We constructed a CRE/lox clonal deletion system named BOB (Brother of Brainbow), which allows tissue-specific elimination of gametophytic essential genes from desired cells. The null sectors, marked by a combination of fluorescent proteins, show that genomic removal of RBR leads to cell-autonomous overproliferation of the quiescent center and distal columella stem cells.
Abstract
Mutations that cause lethality in the gametophyte phase pose a major challenge for studying postfertilization gene function. When both male and female haploid cells require a functional gene copy, null alleles cause developmental arrest before the formation of the zygote, precluding further investigation. The Arabidopsis thaliana Rb homolog RETINOBLASTOMA-RELATED (RBR) has an important function in the stem cell niche, but its requirement in both male and female gametophytes has prevented full loss-of-function studies. To circumvent this obstacle, we designed a clonal deletion system named BOB (Brother of Brainbow) in which null mutant sectors marked by double fluorescence are generated in a fully complemented wild-type background. In this system, both copies of a complementing RBR transgene are eliminated by tissue-specific and inducible CRE expression, and homozygous mutant clones can be distinguished visually. Since mutant sectors can be produced in a homozygous, rather than a heterozygous, background, this system facilitates clonal deletion analysis not only for gametophytic lethal alleles but also for any type of mutation. Using the BOB system, we show that RBR has unique cell-autonomous functions in different cell types within the root stem cell niche.
INTRODUCTION
Gene function in multicellular organisms is often revealed through phenotypes conferred by null alleles. However, early requirements for a functional gene product can block development at early stages and prevent studies of later stages. In addition, subtle molecular and cellular changes may precede clear phenotypic characteristics, creating difficulties in assessing the time of onset of first defects.
Currently, two main strategies are used to bypass these problems, both relying on cell-specific gene knockout or knockdown. First, silencing small RNAs (Schwab et al., 2006; Ossowski et al., 2008) can be driven by tissue-specific promoters to facilitate transcript degradation or block translation but are unlikely to completely abolish gene function (Brummelkamp et al., 2002). In addition, small RNAs can move between cell layers and even systemically (Winston et al., 2002; Yoo et al., 2004); thus, their knockdown effect is not fully constrained. Furthermore, small interfering RNAs can influence nontargeted genes (off-targets; Jackson et al., 2003). A second strategy uses manipulation of the DNA to eliminate the coding sequence of a gene of interest (GOI) in desired regions creating a local loss of function. All currently available methods for generating such genetic null sectors are based on loss of heterozygosity. Cells in heterozygotes are depleted of their single wild-type allele, forming a homozygous mutant clone, which is visibly marked (e.g., by a fluorescent protein) in a cell-autonomous manner. Loss of heterozygosity can be induced by several techniques, such as irradiation or site-specific recombination, using the yeast Flp/FRT or the bacteriophage Cre/lox systems (Hoess et al., 1982; McLeod et al., 1986). In animal systems, recombinase enzymes have been expressed tissue specifically and engineered to be inducible (Metzger et al., 1995). Despite their versatility, these systems cannot be readily implemented for mutations in genes that are essential for the production of both gametes, for which heterozygous progenies are rare.
In Arabidopsis thaliana, stem cells are exquisitely sensitive to the dosage of the Retinoblastoma homolog, RBR. Based on hypomorphic mutants, a specific role for this factor has been proposed in the maintenance of the quiescent center (QC), a slowly dividing organizer cell population within the niche, and in progression from the stem cell state toward differentiation (Wildwater et al., 2005). Analysis of RBR functions in the shoot meristem also reported roles in differentiation (Wyrzykowska et al., 2006). However, female gametophytes strictly require the wild-type RBR allele, while transmission of mutant alleles through the male gametophyte has an efficiency of <10% (Ebel et al., 2004). Hence, generating hetero- or hemizygous progenies for clonal deletion analysis by loss of heterozygosity is extremely inefficient; thus, previously described clonal deletion systems (Muzumdar et al., 2007; Adamski et al., 2009) are not applicable for comprehensive analysis of RBR function in the stem cell niche. To circumvent this limitation, we designed a clonal deletion system named BOB (Brother of Brainbow), which allows for region-specific formation of null mutant cells and their detection by double fluorescence in a background harboring two wild-type gene copies. We used the BOB system for analysis of the gametophytic lethal RBR gene and show that RBR is autonomously required in QC and columella stem cells to limit proliferation and in columella daughters to promote differentiation.
