Abstract
Unlike AU-rich elements (ARes) that are largely present in the 3′UTRs of many unstable mammalian mRNAs, the function and abundance of GU-rich elements (GRes) are poorly understood. We performed a genome-wide analysis and found that at least 5% of human genes contain GRes in their 3′UTRs with functional over-representation in genes involved in transcription, nucleic acid metabolism, developmental processes and neurogenesis. GRes have similar sequence clustering patterns with ARes such as overlapping GUUUG pentamers and enrichment in 3′UTRs. Functional analysis using T-cell mRNA expression microarray data confirms correlation with mRNA destabilization. Reporter assays show that compared with ARes the ability of GRes to destabilize mRNA is modest and does not increase with the increasing number of overlapping pentamers. Naturally occurring GREs within U-rich contexts were more potent in destabilizing GFP reporter mRNAs than synthetic GREs with perfectly overlapping pentamers. Overall, we find that GREs bear a resemblance to AREs in sequence patterns but they regulate a different repertoire of genes and have different dynamics of mRNA decay. A dedicated resource on all GRE-containing genes of the human, mouse and rat genomes can be found at brp.kfshrc.edu.sa/GredOrg.
Keywords: GU-rich elements, AU-rich elements, post-transcriptional regulation of gene expression, mRNA turn-over, T-cell stimulation
Introduction
Post-transcriptional gene regulation is essential for normal function of eukaryotic cells and organisms. The mRNA often contain cis-acting regulatory elements in its 3′ untranslated region (3′UTR) such as adenylates/uridylates (AU)-rich elements (AREs), which destabilize the mRNA and can repress translation. AREs are made up of the core pentamer AUUUA and the minimal functional element is the nonamer UUAUUUA(U/A)(U/A).1,2 AREs often exist in overlapping arrangements of several pentamers in a region that is rich in As and Us.3 The ability of AREs to destabilize mRNA increases with the number of overlapping pentamers.1,2 An updated genome wide search showed that at least 10–15% of human genes contain AREs in the 3′UTR.4 These genes mediate regulatory processes such as transmission of external signals, RNA metabolism and developmental processes, particularly those involving early and transient responses.4,5 AREs have been the subject of intense research for more than 2 decades. They are targets of RNA binding proteins such as TTP, HuR, AUF1, KSRP, TIA1 and others (reviewed in ref. 6 and 7). ARE-containing genes are also responsive to external stimuli like stress and inflammatory mediators transduced by MAPK signaling pathways.8
An element known as a GU-rich element (GRE) with a similar sequence pattern to the ARE was discovered in a subset of unstable transcripts in human T-cells and was shown to destabilize the mRNA.9 The core pentamer of a GRE is GUUUG, and the minimal consensus found in the labile mRNA group is UGUUUGUUUGU.9 GREs are also targets for at least one RNA binding protein; CUG-binding protein 1 (CUG-BP1).9 This was further confirmed recently by immunoprecipitation of mRNAs associated with CUG-BP1.10,11 CUG-BP1 is a member of the highly conserved CELF family of RNA-binding proteins that are post-transcriptional regulators of deadenylation, mRNA decay, translation and pre-mRNA processing.12–14 Considering the remarkable similarity of the GRE consensus motif to the ARE consensus motif and lacking an existing comprehensive resource, we performed a genome-wide analysis for GRE-containing genes (subsequently called GRE-genes) and found that at least 5% of human genes contain GREs. Like AREs, GREs are found in arrangements of 2 to 5 overlapping pentamers. Reporter assays of artificial and natural GREs and T-cell mRNA decay data confirmed the mRNA destabilizing activity of GREs but showed marked differences in the mode of function between AREs and GREs. A direct comparison using bioinformatic tools, GFP reporter assays and functional mRNA half-life measurements in activated cells, revealed that the 2 elements appear to share similar sequence patterns and enrichment in the human genes, but regulate distinct mRNA decay and translational pathways.
Results
Genes with GREs are a rich and abundant class.
We performed a genome-wide analysis of the presence of GRE motifs in the 3′UTRs of the human genes. Overall, we analyzed 31,524 human genes and found at least 5.5% to contain the minimum GRE motif in at least one transcriptional variant of the gene (Table 1). We noted that among the genes with no identified GRE, about 19% of the protein coding genes had no annotated 3′UTR and 27% overall were non-coding and by definition, had no 3′UTR to begin with (i.e., truncated annotation or non-coding genes). Thus our results represent a minimum estimate. The association of the GRE motifs with the 3′UTR was examined, and we found that compared with the other regions of the transcript, the GREs are more abundant in the 3′UTR (Table 1). Owing to the close resemblance to the ARE motifs, we similarly separated GRE-genes into 5 clusters based on the number of GUUUG pentamers. Cluster I contains genes with 5 overlapping pentamers, cluster II genes with 4 overlapping pentamers, cluster III genes with 3 overlapping pentamers, cluter IV genes with 2 overlapping pentamers flanked by 2 Gs or U (K) and cluster V with one pentamer flanked with 3 Ks (Table 1). Specifically, for GRE clusters I, II and III combined, we found an almost 11-fold enrichment in GRE sites compared with the 5′UTR and more than 12-fold enrichment compared with the Coding Region Sequence (CDS) of the transcripts examined (Table 1). This finding is similar to that found with ARE patterns4,5 and suggests that GREs are 3′UTR specific post-transcriptional cis-elements with similar sequences patterns to AREs.
Table 1.
