven assembly of A rings and A patches. (A) Role of the Min system in establishing the midcell in Escherichia coli. An E. coli cell initiating cell division is depicted (inner and outer membranes, yellow; FtsZ ring, purple; MinC, green), wherein the pole-to-pole oscillation of MinC (green arrows) results in a time-averaged MinC concentration gradient that is minimal at midcell, where FtsZ polymerizes. (B) Previous model of the Min system in B. subtilis. A single B. subtilis cell is depicted (plasma membrane, yellow; cell wall, gray). Analogous to the E. coli model, DivIVA (red) was previously thought to primarily localize to the hemispherical poles, where it sequestered MinC in order to create a static MinC concentration gradient that is minimal at midcell. The fainter localization of DivIVA and MinC to midcell was proposed to occur at a time at which the FtsZ ring was resistant to MinC activity, thereby allowing continued division. (C) Revised model of the role of the Min system in B. subtilis. A chain of B. subtilis cells is depicted in which DivIVA localizes at the onset of membrane constriction and assembles into doublet A rings that recruit MinC (not shown for simplicity) to two rings that flank the active division site. This spatially separates MinC and FtsZ, allowing continued cell division, and mediates the transient localization of MinC adjacent to the newly formed septum, where it can prevent formation of an aberrantly positioned division septum. We therefore propose that the curvature-based DivIVA localization mechanism conveys both temporal and spatial regulation on MinC, preventing it from acting on FtsZ at the active division site. As chains of cells separate, A rings collapse into patches, or perhaps ring with small central openings, as flat septa transform into hemispherical poles.
In keeping with this model, we show that deletion of divIVA results in minicell formation next to newly completed septa, as well as filamentation, which is presumably due to the lack of spatial regulation of Min activity. In cells that harbor deletions of both divIVA and minC, the filamentation phenotype caused by unrestricted MinC activity is corrected and only the minicell phenotype at newly completed septa remains (6, 20, 27). It is tempting to speculate that midcell-localized Min proteins might play a role during as well as immediately after cytokinesis, but determining this will require further experiments. Curiously, time-lapse imaging of E. coli has suggested that as MinCD oscillate from pole to pole, they often pause at midcell in dividing cells (39, 43), suggesting that transient localization during constriction might be evolutionarily conserved.
It is worth noting that unlike MinC, which is only transiently associated with active division sites (25), DivIVA rings remained localized at mature septa long after cytokinesis was finished. The mechanism regulating the transient association of MinC with A rings and MinJ rings is presently unknown. Additionally, once cell division completes, MinC is not rerecruited to A rings at mature poles (25), and the mechanism by which MinC discriminates between DivIVA/MinJ at mature and incomplete septa is also unknown. Perhaps the reason underlying the unusual stability of A rings at septa, though, concerns the second reported function of DivIVA (7, 44). In stationary phase, chains of B. subtilis separate into individual cells and stop dividing symmetrically (45) prior to the onset of sporulation. Replicated chromosomes are then tethered to cell poles in a manner dependent on DivIVA (7), which remains localized and is positioned to carry out this task (Fig. 7, bottom).
The recently solved structure of DivIVA revealed an elongated conformation largely composed of coiled coils (18). An N-terminal phenylalanine that upon dimerization of DivIVA was encircled by positively charged residues was predicted to mediate what is likely an extremely tenuous interaction with the plasma membrane, a hypothesis confirmed by mutagenic analysis. Perhaps only such weak membrane interactions are compatible with the extensive reconfiguration of DivIVA that is required as A rings collapse into A patches.
Similar examples of architecture-driven assemblies of large structures may exist elsewhere in biology. The coiled-coil structure of DivIVA resembles a eukaryotic family of cytoskeletal proteins called tropomyosins (18, 46). The tropomyosin Cdc8 in fission yeast Schizosaccharomyces pombe is a cell division machinery component which also assembles into rings at cell division septa and patches at the poles of the rod-shaped fungus (47). Additionally, when the tropomyosin Tpm1 from budding yeast Saccharomyces cerevisiae was produced in S. pombe, not only did it assemble into rings but it also collapsed into polar patches during the course of mitosis (48), suggesting a conserved strategy that somehow ensures the fidelity of cell division. Perhaps factors involved in cellular processes such as cell division in different species exploit changes in cellular architecture in order to exert their function at the right place and at the right time.
