This study examines the lignin biosynthesis pathway in the lycophyte Selaginella moellendorffii. An O-methyltransferase gene was found clustered with the gene encoding a phenylpropanoid meta-hydroxylase known to catalyze the formation of syringyl lignin subunits. The O-methyltransferase is shown to function in the same enzymatic pathway as the phenylpropanoid meta-hydroxylase.
Abstract
Syringyl lignin, an important component of the secondary cell wall, has traditionally been considered to be a hallmark of angiosperms because ferns and gymnosperms in general lack lignin of this type. Interestingly, syringyl lignin was also detected in Selaginella, a genus that represents an extant lineage of the most basal of the vascular plants, the lycophytes. In angiosperms, syringyl lignin biosynthesis requires the activity of ferulate 5-hydroxylase (F5H), a cytochrome P450-dependent monooxygenase, and caffeic acid/5-hydroxyferulic acid O-methyltransferase (COMT). Together, these two enzymes divert metabolic flux from the biosynthesis of guaiacyl lignin, a lignin type common to all vascular plants, toward syringyl lignin. Selaginella has independently evolved an alternative lignin biosynthetic pathway in which syringyl subunits are directly derived from the precursors of p-hydroxyphenyl lignin, through the action of a dual specificity phenylpropanoid meta-hydroxylase, Sm F5H. Here, we report the characterization of an O-methyltransferase from Selaginella moellendorffii, COMT, the coding sequence of which is clustered together with F5H at the adjacent genomic locus. COMT is a bifunctional phenylpropanoid O-methyltransferase that can methylate phenylpropanoid meta-hydroxyls at both the 3- and 5-position and function in concert with F5H in syringyl lignin biosynthesis in S. moellendorffii. Phylogenetic analysis reveals that Sm COMT, like F5H, evolved independently from its angiosperm counterparts.
INTRODUCTION
Paleobotanical and stratigraphic data suggest that the earliest tracheophytes arose during the Late Silurian and Devonian periods (~420 to ~360 million years ago) (Kenrick and Crane, 1997). This group of plants distinguished themselves from primitive bryophytes by the development of vascular tissue that was capable of transporting fluids throughout the plant body. It is thought that the evolution of vasculature in plants involved the recruitment of the plant phenylpropanoid metabolic pathway to synthesize and deposit the heterogeneous aromatic polymer lignin in the xylem cell wall (Boyce et al., 2004; Weng and Chapple, 2010). Lignin physically reinforces the plant cell wall and provides xylem cells with the strength to withstand the tension generated during transpiration, and by stiffening the cell walls of supportive tissues such as fibers, lignin provides plants with the structural support to stand upright. The innovation of lignified vascular tissue marked a significant step of early land plants toward their ultimate adaptation to the terrestrial environment, which consequently facilitated their dominance of the Earth’s flora during the Carboniferous period (~360 to ~300 million years ago) (Friedman and Cook, 2000).
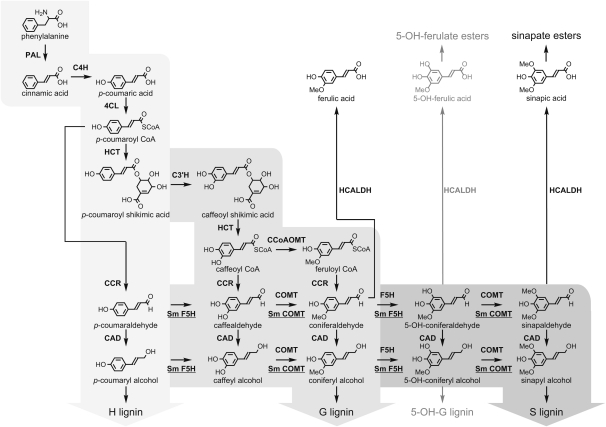
Lignin found in angiosperms is generally comprised of three major types of aromatic units, p-hydroxyphenyl (H), guaiacyl (G), and syringyl (H) units, which derive, following polymerization, from three p-hydroxycinnamyl alcohols: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, also known as the monolignols (Boerjan et al., 2003). Studies on the monolignol biosynthetic pathway over the past two decades have revealed that lignin monomer synthesis requires, among other enzymes, a suite of cytochrome P450-dependent monooxygenases (P450s) and S-adenosyl-l-methionine (SAM)-dependent O-methyltransferases (OMTs) (Boerjan et al., 2003). Whereas three P450s, cinnamic acid 4-hydroxylase (C4H), p-coumaroyl shikimic acid 3′-hydroxylase, and ferulic acid 5-hydroxylase (F5H) catalyze the aromatic hydroxylations para and meta to the sidechain, caffeoyl-CoA O-methyltransferase and caffeic acid O-methyltransferase (COMT) subsequently methylate the free meta-hydroxyls generated by C3′H and F5H, resulting in a set of intermediates with their ring hydroxylation/methoxylation status characteristic of H, G, or S units in lignin (Figure 1).
Figure 1.
The Monolignol Biosynthetic Pathway in Vascular Plants.
Light, medium, and dark-gray areas illustrate three levels of aromatic ring modifications that lead to the biosynthesis of H, G, and S lignin. The metabolic pathway derived from 5-hydroxyguaiacyl (5-OH-G) intermediates is depicted in gray, since it exists only when the COMT step is blocked. The enzymes illustrated on the pathway are by default based on the current knowledge acquired from studying angiosperms, except for the enzymes that define the unique pathway toward S lignin biosynthesis in Selaginella, which are underlined and positioned underneath the arrows. The enzymes and their abbreviations are as follows: PAL, phenylalanine ammonia-lyase; 4CL, 4-hydroxycinnamoyl CoA ligase; HCT, hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase; C3′H, p-coumaroyl shikimic acid 3′-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyl transferase; CCR, (hydroxy)cinnamoyl-CoA reductase; HCALDH, hydroxycinnamaldehyde dehydrogenase; CAD, (hydroxy)cinnamyl alcohol dehydrogenase.
Among the ring modification enzymes mentioned above, F5H and COMT constitute a branch of the phenylpropanoid pathway that is only required for the biosynthesis of S lignin. Downregulation of F5H or COMT in transgenic alfalfa (Medicago sativa) has been shown to result in a reduction of S lignin (Guo et al., 2001; Reddy et al., 2005), and in Arabidopsis thaliana, loss-of-function mutations in either F5H or COMT completely abolish the accumulation of S lignin in their respective mutants (Chapple et al., 1992; Goujon et al., 2003). It has been proposed that F5H and COMT are recent additions to the plant biochemical repertoire, restricted only to angiosperms, because S lignin is absent in most gymnosperms and ferns (Osakabe et al., 1999), although some exceptions exist (Baucher et al., 1998; Weng et al., 2008a). For example, S lignin has also been detected in the lycophyte genus Selaginella (Towers and Gibbs, 1953; White and Towers, 1967; Erickson and Miksche, 1974; Faix et al., 1977; Logan and Thomas, 1985; Jin et al., 2005; Weng et al., 2008b). Lycophytes embody the most ancient lineage of vascular plants living on earth, which diverged from euphyllophytes, including ferns and seed plants, 400 million years ago. The relatively isolated occurrence of S lignin in lycophytes suggests that S lignin could have arisen independently in this lineage (Weng et al., 2008b, 2010b).
We recently reported the identification of a novel P450 (F5H) from Selaginella moellendorffii (Weng et al., 2008b). We showed that Sm F5H not only possesses catalytic activity equivalent to its angiosperm counterparts in mediating 5-hydroxylation of G-substituted intermediates (Weng et al., 2008b), but also possesses additional activity as a phenylpropanoid 3-hydroxylase that defines a novel pathway toward S lignin biosynthesis from p-coumaraldehyde and p-coumaryl alcohol (Weng et al., 2010b) (Figure 1). Phylogenetic analysis suggested that Sm F5H is not orthologous to its angiosperm counterpart, indicating its independent origin in Selaginella (Weng et al., 2008b). The presence of a novel S lignin biosynthetic pathway in Selaginella further suggests that Selaginella must also contain a functional COMT that is capable of methylating each of the meta-hydroxylated intermediates generated by F5H. Although previous kinetic studies have suggested that angiosperm COMTs are relatively promiscuous in their abilities to catalyze transmethylation reactions on both 3-hydroxylated H and 5-hydroxylated G intermediates (Parvathi et al., 2001; Zubieta et al., 2002), data from older literature showed that OMT extracted from gymnosperm species generally have little activity toward 5-hydroxyferulic acid compared with caffeic acid, suggesting that the specific function of COMT for S lignin biosynthesis is probably not fundamental to all tracheophytes but rather a newly evolved feature in angiosperms (Kuroda, 1983).
Here, we report the characterization of a series of OMTs from S. moellendorffii. We provide both in vivo and in vitro evidence to show that Selaginella contains a functional COMT that is capable of mediating specific methylation reactions on meta-hydroxylated lignin biosynthetic intermediates. Similarly to the case of Sm F5H, the COMT in Selaginella also appears to have evolved independently of its angiosperm counterparts. Taken together, our data suggest that independent occurrences of S lignin in phylogenetically divergent angiosperm and lycophyte lineages are due to independent recruitment of biosynthetic enzymes and the pathways defined by them.
