Abstract
RNA polymerase II traverses nucleosomes rapidly and efficiently in the cell but it has not been possible to duplicate this process in the test tube. A single nucleosome has generally been found to provide a strong barrier to transcript elongation in vitro. Recent studies have shown that effective transcript elongation can occur on nucleosomal templates in vitro, but this depends on both facilitated uncoiling of DNA from the octamer surface and the presence of transcription factors that maintain polymerase in the transcriptionally competent state. These findings indicate that the efficiency and rate of transcription through chromatin could be regulated through controlled DNA uncoiling. These studies also demonstrate that nucleosome traversal need not result in nucleosome displacement.
Key words: RNA polymerase II, transcript elongation, nucleosome, chromatin, elongation factors, sin mutations
The study of transcript elongation by RNA polymerase II has generated paradoxical results. In the nucleus pol II transcribes genes packaged in nucleosomes with apparent high efficiency at a rate of 3–4 kb/min.1 On the other hand, many studies have shown that during transcription in vitro pol II is unable to traverse even a single nucleosome efficiently.2–5 In the absence of additional factors, pol II alone tends to pause within the first nucleosome it encounters; recovery from these pauses is inefficient and very slow.3,4,6–11
Considering the structure of the nucleosome, it is not surprising that pol II has difficulty in transcribing chromatin templates. The 146 bp of nucleosomal DNA wrapped around the histone octamer core makes 14 consecutive strong contacts with the underlying histones.12 Packaging the template into nucleosomes does result in a greater tendency of pol II to pause at sites which were difficult for polymerase to cross on pure DNA templates.2,3,5 However, the pausing pattern generated by pol II on nucleosomal templates is not simply the sum of increased pausing at established sites plus a 10 bp periodicity of pausing imposed by the nucleosome. Significant new insight into the mechanism of nucleosome traversal has emerged from studies with mononucleosome templates.3–5,13 The strongest barrier for pol II in these nucleosomes occurred at an interesting location: about 45 bp within the nucleosome, where the polymerase is just beginning to invade the template segment organized by the central H3/H4 tetramer.3 Once the pol II crossed the nucleosomal dyad, there was little pausing observed as transcription proceeded to the end of the nucleosome.3 The barriers to pol II imposed by nucleosomes assembled over three strong positioning elements14 were found to be surprisingly polar; that is, traversal was significantly more difficult in one of the two possible orientations.3 Thus, the nucleosomal blockade is not simply proportional to the overall affinity of the underlying DNA for the histone octamer.
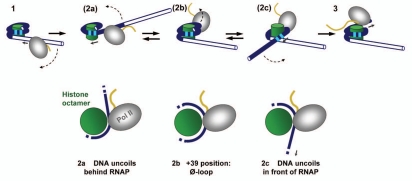
In order to explain all of these observations, the existing crystal structures of the nucleosome and a yeast pol II elongation complex were combined to model the structure of RNA polymerase II paused at various locations within a nucleosome.15 The structure of a complex with pol II halted 39 bp into the nucleosome proved to be particularly informative. This is just upstream of the point that biochemical studies had mapped as the major pause site (See Fig. 1, complex 2a). In the +39 complex, pol II has swung completely away from the octamer surface. Once pol II has transcribed 39 bp into the nucleosome, the upstream DNA can reassociate with the proximal H2A/H2B dimer behind the enzyme. After reassociation, very little DNA is displaced from the histones; thus, this complex was designated a zero-loop (Ø-loop) complex (complex 2b in Fig. 1). The 2b complex showed an unanticipated but critical additional feature: at this position, pol II begins to clash with the DNA binding to the downstream H2A/H2B dimer. For transcription to continue past +39, the template must unwind from the octamer beyond the strong histone-DNA contacts which flank the nucleosome dyad. This downstream unwinding is nucleated by the clash between pol II and the downstream DNA-H2A/H2B dimer interaction (complex 2c, Fig. 1; see ref. 15). Once the downstream template is available, pol II can continue traversal of the nucleosome (step 3 in Fig. 1). This can be accompanied by subsequent formation of a Ø-loop at the position +49 (see Ref. 15).
Figure 1.
Proposed mechanism of transcription through nucleosomes (modified from ref. 15). Pol II enters the nucleosome (1), partially displaces upstream DNA (2a) and initially forms a Ø-loop at +39 (2b), inducing reversible uncoiling of downstream DNA (2c); transcription is accompanied by nucleosome recovery (3). A more detailed description of the uncoiling and traversal pathway is presented in reference 15. Strong DNA-histone interactions which flank the nucleosome dyad are indicated by the squares. Note that the Ø-loop can also be formed at the position +49.
