This work identified two different Arabidopsis mutants that have reduced susceptibility to an infectious biotrophic pathogen due to overaccumulation of the amino acid Thr. This is detrimental for the host plant and the infecting pathogen but does not affect disease caused by some other pathogen species. Therefore, the host metabolic state can influence disease in quite a specific manner.
Abstract
Reliance of biotrophic pathogens on living plant tissues to propagate implies strong interdependence between host metabolism and nutrient uptake by the pathogen. However, factors determining host suitability and establishment of infection are largely unknown. We describe a loss-of-inhibition allele of ASPARTATE KINASE2 and a loss-of-function allele of DIHYDRODIPICOLINATE SYNTHASE2 identified in a screen for Arabidopsis thaliana mutants with increased resistance to the obligate biotrophic oomycete Hyaloperonospora arabidopsidis (Hpa). Through different molecular mechanisms, these mutations perturb amino acid homeostasis leading to overaccumulation of the Asp-derived amino acids Met, Thr, and Ile. Although detrimental for the plant, the mutations do not cause defense activation, and both mutants retain full susceptibility to the adapted obligate biotrophic fungus Golovinomyces orontii (Go). Chemical treatments mimicking the mutants’ metabolic state identified Thr as the amino acid suppressing Hpa but not Go colonization. We conclude that perturbations in amino acid homeostasis render the mutant plants unsuitable as an infection substrate for Hpa. This may be explained by deployment of the same amino acid biosynthetic pathways by oomycetes and plants. Our data show that the plant host metabolic state can, in specific ways, influence the ability of adapted biotrophic strains to cause disease.
INTRODUCTION
Plant-infecting microbes have evolved diverse lifestyles to survive changing environments and host selection pressures. The success of many pathogens depends on a high degree of specialization to a particular plant host which, at the same time, can restrict their host range (Kämper et al., 2006; Tyler et al., 2006). Non-host resistance, defined as immunity of an entire plant species against all genetic variants of a pathogen species, is one major host range-limiting factor. In Arabidopsis thaliana, non-host resistance has been shown to depend on the rapid mobilization of vesicles and defense-associated proteins to attempted invasion sites (Kwon et al., 2008; Wang et al., 2009). Also, chemical responses involving the Trp-derived phytoalexin camalexin and indolic glucosinolates protect cells against attempted infection by biotrophic and necrotrophic pathogens (Bednarek et al., 2009; Sanchez-Vallet et al., 2010; Schlaeppi et al., 2010). As a consequence, mutant plants defective in two redundant CYP79B monooxygenases catalyzing the conversion of Trp to indole-3-acetaldoxime, and thus lacking Trp-derived secondary metabolites, support increased growth of normally nonadapted pathogen isolates (Sanchez-Vallet et al., 2010; Schlaeppi et al., 2010).
Host-adapted pathogens have evolved the ability to overcome non-host defenses and successfully colonize plant tissues. Effector molecules produced by infectious microbes are known to suppress plant defenses or manipulate host development in favor of pathogen growth (reviewed in Bent and Mackey, 2007). In a coevolutionary conflict with the host, some effectors become recognized by intracellular nucleotide binding site–leucine-rich repeat–containing immune sensors (NLRs). As a pathogen effector-driven process, receptor-mediated (race-specific) resistance depends on the genetic repertoire of the host and adapted pathogen (Bent and Mackey, 2007). NLR activation upon direct or indirect recognition of an effector molecule induces a signaling cascade leading to pathogen containment often involving host programmed cell death (the hypersensitive response [HR]) and accumulation of the systemic resistance signaling hormone salicylic acid (SA) (Bent and Mackey, 2007; Vlot et al., 2009). Many NLRs additionally require HSP90 and its cochaperones SGT1 and RAR1 for stability and/or activation (Shirasu, 2009; Zhang et al., 2010).
The obligate biotrophic oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) has become specialized to infect Arabidopsis, causing downy mildew disease. Coadaptation within this natural pathosystem is reflected by the extensive genetic variation in responses of different Arabidopsis accessions to Hpa isolates (Holub et al., 1994). Oomycetes are distinct from true fungi in that they possess a cellulose-based cell wall with little or no chitin and are phylogenetically close to brown algae and diatoms within the Stramenopile (heterokont) lineage (reviewed in Coates and Beynon, 2010). On susceptible Arabidopsis accessions, Hpa asexual conidospores can germinate on leaves to produce a ramifying intercellular network of hyphae with haustorial invaginations of individual host cells believed to serve as specialized feeding structures (Coates and Beynon, 2010). As an obligate biotrophic parasite, Hpa is adept at maintaining host cell viability and integrity, most likely through the collective activities of protein effectors, some of which are translocated across the haustorial and host cell membranes into the plant cytoplasm (Whisson et al., 2007; Kale et al., 2010). The Hpa life cycle is completed by formation of conidiospores on the leaf surface and generation of longer lived sexual oospores inside leaves (Coates and Beynon, 2010). NLR recognition of specific Hpa effectors on resistant Arabidopsis accessions leads to a typical HR and accumulation of SA (Vlot et al., 2009; Coates and Beynon, 2010).
The absolute dependence of obligate biotrophs such as Hpa on their hosts and the intimate physical and molecular interactions between host and pathogen structures implies that additional host factors besides the dedicated defense machinery may be decisive for infection. Plant factors required for host colonization by symbiotic nitrogen-fixing bacteria have been identified (e.g., Hakoyama et al., 2009). Plant susceptibility determinants for biotrophic pathogen colonization have also been described but are not well understood. For example, successful entry of powdery mildew fungal germinating spores into host epidermal cells depends on functional MILDEW RESISTANCE LOCUS O (MLO) protein in barley (Hordeum vulgare) and Arabidopsis, although the precise function of MLO is unclear (Büschges et al., 1997; Consonni et al., 2006). Genetic screens of Arabidopsis identified a range of recessive powdery mildew resistant and downy mildew resistant (dmr) mutants that affect host cell wall composition and/or stress metabolite status (reviewed in O’Connell and Panstruga, 2006). dmr1, mediating strong and specific resistance to Hpa, encodes homoserine kinase, a chloroplast enzyme involved in the biosynthesis of the Asp-derived amino acids Met, Thr, and Ile (van Damme et al., 2009). dmr1 mutants accumulated high levels of homoserine (HS), and treatment of plants with HS induced resistance to Hpa but did not interfere with Hpa spore germination or radial growth of the hemibiotrophic oomycete Phytophthora capsici in vitro (van Damme et al., 2009). The mechanism underlying increased downy mildew resistance through accumulation of HS in the chloroplast was not defined (van Damme et al., 2009).
Here, we describe the characterization of two nonallelic Arabidopsis mutants that display enhanced resistance to host-adapted Hpa. These mutants were identified in a screen for genetic suppressors of susceptibility to Hpa (isolate Noco2) caused by a rar1 mutation disabling RPP5 (NLR) recognition (Muskett et al., 2002). The rar1 suppressor (rsp) mutations cause perturbations of plant amino acid homeostasis by different molecular mechanisms, leading to strong and specific resistance to Hpa. In contrast with the induced defense programs triggered by Hpa effector recognition, we find that suppression of Hpa growth on rsp mutants is a result of Thr accumulation rendering host tissues unsuitable as a growth substrate. The metabolic state provoked by the rsp mutations is also detrimental for the plant, and specific loss of susceptibility to adapted Hpa isolates in the rsp mutants might be explained by deployment of the same metabolic pathway in the phylogenetically related oomycete. Our data suggest that host metabolic status can influence the range of biotrophic pathogens causing disease.
RESULTS
Mutants with Altered Responses to Hpa
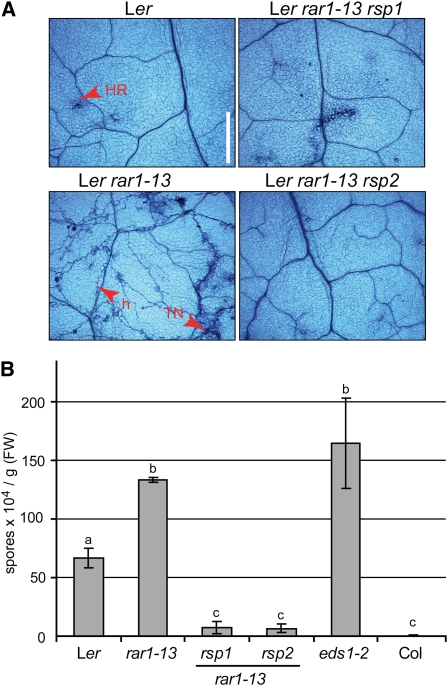
Complete resistance in Arabidopsis accession Landsberg erecta (Ler) to Hpa isolate Noco2 mediated by RPP5 (Parker et al., 1993) depends on RAR1, encoding an HSP90 cochaperone (Muskett et al., 2002; Zhang et al., 2010). We used the intermediate resistance phenotype of a partially defective rar1 mutant, rar1-15, as a sensitized background for a forward genetic screen to identify mutants with an altered response to Noco2 infection. Seeds of rar1-15 were ethyl methanesulfonate mutagenized, and ~2600 M2 families derived from single M1 plants screened for enhanced (rar1 enhancer [ren]) or reduced (rsp) susceptibility to Hpa Noco2. A number of ren and rsp mutants were selected for further characterization. Here, we describe analysis of two nonallelic rsp mutants, rsp1 and rsp2. Increased resistance of rsp1 and rsp2 to Hpa Noco2 was maintained after crossing into the rar1-13 null mutant background, indicating that the resistance does not depend on a partially functional rar1-15 protein. We used the rar1-13 rsp1 and rar1-13 rsp2 mutants for further characterization of rsp1 and rsp2 phenotypes. When infected with Hpa isolate Noco2, pathogen growth and sporulation were seen on Ler rar1-13, contrasting with the complete resistance associated with an HR in wild-type Ler, visualized microscopically after trypan blue (TB) staining of leaves (Figure 1A). The rar1-13 rsp1 and rar1-13 rsp2 double mutants exhibited strong resistance to Hpa. Whereas limited pathogen growth was occasionally observed in leaves of rar1-13 rsp1, there were no symptoms of disease or host cell death in rar1-13 rsp2 plants (Figure 1A). The absence of HR lesions in rar1-13 rsp2 was especially evident when infected plants were observed under UV light (see Supplemental Figure 1A online). To test whether the increased resistance of rar1-13 rsp1 and rar1-13 rsp2 was dependent on RPP5, we infected the mutants with the virulent Hpa isolate Cala2, which is not recognized by RPP5. As expected, enhanced Cala2 sporulation was observed on rar1-13 and eds1-2 compared with wild-type Ler (Figure 1B). eds1-2 is defective in downstream signaling after activation of NLRs containing an N-terminalToll/interleukin-1 receptor domain (Aarts et al., 1998; Wirthmueller et al., 2007) and was included in pathogenicity assays as a mutant with high susceptibility. By contrast, pathogen sporulation was strongly reduced on rar1-13 rsp1 and rar1-13 rsp2 mutant plants that grouped statistically with the fully resistant Columbia-0 (Col-0) accession (Figure 1B). We concluded that the mutations in rsp1 and rsp2 cause reduced susceptibility to Hpa infection independently of RPP5.
