Abstract
A programmed ribosomal frameshift (PRF) in the decoding of APC (adenomatous polyposis coli) mRNA has been identified and characterized in caenorhabditis worms, Drosophila and mosquitoes. The frameshift product lacks the C-terminal approximately one-third of the product of standard decoding and instead has a short sequence encoded by the -1 frame which is just 13 residues in C. elegans, but is 125 in D. melanogaster. The frameshift site is A AAA AAC in Caenorhabditids, fruit flies and the mosquitoes studied while a variant A AAA AAA is found in some other nematodes. The predicted secondary RNA structure of the downstream stimulators varies considerably in the species studied. In the twelve sequenced Drosophila genomes, it is a long stem with a four-way junction in its loop. In the five sequenced Caenorhabditis species, it is a short RNA pseudoknot with an additional stem in loop 1. The efficiency of frameshifting varies significantly, depending on the particular stimulator within the frameshift cassette, when tested with reporter constructs in rabbit reticulocyte lysates. Phylogenetic analysis of the distribution of APC programmed ribosomal frameshifting cassettes suggests it has an ancient origin and raises questions about the possibility of synthesis of alternative protein products during expression of APC in other organisms such as humans. The origin of APC as a PRF candidate emerged from a prior study of evolutionary signatures derived from comparative analysis of the 12 fly genomes. Three other proposed PRF candidates (Xbp1, CG32736, CG14047) with switches in conservation of reading frames are likely explained by mechanisms other than PRF.
Key words: adenomatous polyposis coli, programmed ribosomal frameshifting, recoding, RNA pseudoknot, translational control, alternative decoding
Introduction
Diversion of a proportion of translating ribosomes to an alternative reading frame is utilized in the decoding of a substantial number of mobile genetic elements including viruses.1,2 This frameshifting is generally programmed in the sense that it is evolutionarily conserved and required for the synthesis of a particular functional protein or a variant protein with a distinct function. The efficiency of programmed ribosomal frameshifting (PRF) is often elevated above the background level by stimulatory elements, frequently called recoding signals, embedded in the mRNA. Many examples of programmed ribosomal frameshifting (PRF) are known in the decoding of viral genes and mobile chromosomal elements such as in several families of bacterial insertion sequences,3 in the S. cerevisiae Ty elements,4 as well as in diverse higher eukaryotic retrotransposons.5 However, only few cases of PRF are known in the de coding of non-mobile cellular chromosomal genes.
The characteristics of frameshift sites and recoding signals are relevant to identifying new cases of PRF. Most −1 PRF sites are easier to identify than their +1 counterparts due to commonalities in the frameshift site that usually consists of a heptameric nucleotide sequence that allows dissociation of codon: anticodon pairing in both the ribosomal P- and A-sites and reassociation to mRNA in the −1 frame.6–9 However, some exceptions are known when the shift site involves just a tetrameric10 or a hexameric nucleotide sequence.11
Such patterns are often accompanied by specific 3′ stimulatory signals in the form of secondary RNA structures such as stem loops or pseudoknots, separated from the shift site by 5 to 9 bases.12,13 In some instances, however, a distal component also participates in conjunction with the 3′ proximal sequence.14 A number of computational approaches for the identification of PRF have been developed.15 Those that predict PRF in homologs of known PRF genes achieve high sensitivity.16,17 However, elucidation of novel instances of PRF (where no PRF homologs are known) remains difficult. A direct search for sequence patterns known to be shift-prone may reveal numerous novel sites of ribosomal frameshifting, as has been shown in both prokaryotes18 and eukaryotes.19 While such instances of frameshifting may be biologically relevant (e.g., for fine-tuning mRNA turnover20,21), we use the term ‘programmed ribosomal frameshifting’ to refer to cases where frameshifting has a more specific function (e.g., required for synthesis of a distinct functional product) and is subject to purifying selection as evidenced by phylogenetic conservation of the frameshift site. ‘Unprogrammed’, but nonetheless operational, sites of ribosomal frameshifting have been shown to have destabilizing effects on mRNAs.21 However it remains to be determined whether this is also the case where ribosomal frameshifting is ‘programmed’. Such cases may indeed affect mRNA stability at some level or, alternatively, the corresponding mRNAs may have evolved specific stabilizing elements that negate the potential destabilizing effects of frameshifting.22
Computational identification of PRF events is facilitated when both overlapping ORFs are long. In such cases, both ORFs can be easily identified, and the estimation of their coding potential is feasible. However, several PRF cases involve one very short ORF. Examples where the first ORF is short include the autoregulatory PRF in bacterial release factor 2,23,24 and antizyme25 decoding. Short second ORFs are present in E. coli dnaX,26,27 and mammalian MA3,28 frameshifting. The greatest phylogenetic range of these cases is seen for antizyme +1 frameshifting which acts as a sensor and effector for the control of intracellular polyamine levels from yeast to mammals.
Comparative sequence analysis is important for identifying novel occurrences of PRF as exemplified by recent work on alpha-viruses and flaviviruses.29,30 Purifying selection acts on the evolution of most protein-coding genes and results in characteristic patterns of sequence change, where substitutions to synonymous codons dominate over changes to nonsynonymous codons. This can be used to identify the frame and extent of protein coding sequences.31 Using this principle, Lin and colleagues32 developed a computational approach based on two quantitative metrics: reading frame conservation and codon substitution frequencies, for identifying protein coding regions based on the analysis of multiple nucleotide alignments of closely related sequences. They applied this technique to re-evaluate D. melanogaster genome annotation using genomic alignments of 12 Drosophila species and identified four genes whose evolutionary signatures for protein coding regions switch from one translational frame into another. They proposed that these genes are candidates for PRF.32 They are Xbp1, CG32736, CG14047 and APC (adenomatus polyposis coli). We have computationally re-examined these candidates to gain insight into what mechanisms may be responsible for the apparent transition of evolutionary signatures between different reading frames. This analysis has confirmed the prediction of APC as a gene with PRF, while transitions between frames in the other three cases are more likely due to other mechanisms. Further, we confirmed and experimentally characterized APC PRF on the frameshift cassettes from several species.