RESULTS
The BOB System for Generating Marked Homozygous Deletion Clones
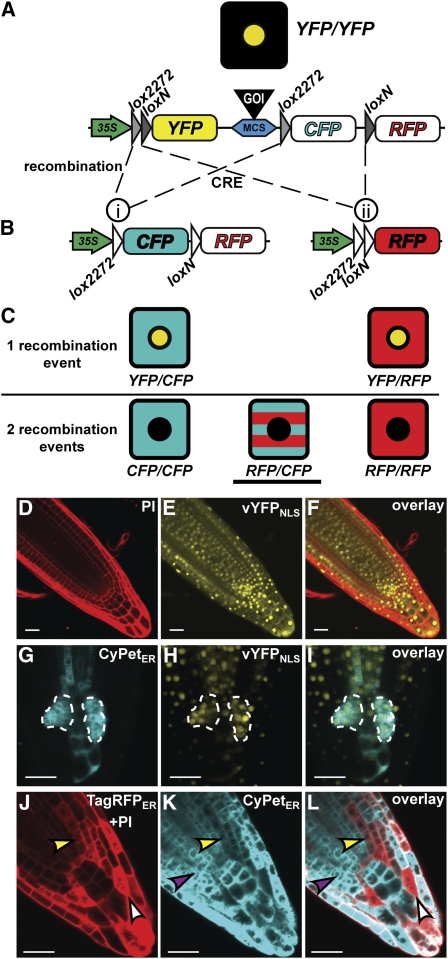
The basic strategy behind the BOB clonal deletion system is that each cell, carrying the homozygous mutant (gametophytic-essential) allele for the GOI, also harbors a single homozygous insertion of the BOB construct, which contains the complementing GOI cloned within it and flanked by lox sites. Induced Cre-dependent recombination mediates the loss of both transgenic copies of the complementing GOI, thus producing null sectors within a wild-type plant. To generate the BOB construct, we combined two different lox site variants with three fluorescent proteins (Figure 1A), a concept that was originally designed to track neuronal networks in brain tissues (Livet et al., 2007). The arrangement of the different lox sites with respect to the GOI and two of the fluorescent proteins is configured such that CRE-mediated deletion of the GOI forms a clone expressing one of two different fluorescent markers. Each deletion of the GOI can generate only one fluorescent signal; hence, double fluorescence can only be the outcome of deletion of both wild-type copies that were located in the BOB T-DNA on the two homologous chromosomes (Figure 1B). These double fluorescence signals thus identify null cells that have lost both wild-type copies of the GOI.
Figure 1.
Induction and Analysis of Clones Using the BOB System.
(A) and (B) The BOB construct before (A) and after (B) CRE-mediated recombination as indicated by dashed lines. Recombination between lox2272 sites (i) fuses the 35S promoter with CypetER. A transcriptional terminator (data not shown) at the 5′ of CyPetER prevents TagRFPER expression. Recombination between loxN sites fuses the 35S promoter to TagRFPER (ii). Colored, filled objects represent active 35S promoter (green), transcribed fluorescent proteins (yellow, cyan, and red), lox sites (gray triangles), and a multicloning site (blue) for cloning a GOI (black). White filled objects represent nonactive modules (i.e., CyPetER and TagRFPER before recombination and lox sites after recombination).
(C) Illustration of possible fluorescently marked cells resulting from one recombination (top) or two recombination events (bottom). Underlined cell depicts the type of clones that were used to identify NHCs in all analyses.