Figure 1.
rsp1 and rsp2 Confer Strong Resistance against Different Hpa Isolates.
(A) Disease symptom formation on rsp mutant leaves. Three-week-old plants were infected with Hpa isolate Noco2, and first true leaves were stained with TB at 7 d after inoculation and examined under a light microscope. h, hyphae; TN, trailing necrosis. Bar = 0.5 mm.
(B) Conidiospore formation 6 d after infection of 3-week-old plants with Hpa isolate Cala2. Accession Col was included as a control expressing RPP2 resistance to Cala2. Standard deviations of three biological replicates are shown. Significantly different classes are indicated by lower-case letters (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each). FW, fresh weight.
rsp1 and rsp2 Are Not Constitutive Resistance Mutants
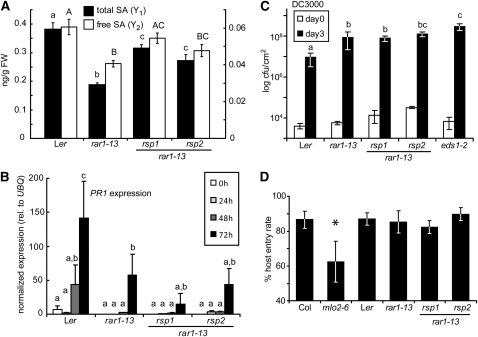
The rar1-13 rsp1 and rar1-13 rsp2 mutants are smaller than wild-type Ler or rar1-13 when grown under normal conditions in soil (see Supplemental Figure 1B online). This feature, together with their reduced susceptibility to a virulent Hpa isolate, suggested they may be expressing constitutive resistance, which is normally accompanied by growth retardation, increased steady state accumulation of SA, and expression of Pathogenesis-Related (PR) genes leading to broad spectrum resistance (e.g., Lu et al., 2003; Zhang et al., 2003). We therefore tested for hallmarks of constitutive resistance. Free and total SA levels were reduced in rar1-13 compared with the wild type. In rar1-13 rsp1 and rar1-13 rsp2, SA accumulation was intermediate between wild-type Ler and rar1-13 (Figure 2A) and therefore not characteristic of constitutive resistance mutants. We then tested whether the enhanced Hpa resistance of the rsp mutants might correlate with increased basal expression or accelerated PR gene induction upon infection that would reflect priming of defenses (Conrath et al., 2002). Plants were infected with Hpa isolate Noco2. Samples were taken up to 72 h after inoculation and expression of the SA-responsive PR gene PR1 measured by quantitative RT-PCR. Individual analysis of variance (ANOVA) tests detected significant PR1 induction over time in all genotypes (Figure 2B). No significant differences in PR1 expression were detected between rar1-13, rar1-13 rsp1, and rar1-13 rsp2 (Figure 2B). These data show that the increased resistance of the rar1-13 rsp mutants cannot be explained by enhanced basal or Hpa-induced SA defenses. We infected plants with a virulent strain (DC3000) of the bacterial pathogen Pseudomonas syringae pv tomato (Pst) to test whether increased resistance of rar1-13 rsp mutants extended to an unrelated hemibiotrophic pathogen. Pst DC3000 grew more in rar1-13 and eds1-2 compared with Ler wild type, as expected (Figure 2C; Muskett et al., 2002). The rar1-13 rsp1 and rar1-13 rsp2 double mutants supported similar (rsp1) or higher (rsp2) bacterial growth compared with rar1-13 (Figure 2C). These data argue against the rsp mutations causing constitutive disease resistance.
Figure 2.
rsp1 and rsp2 Are Not Constitutive Resistance Mutants.
(A) Steady state SA accumulation in 4-week-old plants. Total (black bars, Y1) and free (white bars, Y2) SA was extracted from leaf tissue and measured by GC-MS. Standard deviations of three samples are shown. Letters indicate classes separated by significant differences (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each): lower-case, total SA; upper-case, free SA. FW, fresh weight.
(B) PR1 induction by Hpa. Three-week-old plants were infected with Hpa isolate Noco2 and samples taken at the indicated time points. Expression of PR1 and UBQ10 was determined by quantitative RT-PCR. Relative expression of PR1 normalized to UBQ in each sample is displayed, and error bars indicate standard deviations of three independent samples. Significantly different classes are indicated by lower-case letters (repeated measures ANOVA, Tukey’s post-hoc test, P < 0.05 each).
(C) Growth of Pst DC3000 on rsp mutant plants. The indicated lines were spray infected with bacteria at 4 × 105 colony-forming units (cfu) per mL. Bacterial growth (cfu/cm2) was determined 1 h (white bars) and 3 d (black bars) after infection. Error bars represent standard deviations of four samples. Significantly different classes are indicated by lower-case letters (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each).
(D) Host entry rate of G. orontii. Entry rates on the indicated plant lines were determined from Coomassie blue–stained leaves 2 d after infection of 6-week-old plants. Standard deviations of at least five counts from different leaves are shown. Asterisk indicates significant differences (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each).
To test whether increased resistance to Hpa observed in the rar1-13 rsp mutants extends to a different host-adapted obligate biotrophic pathogen, we inoculated plants with the adapted powdery mildew fungus Golovinomyces orontii (Go). Fungal entry into Arabidopsis epidermal cells is used as an indicator of early infection competence. The host cell entry rate of Go in rar1-13 rsp1 and rar1-13 rsp2 cells was therefore compared with that in wild-type Ler and the rar1-13 single mutant. Accession Col (susceptible) and the Col mlo2-6 (MILDEW RESISTANCE LOCUS O2) mutant, which is more resistant to Go (Consonni et al., 2006), were included as additional controls in the experiment. Fungal host entry rates were similar in Col and Ler and not altered by the rar1-13, rsp1, or rsp2 mutations, but significantly reduced in mlo2-6 (Figure 2D). Development of macroscopic disease symptoms was evaluated between 4 and 7 d after infection. There was enhanced sporulation on rar1-13 rsp2 plants compared with Ler and rar1-13 (see Supplemental Figure 2 online). Symptom formation on rar1-13 rsp1 was accompanied by leaf yellowing but Go sporulation itself was not altered. As expected, sporulation on mlo2-6 was reduced compared with Col (see Supplemental Figure 2 online). Since resistance of the rsp mutants does not extend to another host-adapted biotrophic pathogen, we concluded that the rsp1 and rsp2 defects perturb the Arabidopsis–Hpa interaction in quite a specific manner. This prompted us to clone and characterize the RSP1 and RSP2 genes.
rsp2 Is a Loss-of-Function Allele of DIHYDRODIPICOLINATE SYNTHASE2
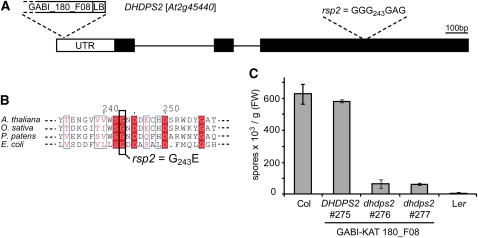
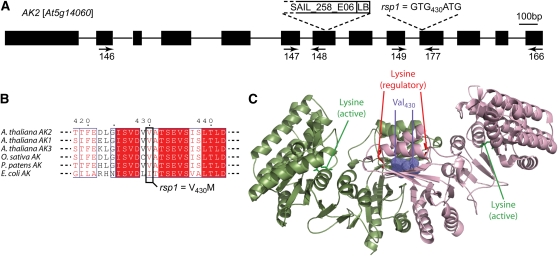
When backcrossed rar1-13 rsp2/RSP2 plants were selfed, resistance to Hpa Noco2 segregated in a 1:3 ratio (29R:108S; χ2 = 1.07) in the progeny indicative of a monogenic recessive trait. rar1-13 rsp2 was crossed to the Col rar1-28 null mutant to generate segregating F2 families for map-based cloning. Although RPP5 also segregates in the progeny (Parker et al., 1997), we observed a clear 1:3 ratio of resistant to susceptible plants in the mapping population consistent with the enhanced resistance of rsp2 being largely independent of RPP5 (Figure 1B). Linkage between rsp2 resistance and the erecta morphology was found and rsp2 was mapped in ~600 phenotyped F2 plants to an 86-kb interval on the lower arm of chromosome 2. This interval contains 26 annotated genes. DNA sequencing revealed a G-to-A exchange in the coding region of DIHYDRODIPICOLINATE SYNTHASE2 (DHDPS2, At2g45440), resulting in a predicted exchange of Gly-243 to Glu in DHDPS2 (Figures 3A and 3B).
Figure 3.
Molecular Cloning of the rsp2 Mutation.
(A) Organization of the DHDPS2 locus. Positions of the rsp2 mutation and T-DNA insertion in GABI_180_F08 are indicated. LB, left border.
(B) Alignment of DHDPS2 proteins and position of the amino acid exchange in rsp2. (At, Q0WSN6; Os, Q7XPB5; Pp, A9SGY8; Ec, C9QPU6).
(C) Conidiospore formation 7 d after infection of 3-week-old plants with Hpa isolate Noco2. Biological replicates were pooled and treated as one sample. Error bars are derived from five technical replicates. FW, fresh weight.
[See online article for color version of this figure.]
DHDPS enzymes catalyze the formation of dihydrodipicolinate from l-aspartate-4-semialdehyde (ASA) as the committing step for biosynthesis of Lys in the biosynthesis of the Asp-derived amino acids Lys, Met, Thr, and Ile, also called the Lys superpathway (Jander and Joshi, 2010; see also scheme in Figure 5A). All reactions of this pathway from Asp phosphorylation to the amino acid end products Lys, Thr, Met, and Ile take place in the chloroplast (Ravanel et al., 2004). The mutated Gly residue in rsp2 is highly conserved among DHDPS proteins (Figure 3B) and has a high bias in the DHDPS motif (http://pfam.sanger.ac.uk/family?acc=DHDPS#tabview=tab3). The recessive nature of rsp2 suggests it is a reduced or loss-of-function (LOF) allele of DHDPS2. To test this, we searched for additional Arabidopsis dhdps2 mutations. From several candidate T-DNA insertions in DHDPS2 (http://signal.salk.edu/cgi-bin/tdnaexpress), one insertion in the 5′ untranslated region of DHDPS2 was confirmed (Figure 3A). Homozygous plants of this line exhibited strong resistance compared with a control line containing the wild-type DHDPS2 locus isolated from the same population (Figure 3C). We therefore concluded that rsp2 is a LOF allele of DHDPS2 and the mutant was renamed dhdps2-2.