GRE motifs and their abundance in human genes
| GRE Motif | Cluster | 3′UTR | 5′UTR | CDS |
| GUUUGUUUGUUUGUUUGUUUG | I | 0.12% | 0.01% | 0.01% |
| GUUUGUUUGUUUGUUUG | II | 0.24% | 0.02% | 0.01% |
| GUUUGUUUGUUUG | III | 0.73% | 0.07% | 0.07% |
| KK[GUUUGUUUG]KK | IV | 0.30% | 0.03% | 0.11% |
| KKKU[GUU UG]UKKK | V | 4.15% | 0.44% | 1.41% |
The total number of genes is 31,524. A mismatch was allowed in each motif (Clusters I–III) or in either of the 2 regions flanking the minimal pattern indicated in brackets (IV and V). Kindicates G or U. We allowed a single mismatch against the motifs to enhance sensitivity (at the expense of specificity) and to accommodate the fact that mRNA and EST sequences are typically prone to sequencing errors.
When we examined the set of genes that contained GREs, AREs or both, we observed that the set containing both was small relative to the sets of either. The numbers of GRE- and ARE-genes in cluster groups I–IV were 438 and 729, respectively, while only 34 transcripts contained both sites simultaneously. Interestingly, this overlap though small, is larger than the number expected by chance (p < 0.001; χ2 test). Closer examination of these genes (data not shown) showed no detectable functional class enrichment, enhancing the likelihood that this was a random event.
GRE-genes and ARE-genes have overlapping functional classes.
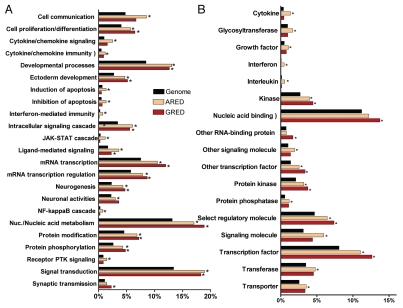
We analyzed GRE-genes for overrepresentation in different functional categories based on the PantherDB (www.pantherdb.org) database annotations, which provides a framework similar to the Gene Ontology but with much simplified structure and statistics. The biological processes that involve the GRE-genes are varying and include nucleic acid metabolism, mRNA transcription, developmental processes, signal transduction and neurogenesis as the most significant processes (Fig. 1A). In terms of molecular pathways, these genes seem to be most closely connected to cytokine-mediated signaling such as the TGFβ signaling pathway. We found, in terms of molecular function, that GRE-genes were mostly transcriptional factors and RNA binding proteins (p < 0.001, χ2 test after multiple testing correction) (Fig. 1). A detailed comparison between ARE- and GRE-genes in terms of over-represented molecular functions and biological processes is shown in Figure 1. In general, GRE-genes follow a pattern similar to AREs, although there are distinct differences. For example, 80% of interferon genes contain AREs, but none have an identifiable GRE. Likewise, many more interleukins, cytokines and glycosyltransferases have AREs when compared with GREs (Fig. 1B). Also, GRE-genes are less prominent than ARE-genes in the JAK-STAT cascade, the NFκB cascade, apoptosis regulation and interferon, cytokine and chemokine mediated immunity and signaling. In contrast, GRE-genes are more often involved in neurogenesis, neuronal activities, synaptic transmission and anterior/posterior patterning (Fig. 1A).
Figure 1.
Differential functional classification of GRE- and ARE-genes. Percentage of genes associated with the annotation terms with respect to the overall size of the data set. RefList is the full human genome gene set of 25,431 genes (in PantherDB); ARED is ARE-genes set of 3,107 genes (that are common with PantherDB); GRED is GRE-genes set of 1,543 genes (that are common with PantherDB). Terms found significantly over-represented (at p < 0.05 after correction for multiple testing) in either of ARED or GRED (with respect to RefList) are shown. *p values (<0.05) indicate over-representation significance compared with RefList: (A) Biological process terms. (B) Molecular function terms.
We assembled an online database of all GRE-genes as a database resource. The website for the database is brp.kfshrc.edu.sa/GredOrg. In the online version, we have extended the GRE-gene search beyond the human genome to include the genomes of the mouse and rat using similar methods. There was significant homology in the GREs among the 3 species, indicating that the GRE represents a conserved posttranscriptional regulatory mechanism (data not published).
Global gene expression analysis suggests GREs are functional post-transcriptional regulators.
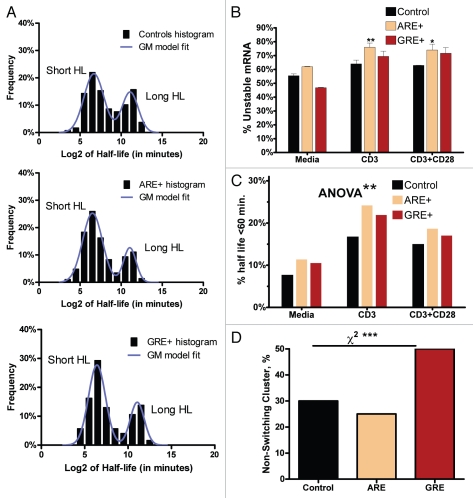
By combining our genome-wide analysis of the GRE or ARE gene content with an existing genome-wide microarray-based profile of T-Cell gene expression15 under various experimental conditions (medium, anti-CD3 or anti-CD+anti-CD28), we were able to observe clear effects of the presence of GRE or ARE sequences on mRNA expression and stability. First, we found that the log2 of half-life values conferred a bimodal mixture of two normal distributions, naturally splitting the transcripts into a short mRNA half-life class and a long mRNA half-life class. This was a consistent characteristic for all groups under all the three experimental conditions (shown with anti-CD3 data, Fig. 2A). We utilized this feature (Fig. 2A) to obtain a data-driven definition of the short-half-life class (also called unstable mRNAs) and then estimate its size relative to the group (i.e., the percentage of unstable mRNAs in controls (no ARE or GRE), in ARE-mRNAs and in GRE-mRNAs; see Fig. 2A). Under all conditions, the percentage of short half-life transcripts in the ARE-mRNAs was always larger than the control. For GRE-mRNA, it was larger than the control at the activated conditions but not with medium only condition. The percentage of short half-life transcripts in ARE-mRNAs was always greater than in GRE-mRNAs (Fig. 2B). Further, the results also suggest that the effects of GREs are weaker than those of AREs. There was an effect of T-cell activation on the overall shift toward a shorter mRNA half life cluster. This is particularly observed with further unstable mRNAs (at half-life <26 = 64 min) (Fig. 2C). Further classification indicates that the genes demonstrated 2 distinct classes: a non-switching class that essentially maintained its stability levels in response to T-cell activation (i.e., short half-lives remained short and long half-lives remained long) and a class that changed its stability patterns. We observe that the non-switching class of GRE-mRNAs constitutes a higher proportion (50%) when compared with the non-switching class of ARE-mRNAs, which constitutes 25% (χ2, p = 0.0004; Fig. 2D).