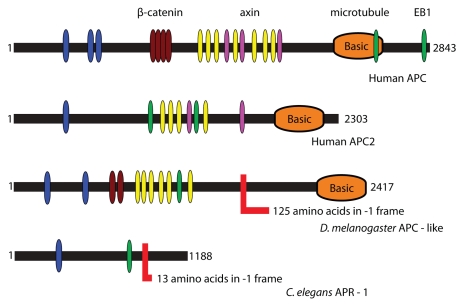
APC functions in the canonical Wnt/Wingless (Wg) signaling pathway in mammals and Drosophila33 (see Figure 1 for APC domain organization). APC homologs are found exclusively and universally in animals, along with other members of the Wnt pathway.34 The Wnt pathway is implicated in many developmental processes, such as cell proliferation, polarity and migration.35 Two paralogs of APC exist in mammals, APC and APC2, while two paralogs also exist in Drosophila, dAPC and dAPC2, although these are not direct orthologs of mammalian APC/APC2.36,37 Mutation of both the mammalian and Drosophila APC genes results in aberrant Wnt/Wg signaling and mutants of mammalian APC are an important direct cause of colon cancer.31 With respect to controlling cell growth and proliferation, however, a role for dAPC has recently been described in regulating the intestinal stem cell proliferation.38 The C. elegans APC homolog apr-1 seems to have a similar role as part of the Wnt pathway in regulating proliferation of the stem cell-like seam cells.39,40
Figure 1.
Domain organization of APC proteins. Highlighted are conserved PFAM domains identified by a PFAM search.76 The color codes are as follows: blue-PF00514 (armadillo), brown-PF05972, yellow-PF05923 (cysteine-rich regions), purple-PF05924 (SAMP), orange-PF05956 (basic) and green-PF05937 (EB1 binding domain). Frameshift sites are shown with an L-like red shape and indicate the length of the C-terminal extension after PRF in each case.
Results
Computational sequence analysis of APC PRF in insects.
One of the four candidates identified by Lin et al.,26 the shift in Xbp1, is due to a conserved nonstandard splicing event. In both yeast and human, ER stress activates the RNase domain of the IRE-1 protein causing the excision of a 23-base non-canonical intron in the Xbp1 transcript. This causes a shift in reading frame, leading to a longer protein encoding an active transcription factor. A pair of stem-loop structures is present at the splice sites in yeast and human and is conserved among insect orthologs (Sup. Fig. 1). Drosophila ESTs encode both spliced and unspliced forms, and splicing is induced by ER stress.41 Details of the analysis of Xbp1 and the following two candidates are in Supplemental material. The mRNA of one of them, CG32736, is probably bicistronic. Both ORFs are well conserved throughout coelomates, but are not well-annotated nor are they members of larger gene families. Although both ORFs are found in the same transcript in most coelomates (including invertebrate chordates and the lamprey, an early branching vertebrate), they are not always in the same reading frame. All other vertebrates have the two ORFs at different chromosomal locations. In contrast, the strong predicted frame-shift in CG14047 is not easily explained and, though CG14047 is conserved throughout insects, its function is unknown. The nucleotide sequence of the shift region is inconsistent with known models of programmed ribosomal frameshifting. Moreover, the behavior cannot be explained by a single frameshifting event.
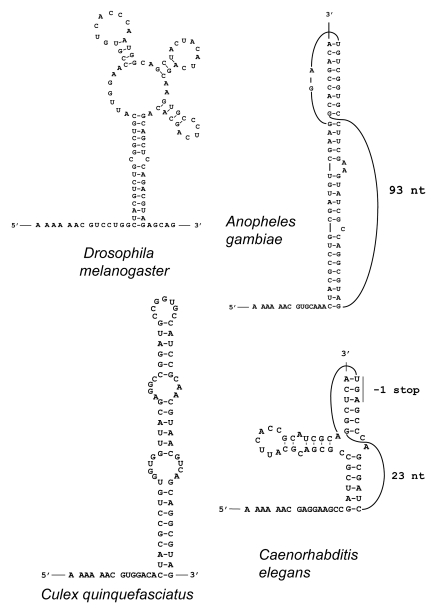
The shift in evolutionary signatures for the APC gene, the fourth candidate identified by Lin et al.,26 is at position chr3R:24,661,410 (here and further genomic coordinates are given for 2006 BDGP R5 assembly). A nucleotide alignment of APC from the 12 sequenced Drosophila species genomes and that of the malaria mosquito Anopheles gambiae, is shown in Figure 2. The organization of the frameshift cassette is very similar to those that are used in many viral genomes to promote −1 frameshifting.12 It consists of a tandem slippery site A_AA.A_AA.C (underscores separate codons in the initial frame and dots separate codons in the new −1 frame) that, following anticodon: codon dissociation, allows two tRNAs to re-pair to the mRNA in the −1 frame. The shift sequence is followed by predicted conserved mRNA secondary structures that may stimulate the frameshifting level. The tandem slippery site also has a conserved CC at its 5′ end (in accordance with the hypothesis that the E-site is involved in modulating −1 frameshifting42) and has a conserved G at its 3′-end (in accordance with an observation that the stacking potential of the third A-site codon base and its 3′ adjacent base affects frameshifting efficiency43). The predicted secondary structure is a four-way junction stem-loop that is conserved across all Drosophila species (Fig. 3A shows the predicted structure for D. melanogaster). Interestingly, mosquitoes have different predicted 3′ stimulatory structures. In the malaria mosquito, Anopheles gambiae, there is a predicted RNA pseudoknot, while in the house mosquito, Culex quinquefasciatus, there is a strong RNA stem loop (Fig. 3). The Culicinae and Anophelinae clades are reported to have diverged about 150 Mya44 and therefore a considerable sequence divergence is not surprising. The location of the shift in coding potential between frames detected by Lin et al.32 is 143 nucleotides downstream from the predicted location of the actual frameshifting site described above—chr3R:24,661,696 genomic coordinate. The reasons for this are easy to understand. Since the likely frameshift cassette is highly conserved, there are few informative codons that can be used for the analysis of protein coding evolutionary signatures. So it is not surprising that Lin et al.32 predicted the shift location 3′ of the bona fide frameshift site. Lin et al.32 reported a 3′ switch of the evolutionary signature back to the zero frame, and this corresponds to the location of the stop codon in the −1 frame segment of the overlap (beyond which, there should be no purifying selection in the −1 frame).
