This work shows that the rice AGAMOUS-LIKE6 MADS box gene MADS6 acts as a master regulator in specifying floral organ identities and meristem determinacy through interaction with B-, C-, D-, and E-class floral homeotic genes.
Abstract
AGAMOUS-LIKE6 (AGL6) genes play essential roles in flower development, but whether and how they work with floral organ identity genes remain less understood. Here, we describe interactions of the rice (Oryza sativa) AGL6 gene MADS6 with other rice floral homeotic genes in flower development. Genetic analyses revealed that MADS6 specifies the identity of the three inner whorls and floral meristem determinacy redundantly with SUPERWOMAN1/MADS16 (B-gene) or MADS3 (C-gene). MADS6 was shown to define carpel/ovule development and floral determinacy by interacting with MADS13 (D-gene) and control the palea and floral meristem identities together with the YABBY gene DROOPING LEAF. Expression analyses revealed that the transcript levels of six B-, C-, and E-class genes were reduced in mads6-1 at the early flower developmental stage, suggesting that MADS6 is a key regulator of early flower development. Moreover, MADS6 can directly bind to a putative regulatory motif on MADS58 (C-gene), and mads6-1 mads58 displayed phenotypes similar to that of mads6-1. These results suggest that MADS6 is a key player in specifying flower development via interacting with other floral homeotic genes in rice, thus providing new insights into the mechanism by which flower development is controlled.
INTRODUCTION
The flowers of angiosperms display a huge diversity in structure (Theissen and Melzer, 2007). The ABCDE model proposed that the combinatorial action of class A, B, C, and E floral homeotic genes determines floral organ identity in eudicot model species, such as Arabidopsis thaliana, Petunia hybrida, and Antirrhinum majus. In particular, A- and E-function genes specify the identity of sepals in the first outer whorl; A-, B-, and E-function genes together determine the identity of petals in the second whorl; B-, C-, and E-function genes coordinately define stamen identity in the third whorl; and C- and E-function genes act together to specify carpels in the fourth whorl (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Theissen, 2001; Theissen and Saedler, 2001; Ditta et al., 2004).
As one of the largest families in higher plants, the grass family (Poaceae) contains many economically important crops, such as rice (Oryza sativa), barley (Hordeum vulgare), and maize (Zea mays) (Clayton and Renvoize, 1986; Linder and Rudall, 2005). These crops evolved floral organization and morphologies distinct from those of eudicots and even other monocots (Grass Phylogeny Working Group, 2001; Rudall et al., 2005; Whipple et al., 2007). Spikelet, the structural unit of grass flowers, has a varied number of bract-like organs and florets. In rice, a spikelet consists of two pairs of sterile glumes (i.e., rudimentary glumes and empty glumes) and one floret that contains a lemma and a palea in whorl 1, two lodicules in whorl 2, six stamens in whorl 3, and a pistil in whorl 4 (Yuan et al., 2009; Zhang and Wilson, 2009). However, the molecular basis underlying grass flower development remains poorly understood (Clifford, 1987; Whipple et al., 2007).
Recently, emerging evidence suggests that the ABCDE genetic model is partially applicable to grasses such as rice and maize (Kyozuka et al., 2000; Kater et al., 2006; Preston and Kellogg, 2006; Prusinkiewicz et al., 2007; Reinheimer and Kellogg, 2009; Thompson and Hake, 2009; Zhang and Wilson, 2009). For instance, mutations in maize Silky1 and rice SUPERWOMAN1 (SPW1) or MADS16, both of which are orthologs of the Arabidopsis B-function gene, AP3, cause homeotic transformations of stamens to carpels and lodicules to lemma- or palea-like structures (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2004), suggesting the conserved role of the B-class genes from grasses to Arabidopsis. In grasses, there are duplicated and subfunctionalized C-class genes (Kramer et al., 2004; Zahn et al., 2006). For example, rice contains two AG homologs, MADS3 and MADS58 (Kramer et al., 2004). MADS3 plays a key role in stamen and ovule identity specification, late anther development, and floral meristem determinacy (Yamaguchi et al., 2006; Hu et al., 2011; Li et al., 2011). Using an RNA interference approach, MADS58 was shown to be required for defining floral meristem determinacy and carpel architecture (Yamaguchi et al., 2006). Similarly, maize has three AG homologs: zag1 (zea agamous1), zmm2 (Zea mays mads2), and zmm23 (Münster et al., 2002). The zag1 gene is required for floral meristem determinacy, but the biological functions of zmm2 and zmm23 have not been elucidated (Mena et al., 1996). Rice contains two D-class genes, MADS13 and MADS21, which are orthologous to the Arabidopsis SEEDSTICK (STK) and petunia FLORAL BINDING PROTEIN7 (FBP7) and FBP11 genes (Colombo et al., 1995). MADS13 was shown to be involved in ovule identity specification and floral meristem termination (Dreni et al., 2007; Li et al., 2011). However, mutants of the Arabidopsis STK gene do not display altered ovule identity (Pinyopich et al., 2003).
Grasses have diversified E-class (SEPALLATA [SEP]) genes. Rice has at least five SEP-like genes: MADS1/LEAFY HULL STERILE1 (LHS1), MADS5, MADS7, MADS8, and MADS34 (Malcomber and Kellogg, 2004, 2005; Zahn et al., 2005; Arora et al., 2007). LHS1 specifies the identity of lemma and palea and the meristem of inner floral organs (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006a). Transgenic plants with reduced expression of both MADS7 and MADS8 exhibit late flowering, homeotic transformations of lodicules, stamens, and carpels into palea/lemma-like organs, and a loss of floral determinacy. Simultaneous reduction of the expression of four rice SEP-like genes (i.e., LHS1, MADS5, MADS7, and MADS8) causes homeotic transformation of all floral organs except the lemma into leaf-like organs (Cui et al., 2010). MADS34 (PANICLE PHYTOMER2) controls the development of inflorescences and spikelets (Gao et al., 2010; Kobayashi et al., 2010). Analysis of mads34 mads1 indicates that MADS34 and LHS1 redundantly specify the identities of floral organs, including lemma/palea, lodicules, stamens, and carpel (Gao et al., 2010).
Sequence and phylogenetic analyses indicated that AGAMOUS-LIKE6 (AGL6)-like and SEP-like genes have high sequence similarities, forming sister clades on the phylogenic tree (Theissen et al., 2000; Becker and Theissen, 2003; Zahn et al., 2005). SEP-like genes are only found in angiosperms, while AGL6 genes are ancient and widely distributed in gymnosperms and angiosperms. Recently, AGL6-like genes in monocots and eudicots were shown to play essential roles in flower development (Schauer et al., 2007; Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Rijpkema et al., 2009; Li et al., 2010). The Arabidopsis genome contains two AGL6 genes, AGL6 and AGL13 (Vandenbussche et al., 2003a), suggesting possible functional redundancy between the two genes. Mutation or knockdown of AGL6 or AGL13 does not result in an abnormal flower phenotype (Schauer et al., 2007; Koo et al., 2010; Yoo et al., 2011). Loss-of-function mutants of the only Petunia AGL6 gene, Ph AGL6, show no morphological abnormalities of floral organs, but Ph AGL6 functions redundantly with the SEP genes FBP2 and FBP5 in petal and anther development, and its protein physically interacts with FBP2 (Vandenbussche et al., 2003b; Rijpkema et al., 2009).
The AGL6-like genes from grasses form two paralogous clades: the MADS17 clade containing only the rice MADS17 gene, and the MADS6 clade, which includes rice MADS6 (also called MOSAIC FLORAL ORGANS1 [MFO1]) and maize ZAG3 and ZAG5 (Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Li et al., 2010). Grass AGL6-like genes were shown to be expressed in paleas, lodicules, ovules, and floral meristems, and each of these expression domains may represent a distinct function of the gene product (Reinheimer and Kellogg, 2009). In rice, the expression level of MADS6 is high in floral meristem at early stages and in the palea and inner floral organ primordia (lodicule, stamen, and pistil) at later stages (Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Li et al., 2010). MADS17 transcripts were detected in the floral meristem at the early stage and in the lemma, palea, lodicule, pistil, and (weakly) in empty glumes and stamens at late stages, with its protein product functioning redundantly with MADS6 in flower development (Ohmori et al., 2009; Reinheimer and Kellogg, 2009). Our previous studies revealed that the palea of mads6-1 flowers develops five to six vascular bundles, which resembles the identity of a wild-type lemma, suggesting the role of MADS6 in specifying the identity of palea. In addition, mads6-1 flowers are retarded in development at the early stage, exhibit homeotic conversion of lodicules and stamens into glume-like and mosaic structures, have defective carpels and ovules, and contain indeterminate meristem at later flower developmental stages. Furthermore, we showed that the MADS6 gene is able to specify floral state by determining floral organ and meristem identities together with LHS1/MADS1 because mads1-z mads6-1 double mutants display severe floral defects, such as no inner floral organs or glume-like structures within flowers and strongly indeterminate floral meristem, phenotypes not observed in the single mutants (Li et al., 2010). A mutation of the maize AGL6 gene zea agamous3 (zag3) results in extra mosaic or fused floral organs in the upper floral meristem and additional floral meristems in the lower floral meristem. zag3 and the maize homolog of AG, zag1, can genetically and physically interact in promoting floral meristem identity (Thompson et al., 2009). These findings suggest that AGL6 genes have SEP-like functions in flower development.
Despite the findings that AGL6-like genes have a role in defining floral organ and meristem identities, whether and how they interact with other floral homeotic genes in these processes remain largely unknown. Here, we report that MADS6 interacts with several known flower homeotic genes in specifying flower development and determining floral meristem fate in rice. We show that MADS6 not only interacts with B-, D-, and E-class proteins but also regulates the expression of these genes, thus providing novel insights into the mechanism by which AGL6-like genes exert their functions in plant flower development.