Figure 2.
GRE- and ARE-motif effect on mRNA stability Microarray-based mRNA half-life data (see Materials and Methods for details) were stratified according to the experimental condition, transcript type and half-life classification (short vs. long half-lives, based upon a Gaussian-mixture model fit. (A) The histograms of log-2 values (columns) of mRNA half-lives in each gene category (Control, ARE-genes and GRE-genes) were obtained and fitted using a bi-modal Gaussian mixture model (line). The control set represent mRNAs with neither ARE nor GRE. A representative model under the anti-CD3 experimental condition is shown. (B) The fraction of short half-life transcripts out of the total within each stratum. Data are Mean ± SEM and represent fitted parameter uncertainties. For each stimulus, ARE-mRNAs and GRE-mRNAs results were compared with the Control using the unpaired t-test with Bonferroni correction for multiple testing. **p < 0.01 and *p < 0.05. Using the 2-way ANOVA test, the effect of the motif (Control, ARE+ or GRE+) was found to be significant at p < 0.01. (C) A simpler classification scheme based on the arbitrary cutoff of 64 (26) minutes to classify short and long half-life transcripts yields essentially the same results as in (A). (D) The proportions of control mRNAs, ARE-mRNAs and GRE-mRNAs that do not change their stability patterns (non-switching class). p < 0.001 using chi-square analysis.
Effects of the length of mRNA decay elements on mRNA stability and steady-state expression.
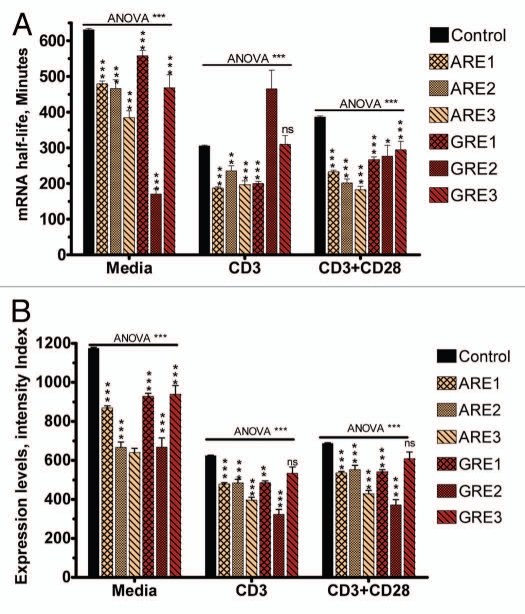
Further examination of the effects of GRE was performed in relationship to the length of the GRE sequences, i.e., the number of overlapping repetitions of the pentamer (GUUUG). Unlike ARE effects, in which longer AREs (3 overlapping pentamers) have consistently stronger effects on mRNA decay than shorter AREs in non-induced and in anti-CD3/anti-CD28 stimulated conditions, there was no similar pattern observed with GREs (Fig. 3A). In general, the effects of GREs on steady-state mRNA expression levels do not appear to be dependent on the number of overlapping GREs, particularly under stimulated conditions (Fig. 3B). In contrast, under both basal and stimulated conditions, AREs consistently appear to have an effect on steady-state mRNA expression levels proportional with the number of overlapping AREs (Fig. 3B). Overall, both AREs and GREs caused a significant reduction in steady-state mRNA expression levels and half-lives, but GREs had a weaker effect. Taken together, these results suggest the two elements have different dynamic effects on mRNA levels.
Figure 3.
GRE and ARE motif-cluster effect on steady-state mRNA expression levels and mRNA decay. Microarray-based mRNA expression and half-life data (see Materials and Methods for details) were stratified according to the experimental condition (media, anti-CD3 and anti-CD3+anti-CD28) and transcript type as indicated. Data are Mean ± SEM for each stratum. Kruskal-Wallis (nonparametric 1-way ANOVA) tests (for different motif strata under each stimulatory condition separately) were all significant (p < 0.005). These were followed by pairwise nonparametric tests against the Control stratum. NS, *, ** and ***indicate p-value after correction for multiple testing (non-significant, <0.05, <0.01 and <0.005 respectively). (A) Effect of motif cluster on average mRNA half life. (B) Effect of motif cluster on steady-state mRNA expression levels.
Effect of natural and synthetic GU-rich elements on reporter mRNA stability.
