Analysis of a series of mutants lacking each of the three isoforms of the Lhcb4 light-harvesting complex in Arabidopsis showed that this complex plays an important role in the macro-organization and photoprotection of photosystem II.
Abstract
The role of the light-harvesting complex Lhcb4 (CP29) in photosynthesis was investigated in Arabidopsis thaliana by characterizing knockout lines for each of the three Lhcb4 isoforms (Lhcb4.1/4.2/4.3). Plants lacking all isoforms (koLhcb4) showed a compensatory increase of Lhcb1 and a slightly reduced photosystem II/I ratio with respect to the wild type. The absence of Lhcb4 did not result in alteration in electron transport rates. However, the kinetic of state transition was faster in the mutant, and nonphotochemical quenching activity was lower in koLhcb4 plants with respect to either wild type or mutants retaining a single Lhcb4 isoform. KoLhcb4 plants were more sensitive to photoinhibition, while this effect was not observed in knockout lines for any other photosystem II antenna subunit. Ultrastructural analysis of thylakoid grana membranes showed a lower density of photosystem II complexes in koLhcb4. Moreover, analysis of isolated supercomplexes showed a different overall shape of the C2S2 particles due to a different binding mode of the S-trimer to the core complex. An empty space was observed within the photosystem II supercomplex at the Lhcb4 position, implying that the missing Lhcb4 was not replaced by other Lhc subunits. This suggests that Lhcb4 is unique among photosystem II antenna proteins and determinant for photosystem II macro-organization and photoprotection.
INTRODUCTION
Oxygenic photosynthesis is performed in the chloroplast by a series of reactions that exploits light as an energy source to fuel ATP and NADPH production for CO2 fixation and synthesis of organic compounds. Light harvesting is the primary process in photosynthesis and consists of absorption of photons by an array of hundreds of chlorophylls (Chls) organized into photosystems. Excitons are transferred among Chls and to the reaction center (RC), where charge separation occurs. In photosystem II (PSII), electrons are transferred to the quinonic acceptor plastoquinone (PQ), leading to charge accumulation in the oxygen evolving complex and water splitting (Nelson and Ben-Shem, 2004). RC of PSII consists of the D1/D2/cytochrome b559 complex carrying the cofactors for electron transport, which forms, together with the nearest subunits CP43 and CP47, a core complex (Ferreira et al., 2004). Core complexes form dimers (C2), which bind an extended system of nuclear-encoded light-harvesting proteins (Lhc), each binding Chl a, Chl b, and xanthophylls. Lhcb4 (CP29) and Lhcb5 (CP26) are located near the core and mediate the binding of a trimeric LHCII antenna complex called LHCII-S (strongly bound). These components form the basic PSII supercomplex structure, called C2S2 (Boekema et al., 1999), which is the major PSII form found in plants grown under high light (HL) (Morosinotto et al., 2006; Frigerio et al., 2007). Under low/moderate light, larger supercomplexes, called C2S2M2, are formed: one additional monomeric subunit Lhcb6 (CP24) and two LHCII trimers (LHCII-M and LHCII-L, moderately and loosely bound, respectively) are accumulated to extend the light-harvesting capacity (Melis 1991; Ballottari et al., 2007). In Arabidopsis thaliana, single genes encode Lhcb5 (CP26) and Lhcb6 (CP24) subunits, while Lhcb4 (CP29) is encoded by three highly conserved genes. Lhcb4.1 and Lhcb4.2 are similarly expressed, while the level of the Lhcb4.3 messenger is 20 times lower under control conditions (Jansson, 1999). The polypeptide encoded by Lhcb4.3 is predicted to lack a large part of the C-terminal domain, a peculiar feature of Lhcb4.1 and Lhcb4.2 isoforms, leading to the suggestion of renaming it as Lhcb8 (Klimmek et al., 2006).
A remarkable property of the antenna system is the ability to actively regulate PSII quantum efficiency to avoid the damaging effects of excess light. Indeed, under constant moderate light conditions, the efficiency of energy conversion is high, due to photochemical reactions. On the other hand, fluctuations of light intensity/temperature/water availability may yield into the excitation of PSII over the capacity for photochemical quenching of Chl singlet excited states (1Chl*). The consequent lifetime increase enhances the probability of Chl triplet (3Chl*) formation by intersystem crossing and yields into single oxygen (1O2) production (Melis, 1999). Since formation of 3Chl* is a constitutive property of Chls (Mozzo et al., 2008), photoprotection mechanisms are activated to prevent damage and improve fitness in the ever-changing environment experienced by plants. Safety systems have evolved to either detoxify the reactive oxygen species (ROS) (Asada, 1999) or to prevent their formation (Niyogi 2000) by (1) downregulating 1Chl* lifetime through the process of nonphotochemical quenching (NPQ) that dissipates excess excited states into heat (Horton 1996); (2) by quenching 3Chl*; and (3) by scavenging ROS. Sustained overexcitation is counteracted by the long-term reduction of PSII antenna size (Anderson, 1986). Additional regulation is activated in limiting light conditions by transferring LHCII complexes between photosystems, which balances excitation delivery to photosystem I (PSI) and PSII (Allen, 1992; Haldrup et al., 2001).
The conservation of the different Lhcb gene products through evolution suggests each has a specific functional role, which, however, is not yet fully clarified. Two major approaches have been used to this aim: namely, the analysis of individual gene products either purified from thylakoids (Bassi et al., 1987; Caffarri et al., 2001) or recombinant (Caffarri et al., 2004; Formaggio et al., 2001) and reverse genetics, by which plants lacking one or more Lhcbs have been produced and characterized (Andersson et al., 2001; Kovács et al., 2006; de Bianchi et al., 2008; Damkjaer et al., 2009). These studies have provided evidence for differential roles of LHCII components (Lhcb1-3) with respect to monomeric subunits (Lhcb4-6). Antisense lines of Arabidopsis devoid of Lhcb1+2 (Ruban et al., 2003) showed a reduced fitness for plants grown in the field, while only minor differences were observed for growth rate, PSII quantum yield, photosynthetic rate, and capacity for NPQ in a controlled environment (Andersson et al., 2003). Similar considerations apply to Lhcb3, the major effect of its depletion consisting of a faster kinetic of state I to state II transitions (Damkjaer et al., 2009). More specific effects have been reported for monomeric Lhcb4 and Lhcb5, which have been shown to (1) carry protonatable (DCCD binding) sites (Pesaresi et al., 1997; Walters et al., 1996); and (2) undergo conformation changes upon exchange of violaxanthin (Viola) to zeaxanthin (Zea) in their L2 xanthophyll binding site (Morosinotto et al., 2002), an important feature since Zea is synthesized in excess light conditions and correlates with NPQ (Demmig-Adams et al., 1989). The Zea-dependent allosteric change was shown to affect the properties of these pigment proteins, including a decrease in fluorescence lifetime (Crimi et al., 2001; Moya et al., 2001) and formation of carotenoid radical cations (Ahn et al., 2008; Avenson et al., 2008), suggesting a major role in NPQ. Consistently, koLhcb6 plants exhibited decreased qE (Kovács et al., 2006; de Bianchi et al., 2008), while koLhcb5 plants were affected in the slower relaxing component of NPQ called qI. Other properties of Lhcb proteins, which are essential for photoprotection, appear to be redundant. In fact, while the total depletion of Lhcb proteins induces photosensitivity in ch1 mutants (Kim et al., 2009; Dall’Osto et al., 2010), no individual knockout (KO) lines have shown major impairment in their capacity to resist HL treatment so far (Ruban et al., 2003; de Bianchi et al., 2008). Lhcb4 antisense lines showed NPQ kinetics and amplitude, photosynthetic electron transport rate, and light sensitivity similar to that of the wild type (Andersson et al., 2001). However, since this work only employed growth in mild conditions, the question is open for the role of Lhcb4 in photoprotection. Indeed, the maintenance of Lhcb4, in HL stress conditions that leads to the reduction of Lhcb6 and LHCII (Ballottari et al., 2007), suggests that Lhcb4 might be a key factor in light harvesting and photoprotection. Lhcb4 phosphorylation in both monocots (Testi et al., 1996) and dicots (Hansson and Vener, 2003; Tikkanen et al., 2006) affects the spectral properties of the protein (Croce et al., 1996) and correlates with higher resistance to HL and cold (Bergantino et al., 1995; Mauro et al., 1997) and water stress (Liu et al., 2009). Although the mechanism of this protective effect is unknown, there is evidence for a link between Lhcb4 and NPQ, since the protein has been identified as an interaction partner of PsbS (Teardo et al., 2007), the pH-dependent trigger of qE (Li et al., 2000), and it is part of a pentameric complex whose dissociation is indispensable for the establishment of NPQ (Betterle et al., 2009). Finally, two Chl ligands in Lhcb4 have been identified as components of the quenching site, which catalyzes the formation of a xanthophyll radical cation (Holt et al., 2005; Avenson et al., 2008).
In this work, we constructed KO mutants for Lhcb4 isoforms in Arabidopsis and analyzed their performance in photosynthesis and photoprotection. PSII quantum efficiency and capacity for NPQ were affected by lack of Lhcb4, and, unlike that of any other Lhcb subunit, the capacity of resisting excess light conditions was affected. Although depletion of Lhcb subunits is usually complemented by the overaccumulation of other members of the subfamily (Ruban et al., 2003; de Bianchi et al., 2008), this is not the case for Lhcb4 and PSII supercomplexes isolated from koLhcb4 that although they retained their C2S2 organization, they lacked Lhcb4. This caused a different mode of binding of LHCII-S trimer within the PSII supercomplex and changed the overall shape of the C2S2 particle. Deletion of either Lhcb4.1 or Lhcb4.2 yielded into a compensatory accumulation of the remaining subunit; instead, the double mutant koLhcb4.1 4.2 was unable to accumulate Lhcb4.3. We conclude that Lhcb4 is a fundamental component of PSII, which is essential for maintenance of both the function and structural organization of this photosystem.
RESULTS
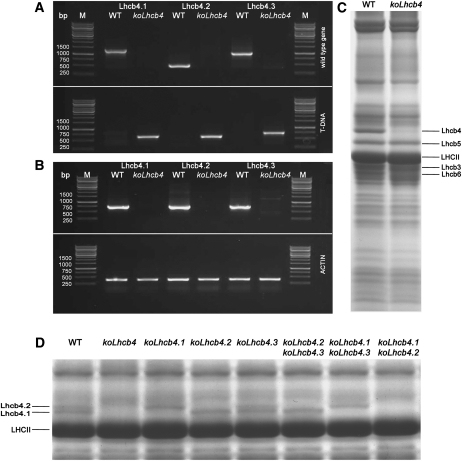
The construction of a plant without a CP29 complex (hereafter named koLhcb4) requires the isolation of KO mutants at three distinct loci, namely, Lhcb4.1, Lhcb4.2, and Lhcb4.3 (Jansson, 1999). We identified koLhcb4.1, koLhcb4.2, and koLhcb4.3 homozygous plants in T-DNA F5 seed pools, obtained from the European Arabidopsis Stock Centre (NASC), by PCR analysis of genomic DNA using specific primers (Figure 1A). The triple KO mutant koLhcb4 was obtained by selection of the progeny from crossing single mutants. PCR analysis confirmed that all Lhcb4 coding regions carried a T-DNA insertion in both alleles (Figure 1A), while RT-PCR showed that mRNAs encoding Lhcb4 isoforms were absent in the mutant (Figure 1B).
Figure 1.
Genetic and Biochemical Characterization of the koLhcb4 Mutant (Triple Mutant for the Three Isoforms of Lhcb4).
(A) Amplification of Lhcb4.1, Lhcb4.2, and Lhcb4.3 loci with allele-specific PCR primers. Top panel: amplification using gene-specific primers. Bands of 1378, 520, and 1046 bp were obtained for the amplification of the Lhcb4.1, Lhcb4.2, and Lhcb4.3 loci, respectively. Bottom panel: amplification using T-DNA–specific primers. Bands of 685, 661, and 773 bp were obtained for the amplification of Lhcb4.1, Lhcb4.2, and Lhcb4.3 KO loci, respectively. Details of primer sequences are reported in Methods. WT, wild type.
(B) RT-PCR measurement of gene-specific transcripts. Sequences of the oligonucleotides used are reported in Methods. Top panel: for each gene, RNA extracted from the wild type and the corresponding mutant was subjected to reverse transcription, followed by 30 cycles of PCR amplification. Bottom panel: amplification of the housekeeping gene actin2 transcript from the same RNAs used as loading control. M, molecular weight marker. The expected sizes of the PCR products are as follows: Lhcb4.1, 724 bp; Lhcb4.2, 715 bp; Lhcb4.3, 730 bp; and actin, 384 bp. Each RT-PCR measurement was repeated three times.
(C) SDS-PAGE analysis of wild-type and koLhcb4 mutant thylakoid proteins performed with the Tris-Tricine buffer system (Schägger and von Jagow, 1987). Selected apoprotein bands are marked. Purified thylakoid sample, corresponding to 15 μg of chlorophylls, was loaded in each lane.
(D) SDS-PAGE analysis performed with the Tris-Tricine buffer system with the addition of 7 M urea to the running gel in order to separate Lhcb4 isoforms in the Lhcb4 KO mutants. Selected apoprotein bands are marked. Fifteen micrograms of chlorophylls were loaded in each lane.
During the course of this work, two additional lines (N124926 and SK32480) with a T-DNA insertion mapped to the Lhcb4.1 and Lhcb4.2 genes, respectively, became available. Both lacked the corresponding protein. The lines showed very similar phenotypic characteristics to N376476 (koLhcb4.1) and N877954 (koLhcb4.2), respectively. Below, we present data from the N376476 and N877954 lines, unless otherwise stated.