RESULTS
Transcriptome Analysis of mads6-1 Flowers
To further elucidate the regulatory role of MADS6, we compared genome-wide mRNA levels in wild-type and mads6-1 flowers at stage Sp6, when stamen primordia are formed, using microarray analyses with an Agilent 4×4 4K oligonucleotide DNA chip. Stage Sp6 flowers were collected according to spikelet length and flower morphology defined by Ikeda et al. (2004), and three independent biological replicates were performed to assess its reproducibility. Data were analyzed by the Empirical Bayes method (Smyth, 2004). Initial filtering of candidate genes was performed using a false discovery rate cutoff of 0.5%, followed by a secondary selection using at least twofold changes in gene expression as the cutoff. Fifty-nine genes were found to have at least twofold changes in expression in mads6-1 flowers compared with the wild type. Among them, 26 were upregulated and 33 were downregulated (see Supplemental Table 1 online).
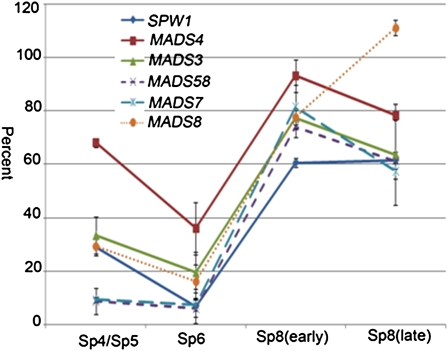
The expression of six MADS box genes, including the B-class genes MADS4 and SPW1, C-class genes MADS3 and MADS58, and E-class genes MADS7 and MADS8, was downregulated in the mutant (Table 1). In particular, MADS58 and MADS7 were downregulated ~22.5-fold (for one MADS58 probe of chip) and 11.6-fold, respectively. The microarray data were further confirmed by serial quantitative RT-PCR (qRT-PCR) analysis, which showed that the mRNA levels of these six genes were reduced from stage 4 to stage 6 and came back up at stage 8 (Figure 1).
Table 1.
Microarray Data Showing the Downregulated Expression of Six MADS Box Genes at Stage Sp6 in mads6-1 Flower
| Gene Name | Fold Change |
| MADS16 | −8.4 |
| MADS4 | −3.7 |
| MADS3 | −7.7 |
| MADS58 | −22.5a |
| MADS58 | −6.1a |
| MADS7 | −11.6 |
| MADS8 | −6.3 |
Different probes for MADS58.
Figure 1.
Expression Analysis of Six MADS Box Genes in mads6-1 Flowers.
qRT-PCR results showing the relative expression levels of SPW1, MADS4, MADS3, MADS58, MADS7, and MADS8 in mads6-1 flowers at stage 4/5, stage 6, early stage 8, and late stage 8. The expression level of each gene in the wild type control was set as 100%, and error bars indicate sd (n = 3).
To clarify the functional relationship between MADS6 and the floral homeotic genes whose expressions were significantly downregulated in mads6-1, we conducted detailed genetic analyses using double mutants between mads6-1 and spw-1, mads3-4, mads58, mads13-3, and dl-sup6. In addition to phenotypic analysis, we also performed in situ analysis to determine the regulatory relationship between MADS6 and SPW1, MADS3, MADS58, MADS13, and DL at the transcription level.
Interaction between MADS6 and SPW1
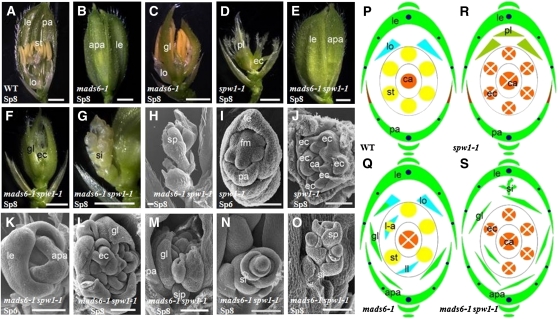
SPW1 is a B-class gene required for determining the identity of lodicules and stamens. In spw1-1 flowers, lodicules are transformed into glume-like structures and stamens into carpel-like organs (Nagasawa et al., 2003) (Figures 2A, 2D, 2P, and 2R). mads6-1 spw1-1 flowers displayed enlarged palea and glume-like organs, which are similar to the phenotypes of mads6-1 at stage Sp8 (Figures 2A to 2C, 2E, 2Q, and 2S) (Ohmori et al., 2009; Li et al., 2010), suggesting that MADS6 specifies palea identity, and this function is independent from SPW1. Unlike spw1-1 or mads6-1, in which the lodicule identity is partially lost, no lodicule-like organs were observed in whorl 2 of mads6-1 spw1-1 flowers (Figure 2F; see Supplemental Table 2 online), suggesting that MADS6 and SPW1 synergistically specify lodicule identity. Similar to spw1-1, mads6-1 spw1-1 flowers displayed the conversion of stamens into carpel-like structures, each of which contains one to four stigmas (Figures 2F and 2S). Consistent with this phenotype, the expression of the carpel marker gene DL was found in the ectopic organs (see Supplemental Figures 1K to 1M online). These results suggest that SPW1 plays a more important role in stamen identity. In the double mutant, the average number of glume-like organs per flower was 5.86 (n = 43) compared with 2.09 in spw1-1 and 4.24 in mads6-1 (see Supplemental Table 2 online). The average number of carpel-like organs per flower was 5.07 (n = 43) in the double mutant, while it was 7.00 for spw1-1 (n = 43) and 1.10 for mads6-1 (n = 43) (see Supplemental Table 2 online). More interestingly, about half of the flowers (22 in 43) in the double mutant developed new inflorescence-like organs at the position of wild-type lodicules (Figures 2G and 2H), a phenotype that was not observed in single mutant flowers, implying that MADS6 and SPW1 can repress the inflorescence primordia in the second whorl in a redundant manner. These observations suggested that both MADS6 and SPW1 function in specifying the identity of floral organs in the three inner whorls and the determinacy of floral meristem. Furthermore, the functions of these two proteins are redundant in some while independent from each other in other aspects of flower development.
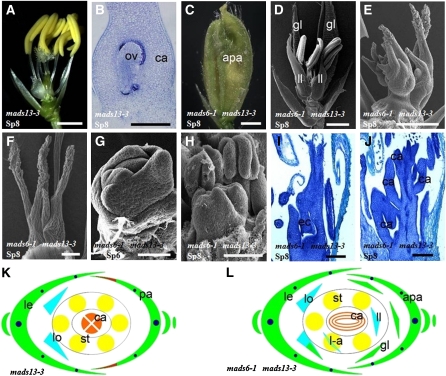
Figure 2.
Phenotypes of spw1-1 and mads6-1 spw1-1 Flowers.
(A) Wild-type (WT) flower with one lemma, one palea, two lodicules, six stamens, and one pistil.
(B) mads6-1 flower with abnormal palea.
(C) and (D) Typical flower of mads6-1 (C) and spw1-1 (D) in which the lemma and the palea were removed to show the inner organs.
(E) mads6-1 spw1-1 flower.
(F) and (G) mads6-1 spw1-1 flowers of the type I (49%, n = 43) (F) and type II phenotype (51%, n = 43) (G). Lemma and palea were removed.
(H) to (O) Scanning electron microscopy analysis.
(H) A mads6-1 spw1-1 flower showing the new inflorescence formed on the lemma side.
(I) and (J) spw1-1 flowers at stage Sp6 (I) and Sp8 (J).
(K) and (L) mads6-1 spw1-1 flower at stage Sp6 (K) and early stage of Sp8 (L).
(M) to (O) mads6-1 spw1-1 flowers showing the initiation and development of additional inflorescence-like primordia at the position of lodicule.
(P) and (Q) Diagrams of wild-type (P) and mads6-1 (Q) flowers.
(R) and (S) Diagrams of spw1-1 (R) and mads6-1 spw1-1 (S) flowers.
apa, abnormal palea; ca, carpel; ec, ectopic carpel; gl, glume-like organ; le, lemma; lo, lodicules; l-a, lodicule-anther mosaic organ; pl, palea-like organ; si, secondary inflorescence; sip, secondary inflorescence primordium; sp, spikelet; st, stamen. Bars = 1 mm in (A) to (G), 100 μm in (H), (J), and (L) to (O), 50 μm in (I) and (K).
To analyze phenotypes of the double mutant in more depth, we performed scanning electron microscopy. No structural differences between the flowers of spw1-1 and the wild type were detected at stage Sp6, when stamen primordia initiate (Figure 2I). However, similar to that of mads6-1 (Li et al., 2010), the inner flower organ primordia of the double mutant displayed retarded growth (Figure 2K). At stage Sp8, spw1-1 displayed ectopic carpel primordia (Figure 2J), and the double mutant flowers developed into carpel- and glume-like structures (Figures 2L and 2M). Also, additional inflorescence primordia were detectable in some mads6-1 spw1-1 flowers (Figures 2N and 2O). Results of the scanning electron microscopy experiment were consistent with the genetic analysis, further substantiating the role MADS6 and SPW1 in regulating flower development.