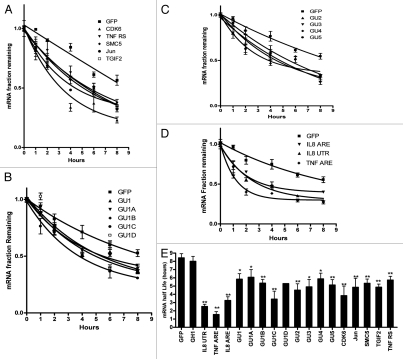
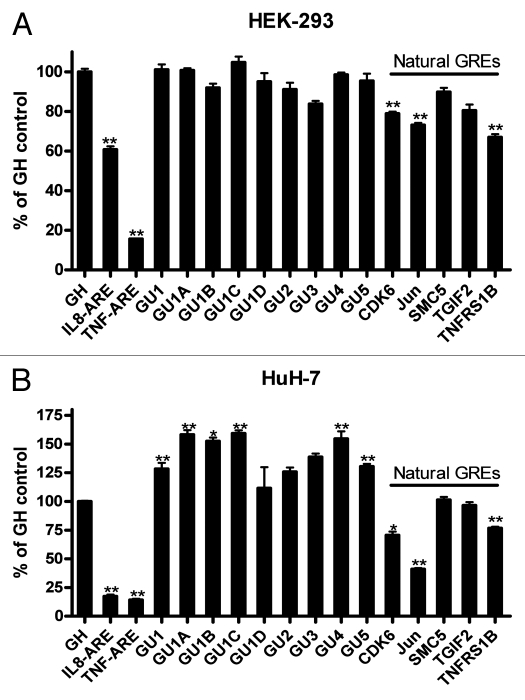
We analyzed the effect of GREs which were introduced in the 3′UTR of the GFP reporter, on mRNA stability. Previously, the effect of GU-rich elements was tested using the Tet-off reporter system in HeLa cells,9 therefore here we used Huh-7 cells and Actinomycin D chase for mRNA half-life determination. This well-differentiated hepatoma cell line responds to a variety of pro-inflammatory cytokines and has been used for hepatitis, cytokine and post-transcriptional studies.16–19 First, GFP reporter constructs fused to 3′UTRs containing natural GREs of the genes TGIF2, SMC5, CDK6, JUN and TNFRS1B were tested and showed enhanced mRNA decay (Fig. 4A). Next we analyzed the effect of sequence context in a single GRE pentamer (Table 2 and Fig. 4B). We also tested the effect of overlapping GRE elements from 2 to 5 elements (Fig. 4C). All synthetic GRE elements had similar effects on mRNA decay; the enhanced decay was largely independent of the sequence context of a single GRE or the number of overlapping pentamers. The destabilizing effects of the AREs from IL8, TNFα or a construct that contains a larger part of the 3′UTR of IL8 (237 bases) were also assessed (Fig. 4D). Two controls were used: the GFP vector with its stable BGH 3′UTR and another control that contains a GU- and AU-free insert derived from the stable human growth hormone (GH1) transcript of comparable size to the GREs and AREs used (Table 2). The mRNA half-life determinations of all the natural and synthetic GREs were performed in three independent experiments (Fig. 4E). All GREs tested had a statistically significant (p < 0.05) impact on mRNA half life (4–6 h compared with 8 h for controls; Fig. 4E). It appeared that GU1C had stronger effect on mRNA stability among other synthetic GREs; however, statistical significance test (one-way ANOVA) showed this is not the case. The AREs had more potent destabilizing effects on GFP mRNA (Fig. 4E).
Figure 4.
The decay patterns for GRE-GFP reporter mRNA. HuH-7 cells were transfected with the indicated constructs (Table 2). Actinomycin D was added for one, 2, 4, 6 and 8 h. Total RNA was extracted and the GFP reporter mRNA was quantified by RT-Real Time-PCR. (A) Decay curves of naturally occurring GREs from the genes TGIF2, SMC5, CDK6, JUN and TNFRS1B. (B) Decay curves of constructs with different context forms of a single GRE. (C) Decay curves of mRNA expressed from constructs with 2 and up to 5 overlapping GREs. (D) Decay curves of mRNA expressed from constructs with the AREs of IL-8 and TNF and a 237 base ARE containing region from IL8 mRNA. Data are shown in fraction of initial mRNA levels. Each time point is Mean ± SEM of 6 real time PCR measurements in 3 independently performed experiments (each experiment is with 2 measurements). (E) Average mRNA halflife of 3 independently performed experiments. * and **p < 0.05 and < 0.01, respectively, using student's t-test.
Table 2.
Synthetic and natural GU-rich and AU-rich inserts and controls used (M, mutant)
| Name | Sequence |
| GH1 | GACCCCUCCCCAGUGCCUCUCCUGGCCCUGGAAGUUGCCACUCCAGUGCCCACCAGCCUUGUCC |
| IL8 ARE | GAUCC GUGUAACUUAUUAACCUAUUUAUUAUUUAUGUAUUUAUUUAAGCAUCAAAUAUUUGUGCAAGAAU |
| TNFα ARE | GAUCCUUGUGAUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUUACAGAUGAAUGUAUUUAUUUGGGAGAU |
| GU1 | GACCCCUCCCCAGUGCCUCUCCUGGCCCUGGGUUUGGCCACUCCAGUGCCCACCAGCCUUGUCC |
| GU1-M | GACCCC UCCCCAGUGCCUCUCCUGGCCCUGGGUCUGGCCACUCCAGUGCCCACCAGCCUUGUCC |
| GU1A | GACCCCUCCCCAGUGCCUCUCCUGGCCCUGUGUUUGUCCACU CCAGUGCCCACCAGCCUUGUCC |
| GU1B | GACCCCUCCCCAGUGCCUCUCCUGGCCCGUUGUUUGUUCACUCCAGUGCCCACCAGCCUUGUCC |
| GU1C | GACCCCUCCCCAGUGCCUCUCCUGGCCCUCUUU GUUUGUUUCCCAGUGCCCACCAGCCUUGUCC |
| GU1D | GACCCCUCCCCAGUGCCUCUCCUGGCCCUGUGUGUUUGUGUGCCAGUGCCCACCAGCCUUGUCC |
| GU1D-M | GACCCCUCCCCAGUGCCUCUCCUGGCCCUGUGUGUCUGUGUGCCAGUGCCCACCAGCCUUGUCC |
| GU2 | GACCCCUCCCCAGUGCCUCUCCUGGCCGUUUGUUUGGCCACUCCAGUGCCCACCAGCCUUGUCC |
| GU3 | GACCCCUCCCCAGUGCCUCUCCUGGGUUUGUUUGUUUGCACUCCAGUGCCCACCAGCCUUGUCC |
| GU3-M | GACCCCUCCCCAGUGCCUCUCCUGGGUCUGUCUGUCUGCACUCCAGUGCCCACCAGCCUUGUCC |