RESULTS
Cell Wall Structure in the Stem of S. moellendorffii
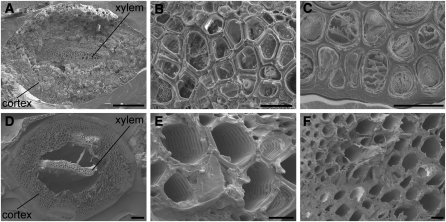
Selaginella possesses a protostele with xylem cells surrounded by a cortical cylinder (Figures 2A and 2D), an anatomy distinct from the eustele typically found in angiosperms. Using the Mäule histochemical staining method, we previously observed that S lignin is predominantly deposited in the cortex, rather than the xylem, in Selaginella (Weng et al., 2008b). To gain more insight into the cell wall secondary thickening and lignification process in these two different tissue types in S. moellendorffii stem, we first investigated the stem cell wall structure by scanning electron microscopy. Sections from both young shoot (~1 cm from the apex) and mature stem (~1 cm from the base) were prepared and examined. In the xylem of both young and mature stem, secondary cell walls with scalariform thickenings were observed in most of the vessel element cells (Figures 2B and 2E). Although many xylem vessel elements from the young shoot were found to contain living contents and were probably not yet mature (Figure 2B), all xylem vessel elements in the mature stem were fully differentiated and had lost their cell contents (Figure 2E). Most of the cortical cells in the young shoot were living cells with obvious cell contents (Figure 2C). The presence of a thick secondary wall could be observed in cortical cells (Figure 2C), reminiscent of the sclerified interfascicular fiber cells found in angiosperms. Like the xylem vessel elements, the cortical cells also die at maturity (Figure 2F).
Figure 2.
Cell Wall Structure of S. moellendorffii Examined by Scanning Electron Microscopy.
Global view of cross sections of Selaginella young (A) and old (D) stems showing a protostelic arrangement with xylem surrounded by cortex. Higher magnification photographs illustrate xylem ([B] and [E]) and cortical cells ([C] and [F]) from young ([B] and [C]) and old ([E] and [F]) regions of the stem. Bar = 100 μm in (A) and (D) and 10 μm in (B), (C), (E), and (F).
Tissue-Specific Lignin Analysis in S. moellendorffii Stem
It has been shown previously using histochemical staining methods and derivatization followed by reductive cleavage (DFRC) lignin analysis that the Selaginella stem cortex contains lignin derived from H, G, and S units, while xylem contains G lignin with only a trace of S lignin (Weng et al., 2008b, 2010b). To further delineate the composition of the whole lignin material and begin to obtain some structural insights regarding the interunit linkage distributions of the lignins in the two different tissue types in Selaginella stem, we took advantage of the unique protostelic structure of Selaginella stem and separated, with the aid of microscopy examination, enough xylem and cortical tissue for NMR analysis. Changes in the S:G:H distribution in the lignins are most readily visualized from the aromatic region of NMR spectra, particularly the two-dimensional 13C–1H correlation (HSQC) spectra correlating protons with their attached carbons (Figure 3A). The Selaginella whole stem lignin is S-rich (S:G:H = 76:22:2). Unlike in angiosperms, however, there is also a substantial H component; this is also seen in species such as kenaf (Hibiscus cannabinus; Kim et al., 2008). There are also some unrecognizable components in the aromatic region of the spectrum, components that appear to be removed by more exhaustive solvent extraction during cell wall isolation. The p-hydroxybenzoates found to acylate some lignins, including those of poplar (Populus sp), are not present. The most striking observations come from examination of spectra from the separated xylem-enriched material versus the residual cortex. Xylem contains essentially pure G lignin, with only ~5% S units in this isolated fraction. The cortex spectrum more closely resembles the whole material, with a high syringyl level and similar S:G ratio. The H-level was not measured due to overlap with other contours but again appears to be associated with only the cortex fraction.
Figure 3.
Tissue-Specific Lignin Analysis in S. moellendorffii.
(A) Aromatic regions of two-dimensional HSQC NMR spectra of acetylated cellulolytic enzyme lignins revealing compositional aspects. An isolated Björkman lignin from poplar was analyzed in parallel for comparison because it has a similar S:G level as that of the Selaginella lignin samples but has p-hydroxybenzoates acylating the γ-positions of some lignin sidechains. S:G:H levels are from uncorrected volume integrals of the analogous S2/6, G2, and H2/6 correlations (with the G2 integral being logically doubled); PB levels are uncorrected integrals and are expressed as a percentage of the total S+G+H level. WT, wild type.
(B) Sidechain regions of two-dimensional HSQC NMR spectra of acetylated cellulolytic enzyme lignins reflecting the resulting structural differences (i.e., the distribution of bonding patterns between the units that result from radical coupling reactions from the differing monomer supplies). Some effects are pronounced; for example, 5-coupling can only occur in H or G units; consequently, units B and D are more pronounced in the G-rich xylem and are relatively minor in the S-rich cortex. Resinols (C) are generally more prevalent in S-rich lignins; their virtual absence in the cortex lignin is most unusual (see Supplemental Figure 1 online). The relatively newly discovered spirodienones S (Ralph et al., 2006; Zhang et al., 2006) are typically more prevalent in S-rich lignins; they are readily seen in the S-rich cortex lignin.
The sidechain region in the NMR spectra peripherally reflects the changes in the S:G:H distribution and is rich in detail regarding the types and distribution of interunit bonding patterns present in the lignin fraction (Figure 3B; see Supplemental Figure 1 online). The xylem lignin spectrum is typical of a G-rich lignin with residual polysaccharides (Ralph et al., 1999). It contains evidence for the major β-ether units A, resinols C, and, due to the availability of the 5-position for radical coupling in guaiacyl units, phenylcoumaran B and dibenzodioxocin D units. The cortex spectrum, as is typical for S-rich lignins, is particularly rich in β-ether units A, has only low levels of 5-linked (B and D) units, and readily shows the spirodienone units S (from β-1-coupling reactions) that have only recently been authenticated and are most prevalently associated with S-rich lignins (Zhang et al., 2006).
Identification of S. moellendorffii COMT Candidate Genes
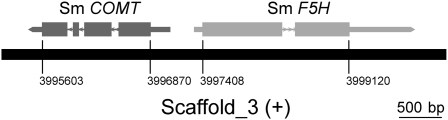
Angiosperm COMTs belong to the SAM-dependent methyltransferase superfamily, which all use SAM as the methyl group donor, yielding S-adenosyl-l-homocysteine and the methylated derivative of the substrate as products (Zubieta et al., 2001). We hypothesized that the COMT from Selaginella may also belong to this family and could be identified based on sequence similarity. We performed a BLAST search using Arabidopsis COMT (At COMT) as the probe against the S. moellendorffii genome. Four putative Selaginella COMT candidate genes were identified, which showed amino acid sequence identity with At COMT ranging from 37 to 51% (Table 1). To our surprise, one Selaginella COMT candidate (51% identical to At COMT) was found directly adjacent to Sm F5H in the genome. Its expression is driven in the opposite direction from a promoter within the same intergenic region (Figure 4). We cloned all four Selaginella COMT candidate genes for further functional analysis.
Table 1.
Amino Acid Sequence Percentage Identity among At COMT and Selaginella COMT Candidates
| Sm COMT | Sm COMT-Like1 | Sm COMT-Like2 | Sm COMT-Like3 | |
| At COMT | 51 | 40 | 43 | 37 |
| Sm COMT | 40 | 55 | 36 | |
| Sm COMT-like1 | 39 | 29 | ||
| Sm COMT-like2 | 36 |
Figure 4.
The Arrangement of COMT and F5H in the S. moellendorffii Genome.
Sm COMT and Sm F5H are clustered and oriented in an opposite direction from each other.
Complementation of the Arabidopsis COMT-Deficient Mutant by Sm COMT
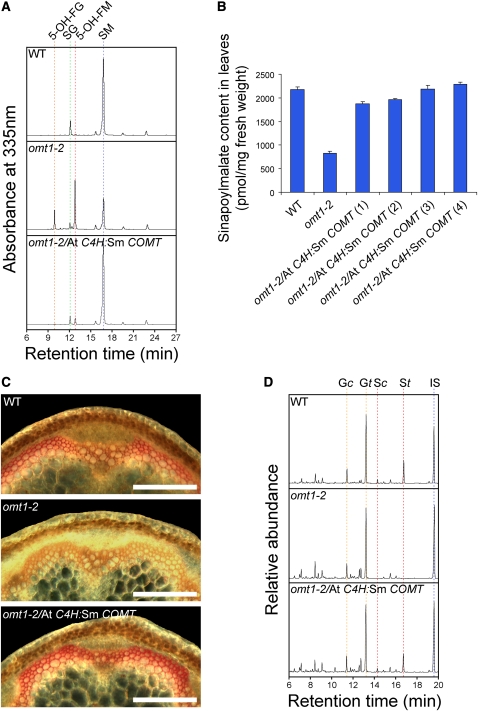
To test the function of Selaginella COMT candidate genes in planta, we transformed them into an Arabidopsis COMT-deficient mutant, omt1-2, under the control of the Arabidopsis C4H promoter and examined the plants for phenotypic complementation. Arabidopsis omt1-2 completely lacks S units in its lignin and accumulates 5-hydroxyferulate esters, which substitute for a large proportion of the sinapate esters normally found in rosette leaves (Figures 1 and 4) (Weng et al., 2010a). These observations are consistent with previous reports of another knockout allele of Arabidopsis omt1 (Goujon et al., 2003).
The leaf methanolic extracts from multiple T1 transformants for each Selaginella COMT candidate construct were analyzed by HPLC. Three of the candidates failed to complement the leaf hydroxycinnamate ester phenotype of omt1-2 (see Supplemental Figure 2 online), but the Selaginella COMT candidate that is adjacent to Sm F5H almost entirely alleviated the accumulation of 5-hydroxyferulate esters and restored normal sinapoyl malate and sinapoyl glucose accumulation in omt1-2 leaves (Figure 5A). More rigorous analysis at the T2 generation further confirmed that the leaf sinapoyl malate content was brought back to wild-type levels in all the independent transgenic lines examined (Figure 5B). This datum suggests that this Selaginella COMT candidate can take the place of At COMT in sinapate ester biosynthesis in vivo and was thus designated as Sm COMT.
Figure 5.
Complementation of the Arabidopsis COMT-Null Mutant by Sm COMT.
