Abstract
Inclusion body myopathy with Paget disease of bone and frontotemporal dementia (IBMPFD) is an autosomal dominant disorder characterized by progressive myopathy that is often accompanied by bone weakening and/or frontotemporal dementia. Although it is known to be caused by mutations in the gene encoding valosin-containing protein (VCP), the underlying disease mechanism remains elusive. Like IBMPFD, neurofibromatosis type 1 (NF1) is an autosomal dominant disorder. Neurofibromin, the protein encoded by the NF1 gene, has been shown to regulate synaptogenesis. Here, we show that neurofibromin and VCP interact and work together to control the density of dendritic spines. Certain mutations identified in IBMPFD and NF1 patients reduced the interaction between VCP and neurofibromin and impaired spinogenesis. The functions of neurofibromin and VCP in spinogenesis were shown to correlate with the learning disability and dementia phenotypes seen in patients with IBMPFD. Consistent with the previous finding that treatment with a statin rescues behavioral defects in Nf1+/– mice and providing further support for our hypothesis that there is crosstalk between neurofibromin and VCP, statin exposure neutralized the effect of VCP knockdown on spinogenesis in cultured hippocampal neurons. The data presented here demonstrate that there is a link between IBMPFD and NF1 and indicate a role for VCP in synapse formation.
Introduction
In the central nervous system of mammals, dendritic spines are the locations of more than 90% of excitatory synapses (1) and therefore constitute the functional subcellular structures for excitatory neurotransmission (2–6). Neurofibromin, a large protein (2818 aa residues) encoded by the human NF1 gene (7, 8), is one regulator of dendritic spine formation (9). Mutations of the gene cause neurofibromatosis type 1 (OMIM 162200), one of the most common autosomal dominant disorders, affecting about one in 3,500 individuals. Neurofibromatosis type 1 (NF1) is characterized by skin pigmentations (café-au-lait spots and freckling) and formations of benign peripheral nerve sheath tumors (neurofibromas). In addition, many other features are frequently found in patients with NF1, including cognitive deficits as well as skeletal lesions and malformations. In children, NF1 is frequently associated with learning difficulty (10) and greater susceptibility to autism (11, 12). The function of neurofibromin in synaptogenesis (9) and formation of barrel cortex (13) may partially explain these neurological symptoms. It is also known that neurofibromin regulates the functions of osteoclast (14, 15) and skeletal muscle development (16). Although the Ras/MAPK pathway, the downstream signaling of neurofibromin, has been implicated in bone resorption (17), the detailed mechanism underlying the bony defects in patients with NF1 remains elusive.
The tumor suppressor activity of neurofibromin is largely dependent on its Ras-specific GTPase-activating protein (RasGap) activity (reviewed in refs. 18, 19). In addition, neurofibromin also regulates adenylate cyclase activity through both Gαs-dependent and -independent pathways, thus controlling the cAMP concentration in cells (20). Our previous study showed that neurofibromin is widely distributed in different subcellular compartments of neurons, including synapses (21). It acts downstream of syndecan-2, a synaptic heparan sulfate proteoglycan, in the regulation of dendritic spine formation (9). Neurofibromin interacts with syndecan-2 (22) and activates the PKA–Enabled/vasodilator-stimulated phosphoprotein (PKA-Ena/VASP) pathway to promote actin polymerization and bundle formation (9). Interestingly, although the PKA pathway is essential for dendritic spine formation, activation of PKA alone is not sufficient for the process (9), possibly due to the involvement of multiple downstream pathways of neurofibromin in spinogenesis.
Valosin-containing protein (VCP), also known as p97, is a multifunctional AAA (ATPases associated with a variety of cellular activities) protein (reviewed in refs. 23, 24) involved in a variety of cellular events, including cell cycle control, membrane fusion, ER-associated protein degradation (ERAD), and autophagy (24–32). VCP is associated with several neurodegenerative disorders (reviewed in refs. 33–35). Mutations in the VCP gene result in inclusion body myopathy with Paget disease of bone and frontotemporal dementia (IBMPFD, ref. 36), a dominant inherited disorder (OMIM 167320). Expression of mutant VCP in transgenic mice or introduction of an IBMPFD mutation into mice through a gene-targeting approach induces degeneration in muscle, bone, and brain (37, 38), recapturing the phenotypes of the patients with IBMPFD. Recently, human genetic analysis also indicated that VCP mutations account for 1%–2% of autosomal dominantly inherited ALS (39). In addition, VCP interacts with the polyglutamine-containing aggregates that are found in patients with Huntington and Machado-Joseph diseases (40).
So far, the mechanism of IBMPFD pathogenesis has not been elucidated. VCP controls polyubiquitin chain turnover (41) and contributes to both formation and clearance of the ubiquitylated inclusion bodies (42). An IBMPFD-associated VCP mutant was shown to induce aggregation of polyubiquitin-conjugated proteins in myoblastoma cells (43). VCP mutations have also been shown to cause the dysfunction of autophagy, which may additionally contribute to the pathogenesis of IBMPFD (31, 44). In addition to defects in protein degradation, dystrophic neurites are frequently found in patients with frontotemporal dementia (FTD) (45–47). Recently, VCP has been shown to regulate remodeling of neuronal morphology in Drosophila (48). It is likely that VCP actively contributes to neuronal morphogenesis and that dysfunction of VCP may therefore result in neurodegeneration.
VCP forms a homohexameric barrel and hydrolyzes ATP to generate the mechanical force for its function as a molecular chaperon (23, 27, 49). It possesses two ATPase domains (D1 and D2). The D2 domain carries the major ATPase activity, while the D1 domain is also the hexamerization domain of VCP. The N-terminal region (N-domain) of VCP is involved in the interaction with various adaptors that direct VCP to various cellular events. The identified IBMPFD mutations are highly clustered in the N- and D1 domains of VCP (36, 47, 50). Interestingly, all mutation hot spots are located at the interface between the N- and D1 domains (51). Therefore, IBMPFD mutations can change the conformation of the catalytic domains, alter ATPase activity, and compromise the function of VCP (52).
In this study, we first examined the number of dendritic spines of CA1 neurons in Nf1+/– mice. Golgi staining showed that the spine number in Nf1+/– mice is lower than that in the WT littermates, consistent with our previous finding that neurofibromin plays a role in controlling spine density of neurons. Using a proteomics approach, we identified VCP as a neurofibromin-associated protein. We further provide evidence that the interaction between neurofibromin and VCP regulates the density of dendritic spines. Our study found a crosstalk between these two proteins, neurofibromin and VCP, each of which is associated with genetic disease and regulates neuronal morphogenesis.
Results
Dendritic spine density is lower in Nf1+/– mice.
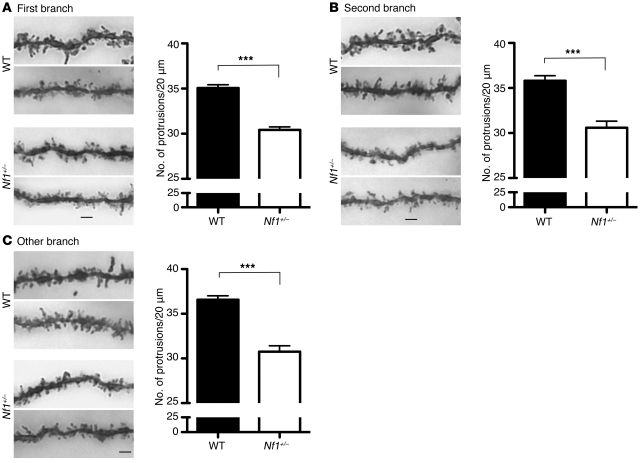
We have previously shown that neurofibromin regulates dendritic spine formation, particularly the initiation step of spinogenesis, namely dendritic filopodia formation, in cultured hippocampal neurons. Because it was shown earlier that the loss of an Nf1 allele results in cognitive deficits and impaired formation of barrel cortex (10, 13), we examined spine density in Nf1+/– mice using Golgi staining to evaluate the effect of Nf1 haploinsufficiency on neuronal morphology in vivo. Compared with WT littermates, the spine densities of CA1 pyramidal neurons in Nf1+/– mice were significantly lower, irrespective of whether first-, second-, or higher-order branches of apical dendrites were analyzed (Figure 1). These results support a role for the Nf1 gene in the regulation of dendritic spine density in mouse brain.
Figure 1. The density of dendritic spines is reduced in Nf1+/– heterozygous mice.
(A) First branches, (B) second branches, and (C) the remaining higher-order branches of apical dendrites of CA1 pyramidal cells. For each group, 2 representative images are shown. The right panels indicate the quantitative results of the spine density. Data represent mean plus SEM. The numbers of analyzed dendrites were (A) WT, 51; Nf1+/–, 55; (B) WT, 21; Nf1+/–, 12; (C) WT, 51; Nf1+/–, 30. Scale bars: 2 μm. ***P < 0.001.
Neurofibromin interacts with VCP.
To further explore the function of neurofibromin, a neurofibromin antibody was employed to identify the neurofibromin-associated proteins by co-immunoprecipitation from rat brain extracts. The precipitates were then separated by 2D gel electrophoresis and analyzed by mass spectrometry. In comparison with the control IgG, the most robust protein precipitated by neurofibromin antibody appeared as a spot at approximately 90 kDa on the 2D gel (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI45677DS1) and was identified as VCP using both MALDI-TOF (Supplemental Figure 1B) and MS/MS analyses (Supplemental Figure 1C). In the MALDI-TOF analysis, a total of 36 peptides matched rat VCP (coverage of 36%). In the MS/MS analysis, 7 peptides were identical to rat VCP protein. The interaction between neurofibromin and VCP was specific, because control IgG did not precipitate VCP (Supplemental Figure 1A). In addition to VCP, p47, a cofactor of VCP, was also present in the precipitate of neurofibromin antibodies (Supplemental Figure 1, A and B), suggesting that neurofibromin forms a complex with VCP and p47. Other protein spots on the 2D gel were also analyzed by MALDI-TOF. However, none of the promising protein candidates were identified. Since VCP was the most promising protein identified from the 2D gel, we focused on VCP in the current study.
To confirm the interaction between neurofibromin and VCP, we performed co-immunoprecipitation-immunoblotting assays using adult rat brain extracts. The result showed that VCP was coprecipitated by neurofibromin antibody (Figure 2A). The reciprocal immunoprecipitation also showed the precipitation of neurofibromin by VCP antibody from rat brain extracts (Figure 2A). Non-immune control IgG was used again as negative control to ensure the specificity of co-immunoprecipitation (Figure 2A). Because neurofibromin and VCP are not neuron-specific proteins, we also examined the interaction between neurofibromin and VCP in non-neuronal cells. Similar to the results obtained with rat brain extracts, neurofibromin antibody precipitated endogenous neurofibromin as well as endogenous VCP from HEK293T cell extract (see below). These immunoblot analyses were consistent with the results obtained by mass spectrometric analyses showing that neurofibromin associates with VCP.