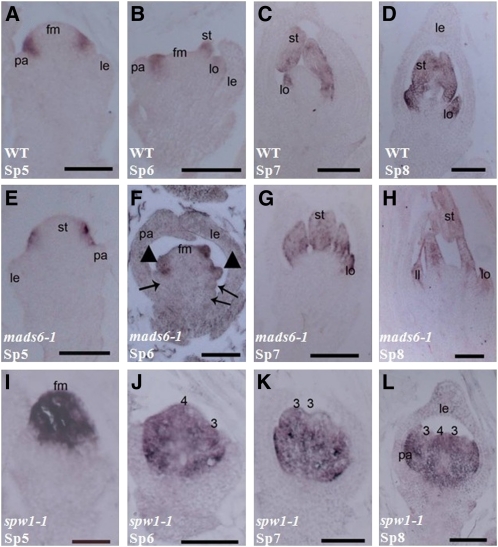
RNA in situ hybridization analysis of the mads6-1 mutant revealed that at stage Sp5, when lodicule primordia are formed, the expression level of SPW1 in the lodicule primodium was slightly weaker than in the wild type (Figures 3A and 3E) (Nagasawa et al., 2003). At stage Sp6, the expression of SPW1 was detected in lodicule and stamen primordia of mads6-1 (Figures 3B and 3F). At stages Sp7 and Sp8, the expression of SPW1 was observed in lodicule and stamen primordia in mads6-1 similar with its expression pattern in the wild type (Figures 3C, 3D, 3G, and 3H). In spw1-1, the expression of MADS6 was detected in paleas, lodicules, carpels, and the receptacle similar to its expression profile in the wild-type plants (Figures 3I to 3L) (Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Li et al., 2010). These in situ analysis results are consistent with the microarray and qRT-PCR experiments, supporting the notion that MADS6 is able to activate the expression of SPW1 at the early flower developmental stage but does not obviously affect its expression at late stages.
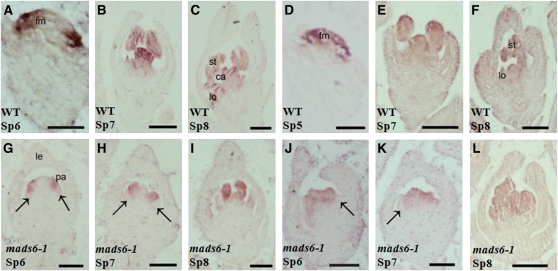
Figure 3.
Expression Pattern of SPW1/MADS16 and MADS6.
(A) to (D) SPW1 expression in wild-type (WT) flowers at stage Sp5 (A), Sp6 (B), Sp7 (C), and Sp8 (D).
(E) to (H) Expression of SPW1 in mads6-1 flowers at Sp5 (E), Sp6 (F), Sp7 (G), and Sp8 (H).
(I) to (L) Expression pattern of MADS6 in spw1-1 flowers at stage Sp5 (I), Sp6 (J), Sp7 (K), and Sp8 (L).
ca, carpel; ec, ectopic carpel; gl, glume-like organ; le, lemma; ll, lodicule-like organ; lo, lodicules; pl, palea-like ogan; pa, palea; si, secondary inflorescence; sip, secondary inflorescence primordium; sp, spikelet; st, stamen. Bars = 50 μm in (A), (E), and (I) and 100 μm in (B) to (D), (F) to (H), and (J) to (L). Numbers in (J) to (L) indicate the whorl number.
MADS6 and DL Redundantly Regulate Floral Meristem Identity
DL, which belongs to the YABBY gene family, determines carpel identity and regulates vascular pattern of the lemma (Nagasawa et al., 2003; Yamaguchi et al., 2004; Li et al., 2011). In the construction of double mutants, we used the null allele dl-sup6, which is allelic to the previously reported dl-2 mutant (Nagasawa et al., 2003; Yamaguchi et al., 2004; Li et al., 2011). dl-sup6 has drooping leaves, ectopic stamens in the fourth whorl at the position of the carpel, and loss of floral meristem determinacy in some of the flowers (Figures 4A, 4B, and 4J) (Li et al., 2011).
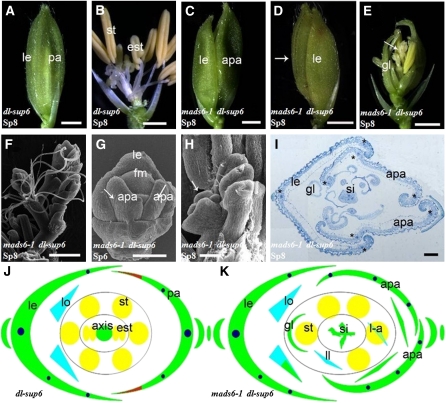
Figure 4.
Flower Phenotypes of mads6-1 dl-sup6.
(A) dl-sup6 flower at stage Sp8.
(B) dl-sup6 flower at stage Sp8, showing inner organs.
(C) and (D) mads6-1 dl-sup6 flowers of the type I (C) and type II (D) phenotypes at stage Sp8. The arrow in (D) indicates the altered palea structure.
(E) mads6-1 dl-sup6 flowers at stage Sp8, after the removal of the lemma and the palea. The arrow indicates one additional inflorescence-like structure in the center.
(F) to (H) Scanning electron microscopy analysis.
(F) mads6-1 dl-sup6 flower at stage Sp8, showing an inflorescence-like structure in the center.
(G) mads6-1dl-sup6 flower at stage Sp6. Arrows indicate two palea-like structures.
(H) mads6-1 dl-sup6 flower with inflorescence-like meristem at early stage Sp8.
(I) Transverse section of a mads6-1 dl-sup6 flower at stage Sp8 showing two abnormal palea-like organs on the palea side.
(J) and (K) Floral diagrams of dl-sup6 (J) and mads6-1 dl-sup6 (K).
apa, abnormal palea; est, ectopic stamen; gl, glume-like organ; le, lemma; lo, lodicules; l-a, lodicule-anther mosaic organ; si, secondary inflorescence; st, stamen. Bars = 1 mm in (A) to (E), 500 μm in (F), 50 μm in (G) and (H), and 100 μm in (I).
Flowers of the mads6-1 dl-sup6 double mutant could be divided into two types based on palea morphology. Type I was found in 58% of the flowers (n = 56). They displayed widened palea, which is similar to that of mads6-1 (Figure 4C). Type II flowers (42%; n = 56) had two palea-like organs (Figures 4D and 4K), which were not observed in the single mutants. Examination of transverse sections showed that these two palea-like organs had the characteristic marginal tissue of the lemma but contained three vascular bundles similar to that of wild-type palea (Figures 4I and 4K). Consistent with this finding, two palea-like primordia were observed at stage Sp6 on the palea side in type II flowers (Figure 4G). These results revealed that MADS6 specifies the palea identity together with DL.
The phenotypes of the mads6-1 dl-sup6 floral organs in whorls 2 and 3 appeared similar to that of mads6-1 (Figure 4E), suggesting that MADS6 is involved in defining the identities of lodicules and stamens, whereas DL does not play a role in it. This finding is consistent with the lack of DL expression in whorls 2 and 3 (see Supplemental Figure 1 online). Interestingly, all flowers in mads6-1 dl-sup6 displayed an inflorescence-like structure in whorl 4 (n = 62) (Figures 4E, 4F, 4H, and 4K), yet this phenotype was rarely observed in mads6-1 (Li et al., 2010). These results suggested that MADS6 and DL act synergistically in terminating floral meristem.
RNA in situ hybridization analysis detected DL expression in the wild-type lemma and carpel, not in the palea (Yamaguchi et al., 2004) (see Supplemental Figures 1A to 1C online). However, ectopic expression of DL was observed in the altered palea organ and ectopic carpels or abnormal ovules in mads6-1 (see Supplemental Figures 1D to 1G online) (Ohmori et al., 2009; Li et al., 2010). By contrast, MADS6 expression was observed in the dl-sup6 floral primordia at stage Sp4, and in palea and carpel primordia as well as the floral meristem at stages Sp7 and Sp8, just like in the wild type (see Supplemental Figures 1H to 1J online). Together, these results suggested that, whereas MADS6 may repress the expression of DL, DL does not have an obvious effect on MADS6 expression.
MADS6 and MADS13 Redundantly Regulate Carpel/Ovule Identity and Floral Determinacy
MADS13 is a D-class gene that functions in specifying ovule identity and floral meristem determinacy (Dreni et al., 2007). We recently identified a strong MADS13 allele, mads13-3, which showed carpelloid structures, indeterminate floral organs, and complete female sterility caused by aborted ovule development (Figures 5A, 5B, and 5K) (Li et al., 2011).
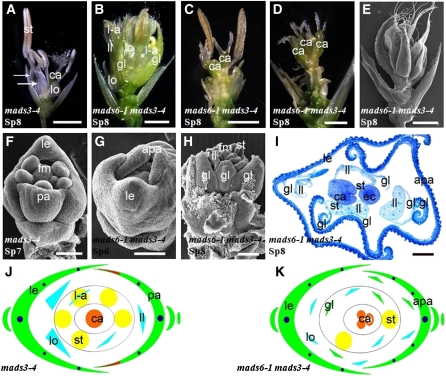
Figure 5.
Flower Phenotypes of mads6-1 mads13-3.
(A) One mads13-3 flower at stage Sp8 showing normal floral morphology.
(B) Longitudinal section of one mads13-3 carpel at stage Sp8 showing abnormal ovule.
(C) One mads6-1 mads13-3 flower at stage Sp8 displaying abnormal palea.
(D) to (H) Scanning electron microscopy analysis.
(D) One mads6-1 mads13-3 flower at stage Sp8 displaying ectopic glume-like structures and abnormal lodicules and stamen development.
(E) One mads6-1 mads13-3 flower at stage Sp8 showing higher-order carpel-like structures in the center.
(F) One mads6-1 mads13-3 flower at stage Sp8 showing the transformation from ovule to carpel-like structure.
(G) One mads6-1 mads13-3 flower at stage Sp6 showing delayed inner floral organ development.
(H) One mads6-1 mads13-3 flower at early stage Sp8 showing new organ primordium in whorl 4.
(I) and (J) Longitudinal sections of mads6-1 mads13-3 flowers showing weak (I) and severe (J) phenotypes in the floral center.
(K) and (L) Floral diagrams of mads13-3 (K) and mads6-1 mads13-3 (L).
apa, abnormal palea; ca, carpel; ec, ectopic carpel; gl, glume-like organ; le, lemma; ll, lodicule-like organ; lo, lodicules; l-a, lodicule-anther mosaic organ; ov, ovule; pa, palea; st, stamen. Bars = 1 mm in (A), (C), and (D), 500 μm in (E), 100 μm in (B), (F), and (H) to (J), and 50 μm in (G).