(A) HPLC profiles of 3-week-old Arabidopsis leaf extracts. 5-OH-FG, 5-hydroxyferuloyl glucose; SG, sinapoyl glucose; 5-OH-FM, 5-hydroxyferuloyl malate; SM, sinapoyl malate.
(B) Restoration of leaf sinapoyl malate production in four independent lines of omt1-2/At C4H:Sm COMT transgenic plants quantified by HPLC. Error bars represent 1 sd of triplicate samples. WT, wild type.
(C) Mäule staining of 6-week-old Arabidopsis inflorescence stem sections. Bars = 200 μm.
(D) Gas chromatograms of the DFRC lignin analysis monomer products from cell wall samples prepared from 3-month-old Arabidopsis inflorescence stems. G/S, guaiacyl/syringyl lignin derivative; c/t, cis/trans; IS, internal standard.
To test whether Sm COMT can complement the lignin phenotype of omt1-2, we first examined the lignin composition of omt1-2/At C4H:Sm COMT transgenic plants using the Mäule histochemical staining reagent. Whereas omt1-2 stem sections stain yellow in both the sclerenchyma and the xylem, a reaction that indicates the absence of S lignin, omt1-2/At C4H:Sm COMT transgenic plants exhibit a staining pattern similar to the wild type with red staining localized in the sclerified parenchyma cells, suggesting S lignin accumulation in these cells (Figure 5C). To definitively determine whether the positive Mäule staining observed in the Sm COMT transgenic plants is due to the presence of S lignin units, we analyzed the cell wall samples from these plants by DFRC lignin analysis, a method specific for β-O-4–linked lignin units (Lu and Ralph, 1998). The results showed that S lignin biosynthesis was restored in the transgenic plants (Figure 5D); all the independent transgenic lines exhibited a lignin S mole percentage similar to that of the wild type (Table 2).
Table 2.
S Lignin Mole Percentage of Columbia Wild Type, omt1-2, and omt1-2/At C4H:Sm COMT T2 Transgenic Plants Quantified by DFRC Lignin Analysis
| Genotype | Lignin Mol % S |
| Wild type | 24 ± 6 |
| omt1-2 | N.D. |
| omt1-2/At C4H:Sm COMT (1) | 16 ± 5 |
| omt1-2/At C4H:Sm COMT (2) | 20 ± 5 |
| omt1-2/At C4H:Sm COMT (3) | 22 ± 4 |
| omt1-2/At C4H:Sm COMT (4) | 24 ± 5 |
N.D., not detectable; ± represents 1 sd for biological triplicates.
The unique structure of the Selaginella genome locus containing Sm COMT and Sm F5H suggests that the expression of these two genes may be coregulated by common cis-regulatory elements located in the same promoter region. To test the hypothesis that these cis-regulatory elements related to S lignin biosynthesis in Selaginella could be conserved in all vascular plants and are recognizable by the lignification-associated transcription factors in angiosperms, we transformed a Selaginella genomic fragment harboring Sm COMT, Sm F5H, and their 3′-downstream regions into the Arabidopsis double mutant of omt1-2 and the F5H-null fah1-2 and looked for complementation. However, none of the T1 transgenic plants exhibited the complemented lignin phenotype (see Supplemental Figure 3 online), suggesting that the cis-regulatory elements in the promoter region of Sm COMT and Sm F5H are probably not conserved between Selaginella and Arabidopsis.
Enzyme Kinetic Analysis of Sm COMT
To assess the substrate specificity of Sm COMT, we expressed N-terminal hexahistidine-tagged Sm COMT in Escherichia coli, purified the tagged protein using nickel affinity chromatography, and performed kinetic assays using a range of meta-hydroxylated phenylpropanoid pathway intermediates, including caffeic acid, caffealdehyde, caffeyl alcohol, 5-hydroxyferulic acid, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol. At COMT was also expressed, purified, and assayed in parallel for comparison. The kinetic constants were inferred from these assays and summarized in Table 3 (see Supplemental Figures 4 and 5 online.
Table 3.
Kinetic Properties of Recombinant Sm COMT and At COMT toward meta-Hydroxylated Phenylpropanoid Intermediates
| At COMT |
Sm COMT |
|||||
| Substrate | Km (μM) | Vmax (nkat·mg−1) | Vmax/Km | Km (μM) | Vmax (nkat·mg−1) | Vmax/Km |
| Caffeic acid | 8.05 × 102 | 16.3 | 2.02 × 10−2 | 5.55 × 103 | 0.422 | 7.60 × 10−5 |
| Caffealdehyde | 0.914 | 4.88 | 5.34 | 12.35 | 1.68 | 0.136 |
| Caffeyl alcohol | 7.70 | 11.5 | 1.49 | 1.17 | 4.45 | 3.80 |
| 5-OH-ferulic acid | 2.25 × 102 | 14.5 | 6.44 × 10−2 | 4.24 × 103 | 0.951 | 2.24 × 10−4 |
| 5-OH-coniferaldehyde | 2.74 | 23.0 | 8.39 | 6.97 | 6.21 | 0.891 |
| 5-OH-coniferyl alcohol | 4.91 | 21.4 | 4.36 | 0.784 | 4.80 | 6.12 |
At COMT shows kinetic properties comparable to those previously reported for M. sativa COMT (Ms COMT) (Parvathi et al., 2001). Caffealdehyde and 5-hydroxyconiferaldehyde are the preferred substrates for At COMT, with their Km values at the lowest among all the tested substrates. Caffeyl alcohol and 5-hydroxyconiferyl alcohol are less preferable substrates, with their Vmax/Km values slightly lower than those for the aldehydes. Caffeic acid and 5-hydroxyferulic acid are poor substrates for At COMT, with their Vmax/Km values at hundreds of times lower than those of their corresponding aldehydes and alcohols.
As they are for At COMT, caffeic acid and 5-hydroxyferulic acid are poor substrates for Sm COMT, but with Vmax/Km values even lower than those values for At COMT. At the aldehyde and alcohol level, Sm COMT shows kinetic constants in a range generally comparable to those of At COMT, although Sm COMT exhibits higher catalytic efficiency with alcohols as substrates, exhibiting both lower Km values and higher Vmax values. These results indicate that Sm COMT has a catalytic capacity comparable to those of its angiosperm counterparts in mediating methylation reactions on 3-hydroxylated or 5-hydroxylated lignin biosynthetic precursors at the aldehyde and alcohol levels.
Interestingly, we also observed that for caffeyl alcohol and 5-hydroxyconiferyl alcohol assays, Sm COMT may exhibit substrate inhibition toward the two alcohols, a phenomenon that was not seen with other substrates or when At COMT was assayed with any substrate (see Supplemental Figures 4 and 5 online).
To test whether Sm COMT can also methylate hydroxycinnamoyl CoA esters, we performed enzyme assays using 5-hydroxyferuloyl CoA, a potential methyl acceptor, with a concentration series ranging from 100 to 1 μM. However, no methyltransferase activity could be detected in these assays, suggesting that hydroxycinnamoyl CoA esters are unlikely to be relevant substrates for Sm COMT in vivo.
To further characterize COMT in Selaginella, we extracted total soluble protein from S. moellendorffii whole-plant tissue and assayed its activity toward a range of potential COMT substrates. Unfortunately, these compounds were degraded by an unknown enzymatic activity present in the crude extract, possibly polyphenol oxidases or diooxygenases, which are known to metabolize catechol-substituted phenolic substrates (Prescott and John, 1996; Marusek et al., 2006). To solve this problem, we fractionated the Selaginella crude protein extract by size exclusion followed by ion exchange chromatography. This procedure yielded a fraction in which the COMT activity was free from the confounding activities described above, facilitating detailed kinetic analysis. The kinetic constants measured using this Selaginella enzyme fraction against caffeic acid, caffealdehyde, caffeyl alcohol, 5-hydroxyferulic acid, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol are comparable to the values determined using recombinant Sm COMT (see Supplemental Table 1 online), consistent with the hypothesis that the protein we have characterized in recombinant form is the dominant COMT in Selaginella.
Tissue-Specific Expression of Sm COMT
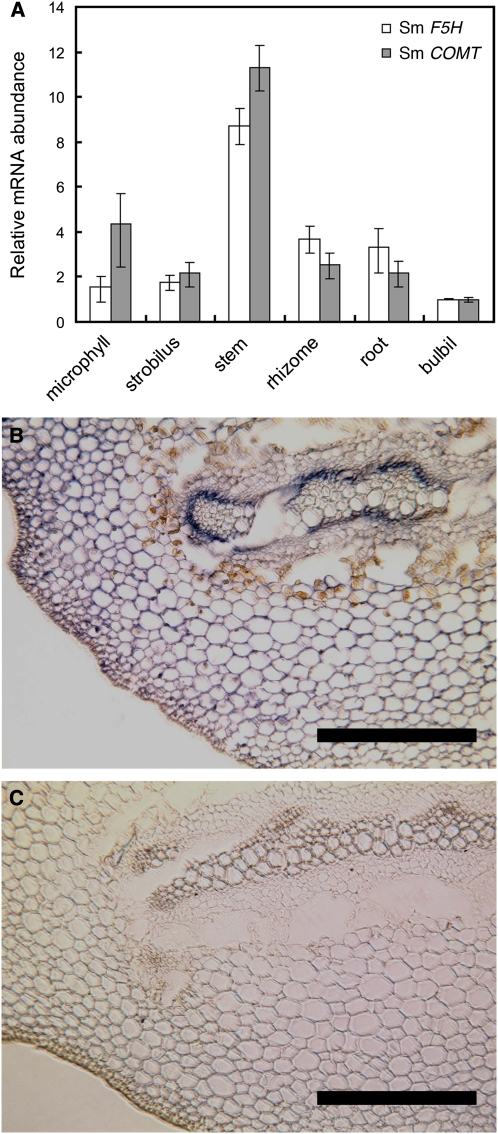
To evaluate the tissue specificity of Sm COMT expression in Selaginella, we first conducted a quantitative RT-PCR (qRT-PCR) experiment using RNA extracted from various tissues, including microphyll, strobilus, stem, rhizome, root, and bulbil. Across the six tissue types, Sm COMT is expressed at the highest level in the stem tissue, consistent with its role in lignin biosynthesis (Figure 6A). We also examined the mRNA abundance of Sm F5H in parallel, which revealed a similar tissue transcript distribution pattern to that of Sm COMT (Figure 6A), consistent with the hypothesis that these two genes are coregulated.
