This work presents the cucumber mitochondrial genome sequence. Its extremely large size reflects the proliferation of dispersed repeats, large introns, and an abundance of chloroplast and nuclear DNA. The genome has an unusual multichromosomal structure, and computational analyses reveal a large number of recombinationally active repeats.
Abstract
Members of the flowering plant family Cucurbitaceae harbor the largest known mitochondrial genomes. Here, we report the 1685-kb mitochondrial genome of cucumber (Cucumis sativus). We help solve a 30-year mystery about the origins of its large size by showing that it mainly reflects the proliferation of dispersed repeats, expansions of existing introns, and the acquisition of sequences from diverse sources, including the cucumber nuclear and chloroplast genomes, viruses, and bacteria. The cucumber genome has a novel structure for plant mitochondria, mapping as three entirely or largely autonomous circular chromosomes (lengths 1556, 84, and 45 kb) that vary in relative abundance over a twofold range. These properties suggest that the three chromosomes replicate independently of one another. The two smaller chromosomes are devoid of known functional genes but nonetheless contain diagnostic mitochondrial features. Paired-end sequencing conflicts reveal differences in recombination dynamics among chromosomes, for which an explanatory model is developed, as well as a large pool of low-frequency genome conformations, many of which may result from asymmetric recombination across intermediate-sized and sometimes highly divergent repeats. These findings highlight the promise of genome sequencing for elucidating the recombinational dynamics of plant mitochondrial genomes.
INTRODUCTION
The mitochondrial genomes of seed plants are notoriously variable in both size and structure. The known size range spans more than an order of magnitude, with estimates ranging from 208 kb in white mustard (Brassica hirta) to 2.9 Mb in muskmelon (Cucumis melo) (Ward et al., 1981; Palmer and Herbon, 1987). It has long been known that much of this variation is captured by just four species within the flowering plant family Cucurbitaceae (Ward et al., 1981). The mitochondrial genomes of two of these species, watermelon (Citrullus lanatus) and zucchini (Cucurbita pepo), have been sequenced, and their 2.6-fold size difference (379 kb in watermelon and 983 kb in zucchini) reflects in large part vast differences in the amounts of repetitive DNA and integrated chloroplast sequences (Alverson et al., 2010). Limited data on the largest two genomes, from cucumber (Cucumis sativus; ~1.8 Mb) and muskmelon (~2.9 Mb), suggest that they do not contain disproportionately more genes (Stern and Newton, 1985) or chloroplast-derived DNA (Stern et al., 1983; Havey et al., 1998). Although neither genome is thought to contain large-scale segmental duplications (Havey et al., 1998), limited sequencing suggests that short dispersed repeats might account for as much as 13% of the cucumber mitochondrial genome (Lilly and Havey, 2001). In short, the factors underlying the growth of these extraordinarily large mitochondrial genomes remain unclear.
Much of the size and structural variation in plant mitochondrial genomes reflects differences in repetitive DNA content. Repeated sequences are also the substrates for intragenomic recombination and so underlie evolutionary changes in mitochondrial genome organization and structural dynamism in vivo (Lonsdale et al., 1984; Palmer and Shields, 1984). Although the patterns of crossing-over and reciprocal exchange across large (>1 kb) repeats have received much of the attention in this area, it is increasingly clear that recombination involving intermediate-sized repeats (0.1 to 1 kb) also plays an important role in the evolution of plant mitochondrial genomes (Arrieta-Montiel et al., 2009). Unlike large repeats, recombination across smaller repeats tends to be asymmetric, resulting in a small number of just one of the two possible recombination products (Shedge et al., 2007; Arrieta-Montiel et al., 2009). These subsequently can be amplified to a much higher frequency in a process known as substoichiometric shifting (Small et al., 1987; Arrieta-Montiel et al., 2009). This process has been well documented in Arabidopsis thaliana, whose mitochondrial genome contains relatively few intermediate-sized repeats. This has provided a tractable model for characterizing the substoichiometric forms derived from a small and specific set of repeats (Arrieta-Montiel et al., 2009). Alternatively, deep genome sequencing offers a promising way to randomly sample from the entire substoichiometric reservoir and is especially attractive for genomes with larger numbers of intermediate-sized repeats. In addition to shedding light on the substoichiometric shifting phenomenon, deep sequencing might also reveal the extent to which interspecies variation in mitochondrial DNA content reflects differences in the main versus substoichiometric fractions of the genome.
We sequenced the mitochondrial genome of cucumber and identified, to the extent possible, the sources of extra DNA in its large 1685-kb genome, taking advantage of the recently reported nuclear genome sequence of cucumber (Huang et al., 2009). The cucumber mitochondrial genome has an unusual multichromosomal architecture that we characterized with a complementary set of laboratory and computational approaches. In addition, the physical coverage of the genome by random shotgun clones was sufficiently high that we were able to quantify the relative abundances of different genomic conformations and peer into the apparently large pool of substoichiometric forms. These data provide insights into the growth of plant mitochondrial genomes and novel perspectives on the pattern and process of intragenomic recombination in plant mitochondria.
RESULTS
Genome Size, Genes, and Introns
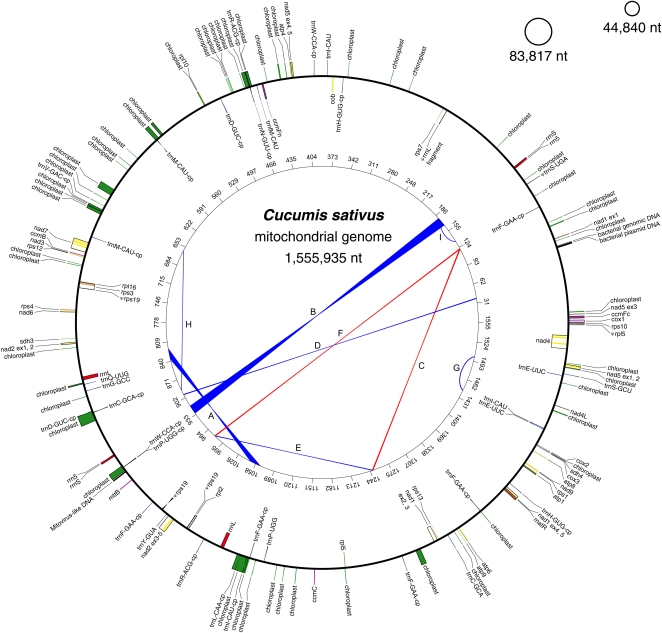
The cucumber mitochondrial genome assembled into three circular-mapping chromosomes of lengths 1,555,935; 83,817; and 44,840 nucleotides (Figure 1). In the “Multichromosomal Genome Structure” section of the Discussion, we discuss the possibility that the two small circles are instead extrachromosomal plasmids as well as the uncertain relationship between circular maps and the in vivo topology of plant mitochondrial genomes. The three chromosomes are nearly identical in nucleotide composition (44.2 to 44.6% G+C content). All of the identifiable, intact mitochondrial genes are on the main 1556-kb chromosome (Figure 1), which is missing just four (rps2, rps11, rps14, and rps19) of the 41 protein-coding genes inferred to have been present in the ancestral seed plant mitochondrial genome (Adams et al., 2002; Sloan et al., 2010) (see Supplemental Figure 1 online). In contrast with rps2 and rps11, whose absence from most eudicot mitochondrial genomes indicates ancient gene losses in this lineage, the phylogenetic distribution of rps14 and rps19 indicates much more recent losses (Adams et al., 2002; Sloan et al., 2010). Although the rps19 loss is specific to the cucumber lineage, the mitochondrial copy of rps14 is a pseudogene in cucumber and other Cucurbitaceae (zucchini and watermelon), suggesting this gene was functionally lost early on in the evolution of the family. Both recently lost genes appear to have been functionally transferred to the cucumber nuclear genome (Huang et al., 2009), which contains an intact mitochondrial rps14 homolog and a mitochondrial rps19 homolog that is split between two contigs. A matching full-length rps19 transcript in the cucumber EST database (http://www.icugi.org/) indicates that the nuclear rps19 is properly transcribed.
Figure 1.
The 1685-kb Mitochondrial Genome of Cucumber.
The genome consists of three circular-mapping chromosomes whose relative abundance varies over a twofold range. For the main 1556-kb chromosome, features on transcriptionally clockwise and counterclockwise strands are drawn on the inside and outside of the circle, respectively. Chloroplast-derived sequences (labeled “chloroplast”) were arbitrarily drawn on the counterclockwise strand. Genes from the same protein complexes are similarly colored, as are rRNAs and tRNAs. The inner circle shows the locations of direct (blue) and inverted (red) repeats (A to I) with the most compelling evidence for recombination activity (see Methods and Supplemental Data Set 2 online). Numbers on the inner circle represent genome coordinates (kb). No features are shown on the two relatively featureless small chromosomes.