(D) to (F) Confocal images of BOB-RBR roots prior to CRE induction. Visualization of PI-marked cell walls in the red channel (D), vYFPNLS expression prior to CRE induction in the yellow channel (E), and their overlay (F).
(G) to (I) Confocal images of a root tip with two highlighted single recombination clones (dashed enclosure) visualized 5 d postinduction. Highlighted clones analyzed in the cyan channel (CyPetER; [G]), the yellow channel (vYFPNLS; [H]), and in the overlay (I).
(J) to (L) Efficient clone formation using the BOB system upon 1-h HS induction. Clones in a HS:CRE;BOB root tip 3 d after HS induction are shown in the red channel (PI + TagRFPER; [J]), the cyan channel (CyPetER; [K]), and overlay (double fluorescence; [L]). Representative CyPetER, TagRFPER, and double fluorescence clones are denoted by purple, white, and yellow arrowheads, respectively. Bars = 20 μm.
Before recombination, cells ubiquitously express a 35S-driven nuclear-localized venus-yellow fluorescent protein (vYFPNLS, a YFP variant with an enhanced bright signal; Figures 1A and 1D to 1F). Induction of CRE expression leads to intrachromosomal recombination and irreversible loss of the complementing transgene (GOI) together with the vYFPNLS. As a result of this recombination, the 35S promoter is fused to and activates expression of CyPetER (a cyan fluorescent protein variant localized to the endoplasmic reticulum [ER]) or TagRFPER (a red fluorescent protein [RFP] variant localized to the ER, visualized together with propidium iodide [PI]–stained cell walls), thereby marking the cells in which a recombination event has occurred (Figure 1B). A single recombination on one of the homologous chromosomes generates expression of either CyPetER or TagRFPER, while vYFPNLS remains expressed from the nonrecombined BOB copy (Figures 1C and 1G to 1I). Null cells that have experienced two recombination events, one on each chromosome, express CyPetER, TagRFPER, or both (Figures 1C and 1J to 1L) and lose vYFPNLS expression.
To test the efficiency of the BOB system, wild-type plants containing a heat shock (HS) promoter driving the CRE recombinase (HS:CRE) were transformed with the empty BOB construct (without any GOI). Subsequently, selected transformants were heat shocked at 37°C to induce TagRFPER and/or CyPetER expressing clones (Figures 1J to 1L). One-hour heat induction was sufficient to elicit formation of at least one recombination event per cell, and we observed all types of clones outlined in Figure 1C, including double CyPetER/TagRFPER clones (Figures 1C, underlined cell, and 1J to 1L, yellow arrowhead). These double color homozygous clones are especially important as they represent absolute markers for independent excisions on both chromosomes. To exclude the possible effects of recombination and/or expression of high levels of fluorescent proteins on root growth, we heat shocked BOB seedlings segregating for the HS:CRE for 1 h. Formation of broad clones spanning almost every cell did not cause any growth defects, and seedlings with clones (carrying the HS:CRE insertion) were indistinguishable from seedlings with no clones (see Supplemental Figures 1A to 1D online).
The BOB System Allows Clonal Analysis of the Gametophytic Lethal RBR Gene
Before detailed analysis of cell type–specific effects of RBR deletion was possible, we needed to establish an rbr line carrying a single insertion of the BOB construct that contains the complementing wild-type RBR allele. We cloned an 8.4-kb genomic region from wild-type Columbia-0 (Col-0) spanning the RBR gene into the BOB construct (BOB-RBR). For clonal deletion analysis, it is crucial to work with a single BOB-RBR insertion line. Therefore, we identified single insertion transformants based on a DNA gel blot experiments, using an RBR-specific probe (see Supplemental Figure 2A online). These were crossed with rbr-3/+ to generate F1 offspring. Based on the sulfadiazin resistance associated with the rbr-3 allele, we noticed that some of rbr-3/+;BOB-RBR+/− F1 plants generated F2 offspring showing non-Mendelian segregation. We tested whether aneuploidy might cause this abnormal segregation pattern (Johnston et al., 2010). Fluorescence-activated cell sorting (FACS) analysis using inflorescence tissue isolated from the respective F2 plants confirmed that these were triploid (see Supplemental Figure 2D online) and they were discarded. The diploid lines (see Supplemental Figure 2C online) were left to self-pollinate and generate the desired rbr-3/rbr-3;BOB-RBR+/+ offspring. Single F2 plants were tested again for a single BOB-RBR insertion as described above, now using a T-DNA–specific probe (see Supplemental Figure 2B online).
