Abstract
Trans-translation is a universal quality-control process eubacteria use to degrade incompletely synthesized proteins and rescue ribosome stalled on defective mRNAs. This process is facilitated by a ribonucleoprotein complex composed of transfer-messenger RNA (tmRNA)—a chimera made of a tRNA-like molecule and a short open reading frame (ORF)—and small protein B (SmpB). Determination of the structure of tmRNA and SmpB in complex with the ribosome, at the stage when translation has resumed on tmRNA, has provided an increased understanding of the structure of tmRNA as it transits the ribosome and unique insights into the complex mechanism of template switching on the ribosome and SmpB-driven selection of the correct reading frame on tmRNA's ORF.
Key words: ribosome, translational regulation, trans-translation, tmRNA, SmpB, pseudoknot
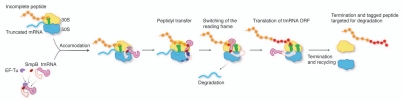
In eubacteria, translation of a truncated mRNA stalls the ribosome at the end of the mRNA, which in the absence of a rescue mechanism would result in an incomplete, potentially toxic polypeptide and render the ribosome unable to recycle. The stalled ribosome is rescued by an intervention by the transfer-messenger RNA (tmRNA) and its protein partners (Fig. 1).
Figure 1.
Schematic of the trans-translation process. The complex formed by tmRNA•SmpB and EF-Tu recognizes ribosome stalled by truncated mRNA, and binds to the empty ribosomal A site. Upon accommodation, EF-Tu leaves the ribosome, and the tmRNA accepts the incomplete polypeptide through the peptidyl transfer reaction. Next, tmRNA•SmpB is translocated to the ribosomal P site, while the reading frame of translation is switched from the truncated mRNA to the open reading frame (ORF) of the tmRNA. Translation resumes until ribosome reaches the stop codon on the ORF. Termination and recycling follow to release the tagged peptide and recycle the ribosome.
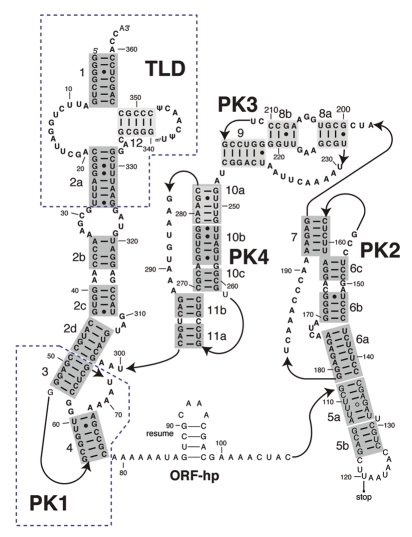
In E. coli, the tmRNA is a 363-nucleotide RNA molecule that consists of several structured domains (Fig. 2). The tRNA-like domain (TLD), which lacks an anti-codon stem-loop when compared to a canonical tRNA, is readily recognized by alanyl-tRNA synthetase and charged with an alanine. It connects to the rest of the tmRNA through the helix 2a. TLD from Aquifex aeolicus and Thermus thermophilus has been co-crystallized with SmpB.1,2 These crystal structures confirmed earlier reports that TLD has an ∼120° inter-helix angle,3 as opposed to ∼90° exhibited in the tRNA. The position of SmpB in these structures suggests that it acts as the anti-codon arm of a tRNA while the linker helix 2a mimics the long variable arm of a class II tRNA. SmpB is indispensable for all activities of tmRNA.4–11
Figure 2.
Secondary structure of the E. coli transfer-messenger RNA (tmRNA). Landmarks: TLD, tRNA-like domain; PK1-4: pseudoknots 1–4; ORF-hp, open reading frame hairpin; resume, resume codon; stop, stop codon. Labels 1–12 next to the helices refer to the standard numbering of the helices.