Figure 2.
Multiple alignment of fly and malaria mosquito APC gene sequences corresponding to the region translated in the −1 frame. Heptameric frameshift sites are shown in yellow. Nucleotides involved in base-pairing interactions within the same double-stranded regions of the predicted secondary structure are differentially colored, e.g., the second stem of the predicted pseudoknot in the malaria mosquito (anoGam1) is shown in red. Secondary structures are also shown in bracket format below the alignment. The first bracket row corresponds to the four-way junction in fruit flies. The second bracket row corresponds to the malaria mosquito pseudoknot.
Figure 3.
Predicted stimulatory PRF secondary RNA structures in flies and mosquitoes. The sequences shown include the A AAA AAC slippery pattern (with codons in the zero frame separated by spaces) at the 5′ end and the entire mRNA regions forming the predicted stimulatory structures 3′ from the shift site. Latin names of the organisms are indicated.
The sequences of APC homologs from various species are highly divergent. This obstructs correct identification of the full APC CDS in some species. A further impediment to constructing correct APC gene models is its low expression level and consequent paucity of EST sequences. We failed to identify frameshifting in APC decoding in three hymenopteran genomes, those of the honey bee Apis mellifera and the ants Harpegnathos saltator and Camponotus floridanus. The genomic region between the two exons that share similarity with dAPC upstream and downstream of the PRF site lack the A AAA AAC motif. Either PRF is not used during expression of hymenopteran APC or the PRF cassettes diverge so much that we are not able to recognize it.
Identification of an APC frameshifting cassette in nematodes.
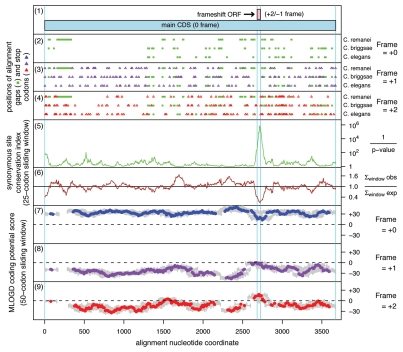
A meaningful comparative sequence analysis similar to the one that revealed APC as a PRF candidate in Drosophila species32 is practical only for sequences that are relatively closely related. Evolutionary distances between APC sequences from nematodes of the Caenorhabditis genus allow such an analysis. To analyze evolutionary behavior of APC genes from Caenorhabditis their protein products were aligned, and then back-translated nucleotide alignments were subjected to analysis by the MLOGD program.45 MLOGD uses nucleotide and amino acid substitution matrices to model sequence evolution and estimate the likelihood, in a sliding window, that an alignment is coding in each possible reading frame. This analysis did not reveal a significantly long region of the APC alignment in which substitutions would indicate evolutionary conservation of protein coding sequence in an alternative reading frame. However, a slight increase in the signal for the −1 frame was observed for a very short fragment of mRNA (Fig. 4). We further analyzed the alignment by examining conservation at synonymous sites using the method described in reference 30. Locally enhanced nucleotide conservation at synonymous positions is indicative of overlapping functional elements such as frameshift-stimulating motifs. This revealed a highly significant conservation peak that coincided with the short MLOGD coding signal for the −1 reading frame (Fig. 4). The sequence alignment around this peak shows a conserved motif of A AAA AAC, identical to the frameshift site of Drosophila APC (alignment, Fig. 5). Conservation of the sequence 3′ from the shift site is apparently due to the existence of an RNA pseudoknot that likely plays a stimulatory role in the frameshifting. However, the secondary structure involved is dramatically different from those used by fruit flies and mosquitoes (Fig. 3). Frameshifting at the A AAA AAC in the C. elegans APC gene would give a product with a C-terminal “tail” of 13 amino acids encoded by the −1 frame, whose last 7 amino acids and stop site are absolutely conserved within the 5 Caenorhabditids.
Figure 4.
Coding potential statistics for Caenorhabditis APC. (1) Map of the APC coding sequence showing the predicted frameshift ORF. (2–4) The positions of stop codons in each of the three forward reading frames. The +0 frame corresponds to APC and is therefore devoid of stop codons. (5 and 6) Conservation at synonymous sites within APC (see ref. 30 for details). (5) depicts the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, while (6) depicts the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model. (7–9) MLOGD sliding window plots (see ref. 61 for details). The null model, in each window, is that the sequence is non-coding, while the alternative model is that the sequence is coding in the given reading frame. Positive scores favor the alternative model and, as expected, in the +0 frame (7) there is a strong coding signature throughout APC . In the +1 and +2 frames (8 and 9), scores are generally negative with some random scatter into low positive scores, except in the region occupied by the predicted frameshift ORF, where there is a strong positive coding signal in the +2/−1 reading frame. (Due to the limited sequence data available, a 50-codon sliding window was used to increase the signal-to-noise ratio throughout the alignment, notwithstanding that this is expected to dilute the signal from the overlapping ORF itself—since, at ∼16 codons, the overlapping ORF is much shorter than the 50-codon window). Note that, regardless of the sign (either positive or negative), the magnitude of MLOGD scores tends to be lower within the overlap region (7–9) due to there being fewer substitutions with which to discriminate the null model from the alternative model in this region of above-average nucleotide conservation.
