The plastid ribosomal release factor-like protein PrfB3 arose from a gene duplication and has subsequently lost the two most important tripeptide motifs required for stop codon recognition and catalysis. This work shows that PrfB3 is essential in vascular plants and has been recruited for binding and environment-dependent stabilization of petB RNA to regulate cytochrome b6f complex levels.
Abstract
Land plant genomes encode four functional ribosomal peptide chain release factors (Prf) of eubacterial origin, two (PrfA and PrfB homologs) for each endosymbiotic organelle. Formerly, we have shown that the Arabidopsis thaliana chloroplast-localized PrfB homolog, PrfB1, is required not only for termination of translation but also for stabilization of UGA stop codon-containing chloroplast transcripts. A previously undiscovered PrfB-like protein, PrfB3, is localized to the chloroplast stroma in a petB RNA-containing complex and found only in vascular plants. Highly conserved positions of introns unequivocally indicate that PrfB3 arose from a duplication of PrfB1. Notably, PrfB3 is lacking the two most important tripeptide motifs characteristic for all eubacterial and organellar PrfB homologs described so far: the stop codon recognition motif SPF and the catalytic center GGQ for peptidyl-tRNA hydrolysis. Complementation studies, as well as functional and molecular analyses of two allelic mutations in Arabidopsis, both of which lead to a specific deficiency of the cytochrome b6f complex, revealed that PrfB3 is essentially required for photoautotrophic growth. Plastid transcript, polysome, and translation analyses indicate that PrfB3 has been recruited in vascular plants for light- and stress-dependent regulation of stability of 3′ processed petB transcripts to adjust cytochrome b6 levels.
INTRODUCTION
The chloroplast has evolved as a result of the endosymbiotic event in which a cyanobacterial ancestor was taken over by a eukaryotic cell. An important characteristic of chloroplast gene expression is the predominance of posttranscriptional control, which is exerted at gene-specific, gene cluster, and genome-wide levels (Cho et al., 2009; del Campo, 2009; Stern et al., 2010). Strategies and regulation of chloroplast gene expression are distinct from those of eubacteria as well as from those in the eukaryotic cytoplasm and reflect a unique chimeric system assembled by multiple origins during endocytobiosis. The cellular genetic system has been embedded into complex circuits that enable an adaptive and developmental regulation of chloroplast biogenesis at various levels (Bollenbach et al., 2004, 2005; Stern et al., 2010). Although the chloroplast has reduced its coding capacity to <100 proteins, a highly sophisticated system of transcript maturation including endo- and exonucleolytic activities, splicing, editing, and modulation of RNA stability has been developed. These activities are not exploited to the same extent in the free-living cyanobacterial ancestor.
Numerous nuclear-encoded factors have been acquired for processing of plastid transcripts (Stern et al., 2010; Barkan, 2011). Whereas most of these factors appear to be newly evolved (e.g., Meurer et al., 1998; Felder et al., 2001) or to have expanded predominantly in vascular plants (Fisk et al., 1999; Meierhoff et al., 2003), others are of eubacterial origin and often gained new functions (Jenkins and Barkan, 2001; Meurer et al., 2002) or possess ancient domains diversified to mediate novel functions (Till et al., 2001; Ostheimer et al., 2003). Some factors act as global players, whereas others have predominantly specific functions in maturation, stabilization, and editing of plastid transcripts like members of the pentatricopeptide repeat (PPR) protein family (Barkan, 2011; Prikryl et al., 2011). Moreover, genetic and molecular approaches demonstrated that higher-order protein complexes are often involved in processing of plastid transcripts (e.g., Fisk et al., 1999; Ossenbühl and Nickelsen, 2000; Kroeger et al., 2009). In summary, the frequent occurrence of plant-specific genes important for chloroplast mRNA homeostasis demonstrates that regulation at the posttranscriptional level represents a fast evolving process during endosymbiosis.
Up to now, little attention has been paid to the functions of translation termination in chloroplasts (Meurer et al., 1996a, 2002). In contrast with the omnipotent eukaryotic and archaebacterial ribosomal release factors (RFs), two peptide chain release factors are present in eubacteria, chloroplasts, and plant mitochondria, prfA (RF1) for UAA and UAG and prfB (RF2) for UAA and UGA (Meurer et al., 2002; Motohashi et al., 2007). Functional and structural comparisons on the basis of a common Gly-Gly-Gln (GGQ) motif present in all release factors described so far revealed its essential function in the hydrolytic activity (Kisselev and Buckingham, 2000) and may reflect a common evolutionary origin of the eukaryotic/archaebacterial and eubacterial proteins, both of which are supposed to mimic tRNAs when bound to ribosomes (Nakamura and Ito, 2003; Loh and Song, 2010). The other highly conserved motif Ser-Pro-Phe (SPF) for recognition of UGA stop codons is present in all eubacterial and related plastid and mitochondrial PrfB proteins (Meurer et al., 2002).
Previously, we identified the essential roles of the GGQ- and SPF-containing plastid RF2 homolog from Arabidopsis thaliana, At-PrfB1 (formerly designated At-PrfB), in translational termination and stabilization of chloroplast UGA stop codon–containing transcripts (Meurer et al., 1996a, 1996b, 2002).
In this study, we report on the essential role of a PrfB1-homologous RF2-like protein of eubacterial origin, PrfB3, which has lost its function as a release factor and has been recruited by vascular plant chloroplasts specifically to regulate the stability of plastid 3′ processed petB transcripts encoding cytochrome b6 in an environmental-dependent and stress-mediated manner.
RESULTS
Identification and Origin of the Nuclear-Encoded Factor PrfB3 in Arabidopsis
The full-length cDNA of At-PrfB3 encodes a protein consisting of 406 amino acids with a deduced size of ~45 kD (see Supplemental Figure 1 online). PrfB3 can be found only in vascular plants and displays 36.5, 37.5, and 31% sequence similarity over the whole length with the corresponding RF2 and only 23.3, 22.6, and 21.7% with the corresponding RF1 homologous proteins of mitochondria, chloroplasts, and Synechocystis, respectively. PrfB3 has so far not been found in proteomic approaches (http://suba.plantenergy.uwa.edu.au; http://ppdb.tc.cornell.edu), indicating that this protein represents a minority factor, as is found for many regulatory proteins. Based on the degree of sequence similarity, it is not possible to determine the origin of PrfB3 with certainty. To address this issue, we compared the position of introns within the genes in rice (Oryza sativa) and Arabidopsis. The At-PrfB3 gene (At3g57190) contains five introns, which are all identical to those in Os-PrfB3 as well as in the At-PrfB1 and Os-PrfB1 genes encoding the functional plastid release factors (Figure 1; see Supplemental Figure 1 online). The intron positions in At-PrfB2 and Os-PrfB2 encoding the mitochondrial proteins are highly conserved but differ from those found in At-PrfB3 and Os-PrfB3, strongly suggesting that At-PrfB3 directly originated from a duplication of At-PrfB1 before the divergence of monocots and dicots. Remarkably, PrfB3 neither contains the otherwise conserved tripeptide anticodon SPF, which determines release factor specificity in vivo in PrfB proteins (Ito et al., 2000; Nakamura et al., 2000), nor the universally conserved GGQ motif, which is essential for the hydrolytic activity and represents a structural counterpart on the CCA-3′ acceptor stem of the tRNA-aminoacyl group (Frolova et al., 2000) (Figure 1). Moreover, the corresponding nucleotides of both motifs were not simply replaced but rather cut out from the genes by deletion events (Figure 1; see Supplemental Figure 1 online). Therefore, PrfB3 must have lost its function to terminate translation and potentially could have been recruited by the chloroplast for a new function that is not directly related to ribosomal release.
Figure 1.
Partial Sequence Alignment of Eubacterial PrfB and Homologous Plant Proteins.
The conserved part containing the essential tripeptide motifs SPF and GGQ (highlighted in red) characteristic of all functional RF2 proteins in eubacteria (Synechocystis and E. coli), plastids (At-PrfB1 and Os-PrfB1), and mitochondria (At-PrfB2 and Os-PrfB2) was compared with the corresponding region in PrfB3 proteins from Arabidopsis and rice. Conserved amino acid residues that are also present in At-PrfB3 and Os-PrfB3 are shaded in gray. Amino acids specific for the functional release factors are shaded in yellow. Conserved amino acids that are present only in the plastid At-PrfB3 and Os-PrfB3 proteins are shaded in green. The deduced positions of introns are underlined and labeled in blue.
At-PrfB3 Is Targeted to the Chloroplast
Several publicly available programs did not predict an amino terminal transit peptide for import of At-PrfB3 into the chloroplast (ChloroP) (Emanuelsson et al., 1999) but instead identified a putative mitochondrial transit peptide (Predotar) (Small et al., 2004); (TargetP) (Emanuelsson et al., 2000); and (MitoProt) (Claros and Vincens, 1996). It has been suggested previously that At-PrfB3 might be involved in termination of translation of transcripts lacking stop codons and might be a mitochondrial protein (Raczynska et al., 2006). However, by transient expression of At-PrfB3-green fluorescent protein (GFP) fusions, we could not find any hints that At-PrfB3 is imported into mitochondria but the fluorescence of GFP was localized exclusively in chloroplasts (Figure 2A). Antibodies raised against At-PrfB3 were used for immunological analysis to strengthen this finding. Analysis of highly purified mitochondria excluded the possibility that even traces of At-PrfB3 are present in mitochondria, and the data confirmed the presence of the 45-kD protein in the soluble fraction of the chloroplast (Figures 2B and 2C), demonstrating that At-PrfB3 represents exclusively a plastid protein.
