Abstract
During recent years, riboswitches have emerged as potential targets for novel antibacterial substances. In this study, we investigated how one flavin analog, roseoflavin, affected the gene-expression, growth and infectivity of the human bacterial pathogen Listeria monocytogenes to determine the potential of this analog to function as an antibacterial substance. The results indicate that roseoflavin has a profound inhibiting effect on the growth of L. monocytogenes at very low concentrations. Also, expression of the gene located downstream of the FMN riboswitch, a riboflavin transporter, was blocked by the addition of roseoflavin. Base-substitution mutations in the FMN riboswitch allowed the bacteria to grow in the presence of roseoflavin, showing that roseoflavin targeted the FMN riboswitch directly. Surprisingly, we found that roseoflavin stimulated L. monocytogenes virulence gene expression and infection abilities in a mechanism independent of the FMN riboswitch. Our results suggest that roseoflavin can block growth but also enhance Listeria virulence.
Key words: riboflavin, roseoflavin, riboswitch, analogs, metabolites, Listeria, virulence
Introduction
The increasing problem of antibiotic resistance calls for new types of antibacterial treatments. Lately, riboswitches have emerged as possible targets for novel antiobiotics.1,2 Riboswitches are metabolite-binding RNA structures usually found in the 5′ untranslated regions (UTRs) of prokaryotic mRNAs.3 After binding their metabolite substrate selectively, riboswitches undergo a conformational change which ultimately results in an altered expression of the downstream gene.4 In most cases, binding of the metabolite to the riboswitch results in an inhibited expression of the downstream gene, either by a termination of the RNA synthesis or by the formation of a hairpin structure that masks the Shine-Dalgarno sequence.1 Moreover, riboswitches are frequently responsible for feedback mechanisms, i.e., riboswitches often regulate the expression of the biosynthetic enzymes or transport proteins which are responsible for maintaining the concentration of the riboswitchbinding metabolite at a sufficient level inside the cell.2
In certain bacteria, such as Bacillus subtilis, flavin mononucleotide (FMN) riboswitches are responsible for regulating the expression of genes involved in the metabolism of riboflavin (vitamin B2).5,6 Riboflavin is the precursor of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), two cofactors essential for several flavoproteins involved in cell metabolism.7 The Gram-positive bacterium Listeria monocytogenes, a close relative to B. subtilis, lacks the ability to synthesize riboflavin de novo and is thus dependent on riboflavin uptake from the environment.6,8
Two FMN riboswitches have previously been identified in B. subtilis, one (ribD) regulates the gene expression from a riboflavin biosynthetic operon and the other (ribU) regulates the expression of a riboflavin transporter, YpaA.5,9 In contrast to B. subtilis, only one FMN riboswitch, Rli96, has been found in L. monocytogenes.10,11 In this case, the downstream gene, lmo1945, encodes a putative riboflavin transporter similar to the riboflavin transporter YpaA of B. subtilis. L. monocytogenes thus lacks the riboflavin biosynthetic genes.8,11–13
The inability of L. monocytogenes to synthesize riboflavin de novo, makes the FMN riboswitch an attractive target for novel antibiotic compounds.14 If the expression of the riboflavin transporter could be inhibited, it is reasonable to assume that this would impair the ability of the bacteria to grow and divide. Analogs of riboflavin have previously been suggested as potential antibacterial compounds.1,2,7,15,16 A possible advantage with this approach is that no riboswitches of any class has yet been found in humans.1
Previously, the antibiotic substance roseoflavin (8-dimethyl-amino-8-demethyl-D-riboflavin), produced by Streptomyces davawensis,7,17 has, as well as riboflavin, been shown to bind the FMN riboswitch of Fusobacterium nucleatum in vitro.18 In addition, it was shown that roseoflavin was able to downregulate the expression of ribD riboswitch-lacZ reporter genes in B. subtilis.16,19
In this study, we investigated how roseoflavin and riboflavin affect the expression of the FMN riboswitch (Rli96) and its downstream gene, lmo1945, in L. monocytogenes and what effect these compounds have on the growth of the bacterium. Our results showed that roseoflavin was able to block the growth of L. monocytogenes and that both riboflavin and roseoflavin could inhibit expression of lmo1945 by binding to the FMN riboswitch directly. By studying the expression of the virulence genes hly, actA, plcA and prfA in roseoflavin- and riboflavin-treated bacteria, we also examined if these compounds were able to alter the virulence properties of L. monocytogenes. Surprisingly, the results indicate that roseoflavin treatment increases listerial virulence while treatment with riboflavin makes L. monocytogenes less virulent.
Results
Roseoflavin inhibits the growth of L. monocytogenes and downregulates expression of lmo1945 in minimal medium.