Figure 5.
Multiple alignment of genomic sequences in the region of the PRF cassette from seven nematodes. Heptameric frameshift sites are shown in yellow. Nucleotides involved in predicted base-pairing interactions within the same double stranded regions of the secondary structure are differentially colored (e.g., the first stem of the pseudoknot is green, the second is red and the stem in the first loop of the pseudoknot is in blue). The secondary structure of the RNA pseudoknot in the C. elegans sequence is also shown in bracket format above the alignment and its diagram is shown in Figure 2.
Among other nematode APC sequences, Pristionchus pacificus (whose last common ancestor with C. elegans lived 200–300 million years ago) appears to have an A AAA AAC shift site followed 6 nts 3′ by a stem loop structure, while Loa loa (“eye worm”, the cause of a filariasis) and Brugia malayi (a cause of elephantiasis or lymphatic filariasis), likely have an A AAA AAA frameshift site (Fig. 5) but cassettes with the relevant sequences have not been tested.
Expression of APC frameshift cassettes in reticulocyte lysates.
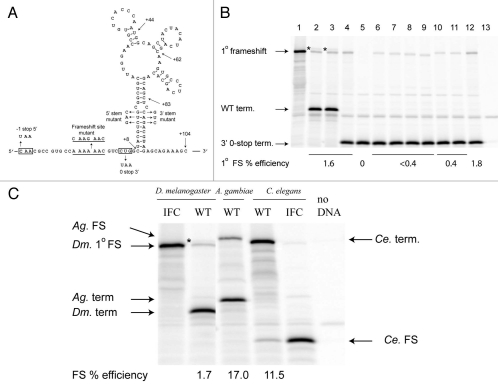
We tested -1 ribosomal frameshifting on the Drosophila APC sequence in vitro. The proposed frameshift heptanucleotide flanked by 15 nt 5′ and 104 nt 3′ (Fig. 6A) was cloned between the renilla and firefly luciferase genes in the pDluc vector46 such that expression of the renilla-APC-firefly luciferase fusion requires −1 frameshifting. Expressing this construct in a rabbit reticulocyte lysate in vitro translation system resulted in two firefly luciferase-containing products (Fig. 6B, lane 2); one co-migrates with the in-frame control for -1 frameshifting at A AAA AAC (Fig. 6B, lane 1), while the other migrates more slowly. The primary frameshift at A AAA AAC occurs 1.7% of the time, and the secondary frameshift occurs at 0.3%. To delimit the origin of the second firefly luciferase-containing product, stop codons were created in the −1 reading frame or in the zero reading frame, 5′ or 3′ of the shifty heptanucleotide, respectively. Occurrence of −1 frameshifting 5′ of the new −1 stop would result in elimination of the secondary product. However, if it were to occur 3′ of the shift site, then the introduced zero frame stop codon should preclude production of the secondary product. The results indicate that the secondary frameshift occurs 3′ of the primary A AAA AAC site (Fig. 6B, lanes 3 and 4). [All subsequent constructs contain the zero frame stop codon to focus on the primary frameshift event; the nature of the secondary frameshift event is currently under investigation.]
Figure 6.
Expression of APC frameshift cassettes in reticulocyte lysates. (A) Drosophila melanogaster APC frameshift cassette. The wild-type sequence with the predicted frameshift site underlined is shown; the sequence of the mutant frameshift site is shown above. Sequences that were mutated to introduce a termination codon in the −1 or the zero frames are boxed. Endpoints of the 3′ deletion series are shown by arrows, as well as nucleotide changes in the stem. (B) In vitro translations of Drosophila APC pDluc constructs. 1° specifies primary frameshift product. (1) The frameshift site was replaced by C AAG AAC C, such that standard translation produces the renilla luciferase-APC-firefly luciferase fusion protein (in-frame control). (2) The wild-type APC sequence ending 104 nt 3′ of the shift site (see A). (3) A C-to-U mutation 5′ of the shift site that results in a UAA stop codon in the −1 frame. (4) CUG was changed to UAA, 3′ of the frameshift site, resulting in a stop codon in the zero reading frame. All subsequent constructs contained the CUG to UAA mutation. (5) Mutation of the frameshift site to C AAG AAC. (6–9) Deletions of the 3′ sequence—endpoints are +83, +62, +44 and +8, respectively (see A). (10) Mutation of the 5′ side of the putative stem. (11) Mutation of the 3′ side of the putative stem. (12) Combination of the 5′ and 3′ stem mutations predicted to restore base pairing (see A). (13) No template added. The positions of the products due to frameshifting at the primary site, A AAA AAC, termination in the WT construct and termination at the zero frame UAA, two codons 3′ of the frameshift site are shown by arrows. The secondary frameshift product of unknown origin is indicated by *. (C) In vitro translations of frameshift cassettes from D. melanogaster, A. gambiae and C. elegans are labelled WT. Lanes with cassettes designed to produce the frameshift product as a result of standard translation are labelled IFC (In-Frame Control). The positions of the respective frameshift and termination products are shown by arrows. Note that with the C. elegans WT cassette, the frameshift product is the smaller of the two products due to a −1 stop codon within the sequence of the predicted 3′ stimulatory element (see Fig. 2). The * signifies the secondary frameshift product from the D. melanogaster APC sequence.