Seed plant mitochondrial genomes contain tRNA genes of both chloroplast and mitochondrial origin. The cucumber mitochondrial genome contains 21 intact chloroplast-derived tRNAs, nine of which fall within recently transferred fragments of chloroplast DNA (see below). Cucumber has experienced recent losses of the mitochondrial-type trnF-GAA, trnK-UUU, and trnS-UGA genes (see Supplemental Figure 1 online), all of which are present in most other eudicot mitochondrial genomes (Sloan et al., 2010), including that of watermelon, the most closely related cucurbit with a fully sequenced mitochondrial genome. Interestingly, trnS-UGA appears to have been lost independently in zucchini, another lineage within this family (see Supplemental Figure 1 online). Although no intact trnK-UUU or trnS-UGA homologs are present in the cucumber mitochondrial genome, it does contain five chloroplast-derived trnF-GAA genes. Three of these occur apart from recently integrated regions of chloroplast DNA, raising the possibility that the mitochondrial trnF-GAA was supplanted recently by one or more chloroplast-derived homologs. Finally, cucumber contains the unusual, bacterial-like trnC-GCA previously characterized from Beta (Kubo et al., 2000) that is also present in both watermelon and Vigna (see Supplemental Figure 1 online). The origin and phylogenetic distribution of this gene are the subjects of ongoing study.
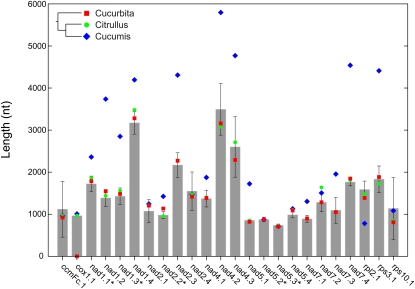
The mitochondrial introns in cucumber reflect a recent history of gain, loss, and, in many cases, major increases in size. The genome contains a highly conserved set of 18 cis- and five trans-spliced group II introns. It also contains a single group I intron, in the cox1 gene, that was acquired by horizontal transfer some 20 million years ago in the common ancestor of watermelon and cucumber (Sanchez-Puerta et al., 2008). Cucumber has experienced the recent loss of a cox2 intron that is still present in both zucchini and watermelon (Alverson et al., 2010). Finally, although three of its introns have shorter than average length, a total of 16 of the 24 introns in the cucumber mitochondrial genome are larger, 14 of them substantially so, than the average length of homologous introns in other seed plants (Figure 2). In total, the 24 introns in cucumber cover 56 kb of the genome (Figure 3). Given minor uncertainties in delimiting the boundaries of trans-spliced introns, this coverage is 35% greater than the corresponding 24 introns in watermelon (Figure 2) (Alverson et al., 2010).
Figure 2.
Many Introns in the Cucumber Mitochondrial Genome Are Substantially Larger Than Homologous Introns in Other Seed Plants.
Mean (±sd) intron lengths were calculated from the set of fully sequenced mitochondrial genomes (except cucumber) (n = 20) for all but the cox1 group I intron, which among the fully sequenced genomes, is present only in watermelon and cucumber. Mean length of the cox1 intron therefore was based on full-length intron sequences in the alignment used by Sanchez-Puerta et al. (2008). Intron sizes for two other cucurbits (Cucurbita [zucchini] and Citrullus [watermelon]) and their phylogenetic relationships to cucumber (Cucumis) are shown to highlight the recency of intron expansions in cucumber. Introns are sequentially numbered beginning from the 5′ end of the gene, and trans-spliced introns in cucumber are marked with an asterisk. Not included are the two introns in the cox2 gene that are widely present in angiosperms (Qiu et al., 1998) but absent from cucumber.
[See online article for color version of this figure.]
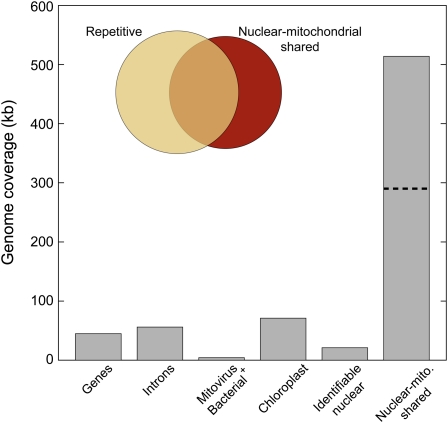
Figure 3.
Coverage of the Cucumber Mitochondrial Genome by Identifiable Coding and Noncoding Features.
Genes includes all protein exons, rRNAs, and tRNAs. Introns include both cis- and trans-spliced forms. The mitochondrial genome contains DNA with identifiable nuclear features (genes and transposable elements) as well as 514 kb of shared nuclear-mitochondrial DNA whose origin and direction of intercompartmental transfer is ambiguous. A little more than half of the nuclear-like sequences are part of the repetitive fraction of the mitochondrial genome, so the dashed line represents the total sequence complexity of the nuclear-like component of the cucumber mitochondrial genome (i.e., each repeated site is counted only once).
[See online article for color version of this figure.]
Using a BLAST e-value cutoff of 1e–6, the cucumber mitochondrial genome shares just 222 kb total sequence with the mitochondrial genomes of two other cucurbit species, zucchini and watermelon. Interestingly, cucumber shares a similar amount of sequence with zucchini (184 kb) and watermelon (168 kb), despite the 2.6-fold difference in size between the mitochondrial genomes of these two species (Alverson et al., 2010). Although many of the shared sequences are located in genic regions that are conserved across all three species, 54 kb of sequence is shared uniquely with zucchini, and 38 kb is shared uniquely with watermelon. Finally, there is little syntenic conservation between cucumber and the two other cucurbits, with most synteny restricted to short regions containing genes and introns (see Supplemental Data Set 1 online).
Horizontal Transfers
As in other seed plants, the large size of the cucumber mitochondrial genome reflects expansions of intergenic regions. Angiosperm mitochondrial genomes commonly contain DNA from diverse extrinsic sources (Stern et al., 1983; Knoop et al., 1996; Bock, 2010), but there have been no reports of recent horizontal sequence acquisitions from bacteria. The main cucumber mitochondrial chromosome contains regions with similarity to β-proteobacterial genomic and plasmid DNA (see Supplemental Table 1 online). The two regions are adjacent, with one of them matching part of a transcriptional regulator gene from the main chromosome of Sideroxydans (BLAST e-value = 6e–29) and the other matching part of a conjugative transfer gene from an Aciodovorax plasmid (BLAST e-value = 0.0) (see Supplemental Table 1 online). The cucumber genome also contains two regions of Mitovirus-derived sequences similar to those found in the mitochondrial genomes of Vitis (Goremykin et al., 2009) and Vigna (Alverson et al., 2011). These bacterial- and viral-derived sequences comprise only a small portion (4.3 kb) of the cucumber mitochondrial genome, especially compared with other categories of sequence of identifiable origin (Figure 3). Aside from the invasive cox1 intron, the cucumber genome shows no evidence for the acquisition via horizontal transfer of whole genes or major portions thereof from other plant mitochondrial genomes.
Intracellular Transfers
The cucumber mitochondrial genome contains an abundance of sequences transferred from the chloroplast and nuclear genomes. We found a total of 71 kb of chloroplast-derived DNA distributed across 53 different regions of the mitochondrial genome, two of which are >10 kb in length (Figures 1 and 3; see Supplemental Data Set 1 online). Each distinct chloroplast fragment is colinear with its corresponding region in the chloroplast genome, though some fragments that are disparately spaced in the chloroplast genome are adjacent in the mitochondrial genome. In absolute terms, only zucchini contains more chloroplast-derived mitochondrial DNA than cucumber (Alverson et al., 2010).
Nuclear-derived sequences are considerably more difficult to detect than chloroplast sequences. Moreover, the large amounts of noncoding sequence in plant mitochondrial and nuclear genomes make it especially difficult to determine the direction of sequence transfer (mitochondrion-to-nucleus or vice versa) for sequences without characteristic features of one or the other genome (Notsu et al., 2002). The cucumber mitochondrial genome contains roughly 21 kb of sequence with identifiable nuclear features, including nuclear pseudogenes (1.2 kb) and transposable elements (20 kb) (Figure 3). The two pseudogenes are similar to the nuclear lectin protein kinase and mandelonitrile lyase pseudogenes found in other angiosperm mitochondrial genomes (Alverson et al., 2010, 2011), suggesting these fragments have been retained in angiosperm mitochondrial genomes for >100 million years (Bell et al., 2010).
We searched the cucumber mitochondrial genome (cv Calypso) against an available cucumber nuclear assembly (cv 9930; Huang et al., 2009) and filtered matches by a relatively stringent set of BLAST e-value cutoffs ranging from 1e–120 to 1e–6. Although the total number of matches varied considerably by e-value (see Supplemental Figure 2 online), overall coverage of the two genomes by shared sequences did not (see Supplemental Figure 3 online). Increasing the e-value stringency dramatically decreased the number of short (<100 nucleotides) matches (see Supplemental Tables 2 and 3 online). In both genomes, many of these short matches overlapped with longer matches, accounting for the apparent disconnect between the number of matches and genome coverage by shared sequences across the range of e-value cutoffs (see Supplemental Figures 2 and 3 online). The following set of results is based on an e-value cutoff of 1e–12.