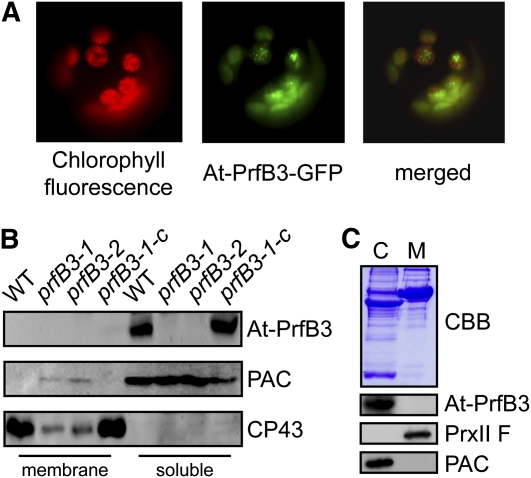
Figure 2.
Localization of At-PrfB3 in the Chloroplast Stroma.
(A) The gene sequence encoding the first 206 N-terminal amino acid residues was fused in frame to the GFP reporter under the control of the cauliflower mosaic virus 35S promoter. The chimeric gene construct was transiently expressed in tobacco protoplasts. Left, chlorophyll a fluorescence; middle, GFP fluorescence; right, merged images. The GFP fluorescence perfectly matched with chlorophyll a fluorescence, confirming chloroplast targeting of At-PrfB3.
(B) Membrane and soluble proteins of the wild type (WT), prfB3 mutants (prfB3-1 and prfB3-2), and complemented lines (prfB3-1-c) were subjected to immunoblot analysis using antibodies raised against At-PrfB3, the membrane protein CP43, and the soluble protein PAC. At-PrfB3 has a size of 45 kD and was present only in the soluble fraction of wild-type and complemented lines. Protein amounts were adjusted according to stained Coomassie blue gels.
(C) Immunoblot analysis of protein fractions from purified wild-type chloroplasts (C) and mitochondria (M) confirmed the presence of PrfB3 in the chloroplasts and excluded targeting into mitochondria. Immunodecoration with the plastid protein PAC and the mitochondrial protein peroxiredoxin (PrxII F) as well as Coomassie blue staining confirms the purity of the organellar fractions.
Distribution of PrfB3 among Photosynthetic Organisms
Database homology searches revealed that At-PrfB3 orthologs are present in the nuclear genomes of monocots and dicots (http://genome.jgi-psf.org). No gene encoding a release factor–like protein that is lacking the SPF and GGQ motif was found in the genomes and EST databases of cyanobacteria, the green algae Chlamydomonas reinhardtii and Ostreococcus tauri, the red algae Cyanidioschyzon merulae and Galdieria sulphuraria, the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum, the moss Physcomitrella patens, or the fern Selaginella moellendorffii (http://genome.jgi-psf.org). No sequence orthologous to PrfB3 is available from coniferophyta. Therefore, it is likely that PrfB3 evolved only recently and is present exclusively in vascular plants, again supporting the high degree of divergence of chloroplast gene expression in photosynthetic organisms.
PrfB3 Is an Essential Chloroplast Protein
To assess the function of PrfB3, we applied a reverse genetics approach and studied a Salk_133921 T-DNA insertion line (prfB3-1) of the At3g57190 gene of Arabidopsis encoding PrfB3. Sequencing of the PCR product obtained with gene-specific and left border T-DNA primers confirmed that the insertion occurred in the fourth exon at position +961 relative to the start codon. Homozygosity of the T-DNA insertion line was confirmed by PCR using adequate control primers (Figures 3A and 3B; see Supplemental Table 1 online). A forward genetics screen identified a mutation, prfB3-2, showing the same phenotype. Sequencing of the PrfB3 locus in prfB3-2 identified a deletion of 273 bp at position +282 relative to the start codon. The deletion includes the entire exon 2 and adjacent intron regions (Figure 3A). Sequencing of RT-PCR products revealed that the deletion produces a premature stop codon and, correspondingly, leads to expression of a truncated protein encoded mainly by exon 1 (Figures 3A and 3C). Both mutations segregated in a Mendelian manner. Test crossings confirmed that prfB3-1 and prfB3-2 are allelic. The stromal PrfB3 protein was lacking in both mutants but was expressed at wild-type-comparable levels in the complemented lines (Figure 2B). Homozygous mutants exhibited a high chlorophyll fluorescence (hcf) phenotype characteristic for plants with deficiencies in the photosynthetic apparatus (Meurer et al., 1996b) and, accordingly, died in the seedling stage and could grow further only under sterile heterotrophic conditions. When grown under a normal light regime (>20 μmol photons m−2 s−1), mutant plants were smaller in size and much paler compared with the wild type; however, their leaf morphology was unchanged (see Supplemental Figure 2 online). Plants grown under very low light of 10 μmol photons m−2 s−1 were less pale and their size was comparable to the wild type (Figure 4A), indicating that the mutants are extremely light sensitive.
Figure 3.
Gene Structure and Mutations of PrfB3.
(A) The structure of the At-PrfB3 gene, composed of six exons (gray boxes), the T-DNA insertion in line prfB3-1, the deletion in prfB3-2 with the resulting splice product and the corresponding sequences (bottom part), and the primers used for PCR analysis (arrows) are indicated. The positions of start (ATG) and stop (TAG) codons of translation are assigned. The resulting premature stop codon in prfB3-2 is labeled in red. LB, left border; RB, right border.
(B) To test homozygosity of the T-DNA insertion, PCR analysis was performed using primers specific for the T-DNA left border (LBb1) and the PrfB3 gene (PrfB3-f and PrfB3-r). For primer sequences, see Supplemental Table 1 online. The gene-specific primers amplified a 454-bp product only in wild-type (WT) and heterozygous plants. The border primer and PrfB3-r amplified a 398-bp fragment in heterozygous and mutant plants. The T-DNA insertion in homozygous lines prevented PCR amplification of the corresponding locus. Sequencing of the PCR product obtained with LBb1 and PrfB3-r primers confirmed that the T-DNA was inserted in the fourth exon at position +961 relative to the start codon. The sizes of PCR products are given in base pairs.
(C) RT-PCR products using gene-specific primers B3-Ex1-f and B3-Ex4-r amplified a 689-bp fragment in the wild type and a 471-bp fragment in the prfB3-2 mutants corresponding to the loss of exon 2 and border regions as revealed by sequencing. Both fragments were amplified in heterozygous plants.
(D) Successful complementation of four lines (numbers 4, 43, 64, and 69) was tested with the same primers as used in (B). The genomic fragment could be amplified only in wild-type and heterozygous plants. In the complemented mutants, only the inserted cDNA gave rise to a PCR product of 258 bp. c, complementing T-DNA.
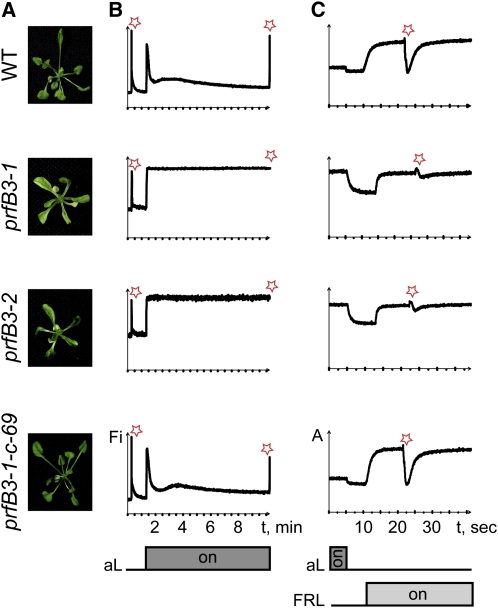
Figure 4.
Phenotype and Functional Analysis of prfB3 Mutants and Complemented Lines.
(A) Phenotype of 25-d-old wild-type (WT) and mutant plants grown on Suc-supplemented medium under low light of 10 μmol photons m−2 s−1. Under these conditions, mutant plants are paler than wild-type plants but similar in size. Knockout line prfB3-1-c-69 complemented with the prfB3 cDNA was undistinguishable from the wild type and able to grow photoautotrophically.
(B) Chlorophyll a fluorescence induction was measured after application of a saturating light pulse (red star) to dark-adapted plants. The ratio Fv/Fm of 0.82 ± 0.01 in the wild type was reduced to 0.54 ± 0.04 in prfB3 mutants. Actinic light (aL) of 50 μmol photons m−2 s−1 was applied, and the photosynthetic parameters qP and NPQ were measured in the steady state after 9 min illumination. Fi, fluorescence intensity (arbitrary units).
(C) Absorbance of P700 at 830 nm is a measure of the P700 redox state. Light-to-dark switches and application of far-red light (FRL) induced redox changes of P700, indicating that PSI is active in prfB3 mutants, although the overall signal is somehow decreased. Saturating light pulses (red stars) in the background of far-red light were sufficient for the complete PSII-dependent reduction of PSI in the wild type. However, these conditions only marginally reduced P700 in the mutants, indicating that the electron transport is interrupted before PSI. A, relative absorbance.
Expression of the full-length cDNA under the control of the constitutive 35S promoter in homozygous prfB3-1 lines fully restored the wild-type phenotype and photoautotrophic growth (Figures 3D, 3E, and 4A to 4C). Functional complementation indicates that the phenotype was exclusively due to the mutations in the PrfB3 gene. Endogenous PrfB1, expressed in the prfB3 mutant background, apparently fails to rescue the mutant defect, supporting the idea that PrfB3 recruited a novel function.
prfB3 Mutations Affect Thylakoid Ultrastructure and Chloroplast Size
Fluorescence imaging revealed that the chlorophyll fluorescence of prfB3-1 mutant grana stacks is not uniformly distributed within the chloroplast but appears to be increased and form distinct spots, indicating major modifications of the thylakoid ultrastructure (see Supplemental Figure 3A online). Electron micrographs showed that the prfB3-1 mutant thylakoid membrane system is rudimentary and that grana stacks are not as densely packed as in the wild type. Furthermore, chloroplast size appeared to be smaller in the mutants (see Supplemental Figures 3B and 3C online).