By screening the F2 generation, it was possible to isolate double mutants expressing single Lhcb4 isoforms, namely, koLhcb4.2 4.3 (retaining Lhcb4.1), koLhcb4.1 4.3 (retaining Lhcb4.2), and koLhcb4.1 4.2 (retaining Lhcb4.3). All of the isolated genotypes did not show a significant reduction in growth with respect to the wild type under control light conditions (100 μmol photons m−2 s−1, 24°C, 8/16 day/night). Electron microscopy analysis of plastids from mesophyll cells of the wild type and mutants was performed to test if the thylakoid structure was changed as an effect of the missing Lhcb (see Supplemental Figure 1 online). All the mutants showed a membrane organization similar to that of wild-type chloroplasts with well-defined grana, containing approximately six discs per granum, as well as similar amounts of grana, stroma lamellae, and end membranes per plastid (see Supplemental Table 1 online).
Thylakoid membranes isolated from the koLhcb4 mutant lacked the corresponding gene product (Figure 1C). A better resolution in the 22 to 35 kD molecular mass range was obtained using a gel incorporating 7 M urea, which split Lhcb4 into a doublet in wild-type thylakoids, the upper band corresponding to Lhcb4.2 as shown by comparison with the pattern from double KO genotypes (Figure 1D). The gel without urea (Figure 1C) revealed that an additional band with lower molecular mass was also missing, which corresponded to Lhcb6, as revealed by immunoblotting (Figures 2A and 2B). The pigment content of the koLhcb4 mutant did not differ from the wild type in either chlorophyll content per leaf area or Chl/Car ratio but showed a significant decrease in the Chl a/Chl b ratio (Table 1).
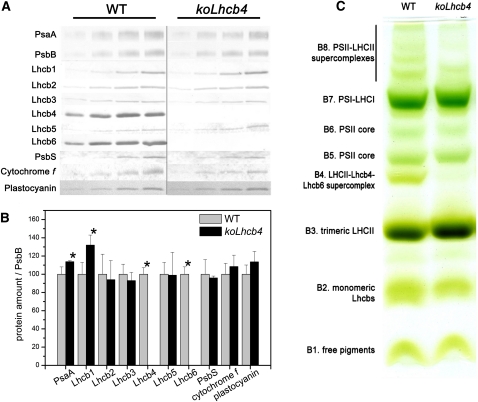
Figure 2.
Polypeptide Composition of Thylakoid Membranes from Wild-Type and koLhcb4 Mutant.
(A) Immunoblotting used for the quantification of photosynthetic subunits in the wild type (WT) and koLhcb4 thylakoids. Immunoblot analysis was performed with antibodies directed against individual gene products: minor antenna proteins, the LHCII subunit, the PSII core subunit PsbB (CP47), the PSI core subunit (PsaA), cytochrome f, and plastocyanin. Thylakoids corresponding to 0.25, 0.5, 0.75, and 1 μg of chlorophyll were loaded for each sample. All samples were loaded on the same SDS-PAGE slab gel.
(B) Results of the immunotitration of thylakoid proteins. Data of PSII antenna subunits were normalized to the core amount, PsbB content (Ballottari et al., 2007), and expressed as a percentage of the corresponding wild-type content. Significantly different values from wild-type membranes are marked with an asterisk.
(C) Thylakoid pigment-protein complexes were separated by nondenaturing Deriphat-PAGE upon solubilization with α-DM. Thylakoids corresponding to 25 μg of chlorophylls were loaded in each lane.
[See online article for color version of this figure.]
Table 1.
Chlorophyll Content and Fluorescence Induction Parameters Determined for Leaves of Arabidopsis Wild-Type and koLhcb4 Plants and Mutants Retaining a Single Lhcb4 Isoform
| Genotype | Chl a/b | Chl/Car | μg Chl/cm2 | Fo | Fv/Fm | t2/3 (ms) | Sm/tFmax (ms−1) |
| Wild type | 3.06 ± 0.07 | 3.63 ± 0.08 | 20.7 ± 3.0 | 0.195 ± 0.002 | 0.790 ± 0.007 | 182 ± 6 | 1.2 ± 0.1 |
| koLhcb4 | 2.83 ± 0.06* | 3.58 ± 0.03 | 19.0 ± 1.1 | 0.253 ± 0.021* | 0.747 ± 0.021* | 176 ± 8 | 1.5 ± 0.3 |
| koLhcb4.2 4.3 | 2.97 ± 0.03 | 3.70 ± 0.13 | 22.5 ± 1.2 | 0.213 ± 0.012 | 0.796 ± 0.004 | 182 ± 5 | 1.1 ± 0.4 |
| koLhcb4.1 4.3 | 2.97 ± 0.03 | 3.61 ± 0.09 | 22.6 ± 4.5 | 0.231 ± 0.012* | 0.777 ± 0.007* | 175 ± 11 | 1.6 ± 0.1 |
| koLhcb4.1 4.2 | 2.83 ± 0.09* | 3.63 ± 0.08 | 22.9 ± 2.5 | 0.266 ± 0.004* | 0.745 ± 0.007* | 184 ± 6 | 1.4 ± 0.2 |
Data are expressed as mean ± sd (n > 5); significantly different values (Student’s t test, P = 0.05) with respect to the wild type are marked with an asterisk.
To determine whether the capacity of the antenna system and its ability to transfer absorbed energy to RCs was affected by the mutation, the functional antenna size of PSII was measured on thylakoids by estimating the rise time of chlorophyll florescence in the presence of DCMU and nigericin. No significant differences were observed in t2/3 Fmax (see Methods for details) between koLhcb4 and the wild type (Table 1; see Supplemental Figure 2 online), suggesting that Lhcb4 depletion did not impair the overall light-harvesting capacity. Analysis of the fluorescence induction in dark-adapted leaves (Butler and Strasser, 1978) revealed a higher F0 value and a significant decrease of maximum quantum efficiency of PSII (Fv/Fm) in koLhcb4 with respect to the wild type. Thus, a larger fraction of absorbed energy is lost as fluorescence in the mutant, implying that the connection between the major LHCII complex and PSII RC is less efficient in the absence of Lhcb4 (Table 1). Nevertheless, parameter Sm/tFmax, which is used for quantifying PSII electron transport (ET) activity, was essentially the same in koLhcb4 and the wild type, suggesting that ET was not limited downstream from QA− in mutant leaves.
Organization and Stoichiometry of Pigment-Protein Complexes
The organization of pigment binding complexes was analyzed by nondenaturing Deriphat-PAGE upon solubilization of wild-type and koLhcb4 thylakoid membranes with 0.7% dodecyl-α-d-maltoside (α-DM). Seven major green bands were resolved (Figure 2C). In the wild type, the PSI-LHCI complex was found as a major band (B7) in the upper part of the gel, while the components of the PSII-LHCII migrated as multiple green bands with different apparent masses, namely, the PSII core dimer and monomer (B6 and B5, respectively) and the antenna moieties, including the Lhcb4-Lhcb6-LHCII-M supercomplex (B4), LHCII trimer (B3), and monomeric Lhcbs (B2). Four faint green bands with high apparent molecular mass, which contained undissociated PSII supercomplexes of different LHCII composition, were detected in the upper part of the gel (B8). The major differences detected in koLhcb4 with respect to the wild type were the lack of B4 and a reduced level of PSII supercomplexes. Densitometric analysis of the Deriphat-PAGE showed a higher PSI/PSII ratio in the koLhcb4 mutant (1.38 ± 0.11) with respect to the wild type (1.04 ± 0.03).
To detect possible alterations in the relative amount of protein components of the photosynthetic apparatus, we determined the stoichiometry of several subunits by immunoblotting titration using CP47 (PsbB) as an internal standard (Ballottari et al., 2007). KoLhcb4 plants lacked both Lhcb4 and Lhcb6 proteins (Figure 2A), while there was a 30% increase in Lhcb1 (Figure 2B) with respect to the wild-type level. The other pigment-protein complexes, as well as cytochrome (cyt) f and plastocyanin, were present in wild-type amounts. A slight but significant increase in PsaA content in koLhcb4 compared with the wild type (Figure 2A) was detected.
We also analyzed the Lhc protein composition in double mutants expressing individual Lhcb4 isoforms (Figure 1D; see Supplemental Table 2 and Supplemental Figure 3 online). The koLhcb4.2 4.3 mutant retained 87% of wild-type Lhcb4 levels, while koLhcb4.1 4.3 had a significantly lower amount of Lhcb4 (59%) with respect to the wild type (see Supplemental Table 2 online). The koLhcb4.1 4.2 mutant did not show any band with mobility similar to Lhcb4.1 and Lhcb4.2 (Figure 1D). Nevertheless, the Lhcb4.3 gene product is expected to have a molecular mass significantly lower than the 0.1 and 0.2 isoforms. Therefore, we cannot exclude that it migrates together with the bulky LHCII band. Immunoblotting analysis also revealed that the level of Lhcb6 polypeptide underwent changes in agreement with the amounts of Lhcb4.1 + Lhcb4.2 protein. In the koLhcb4.1 4.2 mutant, Lhcb6 was not detectable (see Supplemental Table 2 online).
Photosynthesis-Related Functions: ET Rate, State Transition, and P700 Redox State
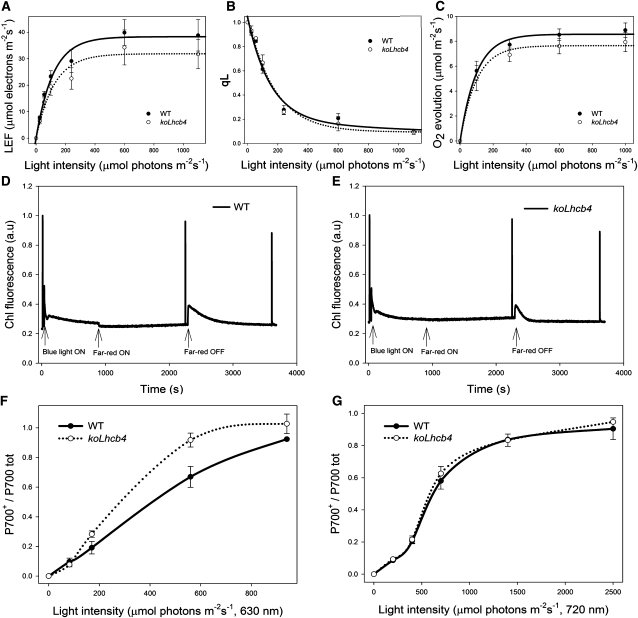
Since pigment-protein complexes participate in modulating ET between photosystems, PSII and PSI function during photosynthesis was further analyzed by chlorophyll fluorometry. KoLhcb4 showed no significant differences with respect to wild-type plants either in the linear electron flow (LEF) or in the QA redox state (qL), as measured at different light intensities in the presence of saturating CO2 on leaves (Figures 3A and 3B), thus indicating no limitations in photosynthesis downstream of PSII. This was further confirmed by measuring rates of O2 evolution in CO2-saturating conditions: no significant reduction in the quantum yield of photosynthesis was observed in koLhcb4 versus wild-type plants at any of the light intensities used (Figure 3C).
Figure 3.
Characterization of Photosynthetic Electron Flow in Wild-Type and koLhcb4 Plants.
(A) Dependence of the LEF rate on light intensity in wild-type (WT) and koLhcb4 leaves. LEF rate is calculated as ϕPSIIPSII·PAR·Aleaf·fractionPSII (see Methods for details).
(B) Photosynthetic oxygen evolution in saturating CO2. Rate of oxygen evolution was measured on whole leaves during illumination with various levels of actinic red light.
(C) Amplitude of qL measured at different light intensities on wild-type and koLhcb4 leaves. qL reflects the redox state of the primary electron acceptor QA, thus, the fraction of open PSII centers.
(D) and (E) Measurement of state I–state II transition in the wild type (D) and koLhcb4 (E). Upon 1-h dark adaptation, plants were illuminated with blue light (40 μmol photons m−2 s−1, wavelength <500 nm) for 15 min to reach state II. A far-red light source was then superimposed on the blue light to induce a transition to state I. Values of Fm, Fm′, and Fm′′ were determined using light saturation pulses (4500 μmol photons m−2 s−1, 0.6 s). a.u., arbitrary units.
(F) and (G) Dependence of the P700 oxidation ratio (ΔA/ΔAmax) on light intensity. ΔA/ΔAmax was measured on wild-type and koLhcb4 leaves at varying actinic intensities, exciting either PSII + PSI (λ = 630 nm; [F]) or PSI only (λ = 730 nm; [G]).
The capacity for state transitions (Allen, 1992) was measured from the changes in chlorophyll fluorescence on leaves (Jensen et al., 2000). The final amplitude of the state transitions (qT) after a 15-min illumination was the same in both genotypes (0.091 ± 0.002 and 0.091 ± 0.006, respectively, in the wild type versus koLhcb4) (Figures 3D and 3E). However, the kinetic of the transition from state I to state II upon switching off far-red light was 3 times faster in koLhcb4 (half-time = 71 ± 5 s) with respect to the wild type (half-time = 204 ± 17 s) (Figures 3D and 3E). Furthermore, it was observed that the switching on of far-red light produced a sudden decrease of fluorescence intensity in the wild type but not in koLhcb4. In fact, fluorescence level in the latter was already very low, consistent with a faster reoxidation of the PQ pool in koLhcb4 with respect to the wild type under low-intensity blue light.