Involvement of the predicted shift site in frameshifting was tested by changing the nucleotide sequence to disrupt tRNA:mRNA base pairing in the −1 frame (C AAG AAC) while maintaining the same amino acid sequence. As expected, this abolished the observed frame-shift (Fig. 6B, lane 5). Several 3′ deletions were designed to determine the importance of particular features of the proposed secondary structure (Fig. 6A). When the 3′ sequence was limited to +83 nt, +62 nt, +44 nt, or +8 nt (Fig. 6B, lanes 6–9), frameshifting was decreased to less than 0.4% in all cases. These results are consistent with the importance of the 16 base-pair stem for frameshifting (it has one mismatch and its 3′ end is 94 nts 3′ of the shift site, Fig. 3). [A construct including ∼800 additional nt of APC sequence 3′ of the proposed stem-loop structure did not affect the level of frameshifting (data not shown) indicating the lack of long-range stimulatory signals.] Disruption of potential base pairing by mutations in the 5′ or 3′ part of the stem decreased frameshifting to 0.4% (Fig. 6B, lanes 10 and 11). However, the combination of the complementary 5′ and 3′ stem mutations, predicted to restore base-pairing (Fig. 6B, lane 12), restored frameshifting to 1.7%, the same level as the parent construct containing the zero frame stop codon 3′ of the shift site.
The predicted frameshift signals in A. gambiae and C. elegans APCs (Fig. 3) were tested as described above for D. melanogaster. The sequences encompassing the proposed structures promoted 17% frameshifting in Anopheles gambiae APC and 11.5% frameshifting in C. elegans APC, both appreciably higher than their counterpart in Drosophila melanogaster (Fig. 6C). In the C. elegans sequence, the first stop codon in the −1 frame after the shift site is involved in base pairing in a stem of a proposed pseudoknot structure so that −1 frameshifting results in the production of a renilla luciferase-APC fusion (smaller product) while translation in the zero frame results in the renilla luciferase-APC-firefly luciferase fusion (larger product).
Discussion
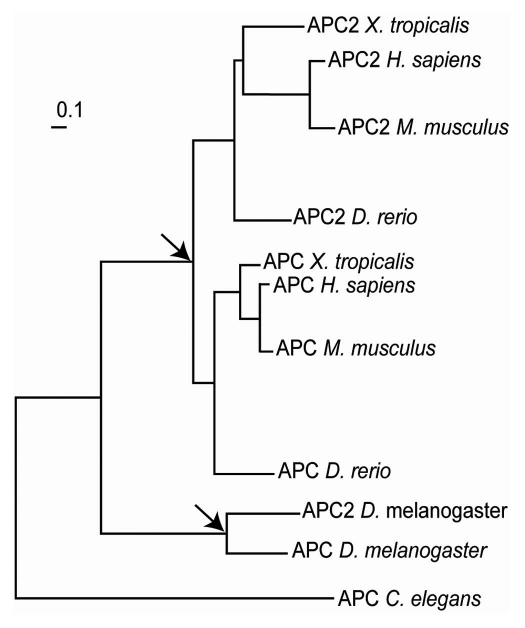
Programmed ribosomal frameshifting is used in the decoding of the APC gene in Caenorhabditis nematodes, mosquitoes and one of the two paralogs in Drosophila. Phylogenetic analysis shows that the APC gene has undergone two independent duplications, in vertebrates and in insects (Fig. 7). The second Drosophila copy is genomically truncated at almost exactly the frameshift region and shows no sign of a frameshift signal. While we did not find the frameshift motif in some other insects, its conservation in nematodes suggests that it had an ancient origin. The poor sequence conservation downstream of the shift site makes it difficult to detect frameshift signals in other metazoan genomes, suggesting that directed cDNA cloning of APC homologs may be required to fully address the origin and extent of this mechanism. However, its existence in flies and nematodes implies that exploitation of this mechanism for the expression of APC protein variants is beneficial. Therefore, its existence in other animal lineages is not unexpected, but its location within APC and its sequence organization could vary considerably, thus identification of PRF in distant sequences may not be an ordinary task.
Figure 7.
Phylogenetic tree of APC genes extracted from TreeFam.70 The nodes corresponding to the events of independent APC gene duplications are indicated with arrows.
The PRF product is shorter than the standard product and shares about two thirds of the same sequence at its N-terminal region (Fig. 1). The proximal gene segment encoding the shared N-terminal part is significantly more conserved than the distal part of the gene. While APC is a multifunctional protein, it is the central region of the protein that is most directly implicated in Wingless signaling. Interestingly, there is a strong tendency for removal of the carboxyl terminus of the protein in mutations that lead to colorectal cancer in humans, mostly (60%) by point mutations leading to frameshift or nonsense mutations. Based on these findings, it is thought that a truncated form of APC, which lacks axin/conductin interaction sites, leads to the constitutive Wg/Wnt signaling and tumorigenesis.47 This suggests that the frameshifted products may have distinct functions to the full length protein.
Multiple functions have been attributed to APC and its interacting partners. While several domains are required for its best characterized function as a protein that binds to β-catenin and thus downregulates Wnt/Wg signaling, APC also interacts with several proteins with established roles in cytoskeleton dynamics, in particular those associated with either cell migration (ASEF, APC-stimulated guanine nucleotide exchange factor; IQGAPI, IQ domain-containing GTP-ase activating protein; KAP3, kinesin super-family associated protein 3; mDIA, diaphanous) or polarized cell division (EB1, end binding 1; hDLG, human Disc Large; Scribble) (reviewed in ref. 48). The C-terminal part of mammalian APC has two domains involved in microtubule binding: a large region containing positively charged amino acids (basic domain),49 and in APC1, at least, an EB1 binding domain.50 Though not sufficient on its own, the core sequence of an EB1 binding site is SxIP.51 Curiously, the Drosophila APC and C. elegans APC studied here have just one SxIP sequence, and it is encoded 5′ of the frameshift site while human APC-2 has two SxIP sequences both of which are in the middle of the protein. The possible significance of these, including juxtaposition to the frameshift site, is unknown. It is notable that interactions specific for polarized cell division are confined to the conserved region that is lost in the ribosomal frameshift derived dAPC products. Considering these data, along with the knowledge that a truncated form of APC is functional in tumorigenesis, we suspect that ribosomal frameshifting limits the way in which APC functions in vivo. Both fly and worm systems will provide good genetic models in which to test the physiologically relevant role(s) of frameshifted APC. This being said, our initial attempts to generate isoform-specific peptide antibodies to delimit the expression and roles of frameshifted and unframeshifted APC isoforms in Drosophila were not successful (data not shown).