Flowers of the mads6-1 mad13-3 double mutant displayed defects in the outer three whorls, a phenotype that is similar to that of mads6-1 (Figures 5C and 5D). Given that the expression of MADS13 is limited to ovules (Dreni et al., 2007) and that the mads13 phenotype is exclusively found in the fourth whorl, we conclude that the function of MADS6 in controlling the organ identities of the outer three whorls is independent of MADS13. However, the flowers of mads6-1 mads13-3 displayed more severe defects in carpel/ovule development and floral meristem determinacy than the single mutants. We grouped these phenotypes into two categories: weak and strong. Fifty-seven percent (n = 55) of the double mutants had a weak phenotype, in which the ovule is transformed into carpelloid structures, a phenotype that is close to the severe phenotype shown in mads13-3 or mads6-1 (Figures 5F and 5I). Forty-three percent (n = 55) of the double mutants had the strong phenotype, in which floral meristem determinacy is largely lost due to the generation of higher order carpel-like organs along the floral axis (Figures 5E, 5J, and 5L). Consistent with these results was the detection of ectopic expression of the carpel marker gene DL in these ectopic carpel-like organs (see Supplemental Figures 1N and 1O online). Further scanning electron microscopy analysis showed that at stage Sp6, the phenotype of the mads6-1 mads13-3 floral primordia was close to that of mads6-1 (e.g., delayed stamen primordial development) (Figure 5G). However, after the carpel is formed at early Sp8 stage, the double mutant developed much stronger defects than the single mutants and had indeterminate floral meristems (Figure 5H). These results suggested that MADS6 and MADS13 have partial functional redundancy in specifying carpel/ovule identity and terminating floral stem cell activity.
In situ hybridization analysis detected the expression of MADS13 in wild-type ovules at stage Sp8 (see Supplemental Figures 2A and 2B online), which is consistent with a previous report (Dreni et al., 2007). In mads6-1, MADS13 expression was detected in ovule primordia at early Sp8 stage, similar to its expression in the wild type (see Supplemental Figure 2C online), and in ectopic ovules at late Sp8 stage (see Supplemental Figures 2D and 2E online). In mads13-3, the expression of MADS6 was observed in the floral meristem and primordia of the palea, lodicules, and the carpel at stages Sp7 to Sp8 (see Supplemental Figures 2F to 2H online), which is similar to its expression pattern in the wild type. These results suggested that MADS6 and MADS13 do not obviously regulate the expression of each other at the transcriptional level during early stages of ovule specification.
MADS6 Positively Regulates the Expression of MADS7 and MADS8
MADS7 and MADS8 are the two closest rice homologs of Arabidopsis SEP3 (Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007). Like SEP3, MADS7 and MADS8 are expressed in the three inner whorls (Cui et al., 2010). MADS6 was shown to interact with MADS7 and MADS8 in a yeast two-hybrid analysis (Moon et al., 1999). Moreover, Cui et al. (2010) revealed that plants in which both MADS7 and MADS8 were silenced displayed strong defects in flower development, where stamens were transformed into lodicules or glume-like organs and flowers appeared to lose determinacy by generating higher order carpels, suggesting functional redundancy between MADS7 and MADS8 in specifying flower development.
In situ hybridization detected the expression of MADS7 in the primodia of lodicules, stamens, and carpels in the wild type (Figures 6A to 6C), which agrees with results from the previous report (Cui et al., 2010). In addition, the expression of MADS8 was detected in the floral meristem (Figure 6D) and whorls 2, 3, and 4 (Figures 6E and 6F) in the wild type. In mads6-1, however, the expression of MADS7 and MADS8 was markedly reduced in floral meristem in the early stage of flower development (Figures 6G to 6L), which is consistent with the microarray data and qRT-PCR analysis (Table 1, Figure 1). Together, these results supported the notion that MADS6 positively regulates the expression of MADS7 and MADS8 during rice flower development.
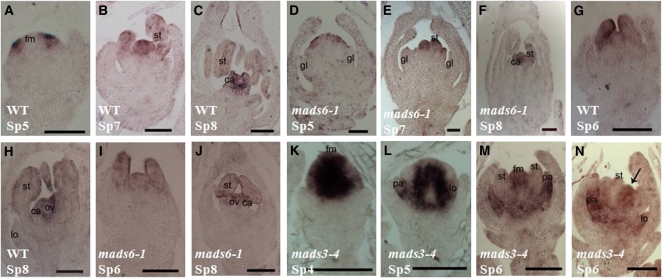
Figure 6.
Expression Pattern of MADS7 and MADS8 in Wild-Type and mads6-1 Flowers.
(A) Transcripts of MADS7 in the primordia of the second and third whorls in the wild-type (WT) flower at early stage Sp6.
(B) and (C) MADS7 expression signals in whorls 2, 3, and 4 of the wild-type flower at stage Sp7 (B) and Sp8 (C).
(D) to (F) MADS8 expression in the wild-type flower at early stage Sp5 (D), Sp7 (E), and Sp8 (F).
(G) to (I) Expression of MADS7 in mads6-1 flowers at stage Sp6 (G), Sp7 (H), and Sp8 (I).
(J) to (L) Expression of MADS8 in mads6-1 flowers at stage Sp6 (J), Sp7 (K), and Sp8 (L).
le, lemma; pa, palea; lo, lodicules; st, stamen; ca, carpel. Arrows indicate lodicule primordia. Bars= 50 μm in (A), (D), (G), and (J), and 100 μm in (B), (C), (E), (F), (H), (I), (K), and (L).
MADS6 Positively Regulates the Expression of MADS3 and MADS58
In mads6-1 flowers, the initiation of stamen development is much delayed and stamens are partially converted into lodicule-like or lodicule-anther mosaic organs during early flower development (Ohmori et al., 2009; Li et al., 2010). MADS3, a C-class gene in rice, has been shown to play a key role in controlling the development of lodicules and stamens (Yamaguchi et al., 2006; Hu et al., 2011). More recently we showed that MADS3 also defines carpel development and floral determinacy redundantly with MADS13 (Li et al., 2011). In addition, two intermediate alleles, mads3-2 and mads3-4, displayed mild homeotic transformation from stamens into lodicule-like or lodicule-anther mosaic organs (Yamaguchi et al., 2006; Hu et al., 2011) (Figures 7A, 7F, and 7J).
Figure 7.
Flower Phenotypes of mads6-1 mads3-4.
(A) mads3-4 flower at stage Sp8. Lodicule-anther mosaic organs are indicated by arrows.
(B) to (D) Flower phenotypes of mads6-1 mads3-4 at stage Sp8, including many glume- or lodicule-like and lodicule-anther mosaic organs (B), two carpels in the center (C), and elongated flower axis or inflorescence-like organ and several carpel-like organs on the top (D).
(E) to (H) Scanning electron microscopy analysis.
(E) One mads6-1 mads3-4 flower at stage Sp8 showing lodicule- and glume-like organs.
(F) One mads3-4 flower at Sp7 showing the primordium of one ectopic organ in the second whorl.
(G) One mads6-1 mads3-4 flower at stage Sp6 showing delayed development of inner floral organs, a similar phenotype to the mads6-1 flowers.
(H) One mads6-1 mads3-4 flower at stage Sp8 showing primordia of lodicule- or glume-like organs.
(I) Transverse section analysis of a stage-Sp8 mads6-1 mads3-4 flower, which has six glume-like organs, four lodicule-like organs, two stamens, and two carpels.
(J) and (K) Floral diagrams of mads3-4 (J) and mads6-1 mads3-4 (K).
ca, carpel; ec, ectopic carpel; fm, floral meristem; gl, glume-like organ; l-a, lodicule-anther mosaic organ; ll, lodicules-like organ; lo, lodicules; st, stamen. Bars = 1 mm in (A) to (C), 500 μm in (D) and (E), 50 μm in (F) and (G), and 100 μm in (H) and (I).
The morphology of palea in the mads6-1 mads3-4 double mutant appeared similar to mads6-1, suggesting that MADS6’s function in regulating palea identity is independent of MADS3. However, more severe defects were observed in whorls 2 to 4. In the double mutant, the average number of ectopic lodicule- or glume-like organs increased to 7.25 (n = 91), compared with 5.34 (n = 79) in mads6-1, and the number of stamens in the double mutant decreased to 2.45 (n = 91) from 6.00 in the wild type (Figures 7B, 7E, 7I, and 7K; see Supplemental Table 3 online). Defects in the double mutant also seemed to be intensified in whorl 4, where elongated floral axis or an inflorescence-like structure was observed in the floral center, and the average number of carpels was 2.2 (n = 91) (Figures 7C and 7D; see Supplemental Table 3 online). Scanning electron microscopy observation indicated that mads6-1 mads3-4 had delayed inner floral organ development at stage Sp6 (Figure 7G), which is similar to that of mads6-1. At stage Sp8, the mads6-1 mads3-4 double mutant displayed lodicule- or glume-like organs compared with the wild type or mads3-4 (Figure 7H). These results suggested that MADS6 and MADS3 synergistically determine the identity of the three inner floral organs and floral determinacy.
In situ analysis detected the expression of MADS3 at stage Sp5 in the wild type at the position where the stamen primordium was to be formed (Figure 8A). At stages Sp7 and Sp8, the mRNA of MADS3 was detectable in whorls 3 and 4 of the wild-type flowers (Figures 8B and 8C) (Yamaguchi et al., 2006). However, in mads6-1, the expression of MADS3 seemed much weaker and delayed (Figures 8D to 8F). Similarly, reduced expression of MADS58 was observed in mads6-1 at the early stage (Figures 8G to 8J). This result is consistent with the microarray data and qRT-PCR analysis, suggesting that MADS6 may promote the expression of MADS3 and MADS58 during early flower development. Conversely, we observed no obvious change in MADS6 expression between mads3-4 and the wild type, with the exception of some signals in ectopic lodicule-like primodia (Figures 8K to 8N), suggesting that MADS3 does not affect the expression of MADS6 at the transcriptional level to significant degrees.