The ORF constitutes an essential segment of the mRNA-like domain (MLD) and encodes 10 residues of a degradation tag in E. coli. The latter connects to the TLD through four pseudoknots, designated PK1 through PK4. Only the three-dimensional structure of PK1 is presently known at atomic resolution.12
Upon recognizing the stalled ribosome, the tmRNA•SmpB complex, with the help of EF-Tu and GTP, binds to the ribosome to occupy the empty A site on the 30S subunit.13,14 The structure of this so-called pre-accommodation complex has been solved by cryo-EM.14–16 In these studies, the complex of tmRNA•SmpB and the ribosome was formed in vitro in the presence of kirromycin, an antibiotic that inhibits hydrolysis of GTP and stalls the EF-Tu•GDP on the ribosome. These maps show a striking arc structure, which is composed of the pseudoknots, curving around the beak of the 30S subunit and the mRNA entry channel. As in the pre-accommodation state in canonical translation, the complex formed by TLD, SmpB and EF-Tu•GTP adopts the A/T configuration, and EF-Tu interacts both with the GTPase-associated center (GAC) of the ribosome and the elbow region of the TLD.17
Next, the complex enters the accommodation state, in which the EF-Tu has left the ribosome. The structure of this accommodated complex has been solved by cryo-EM,15,16,18,19 as well. While in the earlier studies only part of the tmRNA was visualized,15,16 the recent studies used more sophisticated classification methods and obtained structures with well-resolved density of the entire tmRNA molecule.18,19 Compared to the pre-accommodation structure, the TLD•SmpB is completely settled into the ribosomal A site, and the ORF has moved closer to the neck of the 30S, apparently ready to be inserted into the mRNA entry channel.
Subsequently, TLD•SmpB is translocated to the ribosomal P site, and the ORF of the tmRNA enters the mRNA entry channel to displace the defective mRNA molecule.20,21 Translation resumes on the ORF and continues until the ribosome reaches the stop codons on the ORF. The nascent polypeptide chain is then released and the ribosome dissociated,22 probably following the normal course of termination and recycling. Through the addition of the tag encoded by the ORF, the released polypeptide is marked for degradation.
The stages described in the last paragraph have been studied extensively using genetic and biochemical approaches. How the correct reading frame is established, at the point where the ribosome switches from the stalled mRNA to the ORF of the tmRNA, has remained a crucial question in the study of trans-translation. Over the last few years, researchers have focused on the interaction between SmpB and the region upstream of the tmRNA resume codon. This region of an eleven nucleotide-long single-stranded RNA has been shown to bind to E. coli SmpB in vitro.23,24 Several studies have demonstrated that mutations of the five nucleotides preceding the resume codon—the -1 triplet and the U85 and A86 nucleotides—induced frame-shifting in the translation of the ORF, and reduced or even completely abolished tmRNA tagging activity.21,24–28 Moreover, it has been shown that certain mutations in SmpB restore tagging activity of tmRNA mutated in the A86 nucleotide.29,30 These studies suggested that the interaction of the SmpB with the upstream nucleotides plays a critical role in the selection of the correct reading frame.
Another major question to be answered in the studies of trans-translation is how tmRNA passes through the ribosome despite its complex topology and the constraints imposed by the tight intersubunit space. The cryo-EM structure of the tmRNA-ribosome complex in the resume state (the state in which the TLD has moved to the ribosomal P site and translation is ready to resume on the ORF of the tmRNA), obtained independently by our group and the Gillet group, addressed both of these questions.19,31
The two structures, published back-to-back, are from two different organisms, E. coli (our group) and T. thermophilus (the Gillet group). Our complex was formed in vivo. In order to facilitate its purification, we inserted a MS2 hairpin into the helix 5 of the tmRNA. In contrast, Weis and his coworkers assembled the tmRNA:ribosome complex in vitro. By binding EF-G to the accommodated state complex, they induced translocation of the A site-bound tmRNA•SmpB to the P site. These approaches produced complexes with the TLD•SmpB accommodated in the P site but evidently illustrate different states of the resume complex. In our structure the A site is occupied by a tRNAAla, while it is empty in the Weis complex. The opposite is true for the E site, which is empty in our structure but occupied in the complex created by Weis and coworkers. Thus, the in vitro-formed complex might be depicting an early state of the resume complex, where translocation of TLD•SmpB has been completed and the A site is ready to accept the aminoacylated tRNA. In the in vivo-formed complex, apparently, the presence of the A site-bound tRNA implies that the E-site tRNA had already dissociated from the ribosome.
In comparing the results of the two studies, we note that there is evidence of greater residual heterogeneity in the study of Weis and coworkers, which may be linked to differences in the purification and classification methods employed. A telltale of heterogeneity is resolution achieved with a given number of particles [13.6 Å with 20,873 particles, as opposed to 18.4 Å (FSC = 0.5 applied to the published19 FSC curve) with 70,761]. As a consequence, the cryo-EM density maps differ somewhat in the degree of detail, especially in the density attributable to the tmRNA, but the atomic structures inferred for the linked complex of tmRNA and SmpB (PDBID: 3IZ4;29 PDBID: 3IYR27) are nevertheless in remarkable agreement.