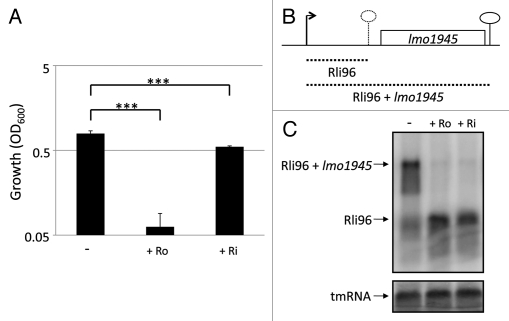
Riboflavin as well as roseoflavin (Fig. 1), a naturally occurring antibacterial analog of riboflavin, were examined for their abilities to inhibit listerial growth in minimal medium. It should be noted that L. monocytogenes is auxotrophic for riboflavin and hence requires a small amount of riboflavin in the medium (13.3 µM20). Roseoflavin, but also riboflavin to some extent, was found to significantly impair the growth of L. monocytogenes at a concentration of 100 µM (Fig. 2A). In B. subtilis, addition of roseoflavin reduced expression of the ribD-operon located downstream of a FMN-riboswitch after either the riboflavin transporter was overexpressed,16 or the ribD-promoter was replaced by the constitutive lysC promoter.19 In the latter study, promoter mutations and in-line probing experiments showed that roseoflavin indeed acted at the ribD-FMN riboswitch.19 To test the effect of riboflavin and roseoflavin on lmo1945 expression in L. monocytogenes (the gene downstream of the FMN riboswitch Rli96, Fig. 2B), these compounds were added to a growing culture (OD600 = 0.25) of L. monocytogenes for approximately 1.5 generations at a concentration of 100 µM. As seen in the northern blot, treatment with roseoflavin, as well as riboflavin, completely inhibited the expression of lmo1945 (Fig. 2C). Instead, transcription terminated at the FMN riboswitch, Rli96. These results clearly indicated that roseoflavin and riboflavin (or more likely, their phosphorylated derivatives16,19) are able to bind Rli96 and cause a structural alteration in the FMN riboswitch leading to transcriptional termination.
Figure 1.
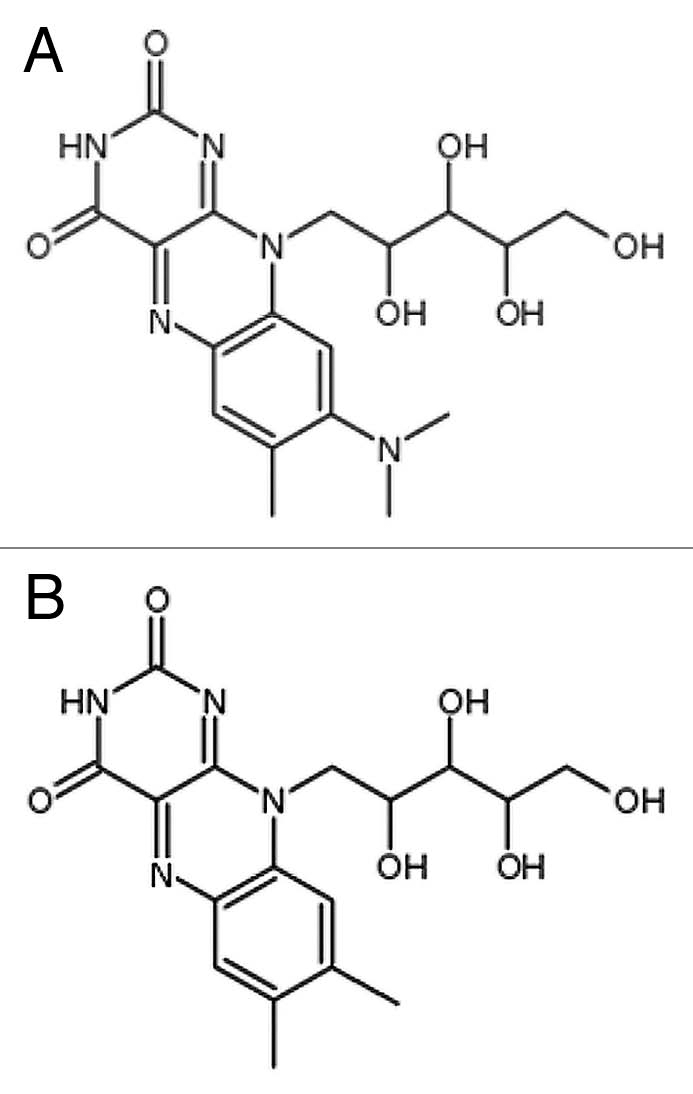
Chemical structures of natural metabolites and analogs. (A) Roseoflavin (B) Riboflavin.
Figure 2.