| GU4 | GACCCCUCCCCAGUGCCUCUCCUGGGUUUGUUUGUUUGUUUGCCAGUGCCCACCAGCCUUGUCC |
| GU5 | GACCCCUCCCCAGUGCCUCUCGUUUGUUUGUUUGUUUGUUUGCCAGUGCCCACCAGCCUUGUCC |
| CDK6 | UAGUUUACUGUUUUGAAAUCAAUGCAAGAGUGAUUGCAGCUUUAUGUUCAUUUGUUUGUUUGUUUGUCUGUUUGUUU |
| CDK6-M | TAGUUUACUGUUUUGAAAUCAAUGCAAGAGUGAUUGCAGCUUUAUGUUCAUUUGUCUGUCUGUCUGUCUGUCUGUUU |
| JUN | UUUGUAAGUUAUUUCUUGUUUGUUUGUUUGGGUAUCCUGCCCAGUGUUGUUUGUAAAUAAGAGAUUUG |
| SMC5 | ACUUAAAACAAAAGUUUUUUUGUUUGUUUGUUUGUUUGUUUUUUUGAGAUGGAGUCUCACUCUG |
| TGIF2 | CCCAUUUUUUCUUUUGCUGUUUUGUUUUUUGUUUUUUGUUUGUUUGUUUGUUUUUUUGAGACAG |
| TGIF2-M | CCCAUUUUUUCUUUUGCUGUUUUGUUUUUUGUUUUUUGUCUGUCUGUCUGUUUUUUUGAGACAG |
| TNFRSF1B | CUGGCUUCUGGAGCCCUUGGGUUUUUUGUUUGUUUGUUUGUUUGUUUGUUUGUUUCUCCCCCUG |
GFP reporter assays on GU-rich and AU-rich elements.
We used the same constructs as above (Table 2) in cell-based fluorescence assays in HEK 293 cells and HuH-7 cells. The synthetic GREs in HEK-293 cells did not have a detectable effect on GFP protein expression as measured by fluorescence readout, while the natural GREs of CDK6, JUN, TGIF2 and TNFRS1B had weak to moderate (although statistically significant) downregulatory effect (Fig. 5A). Also, the AREs were more potent in blocking GFP protein expression than GREs in the 2 cell lines tested here. Surprisingly, in HuH-7 cells, all synthetic GREs led to a detectable upregulation of protein expression while natural GREs in the case of CDK6, JUN and TNFRS1B led to downregulation of GFP expression (Fig. 5B). In both cell lines, the effect of natural GREs was stronger in downregulating protein expression than that of the synthetic elements. Also in both cell lines the GREs of CDK6, JUN and TNFRS1B were more potent than SMC5 and TGIF2 genes.
Figure 5.
Effect of GREs and AREs on GFP reporter expression. HEK-293 (A) or HuH-7 cells (B) were transfected in 96-well microplate with the indicated constructs. Fluorescence due to the activity of the GFP protein was quantified by imaging. The results are shown in percent of the GH1 control (100%). Florescence was measured 24 h post-transfection in HEK293 and 48 h in Huh-7 cells (since they have different transfection efficiencies). Mean ± SEM of 8 replicate microwells from 2 independent experiments (A) or 12 replicate microwells from 3 experiments (B). * and **p < 0.05 and < 0.01, respectively, using student's t-test, compared with control (GH1).
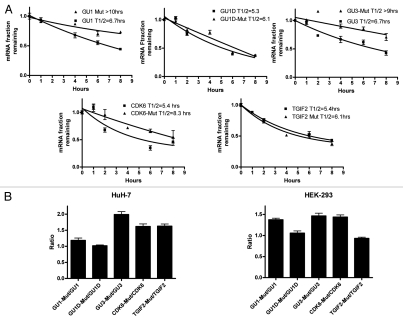
The effect of point mutations on GU-rich element activity.
We introduced U to C substitutions in the central U of all GUUUG pentamers in the reporter constructs for GU1, GU1D, GU3, CDK6 and TGIF2 (Table 2, M-designated constructs). The point mutations lead to significant stabilization in GU1, GU3 and CDK6 but not in GU1D and TGIF2 in HuH7 cells (Fig. 6A). The mutants and corresponding constructs were again transfected into HuH-7 and HEK-293 cells to analyze GFP reporter expression (Fig. 6B). None of the mutations led to reduction in protein expression. Significant mutation-dependent upregulation of expression was observed in GU3 and CDK-6. The mutation in GU1D had no significant effect on expression. The mutation in GU1 led to a modest upregulation in HuH-7 cells and a stronger one in HEK-293 cells. The mutation in TGIF2 led to increased expression in HuH-7 cells with no significant effect in HEK-293 cells.
Figure 6.
Effect of point mutations within GREs on reporter mRNA stability and protein expression. (A) Decay curves of GFP reporter mRNA in HuH-7 cells that were transfected either with the indicated GRE constructs or the corresponding constructs with point mutations (Mut) (Table 2). Actinomycin D was added for one, 2, 4, 6 and 8 h. Total RNA was extracted and the GFP reporter mRNA was quantified by RT-Real Time-PCR (B) HuH-7 and HEK-293 cells were transfected in 96-well microplate with the indicated constructs or the corresponding constructs with point mutations (Table 2). Fluorescence due to the activity of the GFP protein was quantified by imaging 24 h post-transfection. The results are shown in Ratios of the fluorescent intensity of the mutant over non-mutated GU-rich (Table 2). Mean ± SEM of 6 replicate microwells.