Fluorescence induction analysis on intact leaves showed a F0 value up to 40% higher in koLhcb4 versus the wild type, implying the absence of Lhcb4 caused a lower efficiency in energy transfer between LHCII and PSII RC (Table 1). Photosynthetic electron flow through PSI during steady state photosynthesis in vivo was measured from the dependence of the P700+/total P700 ratio on intensity of PSI+PSII light (λ = 630 nm; Figure 3F). In the wild type, this value approached saturation level at >1000 μmol photons m−2 s−1. In koLhcb4, the saturation was observed already at 600 μmol photons m−2 s−1, which implies a higher functional antenna size of PSI in mutant leaves. To investigate the origin of this difference, cyclic electron flow (CEF) was determined by following (1) the rereduction of P700+ at 705 nm, upon far-red saturating flashes, on leaves infiltrated with DCMU (see Supplemental Figure 4A online) and (2) the relationship between LEF and the steady state proton flux across the thylakoid membrane (νH+; see Supplemental Figure 4B online). The fast decay component of P700+, which has been attributed to CEF, was the same in both the wild type and koLhcb4, as well as the dependence of νH+ to LEF. Redox kinetics of cyt b6 reduction and of cyt f oxidation (see Supplemental Figure 4C online) confirmed that both PQ diffusion and cyt b6f activities were not significantly affected in koLhcb4 plants, thus ruling out the possibility that less efficient diffusion of PQ or plastocyanin, or altered activities of the cyt b6f complex, could account for the faster P700 oxidation. Analysis of the functional antenna size of PSI, measured by the rate coefficient of P700 oxidation in steady far-red light following a saturating flash, did not reveal any difference between the two genotypes (see Supplemental Figure 4D online). To further investigate this point, P700 redox state was measured using PSI light (actinic far red, λ = 720 nm) rather than PSI+PSII light. The difference in P700 oxidation rate between wild-type and mutant leaves disappeared once measured with far-red light (Figure 3G), implying that PSII components contribute to increase PSI antenna size in koLhcb4. To verify this suggestion, we investigated the distribution of Chl b and Chl a absorption forms contributing to PSI and PSII excitation by 77K chlorophyll fluorescence spectroscopy. Wild-type and koLkcb4 leaves were either dark adapted or illuminated for 5 min (500 μmol photons m−2 s−1, room temperature), upon which they where rapidly frozen in liquid nitrogen and grounded in a cooled mortar, diluted to 0.05 absorption units with cold buffer containing 5 mM Mg2+, and finally frozen to 77K. Supplemental Figure 5A online shows the emission spectra from dark-adapted versus light-treated wild-type samples. A small increase in the amplitude of the 685-nm emission peak, from PSII versus the 735-nm peak from PSI was obtained upon illumination, a behavior likely due to a completion of the transition to state I. Illumination of koLhcb4 leaves, instead, yielded an opposite effect, with a decreased amplitude of the PSII emission peak (see Supplemental Figure 5B online). Consistently, excitation spectra for the PSI emission (735 nm) clearly show that, while the Chl b contribution is essentially the same in dark- versus light-treated wild type, it is increased in the mutant, implying an enhanced LHCII contribution to PSI (see Supplemental Figures 5C and 5D online).
NPQ of Chlorophyll Fluorescence
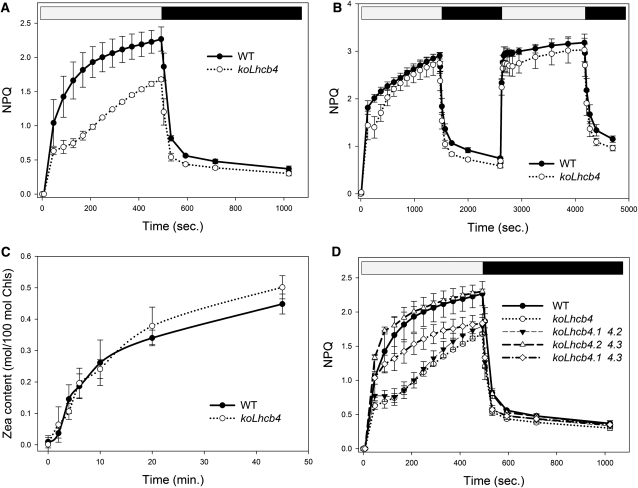
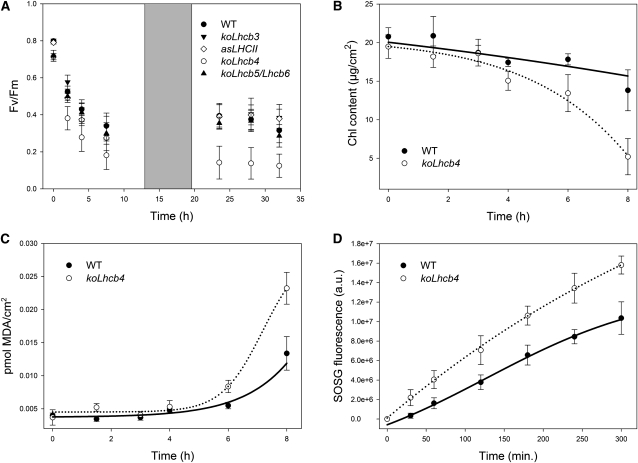
Thermal energy dissipation is a major photoprotection mechanism in plants. Although a quenching component has been localized in PSII core (Finazzi et al., 2004), most quenching activity was associated to the antenna system (Horton and Ruban, 2005; Havaux et al., 2007). In a previous work (Betterle et al., 2009), we observed that the koLhcb4 mutant, among other KO mutants for Lhcb genes, cannot assemble the CP29-CP24-LHCII complex (B4C), whose dissociation is essential for triggering of NPQ. Here, we focus on the elucidation of the mechanistic role of Lhcb4 in NPQ and photoprotection. The NPQ activity of koLhcb4 plants is shown in Figure 4A. Upon exposure of the wild type to saturating light intensity (1200 μmol photons m−2 s−1, 24°C), NPQ showed a rapid rise to 1.4 in the first minute followed by a slower raise, reaching a value of 2.3 after 8 min. Induction of NPQ in koLhcb4 was slower (0.7 at t = 1 min) and reached a lower amplitude (1.6 at t = 8 min). Recovery in the dark was faster and wider in koLhcb4 compared with the wild type (Figure 4A), as previously shown (Betterle et al., 2009). NPQ activity of koLhcb4 reached the wild-type level upon longer light treatment (25 min). When a second illumination period was applied (Figure 4B), the delay in NPQ rise was reduced, while dark recovery was still faster in koLhcb4. It should be noted that Zea synthesis had the same kinetic in the wild type and koLhcb4 (Figure 4C), implying that the delayed NPQ onset was not due to a delayed Zea synthesis.
Figure 4.
Kinetics of the Formation and Relaxation of Photoprotective Energy Dissipation.
(A) Measurements of NPQ kinetics on wild-type (WT) and koLhcb4 leaves illuminated with 1200 μmol photons m−2 s−1, 24°C.
(B) NPQ kinetics of wild-type and koLhcb4 plants during two consecutive periods of illumination with white light (1200 μmol photons m−2 s−1, 25 min, 24°C) with an 18-min period of darkness in between, as indicated by the white and black bars.
(C) Time course of Viola deepoxidation in wild-type and koLhcb4 plants. Leaf discs from dark-adapted leaves were illuminated at 1200 μmol photons m−2 s−1, 24°C (white actinic light). At different times, discs were frozen in liquid nitrogen and total pigments extracted before HPLC analysis.
(D) NPQ kinetics of the wild type, koLhcb4, and mutants retaining single Lhcb4 isoforms. The expression of the isoform Lhcb4.1 (koLhcb4.2 4.3) leads to complete compensation of the NPQ phenotype of koLhcb4; instead, the expression of the Lhcb4.2 isoform partially recovered the quenching ability in the first minutes of induction; nevertheless the mutant failed to fully match the wild type quenching capacity within 8 min of illumination. The presence of the Lhcb4.3 gene did not contribute to NPQ activity. Symbols and error bars show means ± sd (n > 3).
We also investigated whether the three Lhcb4 isoforms had the same activity in NPQ by analyzing the kinetic of NPQ rise in double mutants retaining one single Lhcb4 gene. The results reported in Figure 4D show that mutants conserving the Lhcb4.1 gene had slightly faster onset of NPQ than the wild type, while those retaining Lhcb4.2 gene were unable to reach the final NPQ level of wild-type plants. Mutant plants retaining only Lhcb4.3 behaved like koLhcb4. The faster fluorescence recovery observed in koLhcb4 compared with the wild type is present in the mutants retaining only Lhcb4.2 or Lhcb4.3 genes, whereas mutant retaining Lhcb4.1 recovered with the slower wild-type kinetic.
Photosensitivity under Short- and Long-Term Stress Conditions
Treatment of plants with strong light produces photoxidative stress, whose severity is enhanced by low temperature. Under these conditions, enhanced release of 1O2 leads to bleaching of pigments, lipid oxidation, and PSII photoinhibition, which is accompanied by a decrease in Fv/Fm (Zhang and Scheller, 2004). The sensitivity to photoxidative stress of wild-type and Lhcb4-depleted plants was assessed upon transfer from control conditions to HL + low temperature (500 μmol photons m−2 s−1, 4°C). The level of Fv/Fm was monitored for 2 d (Figure 5A). In wild-type plants, the Fv/Fm parameter gradually decreased from 0.8 to 0.35 during the treatment, while in koLhcb4 plants, the decrease was stronger, reaching a value of 0.15 at the end of the treatment. As a reference, we compared the photosensitivity of different antenna mutants, namely, koLhcb5/Lhcb6, koLhcb3, and LHCII antisense plants (asLHCII). Interestingly, mutants lacking antenna components other than Lhcb4 showed the same level of photoinhibition as wild-type plants, implying that Lhcb4 is the only antenna component indispensable for full level of photoprotection under photoxidative stress (Figure 5A). Measurements of Fv/Fm recovery after photoinhibitory treatment (see Supplemental Figure 6 online) clearly showed that wild-type and koLhcb4 leaves had the same capacity of PSII quantum efficiency recovery, implying that a higher photosensitivity of mutant plants is due to a less effective photoprotection rather than to impaired PSII repair mechanism (Aro et al., 1994). Loss of Lhcb4 caused a decrease in PSII quantum yield (Figure 5A) that could be caused either by damage of the PSII core complex or by an incomplete excitation transfer to PSII RC. To provide a more complete characterization of the photodamage, leaf discs from the wild type and koLhcb4 were submitted to HL + cold stress (1500 μmol photons m−2 s–1, 4°C) and the time course of pigment photobleaching and lipid peroxidation was measured (Figures 5B and 5C). Analysis indicates that chlorophyll bleaching was faster and malondialdehyde (MDA) production was higher in koLhcb4 with respect to wild-type leaves, implying a higher level of lipid peroxidation (Havaux et al., 2005). Photodamage can be caused by the production of ROS, including singlet oxygen (1O2) (Triantaphylidès et al., 2008). To identify if this species was involved in the preferential photodamage of koLhcb4, we quantified production of 1O2 directly in wild-type and koLhcb4 leaves using vacuum-infiltrated 1O2-specific probe (see Methods for details). After illumination with strong light at 4°C, koLhcb4 leaves clearly showed a significantly higher release of singlet oxygen compared with the wild type (Figure 5D).
Figure 5.
Photooxidation of Arabidopsis Wild Type and koLhcb4 Mutant Exposed to HL and Low Temperature.
(A) PSII photoinhibition (Fv/Fm decay) was followed in the wild type (WT), koLhcb4, and antenna-depleted mutant (koLhcb5 Lhcb6, koLhcb3, and LHCII antisense) plants, treated at 550 μmol photons m−2 s−1, 4°C for 30 h with a 6-h period of low light (20 μmol photons m−2 s−1) between the 12 h of HL stress; low-light interval permitted the PSII efficiency recovery.
(B) and (C) Detached leaves floating on water were treated at 1500 μmol photons m−2 s−1 at 4°C, and kinetics of chlorophyll bleaching (B) and MDA formation (C) were recorded.
(D) Wild-type and mutant detached leaves were vacuum infiltrated with 5 μM SOSG, a 1O2-specific fluorogenic probe. SOSG increases its fluorescence emission upon reaction with singlet oxygen. The increase in the probe emission was followed with a fiber optic on the leaf surface during illumination with red actinic light (550 μmol photons m−2 s−1) at 4°C. a.u., arbitrary units.
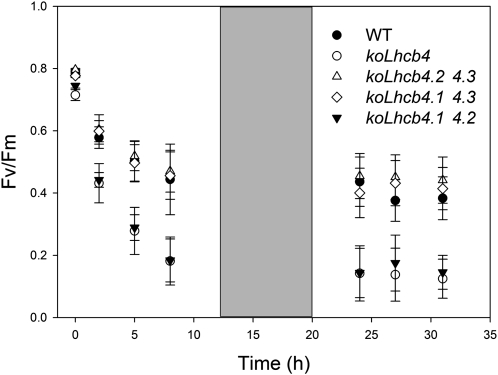
An interesting question is whether the different Lhcb4 isoforms have a specific importance in photoprotection. We measured the level of PSII photoinhibition on KO mutants retaining individual Lhcb4 genes. Plants retaining either Lhcb4.1 or Lhcb4.2 showed a wild-type level of resistance to HL + cold stress, while those retaining the Lhcb4.3 gene (koLhcb4.1 4.2 mutant) were similar to koLhcb4 (Figure 6). More detailed analysis was performed after 3 and 8 d of treatment by determining the leaf chlorophyll content, a target of photoxidative stress. Mutants koLhcb4 and koLhcb4.1 4.2 underwent a significant reduction of leaf chlorophyll content (Table 2), while mutants retaining either Lhcb4.1 or 4.2 did not show this effect. This indicates that a compensatory accumulation of Lhcb4.1 or 4.2 isoforms can restore photoprotection to the wild-type level. Interestingly, the presence of wild-type alleles of Lhcb4.3 as the only Lhcb4 isoform did not restore photoprotection, thus prompting us to get further insight into the Lhcb4.3 expression. We investigated accumulation of the Lhcb4.3 isoform upon several stress conditions, namely, (1) 500 μmol photons m−2 s−1, 4°C for 2 d; (2) 900 μmol photons m−2 s−1, 4°C for 10 d; and (3) 1600 μmol photons m−2 s−1, 24°C for 10 d. At the end of these treatments, thylakoids were isolated from the wild type, koLhcb4, and koLhcb4.1/4.2 and analyzed by SDS-PAGE/immunoblotting using a polyclonal antibody. Under all experimental conditions, no Lhcb4 immune-reactive bands were detected in the koLhcb4.1 4.2 sample from all the tested conditions. In order to verify that our antibody was indeed able to detect Lhcb4.3, the three recombinant apoproteins were assayed by the anti-Lhcb4 antibody and were recognized with the same efficiency (see Supplemental Figure 7 online). Based on these experiments, we conclude that plants retaining the Lhcb4.3 gene only were unable to accumulate the encoded protein to a significant level in the conditions explored in this work.
