Ectopic expression of a MYB transcription factor, ODO1, in tomato fruit altered Phe and phenylpropanoid metabolism, as shown by metabolic profiling and flux analysis. This disruption permitted identification of a set of coregulated genes, including prephenate aminotransferase, which converts prephenate to arogenate. The results establish a fruit arogenate pathway for Phe synthesis.
Abstract
Altering expression of transcription factors can be an effective means to coordinately modulate entire metabolic pathways in plants. It can also provide useful information concerning the identities of genes that constitute metabolic networks. Here, we used ectopic expression of a MYB transcription factor, Petunia hybrida ODORANT1, to alter Phe and phenylpropanoid metabolism in tomato (Solanum lycopersicum) fruits. Despite the importance of Phe and phenylpropanoids to plant and human health, the pathway for Phe synthesis has not been unambiguously determined. Microarray analysis of ripening fruits from transgenic and control plants permitted identification of a suite of coregulated genes involved in synthesis and further metabolism of Phe. The pattern of coregulated gene expression facilitated discovery of the tomato gene encoding prephenate aminotransferase, which converts prephenate to arogenate. The expression and biochemical data establish an arogenate pathway for Phe synthesis in tomato fruits. Metabolic profiling and 13C flux analysis of ripe fruits further revealed large increases in the levels of a specific subset of phenylpropanoid compounds. However, while increased levels of these human nutrition-related phenylpropanoids may be desirable, there were no increases in levels of Phe-derived flavor volatiles.
INTRODUCTION
Targeted manipulation of transcription factors can be a powerful tool to define suites of coordinately regulated genes and discover new genes within regulatory networks. Because of its central role in metabolism of many economically and biologically important products, synthesis and metabolism of Phe is of particular interest. For example, overexpression of an Antirrhinum majus MYB had profound effects on transgenic tobacco (Nicotiana tabacum) phenylpropanoid synthesis, suggesting conservation of function across species (Tamagnone et al., 1998). Ectopic expression of two maize (Zea mays) transcription factors in tomato (Solanum lycopersicum) fruit activated the entire flavonoid biosynthesis pathways, resulting in high fruit flavonol levels (Bovy et al., 2002), while ectopic expression of two A. majus transcription factors resulted in fruits with significantly higher levels of health-promoting anthocyanins (Butelli et al., 2008). The power of integrating transcriptome and metabolome data is illustrated by a study of Arabidopsis thaliana flavonoids. Expression of a MYB transcription factor combined with metabolome analysis led to discovery of several new genes involved in anthocyanin synthesis (Tohge et al., 2005).
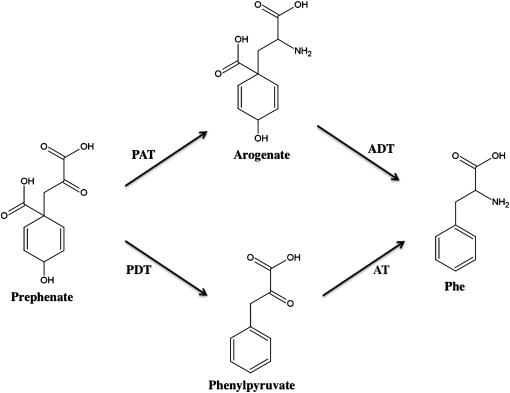
Although Phe synthesis is key to the accumulation of many biologically and economically important secondary metabolites, the biosynthetic pathway is not fully established in plants. Following conversion of chorismic acid to prephenate, two pathways for Phe synthesis have been proposed, and there is experimental support for both (Figure 1). In one branch, prephenate is converted to arogenate by prephenate aminotransferase (PAT) (EC 2.6.1.57) and to Phe by arogenate dehydratase (ADT) (Bonner and Jensen, 1985; Siehl et al., 1986; De-Eknamkul and Ellis, 1988). Alternatively, prephenate is proposed to be converted to phenylpyruvate (PPY) by prephenate dehydratase (E.C. 4.2.1.51) (Tzin et al., 2009). Both of these pathways exist in microbes (Whitaker et al., 1981; Fischer et al., 1993). There is evidence supporting the arogenate branch of the pathway. PAT enzymes have been characterized from Anchusa officinalis (De-Eknamkul and Ellis, 1988) and Nicotiana sylvestris (Bonner and Jensen, 1985). ADT enzymes have been extensively characterized in Arabidopsis (Cho et al., 2007) and petunia (Petunia hybrida; Maeda et al., 2010). Thus, it seems likely that an arogenate pathway exists in plants.
Figure 1.
Proposed Pathways for Phe Synthesis.
Phe is synthesized from prephenate via either arogenate or PPY. PAT, 2.6.1.57; ADT, 4.2.1.91; PDT, prephenate dehydratase; AT, aminotransferase.
The R2R3 MYB transcription factors have essential roles in many aspects of plant secondary metabolism, particularly phenylpropanoid metabolism (Stracke et al., 2001). One MYB, ODO1, was isolated from petunia. Its expression is spatially and temporally coincident with floral volatile synthesis, and RNA interference (RNAi)-mediated suppression causes reduction of multiple Phe-derived volatiles, including 2-phenylethanol, phenylacetaldehyde, and methyl benzoate. RNAi plants exhibit reduced expression of genes in the shikimate and phenylpropanoid pathways, indicating that this MYB is essential for expression of genes encoding key steps in Phe and phenylpropanoid synthesis in petunia flowers (Verdonk et al., 2005). Since ODO1 loss of function reduced expression of genes whose products are essential for Phe synthesis, it is not surprising that synthesis of Phe-derived volatiles was reduced. However, overexpression of ODO1 was not performed. Furthermore, none of the genes whose enzymes are directly involved in synthesis of these volatiles were reported to be downregulated in the RNAi lines. Thus, a direct role for ODO1 in regulating petunia volatile synthesis has not been demonstrated.
We were specifically interested in identifying and altering the expression of key genes regulating both Phe and Phe-derived volatile synthesis. In particular, we sought to increase metabolic flux to Phe and measure the effects on Phe-derived tomato volatiles. The results of Verdonk et al. (2005) indicated that petunia ODO1 is essential for expression of multiple genes responsible for Phe synthesis. Since tomato and petunia are both Solanaceous plants, we overexpressed petunia ODO1 in tomato fruits and assessed the effects of that expression on the metabolome. Using a fruit ripening–specific transcriptional promoter, we measured the primary effects of ODO1 overexpression on the transcriptome and metabolome in a single organ without disruption of primary metabolism in the vegetative organs. We were able to define a set of coordinately regulated genes encoding enzymes of the shikimate and phenylpropanoid pathways that together alter phenylpropanoid levels. We exploited the pattern of coexpression in ODO1-expressing fruits to identify the gene encoding PAT, a gene encoding a critical step in Phe synthesis. This result establishes a pathway to Phe through arogenate in tomato fruits. We further demonstrated significant remodeling of phenylpropanoid flux in transgenic fruits.
RESULTS
Transgenic Expression of Petunia ODO1
To minimize effects on plant growth and development, expression of ODO1 was targeted to fruits using the tomato E8 transcriptional promoter that is expressed throughout fruit tissues beginning at the onset of ripening (Deikman et al., 1992). Five independent transgenic ODO1-expressing lines were obtained (8150, 8117, 7948, 7949, and 7817) (see Supplemental Figure 1 online). As expected, no phenotypic effects were observed on vegetative tissues of transgenic plants. By contrast, transgenic fruits exhibited irregular and delayed ripening (Figure 2). When fruits remained attached to the vine, there was an overall ripening delay of 1 to 2 weeks. Fruits did eventually fully ripen, although the color of the skin was altered in appearance, similar to fruits with modified expression of a tomato MYB transcription factor (Ballester et al., 2010). Although the transgenic fruits did synthesize less ethylene (see Supplemental Figure 2 online), the reductions are probably not sufficient to account for the delayed and irregular ripening phenotype. Furthermore, treatment of detached fruits with propylene did not affect ripening (see Supplemental Figure 3 online). Since the transgenic fruits could be potentially altered in their aromatic amino acid contents, we also examined levels of the Trp-derived indole-3-acetic acid (IAA) in fruits as well as the expression of three potentially auxin-regulated genes. Transgenic fruits were not significantly altered in IAA or expression of these genes (see Supplemental Figure 4 online), eliminating a role for IAA in the delayed ripening phenotype.
Figure 2.
Ripening Phenotypes of Transgenic ODO1 Fruits.
Fruits were tagged at breaker stage and harvested after either 5 (B+5) or 10 (B+10) d. Control is M82.
Consequences of ODO1 Expression on the Ripening Fruit Transcriptome
Petunia ODO1 is required for floral expression of two genes in the shikimate pathway, 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase and 5-enolpyruvylshikimate-3-phosphate synthase, as well as two members of the Phe ammonia lyase gene family (Verdonk et al., 2005). Availability of microarrays containing most of the fruit-expressed genes permitted us to assess the transcriptional network regulated by petunia ODO1 in transgenic tomato fruits. RNAs from two transgenic lines (7949 and 8117) were compared with M82 control RNA using the TOM2 microarray (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/home.cgi). The two transgenic lines exhibited almost identical patterns of altered gene expression (complete data available at http://ted.bti.cornell.edu/cgi-bin/TFGD/miame/home.cgi). Values for genes with functions related to Phe and phenylpropanoid synthesis whose expression was significantly altered are shown in Table 1. The gene identities are based on GenBank BLAST results. In many instances, there are multiple genes encoding these enzymes with only specific family members exhibiting significantly increased expression. For example, one DAHP synthase, corresponding to SGN-U566921, was highly induced in both transgenic lines, while others were not. At least one gene for each step in the shikimate pathway was significantly induced, except for 3-dehydroquinate dehydratase/shikimate dehydrogenase. Notably there was no increased expression of any genes in the branch pathway to Trp. Only a subset of phenylpropanoid-associated genes was induced. No genes related to flavonoid synthesis were upregulated in transgenic fruits. The metabolite data, described below, are also consistent with a lack of flavonoid-related gene induction by ODO1. Expression of a subset of the Phe-associated genes was validated by quantitative RT-PCR (see Supplemental Figure 5 online). The results of these analyses were entirely consistent with the microarray expression results. Taken together, the data indicate that fruit overexpression of ODO1 coordinately regulates expression of a set of genes whose activities are responsible for synthesis and further metabolism of Phe.
Table 1.