A total of 0.9% (2.3 Mb) of the cucumber nuclear assembly (Huang et al., 2009) shows similarity to the mitochondrial genome. We identified 233 scaffolds and contigs in the nuclear assembly with >95% coverage by high identity (>98%) mitochondrial hits. The nuclear assembly also contains a complete set of full- or nearly full-length mitochondrial genes and introns that are identical (or nearly so) to their cognates in the mitochondrial genome. These findings raise the possibility that the nuclear assembly is contaminated with true mitochondrial contigs. Excluding hits to the 233 putative mitochondrial scaffolds and contigs described above reduced the total number of nuclear-mitochondrial matches by more than half to 22,197 hits, which amounts to 0.5% (1.3 Mb) total coverage of the nuclear genome. The following results are based on this reduced data set.
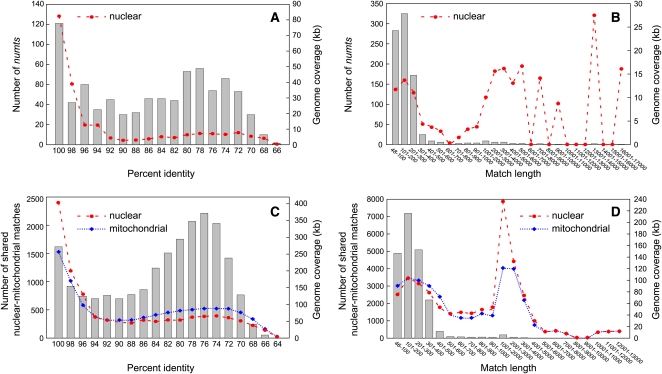
We identified 864 unambiguous mitochondrial transfers to the nuclear genome (numts; Lopez et al., 1994), based on their overlap with mitochondrial genes, introns, or pseudogenes. Numts account for a total of 153 kb (0.1%) of the nuclear assembly. Most of these hits (94%) are <300 nucleotides in length (Figure 4B). The large number of numts with high percent identity to their cognate mitochondrial sequences suggests either a large number of recent transfer events or residual mitochondrial contaminants in the nuclear assembly (Figure 4A). Because of the difficulties in polarizing the direction of sequence transfer for the remaining ~20,000 matches, we treat them as shared nuclear-mitochondrial sequences whose origin and direction of intercompartmental transfer are ambiguous (Notsu et al., 2002). These sequences cover nearly one-third (514 kb) of the cucumber mitochondrial genome (Figure 3), including >90% total combined coverage of the two smaller chromosomes. Most shared nuclear-mitochondrial sequences (95%) are <400 nucleotides in length (Figure 4D), and the distribution of percent identities has a strong peak at 75 to 76% identity (Figure 4C), suggesting that many of the shared sequences date back to a small number of transfer events in the relatively distant past. The secondary peak at 99 to 100% identity represents either numerous recent transfers or residual unfiltered mitochondrial contaminants in the nuclear assembly (Figure 4C).
Figure 4.
Characteristics of Numts and Other Shared Nuclear-Mitochondrial Sequences in Cucumber.
Distributions of lengths and percent identities between nuclear-mitochondrial matches corresponding to identifiable numts ([A] and [B]) and to shared sequences whose origin and direction of intercompartmental transfer are ambiguous ([C] and [D]). Results are based on a BLAST e-value cutoff of 1e–12. The number of matches is shown by gray boxes and plotted on the left ordinate of each panel, whereas nuclear and mitochondrial genome coverage by sequences in each bin is plotted on the right ordinate.
[See online article for color version of this figure.]
More than half of the unpolarized nuclear-mitochondrial sequences are also part of the repetitive fraction of the cucumber mitochondrial genome (Figure 3, inset), which suggests a more limited number of intercompartmental transfers followed by repeated duplication and reshuffling in one or both genomes. Counting once each repeated nuclear-like site in the cucumber mitochondrial genome, the 275 kb of repetitive nuclear-like sequences (Figure 3, inset) reduces to just 51 kb of total sequence complexity. Assuming all 51 kb derives from the nucleus, each nucleotide was duplicated inside the mitochondrial genome an average of five times following the original transfer (or transfers) from the nucleus. Thus, for the entire set of shared nuclear-mitochondrial sequences, counting each nonrepetitive nucleotide site once (239 kb) and just the single-copy fraction (51 kb) of the repetitive nuclear-like fraction gives a total nuclear-like sequence complexity of 290 kb, which represents the maximum amount of recent nuclear-to-mitochondrial sequence transfer in cucumber (Figure 3, dashed line).
Size, Structure, and Content of the Small Chromosomes
Unlike other angiosperm mitochondrial genomes, the cucumber mitochondrial genome has an unusual multichromosomal structure that is not simply the result of ongoing direct-repeat–mediated intragenomic recombination. With physical coverage values in our shotgun libraries of 91 ± 16 clones (1556 kb chromosome), 66 ± 7 clones (84 kb chromosome), and 41 ± 3 clones (45 kb chromosome), the three chromosomes vary in relative abundance by about a factor of two in the DNA sample (made from etiolated seedlings) that we sequenced. There are no large perfect repeats shared between the main and either of the two small chromosomes, and we found no evidence in the genome assembly for recombinational interaction of the main chromosome with either of the smaller chromosomes. However, the smaller chromosomes do share a perfect 3.6-kb repeat that is recombinationally active, as evidenced by conflicts in our clone libraries (Figure 5). Of the 69 clones that span the repeat, 62 (90%) support the split conformation (Figure 5D), which corresponds to a predicted 9:1 (integrated:split) conformation ratio in vivo. Of these 62 clones, 36 derived from the 84-kb chromosome and 26 from the 45-kb chromosome, which is consistent with the relative abundance of these two chromosomes as measured by the overall clone coverage (see above).
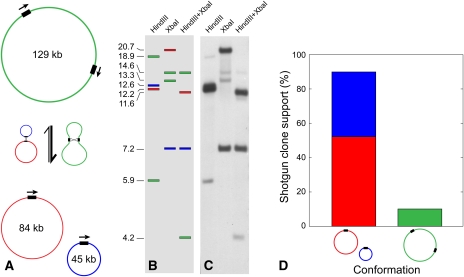
Figure 5.
Homologous Recombination across a Direct Repeat Strongly Favors the Subdivision of a 129-kb Mitochondrial Chromosome into Two Smaller Chromosomes in Cucumber.
Recombination across a 3.6-kb direct repeat (black rectangles) splits a 129-kb (green) mitochondrial chromosome into two subgenomes of lengths 45 kb (blue) and 84 kb (red), and recombination between the latter two reconstitutes the 129-kb chromosome (A). Under the recombination model in (A), the expected (B) and observed (C) patterns from DNA gel blot hybridization of a probe located within the 3.6-kb repeat to purified mitochondrial DNA digested with HindIII, XbaI, and HindIII+XbaI, respectively. The sizes of the bands (kb) are shown to the left of (B). Results show that the 3.6-kb direct repeat mediates homologous recombination. The much stronger hybridization intensity of fragments diagnostic of the 45-kb ([B], blue) and 84-kb ([B], red) chromosomes relative to those of the cointegrated 129-kb chromosome ([B], green) shows that the split conformation predominates (C). Sixty-two (90%) of the 69 informative shotgun clones sequenced for this repeat support the split (45 kb + 84 kb) conformation, whereas just seven of the clones (10%) support the integrated conformation (D). A total of 36 of the 62 clones (58%) supporting the split conformation derived from the 84-kb chromosome (D).
To validate these findings, we performed a DNA gel blot hybridization experiment in which a probe internal to the repeat was hybridized to mitochondrial DNA digested with restriction enzymes that lack cleavage sites within the repeat. This produced a four-band pattern consistent with the expectation (e.g., Palmer and Shields, 1984) for a recombinationally active repeat (Figures 5B and 5C). Consistent with the clone data, the relatively weak hybridization to fragments from the integrated 129-kb chromosome (containing two copies of the repeat in direct orientation) suggests that the split 45 kb + 84 kb conformation substantially outnumbers the integrated conformation (Figures 5B and 5C).
As detailed above, the large 1556-kb chromosome contains all of the intact mitochondrial genes, whereas the 45- and 84-kb chromosomes are devoid of intact known genes. The 45-kb chromosome contains some matches to other sequenced plant mitochondrial genomes and just two open reading frames (ORFs) >150 amino acids in length. It also contains as many as four putatively transcriptionally active regions, based on the comparatively stronger matches (BLAST e-value, percent identity, and/or hit length) of four cucumber unigenes to the 45-kb chromosome than to the nuclear assembly. The 84-kb chromosome is decidedly more plant mitochondrial like in sequence content. It contains a 2-kb region that strongly matches other eudicot mitochondrial genomes, including an rpl5-rps14 pseudogene cluster, which is a common syntenic arrangement in land plants (Takemura et al., 1992). The 84-kb chromosome also contains a 1.1-kb fragment matching the chloroplast inverted repeat and a 1.2-kb fragment of Mitovirus-derived DNA. The 84-kb chromosome contains at least five regions that appear to be transcriptionally active, again based on stronger matches (BLAST e-value, percent identity, and/or hit length) of five cucumber unigenes to the 84-kb chromosome than to the nuclear assembly. One of these five regions contains an ORF >150 amino acids in length. The 84-kb chromosome also contains two ORFs that are 205 and 328 amino acids in length and have overlapping matches to uncharacterized nuclear hypothetical proteins in Ricinus and moderate similarity matches to ESTs from muskmelon.