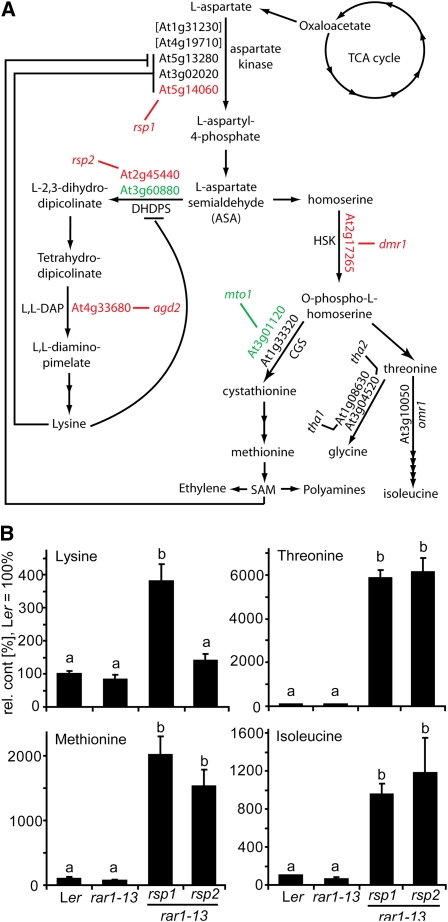
Figure 5.
The Lys Superpathway and Accumulation of Its End Products in rsp Mutants.
(A) Scheme showing the biosynthesis of Asp-derived amino acids in Arabidopsis. Bifunctional AK-HSDH enzymes are shown in brackets. Allosteric inhibition mechanisms mentioned in the text are indicated with negative impacting lines. Mutants identified in this or previous studies are shown in black (not tested), red (resistant to Hpa), or green (no resistance phenotype). TCA, tricarboxylic acid.
(B) Accumulation of the Asp-derived amino acids Lys, Met, Thr, and Ile in rsp mutant and control plants. Polar metabolites were extracted from aerial tissue of 4-week-old soil-grown plants and analyzed by GC-MS. Values were normalized to Ler = 100%, and standard deviations from three independent samples are shown. Significantly different classes are indicated by lower-case letters (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each). Trends were confirmed in an independent experiment.
The Arabidopsis genome contains two genes encoding DHDPS enzymes (http://www.Arabidopsis.org). To test whether DHDPS1 also contributes to Hpa resistance, a T-DNA insertion mutant in DHDPS1 (At3g60880) was isolated (see Supplemental Figure 3 online). Insertion of the T-DNA in an exon strongly reduced DHDPS1 transcript abundance measured by RT-PCR, suggesting that this line is a null dhdps1 mutant. In contrast with dhdps2-2 (rsp2), homozygous dhdps1-1 mutant plants did not exhibit reduced susceptibility to Hpa Noco2 (see Supplemental Figure 3 online).
rsp1 Carries a Mutation in ASPARTATE KINASE2
The rsp1 phenotype was initially scored as a recessive monogenic trait and crossed to Col rar1-28 for mapping. However, scoring of mapping populations infected with Hpa Noco2 revealed that resistance was not strictly recessive and, depending on the infection conditions could appear dominant, suggesting dosage effects. rsp1 was placed on the upper arm of chromosome 5 and a high-confidence interval of 360 kb containing 143 annotated loci was defined using a mapping population of ~500 phenotyped F2 plants. From this interval, a locus encoding ASPARTATE KINASE2 (AK2, At5g14060) was sequenced because rsp1 and rsp2/dhdps2-2 share common developmental defects (see Supplemental Figures 1 and 4 online), and we therefore suspected the mutations may affect the same pathway (see Figure 5A). A G-to-A exchange resulting in a predicted change of Val-430 to Met in AK2 was detected (Figures 4A and 4B). Asp kinases catalyze the conversion of Asp to l-aspartyl-4-phosphate as the first step of the Lys superpathway. The Arabidopsis genome contains three genes encoding monofunctional Asp kinases and two genes encoding bifunctional aspartate kinase/homoserine dehydrogenase (HSDH) enzymes (Figure 5A). Little is known about the in vivo roles of the different isoenzymes, but kinetic parameters in vitro are well described (Curien et al., 2005, 2007). The monofunctional AK2 and AK3 enzymes are allosterically inhibited by Lys, while AK1 is synergistically inhibited by Lys and S-adenosylmethionine. The recently reported crystal structure of the Arabidopsis AK1 dimer revealed binding of both effectors to its ACT1 domain (Mas-Droux et al., 2006). We built a structural model for the AK2 dimer using the AK1 crystal structure as a template (Figure 4C). The Val-430 residue mutated in rsp1, which is not completely conserved among AK enzymes from phylogenetically distant organisms, is located at a homodimer interface built by the two ACT1 domains, and contacts the Val-430 in the other monomer (Figure 4C). Hence, the rsp1 mutation might interfere with AK2 protein function by impairing homodimerization, although the mutation V430M could be modeled without provoking major steric clashes. We isolated a putative AK2 LOF allele from the T-DNA collections in accession Col in which the T-DNA insertion is located in an exon upstream of the mutation detected in rsp1 (Figure 4B; see Supplemental Figure 5 online). Homozygous Col ak2 plants were as susceptible as wild-type Col when tested with Hpa Noco2 (see Supplemental Figure 5B online). Therefore, we reasoned that rsp1 is not a LOF allele of AK2. This is further supported by the presence of a Met residue at the same position in an AK from Streptococcus equi (YP_002743803). Val-430 is also very close to the binding site of the allosteric Lys molecule in AK1, which is conserved with that of the Lys-inhibited Escherichia coli enzyme (Mas-Droux et al., 2006). We therefore reasoned that the V430M mutation might instead affect feedback inhibition of the enzyme in vivo in accordance with the nonrecessivity of rsp1 resistance.
Figure 4.
rsp1 Carries a Mutation in AK2.
(A) Organization of the AK2 locus. Positions of the mutation detected in rsp1, the T-DNA insertion in SAIL_258_06, and primers used for the RT-PCR analysis shown in Supplemental Figure 5 online are indicated. LB, left border.
(B) Alignment of AK2 proteins and position of the amino acid exchange in rsp1 (At AK1, NP_196832; At AK2, O23653; At AK3, NP_186851; Os, Q6YS33; Pp, A9T456; Ec, D3QK31).
(C) Structural model of AK2 generated with AK1 as template. Homodimer subunits are shown in light green and pink. A Lys residue that was cocrystallized in the active site of the original AK1 structure and the inhibitory Lys residue are shown in green and red in each subunit (sticks depiction). The Val-430 residue is highlighted in violet as dotted spheres.
Both rsp1 and rsp2/dhdps2-2 Strongly Perturb Amino Acid Homeostasis
Large differences in accumulation of products of the Lys superpathway have been reported for the previously characterized dhdps2-1 mutant (Craciun et al., 2000; Sarrobert et al., 2000). We investigated whether this is also the case for our newly identified rsp2/dhdps2-2 mutant and possibly rsp1. Polar metabolite extracts from aerial tissue of soil-grown plants were therefore prepared and analyzed by gas chromatography–mass spectrometry (GC-MS). Relative peak areas for signals corresponding to Lys, Ile, Met, and Thr were determined and normalized to Ler wild type, which was set at 100% (Figure 5B). No significant changes were detected between the wild type and rar1-13. In rsp2/dhdps2-2, higher accumulation of all pathway end products was observed, except Lys, which remained unchanged. Remarkably, a 60-fold increase in Thr levels was detected in rsp2/dhdps2-2 compared with the wild type. The amino acid profile of rsp1 was similar to rsp2/dhdps2-2 except that Lys levels increased three- to fourfold (Figure 5B). Altered amino acid levels, including Lys in rsp1 mutant plants, are consistent with our hypothesis that the rsp1 mutant protein has altered inhibition properties leading to deregulated flux of Asp, since the two DHDPS enzymes are normally tightly regulated by feedback inhibition through Lys (Craciun et al., 2000; Vauterin et al., 2000).
rsp1 Is a Loss-of-Inhibition Allele of AK2
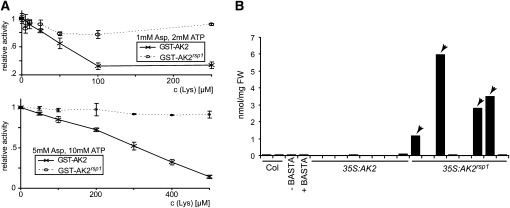
We tested whether the AK2 V430M mutation in rsp1 could indeed give rise to an enzyme refractive to Lys inhibition by expressing wild-type AK2 and mutant AK2rsp1 proteins lacking the chloroplast targeting peptide in E. coli as C-terminal fusions to glutathione S-transferase (GST). Affinity-purified GST-AK2 exhibited strong AK activity when tested in the presence of saturating concentrations of Asp and ATP (80 and 20 mM, respectively), whereas reactions containing a control protein purified under the same conditions did not show activity. The observed activity was fully dependent on the presence of Asp in the reaction mixture (data not shown). Hence, the GST-tagged protein is active and could be used for inhibition assays. GST-AK2 and GST-AK2rsp1 activities were compared under physiological conditions (2 mM ATP and 1 mM Asp) for inhibition by Lys (Figure 6A, top graph). A decrease in activity of the wild-type enzyme was observed with low concentrations of 10 to 25 μM Lys, as previously described (Curien et al., 2007). By contrast, even high Lys concentrations (up to 250 μM) did not lead to appreciable inhibition of AK2rsp1 in our assays. We confirmed this result under conditions of higher substrate availability (5 mM Asp and 10 mM ATP). Although the Lys concentration range for efficient inhibition of GST-AK2 shifted, differential inhibition properties of GST-AK2rsp1 remained (Figure 6A, bottom graph). Thus, a considerably higher activity of the AK2rsp1enzyme would be expected at physiological levels of Lys sufficient to inhibit the wild-type enzyme. We also tested whether the AK2rsp1 mutation causes accumulation of amino acids generated by the Lys superpathway in Arabidopsis aerial tissues by transforming Col with constructs of AK2 or AK2rsp1driven by the constitutive cauliflower mosaic virus 35S promoter. In T1 plants (selected for the cotransformed BASTA resistance marker and confirmed by PCR), 4/30 35S:AK2rsp1 transformants exhibited abnormal growth reminiscent of rsp1, whereas all 35S:AK2 transformants (>30) grew normally (see Supplemental Figure 6 online). Thr accumulation in aerial tissues of individual 35S:AK2 and 35S:AK2rsp1 transformants was measured by HPLC (Figure 6B). Analysis was restricted to the T1 generation because most of the abnormal 35S:AK2rsp1 plants failed to produce seed. Two biological replicates of Col tissue produced highly similar Thr values (37.9 ± 1.2 pmol/mg fresh weight), indicating that major differences could be detected by single measurements. Also, the effect of BASTA pretreatment was evaluated with a control transgenic line not expressing AK2. Thr content measured for this line without treatment was highly similar to Col wild type and decreased by ~25% after BASTA treatment. The Thr content of 35S:AK2 transformants ranged between 13 and 32 pmol/mg (Figure 6B). Similar values were obtained from the phenotypically normal 35S:AK2rsp1 transformants. By contrast, massive accumulation of Thr was detected in the rsp1-like transformants, rising to ~150-fold higher Thr content than the wild type. When 35S:AK2 and 35S:AK2rsp1 T1 plants were inoculated with Hpa Noco2, pathogen sporulation was observed on all plants except rsp1-like 35S:AK2rsp1 transformants included in the experiment, consistent with loss of susceptibility caused by AK2rsp1. The increased accumulation of Lys, Thr, Ile, and Met in rsp1 (Figure 5), the reduced Lys sensitivity of recombinant AK2rsp1 protein (Figure 6A), and the high Thr content of 35S:AK2rsp1 transformants phenotypically resembling the original rsp1 mutant lead us to conclude that rsp1 is a loss-of-inhibition allele of AK2. rsp1 was therefore renamed AK2rsp1 to differentiate it from LOF alleles.