We then sought to determine whether the vYFPNLS expression can be used as an indicator for deletion of the vYFPNLS and RBR genomic sequences by monitoring YFP fluorescence after the formation of clones. Analysis of the root tip, 2 d after 1-h HS, reveals that clones are formed in almost all cells and tissues (see Supplemental Figures 3A and 3C online). Although there is a sharp reduction in vYFPNLS fluorescence upon induction of clones by prolonged HS (see Supplemental Figures 3B, 3E, and 3F online), vYFPNLS expression remained visible up to several days in small null homozygous clones (NHCs) expressing both TagRFPER and CyPetER (Figure 1C, underlined cell). Thus, only cells expressing both CyPetER and TagRFPER were used to unequivocally identify NHCs regardless of the remaining nuclear YFP.
After establishing a complemented rbr-3/rbr-3;BOB-RBR+/+;HS:CRE line, we compared the phenotypes of roots with broad deletion clones (2-h HS) to those having a reduction in RBR levels as previously described (Wildwater et al., 2005). Indeed, using the BOB system, we observed similar phenotypes (i.e., proliferation in the stem cell niche, inhibition of differentiation in the columella, and cell death in columella and vascular tissues) (see Supplemental Figure 4 online; see below). Together with the reduction in vYFPNLS expression, the similarity between these phenotypes indicates that BOB clones truly represent excision of the genomic RBR sequence.
Tissue-Specific Generation of BOB Clones
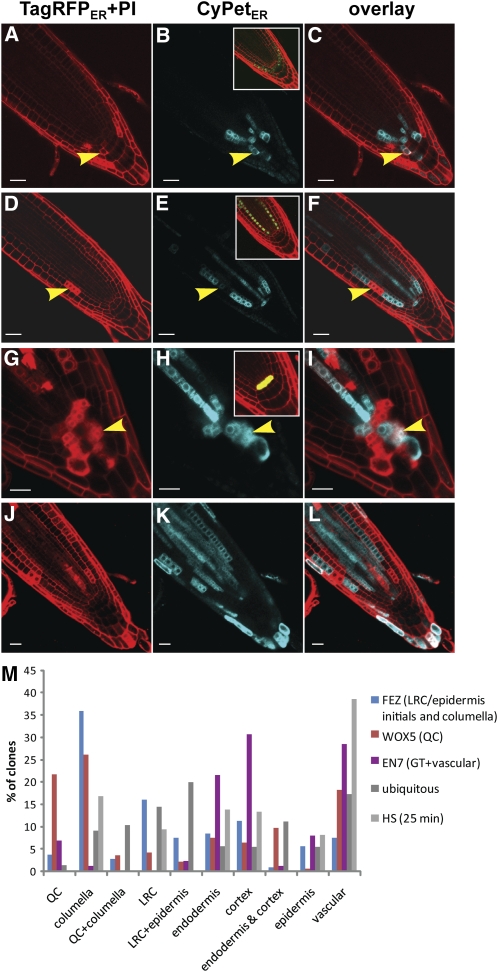
Our previous experiences indicated that it is difficult to obtain specific and small clones in the QC and stem cells upon HS induction using the HS:CRE construct (Heidstra et al., 2004). We applied a two-step strategy to overcome these difficulties. First, we used a CRE-GR (for CRE recombinase, fused to the ligand binding domain of a mutated human glucocorticoid receptor) protein fusion (Brocard et al., 1998), which allows activation of CRE by inducing its nuclear translocation upon dexamethasone (dex) application. Second, we combined this fusion with several tissue-specific promoters to drive transcription of CRE-GR in the particular tissues of interest. We then transformed rbr-3/rbr-3;BOB-RBR+/+ lines with these constructs. This system for tissue-specific CRE-GR activation allowed significant enrichment of clones in desired regions (Table 1). We applied several promoters, among which the FEZ promoter driving expression in the columella and epidermis/lateral root cap stem cells and their daughters (Figure 