Repeats and Recombinant Genome Conformations
As foreshadowed by previous limited sequencing of cucumber mitochondrial DNA (Lilly and Havey, 2001), the cucumber mitochondrial genome is rich in repetitive DNA. In total, repetitive sequences cover 605 kb (36%) of the genome. In absolute terms, this is greater than all other sequenced seed plant mitochondrial genomes but is proportionally similar to what was found in zucchini (Alverson et al., 2010, 2011). Although most repeats are <50 nucleotides in length, the genome has a large number of imperfect and overlapping intermediate-sized repeats that are 100 to 400 nucleotides in length (see Supplemental Table 4 online). The main chromosome has just four perfect repeats >1 kb in length (see Supplemental Data Set 2 online). The two largest repeats create duplicate copies of all three rRNA genes (Figure 1). As described above, the small chromosomes share a 3.6-kb perfect repeat (Figure 5).
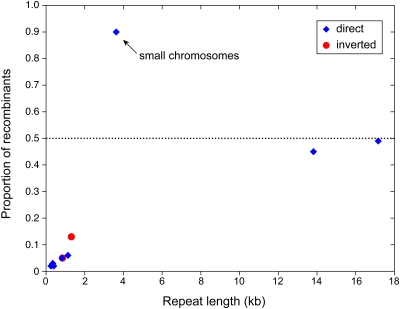
Our computational approach revealed numerous repeats with evidence of recombinational activity. We found 141 repeats in the main chromosome with at least one reconcilable clone, a shotgun clone whose inconsistency with the reference assembly can be reconciled by positing a repeat-mediated recombination event (see Supplemental Data Set 2 online). Nine of these repeats had three or more reconcilable clones, for a combined total of 115 clones, and so showed the strongest support for recombinational activity (Figure 1; see Supplemental Data Set 2 online, repeats A to I). For the largest, 13.8- and 17-kb repeats (both of them direct repeats), the reference and recombinant conformations have a similar abundance (Figure 6), suggesting active and ongoing recombination. Recombinant forms are less abundant for shorter repeats (Figure 6).
Figure 6.
Variation in the Recombinational Equilibria of Repeats on the Large and Small Mitochondrial Chromosomes in Cucumber.
Relative proportion of shotgun clones supporting recombinant genome conformations for 10 repeats with compelling evidence of recombinational activity, based on the support of at least three shotgun clones. In this graph, a repeat with 60 clones supporting the configuration of the reference assembly and 40 clones supporting the recombinant conformation would have a value of 0.4. All repeats shown are on the main chromosome (Figure 1) except for the labeled 3.6-kb repeat on the small chromosomes (Figure 5). As analyzed, direct repeats with points above the dashed line exist predominantly in the split conformation, whereas points below the line favor the integrated conformation.
[See online article for color version of this figure.]
A total of 132 repeats had just one (114) or two (18) reconcilable clones. Both reciprocal recombination products were detected for 5/18 repeats with two reconcilable clones, whereas by definition just one of the two predicted products was detected for the 114 repeats with a single reconcilable clone. To determine the extent to which these 132 repeats represent low-level noise introduced by random chimeric clones, we generated 1000 sets of random clones and reran our analyses on each of these data sets. We found an average of 48.4 (± 6.0) repeats with one or more reconcilable random clones per data set, which corresponds to the expected background number of repeats that will appear recombinationally active based on their support by one or more randomly chimeric clones. Our observed number of repeats with one or two reconcilable clones (132) corresponds to a z score of 13.9 standard deviations from the mean of the null distribution (48.4) and is therefore significantly greater (P < 0.001) than the random expectation under our null model. Although we cannot make conclusions about the recombination activity of any particular one of the 132 repeats with one or two reconcilable clones, these data suggest that a large fraction of them recombine or have done so in the past. The 3.6-kb repeat described above was the only recombinant repeat found on the small chromosomes (Figures 5 and 6).
In addition to recombinant clones, we also looked for sequencing reads that had better matches to hypothetical recombinants than to the reference assembly. These analyses were performed for the main chromosome only. We found 205 reads with a stronger match to a recombinant conformation and a score ≥ 1 (see Methods). Although some of these may represent false positives, we show an example of a read that is the product of recombination across an inverted repeat whose two copies (238 and 248 nucleotides in length) share just 85.6% sequence identity and differ by a 10-nucleotide indel (see Supplemental Figure 4 online). The beginning of the chimeric read perfectly matches the unique upstream flanking region of one copy of the repeat (see Supplemental Figure 4 online, “flanking 1”), and the end of the read perfectly matches the complement of the upstream flanking region of the second copy of the repeat (see Supplemental Figure 4 online, “flanking 3”). The middle repeat–containing portion of the read is a clean chimera of the two divergent repeats (see Supplemental Figure 4C online), and the recombination breakpoint can be narrowed down to a 9-nucleotide window between positions 40 and 48 of the repeat (see Supplemental Figure 4C online). In addition, recombining the genome across this repeat reconciles this clone with the main assembly, based on the criteria outlined above for the analysis of chimeric clones. Thus, both an end-read and the clone from which it derives are entirely consistent with the predicted outcome of recombination across this highly divergent inverted repeat.
DISCUSSION
Foreign Sequences Contribute Significantly to Mitochondrial Genome Expansion in Cucumber
The discovery of bacterially derived DNA in the cucumber mitochondrial genome expands the already extensive list of foreign sequence donors to angiosperm mitochondrial genomes. The bacterial plasmid-like sequence appears to have been transferred from Acidovorax, a destructive seed-borne pathogen and the cause of bacterial fruit blotch in cucumber (Martin et al., 1999) and other cucurbit crops (Makizumi et al., 2011). The mitochondrial genome of Vitis (grapevine) also contains foreign pathogenic sequence, in this case acquired from a grapevine leafroll-associated virus (Goremykin et al., 2009). Although the exact means by which these bacterial sequences were incorporated into the cucumber mitochondrial genome is unclear, they further underscore the amenability of angiosperm mitochondrial genomes to accepting DNA from diverse foreign sources (Bock, 2010).
Chloroplast- and nuclear-derived sequences are common constituents of vascular plant mitochondrial genomes (Stern and Lonsdale, 1982; Knoop et al., 1996; Grewe et al., 2009). The zucchini mitochondrial genome contains a record amount (>113 kb) of transferred chloroplast sequence, accounting for >11% of its nearly 1-Mb mitochondrial genome (Alverson et al., 2010). In absolute terms, the 71 kb of chloroplast sequence in cucumber, though high by comparison to other seed plants, illustrates that the amount of transferred chloroplast DNA does not scale proportionally with genome size.
Plant mitochondrial and nuclear genomes each contain large amounts of noncoding and seemingly featureless sequence, making it difficult to determine the direction of transfer for sequences shared between the two genomes (Notsu et al., 2002). Cucumber is one of only a few plants with fully sequenced chloroplast (Kim et al., 2006), nuclear (Huang et al., 2009), and mitochondrial genomes (this article), which allowed us to characterize patterns of intercompartmental sequence exchange to an extent that is usually not possible. An important caveat is that we have compared the mitochondrial genome of one genetic line (Calypso) to the nuclear genome of a different genetic line (9930), and it is well documented that conspecific genetic lines can vary greatly in mitochondrial genome size and sequence content (Satoh et al., 2004; Allen et al., 2007) as well as numt content (Lough et al., 2008; Roark et al., 2010).
Regardless of how one polarizes the observed transfers, our analyses suggest a relatively high degree of sequence exchange between the mitochondrial and nuclear genomes in cucumber. Based on their matches to identifiable plant mitochondrial features, we identified 864 numts in the cucumber nuclear assembly (Figure 4). This number does not account for the possibility of numt duplications following the initial insertion into the nuclear genome, which is common (and considerably more easy to reconstruct) in human (Hazkani-Covo et al., 2003). The 864 identifiable numts account for 0.1% of the cucumber nuclear genome, which is within the range of what has been found in other angiosperms (Richly and Leister, 2004b; Hazkani-Covo et al., 2010). Some of the ~20,000 unpolarized nuclear-mitochondrial matches likely represent numts, so our estimate of numt content in cucumber is almost certainly a conservative one. Similar to other plants (Notsu et al., 2002; Richly and Leister, 2004a), most numts in cucumber are short, reflecting mitochondrial sequence transfer in small bits and/or rapid fragmentation and decay following insertion of larger fragments.
Nearly one-third of the 1685-kb cucumber mitochondrial genome consists of shared nuclear-mitochondrial sequences whose direction of transfer could not be polarized with confidence. For both numts and the unpolarized class of shared sequences, the continuous distribution of percent identities suggests a history of ongoing sequence exchange (Figure 4). For the shared sequences, however, the strong peak at 73 to 78% identity suggests that many of these sequences are the result of one or a few large-scale transfer events that occurred in the relatively distant past. The secondary peak at 98 to 100% is more difficult to interpret in light of concerns about mitochondrial contaminants in the nuclear assembly. Resolving the history of these sequences is confounded by the highly repetitive nature of the cucumber mitochondrial genome, which has grown, at least in part, through a process of continuous sequence duplication and reshuffling (this study; Lilly and Havey, 2001). This hypothesis is supported by the nuclear-like sequences, some of which appear to have been duplicated an average of five times since their arrival in the mitochondrial genome.