Roseoflavin inhibits the growth of L. monocytogenes and expression of Imo1945 in minimal medium. (A) Bacteria were grown for 10 hours in minimal medium in the absence or presence of roseoflavin (Ro) or riboflavin (Ri) at a concentration of 100 µM. Growth was measured by optical density (OD600). Samples were compared to the control without added compound and shown as mean values with standard deviations (n = 4) and statistical analysis [Student t-test (two-tailed) (***p < 0.001)]. (B) Genetic context of the FMN riboswitch Rli96 and the downstream gene Imo1945. Transcription initiates at a promoter generating Rli96. In presence of a functional metabolite binding Rli96, a structural re-arrangement causes transcription termination at the hatched lollipop, generating Rli96 (hatched short line). In absence of such metabolite, transcription proceeds and an Rli96 + Imo1945 transcript is formed (hatched longer line). (C) L. monocytogenes was grown in minimal medium to an OD600 = 0.25 when roseoflavin (Ro) or riboflavin (Ri) (100 µM) was added for ∼1.5 generations before RNA extraction. Northern blot was hybridized with PCR-generated, radioactively labeled DNA probes complementary to Rli96 and tmRNA (control), respectively.
Roseoflavin can inhibit L. monocytogenes growth and lmo1945 expression also in BHI.
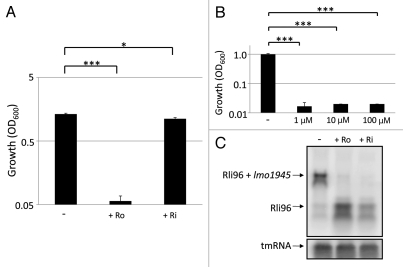
During an infection, it is more likely that L. monocytogenes encounters a milieu rich in nutrients and vitamins inside the host. We therefore examined if the effect exerted by roseoflavin and riboflavin in minimal medium could be observed in BHI (Brain Heart Infusion), a rich growth medium, as well. Presence of especially roseoflavin significantly reduced growth of L. monocytogenes in BHI (Fig. 3A). Interestingly, the growth of L. monocytogenes was inhibited at concentrations as low as 1 µM of roseoflavin (Fig. 3B). Moreover, the addition of roseoflavin and riboflavin to L. monocytogenes dramatically decreased expression of lmo1945, in a manner similar to the effect observed in minimal medium (Fig. 3C). Overall, our results show that roseoflavin is a very potent FMN analog blocking L. monocytogenes growth and expression of lmo1945 also in rich growth media.
Figure 3.
Roseoflavin inhibits the growth of L. monocytogenes and the expression of Imo1945 in BHI. Bacteria were grown for 10 hours in BHI in the absence or presence of roseoflavin (Ro) or riboflavin (Ri) at a concentration of 100 µM. Growth was measured by optical density (OD600). Samples were compared to the control without added compound and shown as mean values with standard deviations (n = 3) and statistical analysis [Student t-test (two-tailed) (***p < 0.001) or (*p < 0.05)]. (B) Bacteria were grown for 10 hours in BHI in the absence or presence of roseoflavin at a concentration of 1, 10 or 100 µM, respectively. Growth was measured by optical density (OD600). Samples were compared to the control without added compound and shown as mean values with standard deviations (n = 3) and statistical analysis [Student t-test (two-tailed) (***p < 0.001)]. (C) L. monocytogenes was grown in BHI to an OD600 = 0.25 when roseoflavin (Ro) or riboflavin (Ri) (100 µM) was added for ∼1.5 generations before RNA extraction. Northern blot was hybridized with PCR-generated, radioactively labeled DNA probes complementary to Rli96 and tmRNA (control), respectively.
Mutations in the FMN riboswitch allow Listeria to grow in the presence of roseoflavin.
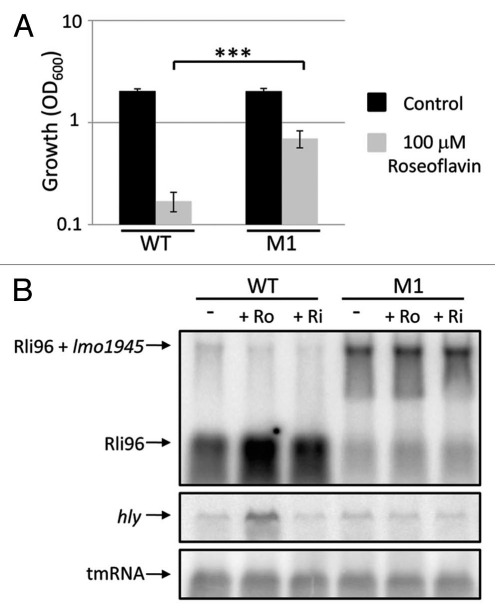
To examine if the interaction between riboflavin/roseoflavin and the FMN riboswitch was direct, two bases were altered (GG to AA) at position 51–52 (M1) in the FMN recognition domain of the FMN riboswitch and these altered bases were introduced into the native chromosomal locus. A base-substitution (G to A) at the corresponding region of the B. subtilis ribD riboswitch allowed expression of the gene downstream of the FMN riboswitch also in presence of riboflavin/roseoflavin.19 The M1 strain was able to grow significantly better than the wild-type strain in the presence of 100 µM of roseoflavin (Fig. 4A). To test whether the expression of the downstream gene lmo1945 was affected in the M1 strain in presence or absence of riboflavin/roseoflavin, a northern blot experiment was performed and in the M1 strain, lmo1945 was expressed in the presence of 100 µM of roseoflavin or riboflavin (Fig. 4B). This was in contrast to the wild-type strain where only a terminated riboswitch fragment could be observed in presence of riboflavin/roseoflavin (Fig. 4B). These data clearly show that roseoflavin directly targets the FMN riboswitch, repressing expression of lmo1945 and hence inhibiting growth of L. monocytogenes.