Figure 6.
Expression Pattern of COMT in S. moellendorffii.
(A) qRT-PCR analysis of transcript abundance of Sm COMT and Sm F5H in various tissue types. Error bars represent 1 sd of biological triplicates.
(B) and (C) In situ hybridization of Sm COMT mRNAs in Selaginella transverse sections using antisense (B) and sense (C) Sm COMT probes. Bars = 200 μm.
To further investigate the expression pattern of Sm COMT in stem, we preformed in situ hybridization experiments on Selaginella stem cross sections. While the antisense probe gave hybridization signals in the cortical cells where S lignin is predominantly deposited (Figure 6B), the sense control probe did not give any hybridization signal (Figure 6C). Notably, the hybridization signal was also observed in the phloem cells surrounding the xylem. The transcript localization pattern of Sm COMT in stem observed in this study is almost identical to that of Sm F5H as previously reported (Weng et al., 2008b), which provides further evidence to support the contention that these two genes may be coregulated.
Phylogenetic Analysis of Sm COMT
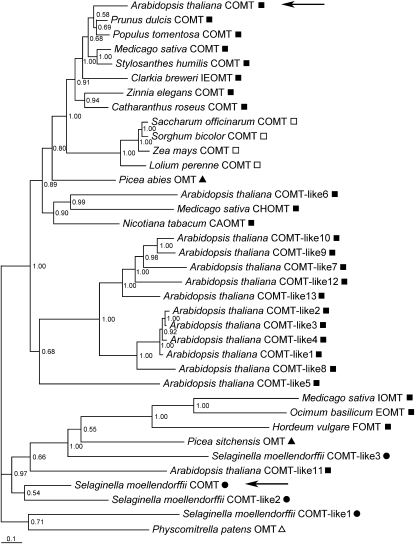
To infer the phylogeny of Sm COMT within the plant OMT family, we performed Bayesian phylogenetic analysis using Sm COMT, angiosperm COMTs, angiosperm OMTs with known functions that can methylate various types of phenylpropanoids, and some OMTs from different land plant lineages with unknown functions (Figure 7; see Supplemental Data Set 1 online). Although all the angiosperm COMTs are clustered together into a clade, sister to a group containing chalcone OMT from alfalfa and catechol OMT from tobacco (Nicotiana tabacum), Sm COMT falls into another clade only distantly related to the angiosperm COMT clade. The OMTs related to Sm COMT in this clade include flavone/isoflavone 7-OMTs from barley (Hordeum vulgare) and alfalfa, eugenol OMT from sweet basil (Ocimum basilicum), two unknown OMTs from Sitka spruce (Picea sitchensis) and Arabidopsis, as well as Sm COMT-like2 and Sm COMT-like3. An unknown OMT from Physcomitrella patens, being the only COMT homolog found in its genome, clustered together with Sm COMT-like1, which could represent the ancestral form of this enzyme family that is highly diversified in higher plants. The observations suggest that COMT may have been recruited for S lignin biosynthesis independently in angiosperms and Selaginella.
Figure 7.
Bayesian Phylogenetic Tree of Sm COMT, Angiosperm COMTs, and Related OMTs from Various Plant Lineages.
Bayesian posterior probabilities are indicated at each node. The scale measures evolutionary distance in substitutions per amino acid. The taxonomy information of the sequences is indicated by the symbols at the right of the gene names (filled square, dicot; open square, monocot; filled triangle, gymnosperm; filled circle, lycophyte; open triangle, bryophyte). CAOMT, catechol O-methyltransferase; CHOMT, chalcone O-methyltransferase; EOMT, eugenol O-methyltransferase; FOMT, flavonoid 7-O-methyltransferase; IEOMT, (iso)eugenol O-methyltransferase; IOMT, isoflavone O-methyltransferase.
Sequence Analysis and Modeling of Sm COMT
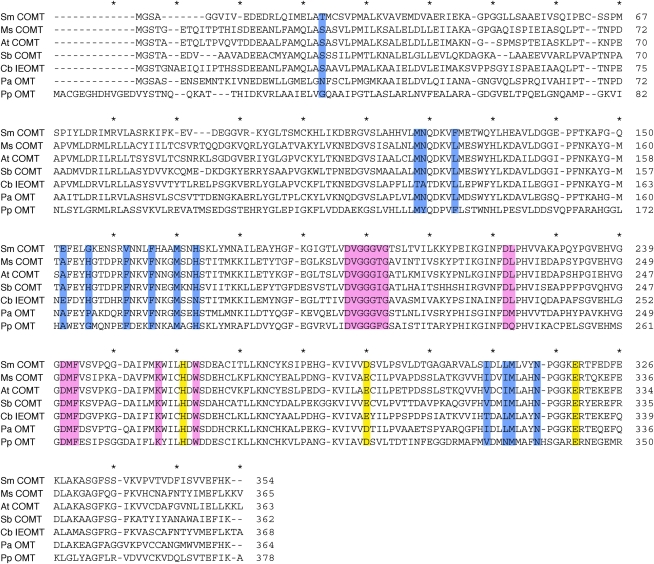
To gain more insight into the evolution and structure-function relationships within the COMT family, we compared Sm COMT with other related proteins in a multiple sequence alignment (Figure 8). This analysis showed that the three catalytic residues, His-269, Glu-297, and Glu-329, previously identified in Ms COMT were highly conserved in Sm COMT (Zubieta et al., 2002), except that the E297 in Ms COMT was substituted with an Asp residue. The residues for SAM binding are also highly conserved between Sm COMT and other OMTs. Whereas angiosperm COMT orthologs are almost entirely conserved in the residues that constitute the methyl acceptor binding pocket, Clarkia breweri IEOMT, a derived OMT paralogous to COMT, shows many nonconservative substitutions at these residues, consistent with its distinct activity as a phenylpropanoid para-hydroxyl O-methyltransferase (Wang and Pichersky, 1998). Although Sm COMT is catalytically equivalent to angiosperm COMTs, major nonconservative substitutions (Leu-136, Ala-162, His-166, and Phe-172 in Ms COMT substituted to Phe-126, Glu-152, Gly-156, and Val-162 in Sm COMT) were found in the corresponding substrate binding residues in Sm COMT, which further supports the notion that the substrate specificity of Sm COMT arose independently from that of angiosperm COMTs.
Figure 8.
Sequence Alignment Analysis of Sm COMT, Together with Two Angiosperm COMTs, and Related OMTs.
Based on the crystal structure of Ms COMT, the residues involved in different aspects of the enzymatic process are highlighted in color. Yellow, catalytic residues; pink, SAM binding residues; blue, substrate binding residues at the active site. Asterisks are placed on top of every tenth site in the alignment. At, Arabidopsis thaliana; Cb, Clarkia breweri; Ms, Medicago sativa; Sb, Sorghum bicolor; Sm, Selaginella moellendorffii; Pa, Picea abies; Pp, Physcomitrella patens.
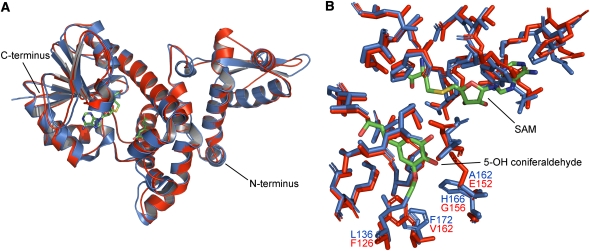
We then generated a Sm COMT structural model based on the Ms COMT crystal structure (Zubieta et al., 2002) and compared the two structures in a spatial manner (Figure 9). The Sm COMT model shares a similar tertiary structure with Ms COMT, including the N-terminal domain responsible for dimerization in Ms COMT (Figure 9A), suggesting that Sm COMT is likely to exist as dimer in solution like other previously reported plant OMTs (Gang et al., 2002; Zubieta et al., 2002). The Sm COMT model contains a methyl acceptor binding pocket similar to that of Ms COMT in overall geometry (Figure 9B). Interestingly, the four nonconservative substitutions in the Sm COMT active site mentioned above form two pairs of spatially contacting residues (F126-V162 and E152-G156), each of which exchange two residues of similar character in reverse orientation, likely resulting in similar spatial filling (Figure 9B). This observation further suggests that, at the molecular level, angiosperms and Selaginella have adopted independent evolutionary solutions toward convergent COMT enzymatic activity geared for lignin biosynthesis.
Figure 9.
Molecular Modeling of Sm COMT.
(A) Tertiary structure of the Sm COMT model (red) superimposed on Ms COMT (blue). The ligands of the template Ms COMT structure, S-adenosyl-l-homocysteine and 5-OH coniferaldehyde, are shown as sticks (green, carbon; red, oxygen; blue, nitrogen; yellow, sulfur).
(B) Putative Sm COMT active site (red) overlaid with Ms COMT active site. Nonconservative differences in those potential substrate binding residues are emphasized in text.