Figure 6.
Photoinhibition of the Wild Type and Mutants Retaining a Single Lhcb4 Isoform Exposed to HL and Low Temperature.
Kinetics of Fv/Fm decay were measured on the wild type (WT), koLhcb4, koLhcb4.1 4.2, koLhcb4.1 4.3, and koLhcb4.2 4.3. Whole plants were treated as described for Figure 5A.
Table 2.
Chlorophyll Content of the Wild Type and Lhcb4 Mutants upon 3 and 8 d of Stress (900 μmol Photons m−2 s−1, 4°C)
| Days of Stress | Wild Type | koLchb4 | koLhcb4.2 4.3 | koLhcb4.1 4.3 | koLhcb4.1 4.2 |
| 0 | 21.7 ± 2.86 | 19.5 ± 0.16 | 22.5 ± 1.2 | 21.5 ± 2.1 | 22.2 ± 2.6 |
| 3 | 19.1 ± 0.7 | 16.5 ± 1.6* | 18.9 ± 2.6* | 17.7 ± 0.9* | 17.2 ± 1.4* |
| 8 | 20.3 ± 2.2 | 13.7 ± 1.7* | 19.7 ± 2.0 | 18.1 ± 2.3 | 15.8 ± 1.8* |
Data are expressed as mean ± sd (n = 5); for each genotype, significantly different values (Student’s t test, P = 0.05) with respect to t0 are marked with an asterisk.
Role of the Zea–Lhcb4 Interaction in Long-Term Membrane Lipid Photoprotection
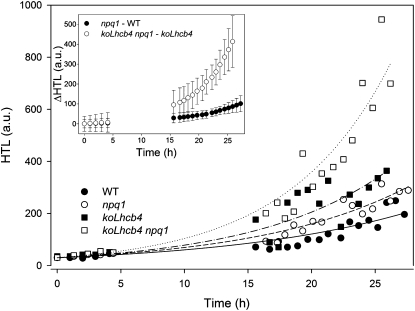
Earlier work has shown that Lhcb4 exchanges Viola with Zea both in vitro (Bassi et al., 1997; Morosinotto et al., 2002) and in vivo upon HL treatment (Bassi et al., 1993), which leads to an increased activity of several photoprotection mechanisms, including ROS scavenging, improved Chl triplet quenching (Mozzo et al., 2008), and formation of carotenoid radical cations (Holt et al., 2005; Avenson et al., 2008). In order to investigate the role of Zea on Lhcb4-dependent photoprotection, koLhcb4 was crossed with npq1 and selected a genotype lacking both the capacity of producing Zea and Lhcb4. The level of stress caused by HL + cold treatment in the wild type, npq1, koLhcb4, and koLhcb4 npq1 was measured from the extent of lipid peroxidation detected by thermoluminescence (TL) (Ducruet and Vavilin, 1999). Figure 7 shows plots of TL amplitudes at different time points during exposure of leaf discs to HL stress (800 μmol photons m−2 s−1, 4°C). In koLhcb4 genotypes, HL treatment produced higher levels of lipid peroxidation with respect to the wild type, while npq1 behavior was intermediate. Experimental points were fitted using a first-order exponential function (Y = A ebx), and the resulting equations were used to obtain the differential effect of Zea in the presence or absence of Lhcb4 (Figure 7, inset). Clearly, a much stronger differential effect of the npq1 mutation was observed in koLhcb4 versus the wild type background, implying that photoprotection mediated by Lhcb4 is enhanced in the presence of Zea.
Figure 7.
Kinetics of Lipid Peroxidation of Arabidopsis Detached Leaves Exposed to HL Stress.
Wild-type, npq1, koLhcb4, and koLhcb4 npq1 mutant leaves floating on water were exposed to HL (800 μmol photons m−2 s−1, 4°C), and photooxidation was estimated from the extent of lipid peroxidation measured by high-temperature TL. Each experimental point corresponds to a different sample. The kinetic of oxidized lipid accumulation was described by fitting the data set with first-order exponential functions: differential kinetics npq1–wild type (WT) and koLhcb4 npq1–koLhcb4 were calculated on the basis of the first-order exponential functions obtained by fitting experimental points (error bars, 95% confidence level). a.u., arbitrary units.
Structural Analysis of Isolated Grana Membranes
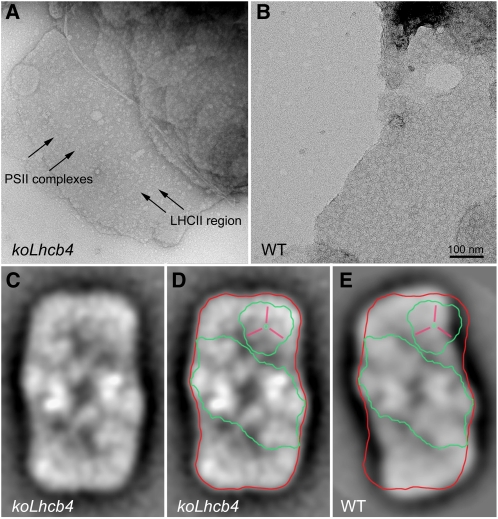
The above results suggest that the koLhcb4 mutant was more strongly affected than genotypes lacking other components of the PSII antenna system. Previous work with koLhcb6 (koCP24) (Kovács et al., 2006; de Bianchi et al., 2008) and koLhcb3 (Damkjaer et al., 2009) has shown that mutations in members of the Lhcb subfamily can affect the macro-organization of PSII supercomplex and PSII-associated regulatory functions. To verify whether a structural effect was induced by lack of Lhcb4, we analyzed both the organization of PSII supercomplexes in grana membranes and isolated PSII supercomplexes by transmission electron microscopy and single particle analysis. Grana membranes from wild-type and koLhcb4 plants exhibit randomly distributed tetrameric stain-excluding particles, corresponding to the PSII-OEC complexes exposed on the luminal surface of dimeric PSII core complexes (Figures 8A and 8B) (Boekema et al., 2000; Betterle et al., 2009). In case of the wild type, a density of PSII complexes in the grana membrane was ~1.5 PSII per 1000 nm2 (Figure 8B). Samples from koLhcb4 were clearly different (Figure 8A), being characterized by a wider spacing of the tetrameric particles (0.9 PSII particles per 1000 nm2). To further investigate how the lack of Lhcb4 affects the structure of PSII supercomplex, purified PSII supercomplexes were analyzed by single-particle analysis. To this aim, freshly isolated thylakoids were mildly solubilized with 0.3% α-DM followed by rapid fractionation by gel filtration (Damkjaer et al., 2009). The first eluted fractions, containing membrane fragments and PSII supercomplexes, were analyzed by electron microscopy. Single projections of PSII supercomplexes images, identified on the basis of their shape as determined by earlier work (Caffarri et al., 2009), were collected and subjected to image analysis, including translational and rotational alignments, multivariate statistical analysis, and averaging of homogeneous classes. Figure 8C shows the average projection map from a set of 1024 projections of C2S2 supercomplexes from plants lacking Lhcb4. Clearly, the projection map is rather similar to the projection map of the wild-type C2S2 supercomplex (Figure 8E). In both genotypes, typical features of the core complex were resolved, together with the S-trimer within the peripheral antenna. However, a detailed comparison between the complexes from koLhcb4 (Figures 8C and 8D) and from the wild type (Figure 8E) (adopted from Boekema et al., 2000) showed a different mode of binding of the S-trimer to the core complex and, thus, a different overall shape of the C2S2 particles. In addition, electron density, at the position where Lhcb4 binds in wild-type C2S2 particles, is less evident but still present in koLhcb4; however, the space between the core complex and LHCII-S seems to be too small to accommodate an Lhcb subunit, suggesting it could represent a lipid-filled area. The simplest way to interpret this result is that PSII supercomplex associates in the absence of Lhcb4 leading to a reorganization of the overall shape of the particle with partial occlusion of the Lhcb4 area and bending of the LHCII-S mass toward the CP47 complex of the core. To verify this hypothesis, we determined the polypeptide composition and relative amounts in the PSII particles from the wild type versus koLhcb4. Grana membranes from wild-type and mutant plants were solubilized with low α-DM concentration (0.3%) and fractionated by nondenaturing Deriphat-PAGE (see Supplemental Figure 8A online), yielding a pattern superimposable to that previously reported (Caffarri et al., 2009). Fractions containing the C2S2M (Band 10) and C2S2M2 (Band 11) supercomplexes were still detectable in koLhcb4, albeit their amounts were reduced compared with the wild type (14 and 9% of C2S2M and C2S2M2). We chose to analyze C2S2M supercomplexes (Band 10) since this fraction is homogeneous while fractions with higher mobility contain more comigrating complexes (Caffarri et al., 2009) (see Supplemental Figure 9 online). Band 10, and other bands used as a reference, were eluted from the gel and their protein composition was determined by SDS-PAGE, quantitative protein gel blotting analysis, and densitometry (see Supplemental Figures 8B to 8D online). In the C2S2M supercomplex (Band 10) from the wild type, polypeptides of PSII core were found along with Lhcb4, Lhcb5, and LHCII. In the same fraction from koLhcb4, the amounts of Lhcb5 and LHCII with respect to PSII core subunits were the same despite the lack of Lhcb4, implying that this C2S2M supercomplex, although exhibiting a similar overall organization and mobility in green gels, lacked one of its inner subunits. Lhcb4 was not substituted by any other protein component that we could confirm by Coomassie blue staining or immunoblotting (see Supplemental Figure 8 online for details).
Figure 8.
Electron Microscopy of Grana Membranes and PSII Particles.
(A) and (B) Electron microscopy of negative staining grana partition membranes were obtained by partial solubilization of thylakoids from koLhcb4 (A) or wild-type (WT) plants (B) with α-DM. High-resolution micrographs show the distribution of stain-excluding tetrameric particles (arrows). Grana partitions from koLhcb4 (A) were characterized by the presence of tetrameric particles more widely spaced than wild-type membranes (B). Arrows indicate PSII core complexes (magenta) and LHCII region (violet).
(C) to (E) Average projection map of a set of 1024 projections of C2S2 supercomplexes from plants lacking Lhcb4 ([C] and [D]) and the wild type (E). Contours representing PSII dimeric core (green), Lhcb4 (yellow), and whole wild-type C2S2 supercomplex (red) are superimposed.
[See online article for color version of this figure.]
DISCUSSION
The Role of Lhcb4/Lhcb6 in the Topology of Grana Membranes and in the Assembly of the PSII-LHCII Supercomplex
Lhcb4 is one of the six homologous Lhc proteins composing the PSII antenna system. These pigment-protein complexes are expected to share a common three-dimensional organization on the basis of the structural data available (Ben Shem et al., 2003; Liu et al., 2004). Previous work with antisense lines has shown a high degree of redundancy among Lhcb subunits; in fact, the PSII supercomplex organization was maintained in the absence of Lhcb1+2 components by overaccumulating Lhcb5 (Ruban et al., 2003). Similarly, plants lacking Lhcb5 and/or Lhcb6 overaccumulated Lhcb1 or Lhcb4 gene products, leading to maintenance of the C2S2 central architecture of the PSII supercomplex (de Bianchi et al., 2008). In the case of koLhcb4, structural redundancy is broken as shown by the destabilization of all supercomplex bands in green gels (Figure 2C; see Supplemental Figure 3 online) and by the differences in the structure of C2S2 particles (Figure 8). These results are consistent with Lhcb4 being the only Lhc subunit that can occupy the position between CP47 and LHCII-M building blocks within the PSII supercomplex (Figures 8C to 8E). This conclusion is supported by the lack of compensation by other gene products in the C2S2M of the mutant (see Supplemental Figures 8B to 8D online).
Isolation of the C2S2 supercomplex from koLhcb4 grana membranes shows that antenna proteins can be associated with the core complex in the absence of Lhcb4 as a docking subunit, which is consistent with the recent isolation of a stable monomeric core with CP26 and the LHCII-S trimer complex (Caffarri et al., 2009). Supercomplexes with a similar shape around the Lhcb4 position were previously found after salt treatment (Boekema et al., 2000). We conclude that in the absence of Lhcb4, PSII does assemble in grana membranes. However, because of the missing subunit, the complex is less stable and assumes a different overall structure with a low-density area located in the position where Lhcb4 is present in the wild type, which is likely occupied by lipids. As for the organization of the outer shell of the PSII antenna system, which is formed by LHCII-M, LHCII-L and Lhcb6, it appears to be strongly affected in koLhcb4. Titration of the different Lhcb proteins with respect to PSII RC showed that the Lhcb6 complex was completely missing in koLhcb4 (Figure 2B). Since the Lhcb6 messenger level was unchanged with respect to the wild type, this implies that removal of Lhcb4 decreases Lhcb6 stability. This is consistent with Lhcb4 being the docking site for Lhcb6 (Andersson et al., 2001; Caffarri et al., 2009), both participating in a pentameric complex called B4C (band 4 complex), which connects inner and outer antenna moieties (Bassi and Dainese, 1992; Betterle et al., 2009). One component of B4C, interacting with Lhcb4 and/or Lhcb6, is Lhcb3 (Betterle et al., 2009), which is still present in koLhcb4 membranes; this result is consistent with the observation that, besides Lhcb4, Lhcb3 can interact with Lhcb1 and Lhcb2 in complexes containing LHCII-M (see Supplemental Figure 8B online, lanes B9 to B11), while it is absent in complexes containing only LHCII-S, such as CS complexes (lanes B6). Less stable C2S2M2 supercomplexes can still form in the koLhcb4 mutant, which lacks both Lhcb4 and Lhcb6, although their molecular interactions are rather weak and the abundance of this complex is only 5% with respect to the wild type. Previous work with koLhcb6 has shown that a large part of the outer antenna formed by LHCII-M and LHCII-L is not directly bound to PSII supercomplexes; rather, trimers L and M form LHCII-only domains that segregate from arrays of C2S2 particles (de Bianchi et al., 2008). No such arrays were observed in koLhcb4 membranes (Figure 8A) nor was PQ diffusion restricted (Figure 3), suggesting that the altered shape of C2S2 particles and/or their instability prevents cooperative interactions with arrays. Besides this, the distance between neighbor PSII centers is higher than in the wild type (Figures 8A and 8B), implying a higher number of LHCII trimers is interposed between PSII centers.