Figure 4.
Mutations in the FMN riboswitch allows Listerial growth and expression of Imo1945 in the presence of roseoflavin. (A) Wild-type or M1 strains were grown for 10 hours in BHI in the absence or presence of roseoflavin at a concentration of 100 µM. Growth was measured by optical density (OD600). Samples were compared to the control without added compound and shown as mean values with standard deviations (n = 6) and statistical analysis [Student t-test (two-tailed) (***p < 0.001)]. (B) L. monocytogenes wild-type or M1 strains were grown in BHI to an OD600 = 0.25 when roseoflavin (Ro) or riboflavin (Ri) (100 µM) was added for ∼1.5 generations before RNA extraction. Northern blot was hybridized with PCR-generated, radioactively labeled DNA probes complementary to Rli96, hly and tmRNA (control), respectively.
The expression of the virulence genes hly, actA and plcA is upregulated in bacteria treated with roseoflavin.
Due to its potential role as an antibacterial drug, we were interested to examine what effect roseoflavin, but also riboflavin, had on virulence gene-expression in L. monocytogenes. RNA was prepared from cultures grown in the presence or absence of roseoflavin and riboflavin (100 µM). Northern membranes were probed against mRNA of the virulence genes hly and actA. hly encodes the virulence factor listeriolysin O, a protein essential for the bacterial escape from the phagosomes inside the host cell,21,22 while ActA, the protein product of actA, is responsible for the intra- and intercellular movement of L. monocytogenes by coordinating actin polymerization.22,23 Surprisingly, the results showed that roseoflavin induced the expression of hly and actA in L. monocytogenes grown in minimal medium, whereas riboflavin dramatically downregulated the expression of these virulence genes (Fig. 5).
Figure 5.
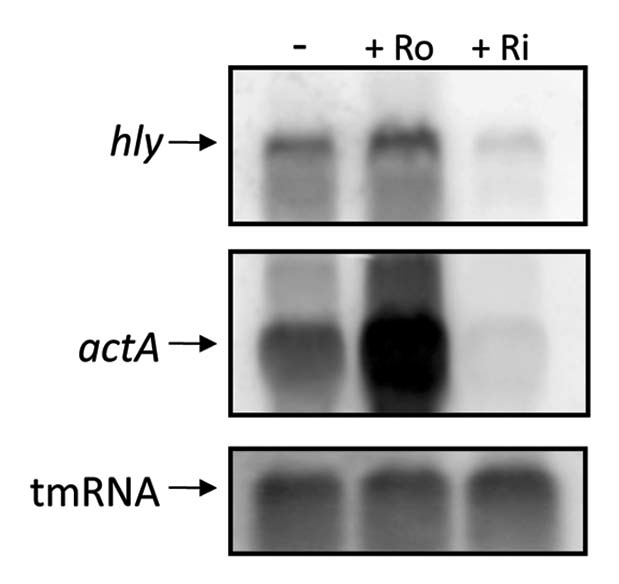
Treatment with roseoflavin results in a decreased expression of the virulence genes hly and actA in minimal medium. L. monocytogenes was grown in minimal medium to an OD600 = 0.25 when roseoflavin (Ro) or riboflavin (Ri) (100 µM) was added for ∼1.5 generations before RNA extraction. Northern blot was hybridized with PCR-generated, radioactively labeled DNA probes complementary to hly, actA and tmRNA (control), respectively.