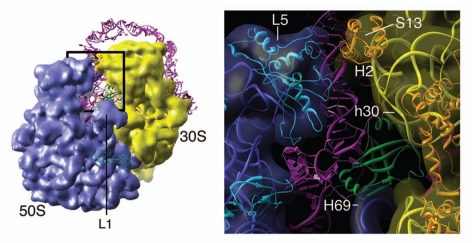
Of particular interest is the region surrounding the ribosomal P site, which is occupied by TLD•SmpB. After fitting all the helices and pseudoknots into the cryo-EM maps, the single-stranded RNA, including the ORF and its upstream region, was modeled taking into account the structural constraints imposed by the ribosomal environment and the presence of the resume codon and tRNA in the A site. Consistent with the biochemical results, the density of SmpB is seen to be in contact with nucleotides upstream of the resume codon, the helix 69 of the 23S rRNA and helix 30 of the 16S rRNA (Fig. 3). Such pattern of interaction is consistent with previous biochemical studies.34 The E. coli SmpB contains a ∼20-amino acids-long C-terminal tail that is not present in the SmpB from T. thermophilus. In our density map, the C-terminal tail of the SmpB appears to interact with the region upstream of the resume codon, as the crystal structure of the T. thermophilus SmpB cannot account for all the density that makes contact with the upstream region's nucleotides. The resolution of our map is not sufficient to infer the structure of the C-terminal tail, but, based on secondary structure prediction, it is very likely that the tail is a long alpha helix. According to our cryo EM map and atomic model, the five nucleotides upstream of the resume codon (-1 triplet, U85 and A86) are likely to interact with the loop preceding the C-terminal tail.
Figure 3.
The interactions between the ribosome, tmRNA and SmpB at the point where the tRNA-like domain/SmpB is bound at the ribosomal P site. Left, cryo-EM density map of the resume state ribosome, superimposed with the atomic model of the ribosome (PDBID: 3FIH and 3FIK) and the modeled structure of tmRNA/SmpB (PDBID: 3IZ4). Right, zoomed-in view of the outlined region on the left. The 23S rRNA is colored in blue, 16S rRNA in yellow, large subunit ribosomal proteins in cyan, small subunit ribosomal proteins in orange, tmRNA in magenta and SmpB in green. Landmarks: L5, ribosomal protein L5; S13, ribosomal protein S13; h30; helix 30 of 16S rRNA; H69, helix 69 of 23S rRNA; H2, helix2 of tmRNA.
Our model also suggests the possible existence of an interaction between the 85UA86 nucleotides and a segment encompassing residues 18 through 24. Because tmRNA activity is relatively tolerant to modifications of the upstream nucleotides,25 we have proposed that these interactions could rely partly on contacts established with their backbone. Actually, the model published by Weis and coworkers suggests a similar mode of interaction between the SmpB and the five nucleotides upstream of the ribosome. However, we noticed that the secondary structure of the SmpB in the Weis model is distorted to some extent. In fact, the proportion of Ramachandran outliers of the Weis model, estimated using MolProbity,35 is ∼12%, while that of ours is less than 1%. This distortion of the SmpB structure, possibly a result of the use of Molecular Dynamics Flexible Fitting on the lower-resolution structure, implies that the interaction pattern suggested in the Weis study may be associated with some uncertainty.
Based on the published structure of the tmRNA-ribosome complex in the accommodated state,18,19 the aforementioned interactions between SmpB and the five nucleotides upstream of the resume codon are unlikely to exist when the TLD•SmpB complex occupies the A site. Thus, they must be established during the translocation of the TLD•SmpB from the A to the P site. The structural alignment analysis of the tmRNA suggests that the resume codon may constitute a part of the loop capping a small hairpin (referred to as ORF-hp in Fig. 2). We speculate that such a structure might exist in both the pre-accommodation and accommodated states, where the arc opening is narrower than in the resume state, so that the single-stranded part of the ORF does not need to be stretched. We believe this hairpin structure may play a role in resuming the stalled translation by initiating the interaction with SmpB.
The other important question, of how tmRNA passes through the ribosome sterically and dynamically, remains largely open. Our cryo-EM studies show that tmRNA maintains a stable structure at least up to the resume state. In both resume complexes, helix 2 of the tmRNA has passed the intersubunit bridge B1a and inserted itself into the space between the central protuberance of the 50S subunit and the head of the 30S subunit, while PK1 remains on the other side of bridge B1a. As for the next step of translocation, for TLD to move from the ribosomal P site to the E site, the biggest hurdle would be for helix 2 to pass through the intersubunit bridge B1b, formed by ribosomal protein S13 and L5. We note that in our resume complex, the head of the 30S subunit tilts slightly toward the solvent side. This displacement breaks bridge B1b to create enough space for the helix 2 to pass through the ribosome (Fig. 3). During its transit through the ribosome, helix 2 is expected to interact with both flanking proteins S13 and L5, with its second internal loop made up by mismatched base pairs. This part of helix 2 is thinner than a regular RNA helix made up by canonical base pairs and thus it is plausible that helix 2 can pass through the bridge B1b without unfolding.