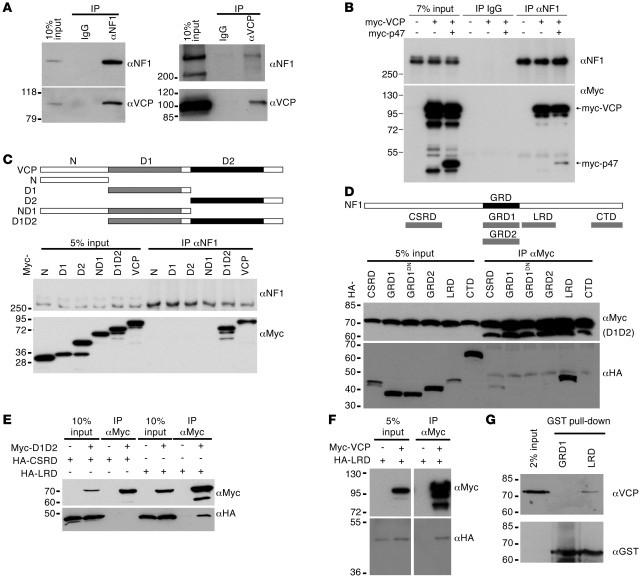
Figure 2. VCP interacts with neurofibromin.
(A) Two-directional immunoprecipitations of neurofibromin and VCP. Adult rat brain extracts were used for immunoprecipitation using the indicated antibodies. Immunoblotting was then performed. (B) Co-immunoprecipitation of p47, VCP, and neurofibromin. HEK293T cells were transfected with Myc-tagged VCP and Myc-tagged p47 or vector control as indicated. One day later, cell extracts were harvested for immunoprecipitation using neurofibromin antibody. Immunoblotting using Myc tag antibody revealed the presence of both VCP and p47. (C–F) Interaction between neurofibromin and VCP in transfected HEK293T cells. Variant Myc-tagged VCP constructs and HA-tagged NF1 constructs were cotransfected into HEK293T cells as indicated, and cell lysates were harvested 24 hours later and immunoprecipitated using antibodies as indicated. (D) Myc-tagged D1D2 construct was cotransfected with variant neurofibromin fragments. GRD1DN, dominant negative mutant of GRD1. (E) 2 mM DTT was added in lysates to reduce oxidation. (G) GST pull-down assay. His-tagged D1D2 of VCP was mixed with LRD or GRD1 GST fusion proteins. The protein complex was then pulled down using glutathione agarose and analyzed by immunoblotting.
The presence of p47 in the neurofibromin protein complex was also confirmed by co-immunoprecipitation. Myc-tagged VCP and Myc-tagged p47 were cotransfected into HEK293T cells. The presence of VCP and p47 in the immunocomplex of neurofibromin can then be examined simultaneously by using a Myc tag antibody for immunoblotting. Indeed, both VCP and p47 were precipitated by neurofibromin antibody (Figure 2B), supporting the association of p47 with the neurofibromin protein complex.
The C-terminal D1D2 region of VCP is required for the interaction with the LRD region of neurofibromin.
To delineate the binding domains of VCP involved in the association with neurofibromin, a series of Myc-tagged constructs containing different domains of VCP (Figure 2C) was expressed in HEK293T cells and immunoprecipitated using neurofibromin antibody. Only full-length VCP and the construct encompassing the D1 and D2 domains of VCP interacted with endogenous neurofibromin (Figure 2C), suggesting that the C-terminal D1 and D2 regions but not the N-terminal region of VCP are involved in the interaction with neurofibromin.
To identify the VCP-interacting domain of neurofibromin, we divided neurofibromin into 7 fragments. Among them, only 4 fragments expressed soluble proteins (Figure 2D). These fragments contained the cysteine/serine-rich domain (CSRD), the GAP-related domain (GRD), the leucine-rich repeat domain (LRD), or the C-terminal domain (CTD) (53). In addition to type I neurofibromin carrying the type I isoform of GRD (GRD1), the alternative splice variant GRD2 was also included in our experiment. These constructs were then tagged with an HA cassette and cotransfected with a Myc-tagged D1D2 construct of VCP. The co-immunoprecipitation experiment conducted with a Myc antibody showed that the LRD of neurofibromin is highly enriched in the precipitates (Figure 2D), suggesting that the LRD is the interaction site for the D1D2 fragment. We noticed that the CSRD fragment, perhaps due to the high content of cysteine residues in the CSRD, had a low solubility and tended to aggregate. Therefore, the trace amount of this protein detected in the Myc antibody precipitate may correspond to aggregates of oxidized CSRD protein generated during antibody and protein A binding. To check this possibility, we reduced the time of binding with antibody and protein A (from 4 to 3 hours) and increased the concentration of DTT (from 1 mM to 2 mM) to reduce oxidation. Indeed, these modifications effectively removed the CSRD fragment from the immunoprecipitates (Figure 2E). By contrast, the LRD still associated with the D1D2 fragment (Figure 2E). These data also support the specific interaction between the LRD and D1D2 constructs.
To further confirm the interaction between the LRD with full-length VCP, we cotransfected HEK293T cells with Myc-tagged VCP and HA-tagged LRD constructs. Similar to the result obtained with the D1D2 fragment, the LRD fragment was coprecipitated with full-length VCP protein using Myc tag antibody (Figure 2F). Note that in addition to the expected full-length protein at approximately 100 kDa, we frequently detected Myc-tag–immunoreactive protein species with an apparent molecular weight less than 95 kDa in precipitates as well as inputs (Figure 2, B, C, and F). Smaller protein fragments were also found for the Myc-tagged D2 and D1D2 truncated mutants (Figure 2, C and D). These faster-migrating protein species are likely C-terminal proteolytic products, because the C-terminal region of VCP has been shown to be sensitive to proteolytic degradation (52).
In addition to co-immunoprecipitation from rodent brain and cultured cells, we performed a GST fusion pull-down assay to validate the direct binding of neurofibromin and VCP. As shown in Figure 2G, purified His-tagged D1D2 of VCP was precipitated by the fusion protein GST-LRD but not GST-GRD1. These data suggest a direct protein-protein interaction between neurofibromin and VCP.
VCP regulates the spine density in cultured hippocampal neurons.
Because neurofibromin regulates dendritic spine formation, we hypothesized that VCP may also be involved in this process. Subcellular distribution of VCP in neurons was first examined. Using GFP to outline cell morphology of neurons, we found that VCP was widely distributed in different compartments of neurons, including soma, dendrites, and dendritic spines (Supplemental Figure 2A). Biochemical fractionation indicated that VCP protein is present in the light membrane (P3), crude synaptosomal (P2), and crude synaptic vesicle (LP2) fractions (Supplemental Figure 2B). The presence of VCP protein in the synaptic fractions supports the possibility that VCP locally regulates synapse morphology or density.
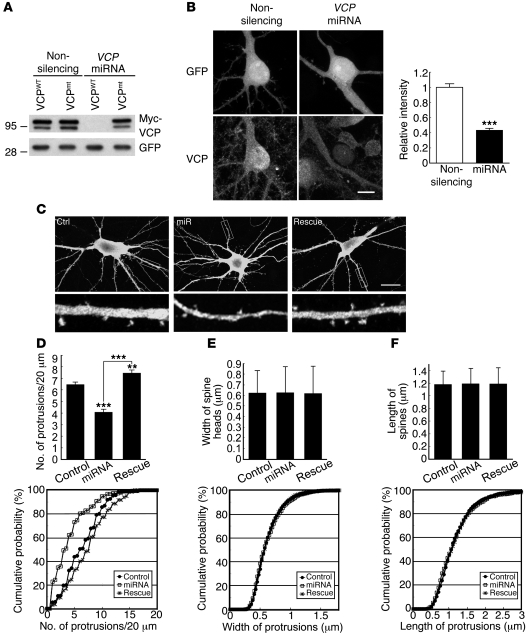
The microRNA (miRNA) knockdown approach was then employed to explore the role of VCP in dendritic spine morphology and density. We generated an artificial miRNA construct coexpressing Emerald GFP (EmGFP) to concurrently label transfected cells and outline the cell morphology. The artificial miRNA was designed to target a site within the VCP gene that is identical in rat and mouse. Therefore, the miRNA construct is expected to reduce the expression of both rat and mouse VCP genes. In addition, a non-silencing control expressing an miRNA sharing no significant homology with mammalian genomes (see Methods for details) was used as a negative control. As expected, the VCP miRNA knockdown clones effectively silenced Myc-tagged mouse VCP expression in HEK293T cells (Figure 3A). In cultured rat hippocampal neurons, the VCP miRNA construct also reduced the expression of endogenous VCP (Figure 3B). In our culture system, it took at least 2 weeks for neurons to be fully differentiated, i.e., to form mature dendritic spines, the location of excitatory synapses. Therefore, in this study, we routinely performed transfection at 12 days in vitro (DIV) and examined neuronal morphology at 18 DIV. Indeed, knockdown of endogenous VCP reduced the density of dendritic spines measured at 18 DIV (Figure 3, C and D; Kolmogorov-Smirnov [KS] test, P < 0.001; t test, P < 0.001). Notably, neither the spine length nor the width of the spine heads was affected as compared with the non-silencing control (Figure 3, E and F). To rule out the possibility of an off-target effect of VCP miRNA, a rescue experiment was performed by cotransfection of a VCP mutant resistant to miRNA. Expression of this VCP silent mutant efficiently increased the spine number (Figure 3, C and D; miRNA vs. rescue, KS test, P < 0.001; t test, P < 0.001), supporting the role of VCP in regulating the density of the dendritic spines.
Figure 3. Reduction of VCP expression impairs dendritic spine formation.
(A) Knockdown effect of the VCP miRNA construct in HEK293T cells. Myc-tagged WT VCP (VCPWT) or a silent mutant of VCP (VCPmt) and non-silencing control or VCP miRNA construct were cotransfected into cells. One day later, cells were harvested for immunoblotting analysis using Myc tag and GFP antibodies. (B) Knockdown of VCP in cultured hippocampal neurons. Transfection of VCP miRNA and non-silencing control was carried out at 12 DIV. Immunostaining using VCP antibody was performed at 18 DIV. The relative VCP protein levels revealed by immunostaining were quantified. Values are presented as means plus SEM. Non-silencing, n = 51; VCP miRNA, n = 35. Scale bar: 10 μm. (C) Cell morphology of VCP-knockdown neurons. Cultured hippocampal neurons were transfected with various plasmids (Control: non-silencing control plus GW1 vector control; miRNA: VCP miRNA plus GW1 vector control; Rescue: VCP miRNA plus miRNA-resistant VCP silent mutant) at 12 DIV and harvested for immunostaining using a GFP antibody at 18 DIV. The lower panels show enlarged images (×5.3) of the boxed regions marked in the upper panels. Scale bar: 20 μm. (D–F) Cumulative probability distributions and means plus SEM of protrusion density (D), protrusion width (E), and protrusion length (F). (D–F) More than 48 neurons, 193 dendrites, and 794 spines for each group from 2 independent experiments were analyzed. In D, P < 0.01, rescue versus control; P < 0.001, miRNA versus control and versus rescue. **P < 0.01, ***P < 0.001.