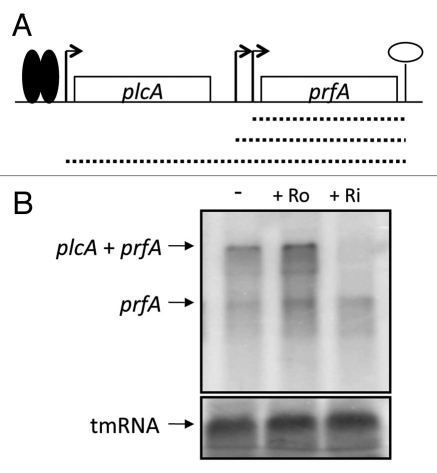
The expression of hly and actA is positively regulated by the transcriptional regulator PrfA22 and to test if the expression of prfA was affected by roseoflavin and riboflavin comparable to hly and actA, the northern membranes were hybridized with DNA probes complementary to prfA. Expression of prfA is initiated at three different promoters.24,25 Expression from two of these promoters results in prfA monocistronic messengers, whereas expression from the third promoter results in a plcA-prfA bi-cistronic messenger (Fig. 6A). Importantly, expression of the plcA-prfA messenger requires a post-translationally active PrfA.22 plcA encodes a phosphoplipase (PlcA) important for the escape from the phagosome.22 Although we detected an upregulation of the prfA expression in roseoflavin-treated bacteria and a downregulation in bacteria treated with riboflavin, this effect was most striking at the plcA-prfA bi-cistronic transcript and almost absent at the prfA monocistronic transcripts (Fig. 6B). Hence, our results indicate that transcription of PrfA is not affected by riboflavin or roseoflavin, but rather its post-translational modification. In the M1 strain, the expression of lmo1945 downregulated hly gene expression also in the presence of riboflavin and roseoflavin (Fig. 4B). This could be explained if an overexpression of the riboflavin transporter (encoded by lmo1945) allows for a maximal uptake of riboflavin from the surrounding media and consequently a downregulated virulence gene expression. Conclusively, our results suggest that the riboflavin/roseoflavin effect on virulence gene expression is uncoupled from the riboswitch regulated expression of lmo1945.
Figure 6.
Roseoflavin does not decrease prfA monocistronic expression. (A) Schematic drawing of the prfA locus. Transcription of prfA initiates at two promoters, generating short prfA transcripts (hatched short lines). Active PrfA protein (solid spheres) can bind to the plcA promoter and stimulate transcription that generates plcA-prfA bi-cistronic transcript (long hatched line). (B) L. monocytogenes was grown in minimal medium to an OD600 = 0.25 when roseoflavin (Ro) or riboflavin (Ri) (100 µM) was added for ∼1.5 generations before RNA extraction. Northern blot was hybridized with PCR-generated, radioactively labeled DNA probes complementary to prfA and tmRNA (control), respectively.
Riboflavin reduces the ability of L. monocytogenes to infect and proliferate within HeLa cells.
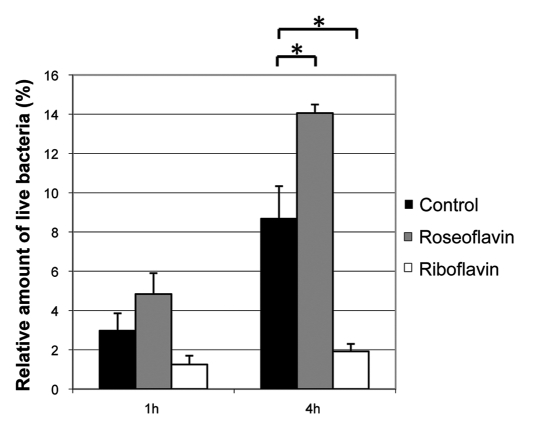
To examine if the changes in virulence gene expression induced by roseoflavin and riboflavin affected the virulence properties of L. monocytogenes, an infection experiment was performed. Bacteria pretreated for one generation with 100 µM roseoflavin or 100 µM riboflavin, respectively, were allowed to infect cells of the cervical cell line HeLa. The result showed that pretreatment with riboflavin reduced the ability of L. monocytogenes to infect and proliferate inside human cells, while pretreatment with roseoflavin increased the infectivity of the bacterium (Fig. 7). These results were in agreement with the virulence gene expression pattern observed in presence of riboflavin and roseoflavin (Figs. 5 and 6). Conclusively, our results indicate that roseoflavin and riboflavin have disparate effects at the regulation of genes controlled by the transcriptional regulator PrfA.
Figure 7.
Riboflavin reduces the ability of L. monocytogenes to infect and proliferate inside HeLa cells. HeLa cells were infected with L. monocytogenes pretreated with roseoflavin or riboflavin (100 µM) for 1 generation at OD600 = 0.2. By viable counting, the amount of live bacteria was measured 1 hour or 4 hour post-infection, respectively, and these quantities were related to the number of live bacteria added to the HeLa cells and shown as mean values with standard deviations (n = 2). The invasivity of the treated bacteria were compared to untreated bacteria using Student t-test (two-tailed) (*p < 0.05).