Discussion
The genome-wide search for GRE-containing genes indicates that like AREs, GREs are present in a significant fraction of the total set of annotated genes and participate in a number of important cellular processes, most of which are transient in nature. We used computational methods to assess the repertoire of GREs in the human genome and analyzed their functional attributes using resting and activated primary human T-cell microarray data as well as synthetic constructs in mRNA and GFP reporter assays. The GREs are functional elements and have different characteristics from AREs, although they share similar overlapping pen-tamer arrangements and both are enriched in the 3′UTR. The GRE- and ARE-genes have both shared and distinct sequence and functional characteristics. Both GREs and AREs are highly specific to the 3′UTR when compared with other regions of the mRNA, supporting their post-transcriptional control role. Our results, namely the differential dynamics of the ARE and GRE effects on mRNA expression and mRNA half life, further signify the sequence complexity of the 3′UTR that may play vital roles in regulation of gene expression.
The higher percentage of non-switching class in GRE-mRNA, i.e., with no change in their stability due to T-cell activation and the fact that longer GREs do not change the mRNA destabilization potency may suggest ON/OFF switching vs. fine-tuning models for GRE and ARE effects; respectively. However, the number of reiterations may be relevant in response to the action of CUG-BP1 which primarily acts on GRE-mRNAs.10,11 Further detailed experimental work is therefore needed to understand these aspects.
Reporter assays in HuH-7 and HEK-293 cells showed that, compared with AREs, most GREs tested had modest effects on mRNA stability (Fig. 4). GREs might not lead to significant destabilization by default, and their effect might be enhanced by cell type and activation status. It has been shown recently that ARE and GRE effects are differentially potent according to the cell type; AREs were more potent in embryonic stem cells while GREs had more impact on mRNA stability in C2C12 muscle cells.11,20 The analysis of synthetic constructs as well as large scale measurements indicate that the ability of AREs to destabilize mRNA increases with the number of overlapping pentamers.1,2,21 This is clearly different in the case of GREs that do not seem to be dependent on the number of overlapping GUUUG pentamers.
Surprisingly, while the synthetic GREs lead to downregulation of mRNA stability in HuH-7 cells, they led to an upregulation of protein expression. The synthetic GREs in HEK-293 cells however, did not lead to such upregulation in protein expression. This unexpected result might be explained by the finding that some GRE binding proteins such as CUG-BP1 may destabilize mRNA but enhance translation.22 CUG-BP2 on the other hand competes with GU-BP1 on the same binding site but can play an opposite role leading to mRNA stabilization and translational shutoff.23 We have quantified the levels of CUG-BP1 and CUG-BP2 mRNAs in HuH-7 and HEK-293 cells by quantitative PCR (data not shown) and found that the levels of CUG-BP1 were comparable in both cell lines while the levels of CUG-BP2 were 10-fold higher in HEK-293 cells. These observations are in agreement with the observed reduced expression of GRE-expression reporters in HEK-293 cells and upregulated expression in HuH-7 cells. Further experiments are needed to assert the potential effects of the differential levels of the RNA binding proteins in the two different cell lines.
Moreover, we found in both cell lines that GREs deriving from natural genes have a stronger ability to downregulate protein expression than all seven synthetic GREs tested. This may indicate that the sequence context of the GRE is important. Natural GREs contain overlapping pentamers but also non-canonical elements like GUUUUUG found in TNFRSF1B, SMC5 and TGIF2. The most effective GREs in downregulating reporter fluorescence were those of CDK6 and JUN mRNAs. Deletion of the GRE region from JUN was previously found to increase JUN mRNA stability despite the presence of other functional ARE-like sequences.24 In this earlier reporter, mutations of the GUUUG to GUAUG or GAUUG in JUN GU-rich region did not significantly affect the mRNA decay indicating the presence of necessary context sequences.24 Mutation to CUUUC results in appreciable reduction of mRNA destabilization, which indicates the importance of the G residues themselves.9 The outcome of these two results may be also due to differences in secondary structures.
An important observation in the functional classification of GRE-genes when compared with the whole genome is over-representation in neurogenesis and neuronal activities. This is in concordance with an important attribute of CUG-BP1, a critical mediator of GRE-mediated mRNA decay, which is involvement in certain RNA mediated neuromuscular diseases such as muscular dystrophy, DM1.25 In DM1, mutant mRNA with extended CUG repeats activate CUG-BP1 hyper-phosphorylation and stabilization resulting in disruption of alternative splicing, mRNA translation and mRNA decay culminating in the symptoms of the disease. CUG-BP1 interacts with MEF2A transcript which acts as a transcription factor involved in myogenesis, and increases its translation.26 Additionally, CUG-BP1 leads to stabilization of TNFα mRNA and subsequently elevation of TNFα in muscle cells contributing to insulin resistance and muscle wasting normally seen in DM1 patients.27 Of particular note, we found that GRE-genes are over-represented (3-fold, p < 0.05) in Huntington's disease pathway. It has been shown that CUG repeat expansion in the mRNA encoding junctophilin-3 is associated with Huntington disease like 2 (HDL2).28 It is likely that this observation is also associated with CUG-BP1 activation. Another molecule, that is indeed a target for CUG-BP1 is GABA-A transporter 4 (GABAR ref. 4) mRNA, and may explain some of the spinocerebellar ataxias 8 syndrome.29
The highly selective abundance of GREs, like AREs, in the 3′UTR and the appreciable number of GRE-genes suggest an important functional role as supported by our expression and reporter data here. The functional representation of GREs in development, mRNA processes and neurogenesis, and the evidence of binding of CUG-BP1 to a number of GRE-mRNAs indicate a significant role for GRE-dependent regulation in health and disease. Thus, analyzing the sequence and functional repertoire of GREs, AREs and miRNAs targets, in the human genome contributes to understanding the molecular systems biology of mRNA decay.
Materials and Methods
Creating a genome-wide GRE database.