Induced Genes in ODO1 Overexpressing Fruits
| Ratio | FDR | Unigene | Description | EC Number |
| 3.94 | 0.004 | SGN-U566921 | DAHP synthase | 2.5.1.54 |
| 4.77 | 0.009 | |||
| 2.13 | 0.005 | SGN-U568781 | Dehydroquinate synthase | 4.2.1.10 |
| 2.43 | 0.015 | |||
| 1.93 | 0.017 | SGN-U582040 | Shikimate kinase | 2.7.1.71 |
| 1.98 | 0.012 | |||
| 1.57 | 0.016 | SGN-U580985 | EPSP synthase | 2.5.1.19 |
| 1.67 | 0.008 | |||
| 3.72 | 0.005 | SGN-U580985 | EPSP synthase | 2.5.1.19 |
| 2.93 | 0.008 | |||
| 3.81 | 0.005 | SGN-U577580 | EPSP synthase | 2.5.1.19 |
| 3.14 | 0.008 | |||
| 2.01 | 0.007 | SGN-U563165 | Chorismate synthase 1 | 4.2.3.5 |
| 2.27 | 0.034 | |||
| 5.44 | 0.005 | SGN-U575627 | Chorismate mutase | 5.4.99.5 |
| 5.05 | 0.008 | |||
| 2.52 | 0.005 | SGN-U567172 | Putative Asp aminotransferase | * |
| 2.29 | 0.008 | |||
| 1.73 | 0.007 | SGN-U567390 | Putative Asp aminotransferase | * |
| 1.27 | 0.018 | |||
| 2.17 | 0.005 | SGN-U573964 | Arogenate/prephenate dehydratase | 4.2.1.91 |
| 1.89 | 0.008 | |||
| 9.73 | 0.005 | SGN-U583010 | Arogenate/prephenate dehydratase | 4.2.1.91 |
| 11.28 | 0.007 | |||
| 2.10 | 0.005 | SGN-U562941 | 4-Coumarate–CoA ligase | 6.2.1.12 |
| 2.40 | 0.008 | |||
| 7.29 | 0.004 | SGN-U580976 | 4-Coumarate–CoA ligase | 6.2.1.12 |
| 8.47 | 0.008 | |||
| 8.83 | 0.005 | SGN-U577586 | 4-Coumarate–CoA ligase | 6.2.1.12 |
| 9.93 | 0.008 | |||
| 15.16 | 0.005 | SGN-U577586/SGN-U593064 | 4-Coumarate–CoA ligase | 6.2.1.12 |
| 17.48 | 0.008 | |||
| 2.09 | 0.006 | SGN-U580050/SGN-U580736 | Phe ammonia-lyase | 4.3.1.24 |
| 1.46* | ||||
| 5.79 | 0.008 | SGN-U572140 | Phe ammonia-lyase | 4.3.1.24 |
| 6.56 | 0.008 | |||
| 6.74 | 0.004 | SGN-U580612 | Phe ammonia-lyase | 4.3.1.24 |
| 7.83 | 0.008 | |||
| 9.05 | 0.005 | SGN-U577677 | Phe ammonia-lyase | 4.3.1.24 |
| 11.58 | 0.008 | |||
| 11.45 | 0.004 | SGN-U577267 | Phe ammonialyase | 4.3.1.24 |
| 18.14 | 0.008 | |||
| 11.55 | 0.006 | SGN-U580050 | Phe ammonia-lyase | 4.3.1.24 |
| 7.83 | 0.008 | |||
| 11.74 | 0.004 | SGN-U590436 | Phe ammonia-lyase | 4.3.1.24 |
| 13.43 | 0.008 | |||
| 13.78 | 0.004 | SGN-U577677 | Phe ammonia-lyase | 4.3.1.24 |
| 16.09 | 0.007 | |||
| 3.83 | 0.006 | SGN-U565006 | Cinnamoyl-CoA reductase | 1.2.1.44 |
| 4.69* | 0.06 | |||
| 5.30 | 0.005 | SGN-U578364/SGN-U578590 | Caffeoyl-CoA O-methyltransferase | 2.1.1.68 |
| 4.78 | 0.008 | |||
| 34.44 | 0.005 | SGN-U581378 | Caffeoyl-CoA O-methyltransferase | 2.1.1.68 |
| 34.79 | 0.008 | |||
| 36.83 | 0.004 | SGN-U590082 | Caffeoyl-CoA O-methyltransferase | 2.1.1.68 |
| 36.67 | 0.008 | |||
| 43.82 | 0.004 | SGN-U577382/SGN-U577729 | Caffeoyl-CoA O-methyltransferase | 2.1.1.68 |
| 33.93 | 0.008 | |||
| 9.61 | 0.005 | SGN-U581122/SGN-U581132 | Cinnamic acid 4-hydroxylase | 1.14.13.11 |
| 9.45 | 0.008 | |||
| 12.39 | 0.005 | SGN-U581122/SGN-U581132 | Cinnamic acid 4-hydroxylase | 1.14.13.11 |
| 11.51 | 0.006 | |||
| 11.31 | 0.005 | SGN-U565500/SGN-U587882 | Tyramine N-feruloyltransferase | 2.3.1.110 |
| 12.73 | 0.008 | |||
| 2.26 | 0.004 | SGN-U578909/SGN-U579184 | Tyramine N-feruloyltransferase | 2.3.1.110 |
| 2.08 | 0.009 | |||
| 2.21 | 0.018 | SGN-U586450/SGN-U586452 | UDP-xylose phenolic glycosyltransferase | 2.4.2.35 |
| 1.70 | 0.044 | |||
| 1.58 | 0.01 | SGN-U562651/SGN-U562652 | Hydroxycinnamoyl CoA quinate transferase | 2.3.1.99 |
| 2.16 | 0.008 | |||
| 0.33 | 0.024 | SGN-U580262 | Chalcone synthase | 2.3.1.74 |
| 0.72* | 0.093 |
Ratios of transgenic to control RNAs for 7949 (top) and 8117 (bottom) as well as the false discovery rate (FDR) are indicated for each unigene. RNA was isolated from fruits at breaker plus 5 d. Asterisk indicates proteins with BLAST identities as Asp aminotransferases tested in this study.
The small set of genes with nonobvious connections to Phe metabolism whose expression was significantly induced supports the conclusion that altered gene expression is a direct consequence of ODO1 expression. Expression of only 28 genes with nonobvious links to Phe metabolism was significantly upregulated 2-fold or greater in both transgenic lines (see Supplemental Table 1 online). One of these genes, S-adenosylmethionine synthetase (SAMS), was also altered in the petunia RNAi lines (Verdonk et al., 2005). Only 12 genes were expressed at significantly lower levels in both transgenic lines, none with obvious links to phenylpropanoid metabolism.
Identification of Tomato PAT
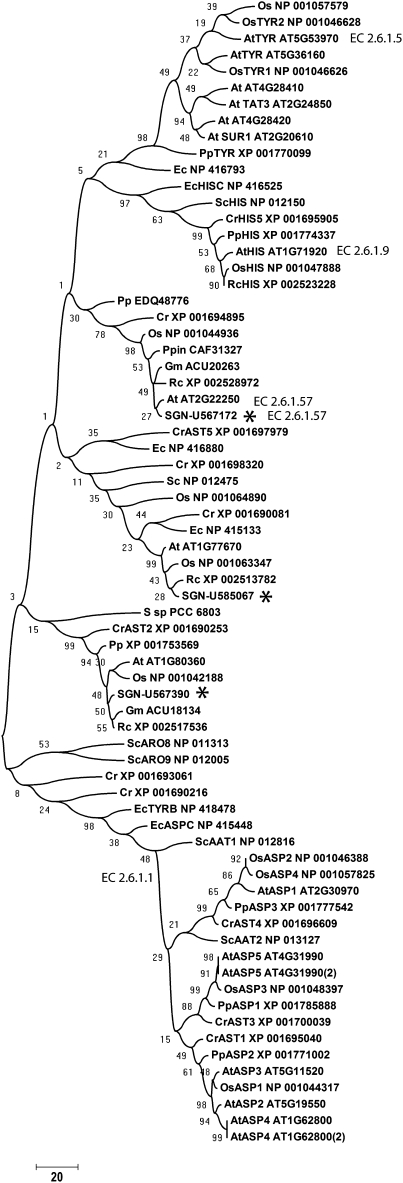
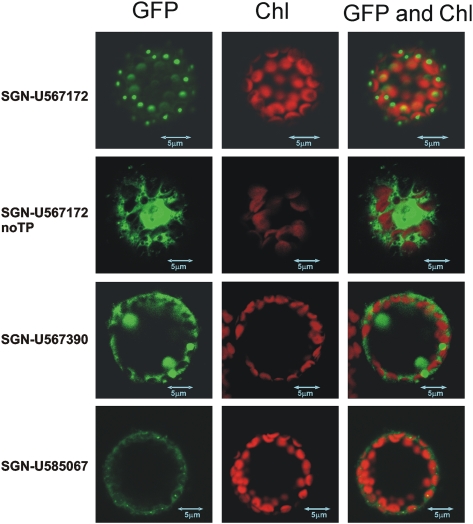
Among the genes that were significantly induced in ODO1-expressing fruits, SGN-U567172 and SGN-U567390 were annotated as putative Asp aminotransferases (Table 1). A third gene whose expression was not altered in ODO1-expressing fruits, SGN-U585067, was identified by similarity to SGN-U567172. We hypothesized that at least one of these genes might encode a PAT. The phylogenetic analysis indicated that these genes encode class I and II aminotransferases and are more closely related to Tyr aminotransferases than Asp aminotransferases (Figure 3). Full-length cDNAs for all three genes were isolated. The predicted SGN-U567172 protein, as well as its closest homologs from other species, contains a putative chloroplast signal peptide, as would be expected for a Phe biosynthetic enzyme. The SGN-U567390 and SGN-U585067 proteins, as well as their closest homologs from other species, do not have obvious signal peptides. To validate the predicted subcellular locations of the proteins, C-terminal fusions between each peptide and green fluorescent protein (GFP) were constructed and transiently expressed in Nicotiana benthamiana leaves (Figure 4). Consistent with predictions, the SGN-U567172 fusion protein was localized to chloroplasts, while the SGN-U567390 and SGN-U585067 fusion proteins were not imported into chloroplasts and are most likely cytoplasmic. The localization was further evaluated by three-dimensional observation of the protoplasts by the Z-stack option of the confocal microscope software (data not shown). The punctate pattern of localization for SGN-U567172 within chloroplasts is consistent with patterns observed for other amino acid biosynthetic enzymes (Farmaki et al., 2007; Maloney et al., 2010).
Figure 3.
Phylogenetic Relationships among PAT-Related Proteins.
Tomato proteins from this article are indicated with an asterisk. Other proteins are as follows: At (Arabidopsis), Cr (Chlamydomonas reinhardtii), Ec (E. coli K12), Fp (Flavobacterium psychrophilum), Gm (Glycine max), Pp (Physcomitrella patens patens), Ppin (Pinus pinaster), Os (Oryza sativa), Rc (Ricinus communis), Sc (Saccharomyces cerevisiae), and Ssp (Synechocystis sp PCC 6803). AAT, aminotransferase; TAT and TYR, Tyr aminotransferase; HIS, histidinoil aminotransferase; AST and ASP, Asp aminotransferase; ARO, aromatic aminotransferase. The EC numbers and protein nomenclature are consistent with KEGG nomenclature. The bar at the bottom indicates the amino acid distance of the sequences of the tree. Bootstrap values are also reported. Asterisks indicate tomato proteins that were assayed for PAT activity in this work.
Figure 4.
Subcellular Locations of Candidate PATs.
C-terminal GFP fusions were transiently expressed in N. benthamiana protoplasts and visualized with confocal microscopy. SGN-U567172 was localized to chloroplasts (top). The same protein without the predicted transit peptide accumulated in the cytoplasm (middle). SGN-U567390 and SGN-U585067 were also located in the cytoplasm, consistent with the lack of identifiable signal peptides. Chloroplasts were visualized by autofluorescence. Chl, chlorophyll.