Figure 8.
Spatial and Temporal Expression Pattern of MADS3, MADS58, and MADS6.
(A) to (F) Expression pattern of MADS3 in flowers of the wild type (WT) ([A] to [C]) and mads6-1 ([D] to [F]). (A) and (D), (B) and (E), and (C) and (F) are from stage Sp5, Sp7, and Sp8, respectively.
(G) to (J) Expression patterns of MADS58 in flowers of the wild type ([G] and [H]) and mads6-1 ([I] and [J]). (G) and (I) and (H) and (J) are from stages 6 and 8, respectively.
(K) to (N) Expression pattern of MADS6 in mads3-4 flowers at stage Sp4 (K), Sp5 (L), and Sp6 ([M] and [N]). The arrow in (N) indicates a lodicule-like or lodicule-anther mosaic organ.
ca, carpel; fm, floral meristem; gl, glume-like organ; lo, lodicule; pa, palea; st, stamen. Bars = 50 μm in (A), (D), (K), and (L) and 100 μm in (B), (C), (E) to (J), (M), and (N).
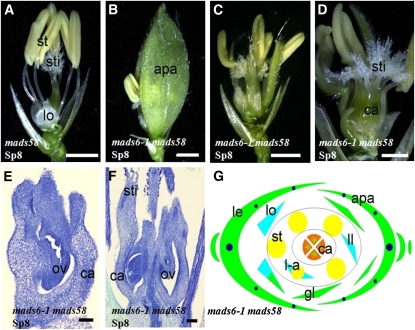
The mads58 allele contains a dSpm element insertion in the second intron of MADS58, which leads to ~35-fold reduction in the expression of MADS58 but no obvious defects in flower development (Figure 9A) (Dreni et al., 2011). The defects of floral organs in the outer three whorls were almost identical in mads6-1 mads58 and mads6-1, except that the number of stigmas and abnormal carpels/ovules was higher in the double mutant (Figures 9B to 9G). The average stigma number per flower was 6.56 (n = 50) in mads6-1 mads58 and 2.77 (n = 44) in mads6-1. About half of the mads6-1 mads58 flowers displayed defective ovule development, each showing two or three carpel/ovule-like structures. The average number of carpel/ovule per flower was 1.5 (n = 50) in mads6-1 mads58 and 1.1 in mads6-1 (n = 44) under the same growth condition (Figures 9E and 9F).
Figure 9.
Floral Phenotypes of mads6-1 mads58.
(A) One mads58 flower at stage Sp8.
(B) to (D) mads6-1 mads58 flowers at stage Sp8 showing the enlarged palea (B), inner floral organs (C), and an increased number of stigmas (D).
(E) and (F) Longitudinal sections of mads6-1 mads58 flowers at stage Sp8 showing abnormal carpel/ovule development.
(G) Diagram of a mads6-1 mads58 flower.
apa, abnormal palea; ca, carpel; gl, glume-like organs; le, lemma; ll, lodicule-like organs; lo, lodicules; l-a, lodicule-anther mosaic organs; ov, ovule; pa, palea; st, stamen; sti, stigmas. Bars = 1 mm in (A) to (C), 500 μm in (D), and 100 μm in (E) and (F).
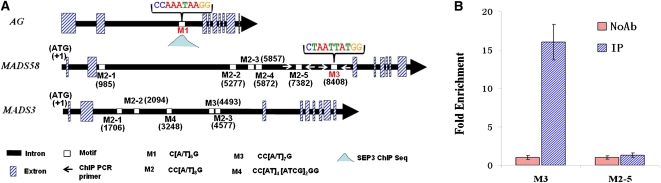
MADS box proteins can regulate gene expression by binding as homo- or heterodimers to sequences containing a consensus core element called the CArG box (Riechmann et al., 1996). For instance, the second intron of the Arabidopsis C-class gene AG contains one CArG-box, M1 [CC(A/T)6G], which is bound by SEP3 and acts as an enhancer sequence required for the proper expression of AG (Deyholos and Sieburth, 2000; Kaufmann et al., 2009) (Figure 10A). To further investigate the regulatory role of MADS6 during rice flower development, we searched the homeotic genes investigated in this study for putative CArG box sequences using the plant CARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002). We found five CArG motifs in the second intron of MADS58 and grouped them into two types: M2 [CC(A/T)8G] and M3 [CC(A/T)7G]. The second intron of MADS3 was also found to contain five CArG motifs, which could be grouped into M2, M3, and M4 [CC(A/T)4(A/T/C/G)2G] types (Figure 10A).
Figure 10.
Analysis of the Association of MADS6 with the Predicted Regulatory Elements in the Three Homeotic Genes.
(A) Gene structures of AG, MADS58, and MADS3 showing the conserved CArG motifs within their second introns.
(B) ChIP-PCR assay showing the specific enrichment of fragments that contain M3 from the second intron of MADS58 by the MADS6 antibody. Error bars indicate sd (n = 3).
M2 to M5 respresent motifs 2 to 5; IP, immunoprecipitated chromatin using MADS6 antibody; NoAb, negative control without MADS6 antibody.
[See online article for color version of this figure.]
To test whether MADS6 is able to bind to the CArG motifs in the second introns of MADS3 and MADS58, we performed chromatin immunoprecipitation (ChIP)-qPCR analysis. First, we developed rabbit polyclonal antibodies against a bacterial-expressed recombinant protein that contains 86 amino acids (amino acid 165 to amino acid 250) from the N terminus of MADS6 (see Methods). Specificity of the antibody was validated using immunoblot analysis, by which we detected, using a protein extract from wild-type flowers, a band of ~30 kD that is close to the expected size (see Supplemental Figure 3 online). ChIP-qPCR analysis using the affinity-purified MADS6 antibody showed specific enrichment of the M3-containing region of MADS58 but not the other CArG motifs in MADS3 and MADS58 (Figure 10B; see Methods and Supplemental Table 4 online). These results suggested that MADS6 may directly regulate the expression of MADS58 through binding to the CArG element in the second intron.
DISCUSSION
MADS6 Is a Key Regulator in Specifying Floral Organ Identities
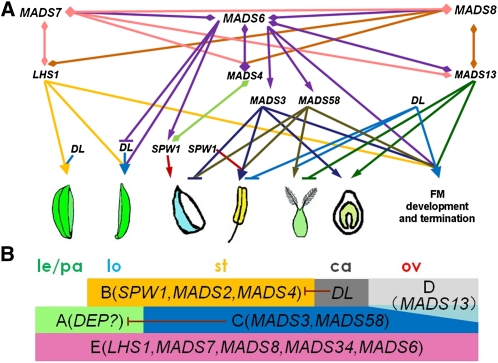
Flowering plants (angiosperms) evolved a tremendous diversity of floral structures (Theissen and Melzer, 2007). In this study, we investigated the genetic interaction of MADS6 with floral homeotic genes SPW1 (B-class), MADS3 and MAD58 (C-class), MADS13 (D-class), and DL and demonstrated that interactions of MADS6 with these floral homeotic genes play essential roles in rice flower development. Expression analyses indicated that the expression of B-, C-, and E-class genes in mads6-1 was reduced at early flower developmental stages (Figure 1, Table 1). This may explain the delayed development of floral organ primordia in the mutant. However, repression of these six MADS box genes at the transcript level was less obvious at late flower developmental stages. Ohmori et al. (2009) reported the upregulation of MADS14, MADS15, MADS3, MADS58, and LHS1 in mfo1-1 elongated lodicules and MADS14, MADS15, and MADS3 in the mfo1-1 carpel during late flower developmental stages, suggesting the developmental stage–dependent control of these floral homeotic genes in rice. In this study, we did not detect obvious changes in the expression pattern of MADS6 in spw1-1, mads3-4, and mads13-3. Based on these results, we propose that MADS6 may act as an upstream regulator that activates the expression of B- (MADS4 and SPW1), C-(MADS3 and MADS58), and E-class (MADS7 and MADS8) genes during early flower development (Figure 11A). Alternatively, the reduced expression of B-, C-, and E-class genes may be a consequence of delayed development of flower organ primordia in mads6-1.
Figure 11.
Proposed Models for Rice Flower Development.
(A) Roles of MADS6 and floral homeotic genes in specifying rice flower development. MADS6 specifies palea identity by repressing DL expression. It regulates lodicule development by activating the expression of SPW1 and MADS4 and physically interacting with the MADS4-SPW1 complex (Seok et al., 2010). MADS6 regulates the stamen, carpel, and meristem identities by activating the expression of MADS3 and MADS58 at early stages and specifies carpel/ovule development and controls floral meristem termination by interacting with MADS13 (Moon et al., 1999). MADS4 or MADS13 interacts with MADS7 and MADS8, respectively (Kater et al., 2006). In addition, MADS6 determines flower development redundantly with LHS1, MADS7, and MADS8 (Ohmori et al., 2009; Li et al., 2010). It also promotes the expression of MADS7 and MADS8 and physically interacts with MADS7 and MADS8 (Moon et al., 1999). Furthermore, MADS6, MADS3, MADS58, DL, and MADS13 redundantly specify floral meristem determinacy (Yamaguchi et al., 2004, 2006; Dreni et al., 2011; Li et al., 2011).