The Intersystem Electron Transport Is Abolished in prfB3-1 and prfB3-2
To gain insights into the primary photosynthetic defect in the mutants, chlorophyll a fluorescence primarily emitted by photosystem II (PSII) at room temperature was measured. The maximum quantum yield of PSII, Fv/Fm, was reduced to 0.54 ± 0.04 in the mutants compared with 0.82 ± 0.01 in the wild type. Under the chosen light conditions, ~85% of the variable fluorescence is lost by photochemical (qP) and nonphotochemical quenching (NPQ) in the wild type (Figure 4B). In prfB3-1 and -2, the actinic light–induced fluorescence remained at its maximum level (qP and NPQ = 0), indicating disruption of the photosynthetic electron transport behind PSII (Meurer et al., 1996b). To estimate further the primary defect in both mutants, the redox state of photosystem I (PSI) was assessed. Light intensities of 50 μmol photons m−2 s−1 induced ~10% oxidation of PSI in the wild type. Although the P700 signal was a bit weaker in the mutants, the actinic light oxidized P700 to 100% (Figure 4C). This indicates that PSI operates but its activity is limited by deficiencies in the intersystemic electron transport chain due to disruption of PrfB3. The photosynthetic characteristics of the complemented lines were comparable to those of the wild type (Figures 4B and 4C).
PrfB3 Is Required for Accumulation of the Cytochrome b6f Complex
Immunological analyses showed that the amounts of the major subunits of the cytochrome b6f complex, such as cytochrome f, cytochrome b6, and the Rieske Fe-S protein, constituted around 4% of wild-type levels in prfB3-1 (Figure 5A). By contrast, PsaF, a representative subunit of PSI as well as PsbI and PsbH from PSII accumulated at wild-type-comparable levels. A similar high degree of reduction was found for the levels of cytochrome b6f complex proteins cytochrome f and subunit IV in prfB3-2. Accumulation of PSII subunits was somewhat reduced in both mutants, for example, CP43 in prfB3-1 and CP47, CP43, and D1 in prfB3-2 (Figures 2B and 5B). PSI subunits PsaD and PsaE were unchanged in prfB3-2 (Figure 5B). Levels of the α- and β-subunits of the ATP synthase were increased in prfB3 mutants (Figure 5A), which is likely to represent a secondary effect of the mutations since cytochrome b6f complex knockout lines in Arabidopsis, Oenothera, and tobacco (Nicotiana tabacum) showed the same phenomenon (Schwenkert et al., 2007) presumably to compensate for the loss of the cytochrome b6f complex.
Figure 5.
Accumulation of Thylakoid Membrane Proteins and Assembly of Complexes in prfB3 Mutants.
(A) Immunoblot analyses of prfB3-1 and the wild type (WT) are shown. Levels of cytochrome b6f complex subunits PetA (Cytf), PetB (Cytb6), and the Rieske protein, the PSII subunits PsbI and PsbH, the PSI subunit PsaF, and α- and β-subunits of the ATP synthase were analyzed. Loading of 100% corresponds to 10 μg of chlorophyll. Cyt, cytochrome.
(B) Immunoblot analyses of prfB3-2 and the wild type are shown. Levels of cytochrome b6f complex subunits PetA (Cytf) and PetD (subunit IV, SU IV), the PSI subunits PsaD and PsaE, and the PSII subunits PsbA (D1), PsbB (CP47), and PsbC (CP43) were analyzed.
(C) Chloroplast protein complexes were solubilized by treatment with 1% (w/v) n-dodecyl-β-d-maltoside and separated by blue native/PAGE (BN-PAGE). After separation in the first dimension, the protein lanes were subjected to a denaturing second dimension gel (SDS-PAGE) followed by silver staining. Macromolecular protein complexes of thylakoid membranes are indicated. The protein spots of the wild type (blue) and prfB3-1 mutant (red) were selectively stained in silico (Photoshop) and subsequently merged. The proteins cytochrome b6, Rieske, and subunit IV of the dimeric cytochrome b6f complex, which appeared only in the wild type, are circled. LHC, light-harvesting complex.
(D) Immunological analysis of the second dimension using cytochrome f antibodies is shown. The blot of the mutant was overexposed compared with the wild type.
(E) Protein complexes were separated in the second dimension after in vivo labeling using [35S]-Met. Arrows indicate positions of monomeric and dimeric cytochrome b6f complexes.
To investigate whether the assembly of higher-order photosynthetic membrane complexes was affected in the mutants, two-dimensional resolution of thylakoid protein complexes was performed by blue native/SDS-PAGE. The spot pattern clearly demonstrates that thylakoid membrane complexes of the ATP synthase, PSI, and PSII were able to assemble efficiently in prfB3-1 (Figure 5C). By contrast, both the dimeric and monomeric cytochrome b6f complexes were below the limits of detection by staining in the mutants (Figure 5C). Therefore, we conclude that the deficiency in prfB3 caused a severe and specific defect in the accumulation of the cytochrome b6f complex.
We asked whether traces of subunits of the cytochrome b6f complex, which accumulate in the mutant, are able to assemble dimeric complexes. Immunoblot analysis of the second dimension demonstrated that the high ratio of dimeric/monomeric complexes was comparable in mutants and the wild type (Figure 5D). This indicates that assembly and stability of the cytochrome b6f complex are not affected in the mutants. Time-solved assembly studies again have shown that higher-order assemblies of thylakoid membrane complexes generally occur efficiently but translation products that belong to monomeric or dimeric cytochrome b6f complexes remained undetectable in prfB3-1 (Figure 5E), indicating that translation of one or more of the proteins of the cytochrome b6f complex is affected. Taken together, the results of the fluorimetric, spectroscopic, and protein analyses allowed the conclusion that accumulation of the cytochrome b6f complex, but not its assembly or stability, is the primary defect in the prfB3 mutations.
3′ Processed petB Transcripts Fail to Accumulate in prfB3 Mutants
To assess whether lack of the cytochrome b6f complex was caused by deficiencies in transcript patterns, we probed numerous plastid transcripts by RNA gel blot hybridizations (Figures 6A to 6C; see Supplemental Figure 4 online). With the exception of mRNAs for the psbB gene cluster, all of the transcripts analyzed accumulated in size and abundance comparable to the wild type in both allelic prfB3 mutants, indicating that global plastid gene expression is not affected by the mutations (see Supplemental Figure 4 online).
Figure 6.
RNA Gel Blot Analysis and S1 Nuclease Mapping of the psbB Gene Cluster.
(A) The structure of the psbB gene cluster and probes A to H used in RNA gel blot analysis in (C) are shown. P, promoter of the plastid encoded polymerase. The gene cluster has been drawn to scale.
(B) The precursor transcript and its most prominent processed and spliced products are numbered from 1 to 13. Transcripts have been drawn to scale.
(C) RNA gel blot analysis of the psbB gene cluster in the wild type (WT) and prfB3 mutants. Probes used (A to I) and labeled transcripts (1 to 13) are shown in (A) and (B), respectively. The size of the transcripts is indicated. Mutant plants were compared with wild-type plants of the same accession.
(D) Transcript termini (arrows) and probes (bold lines) of the psbB gene cluster used for S1 nuclease mapping are shown. The molecular weight of S1 nuclease-resistant probes (thin lines) in nucleotide bases is indicated.
(E) S1 nuclease mapping of processing and splice sites within the psbB polycistron shows that petB-containing transcripts that are processed in the petB-petD intergenic region fail to accumulate in prfB3-2. P, probe.
The plastid pentacistronic psbB-psbT-psbH-petB-petD gene cluster has a promoter for the plastid-encoded polymerase and is conserved among vascular plants. It encodes the subunits CP47 (psbB), T (psbT), and H (psbH) of PSII as well as cytochrome b6 (petB) and subunit IV (petD) of the cytochrome b6f complex (Figure 6A). Each of the petB and petD genes contains an intron. The processing pattern of this gene cluster is rather complex and differs between plant species. Transcript termini of different species have been intensively studied in the past using RNA gel blot analysis and S1-nuclease mapping (Barkan, 1988; Westhoff and Herrmann, 1988; Felder et al., 2001; Meierhoff et al., 2003).
Accumulation of most transcripts derived from the psbB gene cluster was unchanged in both allelic mutants (Figures 6A to 6C). However, both mutants showed the same striking deviation from the wild-type transcript pattern. The 3′ processed and spliced petB-containing transcripts, the monocistronic petB, and the dicistronic psbH-petB transcripts were barely detectable in the mutants. Also, transcript precursors lacking the petB intron (i.e., psbB-psbT-psbH-petB-petD, psbH-petB-petD, and petB-petD) were slightly reduced in both mutants as shown by RNA gel blot analysis (Figure 6C, transcripts 3, 6, and 7). Quantitative RT-PCR analysis revealed that prfB3-1 accumulates 67% ± 2% spliced petB RNA and 105% ± 6% petD RNA compared with the wild type (for oligonucleotides, see Supplemental Table 1 online). This reduction is due to the apparent lack of 3′ processed petB transcripts and reduced levels of spliced precursors. Amounts of all precursor transcripts that include the petB intron were unchanged in the mutants, indicating that splicing takes place efficiently (Figure 6C, transcripts 1, 2, 4, and 5). All this attests that the prfB3 mutation affects only 3′ processed petB transcripts of the psbB gene cluster. Therefore, PrfB3 is involved either in processing of the petB-petD intergenic region or in the stabilization of 3′ processed petB transcripts.