The candidate proteins were further investigated for their enzymatic activities. Recombinant protein containing a C-terminal 6X-His tag with or without the predicted transit peptide were expressed in Escherichia coli. Affinity purified proteins were assayed in vitro for activity with multiple substrates (Table 2). The recombinant SGN-U567172 protein was highly active with prephenate as a substrate. The measured Km for prephenate was similar to that reported for the Anchusa officinalis enzyme (0.08 mM) (De-Eknamkul and Ellis, 1988) and equally efficient with either Asp or Glu as amino group donor, consistent with prior enzyme characterizations (Bonner and Jensen, 1985; De-Eknamkul and Ellis, 1988). The enzyme was also active in the reverse direction with either oxaloacetate and Glu or 2-oxoglutarate and Asp as substrates but was inactive with Phe as substrate. Phe was inhibitory for the forward reaction with a calculated Ki of 0.051 mM. This inhibition is noncompetitive, similar to the Phe inhibition observed with E. coli prephenate dehydratase (Zhang et al., 1998). The protein was equally active with the signal peptide. The enzyme exhibited no activity with PPY, indole-3-pyruvic acid, or 4-hydroxyphenylpyruvate as verified by the assay (data not shown) and by gas chromatography–mass spectrometry (GC-MS) analysis of the reaction products. Arogenate, which is the product of the forward reaction, is not commercially available. To further verify that prephenate was converted to arogenate, the reaction product was acid treated to convert the synthesized arogenate to Phe. This acid-catalyzed conversion of arogenate to Phe is an established validation of arogenate identity as previously described (Zamir et al., 1983; Bonner and Jensen, 1985). Phe was subsequently quantified as described (Chen et al., 2010) (see Supplemental Table 2 online). As further validation of identity, heat stability of the recombinant protein was examined. The N. sylvestris PAT is heat stable with a temperature optimum of 70°C (Bonner and Jensen, 1985). The tomato enzyme showed similar heat stability, being fully active after a 10 min 70°C preincubation. Together, the results indicate that SGN-U567172 encodes a plastid-localized prephenate aminotransferase, hereafter designated Sl PAT (GenBank accession HQ394171). SGN-U567390 and SGN-U585067 exhibited no activity with any of the tested substrates.
Table 2.
Kinetic Parameters of PAT
| Protein | Substrate | Amino Donor | Km mM | Kcat s−1 | Kcat/Km mM−1 s−1 |
| SGN-U567172 -TP | Prephenate | Asp | 0.08 ± 0.01 | 11.35 ± 0.53 | 141.9 |
| SGN-U567172 -TP | Prephenate | Glu | 0.10 ± 0.03 | 11.47 ± 1.73 | 114.7 |
| SGN-U567172 -TP | oxaloacetate | Glu | 0.32 ± 0.06 | 15.06 ± 1.37 | 47.1 |
| SGN-U567172 -TP | 2-oxoglutarate | Asp | 0.28 ± 0.04 | 19.52 ± 2.17 | 69.7 |
| SGN-U567172 | oxaloacetate | Glu | 0.22 ± 0.03 | 10.41 ± 0.51 | 47.3 |
| SGN-U567172 | 2-oxoglutarate | Asp | 0.16 ± 0.05 | 16.61 ± 3.44 | 103.8 |
| AT2G22250 -TP | Prephenate | Asp | 0.2 ± 0.02 23.67 ± 1.66 118.4 | AT2G22250 -TP | Prephenate |
| Glu | 0.10 ± 0.01 | 16.83 ± 0.99 | 168.3 |
Tomato (SGN-U567172) or Arabidopsis (AT2G22250) proteins with or without (-TP) signal peptide were assayed as described in Methods. Values are shown for various substrates ± se. The analysis was performed on 16 to 24 replicates. Nonlinear regressions values for each curve were >95%.
To determine whether the arogenate pathway for Phe synthesis is conserved in other plants, a full-length cDNA from the Arabidopsis gene with greatest similarity to Sl PAT was isolated. At2g22250 encodes a protein with 79% identity and 87% similarity to Sl PAT. BLAST analysis indicates that this is a single copy gene in Arabidopsis, as in tomato. In vitro analysis of recombinant At2g22250 protein, without its transit peptide, indicated similar kinetic properties to the tomato enzyme (Table 2). Thus, At2g22250 is orthologous to Sl PAT. Genetic analysis of T-DNA knockouts of At2g22250 indicated that the gene is essential for embryo development, as the homozygous knockout is lethal at the one-cell stage (Pagnussat et al., 2005).
Arogenate is converted to Phe by action of ADTs. Expression of two genes annotated as prephenate/ADTs was induced in the ODO1-expressing fruits (Table 1). The proteins encoded by these genes are most closely related to a family of Arabidopsis enzymes that specifically convert arogenate to Phe (Cho et al., 2007). As further confirmation of the arogenate pathway for Phe synthesis in tomato fruits, an assay combining recombinant Sl PAT and the putative SGN-U573964 ADT enzymes was performed. The combined recombinant enzymes converted prephenate to Phe in vitro (see Supplemental Figure 6 online). No Phe was produced when the Sl PAT enzyme was heat-inactivated prior to the reaction. These results indicate that Sl PAT and ADT, whose expression was upregulated in ODO1 overexpressing lines, are capable of Phe synthesis.
As final proof of in vivo function, we reconstituted the arogenate pathway in an E. coli Phe auxotrophic mutant. E. coli uses the PPY pathway to synthesize Phe and is incapable of metabolizing arogenate. The pheA18::Tn10 mutant lacks the bifunctional Chorismate mutase-prephenate dehydratase P protein and exhibits an absolute requirement for exogenous Phe (Johnson et al., 1989). The pheA18::Tn10 mutant was transformed with plasmids expressing PAT and ADT (encoded by SGN-U583010). As expected, the PAT expression plasmid alone did not complement the Phe auxotrophy (Figure 5). However, expression of both proteins restored the ability of the mutant to grow on minimal medium lacking Phe (Figure 5). Growth of the mutant expressing both genes was nearly as rapid as that of the mutant in medium supplemented with Phe, indicating reconstitution of a fully functional Phe synthesis pathway. When the ADT was replaced by the other putative ADT that showed induction on the microarray (SGN-U573964), equivalent restoration of growth was observed (data not shown).
Figure 5.
Growth of E. coli Phe Auxotrophs with or without Tomato PAT and ADT.
(A) Growth of E. coli PheA18:Tn10 (DA15) alone, expressing PAT, or PAT and ADT on minimal M9 medium lacking Phe.
(B) Growth of DA15 in minimal liquid medium containing or lacking Phe or plasmids expressing PAT and ADT enzymes.
In summary, Sl PAT exhibits kinetics entirely consistent with those previously reported for plant PAT enzymes. Tomato PAT, combined with ADT, synthesizes Phe both in vitro and in E. coli. The Arabidopsis ortholog exhibits equivalent kinetic properties, and a loss-of-function mutant is lethal at the one-cell stage of embryo development. Taken together, the data indicate that PAT provides the critical and likely essential function to synthesize arogenate that is subsequently converted to Phe by ADT enzymes.
Effects of Petunia ODO1 Expression on Fruit Metabolite Pools
Floral benzenoid volatile emissions were reduced in ODO1 RNAi petunia flowers, making the consequences of overexpression of this transcription factor of interest. Analysis of tomato volatiles in petunia ODO1-overexpressing fruits indicated that there were no statistically significant increases in any Phe-derived tomato fruit volatiles (Table 3). By contrast, all of the lines had significant reductions in benzaldehyde emissions, while four of five lines were significantly reduced in 2-phenylacetonitrile emissions. There were no significant decreases in the unrelated fatty acid–derived volatile, hexanol. We have previously determined that the limiting step to synthesis of several Phe-derived volatiles in tomato is expression of a family of aromatic amino acid decarboxylases (AADCs) (Tieman et al., 2006a). Quantitative RT-PCR analysis indicated that AADC expression was not consistently elevated in the transgenic ODO1-overexpressing lines (data not shown).
Table 3.
Fruit Volatile Emissions
| Volatile | M82 | 7948 | 7949 | 8117 | 8150 | 7817 |
| Hexanol | 55.8 ± 5.4 | 39.1 ± 15.5 | 21.9 ± 15.7 | 27.7 ± 7.0 | 57.5 ± 10.1 | 58.2 ± 16.1 |
| Phenylacetaldehyde | 1.1 ± 0.5 | 0.8 ± 0.6 | 1.0 ± 0.6 | 0.5 ± 0.1 | 0.7 ± 0.3 | 1.8 ± 0.5 |
| Guaiacol | 1.7 ± 0.2 | 1.0 ± 0.2 | 1.1 ± 0.3 | 1.2 ± 0.1 | 1.0 ± 0.2 | 3.0 ± 0.8 |
| 2-Phenylethanol | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 |
| 1-Nitro-2-phenethane | 0.2 ± 0.0 | 0.3 ± 0.1 | 0.2 ± 0.0 | 0.1 ± 0.0* | 0.1 ± 0.0 | 0.2 ± 0.0 |
| Benzaldehyde | 24.0 ± 5.8 | 2.7 ± 0.3* | 1.3 ± 0.7* | 2.7 ± 0.4* | 3.8 ± 0.8* | 4.3 ± 1.7* |
| Benzyl alcohol | 1.3 ± 0.2 | 0.7 ± 0.1 | 0.6 ± 0.4 | 1.0 ± 0.2 | 1.5 ± 0.2 | 0.9 ± 0.2 |
| Guaiacol | 1.7 ± 0.2 | 1.0 ± 0.2 | 0.7 ± 0.3 | 1.0 ± 0.2 | 1.3 ± 0.3 | 3.0 ± 0.9 |
| Methylbenzoate | 3.7 ± 0.4 | 3.5 ± 1.3 | 1.4 ± 0.4 | 2.7 ± 0.4 | 2.8 ± 0.6 | 3.3 ± 0.4 |
| 2-Phenylacetonitrile | 0.42 ± 0.09 | 0.12 ± 0.02* | 0.07 ± 0.03* | 0.25 ± 0.03* | 0.20 ± 0.04* | 0.25 ± 0.07 |
| Methylsalicylate | 0.33 ± 0.11 | 0.38 ± 0.10 | 0.14 ± 0.11 | 0.36 ± 0.07 | 0.40 ± 0.09 | 0.45 ± 0.18 |
| Eugenol | 0.16 ± 0.08 | 0.22 ± 0.11 | 0.05 ± 0.02 | 0.08 ± 0.01 | 0.13 ± 0.03 | 0.28 ± 0.1 |
Average volatile emissions (ng g FW−1 h−1 ± se) from ripe fruits of control (M82) and transgenic fruits harvested when fully ripe. Hexanol was included as a non-Phe-derived volatile. Asterisk denotes significant difference from control (P < 0.05). Statistical analysis was performed by one-way analysis of variance (ANOVA) followed by Dunnett test using GraphPad Prism software.