Characterization of Mutated At COMT and Sm COMT
From the modeling study, we noticed that the position of a pair of residues, His-166 and Ala-162, in Ms COMT in the vicinity of the para-hydroxyl of the phenylpropanoid substrate, may be occupied by Glu-152 and Gly-156 in Sm COMT (Figure 9B). It has been previously suggested that His-166 in Ms COMT could hydrogen bond with the para-hydroxyl moiety of the substrate, contributing to the proper substrate positioning (Zubieta et al., 2002). However, the replacement of this residue with a Glu residue in Sm COMT raises the possibility that His-166 in Ms COMT and/or Glu-152 in Sm COMT may serve as a general base that could deprotonate the substrate para-hydroxyl group to further facilitate the transfer of a methyl group to the meta-hydroxyl position. To test whether these two residues are essential for catalysis, we first generated At COMT-H164A (corresponding to His-166 in Ms COMT), Sm COMT-E152A and Sm COMT-E152Q mutants using site-directed mutagenesis, expressed the mutant proteins in E. coli, and analyzed their kinetic properties against caffeyl alcohol. We found that all three mutants retained OMT catalytic activity, with the catalytic efficiency of At COMT-H164A and Sm COMT-E152Q slightly compromised (Table 4). These results disproved our initial hypothesis and indicated that His-164 in At COMT and Glu-152 in Sm COMT are dispensable for catalysis.
Table 4.
Kinetic Analysis of Wild-Type and Various Mutant Forms of Sm COMT and At COMT toward Caffeyl Alcohol
| Protein | Km (μM) | Vmax (nkat·mg−1) | Vmax/Km |
| At COMT wild type | 7.70 | 11.5 | 1.49 |
| At COMT-H164A | 15.4 | 6.71 | 0.436 |
| At COMT-A160E/H164G | 23.0 | 0.493 | 2.14 × 10−2 |
| At COMT-L134F/F170V | 27.2 | 1.44 | 5.29 × 10−2 |
| Sm COMT wild type | 1.17 | 4.45 | 3.80 |
| Sm COMT-E152A | 1.42 | 7.10 | 5.00 |
| Sm COMT-E152Q | 2.49 | 3.30 | 1.33 |
| Sm COMT-E152A/G156H | 1.49 | 3.77 | 2.53 |
| Sm COMT-F126L/V162F | 1.09 | 5.97 | 5.48 |
The sequence alignment and modeling study suggested four active site residues differ between an angiosperm COMT and Sm COMT. To test whether these residues are interchangeable between an angiosperm COMT and Sm COMT, we generated mutated versions of At COMT carrying A160E/H164G and L134F/F170V substitutions, such that the mutated residues correspond to those in Sm COMT. Similarly reciprocally mutated versions of Sm COMT, Sm COMT-E152A/G156H and Sm COMT-F126L/V162F, were also generated. The kinetic analysis of these mutants toward caffeyl alcohol revealed that the Sm COMT mutants substituted with the At COMT residues are almost unaffected in kinetic constants. On the contrary, replacing At COMT with the corresponding residues from Sm COMT drastically reduced the catalytic efficiency of the enzyme, which could be due to the destabilizing effect of these substitutions in the At COMT sequence context (Table 4).
DISCUSSION
S Lignin Biosynthesis in Angiosperms and Selaginella Evolved through Convergent Evolution
Our understanding of the lignin biosynthetic pathway has been primarily based on decades of research in economically important crop species and trees, as well as the model plant Arabidopsis, all of which are flowering plants (Boerjan et al., 2003; Li et al., 2008). The identification of a bifunctional COMT from Selaginella described in this study, in addition to the previous discovery of the dual meta-hydroxylase F5H from the same species (Weng et al., 2008b, 2010b), indicate that Selaginella has adopted a biochemical pathway distinct from that in angiosperms to synthesize sinapyl alcohol. In contrast with angiosperms, where S lignin biosynthesis is entirely dependent on the availability of G-substituted intermediates, coniferaldehyde and coniferyl alcohol, Selaginella appears to direct H-substituted intermediates, p-coumaraldehyde and p-coumaryl alcohol, toward S lignin biosynthesis via the bifunctional F5H and COMT (Figure 1).
Enzymes that catalyze the same reaction in different organisms are usually encoded by orthologous genes; however, there are cases where the converged enzymatic activities were derived independently (Galperin et al., 1998). For example, fructose 1,6-bisphosphate aldolase in yeast (Saccharomyces cerevisiae) and rabbit (Oryctolagus cuniculus) muscle are capable of catalyzing the same reaction but are only distantly related (Warburg and Christian, 1943; Cooper et al., 1996). In plant secondary metabolism, limonene synthase was found to have arisen independently in angiosperms and gymnosperms (Bohlmann et al., 1998), and of particular relevance to this study, eugenol OMT evolved independently in C. breweri and sweet basil (Gang et al., 2002) (Figure 7). Our phylogenetic analysis suggests that Sm COMT belongs to a plant OMT clade divergent from the clade that contains angiosperm COMTs (Figure 7). The fact that many nonconservative substitutions exist in the substrate binding site of Sm COMT compared with its angiosperm counterparts further supports the independent origins of lignin biosynthetic COMT activities in Selaginella and angiosperms. Unfortunately, gene knockout technology is not currently available in Selaginella, so it is not possible to conduct the loss-of-function experiment to determine whether any OMTs in addition to Sm COMT function in S lignin synthesis in Selaginella. Specifically, we cannot exclude the possibility that other OMTs, widely diverged from angiosperm COMTs, escaped notice during our bioinformatic analysis of the Selaginella genome. Nevertheless, Sm COMT, but not the other three Selaginella OMT homologs, is a functional COMT and is a good candidate for the one that functions in S lignin biosynthesis in Selaginella.
Interestingly, the in vitro enzyme assays revealed that Sm COMT is not entirely identical to its angiosperm counterparts in terms of kinetic properties. Whereas we and others have shown that angiosperm COMTs exhibit higher catalytic efficiency toward meta-hydroxylated phenylpropanoid aldehydes than alcohols (Parvathi et al., 2001; Zubieta et al., 2002), alcohols are superior substrates for Sm COMT, which could be related to the different roles these enzymes have in S lignin biosynthesis, the angiosperm enzymes acting only once in the pathway, and the Selaginella enzyme acting twice. Furthermore, Sm COMT may exhibit substrate inhibition toward caffeyl alcohol and 5-hydroxyconiferyl alcohol, a property not observed in At COMT or in the previously characterized Ms COMT (Parvathi et al., 2001); however, the importance of this phenomenon has yet to be investigated.
The Genes for S Lignin Biosynthesis in S. moellendorffii Are Clustered
The clustering of genes, frequently in operons, is a common feature of prokaryotic genomes (Salgado et al., 2000). By contrast, gene clustering that can produce polycistronic mRNA is generally absent in the genomes of higher eukaryotes (Blumenthal, 1998). There are occasional cases where functionally related genes are arranged closely at a certain genomic locus in higher eukaryotes and are transcriptionally coregulated. For example, the Arabidopsis genome contains an operon-like gene cluster composed of four metabolic genes that are involved in triterpene thalianol biosynthesis (Field and Osbourn, 2008). In maize (Zea mays), five genes required for the biosynthesis of the phytoalexin 2,4-dihydroxy-1,4-benzoxazin-3-one are clustered on chromosome 4 (Frey et al., 1997). Although such operon-like gene clusters do not generate polycistronic mRNA, the individual genes in these clusters are coordinately regulated. In Selaginella, the two genes involved in S lignin biosynthesis, F5H and COMT, are arranged at the same genomic locus in a gene cluster. The two genes share a common upstream region and may be under the control of common cis-regulatory elements. Consistent with this model, the two genes show similar tissue- and cell-specific expression patterns. It is noteworthy that such regulation coordination appears not to exist for angiosperm F5H and COMT. In Arabidopsis, β-glucuronidase expression driven by the COMT promoter is strongly targeted to xylem and is also present in interfascicular fibers and mature phloem in stem (Goujon et al., 2003), whereas β-glucuronidase expression in Arabidopsis stems driven by the F5H promoter together with its 3′ downstream regulatory region (Ruegger et al., 1999) localizes primarily in the interfascicular fiber cells (J. Humphreys and C. Chapple, unpublished data). An alternative explanation for the clustering may come from the field of population genetics: The origin of these operon-like gene clusters could be initially driven by the greater likelihood of fixation of multiple semidominant or dominant traits in populations if they are linked (Barton, 2000).
The discovery of a gene cluster for S lignin biosynthesis in the Selaginella genome raises the question of whether similar gene clustering is common in the Selaginella genome. Indeed, we have identified another gene cluster that contains homologs of C4H and chalcone isomerase, at least if their apparent homology is indicative of their function. These data suggest that there may be opportunities for elucidating gene function in Selaginella based upon clustering of genes of known function with unknown genes that may function in the same pathway.
Involvement of Different Lignin Biosynthetic Pathways in Selaginella Stem Cortex and Xylem
In angiosperms, it has been shown by qualitative UV microscopy, histochemical staining, and cell type–specific lignin analysis that lignin monomer distribution varies in different cell types in vascular tissue (Musha and Goring, 1975; Saka and Goring, 1988; Chapple et al., 1992; Nakashima et al., 2008). In general, lignin in xylem cells is dominated by G units, relative to interfascicular fiber cells where both G and S lignin units are present. Such observations have been attributed to the cell-specific expression of lignin biosynthetic genes and consequently the cell-specific recruitment of different branches of the lignin biosynthetic pathway toward different monomers (Meyer et al., 1998; Chen et al., 2000). The unique protostelic structure of Selaginella stem has allowed us to separate the two lignified cell types, xylem and cortex, with relative ease. We showed that Selaginella xylem, like angiosperm xylem, also contains lignin composed of almost entirely G units, whereas cortical cells, analogous to angiosperms’ interfascicular fiber cells, contain lignin with a high S/G ratio. Such a lignin distribution pattern is consistent with the in situ localization of Sm F5H and Sm COMT transcripts in the cortex, suggesting that the S lignin biosynthetic pathway mediated by Sm F5H and Sm COMT is primarily active in the cortex.