In contrast with maize (Zea mays) and presumably all other monocots (Barkan, 2011), monocistronic and spliced petD transcripts of ~600 nucleotides do not accumulate to significant levels in Arabidopsis and other dicots. In Arabidopsis wild-type plants, petD transcripts, which result from endonucleolytic cleavage in the petB-petD intergenic region, are rapidly degraded irrespective whether they are spliced or not (Figures 6B and 6C) (Felder et al., 2001; Meierhoff et al., 2003). In the prfB3 mutants, intron- and 3′ exon-containing petD transcripts of 1.2 kb (Figure 6C, transcript 8), which lack the first exon, accumulate to even slightly higher amounts presumably due to increased cleavage of the 5′ splice site, indicating that splicing of petD occurs efficiently in the mutants (Figure 6C, transcript 8). Spliced petB transcripts in the wild type are either processed in the petB-petD intergenic region or the petD intron is also missing (Figures 6B and 6C). This indicates that petB splicing depends either on processing of the intergenic region or on petD splicing. The fact that the abundance of these spliced but unprocessed precursors is slightly reduced in both mutants suggests that processing of the petB-petD intergenic region occurs efficiently. The finding that the spliced petB intron of 0.8 kb accumulated to wild-type-comparable levels again suggests that petB splicing is not affected in the mutants (Figure 6C). This all speaks in favor of the idea that PrfB3 evolved to stabilize 3′ processed petB transcripts.
Proposed Function of PrfB3 in Stabilization of 3′ Processed petB Transcripts
Several nuclear and chloroplast mutants have been identified that are affected primarily in splicing or processing of plastid precursor transcripts. In all cases, unspliced or unprocessed precursors accumulated at higher levels (e.g., Jenkins et al., 1997; Meierhoff et al., 2003; Asakura and Barkan, 2007; Petersen et al., 2011). This is not the case for petB precursors in prfB3 mutants. They are instead slightly decreased, again supporting the assumption that spliced and 3′ processed petB transcripts are efficiently generated but are rapidly subjected to degradation in the mutants.
Chloroplast mRNA stability is known to be dependent on the state of transcript termini. Therefore, the mature 3′ and 5′ ends of transcripts from the psbB gene cluster were precisely determined by S1 nuclease mapping in both the wild type and prfB3-2 (Figures 6D and 6E). Both processing sites of the psbH 5′ end and of the psbH-petB intergenic region as well as 5′ and 3′ splicing of petB and petD introns were unchanged in the mutant. Importantly, 3′ processing of petB transcripts, which results in the fragment of 242 bases in the wild type could only barely be detected in the mutant, again indicating that PrfB3 is required for accumulation and/or stabilization of 3′ processed petB transcripts.
Since S1 nuclease mapping is insufficient to detect traces of processed transcripts and to strengthen these data, we mapped transcript ends by sequencing of independently cloned circular RT-PCR products in the wild type and prfB3 mutants. The petB 3′ end always mapped to base position +69 (12 clones) downstream of the petB stop codon in the wild type, precisely confirming the S1 nuclease mapping data. However, in the mutants, the petB 3′ end position was more frequently found at +20 (six clones) and less often at +69 (four clones) or in between at +63 (two clones). This suggests that processing of the petB-petD intergenic region generally takes place in the mutant but that most of the resulting petB transcripts deviate from those in the wild type. The fact that predominantly shorter petB 3′ ends could be found in the mutants again suggests that 3′ processed petB transcripts are rapidly degraded because of the lack of PrfB3. The position of the petB 5′ end −47 bases upstream of the start codon was identical in mutant (12 clones) and wild type (12 clones), indicating that PrfB3 genetically interacts with the petB 3′ end. S1 nuclease mapping failed to detect petD transcripts that were processed in the petB-petD intergenic region. When we used 5′ exon primers in circular RT-PCR experiments (see Supplemental Table 1 online), we could determine the 5′ end at position −31 with respect to the petD start codon, which is 86 nucleotides downstream of the petB 3′ ends. Therefore, petB 3′ and petD 5′ ends do not overlap, indicating that one processing site in between would be sufficient to generate both transcripts in Arabidopsis (see Supplemental Figure 5 online).
Taken together we conclude that PrfB3 is required for stabilizing 3′ processed petB transcripts. Obviously, accumulation of almost unchanged amounts of spliced petB-containing precursors in the mutants is not sufficient to ensure translation of petB. This indicates that either only 3′ processed petB transcripts are translational competent or that PrfB3 plays an additional role in translation of petB or other transcripts encoding subunits of the cytochrome b6f complex.
The Function of PrfB3 in petB RNA Stabilization Is Independent of Translation
Since PrfB3 originated from a duplication of the functional release factor PrfB1, we addressed the question whether the role of PrfB3 is still related to translational events. In contrast with transcripts that are targeted by PrfB1 (Meurer et al., 2002), polysomal loading of petB and petD transcripts is unaltered in prfB3-1 mutants (see Supplemental Figure 6 online). Inhibition of plastid translation in prfB1 induced normal accumulation of transcripts, which are otherwise lacking. By contrast, neither inhibition with lincomycin nor with chloramphenicol induced an increase in petB transcript abundance in prfB3-1 (see Supplemental Figure 7 online). This indicated that the function of PrfB3 in petB RNA stability has been completely uncoupled from translational events.
PrfB3 Is Part of a petB RNA-Containing Complex
To investigate whether PrfB3 acts as part of a protein complex, we analyzed native soluble proteins by size-exclusion chromatography. Collected fractions were subjected to immunological analysis. PrfB3 was found to be part of a 400-kD complex when RNA digestion was inhibited from the beginning of sample extraction (Figure 7A). However, when samples were treated with RNase before they were subjected to chromatography, the 400-kD complex disappeared and most PrfB3 could be found as free protein. In addition, upon RNase treatment, small amounts of a PrfB3-containing complex of ~700 kD were assembled, possibly due to the formation of unspecific aggregation products or a multimeric complex when RNA is lacking (Figure 7A). Electrophoretic mobility shift assays were performed with the purified PrfB3 protein to elucidate whether PrfB3 itself is capable of binding the 3′ untranslated region (UTR) of petB RNA (Figures 7B and 7C). It appeared that PrfB3 efficiently binds to the petB 3′ UTR (Figure 7C). Two binding products were observed, one of lower (B1) and one of higher mobility (B2) (Figure 7C). Presumably the RNA-protein complex becomes more densely packed because a globular three-dimensional structure is formed upon interaction in the B2 product. Addition of unlabeled 3′ UTR RNA to the binding assay led to competition with the labeled probe as revealed by decreasing labeling of binding products B1 and B2. In addition, we used transcripts of the same length (70 bases) from the petB 5′ UTR and petB exon (Figure 7B). Whereas PrfB3 hardly bound the exon region, the 5′ UTR showed a much lower binding affinity than that of the 3′ UTR, indicating the specificity by which PrfB3 binds to its petB 3′ UTR target. A 500-fold excess of transcripts of the exon probe over the 3′ UTR was unable to compete notably. By contrast, the 5′ UTR probe competed with B1, although less efficiently than did the 3′ UTR. However, unlike the 3′ probe, the 5′ UTR was unable to compete with the binding product B2. Moreover, B2 products appeared in larger amounts upon increasing levels of the 5′ UTR probe, indicating that PrfB3 preferentially binds the 3′ UTR (Figure 7C).
Figure 7.
PrfB3 Is Part of an RNA-Containing Complex and Binds to the petB 3′ UTR.
(A) Native soluble proteins were separated by size-exclusion chromatography in the presence of RNase inhibitors, and fractions (1 to 24) were subjected to immunoblot analysis together with wild-type (WT) and prfB3-1 proteins using PrfB3 antibodies (top part). Extracts were RNase treated before subjecting to chromatography (bottom part). The molecular mass of marker proteins is indicated in kilodaltons.
(B) The positions of the petB RNA 70-mers (black lines) of the 5′ UTR, exon and 3′ UTR used for binding, as well as the processing sites (arrows) are shown.
(C) Left: Coomassie blue stain of an SDS-PAGE gel of the purified recombinant At-PrfB3 protein. L, E. coli lysate; E, enriched protein. Right: RNA binding of PrfB3 was analyzed by gel shift assays using increasing concentrations of purified At-PrfB3 protein (0, 6, 60, and 600 nM; indicated by triangles) and the petB RNA 70-mers (indicated below and specified in [B]). Nonlabeled petB RNA 70-mers were used in 10×, 50×, 100×, and 500× molar excess over the radiolabeled 3′ UTR probe for competition experiments using 600 nM protein (black boxes). U, unbound RNA; B1 and B2, bound RNA.