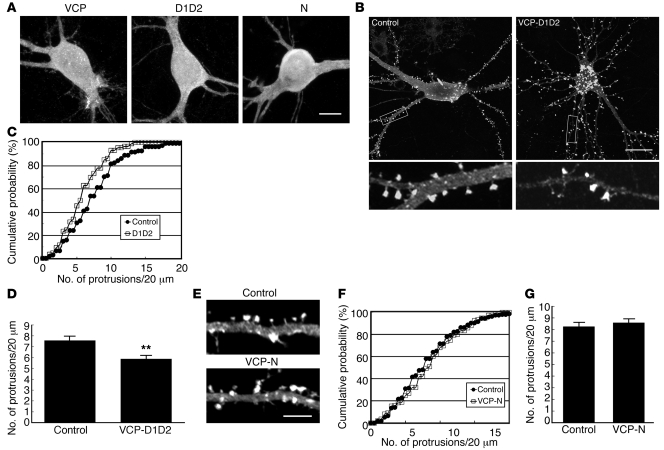
To further confirm the significance of VCP in controlling dendritic spine density, we investigated the effects of the different fragments of VCP in cultured hippocampal neurons. Similar to full-length VCP, the D1D2 and N-domain constructs were widely distributed in neurons (Figure 4A). Compared with the vector control, the presence of the D1D2 fragment reduced the density of dendritic spines (Figure 4, B–D; KS test in Figure 4C, P = 0.013; t test in Figure 4D, P = 0.0014). By contrast, the N-terminal region of VCP, which does not interact with neurofibromin, did not obviously influence the density of dendritic spines (Figure 4, E–G), supporting the specific effect of the D1D2 region on downregulation of the spine density. Moreover, since the D1 and D2 regions possess an ATPase activity, we then investigated whether the ATPase activity of VCP contributes to the effect of the D1D2 fragment on spine density. However, the construct carrying the K524A mutation resulting in ATPase inactivation seemed to have a very strong cytotoxicity to cultured neurons (data not shown). We therefore could not evaluate whether the ATPase activity of VCP is involved in the regulation of dendritic spine density.
Figure 4. Overexpression of the neurofibromin-binding domain of VCP inhibits spine formation.
(A) Immunostaining using Myc-tag antibody reveals the similar expression and distribution of Myc-tagged VCP, D1D2, and N-domain in cultured hippocampal neurons. (B) Myc-tagged VCP D1D2 construct and (E) Myc-tagged N-domain and vector control were cotransfected with GFP-actin, as indicated, into cultured hippocampal neurons at 12 DIV. Six days later, the neuronal morphology was monitored by detection of GFP immunoreactivity. In B, the lower panels show enlarged images (×5.3) of the boxed regions marked in the upper panels. (C and F) Cumulative probability distributions and (D and G) graph of protrusion densities obtained from B and E, respectively. More than 22 neurons and 86 dendrites for each group of experiments were analyzed. P < 0.05, DID2 versus control; **P < 0.01. Scale bars: 10 μm (A); 20 μm (B); 5 μm (E). Values are presented as mean plus SEM.
In conclusion, the above analyses using VCP miRNA and the D1D2 construct suggested a role of VCP in the regulation of dendritic spine density.
Disruption of the interaction between neurofibromin and VCP results in reduced dendritic spine density.
To confirm the role of the interaction between neurofibromin and VCP in spinogenesis, the LRD fragment (Figure 5A), a region of neurofibromin interacting with VCP, was overexpressed in cultured hippocampal neurons. Compared with vector control, expression of the LRD fragment reduced the spine density of cultured hippocampal neurons (Figure 5, B–D; KS test in Figure 5C, control vs. WT LRD, P < 0.001; t test in Figure 5D, control vs. LRD, P < 0.001), supporting the role of the interaction between neurofibromin and VCP in the regulation of dendritic spine density.
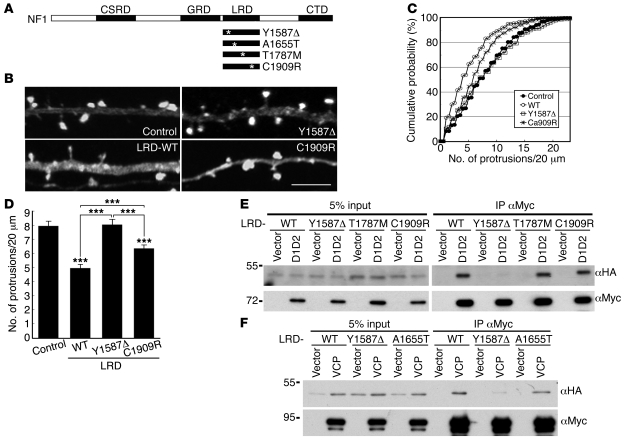
Figure 5. Overexpression of the VCP-binding domain of neurofibromin reduces the number of dendritic spines.
(A) Mutations in the LRD region of the NF1 gene identified from NF1 patients. Locations of mutation sites are indicated by asterisks. (B) Cotransfection of GFP-actin and vector control, HA-tagged WT LRD, or LRD mutants (Y1587Δ and C1909R) into cultured hippocampal neurons at 12 DIV. Six days later, immunostaining was performed using GFP and HA tag antibodies. Only GFP signals are shown. Scale bar: 5 μm. (C) Cumulative probability distributions and (D) graph of protrusion densities. More than 52 neurons and 224 dendrites collected from 2 independent experiments were analyzed. In C, P < 0.01, C1909R versus control and versus Y1587Δ; P < 0.001, C1909R, Y1587Δ, and control versus WT. ***P < 0.001. Values are presented as mean plus SEM. (E) Myc-tagged D1D2 and (F) Myc-tagged VCP were cotransfected with HA-tagged WT and mutant LRD into HEK293T cells as indicated. One day later, total cell extracts were harvested and immunoprecipitated with a Myc antibody. Immunoblotting analysis was then performed using sequentially HA and Myc tag antibodies.
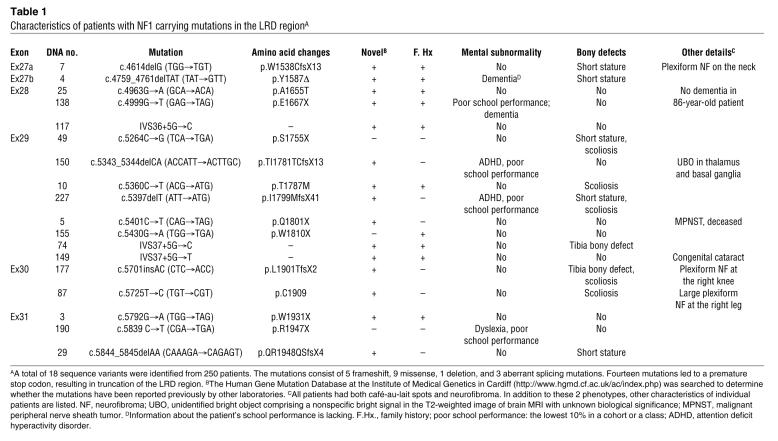
We then asked whether mutations identified in the NF1 gene of patients with NF1 would affect the interaction between neurofibromin and VCP and whether these mutations lead to the reduction in spine density. Mutation screening of 250 Taiwanese NF1 patients fulfilling NIH diagnostic criteria led to the identification of 18 mutations in the LRD region (Table 1), of which only 3 have been identified in previous studies (54–56). Examination of the familial segregation and screening of the control group (>300 Taiwanese participants) suggested that these mutations are pathogenic. Of these 18 mutations, 14 (77.8%) result in a truncated protein, 3 lead to a change of a single amino acid (mutants p.A1655T, p.T1787M, and p.C1909R), and 1 results in the deletion of a single amino acid (residue Y1587, c.4759_4761delTAT; Table 1). In addition to a part of the LRD, the truncated mutant proteins lack more than one-third of the C-terminal residues of neurofibromin. This fact makes it difficult to evaluate the specific role of the interaction between these truncated NF1 mutants with VCP. We therefore focused on the missense and single amino acid deletion mutants (Figure 5A). The interactions between these LRD mutants and the D1D2 fragment or full-length VCP were examined by co-immunoprecipitation. When compared with WT LRD, T1787M, C1909R, and A1655T mutations did not noticeably affect the interaction between the LRD domain and the D1D2 region or full-length VCP (Figure 5, E and F). By contrast, deletion of the residue Y1587 almost completely abolished this interaction (Figure 5, E and F). These data suggest a critical role of this tyrosine residue in the interaction between VCP and neurofibromin LRD.
Table 1 .
Characteristics of patients with NF1 carrying mutations in the LRD regionA
We then investigated the biological significance of the Y1587 deletion. Because the overexpression of WT LRD (Figure 5, B–D) likely disrupted the interaction between endogenous VCP and neurofibromin and thus reduced spine density, we expected that overexpression of the LRD Y1587Δ mutant would not affect spine density, given that this mutant cannot interact with VCP (Figure 5F). Indeed, expression of the LRD Y1587Δ mutant did not reduce spine number (Figure 5, B–D; KS test in Figure 5C, control vs. Y1587Δ, P = 0.83; WT LRD vs. Y1587Δ, P < 0.001; t test in Figure 5D, control vs. Y1587Δ, P = 0.85; WT LRD vs. Y1587Δ, P < 0.001). By contrast, the LRD C1909R mutant, which is capable of interacting with VCP, was able to reduce the spine density (Figure 5, B–D; KS test in Figure 5C, control vs. C1909R, P = 0.002; t test in Figure 5D, control vs. C1909R, P < 0.001), though the inhibitory effect was significantly weaker than that of WT LRD (Figure 5, B–D; KS test in Figure 5C, P < 0.001; t test in Figure 5D, P < 0.001). In conclusion, the results of these analyses suggest that the Y1587Δ mutation in the NF1 gene disrupts the interaction between neurofibromin and VCP and, as a result, affects dendritic spinogenesis.