To summarize, these results suggest that some balance of nuclear sequence acquisition and internal duplication have contributed substantially to the fourfold size difference between the mitochondrial genomes of cucumber and its closest examined relative, watermelon (Alverson et al., 2010), which diverged roughly 20 million years ago (Schaefer et al., 2009). Further refinement of the cucumber nuclear assembly used here (Huang et al., 2009) and analyses of nuclear-mitochondrial genome pairs from the same cucumber cultivar and other cucurbit species should help polarize and date the transfer of these shared sequences, allowing us to better identify those sequences in the cucumber mitochondrial genome that derive from the nucleus.
Gene Loss and Intron Growth in the Cucumber Mitochondrial Genome
Despite its large size, the cucumber mitochondrial genome has experienced several recent gene losses, including two ribosomal protein and three tRNA genes. Ongoing functional transfer of ribosomal protein genes to the nucleus is common in angiosperms (Adams et al., 2002), but the recently reported mitochondrial genomes of Silene latifolia (an extreme case; Sloan et al., 2010), zucchini (Alverson et al., 2010), and now cucumber suggest that tRNA genes might be similarly subject to loss and functional replacement. The recent losses of two mitochondrial trnS variants in zucchini (Alverson et al., 2010) and the mitochondrial trnF in cucumber, along with concomitant acquisitions of chloroplast-derived homologs, suggest that these represent recent functional replacements, making them promising candidates for studies of functional tRNA replacement to address questions such as: How many of the five chloroplast-derived trnF homologs in the cucumber mitochondrial genome function in mitochondrial translation? And, in the early stages of replacement, are multiple copies of a recently co-opted tRNA necessary to satisfy the dosage requirements previously met by a single copy of the displaced native gene?
Although the mitochondrial introns in cucumber have experienced unprecedented levels of expansion compared with other seed plants (Figure 2), introns account for very little of the overall large size of the cucumber mitochondrial genome. Previous sequencing showed that the nad1, nad4, and nad7 introns are substantially larger in cucumber than in other angiosperms (Bartoszewski et al., 2009). Our genome-wide survey showed varying degrees of expansion in 16 of the 24 introns in cucumber. Most of these expansions occurred recently, in the time since the evolutionary split from watermelon some 20 million years ago (Figure 2). Some of the expansions were quite dramatic, resulting in introns that are 250% larger than their counterparts in watermelon. In many cases, the expansions appear to have resulted from just one or a few large sequence insertions. It is unclear whether the forces responsible for the unusual expansion of introns in cucumber mitochondrial DNA are related to those responsible for its major expansion in intergenic spacer content and, thus, overall genome size. The cucumber data also indicate that intron size and number can be decoupled, as the growth in average size of introns in cucumber was accompanied by a decrease in intron number compared with other cucurbits.
Multichromosomal Genome Structure
Most seed plant mitochondrial genomes analyzed to date can be mapped as a single circular master chromosome and a collection of submaster (a.k.a., subgenomic) circular chromosomes arising from active recombination across large direct repeats. The cucumber mitochondrial genome instead maps as three entirely or largely autonomous circular molecules. That is, although the two smaller chromosomes recombine with one another, neither of them was found to recombine with the main chromosome. Our shotgun sequencing libraries also showed that the three cucumber chromosomes exist at different levels in our sample of purified mitochondrial DNA. The main 1556-kb chromosome is the most abundant, occurring at a level 2.2 and 1.5 times that of the 45- and 84-kb chromosomes, respectively. The variation in copy number among the three cucumber chromosomes, together with the physical autonomy of the main chromosome relative to the two smaller ones, suggests that they replicate more or less independently of one another.
It is important to note here that circular maps can reflect any or all of at least three different genomic conformations in vivo: actual genome-sized and subgenomic circular molecules (Palmer, 1988), circular and/or linear head-to-tail concatemers of the genome, and an overlapping series of circularly permuted linear molecules (Bendich, 1993). Observations of plant mitochondrial DNA using pulsed-field gels and electron microscopy have failed to reveal a predominance of circular molecules corresponding to the master chromosome and its predicted recombinationally derived subgenomes, showing instead a more complex assemblage of variably sized (including multimeric) linear and circular molecules and highly branched and sigma-like structures, with the latter perhaps representing actively replicating forms (Bendich, 1993; Oldenburg and Bendich, 1996; Backert et al., 1997; Backert and Börner, 2000; Oldenburg and Bendich, 2001). Importantly, however, the observed stoichiometries of the three cucumber mitochondrial chromosomes and their inferred patterns of recombination should apply regardless of whether the recombining units have a circular or linear topology.
Angiosperm mitochondria often contain one or more circular extrachromosomal plasmids (Lonsdale and Grienenberger, 1992). This, together with the absence of any obvious functional elements on the small cucumber chromosomes, raises the possibility that they should instead be recognized as plasmids. However, there are several key differences between the small cucumber circles and the circular plasmids found in plant mitochondria so far. First, the two small cucumber chromosomes are small only in the exceptional context of the enormous 1556-kb main mitochondrial chromosome of cucumber. The combined size of the two small chromosomes is, in fact, 56 times larger than the largest circular mitochondrial plasmids in angiosperms (Lonsdale and Grienenberger, 1992). Indeed, the smallest 45-kb chromosome itself is about 3 times larger than human and most other animal mitochondrial genomes and about 7 times larger than the smallest known mitochondrial genomes (Burger and Lang, 2003). Second, the two small cucumber chromosomes are virtually identical in base composition to the main mitochondrial chromosome, whereas mitochondrial plasmids rarely have the same base composition as the main genome (Lonsdale and Grienenberger, 1992). Third, the small cucumber circles contain a number of mitochondrial and chloroplast pseudogenes—features typically found in the intergenic regions of mitochondrial genomes but never found in circular mitochondrial plasmids. These observations lead us to conclude that the 45- and 84-kb molecules are genomic chromosomes and not extrachromosomal plasmids, adding angiosperm mitochondria to the growing list of eukaryotes with multichromosomal organelle genomes (Watanabe et al., 1999; Zhang et al., 1999; Armstrong et al., 2000; Burger et al., 2003; Lukes et al., 2005; Shao et al., 2009; Vlcek et al., 2011). Among these, the cucumber mitochondrial genome is unusual for both the extreme size disparity between the main and small chromosomes and the apparent lack of coding sequences on the small chromosomes.
An important part of understanding the origin, evolution, and maintenance of this unusual genomic architecture will be to determine what, if any, function the two small cucumber chromosomes might serve. Because pseudogenes are the only identifiable features on the small chromosomes, they may be nothing more than cemeteries for defunct DNA. If so, then these would represent exceptionally large selfish replicons that would not only impose energetic burdens associated with their replication and maintenance, but also in their recruitment of the functionally important mitochondrial recombination machinery (Maréchal and Brisson, 2010) away from the main chromosome (Figure 5). However, if one or both small cucumber chromosomes contain functional sequences, then a relevant precedent may be found in the mitochondrial mini-circles of kinetoplasts. Although they lack canonical mitochondrial genes and for years seemed to lack an identifiable function, these mini-circles eventually were shown to encode guide RNAs essential for RNA editing of the mitochondrial genes contained within maxi-circles (Pollard et al., 1990; Sturm and Simpson, 1990). This illustrates just one way in which functional elements might be hidden in apparently featureless chromosomes.
Differential Recombinational Equilibria of Large Repeats
The three cucumber chromosomes exhibit striking differences in their recombinational dynamics at large perfect repeats. The main chromosome has two pairs of large direct repeats, both active in recombination leading to near equimolar levels of the reference and recombinant genome conformations (Figure 6). This pattern of recombinational equimolarity appears to be the rule for large recombining repeats that have been investigated semiquantitatively in other angiosperm mitochondrial genomes (Palmer and Shields, 1984; Palmer and Herbon, 1986; Stern and Palmer, 1986; Lejeune et al., 1987; Siculella and Palmer, 1988; Folkerts and Hanson, 1989; Coulthart et al., 1990; Siculella et al., 2001; Sloan et al., 2010). By contrast, the 3.6-kb direct repeat on the small cucumber chromosomes occupies a distinctly different recombinational equilibrium, with the split conformation outnumbering the integrated conformation by 9:1 (Figures 5 and 6). A departure of this magnitude presumably reflects differences in the rates of replication and/or recombination dynamics between the integrated and split conformations. However, these two factors are potentially interrelated, as recombination events may in fact initiate the replication of plant mitochondrial DNA (Oldenburg and Bendich, 1996; reviewed in Maréchal and Brisson, 2010). If the replication rates of the three chromosomes are similar, the apparent recombinational differences might be reconciled under a simple model, which presupposes, for kinetic reasons, that recombination rates are higher within than between chromosomes (Figure 7). If so, recombination across direct repeats should lead to disproportionately higher levels of the split conformation, as seen for the small cucumber chromosomes (Figures 5 and 6). The reaction is effectively counterbalanced when two pairs of interspersed and recombining direct repeats are present, as in the main cucumber chromosome (Figure 1). In this case, recombination at one of the direct repeats will simultaneously increase the frequency of the recombinant conformation for itself along with the reference conformation for the other direct repeat (Figure 7), leading to a state of apparent recombinational equimolarity for both pairs of repeats (Figure 7), as seen in the main cucumber chromosome (Figure 6). If correct, this model appears to be specific to this genome because those examined plant mitochondrial genomes which, in their entirety, are analogous to the two smaller cucumber chromosomes in containing but a single pair of large recombining direct repeats (many of them no larger than the 3.6-kb cucumber repeat) appear to be at or near recombinational equimolarity (Palmer and Shields, 1984; Palmer and Herbon, 1986; Stern and Palmer, 1986; Siculella and Palmer, 1988; Siculella et al., 2001).