Figure 6.
rsp1 Is a Loss-of-Inhibition Allele of AK2.
(A) Asp kinase activity was assayed with wild-type (AK2) and mutant (AK2rsp1) proteins using equal GST fusion protein amounts for enzymatic reactions in the presence of 1 mM Asp and 2 mM ATP (top panel) or 5 mM Asp and 10 mM ATP (bottom panel). Different concentrations of Lys were added to the reactions as indicated, and activity is expressed as percentage of activity without inhibitor. Standard deviations originating from four replicates are shown.
(B) Thr measurements from aerial tissue of AK2 and AK2rsp1-overexpressing plants. Transgenic plants were selected by BASTA resistance and confirmed by PCR. Leaf tissue was harvested from 5-week-old plants and used to determine free Thr content. Measurements derived from plants phenotypically resembling the rsp1 mutant (see Supplemental Figure 6 online) are marked with arrowheads. As the fresh weight of these plants was not sufficient to perform replicate measurements, one single measurement was performed per plant. FW, fresh weight.
AK2rsp1 and rsp2/dhdps2-2 Impede Hpa at an Early Stage of Infection
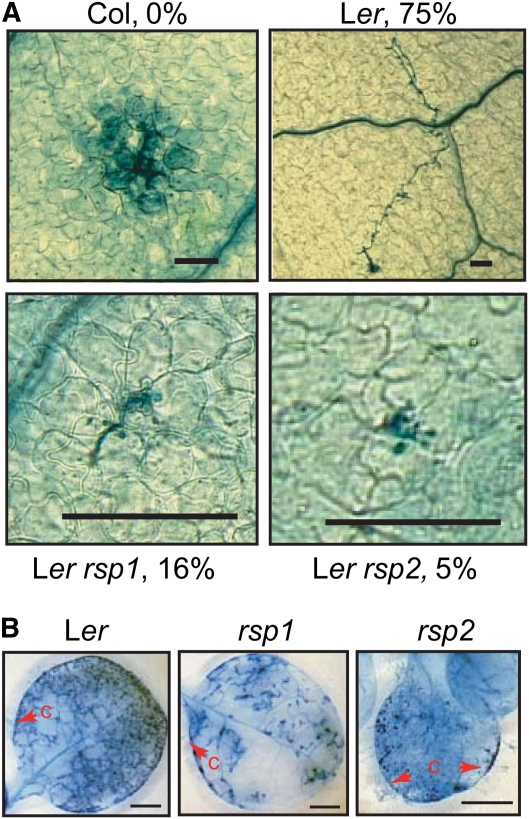
Having identified the molecular lesions underlying the rsp mutant phenotypes, we isolated single AK2rsp1 and rsp2/dhdps2-2 mutants to characterize their impact on Hpa infection in a RAR1 background with a fully functioning innate immune system. When infected with virulent Hpa Cala2, the rsp single mutants exhibited similar levels of resistance as the corresponding rsp rar1-13 double mutant combinations (see Supplemental Figure 7 online), confirming that the rsp phenotype manifests independently of the RAR1 status. We examined early stages of Hpa Cala2 infection (24 h after inoculation) by staining oomycete structures on the leaf surface with the optical brightener calcofluor. At this time point, germinating and fully germinated spores were visible on Ler wild-type leaves, although the germination rate was low (~1%). Spore germination on rsp1 leaves was indistinguishable from the wild type. By contrast, the germination rate appeared lower on rsp2, and we rarely located a germinating spore. This result suggested that Hpa colonization is impeded at a very early time point in rsp2. We then examined infection sites in the first pair of true leaves of wild-type and rsp mutant plants at 48 h after inoculation with Cala2 by TB staining. In the genetically resistant accession Col (due to RPP2 recognition; Sinapidou et al., 2004), each infection site was associated with host cell death and there was no hyphal extension from these sites (Figure 7A). In susceptible Ler wild type, 75% of primary infection sites produced hyphal outgrowth and no host cell death (Figure 7A). In AK2rsp1 and rsp2/dhdps2-2 mutant leaves, the proportion of infection sites producing hyphal outgrowth reduced to 16 and 5%, respectively, but at no site was this associated with host cell death. Therefore, the reduced susceptibility of AK2rsp1 and rsp2/dhdps2-2 mutants to Hpa infection is not due to activation of a classical resistance response. We found that Hpa could grow and form conidiophores at a low level on cotelydons of AK2rsp1 and rsp2/dhdps2-2 and occasionally on true leaves of AK2rsp1 plants (Figure 7B). These phenotypes are in line with a metabolic imbalance or nutrient deficiency in the host-limiting early Hpa colonization of tissues.
Figure 7.
rsp Mutants Do Not Have Hallmarks of Active Resistance to Hpa.
(A) Successful Hpa hyphal outgrowth is reduced in rsp mutants. Three-week-old plants were infected with Hpa Cala2. First true leaves were stained with TB at 48 h after inoculation. Infection sites were analyzed and the fraction of sites with significant hyphal growth expressed as percentage of total infection sites. A representative picture of the predominant reaction is shown for each line. An HR is visible at the interaction site for resistant accession Col. At least 12 leaves were analyzed per genotype, and between 21 (rsp2) and 88 (rsp1) infection sites were evaluated for successful hyphal outgrowth and the occurrence of cell death. Additional pictures of infection structures for comparison of hyphal and haustorial morphologies are shown in Supplemental Figure 9 online. Bars = 62 μm.
(B) Hpa can grow and reproduce on rsp mutant plants. Infections were done as in (A) except TB stainings were performed at 7 d after inoculation on complete plants. Conidiophores (c) are marked with red arrowheads. True leaves are shown for Ler and rsp1, and a cotelydon is shown for rsp2. Bars = 1 mm.
The reduced growth of rsp mutant plants could be partially complemented on synthetic media by addition of Suc but not other osmolytes or signaling sugars to the media (see Supplemental Figures 8A and 8B online). Since this suggested that the rsp mutants might be limited for carbohydrate, we tested whether Suc could also restore susceptibility of rsp mutant plants to Hpa. Plants were initially grown on synthetic media containing Suc and were then transferred to soil at different time points before infection with Hpa and infections monitored by TB staining. Transfer of plants prior to infection increased disease susceptibility of wild-type and rsp mutant plants to a similar extent (see Supplemental Figure 8C online). Nutrient or carbohydrate deficiency is therefore unlikely to underlie rsp resistance to Hpa.
Testing of Candidate Metabolites for Resistance Induction in rsp Mutants
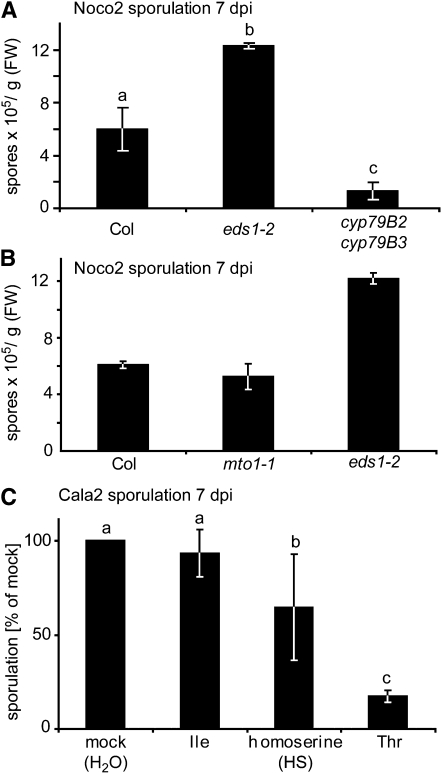
We reasoned that imbalances within the Lys superpathway might lead to increased production of secondary metabolites with resistance properties. For example, camalexin and indolic glucosinolates derived from Trp are important for resistance to adapted and nonadapted fungi (Bednarek et al., 2009, and references therein) and the hemibiotrophic oomycete pathogen Phytophthora brassicae (Schlaeppi et al., 2010). We measured levels of indole-derived secondary metabolites in wild-type and rsp mutant leaves. There were no differences in accumulation of the phytoalexin camalexin, but two- to threefold increased 1-methoxyindol-3-ylmethylglucosinolate levels were detected in rsp1 and rsp2 mutant extracts (see Supplemental Figure 10 online). We therefore tested the importance of indolic secondary metabolites in Hpa resistance by infecting cyp79B2 cyp79B3 (B2B3) double mutant plants that are unable to convert Trp to indol-3-aldoxime (Zhao et al., 2002), which is the precursor of indolic secondary metabolites in Arabidopsis. Compared with wild-type Col, the B2B3 mutant supported lower sporulation of virulent Hpa isolate Noco2 (Figure 8A), suggesting that indolic compounds do not contribute to Arabidopsis resistance to Hpa.
Figure 8.
Thr and HS but Not Indolic Metabolites or Met-Derived Metabolites Affect Arabidopsis Susceptibility to Hpa.
(A) cyp79B2 cyp79B3 double mutant plants are not hypersusceptible to Hpa. Three-week-old plants of the indicated genotypes were infected with Hpa isolate Noco2, and conidiospore formation was quantified at 7 d after inoculation. Error bars indicate standard deviations of three independent samples, and letters indicate significant differences (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each). FW, fresh weight.