Interestingly, we found that H lignin units are present only in the cortex where high levels of S lignin are deposited but are absent in xylem where S lignin exists only in trace amounts. This observation suggests that a pool of H-substituted intermediates may be available in the cortex where it can be readily diverted toward S monolignol biosynthesis by the combined activities of Sm F5H and Sm COMT (Figure 1). Although Sm F5H and Sm COMT can catalyze the hydroxylation reaction and O-methylation on both of the two meta-positions of lignin biosynthetic precursors, the G-rich lignin in xylem suggests that an additional independent phenylpropanoid 3-hydroxylation and 3-O-methylation pathway for G lignin biosynthesis, independent of at least Sm F5H and possibly Sm COMT, exists in the xylem. Presumably, such reactions could be catalyzed by C3′H and caffeoyl-CoA O-methyltransferase in a similar fashion as in angiosperms, as putative orthologs of these genes are present in the Selaginella genome; however, their exact biochemical functions in lignin biosynthesis are still to be elucidated in Selaginella.
The Chemical Characteristics of G and S Lignin May Impact Plant Physiology
We showed that Selaginella xylem, similar to angiosperm xylem, is rich in G lignin, which further supports the importance of G lignin in water transport (Donaldson, 2001). The structural characteristics of the G-rich lignin extracted from Selaginella xylem revealed in this study, together with previous studies (Ralph et al., 2004), indicate that G monomers can form C–C linkages through the 5-position of the aromatic ring, such as in structures B and D shown in Figure 3B, resulting in a more compact and hydrophobic lignin polymer. These properties of G lignin probably imbue the plant cell wall with properties more suitable for water transport than does S lignin. The fact that the transgenic F5H-overexpressing Arabidopsis, with a lignin composed of essentially no G units, shows a modest collapsed xylem phenotype further implicates the unique role of G lignin in water transport (Weng et al., 2010a).
Although S lignin has arisen independently in Selaginella and angiosperms, it is deposited mainly in anatomically analogous tissues, such as cortical and interfascicular fiber cells, but not in xylem cells. Convergent evolution at not only the chemical level but also at the cell-specific distribution level implies that S lignin in Selaginella and angiosperms provides similar selective advantages in each lineage. For example, the ability to synthesize S lignin might confer better mechanical strength to the vascular tissue in angiosperms (Li et al., 2001). It has also been suggested that the enhanced mechanical support provided by S lignin could be important in the maintenance of structural integrity during drought stress (Micco and Aronne, 2007). Interestingly, the occurrences of vessel elements in Selaginella, angiosperms, and the S lignin–containing gymnosperm species under the order of Gnetales are thought to be derived via convergent evolution as well (Duerden, 1934; Logan and Thomas, 1985; Carlquist, 1996). The independent occurrence of S lignin in the Selaginella cortex and angiosperm fiber cells might also have provided the overall surrounding physical strength that allows the development of large water-conducting vessel elements in xylem.
METHODS
Plant Materials
Selaginella moellendorffii was obtained from Plant Delights Nursery and grown in a local greenhouse under 50% shade cloth. Arabidopsis thaliana was grown under a 16-h-light/8-h-dark photoperiod at 100 μE·m−2·s−1 at 22°C. The Arabidopsis COMT T-DNA insertion mutant omt1-2 was ordered from the ABRC under accession number CS25167 (Alonso et al., 2003).
Scanning Electron Microscopy
Selaginella stems were cut into ~3-mm lengths and mounted in a vice cryo holder using cryo-mount adhesive. Samples were plunged into liquid nitrogen and then transferred under vacuum to the cryo prechamber. They were fractured, sublimated for a total of 25 min at −85°C, and then coated with platinum for 120 s prior to moving to the main chamber cryostage. The samples were imaged with an FEI NOVA nanoSEM FESEM operating at 3 or 5 kV, aperture 6, spot size 3, and 4- to 16-mm working distance, and were held at −120°C during imaging.
DFRC Lignin Analysis
Cell wall samples were prepared as previously described by grinding plant stem tissue to a fine powder in liquid nitrogen followed by extraction in neutral phosphate buffer, 80% ethanol, and acetone (Meyer et al., 1998). The DFRC lignin analysis was performed essentially as previously reported (Lu and Ralph, 1998). Briefly, cell wall samples were digested in acetyl bromide/acetic acid solution containing 4,4′-ethylidenebisphenol as an internal standard. The reactions were dried down using N2 gas, dissolved in dioxane/acetic acid/water (5/4/1, v/v/v), subjected to reductive cleavage with Zn dust, purified with C-18 SPE columns (SUPELCO), and acetylated with pyridine/acetic anhydride (2/3, v/v). The lignin derivatives were quantified by gas chromatography with flame ionization detection using response factors relative to the internal standard of 1.30 for coniferyl alcohol diacetate and 1.44 for sinapyl alcohol diacetate.
NMR Lignin Analysis
For NMR lignin analysis, cell wall samples were preground using a Retsch MM301 shaker mill for 3 min at 30 Hz and extracted sequentially with water, 80% methanol, acetone, chloroform-acetone (1/1, v/v), and acetone again. The obtained isolated cell walls were ball-milled for 1.0 h for cortex and for 0.5 h for xylem (in 20 min on/10 min off cycles) using a Retsch PM100 ball mill running at 600 rpm with zirconium dioxide vessels (50 mL) containing ZrO2 ball bearings (10 × 10 mm). The ball milled walls were then digested at 30°C with crude cellulases (Cellulysin; Calbiochem), 30 mg/g of sample, in pH 5.0 acetate buffer for 2 d to generate cellulolytic enzyme lignin (CEL) (Chang et al., 1975). The CEL fraction was then completely dissolved in DMSO/N-methylimidazole (2/1, v/v) (Lu and Ralph, 2003). Following acetic anhydride addition, acetylated enzyme lignins were obtained for NMR spectroscopy.
NMR spectra of acetylated CEL samples in CDCl3 were acquired at 300 K on a 750-MHz (DMX-750) Bruker Biospin instrument equipped with a sensitive cryogenically cooled 5-mm TXI 1H/13C/15N gradient probe with inverse geometry. The central solvent peak was used as an internal (δC 77.0, δH 7.26 ppm). All processing and integration calculations were conducted using Bruker Biospin’s TopSpin v. 2.1 software. An adiabatic HSQC experiment (hsqcetgpsisp) was chosen for its superior phasing and peak shapes as well as uniform reduced J dependence excitation (Kupce and Freeman, 2007). The following parameters were used: 16 transient spectral increments were acquired from 10 to 0 ppm in F2 (1H) using 3002 data points for an acquisition time of 200 ms, an interscan delay of 1 s (for a total scan recycle time of 1.2 s), 170 to 0 ppm in F1 (13C) using 512 increments (F1 acquisition time: 8 ms), with a total acquisition time of 7 h. 13C Decoupling during acquisition was performed by GARP composite pulses from the high-power output decoupling channel. Processing to a final matrix of 2k × 1k points used typical matched Gaussian apodization in F2 (line broadening = −0.15, Gaussian broadening factor = 0.001) and a squared cosine-bell in F1. Lignin assignments were via comparison with previously assigned spectra (Lu and Ralph, 2003; Ralph et al., 2006).
Cloning of Sm COMT and Sm COMT-Like Genes
The Sm COMT cDNA corresponding to the open reading frame (ORF) region was cloned by RT-PCR using a gene-specific primer pair, cc1812-cc1813, and A-T cloned into pGEM T-Easy vector (Promega) to generate pCC0941. The ORF regions of Sm COMT-like1 cDNA and Sm COMT-like2 cDNA were RT-PCR amplified using the gene-specific primer pair cc2139-cc2140 and cc2141-cc2142, respectively. Sm COMT-like3 could not be amplified by RT-PCR, using RNA extracted from Selaginella whole plant, indicating it may not be normally expressed. Therefore, the genomic DNA corresponding to the ORF region of Sm COMT-like3 was PCR amplified with the gene-specific primer pair cc2143-cc2144. The resulting PCR products of the three Sm COMT-like genes were recombined with pCC1155, a Gateway entry vector modified from pDONR 221 (Invitrogen), to generate entry clone pCC1275, pCC1276, and pCC1277, respectively. Detailed information for primers used in this research is summarized in Supplemental Table 2 online.
Transgenic Arabidopsis
To generate the At C4H:Sm COMT construct, pCC0966, the Sm COMT ORF, was PCR amplified from template pCC0941 using primer pair cc1842-cc1841. The amplicon was digested with MfeI and ligated into EcoRI-digested pCC0964. To generate the base binary vector pCC0964 for expressing Sm COMT in planta under the control of the Arabidopsis C4H promoter, the 2977-bp Arabidopsis C4H promoter was SalI and EcoRI released from pCC0916 (Weng et al., 2008b) and ligated into SalI- and EcoRI-digested pCAMBIA1390 (CAMBIA). To generate At C4H:Sm COMT-like1, At C4H:Sm COMT-like2, and At C4H:Sm COMT-like3 constructs, pCC1275, pCC1276, and pCC1277 were recombined with pCC0996, a modified version of pCC0916 with a Gateway cassette and Basta selection marker in planta, to form destination constructs designated as pCC1302, pCC1303, and pCC1304 respectively. To generate a binary vector containing the Sm F5H-Sm COMT genomic locus, a 9050-bp Selaginella genomic fragment, containing Sm F5H, Sm COMT, and their respective 3′-downstream regions, was PCR amplified from the genome using the primer pair cc1931-cc1932. The amplicon was digested with NotI and ligated into NotI-digested pCC1122, a Gateway entry vector modified from pDONR 221 (Invitrogen), to form the entry clone pCC1127. pCC1127 was subsequently recombined with pCC1136, a promoterless Gateway binary vector modified from the backbone of pBI101.2, to generate the destination construct pCC1150. Constructs were introduced into Agrobacterium tumefaciens C58 pGV3850 by electroporation and then transformed into omt1-2 using the floral dip method (Weigel and Glazebrook, 2002).