(B) ABCDE model in rice flower develoment. The putative A-class gene DEP defines palea identity (Wang et al., 2010; Li et al., 2011). The E-function genes LHS1, MADS6, and MADS34 specify lemma/palea identities. The A-class gene (DEP), B-class genes SPW1, MADS2, and MADS4, in combination with the E-class genes LHS1, MADS6, MADS7, MADS8, and MADS34, specify the lodicule identity. The B-class genes, the C-class genes MADS3 and MADS58, together with the E-class genes determine the stamen identity. DL specifies carpel identity. The D-class gene MADS13 and E-class genes determine carpel/ovule identity. Additionally, DL antagonistically regulates the expression of B-class genes, and the C-class gene MADS3 represses the expression of the putative A-class gene DEP (MADS15) (Li et al., 2011).
ca, carpel; le, lemma; lo, lodicule; ov, ovule; pa, palea; st, stamen.
[See online article for color version of this figure.]
Phylogenetic and functional analyses revealed that AGL6 family members are closely related to SEP-like genes (Becker and Theissen, 2003). Arabidopsis SEP and AGL6 genes were shown to activate the expression of B- and C-class genes (Liu et al., 2009a; Koo et al., 2010). Moreover, ChIP sequencing analysis revealed direct association of SEP3 with the regulatory regions of MADS box genes, where peaks of association were found to be located in the promoters of AP1, AP3, SEP1, and SEP2 and the second intron of AG (Kaufmann et al., 2009). In this study, ChIP-qPCR assays revealed direct association of MADS6 with a predicted regulatory motif in the second intron of MADS58 (Figure 10). In addition, dramatically reduced expression of MADS58 was observed in mads6-1 during early flower development (Figures 1 and 8; Table 1), suggesting that MADS6 functions upstream of the C-class gene MADS58 and may directly activate MADS58 expression (Figure 11A).
Similar to SEP proteins (Immink et al., 2009), MADS6 have been shown to form complexes with MADS4, SPW1, MADS13, MADS7, MADS8, MADS14, and MADS15 (Moon et al., 1999; Favaro et al., 2002; Seok et al., 2010) (Figure 11), suggesting that AGL6-like proteins act as integrators that form multimeric complexes with MADS domain proteins from different clades in rice (Figure 11). We hypothesize that these MADS6-containing protein complexes may also be involved in the transcriptional control of floral homeotic genes in rice. This is exemplified by the findings that in Arabidopsis AP3 and PI form functional heterodimers, which can bind DNA in vitro (Riechmann et al., 1996) and regulate their own transcription in vivo (Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996). Further determination of whether such auto/cross-regulatory loops also exist during flower development in grasses will help explain how MADS6 acts both upstream from and in concert with these floral MADS box genes.
The ABCDE Model and Control of Palea Identity in Grasses
Compared with eudicot plants, rice has both conserved and unique genes that act in the regulation of flower development (Figure 11). As a result, the ABCDE genetic model is only partially applicable to the mechanism of flower development in grasses (Thompson and Hake, 2009; Li et al., 2011). For example, B- and C-class function seems to be conserved in grasses, Arabidopsis, and other eudicot species, whereas the function of AGL6 genes, equivalents of the Arabidopsis class E or SEP genes, is partially conserved from eudicots to grasses (Figure 11).
In grasses, AGL6-like genes acquired a new expression domain in the palea, suggesting that their proteins may be key regulators of palea identity (Ohmori et al., 2009; Reinheimer and Kellogg, 2009; Li et al., 2010). It has been hypothesized that during evolution of grass flowers, the palea was a congenital fusion of two perianth parts along with the lemma, which would represent a modified trimerous calyx (Francis, 1920; Zanis, 2007). Congenital fusion (also called phylogenetic fusion and zonal growth) refers to a compound structure that is developed as a homogeneous unit but is potentially derived from separate origins (Cusick, 1966; Verbeke, 1992). According to comparative morphological analysis, the development of compound organs resulting from fusions between individual primordia is thought to play a key role in floral morphogenesis during evolution (Verbeke, 1992). However, the mechanism underlying congenital fusion remains poorly understood.
Our results in this study suggest that both MADS6 and DL are required for the proper establishment of palea identity and morphology. In the wild type, DL was shown to have zygomorphic expression in the lemma, but in mads6 mutants, its expression becomes detectable in the palea as well. In addition, mads6 mutants display loss of the palea identity (Ohmori et al., 2009; Li et al., 2010). DL was recently shown to play a critical role in specifying lemma identity (Figure 11), and dl-sup6 lemmas had altered developmental identity with an increased number of vascular tissues but no obvious change of palea morphology (Li et al., 2011). Intriguingly, some flowers of the mads6-1 dl-sup6 double mutants displayed complete loss of palea identity determination, leading to two separate organs. The complete loss of palea identity may be explained by the idea that congenital fusion of the palea relies on proper establishment of the palea identity. This interpretation is analogous to what was described as the dedoublement phenomenon in Arabidopsis stamens, where a single stamen primordium in the medial position is split in two, producing two pairs of medial stamens (Ronse Decraene and Smets, 1993; Bowman and Smyth, 1998). However, ap3 or pi mutants show the transformation of these organs into carpels, resulting in just single organs in these positions (Bowman et al., 1991; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996). Similarly, this interpretation was used to explain the stamen number reduction in Lepidium (Brassicaceae) (i.e., stamens are reduced either by apparent loss of primordia [in the two lateral positions] or by fusion of two primordia into one [in the two medial positions]) (Endress, 1992; Bowman and Smyth, 1998). Here, we propose that MADS6 may specify the palea identity through repressing the expression of DL in the palea (Figure 11). Consistent with this view, investigations from model dicot plants suggest that the congenital fusion often acts downstream of organ identity specification such that a loss of organ identity alters proper fusion (Alvarez and Smyth, 1999; Vandenbussche et al., 2004; Prunet et al., 2009). In Arabidopsis, analysis of double mutants of crabs claw (crc) and spatula (spt) with homeotic mutants indicated that A and B organ identity genes are capable of negatively regulating the function of CRC and SPT in carpel development (Alvarez and Smyth, 1999). CRC was shown to suppress the radial growth but trigger the longitudinal growth of the developing gynoecium, and SPT is able to promote the development of the carpel margins and the derived tissues (Alvarez and Smyth, 1999).
Previously, we showed that the rice CYCLOIDEA-like homolog RETARDED PALEA1 (REP1) is a palea-specific gene that regulates palea identity and initiation by regulating cell proliferation and expansion, but it does not affect lemma development (Yuan et al., 2009). The rep1 mutants display altered palea identity, such as an increased number of vascular bundles in the palea, which resembles a lemma-like organ (Yuan et al., 2009). Whether and how MADS6 interacts with REP1 in specifying palea identity requires further elucidation.
We have shown that in addition to specifying palea identity, MADS6 also regulates lodicule development by interacting with SPW1, defines the stamen, carpel, and meristem identities with MADS3 and MADS58, and specifies carpel/ovule development and floral meristem determinacy together with MADS13 (Figure 11). Furthermore, MADS6 regulates flower development redundantly with E-class genes, such as LHS1, MADS7, and MADS8. Although no obvious protein–protein interaction or transcriptional regulation between MADS6 and LHS1 was found (Li et al., 2010), MADS6 may promote the expression of MADS7 and MADS8 and physically interact with the products of these two genes (Moon et al., 1999) (Figure 11). Similarly, Arabidopsis AGL6 was shown to interact with several MADS proteins, such as SEP1, SEP3, SHATTERPROOF2, AP1, and FUL (de Folter et al., 2005).
Control of Floral Meristem Determinacy
Floral organs originate from the floral meristem, a pool of pluripotent stem cells (Liu et al., 2009b; Prunet et al., 2009). Mechanisms for the maintenance of floral meristem determinacy seem to be widely conserved among angiosperms (Ferrario et al., 2004; Prunet et al., 2009). Generally, floral meristem activity is abolished after the formation of a fixed pattern of floral organs. It has been shown that Arabidopsis AG is a key regulator in terminating floral meristem by turning WUSCHEL (WUS) off (Lenhard, et al., 2001; Lohmann et al., 2001). Whether grasses use a similar mechanism remains unknown. In the rice genome, there are 13 putative WOX (WUSCHEL-related homeobox gene family) genes; Os WUS was found to be closely related in sequence to the Arabidopsis WUS gene (Nardmann and Werr, 2006; Dai et al., 2007; Zhang et al., 2010b). But the biological function of Os WUS remains unclear. Unlike the WUS gene in Arabidopsis, Os WUS is not expressed in the organizing center of the vegetative shoot apical meristem (Nardmann and Werr, 2006). Grass species have duplicated C-class genes (Mena et al., 1996; Kramer et al., 2004; Yamaguchi et al., 2006; Zahn et al., 2006). In rice, analyses of mads3 and mads58 mutants suggested that MADS3 plays a major role in specifying stamen identity and late anther development (Yamaguchi et al., 2006; Hu et al., 2011), and MADS58 plays a redundant role with MADS3 in specifying floral meristem determinacy and carpel identity (Dreni et al., 2011). More recently, our double mutant analysis revealed that MADS3 and MADS13 redundantly regulate carpel/ovule development and floral meristem determinacy, suggesting that the C-class and D-class genes in rice retain their conserved function even though they had multiple subfunctionalization and/or neofunctionalization events after duplication within AG clade (Li et al., 2011) (Figure 11).
In this study, we have shown that MADS6 regulates floral meristem determinacy together with C-class genes. Mutations in MADS6 cause greatly reduced expression of the AG homologs MADS3 and MADS58, and mads6-1 mads3-4 displayed severe defects in floral meristem determinacy, such as the presence of inflorescence-like structures (Figure 7). Although mads6-1 mads58 and mads6-1 displayed similar defects in the outer three-whorl floral organs, the double mutant flowers had an increased number of stigma and carpel/carpel-like structures compared with mads6-1 (Figure 9), indicating additional defects in floral determinacy. In maize, BDE and ZAG1 were shown to physically interact (Thompson et al., 2009), yet such an interaction was not found between MADS6 and MADS3/MADS58 (Moon et al., 1999). This may suggest that rice MADS6 has conserved and divergent mechanisms from its counterpart in maize in forming protein complexes to specify floral development. Alternatively, MADS6 may have weak interaction with MADS3/58, which was not detected by previous attempts (Moon et al., 1999).