(B) Met or Met-derived metabolites do not confer Hpa resistance. Experiment was performed as in (A). Biological replicates were pooled and treated as one sample. Error bars indicate technical error derived from five counts.
(C) Exogeneous Thr or HS treatments reduce Hpa conidiospore formation. Three-week-old Ler eds1-2 plants were sprayed with 5 mM solutions of the indicated amino acids 3 d and 1 d prior to infection with Hpa isolate Cala2. Conidiospores were quantified at 7 d after inoculation. Data were normalized to the mock-treated control (set at 100%) for seven independent biological replicates, and standard deviations are indicated. Significantly different classes are indicated by lower-case letters (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each).
Both rsp1 and rsp2 accumulate high levels of Met, which is the precursor of S-adenosylmethionine, polyamines, ethylene, and aliphatic glucosinolates (Figure 5A). To test whether high Met content, either directly or through increased production of downstream metabolites, might contribute to resistance in the rsp mutants, we measured Hpa sporulation on mto1-1 mutant plants carrying a mutation in a gene encoding cystathionine γ-synthase (Figure 5A), which leads to high Met accumulation (Inaba et al., 1994; Chiba et al., 1999). The mto1-1 mutant plants supported similar levels of Hpa sporulation as the wild type (Figure 8B). Therefore, increased Met and its downstream metabolites do not explain the resistance in rsp1 and rsp2.
rsp1 and rsp2 also accumulate high levels of Thr and Ile. Arabidopsis mutants with increased levels of Thr and/or Ile in aerial tissue have been reported (Kim and Leustek, 2000; Garcia and Mourad, 2004), but these were unobtainable. Therefore, we spray applied amino acid solutions onto hypersusceptible Ler eds1-2 plants to mimic the mutant metabolic state prior to infection with Hpa Cala2. HS was included in these experiments as induction of resistance to Hpa by this metabolite was recently reported (van Damme et al., 2009). Pretreatment of plants with Ile did not alter Hpa sporulation (Figure 8C). By contrast, sporulation was significantly reduced after HS and, to a stronger extent, Thr pretreatment. We detected only minor HS increases in rsp mutant tissues (see Supplemental Figure 11 online), suggesting that Thr accumulation is more likely to be causal for reduced Hpa growth on rsp mutant plants. Notably, suppression of Hpa growth by Thr application occurred in a dose-dependent manner (see Supplemental Figure 12 online).
Although our experiments indicate that indolic glucosinolates themselves are unlikely to contribute to Hpa resistance (Figure 8A), we examined whether jasmonate (JA)-regulated defenses might underlie the rsp mutant phenotypes since JA application is known to induce indolic glucosinolate metabolites (Mikkelsen et al., 2003). The expression of JA marker genes was not significantly different between wild-type and rsp mutant tissues, and Thr application induced resistance to Hpa to the same extent in plants defective for JA biosynthesis or signaling (see Supplemental Figure 13 online). Therefore, JA-mediated defenses are not responsible for Thr-induced suppression of Hpa infection.
Thr Treatment Recapitulates rsp Mutant Resistance Phenotypes
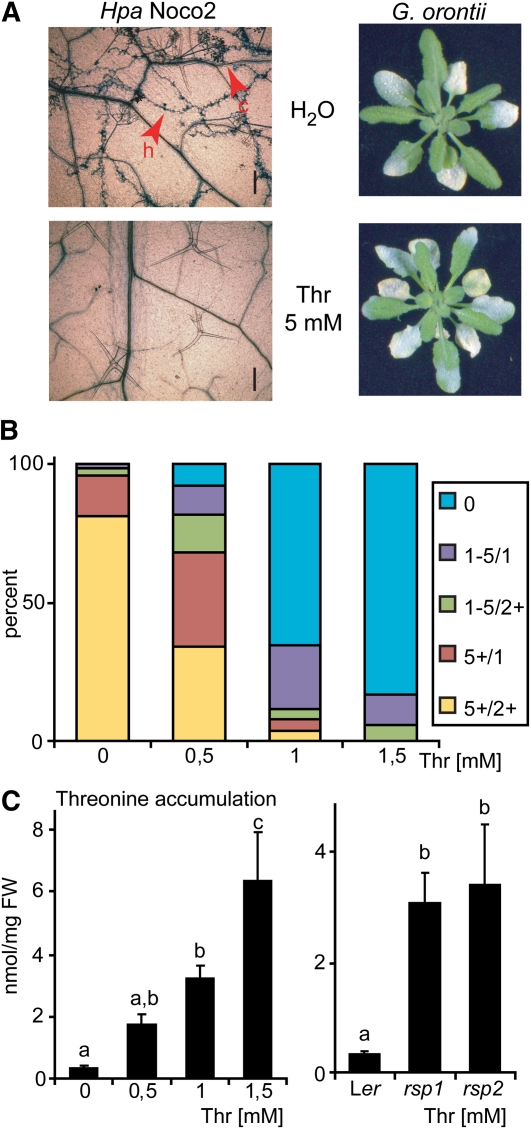
Suppression of Hpa growth by Thr application might result from general toxicity of this metabolite and thus be unrelated to the rsp mutant phenotypes. To exclude this possibility, the same set of plants pretreated with Thr was infected with Hpa or Go. Col plants were used for this assay because Go sporulation was most uniform on this accession. Leaves from plants infected with virulent Hpa isolate Noco2 were stained with TB and examined under the microscope at 7 d after inoculation. Hpa growth was observed in all mock-treated leaves but not in Thr pretreated leaves (Figure 9A, left panel). By contrast, similar levels of Go sporulation were observed on mock- and Thr-treated plants (Figure 9A, right panel). Selective suppression of Hpa growth observed with the rsp mutants can therefore be reproduced by Thr application, excluding broad toxicity of Thr in the chemical treatment experiments.
Figure 9.
Thr Treatment Mimics rsp Mutant Phenotypes.
(A) Thr treatment induces resistance to Hpa but not Go. Four-week-old Col plants were spray treated with Thr as described (Figure 8C). Plants were then infected with either Hpa (isolate Noco2) or Go. Disease symptom formation was evaluated macroscopically at 7 d after inoculation (Go infection) or assessed microscopically after TB staining (Hpa infection). Additional pictures to display the spectrum of pathogen growth are shown in Supplemental Figure 14 online. h, hyphae; c, conidiophores. Bars = 0.2 mm.
(B) Growth of plants on media containing Thr suppresses Hpa growth. Three-week-old Ler eds1-2 plants grown on synthetic media containing different concentrations of Thr were infected with Hpa isolate Cala2. Conidiophore formation was assessed 6 d after inoculation and categorized: 0, no conidiophores; 1-5/1, one to five conidiophores on one leaf; 1-5/2+, one to five conidiophores on two or more leaves; 5+/1, more than five conidiophores on one leaf; 5+/2+, more than five conidiophores on two or more leaves; n ≥ 18. Similar results were obtained in three independent experiments.
(C) Thr accumulation in rsp mutant tissues and plants grown on media containing Thr. Plants from (B) were sampled prior to infection and Thr content measured by HPLC (left graph) in parallel with samples from soil-grown rsp mutant plants (right graph). Data from (B) and (C) are derived from the same experiment. HPLC measurements were performed with ≥4 biological replicates. Significantly different classes are indicated by lower-case letters (one-way ANOVA, Tukey’s post-hoc test, P < 0.05 each). FW, fresh weight.
We then tested whether Thr indeed accumulates in plant tissues after application. Since spray application would not allow us to discriminate between absorbed and surface-deposited Thr, we grew plants on synthetic media containing different concentrations of Thr and measured Thr accumulation in aerial tissues that were not in direct contact with the metabolite. We found that the Hpa infection-suppressing action of Thr could be reproduced under these conditions. Ler eds1-2 plants grown on Murashige and Skoog medium in the presence of 0 to 1.5 mM Thr were infected with Hpa Cala2 and symptom formation scored at 6 d after inoculation. The numbers of conidiophores produced diminished with increasing concentrations of Thr added to the medium (Figure 9B). Thr accumulation was measured by HPLC. Whereas aerial tissues of control plants contained ~0.3 nmol/mg (fresh weight) Thr, this increased to ~3 nmol/mg in tissues grown on 1 mM Thr (Figure 9C, left graph). We concluded that Thr accumulation in aerial tissue upon spray treatment is therefore highly likely. As inclusion of 1 mM Thr to the medium strongly suppressed Hpa infection in the plate assay, we analyzed the extent to which the Thr amounts measured under these conditions relate to rsp mutant Thr contents. Because rsp mutant plants cannot be cultivated on synthetic media lacking Suc (see Supplemental Figure 8 online) leaf samples of soil-grown rsp mutant and control plants were included in the HPLC analysis (Figure 9C, right graph). Thr amounts were similar in the in vitro– and soil-grown control plants, ranging from 0.33 to 0.36 nmol/mg, respectively (Figure 9C). Notably, Thr accumulation in aerial tissues of plants grown on media containing 1 mM Thr was comparable to levels found in rsp mutant tissues (ranging from 3.1 to 3.4 nmol/mg; Figure 9C). Altogether, the results suggest that Thr application closely mirrors the rsp mutant phenotypes, both with regard to Thr accumulation and suppression of Hpa infection.
Effects of Thr on Plant and Hpa Growth Correlate with Deployment of the Diaminopimelate Pathway
Thr toxicity for Arabidopsis grown in vitro has been described (Sarrobert et al., 2000), and expression of an E. coli Thr synthase in Arabidopsis produced Thr overaccumulating plants with wrinkled and thickened rosette leaves and infertility (Lee et al., 2005), phenotypes broadly resembling the rsp1 and rsp2 mutants and AK2rsp1 transgenic plants (see Supplemental Figures 1, 2, and 6 online). Also, lethality of tha2-1 mutant plants defective in THREONINE ALDOLASE2, one of two Arabidopsis enzymes converting Thr to Gly, can be rescued by expression of omr1-5, a feedback-insensitive Thr deaminase (Figure 5A; Garcia and Mourad, 2004; Joshi et al., 2006), suggesting that tha2-1 lethality is due to Thr toxicity (Joshi et al., 2006). Thr overaccumulation therefore appears to be detrimental to Arabidopsis. The Hpa infection phenotypes of rsp mutants (Figure 1) and chemical treatment results (Figure 9) point to a negative effect of Thr on Hpa but not Go infection. We reasoned that the target of Thr interference or a biosynthetic pathway negatively affected by high Thr accumulation might be conserved among plants and oomycetes but not in the fungal ascomycete Go. The presence of genes for biosynthesis of the amino acids Lys, Thr, and Met was therefore compared in a targeted manner between the three different phyla. Thr and Met are derived from Asp in all organisms and their biosynthesis appears to be broadly similar. Ascomycetes use the α-aminoadipate (AAA) pathway with α-ketoglutarate serving as a precursor for biosynthesis of Lys, which thus belongs to the Glu family of amino acids (reviewed in Xu et al., 2006). By contrast, Lys is produced through the diaminopimelate (DAP) pathway in plants and oomycetes and belongs to the Asp family (Figure 5A; Randall et al., 2005; Hudson et al., 2006).