Leaf-Soluble Phenylpropanoid Analysis
Three-week-old Arabidopsis rosette leaves were extracted with 50% methanol and analyzed by reverse-phase HPLC. Leaf extracts were separated on a Microsorb-MV C18 column (Ranin Instruments) using a gradient from 1.5% acetic acid to 35% acetonitrile in 1.5% acetic acid at a flow rate of 1 mL·min−1.
Histochemistry
Mäule’s staining of lignin in Arabidopsis was conducted as described (Chapple et al., 1992). Briefly, hand sections of Arabidopsis stem were fixed in 4% glutaraldehyde, rinsed in water, and treated for 10 min with 0.5% KMnO4. Sections were then rinsed with water, treated for 5 min with 10% HCl, rinsed in water, mounted in concentrated NH4OH, and examined by dark-field microscopy.
qRT-PCR
Total RNA was extracted from various tissue types of Selaginella using the RNeasy plant mini kit (Qiagen). RT reactions were performed using ImProm-II reverse transcriptase (Promega) for each tissue type following the protocol provided by the manufacturer. The resulting cDNA was treated with RNase and used as template for real-time PCR. Quantitative real-time PCR using SYBR Green was performed on the StepOne Real-Time PCR system (Applied Biosystems) using the ΔΔCT method with the default cycling program. Primer pair cc2616-cc2617 was used for Sm F5H and cc2618-cc2619 was used for Sm COMT, whereas cc2620-cc2621 was used for Sm ACTIN as internal standard. All the primer pairs have an amplification efficiency of higher than 90%.
In Situ Hybridization
To examine the localization of Sm COMT mRNA in S. moellendorffii stem, 8-μm sections of paraffin-embedded S. moellendorffii stem were subjected to in situ hybridization as previously described (Vielle-Calzada et al., 1999). To generate Sm COMT antisense or sense probes, pCC0941 was linearized with NcoI or NdeI and transcribed from the SP6 promoter or the T7 promoter, respectively, using the SP6/T7 transcription kit (Roche Applied Science).
COMT Expression, Purification, and Mutagenesis
To generate N-terminally 6×His tagged Sm COMT and At COMT, the ORF of Sm COMT or At COMT was PCR amplified from template pCC0941 or an At COMT cDNA clone 154J19T7 (Zhang et al., 1997) using primer pair cc1822-cc1823 or cc2316-2317. The PCR products were digested with NheI and HindIII and ligated into XbaI and HindIII digested pET28a(+) (Novagen) to generate pCC0956 [Sm COMT-pET28a(+)] and pCC1484 [At COMT-pET28a(+)], respectively.
Various Sm COMT and At COMT mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) following the procedure instructed in the manual. The Sm COMT-E152A, G156H mutant was generated using primer cc2321; the Sm COMT-F126L, V162F mutant was generated using primer pair cc2322-cc2323; the Sm COMT-E152Q mutant was generated using primer cc2324; the Sm COMT-E152A mutant was generated using primer cc2381; the At COMT-A160E/H164G mutant was generated using primer cc2318; the At COMT- L134F/F170V mutant was generated using primer pair cc2319-cc2320; and the At COMT-H164A mutant was generated using primer cc2380.
Constructs containing Sm COMT, At COMT, and their mutants were transformed into Escherichia coli strain BL21 (DE3). Transformed E. coli were grown at 37°C in Luria-Bertani medium containing 100 μg/mL ampicillin until OD600 reached 0.6. After induction with 0.4 mM isopropyl β-d-1-thiogalactopyranoside, the cultures were grown at 25°C for another 10 h. The E. coli cells were pelleted by centrifugation and resuspended in lysis buffer (50 mM NaCl and 20 mM Tris-HCl, pH 8.0), in which lysozyme and DNaseI were added to lyse the cells for half an hour at room temperature. The lysis solution was frozen overnight at −80°C before centrifugation to retrieve the supernatant. The supernatant was passed through a HiTrap chelating HP column (GE Healthcare) fitted on a FPLC system (GE Healthcare) and washed with 10 bed volume of rinsing buffer (50 mM NaCl, 20 mM Tris-HCl, pH 8.0, and 20 mM imidazole), and the His-tagged protein was eluted using elution buffer (50 mM NaCl, 20 mM Tris-HCl, pH 8.0, and 250 mM imidazole). The fractions containing target protein were pooled, desalted over a Zeba Spin Desalting Column (Thermo Fisher Scientific), and stored at −80°C. The protein concentration was determined using the Bradford assay (Bio-Rad). The protein was diluted to an appropriate concentration for kinetic assays in COMT assay buffer (100 mM Tris-HCl, pH 7.5, 0.2 mM MgCl2, and 20% glycerol).
Preparation and Fractionation of Total Soluble Protein from S. moellendorffii
S. moellendorffii whole-plant tissue (10 g fresh weight) was ground to fine powder with mortar and pestle under liquid nitrogen, to which 30 mL extraction buffer (50 mM sodium PIPES, pH 7.0, containing 20% glycerol and 4 mM EDTA) was added. The mixture was incubated at 4°C for 15 min with gentle stirring, filtered through Miracloth (Calbiochem), and centrifuged at 12,000g to obtain clear supernatant. A sample of the total soluble protein extract was desalted using a PD-10 desalting column (GE Healthcare). The total protein extract was first fractionated using gel exclusion chromatography with a Superdex 200 HR26/60 column (Pharmacia Biosystems). The fractions containing specific COMT activity against 5-hydroxyconiferaldehyde were collected, dialyzed, and further fractionated by ion exchange chromatography with a Resource Q column (GE Healthcare). The fractions containing specific COMT activity against 5-hydroxyconiferaldehyde were pooled and used for kinetic assays.
Enzyme Assays
Kinetic assays were performed in COMT assay buffer (100 mM Tris-HCl, pH 7.5, 0.2 mM MgCl2, and 20% glycerol) in the presence of 100 μM SAM and a series of concentrations of phenylpropanoid substrates. The samples were incubated at 30°C for 20 min after addition of the enzyme, terminated by adding glacial acetic acid, extracted with ethyl acetate, dried in vacuo, redissolved in 50% methanol, and analyzed by HPLC, except for the 5-hydroxyferuloyl CoA assays, which were analyzed by HPLC directly after being terminated by acetic acid addition. The kinetic constants, such as Km, Vmax, and Ki, were inferred using the nonlinear regression function integrated in the GraphPad Prism software.
Molecular Modeling of Sm COMT Structure
The Sm COMT model was built by SWISS-MODEL (Arnold et al., 2006) using the Ms COMT structure (PDB ID: 1KYW) as a template. The Sm COMT model was overlaid with the Ms COMT structure and displayed using PyMOL 1.0 (Delano Scientific).
Phylogenetic Analysis
The amino acid alignment was created using the EXPRESSO 3D-coffee function by default settings under the T-COFFEE multiple sequence alignment server (Armougom et al., 2006), and the Bayesian phylogenetic tree was built using MRBAYES 3.1.1 (Huelsenbeck and Ronquist, 2001). The analysis invoked a comparable model (aamodelpr = mixed, nset = 6, rates = invgamma). The Markov chain Monte Carlo (MCMC) analysis was allowed to run for 1,000,000 generations with a sampling frequency of every 1000th generation. The alignment used for phylogenetic tree construction can be found in Supplemental Data Set 1 online.
Accession Numbers
The S. moellendorffii genes identified and characterized in this study have been deposited into GenBank under the following accession numbers: Sm COMT (GQ166949), Sm COMT-like3 (GQ166950), Sm COMT-like3 (GQ166951), and Sm COMT-like3 (GQ166952). The accession numbers for the sequences used in Figure 8 and Figure 9 are as follows: At COMT (NP_200227), At COMT-like1 (NP_173534), At COMT-like2 (NP_173535), At COMT-like3 (NP_173536), At COMT-like4 (NP_849693), At COMT-like5 (NP_174579), At COMT-like6 (NP_974004), At COMT-like7 (NP_974076), At COMT-like8 (NP_177805), At COMT-like9 (NP_177876), At COMT-like10 (NP_177877), At COMT-like11 (NP_190882), At COMT-like12 (NP_198533), At COMT-like13 (NP_200192), Cb IEOMT (AAC01533), Cr COMT (AAK20170), Hv FOMT (CAA54616), Lp COMT (AAD10253), Ms CHOMT (P93324), Ms COMT (AAB46623), Ms IOMT (AAY18582), Nt CAOMT (CAA50561), Ob EOMT (Q93WU3), Pa OMT (CAI30878), Pd COMT (Q43609), Pp OMT (XP_001762717), Ps OMT (ABK24146), Pt COMT (AAF63200), Sb COMT (AAL57301), Sh COMT (2119166A), So COMT (O82054), Ze COMT (Q43239), and Zm COMT (Q06509).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sidechain Regions of 2D HSQC NMR Spectra of Acetylated Lignins.
Supplemental Figure 2. HPLC Chromatograms of 3-Week-Old Rosette Leaf Extract from Arabidopsis Columbia Wild Type, Arabidopsis omt1, omt1-2/At C4H:Sm COMT-like1, omt1-2/At C4H:Sm COMT-like2, and omt1-2/At C4H:Sm COMT-like3 Transgenic Plants.
Supplemental Figure 3. Characterization of the Arabidopsis omt1-2 fah1-2 Double Mutant Transformed with a 9050-bp Selaginella Genomic Fragment Containing COMT, F5H, and Their cis-Regulatory Region.
Supplemental Figure 4. Mechalis-Menten Plots of the Kinetic Assays of Sm COMT and At COMT against Caffeic Acid, 5-Hydroxyferulic Acid, Caffealdehyde, and 5-Hydroxyconiferaldehyde.