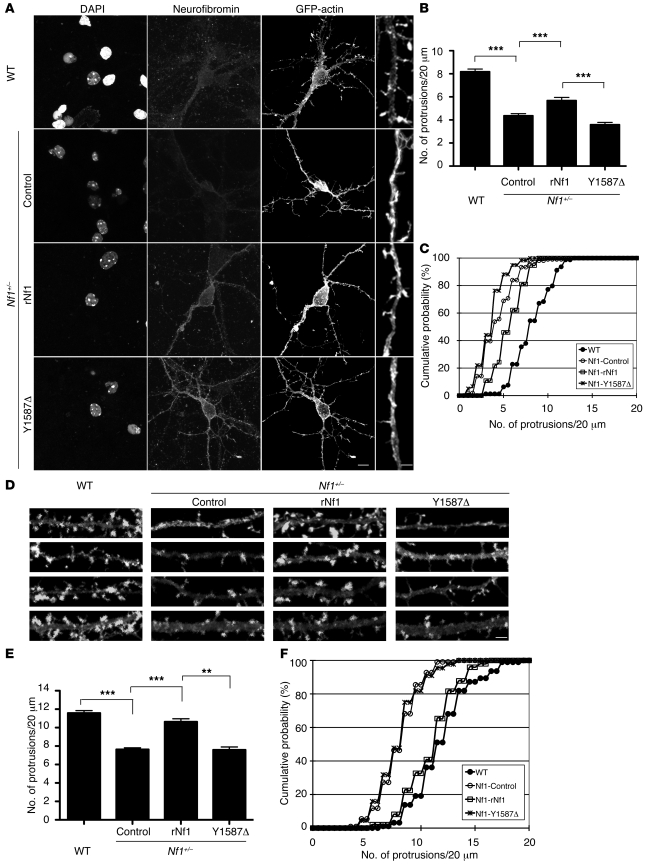
To further confirm the importance of residue Y1587 in the function of neurofibromin, we used cultured cortical neurons prepared from Nf1+/– mice. The aforementioned data suggested that deletion of one copy of the Nf1 gene reduced the spine density of pyramidal neurons in mouse brain. We then examined whether transfection of full-length rNf1 increases the density of dendritic spines in Nf1+/– neurons and whether the full-length Y1587Δ mutant loses this ability. Two sets of experiments with different time points for transfection and immunostaining were performed. In the first set, conditions identical to those in the experiments described above were chosen, i.e., transfection and staining were carried out at 12 DIV and 18 DIV, respectively. In the second set, transfection was done at 7 DIV and staining was carried out at 10 DIV. Our previous study indicated that neurofibromin regulates the initial step of dendritic spinogenesis, namely dendritic filopodia formation (9). We therefore expected that exogenous NF1 should also increase the number of dendritic filopodia in relatively young neurons, such as at 10 DIV. Indeed, compared with vector control, expression of full-length rNf1 increased the density of dendritic protrusion in Nf1+/– cortical neurons at 10 DIV (Figure 6, A–C; Nf1+/– control vs. Nf1+/– rNf1, t test, P < 0.001; KS test, P = 0.007) as well as at 18 DIV (Figure 6, D–F; Nf1+/– control vs. Nf1+/– rNf1, t test, P < 0.001; KS test, P < 0.001). By contrast, expression of the full-length Y1587Δ mutant did not increase the protrusion density in Nf1+/– neurons at 10 DIV (Figure 6, A–C; Nf1+/– rNf1 vs. Nf1+/– Y1587Δ, t test, P < 0.001; KS test, P < 0.001) or at 18 DIV (Figure 6, D–F; Nf1+/– rNf1 vs. Nf1+/– Y1587Δ, t test, P < 0.001; KS test, P < 0.001). These results support the importance of the residue Y1587 for the activity of neurofibromin in controlling spine density.
Figure 6. Overexpression of full-length rNf1, but not Y1587Δ mutant, rescues the dendritic spine phenotype of Nf1+/– neurons in culture.
Mouse cultured cortical neurons were transfected at DIV7 (A) or DIV12 (D) and harvested for staining using neurofibromin and GFP antibodies at DIV10 (A) or DIV18 (D). For WT neurons, GFP-actin and vector control were transfected. For Nf1+/– neurons, GFP-actin was cotransfected with full-length rNf1, full-length Y1587Δ mutant, or vector control, as indicated. B and C, and E and F are quantitative analyses of A and D, respectively. Values in B and E represent the mean plus SEM. The numbers of analyzed dendrites for each group were: (B and C) WT, 79; Nf1+/– control, 106; Nf1+/– rNf1, 37; Nf1+/– Y1587Δ, 59; (E and F) WT, 94; Nf1+/– control, 110; Nf1+/– rNf1, 49; Nf1+/– Y1587Δ, 44. Scale bars: 10 μm in the whole images; 2 μm in the enlarged images (A); 2 μm (D). In C, P < 0.01, Nf1+/– rNf1 versus Nf1+/– control; P < 0.001, Nf1+/– control versus WT and Nf1+/– rNf1 versus Nf1+/– Y1587Δ. In F, P < 0.001, Nf1+/– control versus WT and versus Nf1+/– rNf1, and Nf1+/– Y1587Δ versus Nf1+/– rNf1. **P < 0.01, ***P < 0.001.
Clinically, the proband with the mutation Y1587Δ had multiple café-au-lait spots with numerous cutaneous neurofibroma, which are the typical phenotypes of patients with NF1. This patient was diagnosed with mental subnormality in 2004 at the age of 63 and has been experiencing dementia for the past 3 years. Among the 18 patients with an NF1 mutation in the LRD region, 5 were diagnosed as having mental subnormality characterized by either poor school performance or dementia (27.8%, Table 1). In a previous study, the frequency of mental subnormality of NF1 patients was less than 5% in a Taiwanese cohort (3 of 68, ref. 57). The higher frequency of mental subnormality in NF1 patients carrying mutations in the LRD region may be related to the interaction between neurofibromin and VCP and the role of LRD in the regulation of dendritic spine density.
VCP IBMPFD mutants interact weakly with neurofibromin and impair spinogenesis.
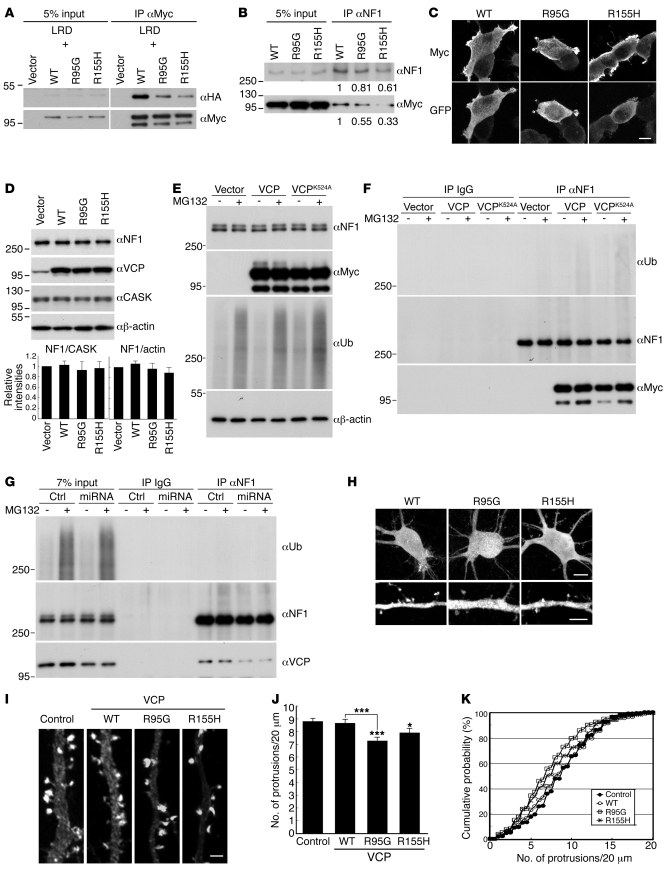
Because the identified IBMPFD mutations are clustered at the interface of the N-domain and the D1 region, it is believed that they can alter the conformation of the hexameric barrel formed by the D1 domain during ATP binding and ATP hydrolysis (51, 52). Because both D1 and D2 domains are required for the interaction with neurofibromin, we speculated that neurofibromin recognizes a special conformation of VCP that may be sensitive to IBMPFD mutations. To investigate this possibility, we conducted co-immunoprecipitation experiments to determine whether IBMPFD mutations would result in a decrease in the interaction of VCP and neurofibromin in HEK293T cells. Compared with WT VCP, both VCP R155H and R95G mutants, which occur most frequently in IBMPFD, interacted weakly with the cotransfected LRD fragment (Figure 7A) or endogenous full-length neurofibromin (Figure 7B), suggesting that IBMPFD mutations in the N-terminal region of VCP reduce the protein-protein interaction mediated through the C-terminal D1 and D2 regions of VCP.
Figure 7. IBMPFD mutations reduce the interaction of neurofibromin and VCP and the density of dendritic spines.
(A) LRD and (B) neurofibromin interact weakly with IBMPFD mutants. Variant Myc-tagged VCP constructs and vector control were cotransfected with HA-tagged LRD or singly transfected into HEK293T cells. One day later, (A) Myc and (B) neurofibromin antibodies were used for immunoprecipitation. Immunoblotting analysis was performed using the antibodies as indicated. The relative intensities to WT lane are shown. Distribution of Myc-tagged VCP and IBMPFD mutants in (C) HEK293T cells and (H) cultured hippocampal neurons was examined by immunostaining using Myc and GFP antibodies. Cotransfection with GFP was performed to outline cell morphology. Expression of (D) IBMPFD mutants, (E and F) VCP K524A mutant, and (G) VCP miRNA construct did not obviously influence the total protein level of neurofibromin. HEK293T cells were transfected with variant VCP constructs, VCP miRNA, or vector control. Immunoblotting was then performed to examine the protein levels of neurofibromin. CASK and/or actin were used as internal controls. In E, F, and G, MG132 was added to cultures 4 hours before harvest or omitted. Ubiquitin antibody FK2 (αUb) was also included in immunoblotting analysis. For F and G, immunoprecipitation was performed using neurofibromin antibody to precipitate endogenous neurofibromin. Immunoblotting using FK2 antibody did not reveal obvious signals in the neurofibromin precipitates. (I) IBMPFD mutants decrease spine density. Cultured hippocampal neurons were cotransfected with GFP-actin and vector control, WT VCP, or IBMPFD mutants (R95G and R155H) at 12 DIV. Cells were fixed at 18 DIV and stained with Myc and GFP antibodies. The morphology of spines was then analyzed based on the GFP signals. (J) Protrusion density. Values are presented as mean plus SEM. (K) Cumulative probability distributions. More than 34 neurons and 146 dendrites for each group collected from 2 independent experiments were analyzed. Scale bars: (C) 10 μm; (H) upper panels, 10 μm; lower panels, 5 μm; (I) 2 μm. In K, P < 0.05, R155H versus control; P < 0.01, R95G versus WT; P < 0.001, R95G versus control. *P < 0.05, ***P < 0.001.
Because previous observations showed that IBMPFD mutant VCP can form aggregates in myoblastoma cells (43), we then wondered whether the reduction of the interaction between an IBMPFD mutant and neurofibromin is caused by a change in the subcellular distribution of mutant VCP protein. However, in HEK293T cells, the subcellular distribution of R155H and R95G mutants was similar to that of WT VCP; each protein was widely distributed, with a tendency to concentrate at the cell cortex (Figure 7C). We did not find any obvious VCP protein aggregates in HEK293T cells. This result indicates that protein aggregation of IBMPFD mutants is cell type–specific and also suggests that the reduced interaction of IBMPFD mutants and neurofibromin is unlikely to be due to an altered subcellular distribution of IBMPFD mutants.