In order to test for deployment of the DAP pathway by Hpa, homologs of DHDPS, dihydrodipicolinate reductase (DHDPR), and diaminopimelate decarboxylase (LysA), which are common to all types of DAP pathway, were searched for in the newly available Hpa genome (see Supplemental Table 1 online; Baxter et al., 2010). Sequences with high similarity to E. coli DHDPS and LysA were detected. For DHDPR, a sequence with moderate similarity to E. coli DapB was found. Notably, the Pfam DHDPR N-terminal (PF01113) and C-terminal (PF05173) domains were identified in the predicted Hpa DHDPR protein. The presence of all three marker genes suggests that Lys biosynthesis occurs via the DAP pathway in Hpa. No sequences supporting the presence of these enzymes were obtained when searching the Go and related Blumeria graminis (Bg) genome assemblies (Spanu et al., 2010). By contrast, there was strong evidence for the presence of the AAA pathway in Go and Bg (see Supplemental Table 1 online). Sequences similar to genes of the AAA pathway were also detected in Hpa. Reciprocal sequence comparisons generally revealed higher similarity to proteins of different function, as previously described for Phytophthora infestans (Randall et al., 2005), arguing against co-option of both DAP and AAA pathways for Lys biosynthesis in oomycetes. We concluded that detrimental effects of Thr on Arabidopsis growth and Hpa infection likely reflect operation of a complex and highly regulated DAP pathway in both systems.
DISCUSSION
Obligate biotrophy implies strong interdependence between host metabolism and nutrient uptake by the pathogen, but the processes allowing establishment and maintenance of a compatible interaction are poorly understood. Here, we show that the primary amino acid metabolic status of a plant can profoundly affect its suitability as an infection substrate for the adapted obligate biotrophic oomycete pathogen, Hpa.
The rsp2 LOF mutant carries a lesion in DHDPS2 (Figure 3), one of two Arabidopsis DHDPS enzymes catalyzing the conversion of ASA to l-2,3-dihydrodipicolinate as the committing step in Lys biosynthesis (Figure 5A). Although expression of DHDPS2 in Arabidopsis was initially reported to be restricted to the root tip (Sarrobert et al., 2000) and mainly vascular tissue in aerial parts (Craciun et al., 2000), analysis of public microarray data (http://www.genevestigator.com) suggests similar levels of DHDPS2 expression in roots and rosette leaves, both of which are colonized by Hpa (Coates and Beynon, 2010). DHDPS1 has a similar expression pattern but with lower overall signal intensity according to microarray data. Since dhdps1 mutant plants did not exhibit altered growth or pathogen resistance (see Supplemental Figure 3 online), DHDPS2 probably accounts for the main DHDPS activity in Arabidopsis. We did not attempt to generate double mutants, as these would be expected to be lethal because no alternative route for Lys biosynthesis is known. Both DHDPS and HSDH, which catalyzes the committing step toward the biosynthesis of Met, Thr, and Ile, use ASA as a common substrate. Due to flux partitioning at the DHDPS/HSDH node, loss of the major DHDPS isoform leads to increased accumulation of products of the Met, Thr, and Ile branch (Figure 5). As Lys is a key regulator of Asp kinases (Curien et al., 2007), flux into the entire pathway is likely increased through compensatory control to reestablish Lys accumulation (for a kinetic model of the pathway, see Curien et al., 2009).
The rsp1 mutation isolated in our study perturbs the Lys superpathway in a different manner to rsp2/dhdps2-2. We show that rsp1 is a loss-of-inhibition allele of AK2, which renders the mutant AK2rsp1 protein refractive to allosteric inhibition by Lys under physiological conditions (Figures 4 and 6). Allosteric transitions deduced from the structures of E. coli AKIII suggest that subtle interdomain movements at the dimer interface are involved in the R- to T-state transition (Kotaka et al., 2006). The exchange of Val-430 located in the dimer interface to Met in AK2rsp1 might impede state transition or directly interfere with inhibitor binding. This molecular characterization of a feedback-insensitive AK variant from Arabidopsis adds to knowledge gained by prior isolation of presumed aspartate kinase mutants with altered regulatory properties (Heremans and Jacobs, 1997). Consistent with a failure in feedback inhibition, expression of AK2rsp1 from its endogenous locus increases flux into the Lys superpathway leading to accumulation of all pathway end products (Figure 5B).
AK2 and DHDPS2 were not obviously linked to plant immunity. This is probably because mutations in these genes do not lead to broad spectrum resistance (Figure 2) but appear to specifically impede colonization by Hpa. Basal innate immunity to adapted Hpa isolates is normally mediated by SA-dependent processes and numerous mutants with constitutively induced SA defenses display enhanced resistance to Hpa (Lu et al., 2003; Zhang et al., 2003). In contrast with these mutants, neither rsp1 nor rsp2 has characteristics of primed or constitutive SA pathway activation (Figure 2). Importantly, in an immune-competent wild-type background, rsp1 and rsp2 suppressed colonization by virulent Hpa isolate Cala2 at an early stage of infection, although the sporadic microcolonies formed in rsp1 did eventually grow and produce some spores (Figure 7). None of the successful or unsuccessful infection sites were associated with host cell death in the rsp mutants. Therefore, the reduced Hpa growth on rsp1 and rsp2 leaves is not through activation of a classical immune response but rather due to a loss of susceptibility. We conclude that perturbations in host metabolism render tissues unsuitable as an Hpa infection substrate.
We performed a number of experiments to elucidate how the host metabolic status might interfere with Hpa infection. While provision of Suc partially restored rsp mutant plant growth in vitro, scarcity of carbohydrates is unlikely to underlie the reduced susceptibility of rsp1 and rsp2 plants because Hpa growth was not appreciably restored by Suc (see Supplemental Figure 8 online). Suppression of Hpa growth through increased accumulation of indolic glucosinolates or Met-derived compounds is also not supported by our data (Figure 8; see Supplemental Figure 10 online). In chemical application experiments, we identified Thr as a potent inhibitor of Hpa growth (Figure 8C). Indeed, the consequences of Thr application compare well with rsp mutant phenotypes both with regard to selective suppression of Hpa growth and to absolute Thr concentrations (Figure 9). It is possible that a toxic metabolite derived from or induced by Thr accumulates upon increased Thr abundance. Since neither rsp mutant metabolic state nor Thr application interfered with growth of the adapted biotrophic ascomycete Go (Figure 9A; see Supplemental Figure 2 online), such a metabolite would have to act specifically on Arabidopsis and Hpa, but not Go. Alternatively, Thr itself could interfere with host and pathogen biosynthetic pathways. Negative effects of Thr by feedback inhibition of the Lys superpathway have been proposed, although failure to rescue lethality of tha2-1 mutant plants accumulating excess Thr by amino acid supplementation suggests otherwise (Joshi et al., 2006). Similarly, we could not restore Hpa growth on rsp mutant plants by providing an amino acid solution upon infection. Surprisingly, however, we also could not rescue the poor growth of the rsp2/dhdps2-2 mutant by Lys supplementation of synthetic media lacking Suc. This suggests an unexpected degree of compartmentalization or additional regulatory mechanisms operating within the Lys superpathway. The precise molecular processes underlying Thr interference remain to be elucidated. We think it likely that Thr overaccumulation interferes with both host and oomycete metabolic processes due to the relatively close phylogeny of the two organisms and the deployment of the DAP pathway in contrast with the more distant ascomycete Go (see Supplemental Table 1 online; Burki et al., 2007).
Two additional loci, AGD2 and DMR1, encoding enzymes of the Lys superpathway have previously been described as having effects on plant immune responses. AGD2 was shown to be a ll-diaminopimelate decarboxylase catalyzing the last step of Lys biosynthesis (Figure 5A; Hudson et al., 2006), and agd2 mutant plants exhibited increased resistance to bacteria and Hpa (Song et al., 2004). We measured the amino acid content of agd2 and detected an approximately twofold increase in Thr content (see Supplemental Figure 15 online), supporting relevance to Lys biosynthesis in vivo. While this small increase in Thr content might contribute to agd2 Hpa resistance, the mutant does not resemble rsp1 and rsp2 since it displays constitutive resistance (Song et al., 2004). By contrast, dmr1 mutant plants defective in homoserine kinase were resistant to Hpa without the hallmarks of constitutive resistance (van Damme et al., 2009). The similar phenotypes of three independent mutants (rsp1, rsp2, and dmr1) affected in enzymes of the Lys superpathway point to a common mechanism leading to impairment of Hpa infection. Our data support Thr as being causal for Hpa growth suppression in rsp mutant tissues, whereas dmr1 mutant plants preferentially accumulate HS (van Damme et al., 2009). It is conceivable that HS taken up by Hpa is subsequently converted to Thr, which might then accumulate in Hpa tissues. In Arabidopsis, HS is rate limiting for the accumulation of downstream metabolites under normal conditions and HS supplementation leads to Thr accumulation (Lee et al., 2005). Genes encoding enzymes of the initial and final reactions of Thr, Ile, and Met biosynthesis and for putative amino acid transporters/permeases are present in Hpa according to primary transcript annotations (http://vmd.vbi.vt.edu/query.php). Also, van Damme et al. (2009) showed HS-induced Hpa growth suppression to be independent from known defense pathways. These data lend support to our hypothesis that incompatibility with Hpa arises by an imbalance in amino acid homeostasis. Here, characterization of the Arabidopsis rsp1 and rsp2 mutants provides a new insight to how plant metabolic status can selectively determine interactions with pathogens. It also prompts a deeper comparative analysis of biotrophic pathogen metabolite uptake and assimilation systems that may also influence host plant selection.