Consequences for Photosynthesis: Light Harvesting and ET
The loss of Lhcb4 in Arabidopsis did not strongly affect growth rate and pigment compositions under control light conditions (Figure 3, Table 1). Indeed, linear and cyclic ET rates were similar to those in wild-type plants, as well as the functional antenna size. However, alterations in the PSII macrostructure in mutant plants did result in differences in chlorophyll fluorescence parameters (Table 1). Increased F0 (Table 1) showed that the efficiency of excitation energy transfer from the antenna to the PSII RC is decreased in mutant plants. In grana membranes from the wild type (Figure 8B), the distribution of PSII particles is homogeneous through the whole surface. This is not the case for koLhcb4, whose grana membranes contained discrete patches of LHCII trimers interspersed by fewer, randomly distributed PSII cores (Figure 8A); therefore, in some discrete areas of koLhcb4 grana membranes, the LHCII/PSII core ratio is increased. In these domains, LHCII fluorescence is probably not efficiently photochemically quenched, thus yielding higher F0 (Table 1). This is consistent with the results obtained in KO mutants for Lhcb5 and Lhcb6 (Kovács et al., 2006; de Bianchi et al., 2008), while no such effect was observed in asLHCII (Andersson et al., 2003). Increased F0 suggests that vacancy of Lhcb4 forces the excitation energy to follow restricted pathways from the peripheral antenna to the core; indeed, the migration time of excitations from antenna to the PSII RC was significantly reduced in koLhcb4 with respect to the wild type (van Oort et al., 2010). Thus, despite the fact that fluorescence induction in the presence of DCMU showed no changes in functional antenna size, the number of LHCII per PSII RC in grana membranes was higher (Figures 8A and 8B). We conclude that in the absence of a well-organized PSII-LHCII supercomplex, the efficiency of excitation energy trapping in koLhcb4 was lower than in the wild type and that the compensatory increase in peripheral LHCII complexes (Figures 2A and 2B) contributed with lower efficiency to light harvesting.
Although functional measurements indicate that there was no major perturbation of PSII and PSI functions, a slight increase in the PSI/PSII ratio was observed in koLhcb4 (Figure 2). The consequences of a lower PSII level were not strong: the parameter Sm/tFmax, expressing the average fraction of open RCs during the time needed to complete closure of centers, was similar in koLhcb4 and wild-type plants, as well as the LEF and QA redox state at several light intensities (Figures 3A and 3B), thus making ET restriction unlikely.
The major effect of the mutation on ET activity was a faster saturation of the P700 oxidation ratio in koLhcb4 with respect to the wild type (Figure 3F). Since both LEF and CEF are not affected in the mutant, we propose that this effect could be related to the higher PSI functional antenna size of the mutant in HL. Indeed, when measurements were performed by changing the quality of actinic light (PSI rather than PSI+PSII light), differences in P700 oxidation rate between wild-type and mutant leaves disappeared (Figure 3G), implying the increase of PSI functional antenna size in HL is due to PSII components. This is consistent with an increased level of spillover in koLhcb4 at higher light intensity. This suggestion was verified by 77K fluorescence spectroscopy (see Supplemental Figure 5 online), showing that HL treatment induces an increased contribution of LHCII to PSI excitation in koLhcb4 but not in the wild type. Since the amplitude of the state I–state II transition is the same in the wild type and in the mutant and, moreover, the stn7 kinase is inhibited in HL (Tikkanen et al., 2006), this spectral change cannot be attributed to state transitions. Instead, we interpret these results as evidence of spillover of excitation from LHCII to PSI. We suggest that this effect ensues from the lower stability of PSII-LHCII supercomplexes in the mutant, with the disappearance of the B4 complex (Figure 2C) and the faster kinetic of state transitions (Figures 3D and 3E). In koLhcb4, an overall weakening of interactions between the PSII core and LHCII would make weakly bound LHCII at the grana margins able to contribute to PSI antenna size particularly in HL, when grana membrane reorganization occurs with segregation of LHCII-rich domains (Kirchhoff, 2008; Betterle et al., 2009; Johnson et al., 2011).
Consequences for Regulation of Light Harvesting: State Transitions
State transition is the mechanism by which the complement of LHCII is balanced between PSII and PSI, depending on the reduction state of the intermediate electron carrier PQ, through its reversible phosphorylation, which induces its disconnection from PSII and binding to PSI (Allen and Nilsson, 1997; Jensen et al., 2000). Lhcb4 can be phosphorylated in monocots (Bergantino et al., 1995; Bergantino et al., 1998) and in Arabidopsis (Hansson and Vener, 2003). In Chlamydomonas reinhardtii, P-Lhcb4 was found to be connected to PSI-LHCI supercomplexes in state II conditions (Takahashi et al., 2006), and Lhcb4-depleted cells showed changes in state transitions (Tokutsu et al., 2009). Thus, changes in state transitions can be expected in Lhcb4-less plants. Results displayed in Figures 3D and 3E show that koLhcb4 was not affected in its state transition total activity, in agreement with the similar reduction state of the PQ pool at all light intensities (Figure 3B) (Bellafiore et al., 2005). We observed that in koLhcb4 the fluorescence changes, upon switching off the far-red light, are faster than in the wild type, implying that the transiently reduced state of the free PQ pool is more promptly relaxed by migration of the LHCII to PSI. The same state transition phenotype has been described for koLhcb6 and koLhcb5 Lhcb6 plants (de Bianchi et al., 2008), and this is a clear indication that the connection between the PSII core and the bulk trimeric LHCII is weaker in these mutants with respect to the wild type. In the case of the koLhcb6 plants, the faster state transition was attributed to the displacement of the M-trimer from the PSII macrostructure, thus enhancing its migration to PSI. We suggest that, in the koLhcb4 mutant, the weaker binding of S trimer (and, possibly, of the M trimer) and the higher amount of weakly connected LHCII trimers available increase the probability of migrating toward PSI upon phosphorylation. In C. reinhardtii, state transitions do not only fulfill the role of balancing light absorption between photosystems, they also increase PSI electron flow at the expense of PSII, acting as a switch between LEF and CEF (Vallon et al., 1991). This was not the case in Arabidopsis since, despite an effect on state transitions (Figures 3D and 3E), no changes in linear (Figure 3A) versus CEF rates (see Supplemental Figures 4A and 4B online) were observed.
Nonphotochemical Fluorescence Quenching
Nonphotochemical dissipation of excess energy is affected in koLhcb4 plants with respect to the wild type: mutant plants showed a delayed rise and a lower NPQ amplitude after 8 min of light (Figure 4A). Upon prolonged illumination and/or upon repeated light treatments (Figure 4B), the amplitude of NPQ was similar to that of the wild type. A similar effect was previously observed in the koLhcb5 Lhcb6 double mutant (de Bianchi et al., 2008). Since Lhcb6 is destabilized in koLhcb4 (Figure 2), this mutant phenocopies a double koLhcb4 Lhcb6 mutant. NPQ kinetic can be modified by changes in the trans-thylakoid ΔpH gradient (de Bianchi et al., 2008), by changes in the level of the pH sensor PsbS (Li et al., 2002, 2004), or by changes in the number/relative abundance of protein subunits hosting quenching sites (Bonente et al., 2008). Changes in luminal pH appear unlikely; in fact, the kinetics of Zea synthesis, catalyzed by the pH-dependent enzyme VDE (Yamamoto and Higashi, 1978), were the same in wild-type and mutant leaves (Figure 4C). Moreover, the level of PsbS was unchanged (Figure 2B), leaving modification in the abundance and identity of quenching sites localized in Lhcb proteins as the most likely cause for the observed phenotype. We interpret our results in the framework of the recently proposed model (Miloslavina et al., 2008; de Bianchi et al., 2010), based on the formation of quenching sites within each of two distinct domains in grana discs: (1) C2S2 particles (containing PSII RC, Lhcb4, Lhcb5, and LHCII-S) and (2) the peripheral antenna, including Lhcb6, LHCII-M, and LHCII-L (Miloslavina et al., 2008; Betterle et al., 2009), which segregate because of the action of PsbS. Zea-dependent quenching activity has been detected within monomeric Lhcb4, b5, and b6 in detergent solution (Avenson et al., 2008), while, in LHCII, it was activated by aggregation and was independent from Zea (Ruban et al., 2007). The similar NPQ kinetics of koLhcb4 and of koLhcb5 Lhcb6 can thus be explained because they both retain a quenching site within the C2S2 domain; the faster quenching kinetic upon repeated illumination is due to the enhancement by Zea of the quenching activity (Niyogi et al., 1998) associated with Lhcb4 (Figure 4) or Lhcb5. This is consistent with the capacity of exchanging Viola with Zea in binding site L2 observed in monomeric Lhcbs (Morosinotto et al., 2002; Wehner et al., 2006) rather than in trimeric LHCII. In koLhcb5 Lhcb6, the recovery of NPQ upon the initial delay is faster and more complete than in koLhcb4, implying that Lhcb4 is more active as a quencher than Lhcb6 and Lhcb5 (de Bianchi et al., 2008). If there are two distinct quenching domains within PSII antenna (Miloslavina et al., 2008) and monomeric Lhcb4-6 proteins are the only sites of quenching in vivo, the peripheral antenna domain should remain unquenched in both koLhcb5 Lhcb6 (de Bianchi et al., 2008) and koLhcb4 because they both lack Lhcb6. Since both genotypes reach NPQ levels similar to the wild type, although with a delayed kinetic, it is likely that some kind of quenching does occur in the peripheral antenna domain disconnected from C2S2 particles in these genotypes. Previous work with koLhcb6 has shown that this component has a quenching effect on the major LHCII antenna (van Oort et al., 2010); here, we observe that, even in the absence of Lhcb6, the overall quenching activity is high, consistent with quenchers being activated in both PSII antenna domains, and conclude that LHCII is likely to contribute to NPQ in vivo, in spite of the fact that it does not exhibit the spectroscopic and structural features of quenching sites (Ahn et al., 2008; Avenson et al., 2008). Thus, it likely activates quenching by a different mechanism (Ruban et al., 2007). Consistent with this view is the finding that a red-shifted fluorescence lifetime component was observed in vivo under NPQ conditions (Müller et al., 2010), which has properties similar to LHCII aggregated in vitro.
While differences in qE were minimized by prolonged/repeated illumination, we observed that the qI component, which is responsible for the slowly relaxing component of quenching, is equally decreased upon short/long or repeated light treatments (Figure 4B). Previous work suggested that qI was specifically due to Zea binding to Lhcb5 (Dall’Osto et al., 2005); instead, removal of Lhcb6 has a negligible effect on qI amplitude, since the double mutant koLhcb5 Lhcb6 did not further decrease its qI level (de Bianchi et al., 2008). Here, we show that removal of Lhcb4 reduces the extent of qI, implying that qI is modulated by both Lhcb4 and Lhcb5.
Consequences for Photoprotection: Resistance to Photooxidative Stress Is Decreased in koLhcb4 Plants with Respect to the Wild Type
Lhcb4-less plants showed a reduced photoprotection capacity when exposed to high irradiance at low temperature. The highest sensitivity to photooxidative stress of koLhcb4, among all other Lhcb KO mutants, is consistent with the higher reduction in fitness of plants lacking Lhcb4 (Andersson et al., 2001; Ganeteg et al., 2004), supporting the importance of this gene product for PSII performance and chloroplast photoprotection. The structural integrity of photosynthetic supramolecular complexes is essential for the resistance to photooxidative stress (Horton and Ruban, 2005), although the reasons are not completely clear. Depletion of light-harvesting antennae has been reported to favor photoinhibition of both PSI (Alboresi et al., 2009) and PSII (Kim et al., 2009; Dall’Osto et al., 2010). KoLhcb4 mutant plants are more sensitive to photooxidative stress than the wild type (Figure 5), and this effect is associated with increased production of 1O2. This effect cannot be ascribed to a pleiotropic effect on photosynthetic ET efficiency, since lack of Lhcb4 does not significantly affect either the rate or the regulation of photosynthetic ET (Figures 3A to 3C). A putative mechanism for photoinhibition is the reduction of 1Chl* dissipation (qE and qI) (Johnson et al., 2007); however, in koLhcb4, qE activity is affected at the onset of illumination, while it is similar to the wild type on a longer timescale. Since our photoinhibitory light treatment was performed at constant light intensity, we conclude that photosensitivity of koLhcb4 plants is not due to differences in qE or qI.
The connection between the PSII core complex and outer LHCII was partially impaired. Indeed, a steady increase in F0 was measured in koLhcb4 plants with respect to the wild type (Table 1); a higher 1Chl* level might lead to 3Chl* formation through intersystem crossing. Although we cannot exclude that the inefficient connection of LHCII to PSII RC could contribute to the higher photosensitivity of koLhcb4, we notice that the koLhcb5 Lhcb6 double mutant was as resistant as wild-type plants (Figure 5A) despite a Fv/Fm reduction comparable to that of Lhcb4-less plants (de Bianchi et al., 2008). Since photoprotection in koLhcb5 Lhcb6 plants was similar to that in the wild type (Figure 5A), neither the increase in F0 per se nor the absence of Lhcb6 can be the cause of higher photosensitivity of koLhcb4 plants. Instead, this effect appears to be specific for the absence of Lhcb4, implying that this gene product is of particular importance in providing PSII photoprotection (Figure 5A). Indeed, Lhcb4 appears to be the most conserved among Lhc proteins associated with PSII (Koziol et al., 2007), and it maintains its stoichiometry with the RC even under stressful growth conditions that lead to a depletion of Lhcb1, 2, and 6 (Ballottari et al., 2007). The most obvious peculiarity of Lhcb4 is that it cannot be replaced in PSII supramolecular architecture by other Lhcbs and is thus required for allowing structural integrity of PSII (Boekema et al., 1999) (Figures 8C to 8E). Thus, the higher photosensitivity in koLhcb4 could be the effect of a less stable PSII supercomplex; indeed, the extreme photosensitivity of Arabidopsis mutants such as ch1 (Dall’Osto et al., 2010) can be ascribed to the absence of light-harvesting complexes surrounding the PSII RC, leaving the PSII core complex exposed to the lipid phase, where radical chain reactions of peroxy-lipids occur during photooxidative stress.