Figure 7.
A Recombination-Based Model to Account for the Apparent Recombinational Equimolarity of Large Direct Repeats on the Main Mitochondrial Chromosome in Cucumber.
The model may account for the apparent recombinational equimolarity of the largest repeats in the main chromosome versus the nonequimolarity of the large 3.6-kb repeat in the small chromosomes (Figures 5 and 6). Repeats in the master circle are represented by gray and white boxes, with letters identifying their unique flanking sequences. Recombinational equimolarity is the state in which active recombination leads to near-equal concentrations of the four possible configurations for a given direct repeat.
Substoichiometric Forms
Although our Sanger sequencing coverage was substantially lower than what is possible with newer technologies, the physical coverage was sufficiently high to detect potentially hundreds of substoichiometric forms present at frequencies roughly two orders of magnitude lower than the main genome. We found 141 repeats in the main genome that showed some evidence for recombination, as evidenced by one or more clones that are inconsistent with the main assembly but can be reconciled by positing a single repeat-mediated recombination event. Although generally found to be infrequent (Korbel et al., 2007), chimeric clones can be created artificially when unrelated DNA fragments are coligated into a single vector (Sambrook and Russell, 2001). However, our conservative null model showed that the number of recombinant repeats we found was significantly higher than expected given a set of random chimeric clones. Most of these repeats were of intermediate size (107 to 636 nucleotides) and were associated with low frequency recombinants, based on their support by just one or two reconcilable clones. Thus, for most repeats, we found just one of the two possible reciprocal products, which occur at a frequency roughly two orders of magnitude lower than the main genome. We therefore expect that many, if not most, of these are the products of rare asymmetric recombination events of the kind described from the mitochondrial genomes of mutant and wild-type Arabidopsis (Shedge et al., 2007; Arrieta-Montiel et al., 2009). Most of this research is based on targeted observations of the modest number of intermediate-sized repeats in the Arabidopsis mitochondrial genome (Arrieta-Montiel et al., 2009), an approach that would be intractable for a mitochondrial genome as large and repeat-rich as in cucumber. Our random sample of substoichiometric molecules was by no means exhaustive, but it nevertheless hints at the large size and complexity of the substoichiometric pool. Deep sequencing will provide a more complete view of the substoichiometric fraction of the cucumber mitochondrial genome and allow for more precise characterization of the rate of asymmetric recombination across intermediate-sized repeats in wild-type plants.
Although the exact length and percent identity requirements for two repeats to engage in recombination are unknown for plant mitochondrial genomes, research in yeast and Escherichia coli has shown that recombination rate scales positively with repeat length (King and Richardson, 1986; Shen and Huang, 1986; Inbar et al., 2000) and can fall dramatically for repeats with nucleotide mismatches (Shen and Huang, 1986) and especially indels (Bucka and Stasiak, 2001). Abundant evidence shows that these general rules also apply to plant mitochondrial genomes (Arrieta-Montiel et al., 2009; Maréchal and Brisson, 2010). Although recombination can be induced for virtually all of the intermediate-sized repeats with >90% sequence identity in Arabidopsis mutants, recombination across a repeat with 80% sequence identity was not inducible, suggesting that it fell below the minimum identity threshold necessary for the mitochondrial recombination machinery (Arrieta-Montiel et al., 2009). Our screen for chimeric sequencing reads allowed us to identify a recombination event mediated by an inverted repeat whose copies (lengths 238 and 248 nucleotides) differ by 35 nucleotide mismatches and a 10-nucleotide indel (see Supplemental Figure 4 online). The longest contiguous stretch of identical sequence was 50 nucleotides in length, but the pattern of mismatches showed that the recombination breakpoint was well outside of this region and is actually flanked on either side by numerous mismatches and is close to the 10-nucleotide indel (see Supplemental Figure 4 online). It therefore appears that recombination can be mediated by repeats with more sequence divergence than previously documented in plant mitochondria. Notably, mitochondrial recombination across sequences with as much as 23% sequence divergence was recently observed in mussels (Ladoukakis et al., 2011). Data from the cucumber shotgun assembly highlight the flexibility of the mitochondrial recombination machinery in plants and further underscore the promise of even deeper next-generation sequencing for providing insights into the process of intragenomic recombination in plant mitochondrial genomes.
METHODS
Mitochondrial DNA Isolation, Genome Sequencing, and Assembly
Mitochondria were isolated from etiolated seedlings of cucumber (Cucumis sativus cv Calypso) using the DNase I procedure (Kolodner and Tewari, 1972), and mitochondrial DNA was purified from lysed mitochondria by CsCl centrifugation (Palmer, 1982). Fosmid and 8-kb clone libraries were constructed and Sanger sequenced by the U.S. Department of Energy Joint Genome Institute. Detailed sequencing protocols are available at http://www.jgi.doe.gov/sequencing/protocols/prots_production.html. Genome assembly and finishing followed Alverson et al. (2010).
Genome Annotation
We annotated protein coding, rRNA, and tRNA genes as described by Alverson et al. (2010). We then searched the cucumber mitochondrial genome against a database of all previously sequenced seed plant mitochondrial genomes using National Center for Biotechnology Information (NCBI)-BLASTN to delimit the boundaries of putatively functional conserved syntenic regions as described by Alverson et al. (2010). Briefly, these are regions that encompass genes and show strong syntenic and sequence conservation to the mitochondrial genomes of a broad range of taxa (e.g., eudicots or all angiosperms) and likely contain promoters, untranslated regions, and trans-spliced introns. We compared the lengths of cucumber introns to those in other fully sequenced seed plant mitochondrial genomes. The lengths of cis-spliced introns and the outermost boundaries of trans-spliced introns were parsed directly from the GenBank files. The internal trans-spliced boundaries were approximated from breaks in sequence conservation across the set of fully sequenced seed plant mitochondrial genomes (see Supplemental Data Set 1 online).
We identified chloroplast-derived sequences by searching the cucumber mitochondrial genome against a database of representative seed plant chloroplast genomes with NCBI-BLASTN. All regions that did not match conserved syntenic regions or chloroplast-derived sequences were extracted and searched against the Repbase repetitive element database (Jurka, 2000) and the following databases maintained by the NCBI: the non-redundant (nr) nucleotide and protein databases, the whole-genome shotgun database (wgs), and the est_others database.
We used the recently sequenced cucumber nuclear genome (Huang et al., 2009) to identify nuclear-derived sequences in the mitochondrial genome by first searching the cucumber mitochondrial genome against the nuclear assembly with NCBI-BLASTN and filtering hits by a range of BLAST e-value cutoffs (see Supplemental Figures 2 and 3 and Supplemental Tables 2 and 3 online). We used megablast to search the mitochondrial genome against a database of 81,401 unigenes downloaded from the cucurbit genomics database (http://www.icugi.org/) and filtered putative mitochondrial and chloroplast gene transcripts by eliminating all hits that matched to chloroplast-derived regions and mitochondrial gene–containing conserved syntenic regions (see above). All NCBI-BLASTN searches used stringent (word_size 9, gapopen 5, gapextend 2, reward 2, penalty –3, dust no) and/or relaxed (word_size 7, gapopen 8, gapextend 6, reward 5, penalty –4, dust no) parameter settings.
DNA Gel Blot Hybridization
We performed a DNA gel blot hybridization experiment to determine whether the two small (45 and 84 kb) circular-mapping chromosomes recombine across a shared 3.6-kb direct repeat. Purified mitochondrial DNA was digested both separately and together with HindIII and XbaI, neither of which has predicted cut sites within the repeat. Digested DNA was electrophoretically separated on a 0.7% agarose gel and then transferred to a positively charged nylon membrane (Roche) following standard protocols. We used PCR to label a 621-nucleotide region internal to the 3.6-kb repeat with DIG-[11]-dUTP (Roche) and performed the DNA gel blot hybridization and chemiluminescent detection protocol as described by Nakazato and Gastony (2006).
Repeats and Intragenomic Recombination Analyses
Repeated sequences were identified by comparing the genome to itself with the megablast algorithm in NCBI-BLAST 2.2.24+ using default parameters except for word_size = 20. All hits with a raw BLAST score ≥ 25, which corresponds to a 25-nucleotide perfect repeat, were considered repeats for calculations of repeat number and genome coverage. We estimated the number of repeats from the number of unique begin–end coordinates of hits from the megablast search, which overestimates the actual number of repeats in the genome (Alverson et al., 2011). Repeats with a raw blast score ≥100 were included in the following recombination analyses.