Although VCP has been shown to associate with the 26S proteasome and to regulate degradation of ubiquitinated proteins (58, 59), the interaction between VCP and neurofibromin did not appear to regulate the protein stability of neurofibromin, as the protein levels of neurofibromin were comparable in HEK293T cells expressing WT VCP and those expressing the IBMPFD mutant (Figure 7D). In addition to IBMPFD mutants, the effects of VCP knockdown and K524A mutation were also examined. Similarly, the levels of neurofibromin protein were not altered by expression of VCP K524A mutant or knockdown of VCP (Figure 7, E and G). In these experiments, MG132, a potent proteasome inhibitor, was either added to the cultures 4 hours before harvesting cell extracts or omitted. Although MG132 increased the signals of protein ubiquitination, it did not increase the total neurofibromin protein levels in either VCP-knockdown cells or K524A mutant–expressing cells (Figure 7, E and G). Moreover, after enrichment by immunoprecipitation, we still could not detect ubiquitinated neurofibromin after alteration of VCP protein level or expression of K524A mutant (Figure 7, F and G). Taken together, all of these analyses suggest that VCP does not influence the protein level or ubiquitination of neurofibromin.
Cultured hippocampal neurons were then used to assess the effect of IBMPFD mutations on spinogenesis. Similar to the results in HEK293T cells, expression of the R95G and R155H mutants was comparable to that of WT VCP in cultured hippocampal neurons. The intensity of immunoreactivity of WT VCP was comparable to that of R95G and R155H mutants (Figure 7H). Besides, we did not find protein aggregates of IBMPFD mutants in neurons (Figure 7H). Like WT VCP, the IBMPFD mutant proteins also entered dendritic spines (Figure 7H). Thus, IBMPFD mutations did not noticeably alter expression levels or the subcellular distribution of VCP. However, the R95G mutant significantly reduced the density of dendritic spines (Figure 7, I–K; KS test in Figure 7K, control vs. R95G, P < 0.001; WT vs. R95G, P = 0.006; t test in Figure 7J, control vs. R95G, P < 0.001; WT vs. R95G, P < 0.001). Although the effect exhibited by the R155H mutant was weaker, it still reduced the density of dendritic spines as compared with the vector control (Figure 7, I–K; KS test in Figure 7K, P = 0.012; t test in Figure 7J, P = 0.028). By contrast, WT VCP did not appear to impair the spine density (Figure 7, I–K). These data suggest that the presence of IBMPFD mutants impairs dendritic spine formation.
VCP acts downstream of neurofibromin in the regulation of dendritic spine density.
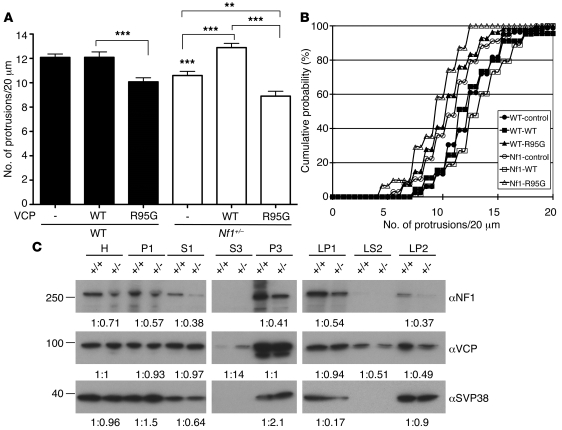
We then used Nf1+/– cortical neurons to investigate whether VCP acts downstream of neurofibromin. In Nf1+/– neurons, expression of WT VCP increased the spine density to a level comparable to that in WT neurons (Figure 8, A and B). By contrast, the VCP R95G mutant did not rescue the density of dendritic spines. In Nf1+/– neurons, the R95G mutant further reduced the spine density compared with vector control (Figure 8, A and B). These data support the conclusion that VCP is located downstream of neurofibromin in controlling spine density.
Figure 8. VCP acts downstream of neurofibromin in the regulation of dendritic spine density.
(A and B) Cultured cortical neurons isolated from Nf1+/– mice and WT littermates were cotransfected with GFP-actin and Myc-tagged WT VCP or R95G mutant or vector control at 12 DIV. The density of dendritic spines was determined at 18 DIV by staining using GFP and Myc antibodies. (A) Mean values plus SEM of protrusion density. **P < 0.01, ***P < 0.001. (B) Cumulative probability of protrusion density. In total, 19–26 neurons and 31–82 dendrites were analyzed. P < 0.01, WT R95G versus WT WT and Nf1+/– control versus WT control; P < 0.001, Nf1+/– WT versus Nf1+/– control and versus Nf1+/– R95G. (C) Subcellular distribution of VCP in Nf1+/– brain. Subcellular fractions of WT and Nf1+/– brains were isolated by a series of centrifugations (95) and analyzed by immunoblotting. Synaptophysin (SVP38) was used as a loading control of LP2. H, total homogenate; P1, crude nuclei, unbroken cells, and debris; S1, supernatant of P1; P3, light membrane fraction (including ER and Golgi body); S3, soluble cytosolic fraction; LP1, lysed synaptosomal membrane; LP2, crude synaptic vesicles; LS2, soluble synaptic cytosol. The signals were quantified using Gel-Pro Analyzer (Media Cybernetics). The intensity ratios of WT to Nf1+/– of each fraction are shown.
Our previous study had shown that neurofibromin was widely distributed in the various membrane fractions purified from rat brains, including the light membrane fraction, lysed synaptosomal membrane fractions, and crude synaptic vesicle fraction (22). Here we found that the neurofibromin protein levels in Nf1+/– brains were lower than those in WT brains regardless of the subcellular fraction examined (Figure 8C). No obvious difference in the VCP protein levels in total homogenates between Nf1+/– mice and WT littermates was detected (Figure 8C). In several subcellular fractions, including light membrane (P3) and lysed synaptosomal membrane (LP1) fractions, the distribution of VCP protein was also comparable in WT and Nf1+/– brains (Figure 8C). However, in Nf1+/– brains, the level of VCP protein was lower in the crude synaptic vesicle (LP2) and synaptic cytosol (LS2) fractions, which contain vesicles and cytosolic fractions at both pre- and post-synaptic sites (Figure 8C). By contrast, VCP protein was more abundant in the total soluble cytosolic fraction (S3) of Nf1+/– mice as compared with WT littermates (Figure 8C). These results suggest that the subcellular distribution of VCP is influenced by neurofibromin.
Taken together, our results suggests that loss of one copy of the Nf1 gene reduces the level of VCP in crude synaptic vesicles and synaptosomal cytosol and may thus impair dendritic spine formation. Overexpression of VCP in Nf1+/– neurons may ectopically increase the VCP level at synapses and rescue the defects in the density of dendritic spines.
Statin treatment rescues the effect of VCP knockdown on spine density.
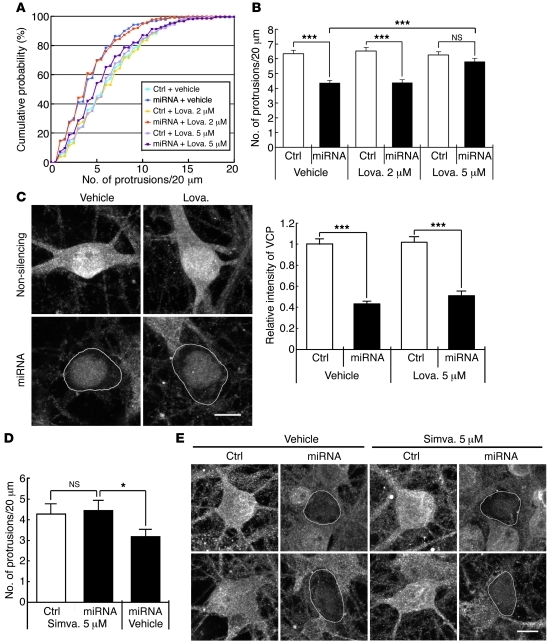
A previous study showed that lovastatin, an inhibitor of the HMG-CoA reductase, rescues the defects of Nf1+/– mice in learning disability (60). We therefore asked whether lovastatin treatment rescues the defect in spinogenesis resulting from silencing expression of VCP. Treatment of cultured hippocampal neurons with lovastatin at a concentration of 2 μM for 3 days did not rescue the effect of VCP knockdown on spine density. However, at a higher concentration (5 μM), the spine density of lovastatin-treated VCP-knockdown neurons was comparable to that of neurons treated with lovastatin alone (Figure 9, A and B). In VCP-knockdown neurons, 5 μM lovastatin treatment increased the density of dendritic spines compared with vehicle control (Figure 9, A and B). The effect of lovastatin was unlikely due to interference with Vcp miRNA knockdown, because the Vcp miRNA construct still effectively knocked down the expression of VCP in the presence of this statin (Figure 9C). Similar results were obtained with simvastatin, with regard to both rescuing the effect of VCP knockdown on spine density (Figure 9D) and absence of interference with VCP knockdown (Figure 9E). Taken together, these data suggest that a statin-sensitive pathway is also involved in VCP-mediated spinogenesis. It is consistent with our hypothesis that VCP and neurofibromin work together to regulate neural function.
Figure 9. Statin treatment abates the effect of VCP knockdown on dendritic spine density.
Cultured hippocampal neurons were cotransfected with GFP-actin and vector control or VCP miRNA at 12 DIV and treated with lovastatin (Lova.) at 15 DIV for 3 days or simvastatin (Simva.) at 14 DIV for 4 days. Quantitative measurements of spine density are shown as (A) cumulative probability distributions and (B and D) graph of protrusion densities. For A and B, more than 53 neurons and 213 dendrites for each group were analyzed. For D, more than 30 dendrites for each group were analyzed. In the presence of statins, knockdown efficiency of VCP miRNA assayed by immunostaining using VCP antibody was also evaluated in C and E. For VCP-knockdown neurons, somas were outlined based on the GFP signal. In C, sample sizes of each group were as follows: vehicle plus control, 51; vehicle plus miRNA, 35; lovastatin plus control, 59; lovastatin plus miRNA, 26. In A, P < 0.001, miRNA vehicle versus control vehicle and miRNA lovastatin 2 μM versus control lovastatin 2 μM. *P < 0.05, ***P < 0.001. Scale bars: 10 μm (C and E).
Discussion
VCP and dendritic spines.