We next examined whether ODO1 overexpression affected the Phe level in fruits. Amino acid analysis indicated that levels of Phe were actually reduced in the three lines examined, two of them significantly (Table 4). No significant differences were observed for Tyr or Trp in the transgenic plants. Significant reductions in Met levels were observed in all three lines. This Met reduction may be related to the higher expression of SAMS observed in the transgenic lines.
Table 4.
Amino Acid Levels in Control (M82) and Transgenic Ripe Fruit Pericarp Tissue
| Amino Acid | M82 | 8117 | 7817 | 7949 |
| Asp | 6.62 ± 0.56 | 5.36 ± 0.58 | 5.88 ± 0.65 | 5.32 ± 0.31 |
| Glu | 10.10 ± 1.05 | 6.59 ± 1.00 | 9.24 ± 1.64 | 7.11 ± 1.01 |
| Asn | 3.30 ± 0.14 | 3.79 ± 0.15 | 2.66 ± 0.37 | 3.41 ± 0.15 |
| Ser | 1.05 ± 0.08 | 1.41 ± 0.41 | 1.58 ± 0.15 | 1.40 ± 0.14 |
| Gln | 17.02 ± 0.81 | 19.81 ± 0.92 | 16.78 ± 1.50 | 18.07 ± 0.85 |
| Gly | 0.21 ± 0.04 | 0.48 ± 0.07 | 0.56 ± 0.07* | 0.50 ± 0.10* |
| Thr | 1.99 ± 0.16 | 1.94 ± 0.10 | 1.77 ± 0.10 | 1.81 ± 0.11 |
| His | 0.13 ± 0.01 | 0.26 ± 0.04 | 0.12 ± 0.01 | 0.15 ± 0.02 |
| Ala | 1.96 ± 0.31 | 3.17 ± 0.47 | 2.83 ± 0.64 | 1.85 ± 0.23 |
| Arg | 0.86 ± 0.10 | 0.43 ± 0.14* | 0.52 ± 0.04 | 0.80 ± 0.10 |
| Tyr | 0.27 ± 0.06 | 0.25 ± 0.03 | 0.21 ± 0.03 | 0.21 ± 0.01 |
| Val | 1.17 ± 0.17 | 1.13 ± 0.12 | 0.89 ± 0.07 | 1.21 ± 0.14 |
| Met | 0.19 ± 0.02 | 0.03 ± 0.01* | 0.06 ± 0.02* | 0.08 ± 0.02* |
| Trp | 0.57 ± 0.11 | 0.49 ± 0.11 | 0.44 ± 0.07 | 0.65 ± 0.08 |
| Phe | 0.88 ± 0.09 | 0.29 ± 0.04* | 0.14 ± 0.06* | 0.67 ± 0.14 |
| Ile | 0.72 ± 0.12 | 0.70 ± 0.09 | 0.61 ± 0.11 | 0.89 ± 0.12 |
| Leu | 0.63 ± 0.07 | 0.37 ± 0.04* | 0.51 ± 0.06 | 0.56 ± 0.07 |
| Lys | 0.59 ± 0.04 | 0.31 ± 0.03* | 0.44 ± 0.04 | 0.51 ± 0.06 |
Each point (μg/g FW ± se) is an average of 6 to 12 independent fruits samples. Asterisk indicates significant difference from M82 (P < 0.05). Statistical analysis was performed by one-way ANOVA followed by Dunnett test.
To validate that there is increased metabolic flux through Phe and downstream phenylpropanoids, untargeted comparative metabolomics was performed on fruit tissue. Results indicated that specific phenylpropanoid conjugates were markedly altered by ODO1 expression (see Supplemental Table 3 online). Various feruloyl and coniferyl esters, some of which are not present at detectable levels in wild-type fruits, were significantly increased in all transgenic lines. On the other hand, dicaffeoyl and tricaffeoyl quinic acid esters as well as several flavonoid species were decreased. These results were further validated by quantitative measurement of phenylpropanoids following acid hydrolysis to remove sugars (Table 5). The changes in levels of various phenylpropanoids in transgenic plants may be related to their dependency on fruit ripening stage (Moco et al., 2006; Iijima et al., 2008), which is slower in ODO1-expressing fruits. Quantitative analyses of phenolic compounds indicated that there were very large increases in the pools of a subset of phenylpropanoids and no increase or reductions in the pools of others (Table 6). Large increases were observed in the levels of ferulic acid as well as coniferaldehyde, which increased from nondetectable levels in wild-type fruit to ~50 mg/kg fresh weight (FW) in transgenic line 7817 (i.e., 2-fold more than caffeic acid, the most common phenylpropanoid in wild-type fruit). There were no consistent significant differences between ODO1 and wild-type fruits in cinnamic, p-coumaric, and caffeic acids. There were also no differences in lignin observed in transgenic fruits by phloroglucinol staining (data not shown). Levels of the major flavonoids naringenin and quercetin were significantly lower than controls. Together, the data indicate that there were substantially elevated levels of certain phenylpropanoids, indicating increased metabolic activity as a consequence of ODO1 overexpression. However, increased metabolic activity was limited to specific branches of the phenylpropanoid pathway.
Table 5.
Phenylpropanoids in M82 and Transgenic ODO1 Fruits
| Compound mg/g FW | M82 n = 5 | PhODO1 7817 n = 9 | PhODO1 7948 n = 9 | PhODO1 7949 n = 3 |
| Cinnamic acid | 2.2 ± 0.1 | 1.6 ± 0.8 | 0.9 ± 0.5* | 3.5 ± 0.6* |
| p-Coumaric acid | 3.4 ± 0.5 | 2.4 ± 0.9 | 1.1 ± 0.6* | 3.0 ± 0.4 |
| Caffeic acid | 30.9 ± 2.7 | 26.1 ± 7.3 | 30.3 ± 9.1 | 27.4 ± 1.2 |
| Ferulic acid | 1.2 ± 0.2 | 3.8 ± 1.2* | 3.0 ± 1.3* | 4.4 ± 1.1* |
| Sinapic acid | ND | ND | ND | ND |
| Coniferylaldehyde | ND | 51.1 ± 13.1* | 40.7 ± 2.1* | 32.2 ± 13.8* |
| Kaempferol | 1.6 ± 0.6 | 1.9 ± 0.8 | 1.7 ± 0.6 | 1.9 ± 0.7 |
| Naringenin | 25.5 ± 10.5 | 4.2 ± 4* | ND* | ND* |
| Quercetin | 20.8 ± 11.9 | 19.5 ± 11.3 | 4.7 ± 1.6* | 4.8 ± 2.0* |
Average (n ≥ 3; ± sd) concentration (μg g−1 fresh weight) of acid hydrolyzable pools of selected phenylpropanoids and flavonoids in fruits harvested at breaker plus 7 d. ND, not detectable (<0.5 μg g−1 FW). Asterisk denotes significant differences compared to the wild type (M82) with one-way ANOVA and Dunnett’s test (P < 0.05).
Table 6.
Untargeted LC-MS Analysis of Tomato Fruit Extracts
| Change in Ph ODO1Lines | Putative ID | M82 | 7817 | 7948 | 7949 | t Test 7817 | t Test 7948 | t Test 7949 |
| n = 5 | n = 9 | n = 9 | n = 3 | |||||
| UP | Feruloyl-dihexoside-deoxyhexoside | 52 | 150 | 993 | 320 | 0.088671 | 0.000005 | 0.000007 |
| UP | 3-O-Feruloylquinic acid | 31 | 529 | 310 | 293 | 0.000001 | 0.000014 | 0.000000 |
| UP | Ferulic acid-hexose I | 229 | 904 | 623 | 806 | 0.000089 | 0.019063 | 0.000016 |
| UP | Ferulic acid-hexose II | 305 | 1,828 | 1,512 | 2,332 | 0.000654 | 0.001690 | 0.000003 |
| UP | 2-(3,4-Dihydroxyphenyl)ethanol; 3′-Me ether, hexoside deoxyhexoside | 38 | 383 | 1,620 | 163 | 0.049449 | 0.000110 | 0.000001 |
| UP | 3-(3,4,5-Trihydroxyphenyl)-1,2-propanediol; 3′,4′-Di-Me ether, hexoside | 33 | 1,141 | 2,090 | 1,436 | 0.000124 | 0.000000 | 0.000003 |
| UP | 3-(3,4,5-Trihydroxyphenyl)-2-propen-1-ol, 3′-Me ether, hexoside | 216 | 1,383 | 768 | 1,036 | 0.000015 | 0.000682 | 0.000001 |
| UP | Acetoxy-tomatine + FA | 1,550 | 23,046 | 3,438 | 2,430 | 0.000791 | 0.004964 | 0.023984 |
| UP | α-Tomatin + FA | 3,575 | 8,670 | 5,333 | 4,432 | 0.076583 | 0.060116 | 0.438996 |
| UP | Benzyl alcohol-hexose-pentose | 39 | 965 | 422 | 774 | 0.000137 | 0.002707 | 0.000177 |
| UP | β-Tomatine | 40 | 1,196 | 1,239 | 1,255 | 0.000023 | 0.000209 | 0.000000 |
| UP | C10H12O3-glutathione ester | 35 | 1,542 | 1,961 | 2,474 | 0.000178 | 0.000086 | 0.000000 |
| UP | Coniferyl aldehyde-C7H12O7 ester | 29 | 4,885 | 8,190 | 6,007 | 0.000020 | 0.000000 | 0.000000 |
| UP | Dihydroconiferyl alcohol-hexoside | 51 | 3,458 | 4,914 | 3,183 | 0.000001 | 0.000000 | 0.000000 |
| UP | Glycoalkaloid | 1,437 | 3,508 | 1,011 | 1,205 | 0.022306 | 0.017561 | 0.338359 |
| UP | Kaempferol-(coumaroyl)dihexose-deoxyhexose | 30 | 211 | 150 | 114 | 0.001640 | 0.002464 | 0.000981 |
| UP | Lycoperoside H or hydroxytomatine II + FA | 588 | 1,507 | 776 | 695 | 0.014544 | 0.027100 | 0.077155 |
| UP | Quercetin-dihexose-deoxyhexose | 654 | 2,577 | 1,729 | 1,683 | 0.000050 | 0.000266 | 0.001391 |
| UNCHANGED | UDP-glucose | 9,128 | 8,675 | 10,927 | 9,510 | 0.819235 | 0.119371 | 0.804161 |
| DOWN | 3-Caffeoylquinic acid (chlorogenic acid) | 1,682 | 1,150 | 350 | 433 | 0.036787 | 0.000001 | 0.001936 |
| DOWN | 4-Caffeoylquinic acid | 369 | 298 | 149 | 180 | 0.217157 | 0.000061 | 0.034939 |
| DOWN | 5-Caffeoylquinic acid | 4,986 | 5,664 | 1,491 | 1,196 | 0.696645 | 0.000005 | 0.002737 |
| DOWN | Tricaffeoyl quinic acid I | 1,981 | 962 | 240 | 575 | 0.000432 | 0.000002 | 0.004658 |
| DOWN | Tricaffeoyl quinic acid II | 1,934 | 1,094 | 165 | 316 | 0.025902 | 0.000000 | 0.002046 |
| DOWN | Dicaffeoylquinic acid II | 654 | 396 | 52 | 104 | 0.013309 | 0.000000 | 0.000601 |
| DOWN | Dicaffeoylquinic acid III | 2,624 | 1,600 | 385 | 458 | 0.055148 | 0.000002 | 0.003025 |
| DOWN | Caffeic acid-hexose III | 327 | 180 | 103 | 222 | 0.023263 | 0.000088 | 0.152436 |
| DOWN | Coumaric acid-hexose I | 321 | 235 | 95 | 604 | 0.679422 | 0.000903 | 0.005196 |
| DOWN | DehydroesculeosideA + FA | 1,854 | 955 | 953 | 966 | 0.000669 | 0.000201 | 0.017773 |
| DOWN | EsculeosideA+ FA | 704 | 392 | 412 | 344 | 0.000811 | 0.000740 | 0.010312 |
| DOWN | Glycoalkaloid | 260 | 29 | 49 | 74 | 0.000000 | 0.000001 | 0.001894 |
| DOWN | Hydroxyphenylethanol-rutinoside | 394 | 115 | 79 | 151 | 0.000176 | 0.000086 | 0.044684 |
| DOWN | Kaempferol-3-O-rutinoside | 711 | 660 | 158 | 63 | 0.693256 | 0.000037 | 0.001177 |
| DOWN | Kaempferol-hexose-deoxyhexose, -pentose | 649 | 523 | 126 | 41 | 0.369613 | 0.000009 | 0.001515 |
| DOWN | Lycoperoside F or G I (+ FA) | 2,819 | 1,350 | 1,593 | 1,547 | 0.000059 | 0.000187 | 0.012161 |
| DOWN | Lycoperoside F or G II + FA | 26,392 | 23,220 | 23,937 | 23,781 | 0.012049 | 0.003229 | 0.021076 |
| DOWN | Naringenin | 1,077 | 258 | 61 | 81 | 0.000035 | 0.000000 | 0.001677 |
| DOWN | Naringenin chalcone | 9,742 | 2,776 | 821 | 1,346 | 0.000001 | 0.000000 | 0.000026 |
| DOWN | Naringenin-chalcone glucoside | 214 | 57 | 40 | 46 | 0.000008 | 0.000000 | 0.001140 |
| DOWN | Phloretin-dihexoside | 732 | 245 | 76 | 22 | 0.000169 | 0.000000 | 0.000112 |
| DOWN | Quercetin-3-O-rutinoside | 26,110 | 23,176 | 5,140 | 5,954 | 0.253163 | 0.000000 | 0.000179 |
| DOWN | Quercetin-hexose-deoxyhexose, -pentose | 8,941 | 6,747 | 582 | 178 | 0.410582 | 0.000823 | 0.041050 |
Peak height (ion count) of (putatively) identified compounds in M82 and ODO1 transgenic fruits. Values are average of the indicated number of fruits harvested at breaker plus 7 d. The amount of all compounds differ significantly (P < 0.05) from wild-type M82 in at least two out of three transgenic lines as indicated in the status row: UP, upregulated; DOWN, downregulated in transgenic lines compared to M82. One unaltered metabolite is labeled “UNCHANGED.”