Measurement of lipid peroxidative damage by thermoluminescence in intact leaves of wild type versus koLhcb4 and koLhcb4 npq1 plants showed that Zea and Lhcb4 are synergistically active in protection from photodamage (Figure 7). Therefore, it appears that a specific effect for Zea in providing efficient photoprotection is preferentially amplified when bound to Lhcb4. This observation is consistent with a previous report of a protective effect of Zea (Dall’Osto et al., 2010) in addition to qE enhancement (Niyogi et al., 1998) and to ROS scavenging in the lipid phase (Havaux and Niyogi, 1999; Baroli et al., 2003, 2004; Havaux et al., 2007). Upon light-induced synthesis, Zea enters the protein-bound xanthophyll pool by binding to the L2 site of minor Lhc (Avenson et al., 2008; Jahns et al., 2009) and to the V1 site of LHCII (Caffarri et al., 2001), in addition to the accumulating in the lipid phase. The molecular mechanism of enhanced resistance to photooxidation by Zea is not yet clear. The hypotheses include (1) draining excitation energy from the PS core by xanthophylls bound at the interface with Lhcbs; (2) quenching of 3Chl* energy from RC, possibly by the Dexter exchange mechanism (Nayak et al., 2002) by Zea located in site L2 on specific Lhcs; and (3) preferential scavenging of 1O2 by Zea into site V1 of LHCII (Johnson et al., 2007). Although all of these mechanisms are plausible for explaining the higher photosensitivity of koLhcb4, their relative contribution needs further investigation.
Effect of the Accumulation of the Individual Isoforms on koLhcb4 Phenotype
The attenuation of the F0 and NPQ phenotypes produced by the triple koLhcb4 mutations is different in the three double koLhcb4 mutants. Retention of Lhcb4.1 restores Fv/Fm and NPQ to wild-type levels, while Lhcb4.2 is only partially effective in this function, Lhcb4.3 is completely inefficient (Figures 4D and Figure 6, Table 1). Indeed, plants retaining only Lhcb4.3 show the same reduction in Chl a/b ratio (Table 1), increased photosensitivity (Figure 6), and delayed NPQ kinetic as the triple koLhcb4. Although we cannot exclude that Lhcb4.1, 4.2, and 4.3 gene products are intrinsically different, the penetration of the phenotype appears to be essentially due to the level of the gene product. Indeed, levels of Lhcb4.1, Lhcb4.2, and Lhcb4.3 in the three double mutant are 1, 0.6, and 0, respectively, per PSII RC (see Supplemental Table 2 online). In fact, although Lhcb4.3 transcription is lightly increased under stress conditions (Alboresi et al., 2011), we were unable to detect the corresponding protein even by keeping plants under a variety of stress conditions, implying it is either not translated or rapidly turned over.
The Lhcb4.3 gene appeared recently in plant genome. Its sequence is not present in algal genomes, and no orthologs were found in the moss Physcomitrella patens; therefore, it appeared only at later stages in the evolution of the green lineage (Alboresi et al., 2008). It seems unlikely that a new gene conserved in higher plants has no function or is not translated at all. EST data from Arabidopsis show that Lhcb4.3 is expressed at low levels (Jansson, 1999), and its transcription seems to be confined to dicots (Goff et al., 2002; Yu et al., 2002); furthermore, clustering analysis of Lhc superfamily expression data (Klimmek et al., 2006) confirmed that Lhcb4.3 is regulated in an opposite way with respect to Lhcb4.1 and Lhcb4.2. The putative Lhcb4.3 protein is shorter than other Lhcb4 isoforms, and its sequence is considerably different from both Lhcb4.1 and Lhcb4.2 isoforms, suggesting it might have a different function in the stress conditions in which this gene is actively transcribed. However, we cannot exclude that the Lhcb4.3 isoform might accumulate in special and still uninvestigated environmental/developmental conditions; so far, the role of this isoform remains elusive.
We have shown that Lhcb4 has a specific function in protecting PSII from photoinhibition, which accounts for the increased sensitivity of koLhcb4 to HL stress. This gene product, different from any other PSII antenna component, cannot be replaced by homologous subunits in the PSII-LHCII supramolecular complex. This leads to the formation of incomplete PSII particles with a gap in the position normally occupied by Lhcb4, a modified shape and a lower stability. This is consistent with evidence that a core group of antenna proteins developed prior to green algal diversification (Koziol et al., 2007) and included Lhcb4 protein associated with PSII RC. The weaker interaction between PSII-LHCII components leads to a faster migration of LHCII complexes to the stroma membrane upon phosphorylation, similar to the case of koLhcb6 and koLhcb3. Although PSII activity is not impaired in moderate light conditions, there is a lower level of PSII RC in grana membranes, probably because the PSII core complex is less strongly retained within grana discs, because one of the two monomeric Lhcbs that bridges it to LHCII is missing. Under stress conditions, koLhcb4 is photoinhibited, and this effect is selectively enhanced in the absence of Zea, which is active in preventing the synthesis of ROS species and promoting their scavenging. By binding in between LHCII trimers and the CP47 subunit of PSII, Lhcb4 is crucial for the protection of PSII RC from ROS produced during photosynthesis either by neighbor damaged PSII complexes (Krieger-Liszkay et al., 2008) or by overexcited antennae (Santabarbara et al., 2002; Mozzo et al., 2008). Thus, the absence of a specific antenna subunit, although it does not restrict light harvesting and photosynthetic ET rate, impairs PSII photoprotection capacity through its effect on the assembly of supercomplexes. This is an example of optimization of the building blocks of photosynthetic complexes and the tuning of their interactions with each other toward overcoming the inhibitory effect of increasing oxygen concentration over evolution time. It is worth noting that functional alterations described in koLhcb4 plants likely result from the reduced ability of PSII macroassemblies to undergo proper organization in low versus high light, thus confirming the crucial role of Lhcb4 in modulating structural and functional flexibility of grana membranes.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana T-DNA insertion mutants (Columbia ecotype) GK Line ID282A07 (insertion into the Lhcb4.1 gene), SAIL_910_D12 (insertion into the Lhcb4.2 gene), and SALK_032779 (insertion into the Lhcb4.3 gene) were obtained from NASC collections (Alonso et al., 2003). Homozygous plants (Betterle et al., 2009) were identified by PCR analysis using the following primers: forward 5′-TCACCAGATAACGCAGAGTTTAATAG-3′ and reverse 5′-CACATGATAATGATTTTAAGATGAGGAG-3′ for the Lhcb4.1 sequence, 5′-CATCATACTCATTGCTGATCCATG-3′ for the insertion; forward 5′-GCGTTTGTGTTTAGCGTTTCGACATCTGTCTG-3′ and reverse 5′-GGTACCCGGGTGGTTTCCGACATTAGC-3′ for the Lhcb4.2 sequence, 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′ for the insertion; forward 5′-GTGAGCTGATCCATGGAAGGTGG-3′ and reverse 5′-GGCCGGTTTTGAACGATTGATGTGAC-3′ for the Lhcb4.3 sequence, and 5′-GCGTGGACCGCTTGCTGCAACT-3′ for the insertion. Genotypes koLhcb4, koLhcb4.1 4.2, koLhcb4.2, koLhcb4.3, and koLhcb4.1 4.3 were obtained by crossing single mutant plants and selecting progeny by PCR analysis. For RT-PCR, total RNA was isolated from 4-week-old plants with the Trizol protocol. Reverse transcription was performed using M-MLV reverse transcriptase with oligo(dT) primer and 1.5 μg of total RNA. For normalization purposes, actin2 was chosen as an endogenous control. The primers used were as follows: 5′-GTGGCTCCCGGTATCCATCC-3′ and 5′-TTGAACCGCGAATCCCAAGAAGG-3′ for Lhcb4.1 cDNA; 5′-GGTTTCCGACATTAGCTCCAATTC-3′ and 5′-CTGAACCGCAAAACCCAAGAATC-3′ for Lhcb4.2 cDNA; 5′-CCGGTTCGGGTTCAGTTTCGG-3′ and 5′-GGCAAGGAAGCTGACAGGGC-3′ for Lhcb4.3 cDNA; and 5′-CCTCATGCCATCCTCCGTCTTG-3′ and 5′-GAGCACAATGTTACCGTACAGATCC-3′ for actin2 cDNA.
The Arabidopsis T-DNA insertion mutant koLhcb3 (SALK_020342) from NASC was obtained by selecting progeny by immunoblotting using a specific antibody against the Lhcb3 subunit; asLHCII (Andersson et al., 2003) was obtained from the NASC. The double mutant koLhcb4 npq1 was obtained by crossing single mutants and selecting progeny by immunoblotting (with the α-Lhcb4 antibody) and HPLC upon HL treatment of leaves. Insertion mutants koLhcb6 and koLhcb5 Lhcb6 were isolated as previously described (de Bianchi et al., 2008). Mutants were grown for 5 weeks at 100 μmol photons m−2 s−1, 23°C, 70% humidity, and 8 h of daylight.
Stress Conditions
Short-term HL treatment was performed at 1200 μmol photons m−2 s−1, 45 min, and room temperature (24°C), to measure the kinetics of Zea accumulation, on detached leaves floating on water. Samples were rapidly frozen in liquid nitrogen prior to pigment extraction. Longer photooxidative stress was induced by exposing whole plants to either 550 μmol photons m−2 s−1 at 4°C for 2 d, 900 μmol photons m−2 s−1 at 4°C for 10 d, or 1600 μmol photons m−2 s−1 at 24°C for 10 d. Light was provided by halogen lamps (Focus 3; Prisma) and filtered through a 2-cm recirculation water layer to remove infrared radiation.
Pigment Analysis
Pigments were extracted from leaf discs, either dark adapted or HL treated, with 85% acetone buffered with Na2CO3, and then separated and quantified by HPLC (Gilmore and Yamamoto, 1991).
Membrane Isolation
Functional thylakoids were isolated from dark-adapted or HL-treated leaves as previously described (Casazza et al., 2001).
Gel Electrophoresis and Immunoblotting
SDS-PAGE analysis was performed with the Tris-Tricine buffer system (Schägger and von Jagow, 1987), with the addition of 7 M urea to the running gel to separate Lhcb4 isoforms. Nondenaturing Deriphat-PAGE was performed following the method developed by Peter et al. (1991) with modification described by Havaux et al. (2004). Thylakoids concentrated at 1 mg/mL chlorophylls were solubilized in a final concentration of 1% α/β-DM, and 25 mg of chlorophyll were loaded in each lane. For immunotitration, thylakoid samples corresponding to 0.25, 0.5, 0.75, and 1 μg of chlorophyll were loaded for each sample and electroblotted on nitrocellulose membranes, and proteins were detected with alkaline phosphatase-conjugated antibody according to Towbin et al. (1979). Signal amplitude was quantified (n = 4) using GelPro 3.2 software (Bio-Rad). To avoid any deviation between different immunoblots, samples were compared only when loaded in the same slab gel.
In Vivo Fluorescence and NPQ Measurements
PSII function during photosynthesis was measured through chlorophyll fluorescence on whole leaves at room temperature with a PAM 101 fluorimeter (Heinz-Walz) (Andersson et al., 2001), a saturating light pulse of 4500 μmol photons m−2 s−1, 0.6 s, and white actinic light of 100 to 1100 μmol photons m−2 s−1, supplied by a KL1500 halogen lamp (Schott). NPQ, ϕPSII, qP, qL, and LET were calculated according to the following equations (Van Kooten and Snel, 1990; Baker, 2008): NPQ = (Fm − Fm′)/Fm′, ϕPSI I= (Fm′ − Fs)/Fm′, qP = (Fm′ − Fs)/(Fm′ − F0′), qL = qP·F0′/Fs, LET = ϕPSII·PAR·Aleaf·fractionPSII, where F0/F0′ is the minimal fluorescence from dark/light-adapted leaf, Fm/Fm′ is the maximal fluorescence from dark/light-adapted leaves measured after the application of a saturating flash, Fs the stationary fluorescence during illumination, and PAR the photosynthetic active radiation; Aleaf (leaf absorptivity) was 0.67 ± 0.05 for the wild type, 0.62 ± 0.04 for koLhcb4; fractionPSII was measured by quantitative immunoblot: wild type, 0.50 ± 0.01; koLhcb4, 0.47 ± 0.06.
State transition experiments were performed using whole plants according to established protocols (Jensen et al., 2000). Preferential PSII excitation was provided by illumination with blue light (40 μmol photons m−2 s−1), excitation of PSI was achieved using far-red light from an LED light source (Heinz-Walz; 102-FR) applied for 15 min simultaneously with blue light. Fm level in state I (Fm′) and state II (Fm′′) was determined at the end of each cycle by the application of a saturating light pulse. The parameter qT (PSII cross-section changes) was calculated as (Fm1 − Fm2)/Fm1 (where Fm1/2 is the maximal fluorescence yield in state I/II).