Reduced PrfB3 Amounts Effect Stabilization of petB mRNA
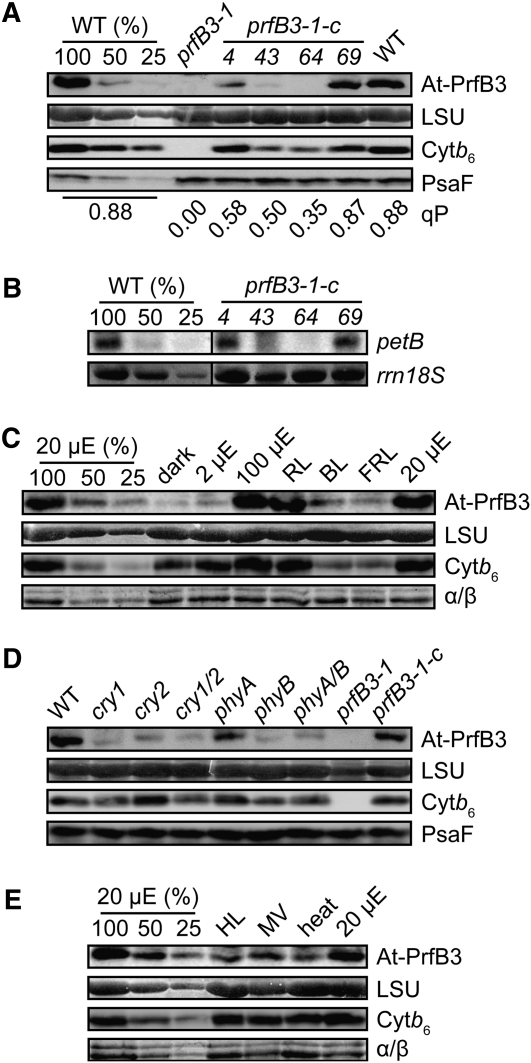
The potentially regulatory function of PrfB3 in stabilizing petB transcripts was first investigated by selecting partially complemented mutant lines (see Supplemental Figure 2 online). Three lines, numbers 4, 43, and 64, showed increased steady state chlorophyll fluorescence levels and a lowered qP (see Supplemental Figure 6 online; Figure 8A) but were able to grow photoautotrophically. Line number 69 showed fluorescence characteristics that are typical for the wild type. It appeared that expression of PrfB3 is reduced in lines number 4, 43, and 64. Only those lines (numbers 43 and 64) in which PrfB3 accumulated below a threshold of ~25% of wild-type levels also accumulated <25% of cytochrome b6 (Figure 8A). Downregulation of PrfB3 levels to >50% of wild-type levels in line 4 was insufficient to reduce amounts of cytochrome b6 comparably. Levels of the PSI protein PsaF were unaffected by the lower accumulation of PrfB3, indicating that PrfB3 levels are crucial for determining levels of cytochrome b6 and accordingly of the entire cytochrome b6f complex. The effect of reduced PrfB3 levels in the partially complemented lines on petB mRNA stability was tested by RNA gel blot analysis. Only lines number 43 and 64 had significantly lower levels of the monocistronic petB mRNA, indicating that accumulation of PrfB3 above 50% had no effect on petB mRNA levels (Figure 8B).
Figure 8.
Regulatory role of PrfB3 in petB RNA Stability and Cytochrome b6 Accumulation.
(A) Levels of the soluble protein PrfB3 and the membrane proteins cytochrome b6 and PsaF were investigated in the wild type (WT), mutant, and partially (numbers 4, 43, and 64) as well as completely complemented (number 69) lines by immunoblot analysis. For the wild type, dilution series of membrane proteins corresponding to 10 (100%), 5, and 2.5 μg chlorophyll and to 100 (100%), 50, and 25 μg of soluble proteins were loaded. As loading control for the soluble proteins, an imidazole stain of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (LSU) is shown. The qP value for each line is indicated below.
(B) Accumulation of monocistronic petB transcripts in wild-type and complemented lines. A dilution series of 8 (100%), 4, and 2 μg of total wild-type RNA was loaded. As loading control the 18S rRNA is shown.
(C) The effect of light quantities and qualities on PrfB3 and cytochrome b6 expression was investigated by immunoblot analysis. As loading control, an imidazole stain of the LSU (soluble protein) and the α/β-subunits of the ATP synthase (membrane proteins) are shown. Plants were grown for 14 d under heterochromatic light (100 μmol photons m−2 s−1) on soil and were then adapted for 3 d to different light regimes. The intensities of heterochromatic light are indicated. μE, μmol photons m−2 s−1; RL, red light (70 μmol photons m−2 s−1); BL, blue light (5 μmol photons m−2 s−1); FRL, far-red light (0.1 μmol photons m−2 s−1).
(D) Expression of PrfB3, cytochrome b6, and PsaF was analyzed in photoreceptor mutants. Levels of LSU are shown as loading control. Plants were grown on soil under heterochromatic light (100 μmol photons m−2 s−1) for 3 weeks. c, complemented
(E) Stress-dependent expression of PrfB3 and cytochrome b6 was analyzed. Controls are the same as in (C). HL, high light (800 μmol photons m−2 s−1, 3 h); MV, methylviologen (50 μM in 0.1% Tween, 2 × 4 h incubation); heat (38°C, 3 h).
Light- and Stress-Dependent Regulation of PrfB3 Levels
The general increase in chloroplast transcript abundance upon illumination of dark-adapted plants is known to depend highly on increased RNA stability (Monde et al., 2000). Therefore, a possible role of light in PrfB3 abundance was investigated. Interestingly, levels of PrfB3 were severely reduced in light-grown plants adapted to darkness, 2 μmol photons m−2 s−1 heterochromatic light, blue (5 μmol photons m−2 s−1), or far-red light (0.1 μmol photons m−2 s−1) for 3 d (Figure 8C). Reduced expression of PrfB3 correlated with reduced amounts of cytochrome b6 but levels of the large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase as well as of the α- and β-subunits of the plastid ATP synthase remained unchanged, reflecting the specificity with which PrfB3 regulates cytochrome b6 levels. Expression of PrfB3 and, thus, cytochrome b6 was comparable to the wild type under red light illumination (70 μmol photons m−2 s−1) and under heterochromatic light of 100 and 20 μmol photons m−2 s−1 (Figure 8C).
The light intensities used differed and were quite weak for blue and far-red light. Therefore, we could not differentiate between light quantity and quality. To address this issue, we performed immunoblot analyses using mutants of several photoreceptors. PrfB3 is severely downregulated in cryptochrome mutants cry1, cry2, and cry1 cry2. Additionally, expression of PrfB3 is severely reduced in phyB and phyA phyB double mutants. This is consistent with normal expression of PrfB3 under red light, which is sensed only by phytochrome B (Quail et al., 1995), and in phyA mutants. Reduced PrfB3 expression below a certain threshold is apparently accompanied by lower cytochrome b6 levels. These data indicate that expression of PrfB3 highly depends on light receptors and accordingly on light quality in addition to light intensity (Figure 8D).
Since the cytochrome b6f complex is known to be responsible for redox signaling (e.g., to balance light absorbance of the two photosystems by triggering phosphorylation of the LHCII kinase; Lemeille and Rochaix, 2010), a possible role of PrfB3 in stress responses was also investigated (Figure 8E). It appeared that application of high light (3 h), oxidative stress as induced by methylviologen treatment (H2O2 production) (8 h), and heat stress (3 h) caused a decrease in PrfB3 amounts to ≤50% of wild-type levels. However, no simultaneous loss of cytochrome b6 could be observed, again indicating that a certain threshold of PrfB3 levels is needed to mediate regulation of cytochrome b6 levels. In conclusion, expression of PrfB3 is quite sensitive to environmental changes, indicating an important regulatory role in adjusting petB RNA and consequently cytochrome b6 protein levels.
DISCUSSION
Phylogenetic Origin, Divergence, and Structure of PrfB3
In this study, we present a previously uncharacterized nuclear-encoded ribosomal release factor-like protein, PrfB3, required for regulation of chloroplast petB transcript stability in Arabidopsis. In contrast with PrfB3, the other four functional organellar release factors of eubacterial origin in Arabidopsis retained the highly conserved stop codon recognition and peptidyl-tRNA hydrolysis motifs, indicating the similarity between termination of translation in eubacteria and organelles. We have shown that At-PrfB1 in addition to its involvement in ribosomal release, also affects stability of transcripts containing UGA stop codons (Meurer et al., 2002). This phenomenon has not been described in eubacteria to date. Therefore, it appears that the additional regulatory functions of At-PrfB1 have been newly acquired by land plants and that they could represent the evolutionary constraints keeping the number of TGA stop codons high in plastids of land plants (Meurer et al., 2002). Several algae, like C. reinhardtii, have lost the TGA stop codon in their plastid genomes and accordingly a gene encoding for the corresponding RF2 (Meurer et al., 2002). The PrfB3 gene arose from PrfB1, but its product does not complement the deficiencies caused by the mutation of the PrfB1 gene, indicating that PrfB3 lost its function as a release factor. In spite of the comparable similarity between PrfB3 and the plastid (37.5%) and mitochondrial (36.5%) RF2, we unequivocally demonstrate that PrfB3 only recently arose from the plastid-localized form, indicating a very fast divergence of this gene. However, the predicted three-dimensional structure appears to be almost identical to that of the crystallized PrfB protein in Escherichia coli (Vestergaard et al., 2001) with the exception of two loops harboring the SPF and GGQ tripeptide motifs in E. coli (see Supplemental Figure 8 online). Here, we provide evidence that PrfB1 and PrfB3 are phylogenetically closely related but functionally distinct plastid proteins in vascular plants.
The high degree of divergence as well as the fluctuation of the gene and intron composition of the psbB gene cluster in plants also attests to the fast evolving gene cluster organization accompanied by the recent acquisition of many plant-specific factors involved in processing of the primary transcript (Barkan, 2011). This is consistent with the higher divergence (even between closely related species) of the target sequence elements in plastid UTRs and intergenic regions compared with that in coding regions, which are by far not targeted as extensively for the control of gene expression (Greiner et al., 2008).
The absence of the PrfB3 gene in the sequenced genomes of cyanobacteria, red, green, and diatom algae, the moss P. patens, and the fern S. moellendorffii (http://www.plantgdb.org) again suggests that PrfB3 evolved in vascular plants just before divergence of monocots and dicots, as a result of a duplication of the ancestral PrfB1 gene and subsequent loss of the peptide chain release function followed by the loss of the two conserved tripeptide motifs (Figure 1).
PrfB3 Protects 3′ Processed petB Transcripts against 3′→5′ Exonucleolytic Degradation
Mapping of transcript termini demonstrated that only spliced and 3′ processed petB transcripts are missing almost completely in both allelic prfB3 mutants (Figure 6E). Several effects potentially could cause reduction of the amounts of these RNAs: reduced splicing, loss of processing, or increased 3′ exonucleolytic attack. Three lines of evidence indicate that there are no primary splicing defects in prfB3: (1) RNA gel blot, S1 nuclease mapping, and quantitative RT-PCR analyses showed that prfB3 was generally able to splice petB transcripts efficiently (Figures 6C to 6E); (2) in contrast with nuclear and chloroplast mutants affected in splicing precursor transcripts (Jenkins et al., 1997; Ostheimer et al., 2003; Barkan, 2011), unspliced forms of petB did not accumulate at higher levels in prfB3 (Figure 6); (3) the pattern and abundance of all petB intron-containing transcripts was identical in the mutant and wild type (Figure 6, intron probe D).