To further characterize these changes, we conducted a stable isotope labeling experiments wherein we incubated pericarp discs isolated from wild-type or ODO1-expressing red ripe fruits and incubated them for 6 h in buffered solution containing 10 mM [2-13C]-phosphoenolpyruvate, a metabolite upstream of Phe biosynthesis. Following this incubation, the samples were washed and snap-frozen in liquid nitrogen, and subsequently the isotope redistribution was quantified by GC-MS as described by Tieman et al. (2006a) as well as by liquid chromatography–mass spectrometry (LC-MS) using a modified version of the protocol of Tohge and Fernie (2010). The total label detected in Phe, the volatile phenethylamine, and 14 phenylpropanoid compounds are presented in Table 7. To our knowledge, there are very few studies to date that directly characterize metabolic fluxes through this pathway in plants. However, in cases in which it was possible to compare the values obtained for the wild type in our study with those reported previously for potato tuber (Matsuda et al., 2003; Heinzle et al., 2007), results are broadly similar. Comparing the label accumulation in these compounds in the ODO-expressing fruit to that in the wild-type control revealed that the vast majority of the compounds were substantially increased in the overexpressor with the changes in isotope accumulation in 2-(3,4-dihydroxyphenyl)ethanol 3′-Me ether, hexoside deoxyhexoside, 3-O-feruloylquinic acid, 3-(3,4,5-trihydroxyphenyl)-1,2-propanediol;3′,4'-Di-Me ether, hexose, and dihydroconiferyl alcohol-hexoside by and large mirroring those of the steady state levels of these compounds reported in Table 6. By contrast, the label accumulating in chlorogenic acid and in Phe itself was significantly lower in the ODO1 fruits. However, in the former case as well as for the volatile phenylethylamine, changes in the isotope accumulation did not translate into a change in flux due to the differential labeling of the precursor pools between the genotypes (see legend of Table 7 for details). By contrast, flux to the phenylpropanoids was substantially upregulated in the ODO line. Indeed, absolute fluxes could be calculated for all 14 phenylpropanoids in this line, but absolute values could only be determined for seven of these in addition to chlorogenic acid in the wild type. Of these seven absolute fluxes that were determined in both genotypes, all were between 7-fold and almost 1000-fold higher in the ODO-overexpressing fruits. We additionally were able to detect the unlabeled peak only in two of the compounds in the wild type, and since it is apparent that this is <100% labeled, we are able to state that the fluxes to both peaks 3 and 4 were at least 3- and 14-fold higher in the ODO fruits, respectively. Similarly, if we set the limit of detection as the upper value for labeling of peaks 6, 8, and 14, the fluxes to these phenylpropanoids are at least 50-, 30-, and 12-fold higher in the ODO fruits than the control. Taken together, these data suggest that the overexpression of PhODO results in upregulating flux through specific branches of the phenylpropanoid network.
Table 7.
Metabolic Flux through into Phenylpropanoids in Tomato
| Method | Peak No. | Peak | m/z | 13C Accumulation (μmol g FW−1) | Flux from Phe (μmol g FW−1 h−1) | ||
| M82 | ODO | M82 | ODO | ||||
| GC-MS | Phe | 165 | 0.0036±0.0004 | 0.0014 ± 0.0002 | |||
| GC-MS | Phenethylamine (Pea) | 121 | 0.204±0.019 | 0.4907 ± 0.0385 | 0.0067 ± 0.0005 | 0.0094 ± 0.0013 | |
| μmol CGA equivalents g FW−1 | Flux from Phe (μmol CGA equivalents g FW−1 h−1) | ||||||
| LC-MS | peak_1 | 3-CGA | 353 | 1.877±0.616 | 0.481 ± 0.347 | 2.393 ± 0.837 | 1.058 ± 0.791 |
| LC-MS | peak_2 | Putative phenolic compound | 427 | 0.609±0.232 | 4.699 ± 0.781 | 0.77 ± 0.28 | 10.217 ± 1.767 |
| LC-MS | peak_3 | Putative phenolic compound | 463 | 0.013a ± 0.004 | 8.015 ± 3.182 | 5.599 ± 3.418 | 17.353 ± 6.575 |
| LC-MS | peak_4 | Putative phenolic compound | 486 | 0.036a ±0.012 | 4.763 ± 1.267 | 3.015 ± 2.472 | 10.396 ± 3.05 |
| LC-MS | peak_5 | Putative phenolic compound | 387 | 0.031±0.012 | 5.307 ± 1.386 | 0.039 ± 0.014 | 11.488 ± 2.733 |
| LC-MS | peak_6 | Putative phenolic compound | 521 | <0.3b | 4.748 ± 1.628 | <0.195b | 10.367 ± 3.724 |
| LC-MS | peak_7 | Putative phenolic compound | 389 | 0.016±0.001 | 15.01 ± 4.478 | 0.0.20 ± 0.002 | 32.476 ± 8.943 |
| LC-MS | peak_8 | Putative phenolic compound | 445 | <0.3b | 2.673 ± 0.595 | <0.195b | 5.805 ± 1.269 |
| LC-MS | peak_9 | 2-(3,4-Dihydroxyphenyl)ethanol 3′-Me ether, hexoside deoxyhexoside | 475 | 0.01±0.008 | 3.326 ± 1.543 | 0.013 ± 0.01 | 7.177 ± 3.164 |
| LC-MS | peak_10 | Putative phenolic compound | 521 | 0.075±0.031 | 2.672 ± 1.245 | 0.096 ± 0.04 | 5.766 ± 2.554 |
| LC-MS | peak_11 | 3-O-Feruloylquinic acid | 385 | 0.083±0.032 | 8.051 ± 1.494 | 0.106 ± 0.042 | 17.447 ± 2.833 |
| LC-MS | peak_12 | 3-(3,4,5-Trihydroxyphenyl)-1,2-propanediol;3′,4′-Di-Me ether, hexose | 389 | 0.017±0.007 | 5.559 ± 0.75 | 0.022 ± 0.01 | 12.067 ± 1.5 |
| LC-MS | peak_13 | Dihydroconiferyl alcohol-hexoside | 343 | 0.0270.009± | 5.182 ± 1.859 | 0.034 ± 0.012 | 11.212 ± 3.79 |
| LC-MS | peak_14 | Putative phenolic compound | 429 | <0.3b | 1.109 ± 0.231 | <0.195b | 2.409 ± 0.489 |
Amount of labeled Phe, phenylethylamine, chlorogenic acid (3-CGA), and phenylpropanoids and derived fluxes from the Phe pool (estimated by division of the isotope accumulation in a compound by the molar fractional enrichment of the Phe pool, which were determined to be 13.1 ± 1.4 and 7.7 ± 1.0 for M82 and ODO, respectively) after [13C]phosphoenolpyruvate feeding for 6 h. Data are means of three replicates ± se. m/z, mass-to-charge ratio.
These compounds are considered as fully labeled.
Detection limit.
DISCUSSION
The factors that regulate synthesis of plant volatile compounds from their precursors are not well understood. In an effort to better understand the relationship between Phe and its volatile metabolites in tomato, we overexpressed an R2R3-type MYB transcription factor from petunia, ODO1, which coordinately alters expression of a set of genes in primary and secondary metabolic pathways related to aromatic amino acids. Ectopic ODO1 expression resulted in elevated levels of a subset of phenylpropanoid compounds but did not significantly increase related compounds, including Phe (Table 4), Phe-derived volatiles, lignin, and flavonoids. Such specific metabolite alterations have been observed previously within phenylpropanoid pathways (Liu et al., 2002; Winkel, 2004).