To create a database of the GRE content of the human genome, we first downloaded the genome annotation (including GO annotations) from ENSEMBL via BioMart (www.biomart.org). We used ENSEMBL release 44 and matched it to NCBI human genome build 36 to computationally extract the 3′UTR regions for each transcript. We then used degenerate pattern matching tools first to scan for the polyA signal (AWTAAA) in the last 50 bases of each sequence. We found 23,539 transcripts with an annotated 3′UTR and a recognizable Poly-A signal in the last 50 bases (strongly suggesting the completeness of the 3′UTR). Each sequence was scanned for the motifs as listed in Table 1. For each transcript, the longest matching motif was assigned as the best match. For each gene and particularly for genes with multiple transcripts, the best match among the transcripts was assigned as the gene_s GRE motif match. If all transcripts of the gene failed to contain a GRE motif (or failed to have an annotated 3′UTR altogether), the gene was considered non-GRE. The core data set was augmented with GO-term associations and gene annotations and stored in a database accessible online via the web with a convenient search facility and interactive presentation.
Functional class enrichment analysis.
The Transcript IDs for GRE-transcripts were converted into Entrez Gene IDs and then uploaded into PantherDB for functional classification. The set was compared against the entire human gene set as a control list. Bonferroni p-value correction for multiple testing was used. About 45% of the genes in the database are functionally unclassified. This information is necessary to attain a sense of scale when assessing Figure 1.
Global analysis of mRNA half-lives.
We combined our genome wide GRE analysis with microarray data that contains T-Cell mRNA expression profiles.15 Briefly, T cells were stimulated under three different conditions (growth media, antiCD3 or anti-CD3+anti-CD28) for 3 h. Following stimulation, Actinomycin D was added and mRNA levels measured on Affymetrix microarrays over a period of 2 h. A 2-step filtering process was devised in order to reduce the noise inherent in the microarray data and the computational pattern matching process. First, we removed from the nearly 6,000 probeset results all those probesets that are known to match more than one transcript, thus reducing the risk of analyzing phantoms, which are averaged expression values resulting from similar but distinct transcripts. For transcripts that were matched by more than one probe set (often 2 probe sets and rarely more), the averages were used. Thus each transcript was unique in the remaining filtered set. The microarray transcripts were matched to GRE-annotated transcripts from the above analysis. The percentages of each GRE-cluster group did not change significantly after the filtering process compared with the values reported in Table 1; thus confirming that no bias was introduced by this step. Analysis of AREs followed exactly the same method described above utilizing ARE-mRNA database data from.4
To detect and compare the effects of the GRE and ARE motifs we stratified the gene set into a control set (genes with no detectable ARE or GRE), an ARE set (genes with the ARE motif but not the GRE motif) and a GRE set (genes with GRE only). We did not analyze the set containing both motifs because of its very small size, which was not sufficient for reliable analysis. In all cases, we compared the three sets within the same cellular condition.
After filtering and stratification, we analyzed the half-life data in two ways:
We obtained a histogram of the base-2 log of the values. In all strata under all conditions and under various different binning strategies the histograms were clearly bimodal, i.e., naturally classifying transcript half-lives into short and long half life mRNA classes. We proceeded to estimate the ratio of the short set to the long set by fitting the histogram data with a bimodal Gaussian Mixture model using GraphPad Prism software functions. The data fitted the mixture very well with R2 at least 0.95 in all cases. This scheme allowed us to estimate the fraction of short half-life transcripts for each stratum and condition in a data-natural way without any arbitrary cutoffs. Interestingly, the Gaussian centroids and intersection points were highly consistent across all strata. We also used a manual cutoff at 64 (26) minutes to identify short half-life transcripts and obtained essentially the same results.
We also directly compared both half-life mRNA data and steady-state mRNA expression levels (at zero minutes from transcriptional arrest) using non-parametric tests and also obtained the usual mean and SEM statistics for each stratum and condition. We sub-stratified the ARE and GRE sets by motif cluster group to determine the effects of the number of overlapping pen-tamer repeats.
Vectors.
The gWIZ GFP vector was purchased from Genelantis (San Diego, CA). GU-rich, AU-rich, mutants and control inserts were made using two annealed synthetic complementary oligonucleotides with BamHI and XbaI overhangs and cloned into the same sites in the BGH 3′UTR of the vector. Constructs with five natural GU-rich elements from the genes TGIF2 (NM_021809), SMC5 (NM_015110), CDK6 (NM_001259), JUN (NM_002228) and TNFRS1B (NM_001066) and the corresponding mutants were cloned (Table 2). For the control, we introduced a 64 bp GU- and AU-free sequence from the 3′UTR of human growth hormone GH1. This size is comparable to the inserted GU- and AU-rich elements (Table 2). In addition, we designed constructs where artificial GU-rich elements were introduced into the GH1 3′UTR sequence while keeping the overall size of the insert constant (Table 2). For comparison with AREs we used constructs where the AREs of IL8 and TNFα± were cloned into the same site (Table 2). The AU-rich region of 237 bp (bases 972_1209 of NM_000584) that belongs to the 1,250 bp IL-8 3′UTR was previously cloned as described in reference ref-type="bibr" rid="R30">30.
Cell lines and transfection.
The cell lines used were HEK293 American Type Culture Collection (ATCC; Rockville, MD) and cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and antibiotics. Huh-7 cell line was obtained from Dr. Stephen J. Polyak (University of Washington, Seattle, WA USA) and was propagated in DMEM medium with 10% FBS and antibiotics. It is hepatoma cell line that has been previously used for hepatitis, cytokine and post-transcriptional studies.16–19 Transfection was performed with GFP reporter constructs with the indicated 3′UTRs using Lipofectamine 2000 reagent (Invitrogen). For RNA analysis experiments, transfections were performed in 10-cm plates using 4 ug vector DNA and 24 h later the cells were split into 43-cm 6-well plates to ensure equal proportions of transfected cells in each plate.
GFP based reporter assay.