The traces of 3′ processed petB transcripts that accumulate in the mutant are mostly shorter at the 3′ end than in the wild type and could represent a metastable intermediate produced by exonucleolytic attack. The 5′ terminus maps at the same position in the mutant and wild type, strongly supporting the idea that the petB 3′ end represents the target of PrfB3. Levels of all petB intron-containing precursors are unaffected, and spliced petB precursors are rather slightly reduced in the mutant, indicating efficient processing of the petB-petD intergenic region. Therefore, we hypothesize that PrfB3 stabilizes 3′ processed petB transcripts.
The finding that PrfB3 is part of an RNA-containing complex also supports the role of the protein in petB RNA protection (Figure 7A). The complex fell apart upon RNase treatment, but a shift toward heavier fractions was also observed. This indicates that the bound RNA is responsible for the formation of the complex running at ~400 kD. Either the complex is shifted to a higher molecular weight because its structure is more relaxed when RNA is absent or, although less likely, a new association partner that replaces the RNA has been assembled. Alternatively, the RNA-free complex tends to form a multimeric assembly of 700 kD. This may indicate that different transient assembly products are also present in vivo. A similar phenomenon has been described recently for MCA1 in C. reinhardtii (Boulouis et al., 2011). As the RNA-containing complex is found in only a few fractions, we assume that it binds exclusively transcripts of a distinct and similar size like the 3′ processed psbH-petB and monocistronic petB RNAs.
The slight reduction in the amounts of spliced petB precursors in the mutants indicates an increased cleavage of the petB-petD intergenic region and hints of a feedback regulation, when either cytochrome b6 proteins or petB transcripts are of low abundance. Presumably, large amounts of accumulating precursor transcripts are translationally incompetent with respect to petB and are rapidly cleaved upon signaling to speed up cytochrome b6 synthesis. This is supported by the fact that the cytochrome b6f complex is virtually absent, although amounts of spliced petB precursors are only marginally reduced in the mutants.
Given that PrfB3 originated from the functional release factor PrfB1, it is conceivable that PrfB3 exerts its function through binding ribosomes. To check this assumption, the effect of two inhibitors, chloramphenicol and lincomycin, known to hinder chloroplast translation in different ways, was analyzed. In prfB1, inhibition of translation stabilizes specific transcripts, which are otherwise unstable (Meurer et al., 2002). However, in prfB3, translation inhibition had obviously no effect on RNA abundance, indicating that translational events are not responsible for rapid degradation of 3′ processed petB RNA (see Supplemental Figure 7 online). Nevertheless, these experiments do not exclude that PrfB3 is also involved in translation, as it was shown that all spliced petB and petD precursors that are associated with ribosomes are subjected to translation in maize (Barkan, 1988, 2011). In addition, a maize mutant lacking the monocistronic petB transcript is still able to produce cytochrome b6 at almost normal rates (Barkan et al., 1994).
However, it should be mentioned here that regulation of petB-petD transcript maturation and presumably petB translation seems to be quite different in maize and Arabidopsis. In contrast with maize, monocistronic petD transcripts in Arabidopsis are below the limits of detection in RNA gel blots. This indicates that petD translation also takes place on precursor transcripts in Arabidopsis but presumably more efficiently than in maize (Barkan et al., 1994). Furthermore, petB 3′ ends overlap with petD 5′ ends in maize (see Supplemental Figure 5 online). This overlapping stretch is presumably masked by PrfB3, CRP1, and/or other factors for stabilization of both processing products in accordance with the function of PPR10 and other PPR proteins in chloroplasts (Barkan, 2011; Prikryl et al., 2011). Only one example from all studies described in the literature shows that processed 5′ and 3′ transcript termini from an intergenic region do not overlap (Hashimoto et al., 2003, Barkan, 2011). In Arabidopsis, the petB 3′ and petD 5′ transcript termini are 86 nucleotides apart and also do not overlap (see Supplemental Figure 5 online). Therefore, only one processing site in the petB-petD intergenic region is sufficient to explain the transcript pattern in Arabidopsis. Either the petD 5′ processing site is of ancient origin as is the cotranscription of petB and petD and it has been lost secondarily in Arabidopsis and other dicots or the processing site has been newly evolved in maize and other monocots. Taken together, the data indicate that petB-petD intercistronic processing occurs efficiently in the prfB3 mutants. The resulting petD transcripts are rapidly subjected to degradation in both the wild type and prfB3, whereas processed petB transcripts are degraded only in the mutant. Thus, we strongly suggest that PrfB3 has been recruited to specifically protect petB transcripts against 3′→5′ exonucleolytic attack by masking the 3′ ends.
It has been proposed that the eubacterial release factor possesses the capability to bind the 23S rRNA already before recognition of the stop codon (Frolova et al., 2000; Korostelev et al., 2010). The RNA binding activity has presumably been conserved in PrfB3 and ultimately been specified for petB transcripts. Indeed, we have shown that PrfB3 predominantly interacts with the petB 3′ UTR region, substantiating the idea that PrfB3 masks the petB 3′ UTR against exonucleolytic attack. No significant binding and competition capability could be observed for a randomly chosen petB exon region of the same length, indicating that transcripts are not generally bound by PrfB3 and that binding to the petB 3′ region is rather specific (Figure 7C). Although the petB UTRs do not share significant sequence or structural similarities, a weak physical interaction of PrfB3 with the 5′ UTR was also observed, raising the possibility of an involvement of PrfB3 in petB translation and stabilization. So far, such dual functions have rather been attributed to factors that bind to the 5′ end of plastid transcripts, presumably allowing access of ribosomes to their binding site (Barkan et al., 1994; Ossenbühl and Nickelsen, 2000; Barkan, 2011).
In addition to PrfB3, several more nuclear-encoded factors required for processing and splicing of the polycistronic psbB-psbT-psbH-petB-petD transcript were identified (Barkan, 2011). Thus, increase of the structural complexity of the processing pattern of the psbB gene cluster was accompanied by the acquisition of a number of newly evolved nuclear-encoded protein factors. This may allow individual regulation of gene expression despite of their presence in a polycistronic context. PrfB3 is a further example of proteins that originally had a housekeeping function and subsequently have been recruited during evolution for fine-tuning of the fast-evolving plastid RNA metabolism. PrfB3 represents the first known factor in plants that is involved in regulation of the stability of a single chloroplast RNA without having PPR, TPR, or OPR motifs.
PrfB3 Expression Is Highly Responsive to Stress and Environmental Changes
Recently, it was hypothesized that most factors involved in chloroplast RNA maturation and complexity evolved to ensure the functionality of chloroplast genetic information and have no authentic regulatory function (Maier et al., 2008). Although expression of several chloroplast RNA binding proteins was shown to depend on development, tissues, and environmental conditions, a truly regulatory function has been shown, to the best of our knowledge, only for the PPR protein MCA1, a factor in C. reinhardtii (Raynaud et al., 2007; Tillich et al., 2010; Ruwe et al., 2011). It appeared that the abundance of MCA1 limits the amount of petA mRNA and accordingly levels of the encoded protein, cytochrome f (Raynaud et al., 2007). This and the fact that the activity of the cytochrome b6f complex is rate limiting for the light reaction in photosynthesis (Yamori et al., 2011) prompted us to investigate whether PrfB3 also has a regulatory function in petB gene expression. First, we have shown that downregulation of PrfB3 below a threshold of ~25% goes along with a severe reduction in the amount of petB RNA and consequently in the abundance of cytochrome b6 protein levels (Figure 8A). Furthermore, we show that expression of PrfB3 is highly responsive to changes in light quantities and qualities (Figures 8C to 8D) as well as to the redox status and stress factors like heat and high light (Figure 8E), which often appear simultaneously and in combination with drought stress in nature. This and the application of the individual stressors for a limited time period may explain why we did not detect a concomitant reduction of cytochrome b6 upon stress induction. Similarly, ATAB2 was shown to be involved in the signaling pathway of light-regulated translation of PSI and PSII proteins during early plant development (Barneche et al., 2006). PrfB3 levels rapidly dropped to ~25 to 50% when we applied only one stress factor under otherwise constant conditions. Presumably, under natural conditions, the response of PrfB3 to combined and more intense stressors may be even more pronounced. We propose that PrfB3 expression is tightly regulated and presumably rate limiting for levels of petB mRNA as well as cytochrome b6 under changing and/or adverse environmental conditions. For example, PrfB3 could exert its function when a selective reduction or increase in the amount of the cytochrome b6f complex is required (Schöttler et al., 2007). Physiologically it would also make sense to regulate primarily the rate-limiting step of photosynthetic electron transport. The complex endonucleolytic processing of RNA precursors in chloroplasts presumably evolved not only to allow subsequent adjustment of processed transcript amounts but also to fine-tune translation rates. This regulation also represents an adaptation to the eukaryotic regulatory system.
METHODS
Source and Growth Conditions of Mutants
The Arabidopsis thaliana T-DNA insertion line prfB3-1 (SALK_133921) accession Columbia-0 was obtained from the SALK collection (http://signal.salk.edu). The mutant line prfB3-2, accession Wassilewskija, was selected from the Versailles mutant collection (Bechtold et al., 1993). Unless stated otherwise, all experiments were performed using 3-week-old wild-type and mutant plants grown on 1× Murashige and Skoog (MS) agar medium supplemented with 1.5% Suc under continuous light at a PFD of 20 μmol photons m−2 s−1 at a constant temperature of 21°C. For regulation experiments wild-type plants, complemented lines, and photoreceptor mutants were grown on soil in a 12-h-light (20°C)/12-h-dark (18°C) cycle with a PFD of 100 μmol photons m−2 s−1 for 2 weeks. When mentioned, plants were transferred to different light qualities for 3 d or subjected to high light (800 μmol photons m−2 s−1, 3 h), methylviologen (50 μM in 0.1% Tween, 2 × 4 h incubation to refresh the solution), or heat (38°C, 3 h) treatment. Since At-PrfB3 is essential for photoautotrophic growth, mutant lines had to be maintained in the heterozygous state. Photomorphogenetic mutants phyA-211, phyB-9 (Chen and Ni, 2006), cry1 (hy4-1), and cry2-1 (Ahmad et al., 1998) and corresponding double mutants were obtained from the Nottingham Arabidopsis Stock Centre (http://Arabidopsis.info).