MATERIALS AND METHODS
Strain construction.
Strains, unless otherwise indicated, are isogenic derivatives of B. subtilis PY79 (49). B. subtilis competent cells were prepared as described previously (50). Strain KR515 (amyE::divIVA-gfp cat) was constructed by transforming PY79 with plasmid pKR179 (19). Construction of strains KR528 (ΔsinI::spc amyE::divIVA-gfp cat) and KR541 (amyE::Phyperspank-divIVA-gfp spc) has been described previously (19). KR546 (ΔdivIVA::erm) was created by allelic replacement of the divIVA open reading frame with an erythromycin resistance cassette (51) constructed by the long-flanking PCR method (52). Strain AD3007 expresses FtsZ-GFP from its native promoter in addition to an untagged copy of FtsZ and has been described previously (25). Strain PE97 (divIVAΩ-CFP cat thrC::Pspac-yfp-ftsZ erm) was assembled by transformation of chromosomal DNA from strains DS4152 (21), and AH175 (53) into PY79. Strain PE99 was constructed by transforming PE97 with chromosomal DNA from strain FG345 (54). Construction of minJ fused to yfp (strain DS3609, an isogenic derivative of B. subtilis 3610) has been described previously (21).
Microscopy.
Overnight cultures grown at 22°C in CH medium (55) were diluted 1:20 into fresh CH medium and grown for 2.5 h at 37°C. If necessary, isopropyl-β-d-thioagalactopyranoside (IPTG) was added (1 mM, final concentration) to induce expression of divIVA-gfp, and medium was supplemented with 0.01 mg/ml (final concentration) of the fluorescent dye FM4-64 to visualize membranes. Cells were harvested, resuspended in phosphate-buffered saline (PBS), and placed on a glass-bottom culture dish (MatTek Corp.). A 1% agarose pad made with distilled water (for static images) or CH medium (for time lapse) was cut to size and placed on top of the cell suspension. Cells were viewed with a DeltaVision Core microscope system (Applied Precision) equipped with an environmental control chamber. Images were captured with a Photometrics CoolSnap HQ2 camera. For static images, 17 planes were acquired every 0.2 µm at 22°C and the data were deconvolved using the SoftWorx software program. Three-dimensional surface representations of fluorescence data were made using the Imaris software program. For time-lapse experiments, cells were imaged at 32°C. Entire fields were photobleached by exposure to fluorescein isothiocyanate (FITC) light for 40 s. Cell growth was typically stalled after photobleaching, with a normal growth rate resuming after approximately 60 min. For photobleaching DivIVA-GFP rings, KR541 cells were grown on an agarose pad at 30°C with 500 µM IPTG. Three prebleach images were collected, after which cells were exposed to 0.5 s of 488-nm laser light, and postbleach images were collected for 1 min. For 3D-SIM, cells were grown on an agarose pad and imaged using a Delta Vision OMX version 3 prototype (Applied Precision) (56, 57).
SUPPLEMENTAL MATERIAL
Localization of overproduced DivIVA-GFP. (A and B) Fluorescence from DivIVA-GFP. Membrane stain FM4-64 (C and D) or overlay of GFP and membrane fluorescence (E and F) in strains expressing divIVA-gfp from either the native promoter (A, C, and E) (strain KR515) or the hyperspank promoter induced with 1 mM IPTG (B, D, and F) (strain KR541). Quantification of DivIVA-GFP fluorescence signal at 25 different septa in each strain revealed a 75.5-fold increase for strain KR541 compared to results for strain KR515. Download Figure S1, PDF file, 0.1 MB.
ACKNOWLEDGMENTS
We thank S. Gottesman, R. Losick, and members of our labs for comments on the manuscript and D. Kearns for strain DS3609.
This work was funded by NIH grants R01GM073898 (to J.P.), R01GM084334 (to J.P.), and R01GM57045 (to K.P.) and by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research (to K.S.R.). Support for the OMX microscope was provided by UCSD Neuroscience Core grant P30 NS047101.
Footnotes
Citation Eswaramoorthy P, et al. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. mBio 2(6):e00257-11. doi:10.1128/mBio.00257-11.