IBMPFD is characterized by three clinical features: myopathy, osteolytic bone lesions, and FTD (44). Using a knockdown approach and IBMPFD mutants, we provide several lines of evidence that VCP plays a role in dendritic spine formation. Expression of the IBMPFD mutants reduces the number of dendritic spines in cultured hippocampal neurons by 10%–18%. Since synapse loss has been suggested in various neurological disorders associated with dementia (61–67), the ability of VCP in the regulation of spinogenesis may be relevant to clinical features of dementia in patients. VCP does not represent the only example where a gene responsible for FTD is involved in neuronal morphogenesis. Chromatin-modifying protein 2B (CHMP2B), the causative gene for FTD3 (68), also regulates dendritic morphology and the number of dendritic spines (69–71). These studies imply that FTD may be caused, at least partially, by cumulative alterations in neuronal morphology, including dendritic arborization and dendritic spinogenesis. To confirm this hypothesis, characterization of genetic mouse models is required in the future.
VCP is a multifunctional protein involved in ERAD, autophagy, and membrane fusion. It is still unclear how IBMPFD mutants reduce the density of dendritic spines. Protein degradation through ERAD and autophagy pathways is certainly a possibility. Furthermore, membrane fusion is likely involved in the process, because vesicle trafficking is actively involved in the regulation of synapse formation and activity. In agreement with this assumption, we found that p47, a cofactor of VCP playing an important role in membrane fusion, was co-immunoprecipitated with neurofibromin and VCP. p47 interacts with the N-terminal region of VCP; the C-terminal D1 and D2 domains of VCP are involved in the interaction with neurofibromin. Neurofibromin, VCP, and p47 likely form a protein complex to regulate neuronal morphology. However, it is possible that all of these events, including ERAD, autophagy, and membrane trafficking, contribute to dendritic spine formation. More investigations need to be carried out to further elucidate the molecular role of VCP in synapse formation.
VCP forms a hexameric barrel. ATP hydrolysis provides the energy to change the conformation of the hexameric barrel and achieve its function as a molecular chaperon (49). IBMPFD mutations cluster at the interface of the N- and D1 domains and modulate ATPase activity and D2 ring conformation of VCP (52). These mutations do not hit the hexamerization site of the D1 region. Therefore, these mutant proteins still form hexameric barrels in solution (52). Mutant VCP may form heterohexamers with WT VCP, thus impairing the function of WT VCP. This would explain, at least partially, why the IBMPFD mutations have a dominant effect of impairing the function of VCP in cells. It may also explain why the IBMPFD mutants that interact weakly with neurofibromin have a dominant-negative effect on the control of dendritic spine density.
Although IBMPFD mutations induce protein aggregates of VCP in C2C12 myoblastoma cells (43), we did not observe such aggregates in transiently transfected HEK293T cells or in cultured neurons. These data suggest that muscle cells are more susceptible to IBMPFD mutations, which may be relevant to the observation that, in patients with IBMPFD, the mean age of presentation for inclusion body myopathy was 42 years, whereas that for FTD was 53 years (36). In addition, since the IBMPFD mutants reduced the spine density without obviously causing VCP aggregation in neurons, our study also suggests that protein aggregation is unlikely a cause of neuronal morphological defects induced by IBMPFD mutations.
The molecular basis of NF1.
NF1 is also an autosomal dominant disorder. Although there are four major features of NF1 (i.e., café-au-lait spots, peripheral neurofibromas, skinfold freckling, and Lisch nodules), many minor features and medical complications are present in a substantial number of patients with NF1 (72), increasing the difficulty of effective therapeutic treatment. So far, it is unclear why clinical features of NF1 patients vary markedly. One reason may be the lack of a mutational hot spot in the NF1 gene. Mutations identified in patients with NF1 are widely spread over the entire gene. Different domains of neurofibromin are expected to carry out specific biological functions; however, this issue has not yet been fully addressed. With the exception of the GRD and LRD regions, the remaining parts of neurofibromin lack recognizable protein domains. Therefore, numerous efforts have been directed to identify neurofibromin-interacting proteins (22, 73–78). Identification of candidate proteins that interact with neurofibromin may provide better and more effective treatments for individual NF1 patients who carry a mutation in a specific domain. In the present study, we identified and characterized the interaction of VCP and neurofibromin. We also found that the reduced spinogenesis in VCP-knockdown neurons was rescued by lovastatin and simvastatin. The results support the notion that neurofibromin and VCP contribute to the same pathway to control spinogenesis and also imply that, for patients carrying a mutation in the LRD region of the NF1 gene, statins may be good candidates as therapeutic agents. In fact, a previous study has shown that lovastatin treatment reverses the learning deficits of Nf1+/– mice (60, 79). Furthermore, lovastatin improves osteoblast activity and rescues tibial dysplasia and the bone healing defect caused by loss of Nf1 (79, 80). However, a randomized controlled trial showed that simvastatin treatment does not improve cognitive function in patients with NF1 (81). A possible explanation for this conflict might be that statin may be only effective in a fraction of NF1 patients, such as patients carrying mutations that alter the interaction between neurofibromin and VCP. More investigations have to be carried out to evaluate this possibility.
The effects of statins.
Statins are well known to inhibit HMG-CoA reductase, the rate-limiting enzyme of the mevalonate pathway. The mevalonate pathway produces cholesterol and other isoprenoids. Therefore, statins are able to reduce the cholesterol level and lipid modification of proteins in cells. It has been shown that lipid modification is critical for the activity of Ras and that lovastatin inhibits Ras isoprenylation and activities in controlling cell growth and neuronal differentiation (82, 83). Since neurofibromin also inhibits Ras activity through its GAP domain, Li and colleagues used statins to rescue the learning defects of Nf1+/– mice (60). In addition to protein isoprenylation, statins may also influence the density of dendritic spines through regulation of the cholesterol level, since cholesterol has been shown to be involved in spinogenesis (84). Although cholesterol is required for spinogenesis, the cholesterol/glycerophospholipid molar ratio gradually decreased during neuronal differentiation (85). Therefore, it is likely that in mature neurons, even though the total cholesterol level is lower, cholesterol is enriched at dendritic spines to maintain spine morphology. Taken together, these studies also suggest that cholesterol homeostasis is important for neuronal development. Interestingly, VCP has been shown to be involved in sterol-induced dislocation of HMG-CoA reductase from the ER, which leads to ubiquitination and degradation of HMG-CoA reductase (86–88). Therefore, treatment with statins may rescue an imbalance of cholesterol synthesis and thus reverse the spine phenotype in VCP-deficient neurons. It will be interesting to further confirm the role of statins in the VCP-regulated pathway.
The crosstalk between neurofibromin and VCP.
Mutations of NF1 and VCP result in different genetic disorders. However, when the phenotypes of NF1 and IBMPFD are analyzed in detail, some similar, though not identical, features can be found. NF1 patients are prone to learning disability, whereas some IBMPFD patients display the features of dementia. The function of neurofibromin and VCP in the regulation of dendritic spine density may account, at least in part, for these phenotypes. In our 250 NF1 patients, 18 mutations occurred in the LRD region (Table 1). Among these patients, 5 of 18 (27.8%) developed mental subnormality. The percentage of NF1 patients with mental subnormality, including poor school performance and attention deficit hyperactivity disorder, was as high as 60% in previous studies (89), while in a study focusing on a Taiwanese cohort, the total was less than 5% (3 of 68). The evidence provided in this report suggests that the learning disability in patients with an NF1 mutation in the LRD region is relatively common. This clinical feature supports a genetic interaction between VCP and neurofibromin.
In addition to learning disability, 9 of 18 patients (50%) with mutations in the LRD region of NF1 had problems in the skeletal system, including short stature, tibia bone defects with pseudoarthrosis, and scoliosis. Abnormal skeletal development occurs in 10%–20% of NF1 patients (90). Multiple forms of scoliosis occur in at least 10% of NF1 patients (91); our experience suggests that the frequency of bone defects in NF1 patients might be as high as 25% (17 of 68) (57). Thus, patients with a mutation in the LRD region of the NF1 gene are also susceptible to bone dysplasia and defects. This correlation also favors a genetic interaction between neurofibromin and VCP, since IBMPFD mutations also have an impact on bone metabolism.
So far, it is not clear why the phenotype of a mutation in the NF1 gene occurs at an early stage, while that of a mutation in the VCP gene occurs at a later stage of life. This discrepancy may indicate that another regulation, such as other reacting protein or posttranslational modification, is involved in regulation of the role of neurofibromin and VCP in spinogenesis. It is also likely that neurofibromin and VCP have other functions in neuronal morphogenesis and therefore display differential phenotypes. For instance, the C1909R mutation of neurofibromin did not obviously affect the interaction between the LRD fragment and VCP. However, overexpression of the LRD C1909R mutant still had a weaker impact on spinogenesis in cultured hippocampal neurons, suggesting that there is an alternative pathway downstream of neurofibromin to control spinogenesis that does not require an interaction with VCP.
In conclusion, our study provides the first evidence to our knowledge that VCP regulates dendritic spine formation, which may correlate with the dementia phenotype in patients with IBMPFD. Moreover, VCP interacts with neurofibromin to regulate dendritic spine formation, implying an interconnection between NF1 and IBMPFD.
Methods
Human research subjects
Patients with NF1 were clinically assessed by clinical geneticists and/or neurologists. All the clinical information pertinent to the diagnosis fulfilled the NIH diagnostic criteria of NF1, and, if applicable, laboratory test results from patients with a NF1 mutation were interpreted by a member of our research team as well as referral physicians. The mental subnormalities and school performance were reported by either the patients themselves or their accompanying family members.
Mutation analysis of the NF1 gene
Two hundred and forty-six Taiwanese patients with NF1 were recruited. Genomic DNA from peripheral blood samples of patients was extracted for screening. Sequencing reactions of the region containing 6 exons (exons 27a, 27b, 28, 29, 30, and 31), which encompasses the LRD of neurofibromin, were performed. In total, 18 sequence variants were identified, comprising 5 frameshift, 9 missense, 1 deletion, and 3 splicing aberrant mutations (Table 1). Fourteen mutations (77.8%) led to a premature stop codon truncating the LRD region. The genetics study was approved by Institutional Review Board of the National Taiwan University Hospital, Taipei, Taiwan.
Antibodies and chemicals
The following antibodies were used in this report: neurofibromin NF1(N) and GST (Santa Cruz Biotechnology Inc.), VCP (for immunoblotting, BD Biosciences; for staining, Bethyl), GFP (Invitrogen), ubiquitin FK2 (Assay Designs), HA (both mouse monoclonal 12CA5 and rat monoclonal, Roche), and Myc 9B10 (Cell Signaling Technology). Simvastatin and lovastatin were obtained from Sigma-Aldrich. MG132 was purchased from Calbiochem.
Immunoprecipitation
Rat brain or cells were homogenized in lysis buffer (1× PBS buffer containing 1% Triton X-100, 1–2 mM DTT, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 10 μg/ml pepstatin, and 1 mM N-tosyl-l-phenylalanylchloromethyl ketone) and centrifuged at 100,000 g at 4°C to remove cell debris. The solubilized extract was subjected to immunoprecipitation using specific antibodies and control non-immune IgG. After mixing by rotation for 4–5 hours at 4°C, the precipitates were sequentially washed 5 times with PBS containing 0.2% Triton X-100, twice with PBS, and once in 10 mM Tris-HCl buffer (pH 7.4). The proteins in the washed precipitates were then analyzed by 2D gel electrophoresis or immunoblotting.