2B, inset; Willemsen et al., 2008), the EN7 promoter driving expression mainly in the endodermis (Figure 2E, inset; Heidstra et al., 2004), and the WOX5 promoter driving expression in the QC (Figure 2H, inset; Blilou et al., 2005). Almost 60% of the clones induced upon dex treatment of FEZ-driven CRE-GR were formed in the columella, lateral root cap, or distal epidermis, as expected (Figures 2A to 2C, Table 1). Dex induction of CRE-GR expressed from the EN7 promoter facilitated selection of clones in the meristematic ground tissue and vascular tissue (Figures 2D to 2F, Table 1). Upon dex application, WOX5 promoter–driven CRE-GR resulted in recombination and generation of deletion clones in the QC (22% of the clones) and in the surrounding stem cells (Figures 2G to 2I, Table 1).
Table 1.
Correlation between Tissue Distribution of Clones and Promoter-Specific CRE-GR Activity
| Position | FEZ (LRC/Epidermis Initials and Columella) n = 12 | WOX5 (QC) n = 82 | EN7 (GT) n = 21 | Theoretical Ubiqiutous; n = 3 | HS (25 min); n = 11 |
| QC | 4 (3.8) | 79 (21.8) | 6 (6.8) | 3 (1.3) | 0 (0) |
| Columella | 38 (35.8) | 95 (26.2) | 1 (1.1) | 19 (9) | 39 (16.7) |
| QC + Columella | 3 (2.8) | 13 (3.6) | 0 (0) | 21 (10.3) | 0 (0) |
| LRC | 17 (16) | 15 (4.1) | 0 (0) | 30 (14.5) | 22 (9.4) |
| LRC + Epidermis | 8 (7.5) | 8 (2.2) | 2 (2.3) | 41 (20) | 0 (0) |
| Endodermis | 9 (8.5) | 27 (7.4) | 19 (21.6) | 11 (5.5) | 32 (13.7) |
| Cortex | 12 (11.3) | 23 (6.3) | 27 (30.7) | 11 (5.5) | 31 (13.3) |
| Endodermis and cortex | 1 (0.9) | 35 (9.6) | 1 (1.1) | 23 (11.1) | 0 (0) |
| Epidermis | 6 (5.7) | 2 (0.6) | 7 (8) | 11 (5.5) | 19 (8.2) |
| Vascular | 8 (7.5) | 66 (18.2) | 25 (28.4) | 36 (17.2) | 90 (38.6) |
| Total | 106 | 363 | 88 | 207 | 233 |
Each number represents the amount of clones visualized early after induction (2 to 3 dpg) and its percentage (in parentheses). We counted clones that were likely to arise by a single recombination event based on the shape and the fluorescence of the clone. LRC, lateral root cap; GT, ground tissue.
Figure 2.
Induction of BOB Clones by Tissue-Specific and HS Promoters.
(A) to (C) Confocal images of clones generated in FEZ:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root 3 dpg on dex-containing medium. The inset in (B) shows the expression domain of the FEZ promoter driving GFP.
(D) to (F) Confocal images of clones generated in EN7:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root 3 dpg on dex-containing medium. The inset in (E) shows the expression of H2B:YFP driven by the EN7 promoter (false green).