Supplemental Figure 5. Kinetic Assays of Sm COMT and At COMT against Caffeyl Alcohol and 5-Hydroxyconiferyl Alcohol.
Supplemental Table 1. Kinetic Properties of Fractionated Selaginella Soluble Protein Prep toward meta-Hydroxylated Phenylpropanoid Intermediates.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Amino Acid Sequence Alignment in Fasta Format Used for the Phylogenetic Analysis Presented in Figure 7.
Acknowledgments
We thank J.A. Banks for providing S. moellendorffii plant materials, D. Sherman for the technical assistance with the scanning electron microscopy, F. Lu and R. Dixon for providing chemicals for enzyme assays, and G.V. Louie and J.P. Noel for insightful discussion. This work is supported by the National Science Foundation (Grant IOB-0450289). Partial funding to J.R. was via the Department of Energy Office of Science (Grant DE-AI02-06ER64299) and the Department of Energy Great Lakes Bioenergy Research Center (Department of Energy Office of Science BER DE-FC02-07ER64494).
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. (2006). Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 34(Web Server issue): W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Barton N.H. (2000). Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M., Monties B., Van Montagu M., Boerjan W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17: 125–197 [Google Scholar]
- Blumenthal T. (1998). Gene clusters and polycistronic transcription in eukaryotes. Bioessays 20: 480–487 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bohlmann J., Meyer-Gauen G., Croteau R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce C.K., Zwieniecki M.A., Cody G.D., Jacobsen C., Wirick S., Knoll A.H., Holbrook N.M. (2004). Evolution of xylem lignification and hydrogel transport regulation. Proc. Natl. Acad. Sci. USA 101: 17555–17558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlquist S. (1996). Wood, bark, and stem anatomy of gnetales: A summary. Int. J. Plant Sci. 157: S58–S76 [Google Scholar]
- Chang H.M., Cowling E.B., Brown W., Adler E., Miksche G. (1975). Comparative studies on cellulolytic enzyme lignin and milled wood lignin of sweetgum and spruce. Holzforschung 29: 153–159 [Google Scholar]
- Chapple C.C., Vogt T., Ellis B.E., Somerville C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Meyermans H., Burggraeve B., De Rycke R.M., Inoue K., De Vleesschauwer V., Steenackers M., Van Montagu M.C., Engler G.J., Boerjan W.A. (2000). Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar. Plant Physiol. 123: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S.J., Leonard G.A., McSweeney S.M., Thompson A.W., Naismith J.H., Qamar S., Plater A., Berry A., Hunter W.N. (1996). The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. Structure 4: 1303–1315 [DOI] [PubMed] [Google Scholar]
- Donaldson L.A. (2001). Lignification and lignin topochemistry—An ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Duerden H. (1934). On the occurrence of vessels in selaginella. Ann. Bot. (Lond.) 48: 459–465 [Google Scholar]
- Erickson M., Miksche G.E. (1974). Characterization of pteridophyta lignins by oxidative-degradation. Holzforschung 28: 157–159 [Google Scholar]
- Faix O., Gyzas E., Schweers W. (1977). Comparative investigations on different fern lignins. Holzforschung 31: 137–144 [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Friedman W.E., Cook M.E. (2000). The origin and early evolution of tracheids in vascular plants: Integration of palaeobotanical and neobotanical data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y., Walker D.R., Koonin E.V. (1998). Analogous enzymes: Independent inventions in enzyme evolution. Genome Res. 8: 779–790 [DOI] [PubMed] [Google Scholar]
- Gang D.R., Lavid N., Zubieta C., Chen F., Beuerle T., Lewinsohn E., Noel J.P., Pichersky E. (2002). Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon T., Sibout R., Pollet B., Maba B., Nussaume L., Bechtold N., Lu F., Ralph J., Mila I., Barrière Y., Lapierre C., Jouanin L. (2003). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Jin Z.F., Matsumoto Y., Tange T., Akiyama T., Higuchi M., Ishii T., Iiyama K. (2005). Proof of the presence of guaiacyl-syringyl lignin in Selaginella tamariscina. J. Wood Sci. 51: 424–426 [Google Scholar]
- Kenrick P., Crane P.R. (1997). The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kim H., Ralph J., Akiyama T. (2008). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. Bioenerg. Res. 1: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupce E., Freeman R. (2007). Compensated adiabatic inversion pulses: Broadband INEPT and HSQC. J. Magn. Reson. 187: 258–265 [DOI] [PubMed] [Google Scholar]
- Kuroda H. (1983). Comparative studies on O-methyltransferases involved in lignin biosynthesis. Wood Res. 69: 91–135 [Google Scholar]
- Li L., Cheng X.F., Leshkevich J., Umezawa T., Harding S.A., Chiang V.L. (2001). The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Weng J.K., Chapple C. (2008). Improvement of biomass through lignin modification. Plant J. 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Logan K.J., Thomas B.A. (1985). Distribution of lignin derivatives in plants. New Phytol. 99: 571–585 [DOI] [PubMed] [Google Scholar]
- Lu F., Ralph J. (1998). The DFRC method for lignin analysis. 2. Monomers from isolated lignins. J. Agric. Food Chem. 46: 547–552 [DOI] [PubMed] [Google Scholar]
- Lu F., Ralph J. (2003). Non-degradative dissolution and acetylation of ball-milled plant cell walls: High-resolution solution-state NMR. Plant J. 35: 535–544 [DOI] [PubMed] [Google Scholar]
- Marusek C.M., Trobaugh N.M., Flurkey W.H., Inlow J.K. (2006). Comparative analysis of polyphenol oxidase from plant and fungal species. J. Inorg. Biochem. 100: 108–123 [DOI] [PubMed] [Google Scholar]
- Meyer K., Shirley A.M., Cusumano J.C., Bell-Lelong D.A., Chapple C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micco V.D., Aronne G. (2007). Anatomical features, monomer lignin composition and accumulation of phenolics in 1-year-old branches of the Mediterranean Cistus ladanifer L. Bot. J. Linn. Soc. 155: 361–371 [Google Scholar]
- Musha Y., Goring D.A.I. (1975). Distribution of syringyl and guaiacyl moieties in hardwoods as indicated by ultraviolet microscopy. Wood Sci. Technol. 9: 45–58 [Google Scholar]
- Nakashima J., Chen F., Jackson L., Shadle G., Dixon R.A. (2008). Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): Effects on lignin composition in specific cell types. New Phytol. 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Osakabe K., Tsao C.C., Li L., Popko J.L., Umezawa T., Carraway D.T., Smeltzer R.H., Joshi C.P., Chiang V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96: 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K., Chen F., Guo D., Blount J.W., Dixon R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Prescott A.G., John P. (1996). DIOXYGENASES: Molecular structure and role in plant metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 245–271 [DOI] [PubMed] [Google Scholar]
- Ralph J., Akiyama T., Kim H., Lu F., Schatz P.F., Marita J.M., Ralph S.A., Reddy M.S., Chen F., Dixon R.A. (2006). Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ralph J., Kim H., Peng J., Lu F. (1999). Arylpropane-1,3-diols in lignins from normal and CAD-deficient pines. Org. Lett. 1: 323–326 [DOI] [PubMed] [Google Scholar]
- Ralph J., Lundquist K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Ralph S.A., Christensen J.H., Boerjan W. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem. Rev. 3: 29–60 [Google Scholar]
- Reddy M.S., Chen F., Shadle G., Jackson L., Aljoe H., Dixon R.A. (2005). Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L:). Proc. Natl. Acad. Sci. USA 102: 16573–16578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Meyer K., Cusumano J.C., Chapple C. (1999). Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 119: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka S., Goring D.A.I. (1988). The distribution of lignin in white birch wood as determined by bromination with Tem-Edxa. Holzforschung 42: 149–153 [Google Scholar]
- Salgado H., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J. (2000). Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. USA 97: 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G.H.N., Gibbs R.D. (1953). Lignin chemistry and the taxonomy of higher plants. Nature 172: 25–26 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Thomas J., Spillane C., Coluccio A., Hoeppner M.A., Grossniklaus U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pichersky E. (1998). Characterization of S-adenosyl-L-methionine:(iso)eugenol O-methyltransferase involved in floral scent production in Clarkia breweri. Arch. Biochem. Biophys. 349: 153–160 [DOI] [PubMed] [Google Scholar]
- Warburg O., Christian W. (1943). Isolation and cristilisation of the fermentative enzyme zymohexase. Biochem. Z. 314: 149–176 [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Weng J.K., Akiyama T., Bonawitz N.D., Li X., Ralph J., Chapple C. (2010b). Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Banks J.A., Chapple C. (2008a). Parallels in lignin biosynthesis: A study in Selaginella moellendorffii reveals convergence across 400 million years of evolution. Commun. Integr. Biol. 1: 20–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Weng J.K., Li X., Stout J., Chapple C. (2008b). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Mo H., Chapple C. (2010a). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64: 898–911 [DOI] [PubMed] [Google Scholar]
- White E., Towers G.H.N. (1967). Comparative biochemistry of lycopods. Phytochemistry 6: 663–667 [Google Scholar]
- Zhang H., Wang J., Goodman H.M. (1997). An Arabidopsis gene encoding a putative 14-3-3-interacting protein, caffeic acid/5-hydroxyferulic acid O-methyltransferase. Biochim. Biophys. Acta 1353: 199–202 [DOI] [PubMed] [Google Scholar]
- Zhang L.M., Gellerstedt G., Ralph J., Lu F.C. (2006). NMR studies on the occurrence of spirodienone structures in lignins. J. Wood Chem. Technol. 26: 65–79 [Google Scholar]
- Zubieta C., He X.Z., Dixon R.A., Noel J.P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C., Kota P., Ferrer J.L., Dixon R.A., Noel J.P. (2002). Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]