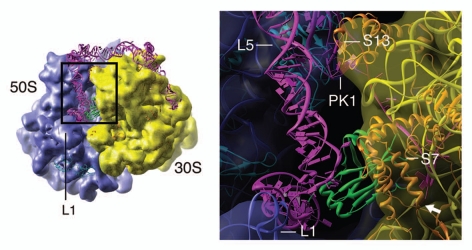
To examine the potential interactions past this point, we moved the tmRNA model to make its TLD•SmpB structure fit into the canonical E-site tRNA36 (Fig. 4). We note that in the resulting position of tmRNA, SmpB presents the -1 triplet right at the intersubunit side of the mRNA exit channel, allowing the insertion of the single-stranded region of the tmRNA into this channel. In fact, SmpB comes to lie very close to ribosomal protein S7, which surrounds the mRNA exit channel, suggesting that SmpB might interact with S7 to facilitate the insertion. The TLD, just like a canonical E-site tRNA,37 would interact with the L1 stalk through the elbow region. This interaction could facilitate the exit of TLD•SmpB, presumably in the same manner as in canonical translation.37 We also note that in the proposed model, PK1 would experience a number of clashes with ribosomal proteins S13 and L5, even if it could pass through the B1a bridge in its entirety. Although we cannot exclude the possibility that the head of the 30S subunit has enough flexibility to open up more, we believe that at this point PK1 unfolds at least partially. This suggestion is consistent with recent observations that PK1 can be replaced by a hairpin without affecting tmRNA tagging activity.38,39 The unfolding could also make the insertion of the single-stranded region of tmRNA into the mRNA exit channel easier.
Figure 4.
Interaction between the ribosome, tmRNA and SmpB inferred from a fitting of the tRNA-like domain/SmpB into the ribosomal E site according to the positioning of the canonical E-site tRNA (PDBID: 3FIH34). Left, cryo-EM density map of the resume state ribosome, superimposed with ribosome model (PDBID: 3FIH, 3FIK34) and the proposed tmRNA/SmpB model that binds at the ribosomal E site. Right, zoomed-in view of the region outlined on the left. The various components of the structure are colored in the same way as in Figure 3. The white arrow, in the lower right corner of the right panel, indicates the mRNA exit channel. Landmarks: L1, L1 stalk of the large subunit; L5, ribosomal protein L5; S13, ribosomal protein S13; S7, ribosomal protein S7; PK1, pseudoknot PK1 of the tmRNA.
In future studies, our model could be used to guide mutation studies to further investigate the mechanism of reading frame selection. In addition, improving resolution of this complex could help in building more precise models, and would be particularly beneficial for studying the structure and function of the C-terminal tail of the E. coli SmpB. To investigate the question of how tmRNA passes through the intersubunit space, we need to obtain the structure of the ribosome-tmRNA complex in the later stages of trans-translation. These complexes are perhaps easier to form in vitro. However, as we discussed in our original publication,31 the insertion of the MS2 hairpin in our tmRNA construct might have over-stabilized the helix 5 that contains part of the ORF, resulting in the vast majority of our in vivo-formed complexes being stalled at the resume state. Thus, to form the complex that depicts later stages of trans-translation in vivo, we could probably insert one or more codons into the single-stranded part of the ORF. This might just allow the TLD to translocate to the ribosomal E site, or any later stages of interest.
Acknowledgements
We thank Melissa Thomas for assistance in the preparation of the illustrations. This work was supported by HHMI, NIH R01 GM29169 and R01 GM55440 (to J.F.), P01 GM064692 (to Robert M. Glaeser) and R01 GM58267 (to J.W.).