The significantly increased accumulation of multiple phenylpropanoids, including coniferaldehyde and ferulic acid as well as their hexose esters, indicates that the entire pathway for Phe synthesis was upregulated in the transgenic plants. The small number of genes with significant induction in both transgenic lines suggests specificity for functions related to synthesis and metabolism of aromatic amino acids. This coordinated upregulation of the Phe biosynthetic pathway in turn led us to examine the set of induced genes for clues regarding the as yet unidentified step in that pathway. We hypothesized that one or more of the coordinately regulated transcripts would encode an enzyme that converts prephenate to either arogenate or PPY. There is strong evidence that plants synthesize Phe via arogenate (Bonner and Jensen, 1985; Siehl et al., 1986; De-Eknamkul and Ellis, 1988). One candidate for the gene encoding PAT, SGN-U567172, was annotated as an Asp aminotransferase. Since Asp or Glu can act as amino group donors for prephenate (Bonner and Jensen, 1985; De-Eknamkul and Ellis, 1988), a genuine PAT might be annotated as an Asp aminotransferase. Assays with recombinant protein as well as complementation of an E. coli Phe auxotroph indicated that SGN-U567172 does encode PAT. Analysis of public microarray results indicate that tomato PAT is constitutively expressed throughout fruit development at a constant level from fertilization to red ripe (http://ted.bti.cornell.edu/cgi-bin/TFGD/array_data spot ID 1-1-3.1.17.4). Phylogenetic analysis of related genes in the tomato genome indicates that there is only a single gene providing this function in tomato (Figure 3). The most closely related sequence in the tomato genome, SGN-U585067, is cytoplasmic (Figure 4) and did not exhibit activity toward prephenate or PPY. The next closest upregulated tomato sequence, SGN-U567390, lacks an obvious transit peptide and clusters closer to an Arabidopsis protein, At1G71920, with demonstrated histidinol-phosphate transaminase activity (Mo et al., 2006). We were unable to demonstrate activity with any of the tested substrates with this enzyme. PAT is evolutionarily conserved, as closely related proteins from pine (Pinus pinaster), rice (Oryza sativa), Chlamydomonas, and Physcomitrella can be identified (Figure 3). Thus, the arogenate pathway is likely to be evolutionarily conserved in plants. A subset of bacteria, most notably cyanobacteria, also synthesize Phe exclusively via arogenate (Whitaker et al., 1981). However, there are currently no sequences available for bacterial aminotransferase enzymes that exclusively produce arogenate.
We demonstrated that the closest homolog of Sl PAT in Arabidopsis (At2g22250) also encodes a PAT. Like tomato, Arabidopsis appears to contain only a single copy of PAT. An At2g22250 knockout is embryo lethal, arresting at the one-cell stage of development (Pagnussat et al., 2005). This lethality is consistent with an essential, nonredundant function in amino acid synthesis and supports the hypothesis that the arogenate pathway is the main route to Phe. Sl PAT, together with ADT, can synthesize Phe (Figure 5; see Supplemental Figure 6 online). It is likely that At PAT also acts in concert with a family of previously characterized ADTs (Cho et al., 2007). Maeda et al. (2011) recently reported that a highly homologous PAT from petunia also performs the same function. Thus, the arogenate pathway is conserved across multiple plant species.
That only 28 genes without obvious connection to Phe metabolism were significantly upregulated (Supplemental Table 1 online) in both transgenic lines indicates that most, if not all, of the metabolic and transcriptional differences are likely to be directly related to ODO1 action. These genes are good candidates for additional factors influencing phenylpropanoid metabolism. For example, the putative glucosyltransferase (SGN-U217248) is an obvious candidate for ferulic and sinapic acid glycosylation. It is likely that the increased SAMS transcription is related to increased demand for C1 groups associated with synthesis of ferulic and sinapic acids. This increased C1 demand may also explain the significant Met and ethylene reductions observed in transgenic fruits. Notably, there is one MYB gene, SGN-U563435, whose expression is significantly increased in transgenic fruits. The closest homologs of this gene in Arabidopsis (Jin et al., 2000) and Vitis vinifera (Matus et al., 2009) are associated with repression of anthocyanin gene expression, consistent with the reductions we observed in flavonoid accumulation.
One of our goals was to understand the relationship between Phe pools and Phe-related volatile synthesis; specifically, if more Phe is synthesized, would that translate into more phenylpropanoid volatiles? While reducing Phe synthesis clearly reduced phenylpropanoid and benzenoid volatile synthesis in petunia flowers, a specific volatile-associated function for ODO1 was not demonstrated. The effects of reduced ODO1 expression on nonvolatile petunia phenylpropanoids was also not previously examined (Verdonk et al., 2005). Thus, the effects of ODO1 overexpression in tomato fruits could not be predicted. Our results indicate that only a subset of tomato phenylpropanoids was highly altered. There were no observable effects on lignin accumulation in fruits, despite the significant increases in precursors. Steady state levels of Phe were actually reduced in the transgenic plants. The smaller Phe pool, combined with no significant increase of AADC expression, explains the lack of effect on an important subset of the Phe-derived volatiles. Chalcone synthase and chalcone isomerase expression was not altered, and significantly lower levels of naringenin (chalcone) and quercetin were observed. Reduced levels of these flavonoids may be the consequence of higher expression of genes directing synthesis of an alternate set of phenylpropanoids, most notably glycosides of ferulic acid and coniferyl aldehyde that exhibited massive increases. Higher ferulic acid appears to stimulate formation of hexose conjugates of ferulic acid (and sinapic acid). It will be interesting to determine whether the coordinately induced glucosyltransferase (SGN-U217248) is responsible for synthesis of these hexose esters. The massive increases in a subset of phenylpropanoids clearly demonstrate increased metabolic flux in tomato fruit tissues. This increased activity of the Phe synthetic pathway was insufficient to increase synthesis of the most important flavor-associated volatiles (phenylacetaldehyde, 2-phenylethanol, and 1-nitro-2-phenethane), possibly because Phe was rapidly converted to other downstream nonvolatile phenylpropanoids. We have also observed a lack of correlation between branched-chain amino acids and their corresponding volatiles in a Solanum pennellii introgression population (Schauer et al., 2006; Tieman et al., 2006b; Maloney et al., 2010). The first committed step in synthesis of the major flavor-associated volatiles is encoded by a small family of AADCs (Tieman et al., 2006a). Based on previous analysis of transgenic plants with altered AADC expression, we determined that the AADC enzymes are rate-limiting to synthesis of these volatiles. Taken together, we conclude that substrate (i.e., Phe) is not a major control point for synthesis of these volatiles. The benzenoid volatiles, synthesized from phenylpropanoids, were also not increased in ODO1-expressing fruits. In fact, synthesis of benzaldehyde was actually significantly lower in the transgenic fruits.
The delayed and irregular fruit ripening phenotype observed in the transgenic lines (Figure 1) cannot be explained at this time, although the low levels of Phe and Met could potentially slow down metabolism. The nonresponsiveness to exogenous propylene treatment indicates that the phenotype is unrelated to ethylene, the most obvious candidate metabolite. Another candidate hormone is IAA, since it is derived from Trp. However, there were no significant differences in either the Trp or IAA contents of the transgenic fruits or in the expression of several genes known to be auxin regulated (see Supplemental Figure 4 online).
In conclusion, we used ectopic expression of a MYB transcription factor to alter a set of genes related to Phe metabolism in tomato fruits. The coordinated upregulation of genes whose products direct synthesis of Phe led to identification of the tomato PAT gene. This increased metabolic activity, which was coupled to a considerably enhanced flux through part of the phenylpropanoid pathway, did not result in higher levels of Phe-derived flavor volatiles, indicating that factors beyond substrate availability limit their synthesis. The specific accumulation of a subset of phenylpropanoid metabolites is in accordance with previous observations in potato tuber discs following wounding (Matsuda et al., 2003) and as such may represent a common regulatory motif in plant secondary metabolism. These data therefore illustrate both the potential for and challenges presented by metabolic engineering fruits for improved flavor and nutrition.
METHODS
Plant Material
Transgenic Solanum lycopersicum cv M82 plants were produced as described (McCormick et al., 1986). Plants were grown in University of Florida greenhouses or in the field (Live Oak, Florida) according to standard agronomic practices. Ethylene measurements were performed as previously reported (Dal Cin et al., 2009). For propylene treatments, fruits were harvested at breaker stage, treated with 500 ppm of propylene for 24 h, and monitored for ripening.
Construct Preparation
A full-length petunia (Petunia hybrida) ODO1 cDNA was cloned into a previously described E8 promoter vector (Díaz de la Garza et al., 2004). This construct was used for production of transgenic plants with kanamycin resistance as a selectable marker. For recombinant protein production, full-length open reading frames with or without the predicted transit peptide of SGN-U567172, SGN-567390, SGN-U585067, and At2g22250 from leaf cDNA were obtained with High Fidelity Phusion DNA polymerase (FINNZYMES, purchased through Fisher Scientific). Leaf cDNAs were prepared from tomato M82 or Arabidopsis thaliana RNA prepared as previously described (Dal Cin et al., 2007). Primers for amplification of the individual constructs are shown in Supplemental Table 4 online. Protein sequences were screened for predicted signal peptides, cleavage sites, and subcellular locations using TargetP (http://www.cbs.dtu.dk/services/TargetP/), ChloroP (http://www.cbs.dtu.dk/services/ChloroP/), and SignalP (http://www.cbs.dtu.dk/services/SignalP/).
Microarray Hybridization and Analysis
Fruit samples were collected at Stage 2 and Stage 2 plus 5 d from lines 7949 and 8117 and control M82. RNA was prepared from four independent biological replicates, each consisting of at least 10 fruits for each time point. RNA was extracted using the methods described by Griffiths et al. (1999). Four biological replicates of each genotype/stage were used in microarray comparisons between the two stages of each genotype. For each biological replicate, two independently synthesized cDNA samples were created and labeled according to the protocol described (Alba et al., 2004) and modified as described in the Tomato Functional Genomics Database (www.ted.bti.cornell.edu) using Cy5 or Cy3 dyes (CyTM Dye Post-labeling Reactive Dye Pack; GE Amersham). Quality and quantity of Cy-labeled cDNA were determined as described (Alba et al., 2004). The result was eight independent probes and hybridizations per comparison (four biological replicates, each labeled with two different dyes) resulting in four biological and four technical replicates for each probe/hybridization.
Transcriptome analyses were performed using the TOM2 long oligonucleotides microarray. Hybridizations and washing regimes were performed using the protocol described (Alba et al., 2004) modified to include the addition of a second wash with wash solution 1 (1× SSC, 0.2% SDS; preheated to 43°C) as well as an additional rinse with wash solution 3 (0.1× SSC; room temperature) when transferring the arrays from wash solution 2 (0.1× SSC, 0.2% SDS; room temperature) to wash solution 3. The full modified protocol can be found at http://ted.bti.cornell.edu/cgi-bin/TFGD/array/home.cgi. The arrays were probed with 200 pmol of Cy5- and Cy3-labeled probes simultaneously. Numerical images were generated from the raw microarray scans using ImaGene software (version 5.5; BioDiscovery). Spots with low intensities (i.e., less than the local background) and of poor quality were flagged by the software and not included in subsequent statistical analysis. Only spots with at least four replicate data points were included in subsequent analysis. Normalization used the print-tip strategy and was applied to ratio values for each array using the marray package in Bioconductor (Yang et al., 2002). Differentially expressed genes were identified using Patterns from Gene Expression (Grant et al., 2005). Genes with false discovery rates of <0.05 and fold change greater than or equal to 2-fold were identified as differentially expressed genes.