The recent demonstration that Drosophila APC plays an important role in maintenance of Drosophila intestinal stem cells is particularly intriguing with respect to our understanding of the role of human APC as a tumor suppressor.38 Best known for its role in colorectal cancer, several in vivo studies in mouse have shown that properly regulated Wnt/Wg signaling is essential for normal proliferation of the precursor cells of the intestinal epithelium.52–54 Thus, the functions of fly and human APC in regulating cell growth and proliferation are likely conserved. Consistent with this idea is the demonstration that Drosophila APC can interact with the Scribbles and Dlg proteins (which result in disc overgrowth phenotypes when mutated), even though Drosophila APC has not itself been shown in genetic studies to function as a tumor suppressor. Notably, the Scribbles and Dlg interaction sites are not present in the frameshifted dAPC isoform.48
Since the focus of selection in the region immediately 3′ of what is now known to be the frameshifting site in Drosophila is the −1 rather than the zero frame, presumably the segment of the shorter protein encoded by the −1 frame (the C-terminal segment) is likely functional also. However, the corresponding region in C. elegans APC is very short, and we cannot exclude the possibility that selection there has only been operating to synthesize a truncated version of the product of standard decoding—one that lacks functions associated with the full-length product.
Whether decoding of either human APC gene yields a frameshift-derived product is unknown, but alternative forms of APC are known to be synthesized. Three alternative transcripts have been identified. (Internal IRES-mediated initiation has also been reported in ref. 55 and ref. 56.) One of the alternative transcripts (Refseq: NM_001127511) produces short protein isoform A (Refseq: NP_001120983), and two others (NM_000038 and NM_001127510) produce long isoform B (NP_001120983). Isoform A and B differ by a single exon encoding 27 amino acid residues in the N-terminal part of APC.57
Interestingly, while the frameshift site is A AAA AAC in the C. elegans, D. melanogaster and mosquito cassettes tested here, the stimulatory secondary RNA structures are highly variable (see Fig. 3). The predicted structures in worms and the malaria mosquito (A. gambiae) are H-type pseudoknots and there is no significant sequence similarity between the worm and mosquito versions of the pseudoknot. Moreover, different worms appear to have strikingly different pseudoknots (see Fig. 5). In flies and the house mosquito, the structures are stem-loops, but of different organization. The Drosophila and mosquito structures share sequence similarity in the area at the base of the structures. For the D. melanogaster stem loop, the 3′ deletion, stem disruption and stem restoration mutations demonstrate the importance of the structure for frameshifting at the primary A AAA AAC site. For C. elegans and A. gambiae, we have shown that the sequences encompassing the proposed RNA structures are sufficient to promote frameshifting, but further mutational analysis is required to establish the exact nature of the structures. Variability in PRF stimulatory structures has been observed previously for −1 PRF in viral genes where, even in closely related species, stimulatory structures could be substantially different, e.g., the existence of either an H-type pseudoknot or kissing loops as stimulatory structures in coronavirus PRF.58
In our experiments with frameshift cassettes from different species, the efficiency of the frameshifting varied considerably—from 1.7% in D. melanogaster to 17% in A. gambiae. Since the efficiency was tested in a heterologous mammalian in vitro system, these efficiencies may not correctly reflect the natural level of the frameshifting. Despite the low efficiency of 1.7% found with the D. melanogaster cassette, the strong evolutionary conservation suggests the frameshift product plays an important role. Presumably either only a very small amount is required or the endogenous levels in the cells where it is normally expressed is higher, conceivably due to a trans-acting factor. Also, it is important to keep in mind that the efficiency of frameshifting could be affected by expression levels59 and translation initiation rates60 of the corresponding mRNA, so that the frameshifting efficiencies determined by the use of artificial genetic constructs may not be truly representative of endogenous frameshifting levels.
The organization of ORFs involved in the production of the alternative PRF APC protein product is unusual and suggests that there might be similar instances of PRF in other chromosomal genes that cannot be readily identified by computational methods. The main reason why PRF was identified in fruit flies was the existence of strong evolutionary selection acting on the protein sequence encoded by the alternative frame while the selection on the standard frame was more relaxed. This created the sequence region with the major protein-coding evolutionary signature corresponding to the alternative frame, as detected by Kellis and colleagues32 and MLOGD.61 Interestingly the selection is much stronger in the −1 frame even though the product of the frameshifting is minor in comparison with the product of standard translation. Although in worms the evolutionary signatures corresponding to the alternative frame are also detectable, the additional peptide generated by the sequence downstream of the frameshift site is much shorter than its fly counterpart. In principle, the situation with APC may well be a fortunate exception. In general, when the functional importance of a frameshift product depends simply on its truncated nature rather than the actual peptide encoded by the new frame, or where selection acting on the protein sequence encoded in the zero frame is similar or stronger than in the alternative frame, frameshifting would not be easily detectable by methods based on identifying protein coding evolutionary signatures. This applies in decoding the E. coli dnaX gene, where the 50% of ribosomes that shift frame at the programmed frameshift site only translate one codon in the new frame before terminating.26 In fact, we were able to identify PRF in Caenorhabditis nematodes foremost because we expected it to be there and specifically searched for it using different methods. Most likely the frame switch between evolutionary signatures would not be detectable in a genome-wide analysis similar to the one performed by Lin et al.32 for fruit flies. Thus, on one hand, identification of PRF in APC illustrates the applicability of comparative sequence analysis for the identification of novel instances of programmed ribosomal frameshifting. On the other hand, it suggests that many instances of PRF would not be detectable by such methods. To accurately investigate the occurrence and utilization of ribosomal frameshifting on a whole genome scale, it is necessary to develop high-throughput techniques for the identification of non-standard decoding. One possibility for the development of such methods is utilization of the ribosomal profiling technique that allows measurement of the density and location of ribosomes on mRNA with sub-codon precision.62
Materials and Methods
Software, computational methods and databases.