Fluorescence induction kinetics was recorded with a home-built apparatus to measure functional antenna size on leaves. Sm/tFmax was calculated from variable fluorescence curves induced with green light (1100 μmol photons m−2 s−1) (Strasser et al., 1995). For measurements of PSII functional antenna size, variable fluorescence was induced with a green light of 15 μmol photons m−2 s−1, on thylakoids (10 μg Chls/mL) in a measuring buffer containing 10 mM HEPES, pH 7.8, 5 mM MgCl2, 30 μM DCMU, and 50 μM nigericin. The reciprocal of time corresponding to two-thirds of the fluorescence rise (T2/3) was taken as a measure of the PSII functional antenna size (Malkin et al., 1981). For measurements of the PSII repair process, whole plants were illuminated at 900 μmol photons m−2 s−1, 4°C for 4 h to induce photoinhibition of PSII, and restoration of the Fv/Fm ratios was subsequently followed at irradiances of 20 μmol photons m−2 s−1, 4°C (Aro et al., 1994).
Analysis of P700 Redox State
All in vivo spectroscopic measurements were performed using a LED spectrophotometer (JTS10; Biologic Science Instruments) in which absorption changes are sampled by weak monochromatic flashes (10-nm bandwidth) provided by LEDs. P700+ reduction following a flash was assayed as detailed by Golding et al. (2004). Leaves were infiltrated with 50 μM DCMU in 150 mM sorbitol. A 400-ms saturating flash of red light (λmax = 630 nm; 3000 μmol photons m−2 s−1) was delivered to the leaf, and changes in absorbance at 705 nm were used to measure the kinetics of P700+ reduction. Oxidized P700 (ΔAmax) was recorded during far-red light illumination (2500 μmol photons m−2 s−1, λmax = 720 nm). The level of oxidized P700 in the leaf (ΔA) was determined during illumination with either orange light (630 nm, from 50 to 980 μmol photons m−2 s−1, λmax = 720 nm) or far-red light (720 nm, from 200 to 2500 μmol photons m−2 s−1) (Zygadlo et al., 2005). Antenna size of PSI was estimated according to Kim et al. (2009).
Analysis of Cytochrome b6f Redox Kinetics
Absorption transients were measured on thylakoids (30 μg Chls/mL) in a measuring buffer containing 10 mM HEPES, pH 7.8, 5 mM MgCl2, 1 mM sodium ascorbate, 20 μM nigericin, and 50 μM methylviologen (Kirchhoff et al., 2000). Cytochrome redox changes were measured in continuous light (630 nm, 300-ms pulse, 560 μmol photons m−2 s−1) at four wavelengths: 548, 554, 563, and 573 nm; redox changes of cytochromes b and f were calculated according to Joliot and Joliot (1984).
Analysis of Steady State Proton Flux across Thylakoid Membranes
Light-induced pmf was estimated on intact leaves, from DIRK changes in absorbance associated with the ECS at 520 nm, as described previously (Livingston et al., 2010). ECS was induced with orange (630 nm) actinic intensities ranging from 15 to 960 μmol photons m−2 s−1. Relative values of steady state proton flux across the thylakoid membrane (νH+) was estimated from the initial slope of the ECS decay upon rapid light–dark transitions.
Gas Exchange Measurement
Measurements of O2 evolution in saturating CO2 were performed using an S101 O2 electrode (Qubit System). Light response curves were determined using broadband red light.
Determination of the Sensitivity to Photooxidative Stress
Photooxidative stress was induced in detached leaves by HL treatment at low temperature. Detached leaves floating on water were exposed to 1500 μmol photons m−2 s−1 for 8 h, in a growth chamber at 4°C, and then immediately frozen in liquid nitrogen. Photooxidative stress was assessed by measuring MDA formation as an indirect quantification of lipid peroxidation (Havaux et al., 2005). Lipid peroxidation was measured on whole plants by TL with a custom-made apparatus (Ducruet, 2003). The amplitude of the TL peak at 135°C was used as an index of lipid peroxidation (Havaux, 2003). Measurements of singlet oxygen production from leaves were performed with SOSG (Molecular Probes), a 1O2-specific fluorogenic probe, as described by Dall’Osto et al. (2010).
Electron Microscopy
Samples were negatively stained with 2% uranyl acetate on glow discharged carbon-coated copper grids. Electron microscopy was performed on a Philips CM120 electron microscope equipped with a LaB6 tip operating at 120 kV. Images were recorded with a Gatan 4000 SP 4 K slow-scan CCD camera at 80,000 magnification at a pixel size (after binning the images) of 0.375 nm at the specimen level with GRACE software for semiautomated specimen selection and data acquisition (Oostergetel et al., 1998). Single-particle analysis of a data set of 1350 PSII particles was performed using Groningen Image Processing software, including multireference and no-reference alignments, multivariate statistical analysis, classification, and averaging of homogeneous classes (van Heel et al., 2000).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At5g01530 (Lhcb4.1), At3g08940 (Lhcb4.2), At2g40100 (Lhcb4.3), At5g54270 (Lhcb3), At4g10340 (Lhcb5), At1g15820 (Lhcb6), At1g08550 (npq1), and At3g18780 (actin2). The KO lines mentioned in the article can be obtained from the NASC under the stock numbers N376476 (koLhcb4.1), N877954 (koLhcb4.2), N532779 (koLhcb4.3), N520342 (koLhcb3), N514869 (koLhcb5), N577953 (koLhcb6), and N6363 (asLHCII).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transmission Electron Micrographs of Plastid from Mesophyll Cells of the Wild type and Mutants.
Supplemental Figure 2. Induction Curves of Chl Fluorescence Measured on Thylakoids Treated with DCMU.
Supplemental Figure 3. Biochemical Characterization of koLhcb4 Mutants.
Supplemental Figure 4. P700 Measurements: Cyclic Electron Transport, PSI Functional Antenna Size, and Cytochrome b6f Redox Kinetics.
Supplemental Figure 5. Low-Temperature Fluorescence Spectra of Chloroplasts.
Supplemental Figure 6. PSII Repair Efficiency under Photoxidative Stress.
Supplemental Figure 7. Insights on the Lhcb4.3 Expression and Function.
Supplemental Figure 8. Insights on the PSII Supercomplex Composition.
Supplemental Figure 9. Schematic Panel of Wild-Type and koLhcb4 PSII Supercomplex Composition.
Supplemental Table 1. Quantitative Analysis of Thylakoid Morphological Traits.
Supplemental Table 2. Polypeptide Composition of Thylakoid Membranes from Wild-Type and KO Mutant Expressing Single Lhcb4 Isoform.
Acknowledgments
We thank Giovanni Finazzi (Commissariat à l’Energie Atomique, Grenoble, France) for helpful discussion. Financial support for this work was provided by the Programmi di Ricerca di Interesse Nazionale (2008XB774B) and by a grant of the Italian Ministry of Research and Marie Curie Actions Initial Training Networks HARVEST (Grant 238017).
AUTHOR CONTRIBUTIONS
S.d.B. isolated single KO mutants, carried out the crossings to obtain all the genotypes used, and performed a biochemical and physiological characterization of their photosynthetic apparatus. S.C., N.B., and L.D. were involved in the photoxidative treatments and stress measurements, in the spectroscopy investigation on thylakoids or isolated complexes, and in data analysis. R.K., E.B., and N.B. carried out electron microscopy structural analysis of isolated grana and PSII particles. R.B. and L.D. conceived the study, participated in its design and coordination, and helped to draft the manuscript.
References
- Ahn T.K., Avenson T.J., Ballottari M., Cheng Y.C., Niyogi K.K., Bassi R., Fleming G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320: 794–797 [DOI] [PubMed] [Google Scholar]
- Alboresi A., Ballottari M., Hienerwadel R., Giacometti G.M., Morosinotto T. (2009). Antenna complexes protect Photosystem I from photoinhibition. BMC Plant Biol. 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A., Caffarri S., Nogue F., Bassi R., Morosinotto T. (2008). In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A., Dall’osto L., Aprile A., Carillo P., Roncaglia E., Cattivelli L., Bassi R. (2011). Reactive oxygen species and transcript analysis upon excess light treatment in wild-type Arabidopsis thaliana vs a photosensitive mutant lacking zeaxanthin and lutein. BMC Plant Biol. 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335 [DOI] [PubMed] [Google Scholar]
- Allen J.F., Nilsson A. (1997). Redox signalling and the structural basis of regulation of photosynthesis by protein phosphorylation. Physiol. Plant. 100: 863–868 [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson J.M. (1986). Photoregulation of the composition, function and structure of thylakoid membranes. Annu. Rev. Plant Physiol. 37: 93–136 [Google Scholar]
- Andersson J., Walters R.G., Horton P., Jansson S. (2001). Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 13: 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J., Wentworth M., Walters R.G., Howard C.A., Ruban A.V., Horton P., Jansson S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - Effects on photosynthesis, grana stacking and fitness. Plant J. 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Aro E.M., McCaffery S., Anderson J.M. (1994). Recovery from photoinhibition in peas (Pisum sativum L.) acclimated to varying growth irradiances (role of D1 protein turnover). Plant Physiol. 104: 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Avenson T.J., Ahn T.K., Zigmantas D., Niyogi K.K., Li Z., Ballottari M., Bassi R., Fleming G.R. (2008). Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283: 3550–3558 [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Ballottari M., Dall’Osto L., Morosinotto T., Bassi R. (2007). Contrasting behavior of higher plant photosystem I and II antenna systems during acclimation. J. Biol. Chem. 282: 8947–8958 [DOI] [PubMed] [Google Scholar]
- Baroli I., Do A.D., Yamane T., Niyogi K.K. (2003). Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I., Gutman B.L., Ledford H.K., Shin J.W., Chin B.L., Havaux M., Niyogi K.K. (2004). Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. J. Biol. Chem. 279: 6337–6344 [DOI] [PubMed] [Google Scholar]
- Bassi R., Dainese P. (1992). A supramolecular light-harvesting complex from chloroplast photosystem-II membranes. Eur. J. Biochem. 204: 317–326 [DOI] [PubMed] [Google Scholar]
- Bassi R., Høyer-Hansen G., Barbato R., Giacometti G.M., Simpson D.J. (1987). Chlorophyll-proteins of the photosystem II antenna system. J. Biol. Chem. 262: 13333–13341 [PubMed] [Google Scholar]
- Bassi R., Pineau B., Dainese P., Marquardt J. (1993). Carotenoid-binding proteins of photosystem II. Eur. J. Biochem. 212: 297–303 [DOI] [PubMed] [Google Scholar]
- Bassi R., Sandona D., Croce R. (1997). Novel aspects of chlorophyll a/b-binding proteins. Physiol. Plant. 100: 769–779 [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Nelson N., Frolow F. (2003). Crystallization and initial X-ray diffraction studies of higher plant photosystem I. Acta Crystallogr. D Biol. Crystallogr. 59: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Bergantino E., Dainese P., Cerovic Z., Sechi S., Bassi R. (1995). A post-translational modification of the photosystem II subunit CP29 protects maize from cold stress. J. Biol. Chem. 270: 8474–8481 [DOI] [PubMed] [Google Scholar]
- Bergantino E., Sandonà D., Cugini D., Bassi R. (1998). The photosystem II subunit CP29 can be phosphorylated in both C3 and C4 plants as suggested by sequence analysis. Plant Mol. Biol. 36: 11–22 [DOI] [PubMed] [Google Scholar]
- Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall’osto L., Morosinotto T., Bassi R. (2009). Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284: 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema E.J., van Breemen J.F., van Roon H., Dekker J.P. (2000). Conformational changes in photosystem II supercomplexes upon removal of extrinsic subunits. Biochemistry 39: 12907–12915 [DOI] [PubMed] [Google Scholar]
- Boekema E.J., Van Roon H., Van Breemen J.F., Dekker J.P. (1999). Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur. J. Biochem. 266: 444–452 [DOI] [PubMed] [Google Scholar]
- Bonente G., Howes B.D., Caffarri S., Smulevich G., Bassi R. (2008). Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J. Biol. Chem. 283: 8434–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler W.L., Strasser R.J. (1978). Effect of divalent cations on energy coupling between the light-harvesting chlorophyll a/b complex and photosystem II. Photosynthesis ’77: Proceedings of the Fourth International Congress on Photosynthesis, Hall D.A., Coombs J., Goodwin T.W., (London: The Biochemical Society; ), pp. 11–20 [Google Scholar]
- Caffarri S., Croce R., Breton J., Bassi R. (2001). The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 276: 35924–35933 [DOI] [PubMed] [Google Scholar]
- Caffarri S., Croce R., Cattivelli L., Bassi R. (2004). A look within LHCII: differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry 43: 9467–9476 [DOI] [PubMed] [Google Scholar]
- Caffarri S., Kouril R., Kereïche S., Boekema E.J., Croce R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casazza A.P., Tarantino D., Soave C. (2001). Preparation and functional characterization of thylakoids from Arabidopsis thaliana. Photosynth. Res. 68: 175–180 [DOI] [PubMed] [Google Scholar]
- Crimi M., Dorra D., Bösinger C.S., Giuffra E., Holzwarth A.R., Bassi R. (2001). Time-resolved fluorescence analysis of the recombinant photosystem II antenna complex CP29. Effects of zeaxanthin, pH and phosphorylation. Eur. J. Biochem. 268: 260–267 [DOI] [PubMed] [Google Scholar]
- Croce R., Breton J., Bassi R. (1996). Conformational changes induced by phosphorylation in the CP29 subunit of photosystem II. Biochemistry 35: 11142–11148 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Caffarri S., Bassi R. (2005). A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., Havaux M., Bassi R. (2010). Enhanced photoprotection by protein-bound vs free xanthophyll pools: A comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant 3: 576–593 [DOI] [PubMed] [Google Scholar]
- Damkjaer J.T., Kereïche S., Johnson M.P., Kovacs L., Kiss A.Z., Boekema E.J., Ruban A.V., Horton P., Jansson S. (2009). The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21: 3245–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bianchi S., Ballottari M., Dall’osto L., Bassi R. (2010). Regulation of plant light harvesting by thermal dissipation of excess energy. Biochem. Soc. Trans. 38: 651–660 [DOI] [PubMed] [Google Scholar]