METHODS
Plant Material, Growth Conditions, and Pathogenicity Assays
Wild-type Arabidopsis thaliana accessions used were Col-0 and Ler. The Ler rar1-13, rar1-15 (Muskett et al., 2002), eds1-2 (Aarts et al., 1998), and Col mlo2-6 (Consonni et al., 2006), mto1-1 (Inaba et al., 1994), and cyp79B2 cyp79B3 (Zhao et al., 2002) mutants are published. Col dhdps1-1 (SALK_147470), dhdps2-3 (GABI-KAT_180F08) (Rosso et al., 2003), ak2-1 (SAIL_258_E06, Syngenta), Ler rsp2/dhdps2-2, and rsp1/AK2rsp1 are characterized here. Oligonucleotides used for genotyping are listed in Supplemental Table 2 online. Plants were grown in soil in controlled environment chambers under a 10-h light regime (150 to 200 μE/m2s) at 22°C and 65% relative humidity. Plants were grown in vitro on one-tenth Murashige and Skoog medium, optionally containing different sugars or amino acids, under long-day conditions (18 h light) at 21°C. Pst DC3000 bacteria were grown for 24 h at 28°C on solid NYG medium (0.5% peptone, 0.3% yeast extract, and 2% glycerol) supplemented with the corresponding antibiotics. For bacterial growth assays, 6-week-old plants were spray inoculated with bacterial suspensions at 4 × 108 colony-forming units/mL in 10 mM MgCl2 containing 0.04% (v/v) Silwet L-77 (Lehle Seeds). In planta bacterial titers were determined at the indicated time points as described (Tornero and Dangl, 2001). Hpa isolates Noco2 and Cala2 were inoculated onto 3-week-old plants at 4 × 104 spores/mL unless indicated otherwise. Plant cell death and Hpa infection structures were visualized under a light microscope after staining leaves with lactophenol TB as described (Muskett et al., 2002) or directly using binocular UV illumination and a GFP1 filter. For quantitative assays, three pots of each genotype were infected and treated as biological replicates. Plants were harvested 6 to 7 d after inoculation and their fresh weight determined, and spores were resuspended in 5 to 10 mL of water and spore concentrations counted using a Neubauer counting chamber under the microscope. For Go inoculations, 4- to 6-week-old plants (as indicated) were touch infected with leaves of sporulating host plants. Host entry rate was determined 2 d after inoculation after destaining leaves with ethanol/acetic acid and staining fungal structures with Coomassie Brilliant Blue. Germinating spores with secondary hyphae were considered as having successfully penetrated.
Expression Analyses
Plants were spray infected with Hpa Noco2 as described above, and leaf samples (~50 mg) from different plants were taken at the indicated time points. Total RNA was extracted from leaves using TRI reagent (Sigma-Aldrich), and RNA was reverse transcribed into cDNA using SuperScriptII (Invitrogen) following the manufacturer's instructions. Quantitative RT-PCR experiments were performed in an iQ5 Real-Time PCR detection system (Bio-Rad) using Brilliant SYBR Green QPCR Core Reagent (Stratagene) as dye. Experiments were performed using three independent biological samples. Relative transcript levels were calculated using the iQ5 Optical System Software (version 2.0). Ubiquitin UBQ10 (At4g05320) transcript levels were used as internal reference. Primers used are listed in Supplemental Table 2 online.
GC-MS Analyses
SA quantification was done as previously described (Straus et al., 2010). For amino acid analysis, metabolites were extracted from 100 mg ground leaf tissue in 1 mL CHCl3/CH3OH/water (1:2:0.3). After shaking for 10 min at 70°C, samples were centrifuged and reextracted with 500 μL CHCl3/CH3OH (2:1). Five hundred microliter of water was added to the pooled supernatants, which were then centrifuged for phase separation. The upper phase was collected and dried. To separate polar from semipolar metabolites, the dried extract was resuspended in 0.5 mL of water with 0.1% trifluoroacetic acid and loaded onto a 100 mg DCS-18 solid phase extraction column (Supelco). Columns were washed twice with 0.6 mL of water containing 0.1% trifluoroacetic acid. For analysis of polar metabolites, the flow-through and washes were collected, dried, and resuspended in 400 μL CH3OH. A 100-μL aliquot was dried and derivatized in 40 μL N-methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane (Fluka) and 40 μL pyridine, including a mix of fatty acid methyl ester (Sigma-Aldrich) as internal standards for 30 min at 90°C. One microliter was injected into a GC-MS system equipped with a HP-5MS column (Agilent). Amino acids were identified by running commercial standards under the same conditions and quantified with Chemstation software from Agilent.
HPLC Analysis
Analysis of indolic glucosinolates was performed as previously described (Bednarek et al., 2009). For amino acid analyses, 100 mg leaf material were used for extractions if available, but less tissue was used for severely affected 35S:AK2rsp1 transgenic plants. Free amino acids were determined using a modified protocol from Scheible et al. (1997). Plant material was extracted for 20 min at 4°C with 400 μL 80% (v/v) aqueous ethanol (2.5 mM HEPES-KOH, pH 7.5) and 400 μL 50% (v/v) aqueous ethanol (2.5 mM HEPES-KOH, pH 7.5) and then 200 μL 80% (v/v) aqueous ethanol. Amino acids were measured in the collected supernantants by precolumn derivatization with orthophthaldehyde in combination with fluorescence detection (λex 330/λem 450) as described (Kreft et al., 2003). Elution was achieved on a Hyperclone C18 BDS column (Phenomenex) on a Summit HPLC system (Dionex), applying a solvent gradient with increasing hydrophobicity (buffer A: 0.2% [v/v] THF, 8.5 mM sodium phosphate buffer [NAPI], pH 7.5; buffer B: 32.5% [v/v] methanol, 20.5% [v/v] acetonitrile, and 18.5 mM NAPI, pH 7.5; flow: 0.8 mL/min; 0 to 2 min: 100% A, 16 min: 77% A, 13% B; 23.25 min: 15% A, 85% B; 32.23 min: 50% A, 50% B; 43.30 min: 40% A, 60% B; 49 to 52 min 100% B, 53 to 60 min: 100% A).
Protein Alignments and Modeling
Protein alignments were made using TCoffee (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi) and graphical views generated with ESPript (http://espript.ibcp.fr/ESPript/ESPript/). Comparative modeling of the At AK2 structure was performed with the modeller9v3 program (Sali and Blundell, 1993) using the structure of At AK1 (PDB: 2CDQ) as template. The high sequence identity between At AK1 and At AK2 (77%) ensures that errors in the model should not exceed 1 Å in Cα root mean square deviation. Mutation V375M in each chain of the dimer At AK2 could be modeled using the backrub module of the rosettav2.3 program (Smith and Kortemme, 2008). Figures were generated using Pymol.
Stable Transgenic Lines
The AK2 coding region was amplified from cDNA and cloned into pENTR/D-TOPO (Invitrogen), and the rsp1 mutation was introduced using site-directed mutagenesis by PCR with complementary oligonucleotides. Primers are listed in Supplemental Table 2 online. Wild-type and mutant AK2 sequences were recombined into Gateway-converted pAM-PAT-MCS. Plasmids were transformed into Agrobacterium tumefaciens strain GV3101:pMP90RK for transformation of Arabidopsis plants using the floral dip method (Logemann et al., 2006).
Recombinant Protein Expression and Purification
A cDNA sequence coding for AK2 with the initiating Met introduced at position 61 to remove the chloroplast targeting peptide was amplified by PCR and cloned into pENTR/D-TOPO, yielding pE AK2_STOP. The rsp1 mutation was introduced by site-directed mutagenesis giving rise to pE AK2rsp1_STOP. Sequences were recombined into pDEST15 (Invitrogen) and confirmed plasmids were transformed into Escherichia coli Rosetta (Novagen). For protein expression, bacteria were grown at 37°C in Luria-Bertani media to an OD600 of 0.6, and expression was then induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside, and bacteria were grown for additional 16 h at 15°C. Bacteria were harvested by centrifugation, taken up in buffer A (50 mM Tris, pH 7.4, 50 mM KCl, 2 mM lysine, 2 mM DTT, 1 mM EDTA, and 10% glycerol) supplemented with Complete Protease inhibitor without EDTA (Roche), DNaseI, and Lysozyme, and lysed by sonification. Lysates were cleared by centrifugation (30 min, 4°C, 30,000g), filtered through 0.22-μm PES membrane filters and batch incubated with 1 mL preequilibrated GST-Sepharose (GE Healthcare) for 1 h. Beads were transferred to 10-mL filter columns (Bio-Rad) and washed with buffer A, then with buffer A containing 300 mM NaCl and finally reequilibrated with buffer A. Proteins were eluted with buffer A containing 10 mM reduced glutathione, concentrated using Vivaspin devices (Sartorius), and buffer exchanged on a HiTrap column (GE Healthcare) equilibrated with buffer B (50 mM Tris, pH 7.4, 50 mM KCl, and 10% glycerol). Proteins were reconcentrated, aliquoted, and stored at −80°C. Purity was confirmed by SDS-PAGE followed by Coomassie Brilliant Blue staining and protein concentration determined by Bradford assay according to the manufacturer (Bio-Rad).
Asp Kinase Activity Assay
AK was assayed using the hydroxamate assay as previously described (Ferreira et al., 2006) with minor modifications. Forty microliters of reaction buffer (25 mM Tris, pH 7.4, 1 mM DTT, 5% glycerol, 2 mM ATP, 1 mM Asp, 10 mM MgSO4, and 500 mM hydroxylamine) was made up to 50 μL with buffer B, proteins contained in buffer B, and/or buffer B containing Lys. After incubation at 35°C, 1 volume of STOP solution was added (0.67 M FeCl3, 0.5 M HCl, and 20% [w/v] trichloractetic acid), and absorbance was measured at 490 nm. The assays were repeated with higher substrate concentrations (5 mM Asp and 10 mM ATP), as these conditions yielded more robust data.
Bioinformatic Analysis
The genomes of Hpa (gene models v8.3; http://vmd.vbi.vt.edu/), Bg (https://www.blugen.org/), and Go (local BLAST server) were queried for sequences with similarity to the sequences of enzymes of AAA and DAP pathway listed in Supplemental Table 1 online using TBLASTn with default settings. Returned sequences producing significant alignments (E ≤ 1e-5) were considered as possible presence of a gene, listed in Supplemental Table 1 online, and used for a reciprocal BLAST search. The first iteration of the PSI-BLAST algorithm (http://www.ebi.ac.uk/Tools/sss/psiblast/) was used with default settings against the UniProt Knowledgebase. From this reciprocal BLAST search, the first entry with an informative annotation was listed in Supplemental Table 1 online to support or not the results of first BLAST searches. Presence of a gene was concluded if an alignment with E ≤ 1e-35 was obtained in the first BLAST search. The result of the reciprocal BLAST was used as judgment for returned results with 1e-35 ≤ E ≤ 1e-5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RAR1 (At5g51700), EDS1 (At3g48090), PR1 (At2g14610), MLO2 (At1g11310), DHDPS1 (At3g60880), DHDPS2 (At2g45440), AK2 (At5g14060), PDF1.2 (AT5G44420), VSP2 (AT5G24770), OPR3 (AT2G06050), JAR1 (AT2G46370), JIN1 (AT1G32640), AOS (AT5G42650), THA2 (AT3G04520), and OMR1 (AT3G10050).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1.Phenotypes of rar1-13 rsp1 and rsp2 Double Mutants.