The coordinates for predicted switches of reading frames in the D. melanogaster genome were obtained from the online supplemental materials for Lin et al.32 at www.broadinstitute.org/∼mlin/fly_genes/dmel_frameshifts.dat. Conversions between coordinates from different assemblies were made with the FlyBase LiftOver tool.63 APC homologs were identified in the NCBI NR and dbEST databases by Blast, and were repredicted using GeneWise64 and genomic sequences from the UCSC genome browser,65 using a variety of homology templates. MultiZ alignments66 of genomic sequences used in this study were obtained from the UCSC conservation tracks for 12 Flies, Mosquito, Honeybee and Beetle MultiZ Alignments.65 To generate multiple alignments of CDSs for the analysis by MLOGD61 and synonymous site conservation,30 nucleotide sequences were translated into protein sequences and aligned using MUSCLE67 and then the resulting protein alignments were backtranslated to nucleotide alignments using t-coffee.68 CDS prediction for worm APC genes was made with the help of the GeneWise program.64 Visualization of sequences for the manual analysis was done with the assistance of Gendoc v2.6.002 (www.nrbsc.org/gfx/genedoc/),69 pDraw32 v1.1.107 (www.acaclone.com) and SequenceCruncher v. 1.09 (gesteland.genetics.utah.edu/bmoore/Downloads/SeqCrn.html). Phylogenetic trees of APC genes were obtained from TreeFam70 and visualized using TreeView.71 Initial secondary RNA structure predictions were carried out with MFOLD72 and SFOLD,73 and then manually reevaluated to examine the potential existence of RNA pseudoknots and for verification via compensatory substitutions supporting the structure. Visualization of secondary RNA structures was performed with PseudoViewer 3.74
Analysis of frameshifting efficiency in WT and mutant PRF cassettes.
Construction of plasmids. The sequences encompassing the predicted frameshift site and the 3′ stimulatory structure (frameshift cassette) for Drosophila melanogaster, Anopheles gambiae and C. elegans APCs were generated using overlapping synthetic oligonucleotides and cloned into the XhoI and BglII sites in vector pDluc,46 a derivative of the p2luc vector.75 The firefly luciferase gene is in the −1 reading frame relative to the upstream renilla luciferase gene such that −1 frameshifting within the frameshift cassette results in a renilla luciferase-APC-firefly luciferase fusion product. A Drosophila melanogaster APC insert including ∼800 nt 3− of the predicted stem-loop structure was synthesized by GenScript and cloned into the HindIII and BglII sites of pDluc (due to an internal XhoI site in the APC sequence). All plasmids were verified by DNA sequencing.
Frameshift assays. Plasmid DNAs (0.2 µg) were used as templates in 10 µl reactions of the rabbit reticulocyte lysate TNT® T7 Quick Coupled Transcription/Translation system (Promega).35 S-methionine (Perkin Elmer) was included in the reactions and protein products were separated by SDS-PAGE. Dried gels were analyzed using a Typhoon PhosphorImager (GE Healthcare) and the amount of radioactivity in each product was determined using the ImageQuant 5.2 program (Molecular Dynamics). After normalization for the number of methionine residues in termination and frameshift products (9 and 22, respectively, for Drosophila APC), the frameshift efficiencies were calculated [frameshift/(frameshift + termination)]. Note that in the C. elegans APC sequence, the −1 termination codon is located within the predicted stimulatory structure, so the smaller product represents the frameshift product.
Acknowledgements
The authors would like to thank (1) Dr. Avril Coghlan for useful suggestions related to identification of APC sequences in nematodes and for her help with TreeFam and (2) Dr. Erik Jorgensen for drawing our attention to the location (with respect to the frameshift site) of the SxIP motif in C. elegans and its potential implication in binding EB1.
Abreviations
- APC
adenomatus polypolis coli
- PRF
programmed ribosomal frameshifting (or frameshift)
- ORF
open reading frame
Financial Support
This work was supported by Science Foundation Ireland (grant number 06/IN.1/B81 to P.V.B., 08/IN.1/B1889 to J.F.A.), the Wellcome Trust (grant number 088789 to A.E.F.), and NIH (grant number HG004164) to G.M.