References
- 1.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 2.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, et al. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci USA. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stagg SM, Frazer-Abel AA, Hagerman PJ, Harvey SC. Structural studies of the tRNA domain of tmRNA. J Mol Biol. 2001;309:727–735. doi: 10.1006/jmbi.2001.4632. [DOI] [PubMed] [Google Scholar]
- 4.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wower IK, Zwieb C, Wower J. Contributions of pseudoknots and protein SmpB to the structure and function of tmRNA in trans-translation. J Biol Chem. 2004;279:54202–54209. doi: 10.1074/jbc.M410488200. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Ueda T. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. J Biol Chem. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- 7.Wower J, Zwieb CW, Hoffman DW, Wower IK. SmpB: a protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 8.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci USA. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nameki N, Someya T, Okano S, Suemasa R, Kimoto M, Hanawa-Suetsugu K, et al. Interaction analysis between tmRNA and SmpB from Thermus thermophilus. J Biochem. 2005;138:729–739. doi: 10.1093/jb/mvi180. [DOI] [PubMed] [Google Scholar]
- 10.Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nameki N, Chattopadhyay P, Himeno H, Muto A, Kawai G. An NMR and mutational analysis of an RNA pseudoknot of Escherichia coli tmRNA involved in trans-translation. Nucleic Acids Res. 1999;27:3667–3675. doi: 10.1093/nar/27.18.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudinger-Thirion J, Giege R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 15.Weis F, Bron P, Rolland JP, Thomas D, Felden B, Gillet R. Accommodation of tmRNA-SmpB into stalled ribosomes: a cryo-EM study. RNA. 2010;16:299–306. doi: 10.1261/rna.1757410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc Natl Acad Sci USA. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 18.Cheng K, Ivanova N, Scheres SH, Pavlov MY, Carazo JM, Hebert H, et al. tmRNA•SmpB complex mimics native aminoacyl-tRNAs in the A site of stalled ribosomes. J Struct Biol. 2010;169:342–348. doi: 10.1016/j.jsb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Weis F, Bron P, Giudice E, Rolland JP, Thomas D, Felden B, et al. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO J. 2010;29:3810–3818. doi: 10.1038/emboj.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova N, Pavlov MY, Ehrenberg M. tmRNA-induced release of messenger RNA from stalled ribosomes. J Mol Biol. 2005;350:897–905. doi: 10.1016/j.jmb.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Williams KP, Martindale KA, Bartel DP. Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 23.Metzinger L, Hallier M, Felden B. The highest affinity binding site of small protein B on transfer messenger RNA is outside the tRNA domain. RNA. 2008;14:1761–1772. doi: 10.1261/rna.1185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konno T, Kurita D, Takada K, Muto A, Himeno H. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA. 2007;13:1723–1731. doi: 10.1261/rna.604907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MR, Healey DW, Robison SG, Dewey JD, Buskirk AR. The role of upstream sequences in selecting the reading frame on tmRNA. BMC Biol. 2008;6:29. doi: 10.1186/1741-7007-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Ishii M, Tadaki T, Muto A, Himeno H. Determinants on tmRNA for initiating efficient and precise trans-translation: some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA. 2001;7:999–1012. doi: 10.1017/s1355838201010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim VI, Garber MB. Analysis of recognition of transfer-messenger RNA by the ribosomal decoding center. J Mol Biol. 2005;346:395–398. doi: 10.1016/j.jmb.2004.11.061. [DOI] [PubMed] [Google Scholar]
- 28.Trimble MJ, Minnicus A, Williams KP. tRNA slippage at the tmRNA resume codon. RNA. 2004;10:805–812. doi: 10.1261/rna.7010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts T, Cazier D, Healey D, Buskirk A. SmpB Contributes to reading frame selection in the translation of transfer-messenger RNA. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watts T, Healey D, Jones D, Buskirk A. The role of tmRNA nucleotides and the SmpB protein in setting the translational frame on tmRNA. FASEB J. 2008;22:224. [Google Scholar]
- 31.Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, et al. Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J. 2010;29:3819–3825. doi: 10.1038/emboj.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 33.Fu J, Gao H, Frank J. Unsupervised classification of single particles by cluster tracking in multi-dimensional space. J Struct Biol. 2007;157:226–239. doi: 10.1016/j.jsb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Hallier M, Desreac J, Felden B. Small protein B interacts with the large and the small subunits of a stalled ribosome during trans-translation. Nucleic Acids Res. 2006;34:1935–1943. doi: 10.1093/nar/gkl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 38.Wower IK, Zwieb C, Wower J. Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro. RNA. 2009;15:128–137. doi: 10.1261/rna.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanner DR, Dewey JD, Miller MR, Buskirk AR. Genetic analysis of the structure and function of transfer messenger RNA pseudoknot 1. J Biol Chem. 2006;281:10561–10566. doi: 10.1074/jbc.M600167200. [DOI] [PubMed] [Google Scholar]