This traversal mechanism has been demonstrated for both yeast and human pol II.3,15 It is frequently accompanied by the loss of a single H2A/H2B dimer but the H3/H4 tetramer is not displaced from its original location.4,16 These features duplicate the apparent effect of pol II passage through nucleosomes in vivo. Within lightly transcribed genes, exchange is observed for H2A/H2B but not for H3 and H4.17–19 Note that a considerably different mechanism, used by RNA polymerase III, involves transfer of a complete histone octamer from in front of the transcribing enzyme to behind it through the obligatory formation of a larger DNA loop.20
Important aspects of the transcription pattern displayed by pol II within nucleosomes in vitro are explained by the model in Figure 1. Details of pausing in the proximal half of the nucleosome vary with sequence, but the strongest stop is always near the position where the proposed metastable Ø-loop complexes form.3 Very little pausing is seen downstream of the nucleosome dyad, consistent with the full unwinding of the downstream template once polymerase has passed the major pause site. The reassociation of template with the upstream histones which begins with the +39 and +49 Ø-loop complexes facilitates retention of the nucleosome even though downstream unwinding must occur to complete nucleosome traversal. Physical studies in which DNA was uncoiled from single nucleosomes21,22 support the importance of the major barrier observed in transcription studies as a particularly difficult point in the process of unwinding the template from the octamer surface.
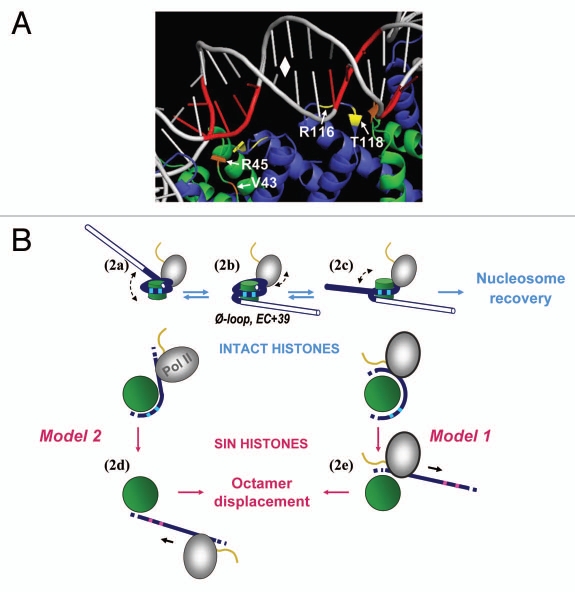
The traversal pathway shown in Figure 1 predicts that alterations in nucleosome structure that facilitate the critical template unwinding event downstream of the Ø-loop should be stimulatory for nucleosome traversal. This idea has recently been tested by substituting H3 or H4 histones with corresponding Sin mutant versions. The Sin mutations are single amino acid changes in H3 or H4, originally identified in yeast because they remove the requirement for chromatin remodelers at certain genes.23 Nucleosomes assembled with vertebrate analogs of Sin mutant histones show specific loss of histone-DNA contacts immediately flanking the nucleosome dyad.24 These contacts (squares in Figs. 1 and 2B) are the strongest of the 14 major histone-DNA interaction points in the nucleosome.22,24 Breaking these interactions is an important part of the overall template unwinding that occurs during traversal (Fig. 1). As predicted, it was found that the incorporation of four different Sin mutant H3 or H4 histones caused significant reduction in the nucleosomal transcription barrier for both yeast and human pol II.25
Figure 2.
Proposed mechanism of transcription through Sin nucleosomes (modified from ref. 25). (A) DNA-histone interactions (from refs. 24 and 44) affected by the particular Sin mutations investigated in reference 25. Histone H3 residues R116 and T118, and H4 residues V43 and R45 are indicated by white arrows. White diamond indicates the nucleosome dyad. (B) Pol II enters the nucleosome, partially displaces upstream DNA (2a) and forms a Ø-loop at +39 (2b), inducing reversible uncoiling of downstream DNA (2c); transcription is accompanied by nucleosome recovery. However, strong DNA-histone interactions (squares) are weakened in Sin nucleosomes, causing a larger downstream DNA region to be displaced (2d and 2e), favoring nucleosome loss.