Plasmid construction
NF1.
Human neurofibromin fragments were amplified by PCR using the following primers: (a) for CSRD (cysteine/serine-rich domain, aa 543–909 of type I protein), 5′-GAAGATCTCAGGAAGCAATGGAGGCTCTG and 5′-GAAGATCTTTACACTTTCTCATGGTTACACACCAT; (b) for GRD1 and GRD2 (GAP-related domain type I, aa 1168–1530 of type I protein, and GAP-related domain type II, aa 1168–1551 of type II protein), 5′-GAAGATCTGGTTACCACAAGGATCTCCAGA and 5′-GAAGATCTTTAGTGCTCTGGAGGACCCAGGTAT; (c) for LRD (leucine repeat domain, aa 1545–1950 of type I protein), 5′-GAAGATCTAGTTCAAAGTTTGAGGAATTTATGACT and 5′-GAAGATCTTTAAACTCTTTGTCGTTTGGCATCATC; (d) for CTD (C-terminal domain, aa 2260–2818 of type I protein), 5′-GAAGATCTGGACCTGACACTTACAACAGTCA and 5′-GAAGATCTTCACACGATCTTCTTAATGCTATT, where underlining denotes the recognition sequence of the restriction endonuclease BglII. The PCR products were then subcloned into the BglII site of the vectors HA-GW1 or pGEX4T1. The full-length rat Nf1 expression construct was a gift of Shigeki Shibahara (Department of Molecular Biology and Applied Physiology, Tohoku University School of Medicine, Tohoku, Japan). Site-directed mutagenesis was carried out using the primer pair 5′-GGGAATCCTATTTTCTATGTTGCACGGAGGTTCAAA-3′ and 5′-TTTGAACCTCCGTGCAACATAGAAAATAGGATTCCC-3′ to introduce the Y1587 deletion mutation into the full-length rNf1 gene.
VCP.
A set of primers — A: 5′-GGGGTACCGCCTCTGGAGCCGATTCAA; B: 5′-GAAGATCTTTAGCCATACAGGTCATCGTCATT; C: 5′-GAAGATCTTTAGCCTACTTCATTCAAGGATTCCTC; D: 5′-GAAGATCTTA TGATGACATCGGTGGTTGCA; E: 5′-GAAGATCTTTAATCCATAGTAACTGC CAGGGAATTC; F: 5′-GAAGATCTGACTTCCGGTGGGCTTTGAG — was used to amplify VCP fragments from EST clone BC043053. The combinations used were as follows: (a) for full-length VCP, primers A and B; (b) for fragment N, primers A and C; (c) for fragment D1, primers D and E; (d) for fragment D2, primers F and B; (e) for fragment ND1, primers A and E; and (f) for fragment D1D2, primers D and B. The PCR products were then subcloned into vectors Myc-GW1 or pET28a. For vector-based miRNA constructs, the target sequence No. 1, 5′-TAGGATAGCAGGATCAATGAT-3′; for mouse and rat Vcp genes and the sequence No. 2, 5′-ATCCATTTCTGTCAGGATCTG-3′; for all human, mouse, and rat VCP genes were determined by using the BLOCK-iT RNAi Designer tool (Invitrogen). The corresponding paired oligonucleotides were then inserted into the vector pcDNA 6.2-GW/EmGFP-miRNA using the BLOCK-iT Pol II miR RNAi Expression Vector Kit with EmGFP (Invitrogen). The plasmid pcDNA 6.2-GW/EmGFP-miR-neg control was employed as a negative control for knockdown experiments; it contained the sequence 5′-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′ that was predicted not to target any known vertebrate gene as an insert. The QuikChange XL Site-Directed Mutagenesis Kit (Agilent) was applied to generate silent mutants resistant to No. 1 miRNA constructs using primers 5′-CATTGATCCTGCTATTTTGAGACCTGGCCGTCTA-3′ and 5′-TAGACGGCCAGGTCTCAAAATAGCAGGATCAATG-3′ (mutated sites are underlined). Plasmid EGFP-actin was purchased from Clontech.
Hippocampal neuronal culture and transfection
Culture and transfection of rat embryonic hippocampal and mouse embryonic cortical neurons were performed as described previously (9, 92).
Analyses of immunofluorescence staining and dendritic spine formation
To study dendritic spine formation, we performed transfection at 12 DIV, and immunofluorescence staining at 18 DIV. Cultured hippocampal neurons were washed twice in PBS and fixed in PBS containing 4% PFA and 4% sucrose for 10 minutes at room temperature. After washing, cells were permeabilized with 10% methanol and 0.2% Triton X-100 in PBS for 5 minutes and then blocked with 3% BSA/0.1% NHS in PBS for 1 hour. The primary antibody was diluted in PBS containing 3% BSA and 0.1% normal horse serum and added to cells for a further overnight incubation at 4°C. After washing, secondary antibody conjugated with Alexa Fluor 488, 555, or 594 (Invitrogen) was added. Following incubation for 1 hour at room temperature and washing to remove unbound antibodies, samples were analyzed with a confocal microscope (LSM510 or LSM700; Zeiss) equipped with a Plan-Apochromat 63× NA 1.4 oil objective lens (Zeiss). Images were captured with LSM or Zen acquisition and analysis software (Zeiss) at 20–22°C and acquired as Z-series of 5–12 sections spaced 0.6–0.8 μm apart. The Z-series were then projected into single images. For publication, the images were processed with Photoshop (Adobe), with minimal adjustment of brightness or contrast applied to the whole images.
The intensities of VCP immunoreactivities were measured using ImageJ software ( http://rsbweb.nih.gov/ij/). The signals of GFP were employed to outline the soma of transfected neurons. The intensities of VCP protein were then compared in control and VCP miRNA–transfected neurons. Morphometric measurement was performed using ImageJ. The number of spines present on the dendrites (along 20 μm of each dendrite starting from a point 20 μm away from the soma) was manually counted and traced. The maximum length from the tip of the spine to the dendritic shaft and the head width of each spine were manually measured by ImageJ region measurement tools. The data were then exported to Excel (Microsoft) for further analysis. The cumulative probability distributions were determined and statistically analyzed by KS test based on the D and corresponding P values. In addition, the mean and SEM of each group were determined and analyzed using a 2-tailed Student’s t test. The experiments were repeated using at least 2–3 independent culture preparations. Because dendritic spine formation is highly sensitive to culture conditions, such as the quality of B27 supplement (93), each experiment was repeated using the same lot of B27 supplement. The data from repeated experiments were pooled for statistical analysis only when the variation of the control group was not significantly different between repeated experiments.
Golgi staining
Two-month-old Nf1+/– mice and WT littermates (94) were obtained from the Jackson Laboratory and sacrificed by cervical dislocation. Whole brains were taken, immediately placed into cold PBS, and then processed for Golgi staining (Rapid GolgiStain Kit, FD NeuroTechnologies). Coronal sections of 150-μm thickness were cut on a vibratome and plated on gelatin-coated microscopy slides. Images were collected using a Zeiss AxioImager-Z1 microscope. Pyramidal cells at the CA1 region of the hippocampus were selected for determination of the number of spines.
Supplementary Material
Acknowledgments
We are grateful to the study participants. We also thank Sue-Ping Lee and Yi-Ling Lin at the Institute of Molecular Biology, Academia Sinica, and the Proteomic Core Facility at the Institute of Biological Chemistry, Academia Sinica, for technical assistance. We further thank Gunnar Johansson for suggestions on the manuscript and Heiko Kuhn for manuscript editing. H.-F. Wang is supported by a postdoctoral fellowship from Academia Sinica. This work was supported by Academia Sinica (to Y.P. Hsueh) and grants from the National Science Council (NSC 98-2321-B-001-002, NSC 99-2321-B-001-032, NSC-100-2321-B-001-032, and NSC 98-2311-B-001-012-MY3 to Y.-P. Hsueh).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(12):4820–4837. doi:10.1172/JCI45677.
See the related Commentary beginning on page 4627.