Our data suggest that in addition to C-class genes, MADS6 has other partners in determining floral meristem identity. MADS6 functions redundantly with SPW1, MADS13, and DL, respectively, in specifying floral meristem identity. Some flowers in mads6-1 spw1-1 exhibited new inflorescence-like organs in lieu of lodicules, suggesting that MADS6 and SPW1 play an important role in floral development, particularly in repressing meristem activity after the establishment of lodicule identity. Their genetic interaction may be explained by their protein–protein interaction observed by Seok et al. (2010). In addition to its role in promoting the expression of MADS4 and SPW1 during early flower development, MADS6 was also shown to physically interact with MADS4 and SPW1, suggesting that MADS6-MADS4-SPW1 complex formation may be essential for flower development (Seok et al., 2010) (Figure 11). Additionally, analysis of mads6-1 mad13-3 suggests the interaction of MADS6 with MADS13 in specifying floral meristem identity. MADS6 can interact with MADS13 at the protein level (Favaro et al., 2002), but they do not seem to regulate each other’s expression as revealed in this study (Figure 11).
Rice DL is different from the well-known ABC genes (Nagasawa et al., 2003; Yamaguchi et al., 2004). Moreover, the role of DL is distinct from the closely related Arabidopsis YABBY gene CRC, which only plays a partial role in carpel identity (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). Our recent analysis of mads3-4 dl-sup6 flowers indicated that DL and MADS3 act redundantly in specifying carpel identity and terminating floral meristem, but they may function in distinct pathways (Li et al., 2011) (Figure 11). Furthermore, previous investigation indicated that DL plays an antagonistic role with class B genes (Yamaguchi et al., 2004; Li et al., 2011). The mads6-1 dl-sup6 double mutant flowers displayed inflorescence-like structures in whorl 4, suggesting that MADS6 and DL may synergistically terminate the floral meristem (Figure 11).
MADS6 is able to specify floral meristem by interacting with E-class genes such as LHS1/MADS1, MADS7, and MADS8 (Figure 11). One possible mechanism is that MADS6 could positively regulate the expression of MADS7 and MADS8. mads1 mads6 double mutants had severe indeterminate floral meristem (Ohmori et al., 2009; Li et al., 2010), while our microarray (this study) and in situ analyses (Li et al., 2010) did not reveal obvious expression changes in LHS1/MADS1 in mads6-1. Although Moon et al. (1999) reported the protein–protein interaction between MADS6 and LHS1/MADS1 in yeast cells, we did not observe this interaction (Li et al., 2010), possibly due to weak protein interaction.
In summary, using double mutant analyses in combination with in situ and ChIP-qPCR experiments, we reveal interactions of MADS6 with floral homeotic genes SPW1, MADS3, MAD58, MADS13, and DL in regulating rice flower development and show that MADS6 positively regulates the expression of the B-, C-, and E-class genes during early flower development. A model (Figure 11) is proposed to illustrate the role of floral homeotic genes in the specification of flower organ identity and meristem determinacy in rice.
METHODS
Plant Materials
The mads6-1, mads13-3, mads3-4, and dl-sup6 mutants were identified previously (Chen et al., 2006b; Hu et al., 2011; Li et al., 2010, 2011). spw1-1 and mads58 were kindly provided by Hajime Sakai, Yasuo Nagato, and Venkatesan Sundaresan. Double mutants were isolated by phenotype observation and verified by genotyping. Methods for genotyping dl-sup6, mads13-3, and mads3-4 were described previously (Li et al., 2011). Primer pairs 6TPF/6TPR, 16TPF/16TPR, and 58TPF/58TPR were used for genotyping mads6-1, spw1-1, and mads58, respectively (see Supplemental Table 4 online). Mutant and wild-type rice (Oryza sativa) plants were planted in the paddy field or greenhouse in Shanghai Jiao Tong University, China.
Histological Analysis and Microscopy Observation
Flower materials were collected at various stages of development according to spikelet size and morphology definitions described by Ikeda et al. (2004), fixed in FAA (10% formalin, 50% ethanol, and 5% acetic acid), and dehydrated in a series of graded ethanol (Chu et al., 2006; Li et al., 2006). For histological analysis, tissues were then infiltrated with xylene and embedded in paraplast plus. Then, materials were sectioned to 8 μm thick and stained with toluidine blue (Bio Basic) and photographed using a Nikon E600 microscope and a Nikon DXM1200 digital camera. Scanning electron microscopy was performed with a JSM-6360LV (Jeol) as described previously (Li et al., 2006).
In Situ Hybridization
Samples were treated as described previously (Li et al., 2006). Constructs of gene-specific probes for MADS13, SPW1, DL, and MADS6 were described previously (Li et al., 2010, 2011). Constructs of MADS3 and MADS58 probes were generated based on descriptions by Yamaguchi et al. (2006) and probes of MADS7 and MADS8 were made as described by Cui et al. (2010). Digoxygenin-labeled antisense and sense probes were transcribed in vitro as described by Li et al. (2006). Images were obtained using an Olympus Nikon E600 microscope.
Microarray Analysis
Microarray experiments were performed as described by Hu et al. (2011). Total RNAs were isolated from three replicates of stage Sp6 flowers from the wild type and mads6-1. Developing flower primordia were collected based on microscopy analysis and spikelet length described by Ikeda et al. (2004).
qRT-PCR
Total RNA was isolated from wild-type and mads6-1 flowers at stage Sp4/SP5, Sp6, early Sp8, and late Sp8. qRT-PCR conditions were the same as those described by Zhang et al. (2010a). Primers for MADS3 and MADS4 were described by Chen et al. (2006a), and primers for SPW1, MADS7, MADS8, and MADS58 were described by Cui et al. (2010). All samples were run with three replicates. Data acquisition and analyses were performed using the Roche Light Cycler software. Sample amounts used were normalized using ACTIN expression.
Preparation of the MADS6 Polyclonal Antibody
The MADS6-specific fragment (493 to 753 bp) was amplified by PCR using the primer pair 6APF/6APR (see Supplemental Table 4 online). PCR products were cloned into pET-32a (Novagen) to produce p32-P-MADS6. The 6XHis-MADS6 fused protein was expressed in Escherichia coli using p32-P-MADS6. Proteins were purified according to the manufacturer’s instructions, and the antibody was prepared as described by Huang et al. (2003) using the specific MADS6 fragment. Nuclear extracts from the wild-type flowers at stage Sp7 were used for immunoblot analysis to test the specificity of the MADS6 antibody, following the protocol used for the ChIP experiments (Zhang et al., 2010b), except that the tissue was not fixed. The glutathione S-tranferase–tagged full-length MADS6 protein was used as the positive control for immunoblot analysis.
ChIP-qPCR and Fold Enrichment Analysis
Rice spikelets at stage Sp7 were treated and sonicated with an Ultrasonic Crasher Noise Isolating Chamber (Scientz). The procedure for ChIP of the MADS6-DNA complexes in wild-type flowers was modified from Haring et al. (2007). For each PCR reaction, 0.5 μL of recovered DNA from immune precipitation (IP) or mock was used as template, three biological replicates were included, and each reaction was repeated three times. Primers used for qPCR analyses are labeled in Figure 10 and listed in Supplemental Table 4 online, and reactions were performed on a Rotor-Gene RG3000A detection system (Corbett Research) using SYBR Green I. The normalized mean cycle threshold (Ct) of each gene was calculated and used to determine fold change according to the method described by Rotor-Gene version 6.0 (Build 38) software and Zhang et al. (2010a). The difference between the Ct of the antibody enrichment and no antibody control (mock) was calculated to obtain the relative enrichment of the fragments containing the putative CArG motifs. Quantification involved normalization of the Ct of each IP or the control sample to obtain a nonspecific adjustment △Ct (△Ct IP − △Ct mock), followed by calculation of relative enrichment of each fragment using the equation 2−(△Ct IP − △Ct control).
Accession Numbers
Sequence data from this article for the cDNAs of MADS6, SPW1, MADS3, MADS58, DL, MADS13, MADS7, and MADS8 can be found in the GenBank/EMBL data libraries under accession numbers AK069103, AK069317, AK108568, AK111723, AK242416, AK070425, AK100263, and AK072867, respectively. Locus identifications in the Rice Genome Annotation Project Database are as follows: MADS6 (Os02g45770), SPW1 (Os06g49840), MADS3 (Os01g10504), MADS58 (Os05g11414), DL (Os03g11600), MADS13 (Os12g10540), MADS7 (Os08g41950), and MADS8 (Os09g32948). Microarray data accession number in NCBI is GSE29349.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Spatial and Temporal Expression Pattern of DL and MADS6 as Revealed by in Situ Hybridization.
Supplemental Figure 2. Spatial and Temporal Expression Pattern of MADS13 and MADS6.
Supplemental Figure 3. Specificity Analysis of MADS6 Polyclonal Antibodies.
Supplemental Table 1. Up- or Downregulated (at Least Twofold Change in Expression, P Value < 0.05) Non-MADS Box Genes in mads6-1 Identified by Bayes Analysis (<0.5% FDR).
Supplemental Table 2. Number of Floral Organs in Wild-Type and mads6-1, spw1-1, and mads6-1 spw1-1 Flowers.
Supplemental Table 3. Number of Floral Organs in Wild-Type and mads6-1, mads3-4, and mads6-1 mads3-4 Flowers.
Supplemental Table 4. Primers Used in This Study.