Oligonucleotides
All oligonucleotides used in this work for PCR, RT-PCR, real-time RT-PCR, circular RT-PCR, probe generation, and other applications are listed in Supplemental Table 1 online.
Complementation of prfB3 and Sequence Analysis
The cDNA of At-PrfB3 was obtained from the RIKEN BioResource Center (Seki et al., 2002) and amplified with Pfu polymerase (Fermentas) using the 5′ phosphorylated PrfB3-P-f and PrfB3-Xba-r oligonucleotide primers. The resulting XbaI-digested PCR product was ligated into the SmaI/XbaI sites of vector pS001-VS (Meurer et al., 1998). The construct obtained, pbinatprfB3, was introduced into Agrobacterium tumefaciens GV3101 (pMP90RK) and subsequently transformed into prfB3-1 mutant segregants using the floral dip method (Clough and Bent, 1998). Complemented homozygous mutants were selected by PCR-based screens using gene-specific primers PrfB3-f and PrfB3-r. Proteins were aligned using the ClustalW program (http://www.clustal.org).
Spectroscopy Analysis
Chlorophyll a fluorescence induction kinetics of wild-type and mutant leaves was measured using a pulse-modulated fluorimeter (PAM101; Walz) as described (Meurer et al., 1996b). Red actinic light (650 nm, 50 μmol photons m−2 s−1) was used for measurements of fluorescence quenching. NPQ and qP parameters were determined by applying repetitive saturation pulses of 800 ms. NPQ and qP were calculated as (Fm − Fm’)/Fm’ and (Fm’ − F)/(Fm’ − Fo), respectively. PSI yield was measured on leaves as absorption changes at 830 nm induced by far-red light (ΔAmax) and in the presence of actinic light (ΔA) (650 nm, 50 μmol photons m−2 s−1) using the PSI attachment of PAM101. The redox state of PSI was expressed as the fraction ΔA/ΔAmax.
Protein Analysis and Antibody Production
Thylakoid membrane proteins were isolated as described (Meurer et al., 1996b). For isolation of soluble proteins, wild-type and mutant leaves were homogenized in isolation medium (0.3 M sorbitol, 20 mM HEPES/KOH, 10 mM NaHCO3, 5 mM EGTA, 5 mM EDTA, and 5 mM MgCl2, pH 8.0) using a Waring blender. The homogenate was filtered through two layers of Miracloth (Merck) and centrifuged at 1500g for 8 min with medium break. The chloroplasts were lysed in lysis buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, and 1% protease inhibitor [Cocktail set VI, Merck]), and after centrifugation at 18,000g, soluble proteins were collected. Proteins were separated by SDS-PAGE, stained with Coomassie Brilliant Blue, or transferred onto nitrocellulose membranes (Protran; Whatman). Blotted proteins were incubated with specific antisera and visualized by chemiluminescence. Antibodies used for detection of PSII, PSI, cytochrome b6f complex, and ATP synthase subunits were described previously (Meurer et al., 1996b). Sera raised against the mitochondrial peroxiredoxin (PrxII F) were described (Finkemeier et al., 2005). For production of antibodies against At-PrfB3, a cDNA fragment corresponding to amino acids 35 to 410, lacking the putative transit peptide, was amplified using Phusion Pfu polymerase (NEB) and primers B3-Topo17-for and B3-Topo17-rev. After cloning the fragment into Gateway pDEST17 vector via Gateway pENTR/D-TOPO vector (Invitrogen), the protein was overexpressed in BL21(DE3)pLysS cells together with an N-terminal 6xHis-tag (AtprfB3-HIS). The antigen was purified from urea-solubilized inclusion bodies to high purity and injected into rabbits at different time intervals (Pineda Antibody Service). As the At-PrfB3 antibody recognizes many unspecific proteins, enrichment of chloroplast soluble proteins is required for detection of At-prfB3. The cloning strategy and antibody production against PAC were as for At-PrfB3 using primers Pac-Topo17-for and Pac-Topo17-rev. Intact chloroplasts and mitochondria were isolated as described (Lezhneva and Meurer, 2004; Sweetlove et al., 2007). Soluble proteins from isolated organelles were isolated by freezing and subsequent centrifugation at 18,000g.
Separation of Radiolabeled or Unlabeled Thylakoid Membrane Complexes by Blue Native/PAGE
Labeling of plastid proteins with [35S]-Met was performed as described (Amann et al., 2004). Blue native/PAGE analysis was performed as reported (Schwenkert et al., 2006). Individual spots were labeled according to their determination by mass spectrometry (Granvogl et al., 2006).
RNA Gel Blot Analysis, Polysome Analysis, Determination of Transcript Termini, and S1 Nuclease Mapping
Isolation of total RNA and RNA gel blot analysis was performed as described (Meurer et al., 2002). Polysomes were isolated from leaf tissue essentially as described (Barkan, 1988). Polysome aliquots (0.5 mL) were layered onto 3.2-mL 15 to 55% Suc gradients and centrifuged for 65 min at 272,000g at 4°C in a SW60Ti rotor (Beckman). Fractions of 0.4 mL were collected, and RNA was purified, denatured with 30% glyoxal, and subjected to RNA gel blot analysis. For determination of transcript termini, ~1 μg of RNA was self-ligated with T4 RNA ligase (Ambion). Resulting products were subjected to RT-PCR analysis using a one-step RT-PCR kit (Roche). This allowed amplification of ligated 3′-5′ boundaries. Enrichment of specific products was achieved by a nested PCR. Products were cloned and sequenced. Alternatively transcript termini were determined by RNase protection analysis (Felder et al., 2001).
Generation of Recombinant At-PrfB3 for RNA Binding Studies
The above-described AtprfB3-HIS fusion protein was expressed in Escherichia coli cells (Rosetta2; Novagen) using standard conditions. Inclusion bodies were purified in buffer A (150 mM NaCl, 50 mM Tris, pH 8, and 0.1% Triton X-100) 150 min after induction of expression and dissolved in 8 M urea, 50 mM Tris, pH 8, 1 mM DTT, and 1 mM EDTA. After dialysis against buffer A, the protein was enriched to high purity using Ni-NTA agarose beads (Qiagen).
Electrophoretic Mobility Shift Assay
Radiolabeled RNA 70-mers (200 pM), generated by in vitro transcription from T7 promoter–containing DNA fragments, and increasing protein concentrations (0, 6, 60, and 600 nM) were used in RNA gel mobility shift assays. For sequence information of the RNA 70-mers of the 3′ UTR, 5′ UTR, and exon region of petB, see Supplemental Table 1 online. RNA–protein interaction took place in a buffer containing 40 mM Tris, pH 7.5, 100 mM NaCl, 0.1 mg/mL BSA, 4 mM DTT, and 0.5 mg/mL Heparin at 25°C for 10 min. As competitors, nonlabeled RNA fragments were used in 10×, 50×, 100×, and 500× molar excess over the labeled petB 3′ UTR probe. Samples were separated on a nondenaturing 8% polyacrylamide (37.5:1 acrylamide:bis-acrylamide) gel prepared in 1× TBE buffer. Gels were fixed, dried, and analyzed with a phosphor imaging system (Typhoon).
Translation Inhibition Experiment
For application of plastid translation inhibitors, 3-week-old plants of prfB3-1 and the wild type were used. To avoid embolism, hypocotyls were clipped in a half-strength MS solution containing 500 mg/L chloramphenicol or 400 mg/L lincomycin, respectively. Leaves were not immersed to ensure increased uptake of the antibiotics by transpiration. Twenty-four hours after incubation in ambient light, leaves of the seedlings were harvested. Control plants were incubated for the same time in a solution containing half-strength MS nutrients. Total RNA was isolated from the harvested material and used for RNA gel blot analysis. The petB and petD probes used for hybridizations were amplified using primers petB-for/petB-rev and petD-for/petD-rev, respectively.
Quantitative Estimates of Spliced petB Transcripts
Quantitative two-step RT-PCR was performed using the LightCycler thermal cycler system (Roche) as described (Lezhneva and Meurer, 2004). Serially diluted samples of Arabidopsis total cDNA, corresponding to 15 ng to 1.5 pg of DNA were used for calibration using primers psaA-f and psaA-r. Quantification of spliced petB and petD transcripts was performed using primers petB-f/petB-r and petD-f/petD-r, respectively.
Subcellular Localization of PrfB3
The cDNA of At-PrfB3 encoding the N-terminal part was PCR amplified using AtprfB3-Sal-f and AtprfB3-Sal-r primers (see Supplemental Table 1 online). The resulting product was digested with SalI and cloned in frame into the SalI site of the GFP expression vector pOL-LT (Mollier et al., 2002). Transient expression was performed in polyethylene glycol–treated tobacco (Nicotiana tabacum) protoplasts (Lyznik et al., 1991), and fluorescence was visualized 18 h after transformation using a Fluorescence Axio Imager microscope in ApoTome mode (Zeiss) (Amann et al., 2004).
Isolation of Arabidopsis protoplasts
Cotyledons of sterile Arabidopsis seedlings were cut into small pieces and incubated at 24°C overnight in the dark in 10 mM MES, 20 mM CaCl2, 0.5 M mannitol, pH 5.8, 5 mg mL−1, macerozyme, and 5 mg mL−1 cellulase (Duchefa). Isolation of protoplasts was as described (Dovzhenko et al., 2003).