To avoid differences in transfection efficiency due to DNA quality, e.g., supercoiling DNA variations and residual endotoxin, linear expression cassettes of the vectors were generated by PCR.31 After purification of the PCR-DNA using Qiagen kit, DNA concentration was determined with the Nanodrop spectrophotometer and the quality of the PCR products was monitored on DNA agarose gels. HEK-293 and HuH-7 cells were seeded in clear bottom black microplates and transfected with 50 ng (HEK-293) or 100 ng (HuH-7) of PCR DNA using Lipofectamine 2000 reagent and left 24 or 48 h. Automated laser-focus image capturing was performed using the 4x magnification with the high-throughput BD Pathway 435 imager (BD Biosciences, San Jose, CA). The variance in GFP fluorescence among replicate microwells was <6%; thus, with this minimum variance, experiments do not warrant transfection normalization.32 Image processing, segmentation and fluorescence quantification was facilitated by the ProXcell program and was previously described in reference 32.
RNA isolation, reverse transcription and real time PCR.
For GFP reporter mRNA expression analysis, total RNA was isolated from reporter transfected cells. The next day after transfection, transcription was blocked with 10 ug/ml Actinomycin D (Sigma) and total RNA was extracted with Tri reagent (Sigma) after one, 2, 4 and 8 h. Reverse transcription was performed using Superscript II and Oligo dT primer (Invitrogen). Real Time PCR was performed using a custom made TaqMan primer set (Applied Biosystems). The primers span the CMV promoter intron A to control for DNA contamination. A 6-carboxyfluorescein (6FAM)-labeled TaqMan probe that targets the CMV exon 1/exon 2 of GFP junction sequence was used. The probe design allowed further control of DNA contamination. The specificity of the cDNA of TaqMan primer was tested on a negative control containing plasmid DNA. The control housekeeping RPL0 mRNA probe was labeled with a 5′ reporter VIC dye and used for normalization. Real time PCR was performed in multiplex form using the Chroma 4 DNA Engine cycler (BioRad). Cell culture experiments for the mRNA half-life determination were performed with HuH-7 cell line that was treated with Actinomycin D (10 µg/ml) to shut-off transcription. Decay curves were derived using GraphPad Prism software using the one phase exponential decay model.30
Acknowledgments
The study and open access charges are supported by intramural funding of the King Faisal Specialist Hospital and Research Center Organization to the BioMolecular Research Program.
References
- 1.Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakheet T, Williams BR, Khabar KS. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:137–140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark A, Dean J, Tudor C, Saklatvala J. Post-transcriptional gene regulation by MAP kinases via AU-rich elements. Front Biosci. 2009;14:847–871. doi: 10.2741/3282. [DOI] [PubMed] [Google Scholar]
- 9.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St. Louis-Vlasova IA, et al. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5:11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timchenko NA, Wang GL, Timchenko LT. RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein beta by interacting with the alpha and beta subunits of eukaryotic initiation translation factor 2. J Biol Chem. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- 14.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, et al. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimazu T, Takada S, Ueno Y, Hayashi Y, Koike K. Post-transcriptional control of the level of mRNA by hepatitis B virus X gene in the transient expression system using human hepatic cells. Genes Cells. 1998;3:477–484. doi: 10.1046/j.1365-2443.1998.00203.x. [DOI] [PubMed] [Google Scholar]
- 17.Raynes JG, Bevan S. Inhibition of the acute-phase response in a human hepatoma cell line. Agents Actions. 1993;38:66–68. doi: 10.1007/BF01991140. [DOI] [PubMed] [Google Scholar]
- 18.Dormoy-Raclet V, Markovits J, Jacquemin-Sablon A, Jacquemin-Sablon H. Regulation of Unr expression by 5′- and 3′-untranslated regions of its mRNA throughmodulation of stability and IRES mediated translation. RNA Biol. 2005;2:27–35. doi: 10.4161/rna.2.3.2203. [DOI] [PubMed] [Google Scholar]
- 19.Hitti E, Al-Yahya S, Al-Saif M, Mohideen P, Mahmoud L, Polyak SJ, et al. A versatile ribosomal protein promoter-based reporter system for selective assessment of RNA stability and post-transcriptional control. RNA. 2010;16:1245–1250. doi: 10.1261/rna.2026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frevel MA, Bakheet T, Silva AM, Hissong JG, Khabar KS, Williams BR. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-36.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barreau C, Watrin T, Beverley Osborne H, Paillard L. Protein expression is increased by a class III AU-rich element and tethered CUG-BP1. Biochem Biophys Res Commun. 2006;347:723–730. doi: 10.1016/j.bbrc.2006.06.177. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay D, Jung J, Murmu N, Houchen CW, Dieckgraefe BK, Anant S. CUGBP2 plays a critical role in apoptosis of breast cancer cells in response to genotoxic injury. Ann NY Acad Sci. 2003;1010:504–509. doi: 10.1196/annals.1299.093. [DOI] [PubMed] [Google Scholar]
- 24.Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 26.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG Triplet Repeat-binding Protein, CUGBP1, in Mice Inhibits Myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding Protein CUGBP1 Regulates Stability of Tumor Necrosis Factor mRNA in Muscle Cells. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes SE, O'Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet. 2001;29:377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- 29.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, et al. RNA Gain-of-Function in Spinocerebellar Ataxia Type 8. PLoS Genet. 2009;5:1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic Acids Res. 2009;37:3612–3624. doi: 10.1093/nar/gkp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.al-Haj L, Al-Ahmadi W, Al-Saif M, Demirkaya O, Khabar KS. Cloning-free regulated monitoring of reporter and gene expression. BMC Mol Biol. 2009;10:20. doi: 10.1186/1471-2199-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Zoghaibi F, Ashour T, Al-Ahmadi W, Abulleef H, Demirkaya O, Khabar KS. Bioinformatics and experimental derivation of an efficient hybrid 3′ untranslated region and use in expression active linear DNA with minimum poly(A) region. Gene. 2007;391:130–139. doi: 10.1016/j.gene.2006.12.013. [DOI] [PubMed] [Google Scholar]