(G) to (I) Confocal images of clones generated in WOX5:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root 3 dpg on dex-containing medium. The inset in (H) shows the expression of GFPER from the WOX5 promoter.
(J) to (L) Confocal images of clones generated in HS:CRE;rbr-3/rbr-3;BOB-RBR+/+ root 3 d after HS. Yellow arrowheads point at double fluorescence clones. Bars = 20 μm.
(M) Percentage of clones in particular regions of the root meristem plotted for the three tissue-specific promoters used for driving CRE-GR (colored bars) and the theoretical ubiquitous promoter and HS promoter (see text) used as controls (gray bars).
To evaluate how efficient these promoters are for generating tissue-specific clones, we compared the percentage of tissue-specific clones to two nonspecific alternatives, based on frequencies given in Table 1 (Figure 2M): (1) A theoretical ubiquitous promoter that is expected to generate clones with similar chance for each cell and (2) the HS promoter (Figures 2J to 2L). The region that was selected for counting the number of cells and the number of clones spans 100 μm shootward of the QC to the distal most columella cells. For example, the QC cells constitute ~1.3% of the cells in the root meristem; thus, a truly ubiquitous promoter would be expected to induce QC clones in 1.3% of the cases. The percentage of QC clones generated using the WOX5 promoter was almost 20 times higher than the percentage of QC cells in the root meristems, while the HS promoter never induced clones in QC cells (P ≤ 0.0001 for both comparisons; Fisher’s exact test). The relative number of clones induced by the FEZ promoter in the columella was 4 times higher (38 out of 106) in comparison to a ubiquitous promoter (19 out of 207; P ≤ 0.0001, Fisher’s exact test) and 2.5 times higher in comparison to the HS-induced columella clones (39 out of 233; P ≤ 0.0002, Fisher’s exact test). More importantly for our analysis, HS induced columella clones were confined to the two outer layers and never observed in columella stem cells and daughter cells (Figures 2J to 2L), while half (19 out of 38) of the columella clones induced by the FEZ promoter were found in columella stem cells and daughter cells (P ≤ 0.0002, Fisher’s exact test). Columella stem cell or daughter cell clones are therefore greatly enriched by use of FEZ promoter–driven CRE. Enrichment of ground tissue (endodermis and cortex) clones using the EN7 promoter was more than twofold compared with a presumed ubiquitous promoter (47 out of 88 versus 45 out of 207; P ≤ 0.0001, Fisher’s exact test) and also twice as efficient compared with HS-induced clones (47 out of 88 versus 63 out of 233; P ≤ 0.0001 for both comparisons; Fisher’s exact test).
The first clones were observed in 3 d postgermination (dpg) seedlings on 5 μM dex. Shorter induction times or concentrations below 1 μM lead to a severe reduction in the number of clones per root. After identification of clones, we transferred seedlings to a dex-free medium to avoid further formation of new clones. Nevertheless, in rare cases, new clones appeared up to 3 d after the removal of dex. We could not exclude that these apparently newly emerging clones might have been present in a slightly different focal plane, only becoming visible during time-lapse analysis when roots are presenting a different median plane. Nevertheless, the shape and fluorescence combination of each clone and its neighboring cells were sufficient to reidentify it in later stages and to discriminate new clones from dividing older ones. Interestingly, regardless of the promoter we used for driving CRE-GR, clones were always induced in a subset of cells within the relevant tissue, while many cells did not undergo deletion/recombination of any of the BOB-RBR copies. We concluded that the CRE-GR fusion was only activated over a considerable dex induction threshold and exploited this feature to generate induction mosaics within a given tissue to compare regions with clones to their juxtaposed wild-type cells.