Weakening of the dyad-proximal histone-DNA contacts can also affect the survival of the nucleosome upon traversal in vitro. While the majority of nucleosomes are not displaced by transcription, an increased tendency for the nucleosome to be released from the DNA was observed when Sin mutant histones were incorporated into the octamer.25 Two related mechanisms, shown schematically in Figure 2B, can be envisioned to explain this effect. Retention of the nucleosome as pol II advances from the Ø-loop position at +39 is dependent on the continuous reassociation of upstream template onto the octamer surface as pol II begins to displace the downstream DNA away from the histones.15 If complete downstream uncoiling occurs at position +39, as may be the case with Sin mutant histones, then the octamer is more likely to be displaced into solution (Model 1, Fig. 2B). This is consistent with footprinting results with bacterial RNA polymerase halted at the relevant location within nucleosomes assembled from wild type or Sin mutant histones.25 An alternative possibility is that full dissociation of the downstream DNA could occur on Sin mutant nucleosomes before any stable reassociation of the upstream DNA with the octamer has occurred (Model 2, Fig. 2B). Transcription-dependent loss of nucleosomes in vivo due to Sin-like mutations has been reported for one Sin mutant.26
The pathway of template uncoiling away from the octamer surface seems to be one key to understanding nucleosome traversal, since facilitating this pathway clearly facilitates transcription through the nucleosome. However, it is also clear that the nucleosome alterations tested to date do not render the nucleosome transparent to pol II.10,25 At physiological ionic strength, traversal of even the most permissive single nucleosomes by pol II remains incomplete over a time course of 5 min; also, the rate at which full traversal is achieved is distinctly slower than elongation rates on the equivalent pure DNA templates.6,10,25 Significantly, those polymerases that fail to complete traversal are often trapped at strong, nucleosome-specific pause sites. At these locations, the initial pause is followed by extensive backtracking along the template,5 which separates the transcript 3′ end from the active site and leaves pol II arrested and transcriptionally inactive. It has been shown that pausing precedes backtracking and arrest,5 which implicates DNA uncoiling from the octamer as the primary part of the barrier. In this context, it is important to note that DNA can “breathe” away from the nucleosome surface.27 This transient disassociation would reveal the downstream template, allowing pol II to avoid pausing and continue transcription. However, the rate at which this “window of transcriptional opportunity” recloses is considerably faster than the bond formation rate for pol II in vitro in the absence of additional factors.6,11 Thus, it seems likely that successful nucleosomal traversal will also depend on transcript elongation factors which assist pol II in exploiting template uncoiling.
It is well established that recovery from backtracking and arrest on pure DNA templates by pol II requires the transcript elongation factor TFIIS.28,29 This factor stimulates the backtracked polymerase to cleave the nascent RNA adjacent to the active site, thereby regenerating an extendable 3′ end.29–31 A number of studies have demonstrated that TFIIS does stimulate nucleosome traversal by pol II.3,5,6,9,11,32 However, even at saturating levels of TFIIS, nucleosome traversal remains slow and incomplete.11 If the primary difficulty for pol II during nucleosome traversal is transcriptional arrest from backtracking, TFIIS should be sufficient to drive pol II completely through nucleosomes as TFIIS is able to do at arrest sites on pure DNA templates.28,33 To appreciate why TFIIS alone might not support effective nucleosome traversal, it is important to recall that the transient disassociation of DNA from the octamer surface which could allow traversal27 reverses more rapidly than the average rate of bond formation by pol II alone.11 Thus, when pol II recovers from nucleosomal arrest through the action of TFIIS, it is likely that it will be unable to continue transcription quickly enough and instead will backtrack and arrest again. The difficulty of resumption of transcription after backtracking is probably increased on nucleosomal templates because of the tendency of the template to reassociate with the octamer core as the polymerase retreats upstream.