The main chromosome was sequenced to an average depth of 10 ± 4 reads and an average physical depth of 91 ± 16 clones. The same shotgun clone libraries resulted in average read and physical depths of 4 ± 2.5 reads and 41 ± 3 clones, respectively, for the 45-kb chromosome and average read and physical depths of 6 ± 3 reads and 66 ± 7 clones, respectively, for the 84-kb chromosome. For the main chromosome, a physical depth of 91 clones means that each nucleotide in the chromosome was contained within an average of 91 different clones that were consistent with the final assembly, whereby a consistent clone is defined as one whose end reads point toward each other and whose separation in the reference assembly is within three standard deviations of the mean insert size for that clone library. Numerous clones failed to satisfy one of these two criteria, possibly because the clone represented an alternate genome conformation that resulted from recombination between genomic repeats in the reference assembly. We identified reconcilable clones (or reconcilable read pairs) as those inconsistent paired-end reads that could be made consistent (i.e., reconciled) by positing a single hypothetical repeat-mediated recombination event, and we used this information to identify and characterize recombinationally active repeats in the cucumber mitochondrial genome. The following analyses were performed on the main chromosome and the integrated (129 kb) conformation of the two small chromosomes.
The initial part of the analysis is similar to the approach used to identify structural variants in the human genome (Korbel et al., 2007). We began by using BLASTN to map all reads to the main reference assembly (e.g., Figure 1) and evaluated the quality of the overall read match as the number of high-quality (Phred score ≥ 20) discrepancies between the read and the reference assembly. Reads with >1 high-quality mismatch in the aligned region or >5 high-quality mismatches outside the aligned region were removed from further consideration, as were all clones in which just one of the two reads mapped to the assembly. Based on these stringent criteria, we mapped 5645 clones to the reference assembly, 577 of which were inconsistent and used to assess recombination. For each repeat, we recombined the genome and reevaluated each inconsistent clone to see whether the rearrangement made that clone consistent as defined above.
The evidence for recombinational activity comes from the support of at least one reconcilable clone. Many repeats had just one or two reconcilable clones, making it necessary to establish a random null model that would give some sense of the expected number of repeats that would appear recombinationally active simply by chance. The size of random chimeric distribution for the null model was based on the total number of inconsistent clones from the main chromosome (513) minus those clones associated with repeats (on the main chromosome) having three or more reconcilable clones (115). Many of the resulting 398 clones are likely associated with genuine low-frequency recombinants involving repeats with just one or two reconcilable clones or, using our assembly as the reference, the products of second- or third-order (or greater) recombination events. Given a set of random clones, our null model therefore should give a conservative estimate of the expected number of repeats with at least one reconcilable clone. The null model was calculated by generating 1000 random sets of read-pair coordinates on the genome using the lengths of the 398 clones that could be random chimerics. For each random data set, we iterated over all the repeats, recombining the genome across each one to determine whether any of the randomly generated chimeric clones was reconcilable. For each set of random clones, we calculated the number of reconcilable clones, and the final expected number of randomly chimeric/reconcilable clones was the mean calculated across the 1000 random data sets.
In addition to looking at entire clones that span hypothetical recombinants, we looked at individual Sanger reads spanning hypothetical recombination points involving shorter repeats (less than the length of a Sanger read). These analyses included all repeats with a raw BLAST raw score >49. For identical repeats, we generated the two possible reciprocal products, and for nonidentical repeats, we generated four possible hypothetical recombination products (2 different repeats × 2 reciprocal recombination products each). In reality, however, recombination across an imperfect repeat can generate many different chimeric products depending on the level of dissimilarity between the repeats and the precise location of the recombination breakpoint. We kept 1000 base pairs up- and downstream of the repeat in the hypothetical products to ensure a full-length match between a recombinant read and the hypothetical product. We then used BLASTN to compare each hypothetical product to all of the sequencing reads, rejecting hits with >10 high-quality mismatches in the aligned portion of the read and >30 high-quality mismatches in the unaligned portion of the read. The aligned portion of the read had to extend beyond both ends of the repeat by at least one base. We scored each potentially recombinant read as the number of bases extending beyond the repeat at the end with the shortest extension. The read was rejected if its match score to the putative recombinant was less than half of its match score to the reference assembly.
Accession Numbers
Genome sequences and annotations from this article can be found in the GenBank data library under accession numbers HQ860792, HQ860793, and HQ860794.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Content in Sequenced Seed Plant Mitochondrial Genomes.
Supplemental Figure 2. Number of numts and Unpolarized Nuclear-Mitochondrial Matches at Different BLAST e-Value Cutoffs.
Supplemental Figure 3. Coverage of the Cucumber Nuclear and Mitochondrial Genomes by Shared Nuclear-Mitochondrial Sequences at Different BLAST e-Value Cutoffs.
Supplemental Figure 4. Recombination across an Imperfect Inverted Repeat Creates a Recombinant Genome Configuration with a Chimeric Copy of the Repeat.
Supplemental Table 1. Details of the Cucumber Mitochondrial BLASTN Matches to Bacterial-Derived Sequences.
Supplemental Table 2. Number of numts, Broken Down by Length, at Different BLAST e-Value Cutoffs.
Supplemental Table 3. Number of Shared Nuclear-Mitochondrial Hits, Broken Down by Length, at Different BLAST e-Value Cutoffs.
Supplemental Table 4. Frequencies of Different Sized Repeats in the Cucumber Mitochondrial Genome.
Supplemental Data Set 1. Linear Map of the Main 1556-kb Cucumber Mitochondrial Chromosome.
Supplemental Data Set 2. Recombinant Clone Data for the 141 Repeats in the Main 1556-kb Cucumber Mitochondrial Chromosome with Evidence for Recombination Activity.
Acknowledgments
We thank Arnie Bendich (University of Washington), Sally Mackenzie (University of Nebraska), Susanne Renner (University of Munich), Dan Sloan (University of Virginia), and two anonymous reviewers for critical comments. We thank Dan Croaker and Nischit Shetty (Seminis Vegetable Seeds) for providing the Calypso seed. This work was supported by the National Institutes of Health (1F32GM080079-01A1 to A.J.A. and RO1-GM-70612 to J.D.P.) and the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment to J.D.P. The U.S. Department of Energy Joint Genome Institute provided sequencing and analyses under the Community Sequencing Program supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231.
AUTHOR CONTRIBUTIONS
A.J.A., D.W.R., K.B., and J.D.P. designed the research. A.J.A. and D.W.R. performed the research and contributed new analytic tools. A.J.A., D.W.R., K.B., S.D., and J.D.P. analyzed the data. A.J.A. wrote the article.