Expression Analysis
Expression of Ph ODO1 in transgenic fruits was assessed by quantitative RT-PCR. A minimum of 10 fruits from each line was collected at breaker and allowed to ripen at room temperature for 7 d. RNA was extracted and cDNA synthesis was performed as previously described (Dal Cin et al., 2005a, 2005b). Expression analysis was performed with SYBR green (Applied Biosystems) as previously reported (Cin et al., 2005) with specific primers (see Supplemental Table 4 online). A titration curve was also prepared for Ph ODO1, Sl UBI, and 18S RNA. For validation of gene expression, RNA was prepared from the same tissue samples used in the microarray experiments described above. Ubiquitin and 18S RNA were used as controls.
Amino Acid Analysis
Amino acids were analyzed using HPLC with o-phthalaldehyde derivitization according to the method of (Kreft et al. 2003). Detection and quantification of products were based on the modification of the primary amino group with o-phthalic acid dialdehyde to a fluorescing derivative, which was measured using a fluorometric detection system. One hundred and sixty microliters of 80% (v/v) aqueous ethanol extract (buffered with 2.5 mM HEPES/KOH, pH 6.8) was mixed with 40 μL of borate buffer (1 M, pH 10.7), incubated for 10 min at room temperature, and centrifuged. Supernatant was mixed with o-phthalaldehyde, and 5 μL of the mixture was injected immediately onto the column (HyperClone ODS [C18], 3 μ 120A; Phenomenex) and eluted with buffer A (water with 0.22% of 0.4 M sodium phosphate buffer and 0.02% THF) and buffer B (water:acetonitrile:methanol, 45:22:35, 2.5% of 0.4 M sodium phosphate) at a flow rate of 1.0 mL/min. Separation conditions were as follows: 0 to 3 min, isocratic elution with 0% B; 3 to 5 min, linear gradient from 0 to 7% B; 5 to 14 min, linear gradient from 7 to 40% B; 14 to 18 min, linear gradient from 40 to 45% B; 18 to 28 min, linear gradient from 45 to 100% B; 28 to 31 min, isocratic elution with 100% B. Equilibration conditions were as follows: 31 to 34 min, isocratic elution with 100% A. Peak areas were integrated using Chromeleon software 6.8 (Dionex) and subjected to quantification by means of calibration curves made from standard mixtures.
Metabolite Analysis
Phenolic and flavonoids were quantified by HPLC-PDA after acid hydrolysis (Muir et al., 2001). Untargeted metabolomics profiling was performed on aqueous-methanol extracts using a HPLC-PDA-QTOF MS system with accurate mass detection (De Vos et al., 2007). Unbiased peak picking and alignment was performed using the accurate mass version of MetAlign (Lommen, 2009) using a signal-to-noise threshold ratio of 5. The resulting accurate mass peak list, containing 2075 unique mass signals, was filtered for signals present in at least three samples (i.e., number of biological replicates per genotype) and then subjected to an in-house developed script (MSClust; Tikunov et al., 2010) to cluster all signals derived from the same metabolite. The resulting 152 centrotypes (=metabolites) were subjected to statistical analysis using a pairwise t test to compare M82 and transgenic lines. Significantly different metabolites (P ≤ 0.05) were manually annotated by matching to the tomato fruit specific databases MotoDB (Moco et al., 2006) and Komics (http://webs2.kazusa.or.jp/komics), and, in the case of novel tomato compounds, to accurate mass databases such as Metabolomics Japan (http://metabolomics.jp) and the Dictionary of Natural Products (http://dnp.chemnetbase.com), using a maximum deviation of observed mass to calculated mass of 5 ppm. Candidate structures were checked for corresponding in-source fragments and UV/Vis absorbance characteristics, if present, and using commercially available authentic standards.
Volatile Analysis
Volatiles were collected from tomato fruits according to (Tieman et al. 2006b). Briefly, air was passed over the samples and volatiles were collected on a SuperQ Resin for 1 h. Volatiles were eluted from the column with methylene chloride and run on a gas chromatograph equipped with a flame ionization detector.
Isotope Labeling Experiments
For [13C]-labeling studies, pericarp disks weighing ~600 mg were supplied with 10 μmol of [2-13C]phosphoenolpyruvate (Sigma-Aldrich) in 50 μL of water for 6 h. Samples were homogenized split into two and were extracted as described previously (Schauer et al., 2006; Tohge and Fernie, 2010). The absolute concentration of metabolites was determined as defined by Tieman et al. (2006a) for primary metabolites and phenethylamine and by comparison to standard concentration curves for chlorogenic acid for secondary metabolites. Analysis of [1-13C]phosphoenolpyruvate–labeled samples was performed exactly as described previously (Tieman et al., 2006a). Uncorrected molar percentage enrichments were evaluated as defined by Roessner-Tunali et al. (2004). The reaction rates from metabolic precursors through intermediates to end-products was estimated by dividing the amount of label accumulating in the product by the calculated average proportional labeling of the precursor pool.
Phylogenetic Analysis
Members of the aminotransferase family were retrieved from KEGG (http://www.genome.jp/kegg-bin/show_pathway?org_name=athandmapno=00400andmapscale=1.0andshow_description=show) and the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (see Supplemental Data Set 1 online). The evolutionary history was inferred using the Maximum Parsimony method (Eck and Dayhoff, 1966). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The Maximum Parsimony tree was obtained using the Close-Neighbor-Interchange algorithm (Nei and Kumar, 2000) with search level 7 in which the initial trees were obtained with the random addition of sequences (10 replicates). The tree is drawn to scale, with branch lengths calculated using the average pathway method (Nei and Kumar, 2000) and are in the units of the number of changes over the whole sequence. All positions containing gaps and missing data were eliminated from the data set (Complete Deletion option). There were a total of 171 positions in the final data set, out of which 169 were parsimony informative. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). An alternative Bayesian approach was also used to determine phylogenetic relationships, with consistent results obtained (see Supplemental Figure 7 online).
Subcellular Protein Localization
Full-length open reading frames were amplified with primers for insertion into the Gateway vector pDONOR221. The open reading frames were then cloned into the expression vector pK7FWG2 (Karimi et al., 2007), resulting in C-terminal GFP fusion proteins. A truncated form of Sl PAT lacking the predicted transit peptide was also created. Constructs were inserted into Agrobacterium tumefaciens ABI and transiently introduced into Nicotiana benthamiana. Mitotrack (Invitrogen) was used to visualize mitochondria and the Mcherry gene (Avisar et al., 2008) to visualize peroxisomes. Protoplasts were prepared as described (Yoo et al., 2007) and observed with a Zeiss confocal microscope at the University of Florida ICBR facility (http://www.biotech.ufl.edu/cellomics/). GFP was excited at 488 nm with an argon laser exciting and visualized between 500 and 530 nm. Chlorophyll autofluorescence was visualized at 633 nm by excitation with a helium-neon laser at 543 nm. Mitochondria and peroxisomes were visualized at 576 nm. Cell size was determined with the program ImageJ
PAT Assays
Full-length open reading frames were amplified as described above using specific primers (see Supplemental Table 4 online) that introduced a C-terminal 6X-His tag. Sl PAT was also engineered without the predicted chloroplast transit peptide by replacing the first 19 amino acids with Met. A control plasmid contained only the His tag from pET28b+. The open reading frames were cloned into pENTR/SD-TOPO and then recombined into pDEST14 (Invitrogen). The resulting expression plasmids along with the pG-KJE8 or the pTf16 chaperone plasmid (Takara Bio) were transformed into Escherichia coli BL21AI (Invitrogen). Protein expression was induced with arabinose and tetracycline or just arabinose (depending on the chaperone plasmid used). Cells were centrifuged and lysed by sonication in PBS, pH 7.4, 0.1 mM pyridoxal phosphate, and Protease Inhibitor Cocktail for His-tagged purification (Sigma-Aldrich) according to the manufacturer’s directions. Proteins were purified from cell lysates with TALON affinity purification resin (Clontech) according to the manufacturer's instructions. His-tagged recombinant proteins were eluted with 150 mM imidazole, PBS buffer, pH 7.0, and 0.1 mM pyridoxal phosphate. Aminotransferase activity was measured using coupled assays with either Glu dehydrogenase (Prohl et al., 2000) or malate dehydrogenase (Bonner and Jensen, 1987) as described. The assay uses several α-keto acids as substrate and Glu or Asp as amino group donor to produce arogenate and either 2-oxoglutarate or oxaloacetate. 4-Hydroxyphenylpyruvate and PPY were also tested as possible Sl PAT substrates with no conversion observed. Reactions were performed at 24°C. Proteins were quantified by the method of Bradford (1976) and by densitometry obtained from the Coomassie Brilliant Blue–stained gels and from images generated from protein blots probed with an anti-His tag antibody. Kinetic parameters were calculated using GraphPad Prism 5 for Windows software (GraphPad Software). Prephenate was converted from the barium salt to the potassium salt with potassium sulfate (Bonner and Jensen, 1985) and quantified by hydrolysis to PPY with HCl and reading the absorbance at 440 nm after the reaction with 2,4-dinitrophenylhydrazine as previously described (Görisch, 1978). The presence of arogenate in the reaction products was determined by acid treatment followed by Phe quantitation (Zamir et al., 1983; Bonner and Jensen, 1985). Conversion of arogenate to Phe was achieved by adding 5 μL 3 n HCl to 100 μL of the reaction mix, incubating at 37° for 15 min, followed by addition of 5 μL 3 n NaOH (Bonner and Jensen, 1985). Phe levels were then determined by derivitization with methyl chloroformate and quantification by GC-MS using the method of Chen et al. (2010). Heat stability of Sl PAT was tested by incubation of enzyme for 10 min at 70°C prior to assay (Bonner and Jensen, 1985).
Complementation of the E. coli Phe Auxotroph
The full-length open reading frames of Sl PAT and ADTs SGN-U583010 and SGN-U573964 were amplified and cloned into vector pENTR/D-TOPO (Invitrogen). Sl PAT was then recombined into the His-tagged expression vector pDEST17. The ADT open reading frames were recombined into a His-tagged expression vector containing the E. coli p15A origin of replication from plasmid pACYC184 (Chang and Cohen, 1978) and a gene for spectinomycin resistance. A third plasmid was constructed for isopropyl β-D-1-thiogalactopyranoside-inducible expression of the Sl PAT and ADT in the E. coli mutant. A BamHI fragment containing the T7 RNA polymerase gene under control of the lac UV5 promoter from pAR1219 (Davanloo et al., 1984) was cloned into vector pCOLADuet-1 (Novagen) containing the ColA origin of replication and kanamycin resistance. The plasmids expressing Sl PAT, ADT, and the T7 RNA polymerase were introduced into E. coli strain DA15 (Johnson et al., 1989) containing the pheA18:Tn10 mutation. The complemented E. coli was grown on minimal medium supplemented with IPTG for induction of the T7 RNA polymerase and production of the recombinant proteins. Production of Sl PAT and ADT proteins in E. coli was confirmed by protein immunoblotting with anti-His tag antibodies.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: S. lycopersicum PAT (SGN-U567172) and HQ394171 Arabidopsis PAT (At2g22250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1 Quantitative RT-PCR Detection of Petunia ODO1 Transcript in Transgenic Tomato Fruits.