Electron Microscopy
The ultrastructural analysis of the chloroplast membrane system was performed as described (Amann et al., 2004).
Size-Exclusion Chromatography
Three-week-old Arabidopsis wild-type and mutant leaves were homogenized in isolation medium as described above. Chloroplasts were lysed in a buffer containing 25 mM Tricine, pH 7.9, 50 mM KCl, 1% protease inhibitors, and 1% RNase inhibitors (Fermentas). The stromal fraction was obtained after centrifugation for 20 min at 18,000g and 70,000g. Supernatants were concentrated in Amicon Ultra filtration devices (Millipore) at 4°C, with or without 15 min RNase treatment (250 units RNaseOne; Promega). Samples (5 mg protein) were loaded onto a Superdex 200 10/300 GL column (GE Healthcare) and eluted with the same buffer. Elution fractions were precipitated with TCA and loaded onto SDS-PAGE gels with subsequent immunoblotting. The column was calibrated with a HMW calibration kit (GE Healthcare).
Structure Prediction
A homology-based model for the structure of At-PrfB3 was predicted using Genesilico (Kurowski and Bujnicki, 2003) taking the x-ray crystal structure of the E. coli PrfB protein as template (pdb 1GQE; Vestergaard et al., 2001). The image was produced using JMol (http://www.jmol.org/). RNA structures were calculated using the mfold algorithm (http://mfold.rna.albany.edu/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative data libraries under the following accession numbers: AT3G57190 (At-PrfB3), AT5G36170 (At-PrfB1), At1g56350 (At-PrfB2), LOC_Os05g31160 (Os-PrfB3), LOC_Os07g36250 (Os-PrfB1), LOC_Os03g19300 (Os-PrfB2), P74476 (Synechocystis sp PCC 6803 PrfB), and AAA24520 (E. coli PrfB).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of the E. coli and Synechocystis PrfB Proteins with Homologs in Plastids of Arabidopsis and Rice.
Supplemental Figure 2. Phenotype and Functional Analysis of prfB3 Mutants and Complemented Lines.
Supplemental Figure 3. Imaging of Chlorophyll a Fluorescence of Arabidopsis Protoplasts and Chloroplast Ultrastructure.
Supplemental Figure 4. RNA Gel Blot Analysis of Plastid Genes in the Wild Type, prfB3-1, and prfB3-2.
Supplemental Figure 5. Alignment of the petB-petD Intergenic Region in Arabidopsis and Maize.
Supplemental Figure 6. Polysome Analysis of the Wild Type and prfB3-1.
Supplemental Figure 7. RNA Gel Blot Analysis of Plants Treated with Plastid Translation Inhibitors.
Supplemental Figure 8. Predicted 3D Structure of At-PrfB3.
Supplemental Table 1. Primer Sequences, Probes, and RNA Fragments Used in This Work.
Acknowledgments
We thank Elli Gerick for excellent technical support. Agata Stêpieñ and Tilmann Abele are acknowledged for help with PCR and cloning. We thank Regina Schweiger for mitochondrial preparations and Iris Finkemeier and Karl-Josef Dietz for providing peroxiredoxin antibodies. Gerhard Wanner is acknowledged for preparing electron micrographs. We also thank Dario Leister for reading the manuscript. This research was supported by the German Science Foundation (Deutsche Forschungsgemeinschaft) to J.M. and P.W. (SFB-TR-1).
AUTHOR CONTRIBUTIONS
J.M. designed the research. R.S., L.L., S.S., S.T., S.F., and J.M. performed the research. R.S., L.L., K.M., P.W., and J.M. analyzed data. L.L. and J.M. wrote the article.
References
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998). Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392: 720–723 [DOI] [PubMed] [Google Scholar]
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. (2007). A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 19: 3864–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Walker M., Nolasco M., Johnson D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13: 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F., Winter V., Crèvecoeur M., Rochaix J.D. (2006). ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 25: 5907–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316: 1194–1199 [Google Scholar]
- Bollenbach T.J., Lange H., Gutierrez R., Erhardt M., Stern D.B., Gagliardi D. (2005). RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach T.J., Schuster G., Stern D.B. (2004). Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 78: 305–337 [DOI] [PubMed] [Google Scholar]
- Boulouis A., Raynaud C., Bujaldon S., Aznar A., Wollman F.A., Choquet Y. (2011). The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ni M. (2006). RED AND FAR-RED INSENSITIVE 2, a RING-domain zinc finger protein, mediates phytochrome-controlled seedling deetiolation responses. Plant Physiol. 140: 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Geimer S., Meurer J. (2009). Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana. DNA Res. 16: 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Claros M.G., Vincens P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779–786 [DOI] [PubMed] [Google Scholar]
- del Campo E.M. (2009). Post-transcriptional control of chloroplast gene expression. Gene Regul. Syst. Bio. 3: 31–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovzhenko A., Dal Bosco C., Meurer J., Koop H.U. (2003). Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma 222: 107–111 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Meierhoff K., Sane A.P., Meurer J., Driemel C., Plücken H., Klaff P., Stein B., Bechtold N., Westhoff P. (2001). The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13: 2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L.J., Dietz K.J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Merkulova T.I., Kisselev L.L. (2000). Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granvogl B., Reisinger V., Eichacker L.A. (2006). Mapping the proteome of thylakoid membranes by de novo sequencing of intermembrane peptide domains. Proteomics 6: 3681–3695 [DOI] [PubMed] [Google Scholar]
- Greiner S., Wang X., Herrmann R.G., Rauwolf U., Mayer K., Haberer G., Meurer J. (2008). The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 25: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Ito K., Uno M., Nakamura Y. (2000). A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403: 680–684 [DOI] [PubMed] [Google Scholar]
- Jenkins B.D., Barkan A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek D.J., Barkan A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L.L., Buckingham R.H. (2000). Translational termination comes of age. Trends Biochem. Sci. 25: 561–566 [DOI] [PubMed] [Google Scholar]
- Korostelev A., Zhu J., Asahara H., Noller H.F. (2010). Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 29: 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger T.S., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski M.A., Bujnicki J.M. (2003). GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 31: 3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S., Rochaix J.D. (2010). State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 106: 33–46 [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Meurer J. (2004). The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 38: 740–753 [DOI] [PubMed] [Google Scholar]
- Loh P.G., Song H. (2010). Structural and mechanistic insights into translation termination. Curr. Opin. Struct. Biol. 20: 98–103 [DOI] [PubMed] [Google Scholar]
- Lyznik L.A., Peng J.Y., Hodges T.K. (1991). Simplified procedure for transient transformation of plant protoplasts using polyethylene glycol treatment. Biotechniques 10: 294–300 [PubMed] [Google Scholar]
- Maier U.G., Bozarth A., Funk H.T., Zauner S., Rensing S.A., Schmitz-Linneweber C., Börner T., Tillich M. (2008). Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol. 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Berger A., Westhoff P. (1996a). A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8: 1193–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Grevelding C., Westhoff P., Reiss B. (1998). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258: 342–351 [DOI] [PubMed] [Google Scholar]
- Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I., Oelmüller R. (2002). A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14: 3255–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996b). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Mollier P., Hoffmann B., Debast C., Small I. (2002). The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 40: 405–409 [DOI] [PubMed] [Google Scholar]
- Monde R.A., Schuster G., Stern D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82: 573–582 [DOI] [PubMed] [Google Scholar]
- Motohashi R., et al. (2007). Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development. Plant Mol. Biol. 64: 481–497 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito K. (2003). Making sense of mimic in translation termination. Trends Biochem. Sci. 28: 99–105 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito K., Ehrenberg M. (2000). Mimicry grasps reality in translation termination. Cell 101: 349–352 [DOI] [PubMed] [Google Scholar]
- Ossenbühl F., Nickelsen J. (2000). cis- and trans-Acting determinants for translation of psbD mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 20: 8134–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer G.J., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K., Schöttler M.A., Karcher D., Thiele W., Bock R. (2011). Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucleic Acids Res. 39: 5181–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H., Boylan M.T., Parks B.M., Short T.W., Xu Y., Wagner D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268: 675–680 [DOI] [PubMed] [Google Scholar]
- Raczynska K.D., Le Ret M., Rurek M., Bonnard G., Augustyniak H., Gualberto J.M. (2006). Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett. 580: 5641–5646 [DOI] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A., Choquet Y. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C. (2011). The RNA-recognition motif in chloroplasts. J. Plant Physiol. 168: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Flügel C., Thiele W., Bock R. (2007). Knock-out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J. Biol. Chem. 282: 976–985 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Legen J., Takami T., Shikanai T., Herrmann R.G., Meurer J. (2007). Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol. 144: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Umate P., Dal Bosco C., Volz S., Mlçochová L., Zoryan M., Eichacker L.A., Ohad I., Herrmann R.G., Meurer J. (2006). PsbI affects the stability, function, and phosphorylation patterns of photosystem II assemblies in tobacco. J. Biol. Chem. 281: 34227–34238 [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Taylor N.L., Leaver C.J. (2007). Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372: 125–136 [DOI] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Beick S., Schmitz-Linneweber C. (2010). Chloroplast RNA-binding proteins: Repair and regulation of chloroplast transcripts. RNA Biol. 7: 172–178 [DOI] [PubMed] [Google Scholar]
- Vestergaard B., Van L.B., Andersen G.R., Nyborg J., Buckingham R.H., Kjeldgaard M. (2001). Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 8: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Westhoff P., Herrmann R.G. (1988). Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur. J. Biochem. 171: 551–564 [DOI] [PubMed] [Google Scholar]
- Yamori W., Takahashi S., Makino A., Price G.D., Badger M.R., von Caemmerer S. (2011). The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 155: 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]