References
- 1.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9(8):2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2(12):880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 3.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75(3):161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28(4):182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Lippman J, Dunaevsky A. Dendritic spine morphogenesis and plasticity. J Neurobiol. 2005;64(1):47–57. doi: 10.1002/neu.20149. [DOI] [PubMed] [Google Scholar]
- 6.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16(1):95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Viskochil D, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62(1):187–192. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 8.Wallace MR, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249(4965):181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y-L, Lei Y-T, Hong C-J, Hsueh YP. Syndecan-2 induces filopodia formation via the neurofibromin-PKA-Ena/VASP pathway. J Cell Biol. 2007;177(5):829–841. doi: 10.1083/jcb.200608121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa RM, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415(6871):526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 11.Mbarek O, Marouillat S, Martineau J, Barthelemy C, Muh JP, Andres C. Association study of the NF1 gene and autistic disorder. Am J Med Genet. 1999;88(6):729–732. doi: 10.1002/(SICI)1096-8628(19991215)88:6<729::AID-AJMG26>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Marui T, et al. Association between the neurofibromatosis-1 (NF1) locus and autism in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2004;131B(1):43–47. doi: 10.1002/ajmg.b.20119. [DOI] [PubMed] [Google Scholar]
- 13.Lush ME, Li Y, Kwon CH, Chen J, Parada LF. Neurofibromin is required for barrel formation in the mouse somatosensory cortex. J Neurosci. 2008;28(7):1580–1587. doi: 10.1523/JNEUROSCI.5236-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevenson DA, et al. Multiple increased osteoclast functions in individuals with neurofibromatosis type 1. Am J Med Genet A. 2011;155A(5):1050–1059. doi: 10.1002/ajmg.a.33965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, et al. Primary osteopathy of vertebrae in a neurofibromatosis type 1 murine model. Bone. 2011;48(6):1378–1387. doi: 10.1016/j.bone.2011.03.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kossler N, et al. Neurofibromin (Nf1) is required for skeletal muscle development. Hum Mol Genet. 2011;20(14):2697–2709. doi: 10.1093/hmg/ddr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson DA, et al. Bone resorption in syndromes of the Ras/MAPK pathway. Clin Genet. 2011;80(6):566–573. doi: 10.1111/j.1399-0004.2010.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/S0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264(1):19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- 20.Hannan F, Ho I, Tong JJ, Zhu Y, Nurnberg P, Zhong Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum Mol Genet. 2006;15(7):1087–1098. doi: 10.1093/hmg/ddl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404(6775):298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 22.Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J Neurosci. 2001;21(11):3764–3770. doi: 10.1523/JNEUROSCI.21-11-03764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunger AT, DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555(1):126–133. doi: 10.1016/S0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
- 24.Woodman PG. p97, a protein coping with multiple identities. J Cell Sci. 2003;116(pt 21):4283–4290. doi: 10.1242/jcs.00817. [DOI] [PubMed] [Google Scholar]
- 25.Vij N. AAA ATPase p97/VCP: cellular functions, disease and therapeutic potential. J Cell Mol Med. 2008;12(6A):2511–2518. doi: 10.1111/j.1582-4934.2008.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarosch E, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4(2):134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Song C, Li CC. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146(1–2):44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. Embo J. 2004;23(5):1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida H. ER stress and diseases. FEBS J. 2007;274(3):630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 30.Uchiyama K, Kondo H. p97/p47-Mediated biogenesis of Golgi and ER. J Biochem. 2005;137(2):115–119. doi: 10.1093/jb/mvi028. [DOI] [PubMed] [Google Scholar]
- 31.Ju JS, Weihl CC. p97/VCP at the intersection of the autophagy and the ubiquitin proteasome system. Autophagy. 2010;6(2):283–285. doi: 10.4161/auto.6.2.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tresse E, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6(2):217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bugiani O. The many ways to frontotemporal degeneration and beyond. Neurol Sci. 2007;28(5):241–244. doi: 10.1007/s10072-007-0829-6. [DOI] [PubMed] [Google Scholar]
- 34.Talbot K, Ansorge O. Recent advances in the genetics of amyotrophic lateral sclerosis and frontotemporal dementia: common pathways in neurodegenerative disease. Hum Mol Genet. 2006;15 spec no 2:R182–R187. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- 35.Kakizuka A. Roles of VCP in human neurodegenerative disorders. Biochem Soc Trans. 2008;36(pt 1):105–108. doi: 10.1042/BST0360105. [DOI] [PubMed] [Google Scholar]
- 36.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 37.Custer SK, Neumann M, Lu H, Wright AC, Taylor JP. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum Mol Genet. 2010;19(9):1741–1755. doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- 38.Badadani M, et al. VCP associated inclusion body myopathy and paget disease of bone knock-in mouse model exhibits tissue pathology typical of human disease. PLoS One. 2010;5(10):e13183. doi: 10.1371/journal.pone.0013183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68(5):857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirabayashi M, et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8(10):977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- 41.Boyault C, et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. Embo J. 2006;25(14):3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, Manno A, Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells. 2007;12(7):889–901. doi: 10.1111/j.1365-2443.2007.01099.x. [DOI] [PubMed] [Google Scholar]
- 43.Weihl CC, Dalal S, Pestronk A, Hanson PI. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum Mol Genet. 2006;15(2):189–199. doi: 10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- 44.Ju JS, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187(6):875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno Y, Hori S, Kakizuka A, Okamoto K. Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci Lett. 2003;343(2):77–80. doi: 10.1016/S0304-3940(03)00280-5. [DOI] [PubMed] [Google Scholar]
- 46.Forman MS, et al. Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J Neuropathol Exp Neurol. 2006;65(6):571–581. doi: 10.1097/00005072-200606000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Kimonis VE, et al. Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. Am J Med Genet A. 2008;146A(6):745–757. doi: 10.1002/ajmg.a.31862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rumpf S, Lee SB, Jan LY, Jan YN. Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development. 2011;138(6):1153–1160. doi: 10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouiller I, et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9(12):950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 50.Schroder R, et al. Mutant valosin-containing protein causes a novel type of frontotemporal dementia. Ann Neurol. 2005;57(3):457–461. doi: 10.1002/ana.20407. [DOI] [PubMed] [Google Scholar]
- 51.Tang WK, et al. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. Embo J. 2010;29(13):2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol Cell Biol. 2009;29(16):4484–4494. doi: 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokuo H, et al. Phosphorylation of neurofibromin by cAMP-dependent protein kinase is regulated via a cellular association of N(G),N(G)-dimethylarginine dimethylaminohydrolase. FEBS Lett. 2001;494(1–2):48–53. doi: 10.1016/S0014-5793(01)02309-2. [DOI] [PubMed] [Google Scholar]
- 54.Messiaen LM, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15(6):541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 55.Girodon-Boulandet E, et al. NF1 gene analysis focused on CpG-rich exons in a cohort of 93 patients with neurofibromatosis type 1. Hum Mutat. 2000;16(3):274–275. doi: 10.1002/1098-1004(200009)16:3<274::aid-humu21>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 56.Cawthon RM, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62(1):193–201. doi: 10.1016/0092-8674(90)90253-B. [DOI] [PubMed] [Google Scholar]
- 57.Lee MJ, et al. Identification of forty-five novel and twenty-three known NF1 mutations in Chinese patients with neurofibromatosis type 1. Hum Mutat. 2006;27(8):832. doi: 10.1002/humu.9446. [DOI] [PubMed] [Google Scholar]
- 58.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273(6):3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 59.Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3(8):740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 60.Li W, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15(21):1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 61.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27(5):457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 62.Terry RD, et al. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 63.Scheff SW, Price DA. Synapse loss in the temporal lobe in Alzheimer’s disease. Ann Neurol. 1993;33(2):190–199. doi: 10.1002/ana.410330209. [DOI] [PubMed] [Google Scholar]
- 64.Liu X, Erikson C, Brun A. Cortical synaptic changes and gliosis in normal aging, Alzheimer’s disease and frontal lobe degeneration. Dementia. 1996;7(3):128–134. doi: 10.1159/000106867. [DOI] [PubMed] [Google Scholar]
- 65.Zhou L, Miller BL, McDaniel CH, Kelly L, Kim OJ, Miller CA. Frontotemporal dementia: neuropil spheroids and presynaptic terminal degeneration. Ann Neurol. 1998;44(1):99–109. doi: 10.1002/ana.410440116. [DOI] [PubMed] [Google Scholar]
- 66.Lipton AM, et al. Contribution of asymmetric synapse loss to lateralizing clinical deficits in frontotemporal dementias. Arch Neurol. 2001;58(8):1233–1239. doi: 10.1001/archneur.58.8.1233. [DOI] [PubMed] [Google Scholar]
- 67.Clare R, King VG, Wirenfeldt M, Vinters HV. Synapse loss in dementias. J Neurosci Res. 2010;88(10):2083–2090. doi: 10.1002/jnr.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37(8):806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 69.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17(18):1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 70.Lee JA, Gao FB. ESCRT, autophagy, and frontotemporal dementia. BMB Rep. 2008;41(12):827–832. doi: 10.5483/BMBRep.2008.41.12.827. [DOI] [PubMed] [Google Scholar]
- 71.Belly A, Bodon G, Blot B, Bouron A, Sadoul R, Goldberg Y. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J Cell Sci. 2010;123(pt 17):2943–2954. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Upadhyaya M, Cooper DN, eds.Neurofibromatosis Type I: From Genotype to Phenotype . Waltham, Massachusetts, USA: Academic Press; 1998. [Google Scholar]
- 73.Lin YL, Hsueh YP. Neurofibromin interacts with CRMP-2 and CRMP-4 in rat brain. Biochem Biophys Res Commun. 2008;369(2):747–752. doi: 10.1016/j.bbrc.2008.02.095. [DOI] [PubMed] [Google Scholar]
- 74.Volta M, Calza S, Roberts AM, Roberts RG. Characterisation of the interaction between syndecan-2, neurofibromin and CASK: dependence of interaction on syndecan dimerization. Biochem Biophys Res Commun. 2010;391(2):1216–1221. doi: 10.1016/j.bbrc.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 75.Kweh F, Zheng M, Kurenova E, Wallace M, Golubovskaya V, Cance WG. Neurofibromin physically interacts with the N-terminal domain of focal adhesion kinase. Mol Carcinog. 2009;48(11):1005–1017. doi: 10.1002/mc.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Schepper S, et al. Neurofibromatosis type 1 protein and amyloid precursor protein interact in normal human melanocytes and colocalize with melanosomes. J Invest Dermatol. 2006;126(3):653–659. doi: 10.1038/sj.jid.5700087. [DOI] [PubMed] [Google Scholar]
- 77.Feng L, et al. PKA phosphorylation and 14-3-3 interaction regulate the function of neurofibromatosis type I tumor suppressor, neurofibromin. FEBS Lett. 2004;557(1–3):275–282. doi: 10.1016/S0014-5793(03)01507-2. [DOI] [PubMed] [Google Scholar]
- 78.Patrakitkomjorn S, et al. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J Biol Chem. 2008;283(14):9399–9413. doi: 10.1074/jbc.M708206200. [DOI] [PubMed] [Google Scholar]
- 79.Kolanczyk M, et al. Modelling neurofibromatosis type 1 tibial dysplasia and its treatment with lovastatin. BMC Med. 2008;6:21. doi: 10.1186/1741-7015-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, et al. Local low-dose lovastatin delivery improves the bone-healing defect caused by Nf1 loss of function in osteoblasts. J Bone Miner Res. 2010;25(7):1658–1667. doi: 10.1002/jbmr.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krab LC, et al. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: a randomized controlled trial. JAMA. 2008;300(3):287–294. doi: 10.1001/jama.300.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mendola CE, Backer JM. Lovastatin blocks N-ras oncogene-induced neuronal differentiation. Cell Growth Differ. 1990;1(10):499–502. [PubMed] [Google Scholar]
- 83.Sebti SM, Tkalcevic GT, Jani JP. Lovastatin, a cholesterol biosynthesis inhibitor, inhibits the growth of human H-ras oncogene transformed cells in nude mice. Cancer Commun. 1991;3(5):141–147. doi: 10.3727/095535491820873371. [DOI] [PubMed] [Google Scholar]
- 84.Hering H, Lin CC, Sheng M. Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J Neurosci. 2003;23(8):3262–3271. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prinetti A, et al. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane domains, in rat cerebellar granule cells developing in vitro. J Biol Chem. 2001;276(24):21136–21145. doi: 10.1074/jbc.M010666200. [DOI] [PubMed] [Google Scholar]
- 86.Cao J, et al. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6(2):115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 87.Leichner GS, Avner R, Harats D, Roitelman J. Dislocation of HMG-CoA reductase and Insig-1, two polytopic endoplasmic reticulum proteins, en route to proteasomal degradation. Mol Biol Cell. 2009;20(14):3330–3341. doi: 10.1091/mbc.E08-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hartman IZ, et al. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285(25):19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tonsgard JH. Clinical manifestations and management of neurofibromatosis type 1. Semin Pediatr Neurol. 2006;13(1):2–7. doi: 10.1016/j.spen.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 90.Crawford AH, Schorry EK. Neurofibromatosis in children: the role of the orthopaedist. J Am Acad Orthop Surg. 1999;7(4):217–230. doi: 10.5435/00124635-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 91.Lee MJ, Stephenson DA. Recent developments in neurofibromatosis type 1. Curr Opin Neurol. 2007;20(2):135–141. doi: 10.1097/WCO.0b013e3280895da8. [DOI] [PubMed] [Google Scholar]
- 92.Chao HW, Hong CJ, Huang TN, Lin YL, Hsueh YP. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol. 2008;182(1):141–155. doi: 10.1083/jcb.200712094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171(2):239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7(3):353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 95.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96(5):1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.