To examine a possible preference of CRE-GR–mediated recombination for either of the two distinct lox variants in the BOB system in planta, we compared the number of CyPetER (lox2272 recombination) and TagRFPER (loxN recombination) expressing clones upon dex induction using the tissue-specific driver lines described above. CyPetER clones, which result by single or double recombination, were almost twice as abundant as the TagRFPER clones (Table 2). This may be caused by the distance between the different lox sites (Coppoolse et al., 2005) or because of differences in their quality as substrates for the CRE recombinase. The frequency of the double fluorescence clones was ~10%, sufficient for efficient detection of NHCs.
Table 2.
Distribution of the Three Clone Types (TagRFPER, CyPetER, and TagRFPER+CyPetER)
| Fluorescence | FEZ; n = 12 | WOX5; n = 82 | EN7; n = 21 | Total |
| CyPetER | 57 | 237 | 68 | 362 (0.63) |
| TagRFPER | 40 | 105 | 19 | 164 (0.29) |
| TagRFPER+CyPetER | 13 | 31 | 4 | 48 (0.08) |
| Total | 110 | 373 | 91 | 574 |
Numbers of each type of fluorescent clone were counted 4 dpg on dex-containing medium in rbr-3/rbr-3;BOB-RBR+/+ carrying tissue-specific promoter driving CRE-GR. The relative proportion of each clone is in parentheses.
Our data demonstrate that tissue-specific activation of CRE-GR is a feasible method in plants to obtain significant enrichment of null clones in desired regions and at preferred time points.
Cell-Autonomous RBR Activity in the QC Constrains Cell Division
RBR has been implicated in several developmental processes, including stem cell maintenance and differentiation (Wildwater et al., 2005; Borghi et al., 2010), but its broad expression domain prevented an accurate functional analysis in specific cells and tissues. The capacity of the BOB system to generate single and isolated null clones allowed us to address three open questions about RBR action in the Arabidopsis root stem cell niche. First, what is the effect of complete RBR removal from different cell types within the stem cell niche? Second, does RBR act in the QC, in the stem cells, or in both? And third, does RBR act cell-autonomously?
To test the effect of RBR depletion from various cell types in the root meristem, we followed clones in this region and compared their behavior to neighbors without excision events. Three-day-old seedlings germinating on dex-containing medium were preselected for the presence of clones under a fluorescence binocular microscope and taken for detailed analysis by confocal microscopy. Relevant NHCs were returned to dex-free growth medium and checked again every 5 to 24 h. Because the WOX5 promoter–driven CRE-GR generated clones in both QC and stem cells upon dex application, we generally used the rbr-3/rbr-3;BOB-RBR+/+;WOX5:CRE-GR line for our analysis. The number and localization of informative clones (clones that originated from a single cell and were followed in more than one time point) used in this study are given in Table 3.
Table 3.
Number and Localization of Informative BOB-RBR Clones
| Location of Clone | NHC | Single Fluorescence Clone | Total |
| QC | 4 | 10 | 14 |
| QC+CSC | 1 | 0 | 1 |
| CSC | 3 | 6(3)a | 9 |
| CSC+CDC | 6 | 5 | 11 |
| CDC | 5 | 3(1)a | 8 |
| QC+CSC+CDC | 2 | 2 | 4 |
| CDiC | 11 | 0 | 11 |
| Endodermis | 3 | 6 | 9 |
| Endodermis+Cortex | 2(1)b | 0 | 2 |
| Cortex | 1 | 3 | 4 |
| Total | 38 | 35 | 73 |
Informative clones are clones that were observed at minimally two time points, usually spanning 24 h. Clones were induced with dex in the rbr-3/rbr-3;BOB-RBR+/+;WOX5:CRE-GR line. CSC, columella stem cell; CDC, columella daughter cell; CDiC, columella differentiated cell. n = 37 roots.
Clones with wild-type phenotype are in parentheses.
A single clone that acquired a second deletion 1 d later is in parentheses.