REFERENCES
- 1. Berg CA. 2005. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 21:346–355 [DOI] [PubMed] [Google Scholar]
- 2. Margolis HC, Beniash E, Fowler CE. 2006. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 85:775–793 [DOI] [PubMed] [Google Scholar]
- 3. Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60:2356–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rudner DZ, Losick R. 2010. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2:a000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cha JH, Stewart GC. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 8. Ben-Yehuda S, et al. 2005. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17:773–782 [DOI] [PubMed] [Google Scholar]
- 9. Lenarcic R, et al. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Boer PA. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74:504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925 [DOI] [PubMed] [Google Scholar]
- 14. Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothfield L, Taghbalout A, Shih YL. 2005. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3:959–968 [DOI] [PubMed] [Google Scholar]
- 16. de Boer PA, Crossley RE, Rothfield LI. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Boer PA, Crossley RE, Hand AR, Rothfield LI. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliva MA, et al. 2010. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 29:1988–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramamurthi KS, Losick R. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U. S. A. 106:13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marston AL, Errington J. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84–96 [DOI] [PubMed] [Google Scholar]
- 21. Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166–1179 [DOI] [PubMed] [Google Scholar]
- 22. Bramkamp M, et al. 2008. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556–1569 [DOI] [PubMed] [Google Scholar]
- 23. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 24. Bramkamp M, van Baarle S. 2009. Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12:683–688 [DOI] [PubMed] [Google Scholar]
- 25. Gregory JA, Becker EC, Pogliano K. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22:3475–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Baarle S, Bramkamp M. 2010. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One 5:e9850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Edwards DH, Errington J. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905–915 [DOI] [PubMed] [Google Scholar]
- 28. Harry EJ, Lewis PJ. 2003. Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol. Microbiol. 47:37–48 [DOI] [PubMed] [Google Scholar]
- 29. Perry SE, Edwards DH. 2004. Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol. Microbiol. 54:1237–1249 [DOI] [PubMed] [Google Scholar]
- 30. Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739–749 [DOI] [PubMed] [Google Scholar]
- 32. Hamoen LW, Errington J. 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J. Bacteriol. 185:693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levin PA, Kurtser IG, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 96:9642–9647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191:4186–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U. S. A. 99:3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nanninga N, Koppes LJ, de Vries-Tijssen FC. 1979. The cell cycle of Bacillus subtilis as studied by electron microscopy. Arch. Microbiol. 123:173–181 [DOI] [PubMed] [Google Scholar]
- 38. Gustafsson MG. 2005. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. U. S. A. 102:13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu Z, Lutkenhaus J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82–90 [DOI] [PubMed] [Google Scholar]
- 40. Raskin DM, de Boer PA. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raskin DM, de Boer PA. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park KT, et al. 2011. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell 146:396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Juarez JR, Margolin W. 2010. Changes in the Min oscillation pattern before and after cell birth. J. Bacteriol. 192:4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomaides HB, Freeman M, El Karoui M, Errington J. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edwards DH, Thomaides HB, Errington J. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balasubramanian MK, Helfman DM, Hemmingsen SM. 1992. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 360:84–87 [DOI] [PubMed] [Google Scholar]
- 48. Skoumpla K, Coulton AT, Lehman W, Geeves MA, Mulvihill DP. 2007. Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 120:1635–1645 [DOI] [PubMed] [Google Scholar]
- 49. Youngman P, Perkins JB, Losick R. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1–9 [DOI] [PubMed] [Google Scholar]
- 50. Wilson GA, Bott KF. 1968. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 95:1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 52. Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265 [DOI] [PubMed] [Google Scholar]
- 53. Handler AA, Lim JE, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol. 68:588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MG. 2009. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6:339–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gustafsson MG, et al. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Localization of overproduced DivIVA-GFP. (A and B) Fluorescence from DivIVA-GFP. Membrane stain FM4-64 (C and D) or overlay of GFP and membrane fluorescence (E and F) in strains expressing divIVA-gfp from either the native promoter (A, C, and E) (strain KR515) or the hyperspank promoter induced with 1 mM IPTG (B, D, and F) (strain KR541). Quantification of DivIVA-GFP fluorescence signal at 25 different septa in each strain revealed a 75.5-fold increase for strain KR541 compared to results for strain KR515. Download Figure S1, PDF file, 0.1 MB.