Supplementary Material
References
- 1.Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, Baranov PV. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 2010;38:D69–D74. doi: 10.1093/nar/gkp788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins JF, Gesteland RF, editors. Recoding Expansion of Decoding Rules Enriches Gene Expression. New York and Heidelberg: Springer; 2010. [Google Scholar]
- 3.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Stahl G, Ben Salem S, Li Z, McCarty G, Raman A, Shah M, et al. Programmed +1 translational frameshifting in the yeast Saccharomyces cerevisiae results from disruption of translational error correction. Cold Spring Harbor Symp Quant Biol. 2001;66:249–258. doi: 10.1101/sqb.2001.66.249. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Havecker ER, Baranov PV, Atkins JF, Voytas DF. Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA. 2003;9:1422–1430. doi: 10.1261/rna.5105503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 7.Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biologist. 1989;1:159–169. [PubMed] [Google Scholar]
- 8.Dinman JD, Icho T, Wickner RB. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 11.Licznar P, Mejlhede N, Prere MF, Wills N, Gesteland RF, Atkins JF, et al. Programmed translational -1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 13.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barry JK, Miller WA. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firth AE, Bekaert M, Baranov PV. Computational resources for studying recoding. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. New York and Heidelberg: Springer; 2010. pp. 281–300. [Google Scholar]
- 16.Bekaert M, Atkins JF, Baranov PV. ARFA a program for annotating bacterial release factor genes, including prediction of programmed ribosomal frameshifting. Bioinformatics. 2006;22:2463–2465. doi: 10.1093/bioinformatics/btl430. [DOI] [PubMed] [Google Scholar]
- 17.Bekaert M, Ivanov IP, Atkins JF, Baranov PV. Ornithine decarboxylase antizyme finder (OAF): fast and reliable detection of antizymes with frameshifts in mRNAs. BMC Bioinformatics. 2008;9:178. doi: 10.1186/1471-2105-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, Atkins JF. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs JL, Belew AT, Rakauskaite R, Dinman JD. Identification of functional, endogenous programmed −1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:165–174. doi: 10.1093/nar/gkl1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plant EP, Wang P, Jacobs JL, Dinman JD. A programmed −1 ribosomal frameshift signal can function as a cis-acting mRNA destabilizing element. Nucleic Acids Res. 2004;32:784–790. doi: 10.1093/nar/gkh256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belew AT, Advani VM, Dinman JD. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 2011;39:2799–2808. doi: 10.1093/nar/gkq1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weil JE, Hadjithomas M, Beemon KL. Structural characterization of the Rous sarcoma virus RNA stability element. J Virol. 2009;83:2119–2129. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 24.Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov IP, Matsufuji S. Autoregulatory frameshifting in antizyme gene expression governs polyamine levels from yeast to mammals. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Vol. 2. New York and Heidelberg: Springer; 2010. pp. 281–300. [Google Scholar]
- 26.Tsuchihashi Z, Brown PO. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 27.Larsen B, Wills NM, Gesteland RF, Atkins JF. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wills NM, Moore B, Hammer A, Gesteland RF, Atkins JF. A functional −1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J Biol Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- 29.Firth AE, Chung BY, Fleeton MN, Atkins JF. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firth AE, Atkins JF. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1' may derive from ribosomal frameshifting. Virol J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nekrutenko A, Makova KD, Li WH. The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12:198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 34.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 36.van Es JH, Kirkpatrick C, van de Wetering M, Molenaar M, Miles A, Kuipers J, et al. Identification of APC2, a homologue of the adenomatous polyposis coli tumour suppressor. Curr Biol. 1999;9:105–108. doi: 10.1016/s0960-9822(99)80024-4. [DOI] [PubMed] [Google Scholar]
- 37.Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- 38.Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development. 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- 39.Huang X, Tian E, Xu Y, Zhang H. The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev Biol. 2009;333:337–347. doi: 10.1016/j.ydbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Ren H, Zhang H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev Biol. 2010;345:144–155. doi: 10.1016/j.ydbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed −1 ribosomal frameshift. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand C, Prere MF, Gesteland RF, Atkins JF, Fayet O. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA. 2002;8:16–28. doi: 10.1017/s1355838202012086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Firth AE, Brown CM. Detecting overlapping coding sequences with pairwise alignments. Bioinformatics. 2005;21:282–292. doi: 10.1093/bioinformatics/bti007. [DOI] [PubMed] [Google Scholar]
- 46.Fixsen SM, Howard MT. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J Mol Biol. 2010;399:385–396. doi: 10.1016/j.jmb.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 48.Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deka J, Kuhlmann J, Muller O. A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur J Biochem. 1998;253:591–597. doi: 10.1046/j.1432-1327.1998.2530591.x. [DOI] [PubMed] [Google Scholar]
- 50.Dikovskaya D, Li Z, Newton IP, Davidson I, Hutchins JR, Kalab P, et al. Microtubule assembly by the Apc protein is regulated by importin-beta--RanGTP. J Cell Sci. 2010;123:736–746. doi: 10.1242/jcs.060806. [DOI] [PubMed] [Google Scholar]
- 51.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 52.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 54.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heppner Goss K, Trzepacz C, Tuohy TM, Groden J. Attenuated APC alleles produce functional protein from internal translation initiation. Proc Natl Acad Sci USA. 2002;99:8161–8166. doi: 10.1073/pnas.112072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozak M. New ways of initiating translation in eukaryotes? Mol Cell Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carson DJ, Santoro IM, Groden J. Isoforms of the APC tumor suppressor and their ability to inhibit cell growth and tumorigenicity. Oncogene. 2004;23:7144–7148. doi: 10.1038/sj.onc.1207954. [DOI] [PubMed] [Google Scholar]
- 58.Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virol. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gurvich OL, Baranov PV, Gesteland RF, Atkins JF. Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli. J Bacteriol. 2005;187:4023–4032. doi: 10.1128/JB.187.12.4023-4032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gendron K, Charbonneau J, Dulude D, Heveker N, Ferbeyre G, Brakier-Gingras L. The presence of the TAR RNA structure alters the programmed −1 ribosomal frameshift efficiency of the human mmunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Firth AE, Brown CM. Detecting overlapping coding sequences in virus genomes. BMC Bioinformatics. 2006;7:75. doi: 10.1186/1471-2105-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drysdale R. Methods in Molecular Biology. Vol. 420. Clifton, NJ: 2008. FlyBase : a database for the Drosophila research community; pp. 45–59. [DOI] [PubMed] [Google Scholar]
- 64.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Notredame C. Computing multiple sequence/structure alignments with the T-coffee package. Curr Protoc Bioinformatics. 2010;8:1–25. doi: 10.1002/0471250953.bi0308s29. Chapter 3:Unit 3. [DOI] [PubMed] [Google Scholar]
- 69.Nicholas KB, Nicholas HB, Deerfield DW. 2nd. GeneDoc: a tool for editing and annotating multiple sequence alignments. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- 70.Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page RD. Visualizing phylogenetic trees using TreeView. Curr Protoc Bioinformatics. 2002;2 doi: 10.1002/0471250953.bi0602s01. Chapter 6:Unit 6. [DOI] [PubMed] [Google Scholar]
- 72.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Byun Y, Han K. PseudoViewer: web application and web service for visualizing RNA pseudoknots and secondary structures. Nucleic Acids Res. 2006;34:W416–W422. doi: 10.1093/nar/gkl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 76.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.