Discussion
In this study, we have investigated the effect of riboflavin and its analog roseoflavin on bacterial growth and gene expression at an FMN riboswitch in the bacterial pathogen Listeria monocytogenes. Roseoflavin inhibited bacterial growth and the expression of the gene located downstream of the target riboswitch, lmo1945, in both minimal medium and in rich medium. The ability of roseoflavin to inhibit the growth of L. monocytogenes lies within its capacity to bind the FMN riboswitch, Rli96, as evidenced by base-substitution mutations within the FMN riboswitch (Fig. 4). Such a mutant (M1) did not terminate transcription in presence of riboflavin or roseoflavin, but instead constitutively expressed the downstream gene lmo1945. In contrast to the wild-type strain, the M1 strain could grow in the presence of roseoflavin (Fig. 4). In agreement with our results, roseoflavin has previously been shown to directly bind to the ribD riboswitch in B. subtilis.16,19
Intriguingly, we observed an increased expression of the virulence genes, plcA, hly and actA as well as an increased ability of roseoflavin-treated bacteria to infect and proliferate inside HeLa cells. In contrast, riboflavin repressed expression of the virulence genes and decreased the cellular infection (Figs. 5–7). Previously, it has been shown that stress conditions such as nutritional starvation results in an increased expression of hly and actA.26 It might therefore be possible that the riboflavin deficiency, which is likely to follow upon treatment with roseoflavin, results in a similar upregulation of these virulence genes. Since the expression of the prfA monocistronic messenger remains essentially unaffected in presence/absence of roseoflavin or riboflavin, it is fair to believe that the virulence effect by roseoflavin and riboflavin lies at the level of post-translational modification of the PrfA protein. Interestingly, the M1 mutant strain carrying a FMN riboswitch being constitutively “ON” (constant expression of lmo1945) did not show any altered expression of hly in presence of riboflavin or roseoflavin (Fig. 4). This indicates that the effect of roseoflavin and riboflavin on PrfA activity is not mediated via the FMN riboswitch per se, but rather through uptake of these metabolites by the riboflavin transporter protein encoded by lmo1945. The mechanism by how PrfA becomes post-translationally modified still remains obscure, but it has been suggested to include uptake of glucose.27,28 Our data indicate that the concentration of roseoflavin affect expression of virulence genes in L. monocytogenes, hence giving an important role for the riboflavin transport protein to control activity of PrfA.
It is likely that roseoflavin employs one or several mechanisms, other than binding to the FMN riboswitch, to affect virulence properties of L. monocytogenes. For example, roseoflavin competes with riboflavin for the transport through YpaA in B. subtilis8 and the ability of roseoflavin to interact with the enzymes catalyzing the conversion of riboflavin to FMN and FAD has not yet been studied in L. monocytogenes. Our results show that listerial growth can be inhibited in both minimal and rich medium by treating bacteria with roseoflavin. Roseoflavin inhibits the growth of L. monocytogenes at very low concentrations (1 µM) in rich medium, indicating a dominant negative effect of roseoflavin over riboflavin (i.e., possible presence of riboflavin in the medium would be outcompeted by roseoflavin), at the FMN riboswitch.
The FMN riboswitch remains an interesting target for the development of novel antibiotics, particularly for treating infections caused by bacteria lacking riboflavin biosynthetic genes. Structural derivatives of roseoflavin and riboflavin might prove to be even more effective. It would be interesting to explore what effect roseoflavin and derivatives of this molecule have on other bacteria, such as Streptococcus pyogenes, also lacking riboflavin biosynthetic genes.6
The capacity of roseoflavin to induce the expression of the virulence genes hly, actA and plcA and the increased ability of roseoflavin-treated bacteria to infect and proliferate inside HeLa cells could prove deleterious for a drug candidate. However, the growth inhibition induced by roseoflavin possibly makes the increased virulence less important in a broader physiological and immunological context.
Experimental Procedures
Strains and cell culture.
Strains, cell culture and growth media. L. monocytogenes EGDe was grown in either Brain Heart Infusion (BHI) broth (Fluka) or minimal medium.20 LB medium solidified with agar (LA plates) was used during the viable count experiments. Cells of the cervical cell line HeLa were grown in RPMI (Sigma) at 37°C in 5% CO2. For knock-out constructions erythromycin was added at a concentration of 7 µgml−.
Chemicals and primers.
Roseoflavin (MP Biomedicals, LLC) was dissolved in water, while dimethyl sulfoxide (DMSO) was used as solvent for riboflavin (Sigma). Dissolved chemicals were stored at +4°C. Table 1 lists all the primers used in this study.
TABLE 1.
Primers used in this study
| Primer | Primer sequence 5′-3′ | |
| Detection primers: | ||
| tmRNA | #2-U | CGG CAC TTA ATA TCT ACG AGC |
| #2-D | CCT CGT TAT CAA CGT CAA AGC C | |
| Rli96 | Rli96-U | GTT CAT CTT CGG GGC AGG |
| Rli96-D | CAA CTA CTT CTC CCA TCC AG | |
| actA | actA–U | TGA TGG TGG TTT TCA TTA CTG C |
| actA–D | CTT ACT TCA CGT GCA GTT TCG | |
| hly | hly-U | GAA GCA AAG GAT GCA TCT GC |
| hly-D | CCA TCT TTG TAA CCT TTT CTT GG | |
| prfA | prfA-3 | GAA ATT GTT TTG GTT TTA TCC CG |
| prfA-4 | TGT AAA AAA CAT CAT TTA GCG | |
| Base-substitution and knock-out primers: | ||
| FMN-RS-MutU: | GCA ATT CCC GAC CGG TAA TTA AAG TCC ACG ATC | |
| FMN-RS-MutD: | GAT CGT GGA CTT TAA TTA CCG GTC GGG AAT TGC | |
| lmo1945-A: | GCA TCG ACA GAC TCA GTG G | |
| lmo1945-D: | GTA CCT GAC ACG ATG TAA AGC | |
Growth assay.