References
- Adams K.L., Qiu Y.-L., Stoutemyer M., Palmer J.D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 99: 9905–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.O., et al. (2007). Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177: 1173–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson A.J., Wei X.X., Rice D.W., Stern D.B., Barry K., Palmer J.D. (2010). Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 27: 1436–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson A.J., Zhuo S., Rice D.W., Sloan D.B., Palmer J.D. (2011). The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS ONE 6: e16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong M.R., Blok V.C., Phillips M.S. (2000). A multipartite mitochondrial genome in the potato cyst nematode Globodera pallida. Genetics 154: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel M.P., Shedge V., Davila J., Christensen A.C., Mackenzie S.A. (2009). Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183: 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Börner T. (2000). Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37: 304–314 [DOI] [PubMed] [Google Scholar]
- Backert S., Lynn Nielsen B., Börner T. (1997). The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 2: 477–483 [Google Scholar]
- Bartoszewski G., Gawronski P., Szklarczyk M., Verbakel H., Havey M.J. (2009). A one-megabase physical map provides insights on gene organization in the enormous mitochondrial genome of cucumber. Genome 52: 299–307 [DOI] [PubMed] [Google Scholar]
- Bell C.D., Soltis D.E., Soltis P.S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97: 1296–1303 [DOI] [PubMed] [Google Scholar]
- Bendich A.J. (1993). Reaching for the ring: The study of mitochondrial genome structure. Curr. Genet. 24: 279–290 [DOI] [PubMed] [Google Scholar]
- Bock R. (2010). The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 15: 11–22 [DOI] [PubMed] [Google Scholar]
- Bucka A., Stasiak A. (2001). RecA-mediated strand exchange traverses substitutional heterologies more easily than deletions or insertions. Nucleic Acids Res. 29: 2464–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Forget L., Zhu Y., Gray M.W., Lang B.F. (2003). Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl. Acad. Sci. USA 100: 892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Lang B.F. (2003). Parallels in genome evolution in mitochondria and bacterial symbionts. IUBMB Life 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Coulthart M.B., Huh G.S., Gray M.W. (1990). Physical organization of the 18S and 5S ribosomal RNA genes in the mitochondrial genome of rye (Secale cereale L.). Curr. Genet. 17: 339–346 [DOI] [PubMed] [Google Scholar]
- Folkerts O., Hanson M.R. (1989). Three copies of a single recombination repeat occur on the 443 kb master circle of the Petunia hybrida 3704 mitochondrial genome. Nucleic Acids Res. 17: 7345–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin V.V., Salamini F., Velasco R., Viola R. (2009). Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 26: 99–110 [DOI] [PubMed] [Google Scholar]
- Grewe F., Viehoever P., Weisshaar B., Knoop V. (2009). A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 37: 5093–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havey M.J., McCreight J.D., Rhodes B., Taurick G. (1998). Differential transmission of the Cucumis organellar genomes. Theor. Appl. Genet. 97: 122–128 [Google Scholar]
- Hazkani-Covo E., Sorek R., Graur D. (2003). Evolutionary dynamics of large numts in the human genome: Rarity of independent insertions and abundance of post-insertion duplications. J. Mol. Evol. 56: 169–174 [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E., Zeller R.M., Martin W. (2010). Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 6: e1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Inbar O., Liefshitz B., Bitan G., Kupiec M. (2000). The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275: 30833–30838 [DOI] [PubMed] [Google Scholar]
- Jurka J. (2000). Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16: 418–420 [DOI] [PubMed] [Google Scholar]
- Kim J.S., Jung J.D., Lee J.A., Park H.W., Oh K.H., Jeong W.J., Choi D.W., Liu J.R., Cho K.Y. (2006). Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 25: 334–340 [DOI] [PubMed] [Google Scholar]
- King S.R., Richardson J.P. (1986). Role of homology and pathway specificity for recombination between plasmids and bacteriophage lambda. Mol. Gen. Genet. 204: 141–147 [DOI] [PubMed] [Google Scholar]
- Knoop V., Unseld M., Marienfeld J., Brandt P., Sünkel S., Ullrich H., Brennicke A. (1996). copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 142: 579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K.K. (1972). Physicochemical characterization of mitochondrial DNA from pea leaves. Proc. Natl. Acad. Sci. USA 69: 1830–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J.O., et al. (2007). Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Nishizawa S., Sugawara A., Itchoda N., Estiati A., Mikami T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 28: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis E.D., Theologidis I., Rodakis G.C., Zouros E. (2011). Homologous recombination between highly diverged mitochondrial sequences: Examples from maternally and paternally transmitted genomes. Mol. Biol. Evol. 28: 1847–1859 [DOI] [PubMed] [Google Scholar]
- Lejeune B., Delorme S., Delcher E., Quetier F. (1987). Recombination in wheat mitochondrial DNA: Occurrence of nine different genomic contexts for the 18 S-5 S genes. Plant Physiol. Biochem. 25: 227–233 [Google Scholar]
- Lilly J.W., Havey M.J. (2001). Small, repetitive DNAs contribute significantly to the expanded mitochondrial genome of cucumber. Genetics 159: 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D.M., Grienenberger J.M. (1992). The mitochondrial genome of plants. Cell Organelles, Herrmann R.G., (Vienna, Austria: Springer-Verlag; ), pp. 183–218 [Google Scholar]
- Lonsdale D.M., Hodge T.P., Fauron C.M. (1984). The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 12: 9249–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.V., Yuhki N., Masuda R., Modi W., O’Brien S.J. (1994). Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J. Mol. Evol. 39: 174–190 [DOI] [PubMed] [Google Scholar]
- Lough A.N., Roark L.M., Kato A., Ream T.S., Lamb J.C., Birchler J.A., Newton K.J. (2008). Mitochondrial DNA transfer to the nucleus generates extensive insertion site variation in maize. Genetics 178: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukes J., Hashimi H., Zíková A. (2005). Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48: 277–299 [DOI] [PubMed] [Google Scholar]
- Makizumi Y., Igarashi M., Gotoh K., Murao K., Yamamoto M., Udonsri N., Ochiai H., Thummabenjapone P., Kaku H. (2011). Genetic diversity and pathogenicity of cucurbit-associated Acidovorax. J. Gen. Plant Pathol. 77: 24–32 [Google Scholar]
- Maréchal A., Brisson N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Martin H.L., O'Brien R.G., Abbott D.V. (1999). First report of Acidovorax avenae subsp. citrulli as a pathogen of cucumber. Plant Dis. 83: 965–965 [DOI] [PubMed] [Google Scholar]
- Nakazato T., Gastony G.J. (2006). High-throughput RFLP genotyping method for large genomes based on a chemiluminescent detection system. Plant Mol. Biol. Rep. 24: 245a–245f [Google Scholar]
- Notsu Y., Masood S., Nishikawa T., Kubo N., Akiduki G., Nakazono M., Hirai A., Kadowaki K. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268: 434–445 [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J., Bendich A.J. (1996). Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell 8: 447–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg D.J., Bendich A.J. (2001). Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. 310: 549–562 [DOI] [PubMed] [Google Scholar]
- Palmer J.D. (1982). Physical and gene mapping of chloroplast DNA from Atriplex triangularis and Cucumis sativa. Nucleic Acids Res. 10: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D. (1988). Intraspecific variation and multicircularity in Brassica mitochondrial DNAs. Genetics 118: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D., Herbon L.A. (1986). Tricircular mitochondrial genomes of Brassica and Raphanus: Reversal of repeat configurations by inversion. Nucleic Acids Res. 14: 9755–9764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D., Herbon L.A. (1987). Unicircular structure of the Brassica hirta mitochondrial genome. Curr. Genet. 11: 565–570 [DOI] [PubMed] [Google Scholar]
- Palmer J.D., Shields C.R. (1984). Tripartite structure of the Brassica campestris mitochondrial genome. Nature 307: 437–440 [Google Scholar]
- Pollard V.W., Rohrer S.P., Michelotti E.F., Hancock K., Hajduk S.L. (1990). Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell 63: 783–790 [DOI] [PubMed] [Google Scholar]
- Qiu Y.-L., Cho Y.R., Cox J.C., Palmer J.D. (1998). The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674 [DOI] [PubMed] [Google Scholar]
- Richly E., Leister D. (2004a). NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol. Biol. Evol. 21: 1972–1980 [DOI] [PubMed] [Google Scholar]
- Richly E., Leister D. (2004b). NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 21: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Roark L.M., Hui A.Y., Donnelly L., Birchler J.A., Newton K.J. (2010). Recent and frequent insertions of chloroplast DNA into maize nuclear chromosomes. Cytogenet. Genome Res. 129: 17–23 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sanchez-Puerta M.V., Cho Y., Mower J.P., Alverson A.J., Palmer J.D. (2008). Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol. Biol. Evol. 25: 1762–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Kubo T., Nishizawa S., Estiati A., Itchoda N., Mikami T. (2004). The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genomics 272: 247–256 [DOI] [PubMed] [Google Scholar]
- Schaefer H., Heibl C., Renner S.S. (2009). Gourds afloat: A dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. Biol. Sci. 276: 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R.F., Kirkness E.F., Barker S.C. (2009). The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 19: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge V., Arrieta-Montiel M., Christensen A.C., Mackenzie S.A. (2007). Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H.V. (1986). Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics 112: 441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siculella L., Damiano F., Cortese M.R., Dassisti E., Rainaldi G., Gallerani R., De Benedetto C. (2001). Gene content and organization of the oat mitochondrial genome. Theor. Appl. Genet. 103: 359–365 [Google Scholar]
- Siculella L., Palmer J.D. (1988). Physical and gene organization of mitochondrial DNA in fertile and male sterile sunflower. CMS-associated alterations in structure and transcription of the atpA gene. Nucleic Acids Res. 16: 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D.B., Alverson A.J., Storchová H., Palmer J.D., Taylor D.R. (2010). Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol. Biol. 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I.D., Isaac P.G., Leaver C.J. (1987). Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial genome diversity in maize. EMBO J. 6: 865–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Lonsdale D.M. (1982). Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature 299: 698–702 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Newton K.J. (1985). Mitochondrial gene expression in Cucurbitaceae: Conserved and variable features. Curr. Genet. 9: 395–404 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Palmer J.D. (1986). Tripartite mitochondrial genome of spinach: Physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 14: 5651–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Palmer J.D., Thompson W.F., Lonsdale D.M. (1983). Mitochondrial DNA sequence evolution and homology to chloroplast DNA in angiosperms. Plant Molecular Biology, ICN-UCLA Symposia on Molecular and Cellular Biology, Goldberg R.B., (New York: Alan R. Liss; ), pp. 467–477 [Google Scholar]
- Sturm N.R., Simpson L. (1990). Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell 61: 879–884 [DOI] [PubMed] [Google Scholar]
- Takemura M., Oda K., Yamato K., Ohta E., Nakamura Y., Nozato N., Akashi K., Ohyama K. (1992). Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort, Marchantia polymorpha. Nucleic Acids Res. 20: 3199–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek C., Marande W., Teijeiro S., Lukes J., Burger G. (2011). Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 39: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B.L., Anderson R.S., Bendich A.J. (1981). The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell 25: 793–803 [DOI] [PubMed] [Google Scholar]
- Watanabe K.I., Bessho Y., Kawasaki M., Hori H. (1999). Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J. Mol. Biol. 286: 645–650 [DOI] [PubMed] [Google Scholar]
- Zhang Z.D., Green B.R., Cavalier-Smith T. (1999). Single gene circles in dinoflagellate chloroplast genomes. Nature 400: 155–159 [DOI] [PubMed] [Google Scholar]