Supplemental Figure 2. Macroscopic Disease Symptom Formation upon Golovinomyces orontii Infection.
Supplemental Figure 3. Isolation and Characterization of a Col dhdps1-1 Mutant.
Supplemental Figure 4.Germination Phenotype of rsp Mutants.
Supplemental Figure 5. Characterization of a Putative ak2 Loss-of-Function Mutant.
Supplemental Figure 6. Macroscopic Phenotypes of T1 Plants Overexpressing Wild-Type AK2 or Mutant AK2rsp1.
Supplemental Figure 7. Hpa Resistanceof rsp Mutants Is Not rar1-13 Dependent.
Supplemental Figure 8. Effects of Sugar on Growth and Hpa Susceptibility of rsp Mutants.
Supplemental Figure 9. Hpa Infection Structures of Virulent Hpa Isolate Cala2 on rsp1 and rsp2 Mutant Plants.
Supplemental Figure 10. Indole Glucosinolate Content of rsp Mutant Plant Tissues.
Supplemental Figure 11. Homoserine Content of rsp Mutant Plants.
Supplemental Figure 12. Dose Dependency of Hpa Growth Suppression by Thr.
Supplemental Figure 13. Expression of JA-Regulated Genes in rsp Mutant Plants and Thr-Induced Hpa Growth Suppression on JA-Signaling Mutants.
Supplemental Figure 14. Growth of Hpa on Mock- or Threonine-Treated Col Plants.
Supplemental Figure 15. Threonine Content of agd2 Mutant Tissues.
Supplemental Table 1. Comparative Genome Analysis for Genes Coding DAP/AAA Pathway Enzymes in Hyaloperonospora arabidopsidis, Golovinomyces orontii, and Blumeria graminis.
Supplemental Table 2. Oligonucleotides Used in This Study.
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Nottingham Arabidopsis Stock Centre for distribution of Arabidopsis lines. We thank G. Jander and P. Schulze-Lefert for helpful discussions, S. Lagauzere, D. Becker, and N. Moret for technical assistance, and Emiel Ver Loren van Themaat for help with bioinformatic analysis. We also thank L. Nussaume, J. Greenberg, G. Van den Ackerveken, R. Ros, and Y. Yoshioka for providing mutant Arabidopsis seed. This work was funded by the Max-Planck Society, Deutsche Forschungsgemeinshaft funding within Collaborative Research Centre (SFB) ‘635’ (J.E.P. and J.S.), and Deutsche Forschungsgemeinshaft Grant PA 917/3-1 (J.E.P. and S.R.).
AUTHOR CONTRIBUTIONS
J.S. and J.E.P. designed research and wrote the article. J.S., S.R., J.K., P.M., and P.B. performed experiments and analyzed data. H.-M.H. and R.H. performed metabolite profiling. R.G. performed structural modeling and interpretation.
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330: 1549–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Burki F., Shalchian-Tabrizi K., Minge M., Skjaeveland A., Nikolaev S.I., Jakobsen K.S., Pawlowski J. (2007). Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2: e790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Chiba Y., Ishikawa M., Kijima F., Tyson R.H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R.M., Naito S. (1999). Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science 286: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Coates M.E., Beynon J.L. (2010). Hyaloperonospora arabidopsidis as a pathogen model. Annu. Rev. Phytopathol. 48: 329–345 [DOI] [PubMed] [Google Scholar]
- Conrath U., Pieterse C.M., Mauch-Mani B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7: 210–216 [DOI] [PubMed] [Google Scholar]
- Consonni C., Humphry M.E., Hartmann H.A., Livaja M., Durner J., Westphal L., Vogel J., Lipka V., Kemmerling B., Schulze-Lefert P., Somerville S.C., Panstruga R. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Craciun A., Jacobs M., Vauterin M. (2000). Arabidopsis loss-of-function mutant in the lysine pathway points out complex regulation mechanisms. FEBS Lett. 487: 234–238 [DOI] [PubMed] [Google Scholar]
- Curien G., Bastien O., Robert-Genthon M., Cornish-Bowden A., Cárdenas M.L., Dumas R. (2009). Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol. Syst. Biol. 5: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G., Laurencin M., Robert-Genthon M., Dumas R. (2007). Allosteric monofunctional aspartate kinases from Arabidopsis. FEBS J. 274: 164–176 [DOI] [PubMed] [Google Scholar]
- Curien G., Ravanel S., Robert M., Dumas R. (2005). Identification of six novel allosteric effectors of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase isoforms. Physiological context sets the specificity. J. Biol. Chem. 280: 41178–41183 [DOI] [PubMed] [Google Scholar]
- Ferreira R.R., Meinhardt L.W., Azevedo R.A. (2006). Lysine and threonine biosynthesis in sorghum seeds: characterisation of aspartate kinase and homoserine dehydrogenase isoenzymes. Ann. Appl. Biol. 149: 77–86 [Google Scholar]
- Garcia E.L., Mourad G.S. (2004). A site-directed mutagenesis interrogation of the carboxy-terminal end of Arabidopsis thaliana threonine dehydratase/deaminase reveals a synergistic interaction between two effector-binding sites and contributes to the development of a novel selectable marker. Plant Mol. Biol. 55: 121–134 [DOI] [PubMed] [Google Scholar]
- Hakoyama T., et al. (2009). Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 462: 514–517 [DOI] [PubMed] [Google Scholar]
- Heremans B., Jacobs M. (1997). A mutant of Arabidopsis thaliana (L.) Heynh. with modified control of aspartate kinase by threonine. Biochem. Genet. 35: 139–153 [DOI] [PubMed] [Google Scholar]
- Holub E.B., Beynon J.L., Crute I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant Microbe Interact. 7: 223–239 [Google Scholar]
- Hudson A.O., Singh B.K., Leustek T., Gilvarg C. (2006). An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 140: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K., Fujiwara T., Hayashi H., Chino M., Komeda Y., Naito S. (1994). Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine (temporal and spatial patterns of soluble methionine accumulation). Plant Physiol. 104: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Joshi V. (2010). Recent progress in deciphering the biosynthesis of aspartate-derived amino acids in plants. Mol. Plant 3: 54–65 [DOI] [PubMed] [Google Scholar]
- Joshi V., Laubengayer K.M., Schauer N., Fernie A.R., Jander G. (2006). Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18: 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295 [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kim J., Leustek T. (2000). Repression of cystathionine γ-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci. 151: 9–18 [Google Scholar]
- Kotaka M., Ren J., Lockyer M., Hawkins A.R., Stammers D.K. (2006). Structures of R- and T-state Escherichia coli aspartokinase III. Mechanisms of the allosteric transition and inhibition by lysine. J. Biol. Chem. 281: 31544–31552 [DOI] [PubMed] [Google Scholar]
- Kreft O., Hoefgen R., Hesse H. (2003). Functional analysis of cystathionine gamma-synthase in genetically engineered potato plants. Plant Physiol. 131: 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Lee M., Martin M.N., Hudson A.O., Lee J., Muhitch M.J., Leustek T. (2005). Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. Plant J. 41: 685–696 [DOI] [PubMed] [Google Scholar]
- Logemann E., Birkenbihl R.P., Ulker B., Somssich I.E. (2006). An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Rate D.N., Song J.T., Greenberg J.T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Droux C., Curien G., Robert-Genthon M., Laurencin M., Ferrer J.L., Dumas R. (2006). A novel organization of ACT domains in allosteric enzymes revealed by the crystal structure of Arabidopsis aspartate kinase. Plant Cell 18: 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Petersen B.L., Glawischnig E., Jensen A.B., Andreasson E., Halkier B.A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett P.R., Kahn K., Austin M.J., Moisan L.J., Sadanandom A., Shirasu K., Jones J.D.G., Parker J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.J., Panstruga R. (2006). Tête à tête inside a plant cell: Establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 171: 699–718 [DOI] [PubMed] [Google Scholar]
- Parker J.E., Coleman M.J., Szabò V., Frost L.N., Schmidt R., van der Biezen E.A., Moores T., Dean C., Daniels M.J., Jones J.D. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9: 879–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.E., Szabo V., Staskawicz B.J., Lister C., Dean C., Daniels M., Jones J.D.G. (1993). Phenotypic characterization and molecular mapping of the Arabidopsis thaliana locus RPP5, determining resistance to Peronospora parasitica. Plant J. 4: 821–831 [Google Scholar]
- Randall T.A., et al. (2005). Large-scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Mol. Plant Microbe Interact. 18: 229–243 [DOI] [PubMed] [Google Scholar]
- Ravanel S., Block M.A., Rippert P., Jabrin S., Curien G., Rébeillé F., Douce R. (2004). Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279: 22548–22557 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A., Ramos B., Bednarek P., López G., Piślewska-Bednarek M., Schulze-Lefert P., Molina A. (2010). Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 63: 115–127 [DOI] [PubMed] [Google Scholar]
- Sarrobert C., Thibaud M.C., Contard-David P., Gineste S., Bechtold N., Robaglia C., Nussaume L. (2000). Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J. 24: 357–367 [DOI] [PubMed] [Google Scholar]
- Scheible W.R., González-Fontes A., Morcuende R., Lauerer M., Geiger M., Glaab J., Gojon A., Schulze E.D., Stitt M. (1997). Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203: 304–319 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F. (2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62: 840–851 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Sinapidou E., Williams K., Nott L., Bahkt S., Tör M., Crute I., Bittner-Eddy P., Beynon J. (2004). Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38: 898–909 [DOI] [PubMed] [Google Scholar]
- Smith C.A., Kortemme T. (2008). Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J. Mol. Biol. 380: 742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.T., Lu H., Greenberg J.T. (2004). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P.D., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Straus M.R., Rietz S., Ver Loren van Themaat E., Bartsch M., Parker J.E. (2010). Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 62: 628–640 [DOI] [PubMed] [Google Scholar]
- Tornero P., Dangl J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28: 475–481 [DOI] [PubMed] [Google Scholar]
- Tyler B.M., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261–1266 [DOI] [PubMed] [Google Scholar]
- van Damme M., Zeilmaker T., Elberse J., Andel A., de Sain-van der Velden M., van den Ackerveken G. (2009). Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell 21: 2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauterin M., Frankard V., Jacobs M. (2000). Functional rescue of a bacterial dapA auxotroph with a plant cDNA library selects for mutant clones encoding a feedback-insensitive dihydrodipicolinate synthase. Plant J. 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R., Xiao S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21: 2898–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Xu H., Andi B., Qian J., West A.H., Cook P.F. (2006). The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem. Biophys. 46: 43–64 [DOI] [PubMed] [Google Scholar]
- Zhang M., Kadota Y., Prodromou C., Shirasu K., Pearl L.H. (2010). Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol. Cell 39: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.L., Goritschnig S., Dong X.N., Li X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]