Over-night cultures of L. monocytogenes (in BHI) were diluted 100-fold in either BHI or minimal medium and the indicated compounds were added to a final concentration of 100 µM, if not otherwise indicated. After growing bacteria for 10 h at 37°C with aeration, OD600 was measured in a spectrophotometer (Shimadzu UV mini 1240) and the obtained values were compared to the control.
Isolation of RNA.
Isolation of RNA was performed essentially as described in reference 29. Listeria monocytogenes over-night cultures (in BHI or LB) were diluted by a factor of 100 in either minimal medium or BHI and incubated at 37°C with shaking. At OD600 = 0.25, metabolites and analogs were added to a final concentration of 100 µM. Subsequently, at OD600 = 0.6, bacteria were pelleted (Beckman Coulter Avanti J-26 XP, JA 25.50 rotor; 11,000 rpm, 3 minutes, 4°C) and frozen at −80°C. Pelleted bacteria were dissolved in a resuspension solution [10% glucose, 12.5 µM Tris (pH 7.5) and 5 µM EDTA] and fresh EDTA (0.5 M). After transferring the samples to bead beater tubes containing approximately 0.4 g glass beads and 0.5 ml acid phenol (pH 4.5), samples were homogenized in a mini bead beater (Biospec products) for 1 minute and 15 seconds. The obtained mixture was centrifuged (Beckman Coulter Microfuge® 22R; 14,000 rpm, 5 minutes, 4°C) before the addition of 1 ml trizol (Ambion) and 100 µl chloroform/isoamylalcohol solution (24:1) to the aqueous (upper) phase. After centrifugation, two more chloroform/IAA extractions followed and RNA was later precipitated in 0.7 volumes of isopropanol at −20°C (20 minutes). RNA was pelleted (14,000 rpm, 20 minutes, 4°C), dried (Savant Integrated SpeedVac System) and dissolved in 200 µl DEPC-treated water. Samples were treated with recombinant DNase I (Roche) for 30 minutes at 37°C and by the addition of phenol/chloroform/IAA (1:24:1) [pH 6.6], the reaction was terminated. Subsequently, a Chloroform/IAA extraction was performed on the centrifuged samples. RNA was precipitated in 1/10 volumes of DEPC-treated NaAc (pH 4.5) and 2.5 volumes of ethanol (99.5%), pelleted and later dissolved in 200 µl DEPC-treated water. The isolated RNA was analyzed on a 1.2% agarose gel and the concentration was measured on a Nanodrop (Nanodrop ND-1000 spectrophotometer). Only RNA isolations showing distinct precursor bands on the agarose gel were used for further experiments.
Northern blotting.
Total RNA was separated on a 1% formaldehyde agarose gel and blotted onto a Hybond-N-membrane (GE Healthcare). The membrane was hybridized with the indicated, PCR-amplified, DNA fragments labeled with α 32P dATP (PerkinElmer) using the GE healthcare Megaprime DNA labeling kit. Oligonucleotides used for PCR amplification are found in Table 1. A STORM device (Molecular Dynamics) was used to develop the blot. Prior to re-hybridizations, membranes were stripped in 0.1% boiling SDS.
Tissue culture invasion experiment with HeLa cells.
An over night culture of L. monocytogenes in LB was diluted 50-fold in minimal medium and incubated at 37°C with aeration. At OD600 = 0.2, the medium was supplemented with either roseoflavin or riboflavin to a final concentration of 100 µM. At OD600 = 0.4, bacteria were pelleted (6,000 rpm, 3 minutes) and resuspended in PBS. Bacteria were then diluted 5-fold in RPMI and added to HeLa cells growing in a 24-well plate (multiplicity of infection 10:1). The infection plate was centrifuged (1,500 rpm, 5 minutes) and incubated for 1 hour at 37°C in 5% CO2. The medium containing non-invaded, extracellular bacteria was removed and cells were washed with PBS. New RPMI containing gentamicin (Invitrogen; 50 µgml−1) was added in order to kill remaining extracellular bacteria and the plate was incubated at 37°C in 5% CO2 for another 4 hours. Samples were collected after 1 and 4 hours (after an additional round of washing with PBS). To lyse the eukaryotic cells, PBS supplemented with 1% Triton X-100 (Sigma) was used. Samples were spread onto LA plates which were incubated at 37°C over night and the number of colony forming units was related to the number of live bacteria added to the HeLa cells.