- de Bianchi S., Dall’Osto L., Tognon G., Morosinotto T., Bassi R. (2008). Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20: 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Winter K., Kruger A., Czygan F.-C. (1989). Light stress and photoprotection related to the carotenoid zeaxanthin in higher plants. Photosynthesis. Plant Biology Vol. 8, Briggs W.R., (New York: Alan R. Liss; ), pp. 375–391 [Google Scholar]
- Ducruet J.M. (2003). Chlorophyll thermoluminescence of leaf discs: Simple instruments and progress in signal interpretation open the way to new ecophysiological indicators. J. Exp. Bot. 54: 2419–2430 [DOI] [PubMed] [Google Scholar]
- Ducruet J.M., Vavilin D. (1999). Chlorophyll high-temperature thermoluminescence emission as an indicator of oxidative stress: Perturbating effects of oxygen and leaf water content. Free Radic. Res. 31 (suppl): S187–S192 [DOI] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Finazzi G., Johnson G.N., Dall’Osto L., Joliot P., Wollman F.A., Bassi R. (2004). A zeaxanthin-independent nonphotochemical quenching mechanism localized in the photosystem II core complex. Proc. Natl. Acad. Sci. USA 101: 12375–12380 Erratum. Proc. Natl. Acad. Sci. USA 101: 17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaggio E., Cinque G., Bassi R. (2001). Functional architecture of the major light-harvesting complex from higher plants. J. Mol. Biol. 314: 1157–1166 [DOI] [PubMed] [Google Scholar]
- Frigerio S., Campoli C., Zorzan S., Fantoni L.I., Crosatti C., Drepper F., Haehnel W., Cattivelli L., Morosinotto T., Bassi R. (2007). Photosynthetic antenna size in higher plants is controlled by the plastoquinone redox state at the post-transcriptional rather than transcriptional level. J. Biol. Chem. 282: 29457–29469 [DOI] [PubMed] [Google Scholar]
- Ganeteg U., Külheim C., Andersson J., Jansson S. (2004). Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 134: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore A.M., Yamamoto H.Y. (1991). Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Golding A.J., Finazzi G., Johnson G.N. (2004). Reduction of the thylakoid electron transport chain by stromal reductants—Evidence for activation of cyclic electron transport upon dark adaptation or under drought. Planta 220: 356–363 [DOI] [PubMed] [Google Scholar]
- Haldrup A., Jensen P.E., Lunde C., Scheller H.V. (2001). Balance of power: A view of the mechanism of photosynthetic state transitions. Trends Plant Sci. 6: 301–305 [DOI] [PubMed] [Google Scholar]
- Hansson M., Vener A.V. (2003). Identification of three previously unknown in vivo protein phosphorylation sites in thylakoid membranes of Arabidopsis thaliana. Mol. Cell. Proteomics 2: 550–559 [DOI] [PubMed] [Google Scholar]
- Havaux M. (2003). Spontaneous and thermoinduced photon emission: New methods to detect and quantify oxidative stress in plants. Trends Plant Sci. 8: 409–413 [DOI] [PubMed] [Google Scholar]
- Havaux M., Dall’osto L., Bassi R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Dall’Osto L., Cuiné S., Giuliano G., Bassi R. (2004). The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. J. Biol. Chem. 279: 13878–13888 [DOI] [PubMed] [Google Scholar]
- Havaux M., Eymery F., Porfirova S., Rey P., Dörmann P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Niyogi K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96: 8762–8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N.E., Zigmantas D., Valkunas L., Li X.P., Niyogi K.K., Fleming G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Horton P. (1996). Nonphotochemical quenching of chlorophyll fluorescence. Light as an Energy Source and Information Carrier in Plant Physiology, Jennings R.C., (New York: Plenum Press; ), pp. 99–111 [Google Scholar]
- Horton P., Ruban A. (2005). Molecular design of the photosystem II light-harvesting antenna: Photosynthesis and photoprotection. J. Exp. Bot. 56: 365–373 [DOI] [PubMed] [Google Scholar]
- Jahns P., Latowski D., Strzalka K. (2009). Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim. Biophys. Acta 1787: 3–14 [DOI] [PubMed] [Google Scholar]
- Jansson S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Jensen P.E., Gilpin M., Knoetzel J., Scheller H.V. (2000). The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275: 24701–24708 [DOI] [PubMed] [Google Scholar]
- Johnson M.P., Goral T.K., Duffy C.D., Brain A.P., Mullineaux C.W., Ruban A.V. (2011). Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.P., Havaux M., Triantaphylidès C., Ksas B., Pascal A.A., Robert B., Davison P.A., Ruban A.V., Horton P. (2007). Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J. Biol. Chem. 282: 22605–22618 [DOI] [PubMed] [Google Scholar]
- Joliot P., Joliot A. (1984). Electron transfer between the two photosystem. I. Flash excitation under oxidizing conditions. Biochim. Biophys. Acta 765: 210–218 [Google Scholar]
- Kim E.H., Li X.P., Razeghifard R., Anderson J.M., Niyogi K.K., Pogson B.J., Chow W.S. (2009). The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: A study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 1787: 973–984 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. (2008). Molecular crowding and order in photosynthetic membranes. Trends Plant Sci. 13: 201–207 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Horstmann S., Weis E. (2000). Control of the photosynthetic electron transport by PQ diffusion microdomains in thylakoids of higher plants. Biochim. Biophys. Acta 1459: 148–168 [DOI] [PubMed] [Google Scholar]
- Klimmek F., Sjödin A., Noutsos C., Leister D., Jansson S. (2006). Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács L., Damkjaer J., Kereïche S., Ilioaia C., Ruban A.V., Boekema E.J., Jansson S., Horton P. (2006). Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18: 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol A.G., Borza T., Ishida K., Keeling P., Lee R.W., Durnford D.G. (2007). Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143: 1802–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Fufezan C., Trebst A. (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Li X.P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li X.P., Gilmore A.M., Caffarri S., Bassi R., Golan T., Kramer D., Niyogi K.K. (2004). Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279: 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li X.P., Gilmore A.M., Niyogi K.K. (2002). Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 277: 33590–33597 [DOI] [PubMed] [Google Scholar]
- Liu W.J., Chen Y.E., Tian W.J., Du J.B., Zhang Z.W., Xu F., Zhang F., Yuan S., Lin H.H. (2009). Dephosphorylation of photosystem II proteins and phosphorylation of CP29 in barley photosynthetic membranes as a response to water stress. Biochim. Biophys. Acta 1787: 1238–1245 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Livingston A.K., Cruz J.A., Kohzuma K., Dhingra A., Kramer D.M. (2010). An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin S., Armond P.A., Mooney H.A., Fork D.C. (1981). Photosystem II photosynthetic unit sizes from fluorescence induction in leaves. Correlation to photosynthetic capacity. Plant Physiol. 67: 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro S., Dainese P., Lannoye R., Bassi R. (1997). Cold-resistant and cold-sensitive maize lines differ in the phosphorylation of the photosystem II subunit, CP29. Plant Physiol. 115: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. (1991). Dynamics of photosynthetic membrane composition and function. Biochim. Biophys. Acta 1058: 87–106 [Google Scholar]
- Melis A. (1999). Photosystem-II damage and repair cycle in chloroplasts: What modulates the rate of photodamage? Trends Plant Sci. 4: 130–135 [DOI] [PubMed] [Google Scholar]
- Miloslavina Y., Wehner A., Lambrev P.H., Wientjes E., Reus M., Garab G., Croce R., Holzwarth A.R. (2008). Far-red fluorescence: A direct spectroscopic marker for LHCII oligomer formation in non-photochemical quenching. FEBS Lett. 582: 3625–3631 [DOI] [PubMed] [Google Scholar]
- Morosinotto T., Baronio R., Bassi R. (2002). Dynamics of chromophore binding to Lhc proteins in vivo and in vitro during operation of the xanthophyll cycle. J. Biol. Chem. 277: 36913–36920 [DOI] [PubMed] [Google Scholar]
- Morosinotto T., Bassi R., Frigerio S., Finazzi G., Morris E., Barber J. (2006). Biochemical and structural analyses of a higher plant photosystem II supercomplex of a photosystem I-less mutant of barley. Consequences of a chronic over-reduction of the plastoquinone pool. FEBS J. 273: 4616–4630 [DOI] [PubMed] [Google Scholar]
- Moya I., Silvestri M., Vallon O., Cinque G., Bassi R. (2001). Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry 40: 12552–12561 [DOI] [PubMed] [Google Scholar]
- Mozzo M., Dall’Osto L., Hienerwadel R., Bassi R., Croce R. (2008). Photoprotection in the antenna complexes of photosystem II: Role of individual xanthophylls in chlorophyll triplet quenching. J. Biol. Chem. 283: 6184–6192 [DOI] [PubMed] [Google Scholar]
- Müller M.G., Lambrev P., Reus M., Wientjes E., Croce R., Holzwarth A.R. (2010). Singlet energy dissipation in the photosystem II light-harvesting complex does not involve energy transfer to carotenoids. ChemPhysChem 11: 1289–1296 [DOI] [PubMed] [Google Scholar]
- Nayak L., Raval M.K., Biswal B., Biswal U.C. (2002). Topology and photoprotective role of carotenoids in photosystem II of chloroplast: A hypothesis. Photochem. Photobiol. Sci. 1: 629–631 [DOI] [PubMed] [Google Scholar]
- Nelson N., Ben-Shem A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5: 971–982 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3: 455–460 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K., Grossman A.R., Björkman O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostergetel G.T., Keegstra W., Brisson A. (1998). Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy 74: 47–59 [Google Scholar]
- Pesaresi P., Sandonà D., Giuffra E., Bassi R. (1997). A single point mutation (E166Q) prevents dicyclohexylcarbodiimide binding to the photosystem II subunit CP29. FEBS Lett. 402: 151–156 [DOI] [PubMed] [Google Scholar]
- Peter G.F., Takeuchi T., Thornber J.P. (1991). Solubilization and two-dimensional electrophoretic procedures for studying the organization and composition of photosynthetic membrane polypeptides. Methods 3: 115–124 [Google Scholar]
- Ruban A.V., Berera R., Ilioaia C., van Stokkum I.H., Kennis J.T., Pascal A.A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007). Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Wentworth M., Yakushevska A.E., Andersson J., Lee P.J., Keegstra W., Dekker J.P., Boekema E.J., Jansson S., Horton P. (2003). Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421: 648–652 [DOI] [PubMed] [Google Scholar]
- Santabarbara S., Bordignon E., Jennings R.C., Carbonera D. (2002). Chlorophyll triplet states associated with photosystem II of thylakoids. Biochemistry 41: 8184–8194 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Strasser R.J., Srivastava A., Govindjee (1995). Polyphasic chlorophyll a fluorescence transient in plants and cyanobacteria. Photochem. Photobiol. 61: 32–42 [Google Scholar]
- Takahashi H., Iwai M., Takahashi Y., Minagawa J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teardo E., de Laureto P.P., Bergantino E., Dalla Vecchia F., Rigoni F., Szabò I., Giacometti G.M. (2007). Evidences for interaction of PsbS with photosynthetic complexes in maize thylakoids. Biochim. Biophys. Acta 1767: 703–711 [DOI] [PubMed] [Google Scholar]
- Testi M.G., Croce R., Polverino-De Laureto P., Bassi R. (1996). A CK2 site is reversibly phosphorylated in the photosystem II subunit CP29. FEBS Lett. 399: 245–250 [DOI] [PubMed] [Google Scholar]
- Tikkanen M., Piippo M., Suorsa M., Sirpiö S., Mulo P., Vainonen J., Vener A.V., Allahverdiyeva Y., Aro E.M. (2006). State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62: 779–793 Erratum. Plant Mol. Biol. 62: 795 [DOI] [PubMed] [Google Scholar]
- Tokutsu R., Iwai M., Minagawa J. (2009). CP29, a monomeric light-harvesting complex II protein, is essential for state transitions in Chlamydomonas reinhardtii. J. Biol. Chem. 284: 7777–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C., Krischke M., Hoeberichts F.A., Ksas B., Gresser G., Havaux M., Van Breusegem F., Mueller M.J. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Bulte L., Dainese P., Olive J., Bassi R., Wollman F.A. (1991). Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88: 8262–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Gowen B., Matadeen R., Orlova E.V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. (2000). Single-particle electron cryo-microscopy: Towards atomic resolution. Q. Rev. Biophys. 33: 307–369 [DOI] [PubMed] [Google Scholar]
- Van Kooten O., Snel J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25: 147–150 [DOI] [PubMed] [Google Scholar]
- van Oort B., Alberts M., de Bianchi S., Dall’Osto L., Bassi R., Trinkunas G., Croce R., van Amerongen H. (2010). Effect of antenna-depletion in Photosystem II on excitation energy transfer in Arabidopsis thaliana. Biophys. J. 98: 922–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters R.G., Ruban A.V., Horton P. (1996). Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. USA 93: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner A., Grasses T., Jahns P. (2006). De-epoxidation of violaxanthin in the minor antenna proteins of photosystem II, LHCB4, LHCB5, and LHCB6. J. Biol. Chem. 281: 21924–21933 [DOI] [PubMed] [Google Scholar]
- Yamamoto H.Y., Higashi R.M. (1978). Violaxanthin de-epoxidase. Lipid composition and substrate specificity. Arch. Biochem. Biophys. 190: 514–522 [DOI] [PubMed] [Google Scholar]
- Yu J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhang S., Scheller H.V. (2004). Photoinhibition of photosystem I at chilling temperature and subsequent recovery in Arabidopsis thaliana. Plant Cell Physiol. 45: 1595–1602 [DOI] [PubMed] [Google Scholar]
- Zygadlo A., Jensen P.E., Leister D., Scheller H.V. (2005). Photosystem I lacking the PSI-G subunit has a higher affinity for plastocyanin and is sensitive to photodamage. Biochim. Biophys. Acta 1708: 154–163 [DOI] [PubMed] [Google Scholar]