These considerations indicate that additional factors, directed at increasing the rate of bond formation by pol II, may be necessary in addition to TFIIS to drive efficient nucleosome traversal. One likely candidate for this role is the general transcript initiation factor TFIIF, which also significantly increases bond formation rates during transcript elongation6,11,34–36 by generally suppressing pausing by pol II.37 Significantly, it has been shown in single bond formation assays that TFIIF and TFIIS cooperate to maximize the rate of transcription.37 Thus, it would be expected that TFIIF will substantially increase the effectiveness of TFIIS in facilitating nucleosome traversal by pol II. In a recent test with two mononucleosomal templates, this has proven to be the case. TFIIS and TFIIF together provided additive increases in traversal and when combined with the presence of a single Sin mutant histone, they supported complete traversal of a nucleosome at rates within a factor of 2 of those seen on the equivalent pure DNA templates.11 Note that in metazoans, both TFIIF and TFIIS have been shown to be associated with the bodies of active genes.32,38,39
Backtracking by pol II at critical nucleosomal arrest sites could also be countered by a factor-independent mechanism involving a second RNA polymerase. The trailing polymerase could directly reverse backtracking as it collides with the stalled downstream polymerase. This should increase traversal by restarting the stalled polymerase, as is seen with TFIIS. It should also lead to nucleosome loss because, unlike the single nucleosome case, the upstream DNA that should reassociate and anchor the template on the nucleosome (Fig. 1 and transition from complex 2a to 2b) is instead taken up by the trailing polymerase. Recent in vitro studies40,41 confirm this prediction, which is consistent with the in vivo observation that nucleosomes are lost within highly transcribed genes but not within less active transcription units.17,42,43
In summary, the nucleosome is not an insurmountable barrier to transcription in the test tube. Investigations of transcription of mononucleosome templates, along with biophysical studies, have shown that the nucleosome is poised for successful traversal through the spontaneous breathing of the template away from the octamer surface and the uncoiling of the DNA in response to invasion by pol II. The nucleosomal barrier can be overcome when template uncoiling is facilitated and factors are available to maximize pol II's catalytic competence. It is important to emphasize that the extent of DNA uncoiling from the octamer during transcription through a nucleosome is large relative to the interactions that remain, so further small destabilizations or more efficient transcription can strongly affect the rate of traversal. For the immediate future, there are several clear challenges in advancing the study of transcription of chromatin templates in vitro. It will be essential to obtain a better understanding of how template uncoiling from the nucleosome surface is driven in the cell. It will also be important to extend these approaches to more physiological templates consisting of arrays of nucleosomes. This will allow us to address the importance of nucleosome-nucleosome interactions in controlling nucleosome traversal by pol II.
Acknowledgements
This work was supported by NSF 0743298 grant to D.L. and Government of the Russian Federation (order #220) and NIH GM58650 grants to V.M.S.
References
- 1.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 3.Bondarenko VA, Steele LM, Ujvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 5.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 6.Izban MG, Luse DS. Factor-Stimulated RNA polymerase-II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 7.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 9.Guermah M, Kim J, Roeder RG. Transcription of in vitro assembled chromatin templates in a highly purified RNA polymerase II system. Methods. 2009;48:353–360. doi: 10.1016/j.ymeth.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ujvari A, Hsieh FK, Luse SW, Studitsky VM, Luse DS. Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J Biol Chem. 2008;283:32236–32243. doi: 10.1074/jbc.M806636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luger K, Mäder A, Sargent DF, Richmond TJ. The atomic structure of the nucleosome core particle. J Biomol Struct Dynam. 2000:185–188. doi: 10.1080/07391102.2000.10506619. [DOI] [PubMed] [Google Scholar]
- 13.Studitsky VM, Clark DJ, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 14.Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 15.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 17.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 19.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 21.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, et al. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/ SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 24.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, et al. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 2010;11:705–710. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung V, Chua G, Batada NN, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome-art. no. e277. Plos Biol. 2008;6:2550–2562. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 28.Reines D, Ghanouni P, Li QQ, Mote J. The RNA polymerase-II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- 29.Izban MG, Luse DS. The RNA Polymerase-II ternary complex cleaves the nascent transcript in a 3′→5′ direction in the presence of elongation factor-SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 30.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/ TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izban MG, Luse DS. The Increment of SII-facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase-IIternary complexes. J Biol Chem. 1993;268:12874–12885. [PubMed] [Google Scholar]
- 34.Price DH, Sluder AE, Greenleaf AL. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kephart DD, Wang BQ, Burton ZF, Price DH. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 36.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFbmediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang CF, Yan HG, Burton ZF. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen and stimulatory factor II. J Biol Chem. 2003;278:50101–50111. doi: 10.1074/jbc.M307590200. [DOI] [PubMed] [Google Scholar]
- 38.Cojocaru M, Jeronimo C, Forget D, et al. Genomic location of the human RNA polymerase II general machinery: evidence for a role of TFIIF and Rpb7 at both early and late stages of transcription. Biochem J. 2008;409:139–147. doi: 10.1042/BJ20070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Fairley JA, Roberts SGE. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci USA. 2010;107:11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, et al. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol. 2010;17:745–752. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 43.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]