Acknowledgments
We thank Zhijing Luo and Mingjiao Chen for mutant screening and generation of mutants, Hajime Sakai and Yasuo Nagato for providing spw1-1, Venkatesan Sundaresan for mads58, Ning Jiang and Zhongchi Liu for comments on the this work, and Jianping Hu for editing this manuscript. We gratefully acknowledge the anonymous reviewers for very helpful comments. This work was supported by Funds from the National Basic Research Program of China (2009CB941500 and 2007CB108700), the National Natural Science Foundation of China (30725022), the Chinese Transgenic Project (2009ZX08009-108B), and the National 863 High-Tech Project (2011AA10A101).
AUTHOR CONTRIBUTIONS
H.L., W.L., Y.H., L.Z., C.Y., J.X., L.D., and M.M.K. performed research and analyzed data. D.Z. and W.L. designed the research and analyzed data. D.Z. wrote the article.
References
- Agrawal G.K., Abe K., Yamazaki M., Miyao A., Hirochika H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Ambrose B.A., Lerner D.R., Ciceri P., Padilla C.M., Yanofsky M.F., Schmidt R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579 [DOI] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R. (1998). Patterns of petal and stamen reduction in reduction in Australian species of Lepidium L. (Brassicaceae). Int. J. Plant Sci. 159: 65–74 [Google Scholar]
- Bowman J.L., Smyth D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Chen L., Chu H.W., Yuan Z., Pan A.H., Liang W.Q., Huang H., Shen M.S., Zhang D.B., Chen L. (2006b). Isolation and genetic analysis for rice mutants treated with 60Co γ-Ray. J. Xiamen Univ. 45: 82–85 [Google Scholar]
- Chen Z.X., Wu J.G., Ding W.N., Chen H.M., Wu P., Shi C.H. (2006a). Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Chu H.W., Qian Q., Liang W.Q., Yin C.S., Tan H.X., Yao X., Yuan Z., Yang J., Huang H., Luo D., Ma H., Zhang D.B. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton W.D., Renvoize S.A. (1986). Genera Graminum: Grasses of the World. (Kew, UK: Kew Publishing; ). [Google Scholar]
- Clifford H. (1987). Spikelet and Floral Morphology. (Washington, DC: Smithsonian Institution Press; ). [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Colombo L., Franken J., Koetje E., van Went J., Dons H.J., Angenent G.C., van Tunen A.J. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., Xu Q., Chong K., Theissen G., Meng Z. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Cusick F. (1966). On phylogenetic and ontogenetic fusions. In Trends in Plant Morphogenesis, Cutter E.G., (London: Longmans, Green & Co; ), pp. 170–183 [Google Scholar]
- Dai M., Hu Y., Zhao Y., Liu H., Zhou D.X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Immink R.G., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos M.K., Sieburth L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P.B., An G., Colombo L., Kater M.M. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D.P., Erreni S., Pajoro A., Caporali E., Zhang D.B., Kater M.M. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. The Plant Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress P.K. (1992). Evolution and floral diversity: The phylogenetic surroundings of Arabidopsis and Antirrhinum. Int. J. Plant Sci. 153(suppl): S106–S122 [Google Scholar]
- Favaro R., Immink R.G., Ferioli V., Bernasconi B., Byzova M., Angenent G.C., Kater M., Colombo L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Ferrario S., Immink R.G., Angenent G.C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7: 84–91 [DOI] [PubMed] [Google Scholar]
- Francis M.E. (1920). Book of Grasses. (Garden City, New York: Doubleday, Page & Co; ). [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., Guo C., Kong H., Xue H., Zhang D. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 88: 373–457 [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.F., Liang W.Q., Yin C.S., Cui X., Zong J., Wang X., Hu J.P., Zhang D.B. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liang W., Pan A., Zhou Z., Huang C., Chen J., Zhang D. (2003). Production of FaeG, the major subunit of K88 fimbriae, in transgenic tobacco plants and its immunogenicity in mice. Infect. Immun. 71: 5436–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Nagasawa N., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54: 147–156 [Google Scholar]
- Immink R.G., Tonaco I.A., de Folter S., Shchennikova A., van Dijk A.D., Busscher-Lange J., Borst J.W., Angenent G.C. (2009). SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., Jang S., Lee S., Nam J., Kim C., Lee S.H., Chung Y.Y., Kim S.R., Lee Y.H., Cho Y.G., An G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater M.M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo S.C., et al. (2010). Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 62: 807–816 [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Jaramillo M.A., Di Stilio V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Kyozuka J., Kobayashi T., Morita M., Shimamoto K. (2000). Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jürgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.F., Liang W.Q., Jia R.D., Yin C.S., Zong J., Kong H.Z., Zhang D.B. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li H.F., Liang W.Q., Yin C.S., Zhu L., Zhang D.B. (2011). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder H., Rudall P. (2005). Evolutionary history of poales. Annu. Rev. Ecol. Evol. Syst. 36: 107–124 [Google Scholar]
- Liu C., Thong Z., Yu H. (2009b). Coming into bloom: The specification of floral meristems. Development 136: 3379–3391 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W.Y., Shen L.S., Tan C.P., Yu H. (2009a). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Malcomber S.T., Kellogg E.A. (2004). Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber S.T., Kellogg E.A. (2005). SEPALLATA gene diversification: Brave new whorls. Trends Plant Sci. 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Mena M., Ambrose B.A., Meeley R.B., Briggs S.P., Yanofsky M.F., Schmidt R.J. (1996). Diversification of C-function activity in maize flower development. Science 274: 1537–1540 [DOI] [PubMed] [Google Scholar]
- Moon Y.H., Kang H.G., Jung J.Y., Jeon J.S., Sung S.K., An G. (1999). Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol. 120: 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münster T., Deleu W., Wingen L.U., Ouzunova M., Cacharrón J., Faigl W., Werth S., Kim J.T.T., Saedler H., Theissen G. (2002). Maize MADS-box genes galore. Maydica 47: 287–301 [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nardmann J., Werr W. (2006). The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 23: 2492–2504 [DOI] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., Nagato Y., Yoshida H. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K., Parameswaran S., Vijayraghavan U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Preston J.C., Kellogg E.A. (2006). Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Morel P., Negrutiu I., Trehin C. (2009). Time to stop: Flower meristem termination. Plant Physiol. 150: 1764–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P., Erasmus Y., Lane B., Harder L.D., Coen E. (2007). Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Reinheimer R., Kellogg E.A. (2009). Evolution of AGL6-like MADS box genes in grasses (Poaceae): Ovule expression is ancient and palea expression is new. Plant Cell 21: 2591–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A.S., Zethof J., Gerats T., Vandenbussche M. (2009). The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 60: 1–9 [DOI] [PubMed] [Google Scholar]
- Ronse Decraene L.P., Smets E.F. (1993). Dedoublement revisited: Towards a renewed interpretation of the androecium of the Magnoliophytina. Bot. J. Linn. Soc. 113: 103–124 [Google Scholar]
- Rudall P.J., Stuppy W., Cunniff J., Kellogg E.A., Briggs B.G. (2005). Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am. J. Bot. 92: 1432–1443 [DOI] [PubMed] [Google Scholar]
- Schauer S.E., Baskar R., Brand L., Bolanos A., Grobei M., Federer M., Jauch U.and Gagliardini, V. (2007). Examination of the role of the Arabidopsis MADS-box transcription factors AGL6 and AGL13 in reproduction. Dev. Biol. 306: 312 [Google Scholar]
- Seok H.Y., Park H.Y., Park J.I., Lee Y.M., Lee S.Y., An G., Moon Y.H. (2010). Rice ternary MADS protein complexes containing class B MADS heterodimer. Biochem. Biophys. Res. Commun. 401: 598–604 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3 [DOI] [PubMed] [Google Scholar]
- Theissen G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J.T., Münster T., Winter K.U., Saedler H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42: 115–149 [PubMed] [Google Scholar]
- Theissen G., Melzer R. (2007). Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. (Lond.) 100: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Thompson B.E., Bartling L., Whipple C., Hall D.H., Sakai H., Schmidt R., Hake S. (2009). bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.E., Hake S. (2009). Translational biology: From Arabidopsis flowers to grass inflorescence architecture. Plant Physiol. 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Theissen G., Van de Peer Y., Gerats T. (2003a). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31: 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Royaert S., Weterings K., Gerats T. (2004). The duplicated B-class heterodimer model: Whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Souer E., Koes R., Tornielli G.B., Pezzotti M., Ferrario S., Angenent G.C., Gerats T. (2003b). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke J.A. (1992). Fusion events during floral morphogenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 583–598 [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., Gu M., Xue Y., Cheng Z. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Ciceri P., Padilla C.M., Ambrose B.A., Bandong S.L., Schmidt R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131: 6083–6091 [DOI] [PubMed] [Google Scholar]
- Whipple C.J., Zanis M.J., Kellogg E.A., Schmidt R.J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 104: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Wu X.L., Lee J.S., Ahn J.H. (2011). AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 65: 62–76 [DOI] [PubMed] [Google Scholar]
- Yuan Z., Gao S., Xue D.W., Luo D., Li L.T., Ding S.Y., Yao X., Wilson Z.A., Qian Q., Zhang D.B. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L.M., Kong H., Leebens-Mack J.H., Kim S., Soltis P.S., Landherr L.L., Soltis D.E., Depamphilis C.W., Ma H. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L.M., Leebens-Mack J.H., Arrington J.M., Hu Y., Landherr L.L., dePamphilis C.W., Becker A., Theissen G., Ma H. (2006). Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol. Dev. 8: 30–45 [DOI] [PubMed] [Google Scholar]
- Zanis M.J. (2007). Grass spikelet genetics and duplicate gene comparisons. Int. J. Plant Sci. 168: 93–110 [Google Scholar]
- Zhang D.B., Wilson Z.A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 1–12 [Google Scholar]
- Zhang H., Liang W.Q., Yang X.J., Luo X., Jiang N., Ma H., Zhang D.B. (2010a). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zong J., Liu J., Yin J.Y., Zhang D.B. (2010b). Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 52: 1016–1026 [DOI] [PubMed] [Google Scholar]