Construction of strain M1.
To replace GG at position 51–52 with AA a two step PCR-strategy was undertaken. First, the primer-pairs: lmo1945-A together with FMN-RS-MutD and lmo1945-D together with FMN-RS-MutD were used to amplify the riboswitch and its surrounding region. These primer-pairs generated a 525 and a 513 base-pair long fragment, respectively. Since with FMN-RS-MutU and with FMN-RS-MutD overlap, the two PCR products could be used as a template for a new PCR using lmo1945-A and lmo1945-B as primers. A 1,003 base-pair long fragment was purified before digestion with SalI (Restriction site in primer lmo1945-A). After purification, the fragment was introduced into SalI/EcoRV digested pMAD30 and the resulting construct was used to transform Escherichia coli strain Novablue (Novagene). The construct was sequenced and to achieve allelic exchange, we followed the protocol of Arnaud et al. 2004. Briefly, the construct was electroporated into L. monocytogenes EGDe at 2,500 V, 200 Ω and 25 µF. Transformants were selected at 30°C on BHI medium containing 7 µgml−1 erythromycin (BHI-Em). One colony was grown in BHI-Em broth at 43.5°C, and the culture was plated onto BHI-Cm agar at 43.5°C. One colony was resuspended in 100 µl of BHI (without erythromycin), and 10 ml of BHI broth was inoculated with 1 µl of this suspension and incubated at 30°C for 6 hours. The temperature was raised to 39°C and after another 3 hours of incubation, dilutions were spread onto LA plates containing X-Gal (50 µgml−1). White erythromycin-sensitive colonies were analyzed by sequencing of PCR amplified fragments generated by using lmo1945-A and lmo1945-D as primers.
Acknowledgements
J.J. is supported by Umeå University, the Swedish Research Council grants (K2011-56X-15144-08-6 and 621-2009-5677) and an ERC starting grant (no 260764-RNAntibiotics).
References
- 1.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 2.Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 4.Soukup JK, Soukup GA. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack M, Grill S. Riboflavin analogs and inhibitors of riboflavin biosynthesis. Appl Microbiol Biotechnol. 2006;71:265–275. doi: 10.1007/s00253-006-0421-7. [DOI] [PubMed] [Google Scholar]
- 8.Vogl C, Grill S, Schilling O, Stülke J, Mack M, Stolz J. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol. 2007;189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. 2005;33:121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 12.Kreneva RA, Gelfand MS, Mironov AA, Iomantas IuA, Kozlov IuI, Mironov AS, Perumov DA. Study of the phenotypic occurence of ypaA gene inactivation in Bacillus subtilis. Genetika. 2000;36:1166–1168. [PubMed] [Google Scholar]
- 13.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, et al. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulhbacher J, St. Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol. 2010;10:551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Bacher A, Eberhardt S, Eisenreich W, Fischer M, Herz S, Illarionov B, et al. Biosynthesis of riboflavin. Vitam Horm. 2001;61:1–49. doi: 10.1016/s0083-6729(01)61001-x. [DOI] [PubMed] [Google Scholar]
- 16.Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- 17.Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Letter: Roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot (Tokyo) 1974;27:86–87. [PubMed] [Google Scholar]
- 18.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan-Thanh L, Gormon T. A chemically defined minimal medium for the optimal culture of Listeria. Int J Food Microbiol. 1997;35:91–95. doi: 10.1016/s0168-1605(96)01205-6. [DOI] [PubMed] [Google Scholar]
- 21.Gedde MM, Higgins DE, Tilney LG, Portnoy DA. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect Immun. 2000;68:999–1003. doi: 10.1128/iai.68.2.999-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dussurget O, Pizarro-Cerda J, Cossart P. Molecular determinants of Listeria monocytogenes virulence. Annu Rev Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- 23.Kocks C, Marchand JB, Gouin E, d'Hauteville H, Sansonetti PJ, Carlier MF, Cossart P. The unrelated surface proteins ActA of Listeria monocytogenes and IcsA of Shigella flexneri are sufficient to confer actinbased motility on Listeria innocua and Escherichia coli respectively. Mol Microbiol. 1995;18:413–423. doi: 10.1111/j.1365-2958.1995.mmi_18030413.x. [DOI] [PubMed] [Google Scholar]
- 24.Freitag NE, Portnoy DA. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 25.Freitag NE, Port GC, Miner MD. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokolovic Z, Riedel J, Wuenscher M, Goebel W. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol Microbiol. 1993;8:219–227. doi: 10.1111/j.1365-2958.1993.tb01566.x. [DOI] [PubMed] [Google Scholar]
- 27.Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, et al. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 29.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 30.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]