Supplemental Figure 2. Ethylene Emissions from M82 and Transgenic Fruits.
Supplemental Figure 3. Propylene Treatments.
Supplemental Figure 4. Auxin and Auxin-Regulated Gene Expression in Transgenic Fruits.
Supplemental Figure 5. Quantitative RT-PCR on Selected Genes.
Supplemental Figure 6.Phe Synthesis by Sl PAT and ADT.
Supplemental Figure 7. Alternative Phylogenetic Tree.
Supplemental Table 1. Expression Patterns of Genes with No Apparent Relations to Phe and Phenylpropanoid Synthesis.
Supplemental Table 2. Acid-Mediated Conversion of Arogenate to Phe.
Supplemental Table 3. Untargeted LC-MS Analysis of Tomato Fruit Extracts.
Supplemental Table 4. List of DNA Primers Used.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 3.
Acknowledgments
This work was supported by National Science Foundation Grant IOS-0923312 (to H.J.K. and J.G.), by the Florida Agricultural Experiment Station, the Centre of Biosystems Genomics (M.T.S.-K., M.A.H., and R.C.H.D.) and the Netherlands Metabolomics Centre that are part of the Netherlands Genomic Initiative (R.C.H.D.), an Alexander von Humboldt scholarship (T.T.), and by the Max-Planck-Society (T.T., S.O., and A.R.F.).
AUTHOR CONTRIBUTIONS
V.D., D.M.T., and H.J.K. designed the research, performed research, analyzed data, and wrote the article. T.T., R.M., and R.C.H.D. performed research and analyzed data. S.O., E.A.S., and M.G.T. performed research. M.T.S.-K., R.C.S., and M.A.H. analyzed data. J.G. designed the research. A.R.F. designed the research, analyzed data, and wrote the article.
References
- Alba R., et al. (2004). ESTs, cDNA microarrays, and gene expression profiling: Tools for dissecting plant physiology and development. Plant J. 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A.I., Makarova K.S., Koonin E.V., Dolja V.V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester A.R., et al. (2010). Biochemical and molecular analysis of pink tomatoes: Deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C.A., Jensen R.A. (1985). Novel features of prephenate aminotransferase from cell cultures of Nicotiana silvestris. Arch. Biochem. Biophys. 238: 237–246 [DOI] [PubMed] [Google Scholar]
- Bonner C.A., Jensen R.A. (1987). Prephenate aminotransferase. Methods Enzymol. 142: 479–487 [DOI] [PubMed] [Google Scholar]
- Bovy A.G., de Vos R., Kemper M., Schijlen E., Almenar Pertejo M., Muir S., Collins G., Robinson S., Verhoeyen M., Hughes S., Santos-Buelga C., van Tunen A. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G.W.M., Hall R.D., Bovy A.G., Luo J., Martin C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chang A.C., Cohen S.N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-P., Yang X.-Y., Hegeman A.D., Gray W.M., Cohen J.D. (2010). Microscale analysis of amino acids using gas chromatography-mass spectrometry after methyl chloroformate derivatization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 2199–2208 [DOI] [PubMed] [Google Scholar]
- Cho M.H., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 282: 30827–30835 [DOI] [PubMed] [Google Scholar]
- Dal Cin V., Danesin M., Boschetti A., Dorigoni A., Ramina A. (2005a). Ethylene biosynthesis and perception in apple fruitlet abscission (Malus domestica L. Borck). J. Exp. Bot. 56: 2995–3005 [DOI] [PubMed] [Google Scholar]
- Dal Cin V., Danesin M., Rizzini F.M., Ramina A. (2005b). RNA extraction from plant tissues: The use of calcium to precipitate contaminating pectic sugars. Mol. Biotechnol. 31: 113–119 [DOI] [PubMed] [Google Scholar]
- Dal Cin V., Galla G., Ramina A. (2007). MdACO expression during abscission: The use of 33P labeled primers in transcript quantitation. Mol. Biotechnol. 36: 9–13 [DOI] [PubMed] [Google Scholar]
- Dal Cin V., Kevany B., Fei Z., Klee H.J. (2009). Identification of Solanum habrochaites loci that quantitatively influence tomato fruit ripening-associated ethylene emissions. Theor. Appl. Genet. 119: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A.H., Dunn J.J., Studier F.W. (1984). Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81: 2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Eknamkul W., Ellis B.E. (1988). Purification and characterization of prephenate aminotransferase from Anchusa officinalis cell cultures. Arch. Biochem. Biophys. 267: 87–94 [DOI] [PubMed] [Google Scholar]
- Deikman J., Kline R., Fischer R.L. (1992). Organization of ripening and ethylene regulatory regions in a fruit-specific promoter from tomato (Lycopersicon esculentum). Plant Physiol. 100: 2013–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos R.C.H., Moco S., Lommen A., Keurentjes J.J.B., Bino R.J., Hall R.D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2: 778–791 [DOI] [PubMed] [Google Scholar]
- Díaz de la Garza R., Quinlivan E.P., Klaus S.M.J., Basset G.J.C., Gregory J.F., III, Hanson A.D. (2004). Folate biofortification in tomatoes by engineering the pteridine branch of folate synthesis. Proc. Natl. Acad. Sci. USA 101: 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck R.V., Dayhoff M.O. (1966). Atlas of Protein Sequence and Structure. (Silver Springs, MD: National Biomedical Research Foundation). [Google Scholar]
- Farmaki T., Sanmartín M., Jiménez P., Paneque M., Sanz C., Vancanneyt G., León J., Sánchez-Serrano J.J. (2007). Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. J. Exp. Bot. 58: 555–568 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fischer R.S., Bonner C.A., Boone D.R., Jensen R.A. (1993). Clues from a halophilic methanogen about aromatic amino-acid biosynthesis in archaebacteria. Arch. Microbiol. 160s: 440–446 [Google Scholar]
- Görisch H. (1978). A new test for chorismate mutase activity. Anal. Biochem. 86: 764–768 [DOI] [PubMed] [Google Scholar]
- Grant G.R., Liu J., Stoeckert C.J., Jr (2005). A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics 21: 2684–2690 [DOI] [PubMed] [Google Scholar]
- Griffiths A., Barry C.S., Alpuche-Solils A., Grierson D. (1999). Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J. Exp. Bot. 50: 793–798 [Google Scholar]
- Heinzle E., Matsuda F., Miyagawa H., Wakasa K., Nishioka T. (2007). Estimation of metabolic fluxes, expression levels and metabolite dynamics of a secondary metabolic pathway in potato using label pulse-feeding experiments combined with kinetic network modelling and simulation. Plant J. 50: 176–187 [DOI] [PubMed] [Google Scholar]
- Iijima Y., et al. (2008). Metabolite annotations based on the integration of mass spectral information. Plant J. 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Chandrasekhar G.N., Georgopoulos C. (1989). Escherichia coli DnaK and GrpE heat shock proteins interact both in vivo and in vitro. J. Bacteriol. 171: 1590–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft O., Hoefgen R., Hesse H. (2003). Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol. 131: 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Blount J.W., Steele C.L., Dixon R.A. (2002). Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Maeda H., Shasany A.K., Schnepp J., Orlova I., Taguchi G., Cooper B.R., Rhodes D., Pichersky E., Dudareva N. (2010). RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Yoo H.J., Dudareva N. (2011). Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat. Chem. Biol. 7: 19–21 [DOI] [PubMed] [Google Scholar]
- Maloney G.S., Kochevenko A., Tieman D.M., Tohge T., Krieger U., Zamir D., Taylor M.G., Fernie A.R., Klee H.J. (2010). Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 153: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F., Morino K., Miyashita M., Miyagawa H. (2003). Metabolic flux analysis of the phenylpropanoid pathway in wound-healing potato tuber tissue using stable isotope-labeled tracer and LC-MS spectroscopy. Plant Cell Physiol. 44: 510–517 [DOI] [PubMed] [Google Scholar]
- Matus J.T., Loyola R., Vega A., Peña-Neira A., Bordeu E., Arce-Johnson P., Alcalde J.A. (2009). Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X., Zhu Q., Li X., Li J., Zeng Q., Rong H., Zhang H., Wu P. (2006). The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol. 141: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S., Bino R.J., Vorst O., Verhoeven H.A., de Groot J., van Beek T.A., Vervoort J., de Vos C.H.R. (2006). A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S., Niedermeyer J., Fry J., Barnason A., Horsch R., Fraley R. (1986). Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 5: 81–84 [DOI] [PubMed] [Google Scholar]
- Muir S.R., Collins G.J., Robinson S., Hughes S., Bovy A., Ric De Vos C.H., van Tunen A.J., Verhoeyen M.E. (2001). Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 19: 470–474 [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. (New York: Oxford University Press). [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Prohl C., Kispal G., Lill R. (2000). Branched-chain-amino-acid transaminases of yeast Saccharomyces cerevisiae. Methods Enzymol. 324: 365–375 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U., Liu J.L., Leisse A., Balbo I., Perez-Melis A., Willmitzer L., Fernie A.R. (2004). Kinetics of labeling of organic and amino acids in potato tubers by gas chromatography following incubation in 13C labeled isotopes. Plant J. 39: 668–679 [DOI] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Siehl D.L., Connelly J.A., Conn E.E. (1986). Tyrosine biosynthesis in Sorghum bicolor: characteristics of prephenate aminotransferase. Z. Naturforsch., C, J. Biosci. 41: 79–86 [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Tamagnone L., Merida A., Parr A., Mackay S., Culianez-Macia F.A., Roberts K., Martin C. (1998). The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tieman D., Taylor M., Schauer N., Fernie A.R., Hanson A.D., Klee H.J. (2006a). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Zeigler M., Schmelz E.A., Taylor M.G., Bliss P., Kirst M., Klee H.J. (2006b). Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Tikunov Y.M., de Vos R.C.H., González Paramás A.M., Hall R.D., Bovy A.G. (2010). A role for differential glycoconjugation in the emission of phenylpropanoid volatiles from tomato fruit discovered using a metabolic data fusion approach. Plant Physiol. 152: 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tzin V., Malitsky S., Aharoni A., Galili G. (2009). Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J. 60: 156–167 [DOI] [PubMed] [Google Scholar]
- Verdonk J.C., Haring M.A., van Tunen A.J., Schuurink R.C. (2005). ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R.J., Byng G.S., Gherna R.L., Jensen R.A. (1981). Diverse enzymological patterns of phenylalanine biosynthesis in pseudomonads are conserved in parallel with deoxyribonucleic acid homology groupings. J. Bacteriol. 147: 526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel B.S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zamir L.O., Tiberio R., Jensen R.A. (1983). Differential acid-catalyzed aromatization of prephenate, arogenate, and spiro-arogenate. Tetrahedron Lett. 24: 2815–2818 [Google Scholar]
- Zhang S., Pohnert G., Kongsaeree P., Wilson D.B., Clardy J., Ganem B. (1998). Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273: 6248–6253 [DOI] [PubMed] [Google Scholar]