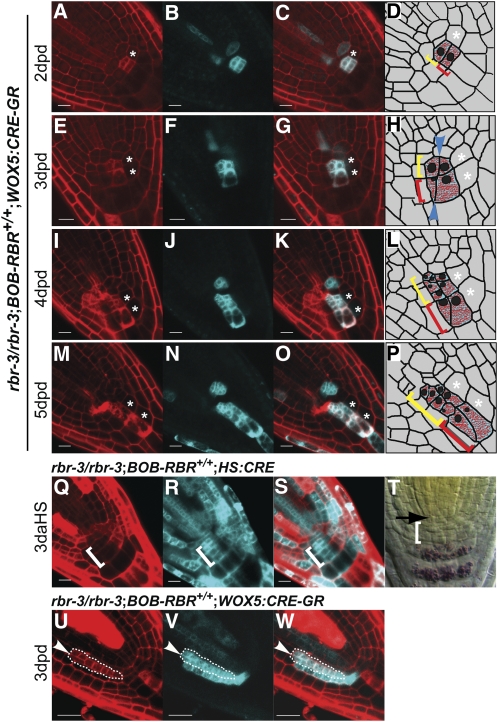
In the case of QC clones, we ensured that each identified clone truly represented a group of cells that originated from a single cell. We followed such clones only if we could visualize a double recombination event at the single-cell stage rapidly after the induction of CRE-GR. We found that null QC cells underwent faster cell divisions with reduced growth periods leading to clusters of small cells originating from the QC and giving rise to new cells within the columella domain (Figures 3A to 3L, white and yellow arrowheads). Figure 3 shows two QC lineages that divide anticlinally toward the columella tissue: a double TagRFPER/CypetER NHC (Figures 3A to 3D, yellow arrowhead) that gives rise to four cells (Figures 3I to 3L, yellow arrowhead) and a single fluorescence TagRFPER clone (Figures 3A, 3C, and 3D, white arrowhead) that gives rise to three cells (Figures 3I, 3K, and 3L, white arrowhead). In contrast with the increased proliferation rate of QC cells lacking RBR, adjacent wild-type QC cells rarely divided (Figures 3C, 3G, and 3K, white asterisk). Since most of the clones expressing single fluorescence marker protein are likely to be heterozygous (Table 2) and the majority of the single fluorescence QC clones showed a similar proliferation phenotype (8 out of 10), we concluded that reduction in RBR levels is sufficient to induce excessive proliferation in the QC. Interestingly, all rbr clones that originated in the QC exclusively populated later columella regions (n = 14).
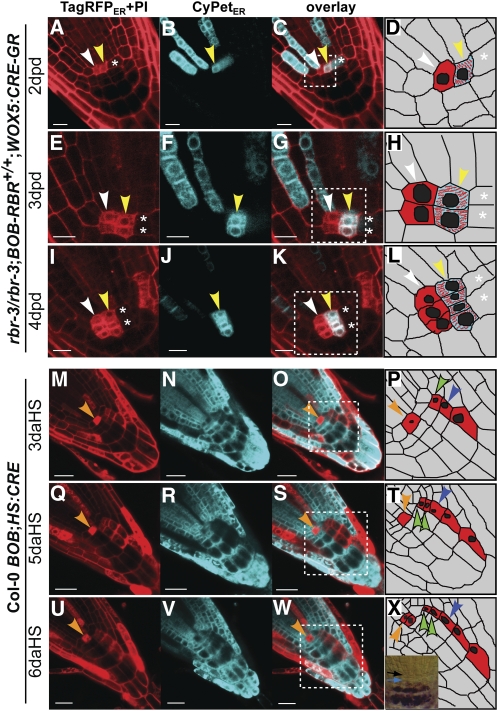
Figure 3.
Loss of RBR in QC Cells Leads to Ectopic Proliferation.
(A) to (L) Confocal images of clones induced in WOX5:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root meristem, analyzed at 2 ([A] to [D]), 3 ([E] to [H]), and 4 ([I] to [L]) d post CRE-GR activation by dex (dpd). White and yellow arrowheads point to ectopically proliferating TagRFPER single fluorescence clone and double fluorescence rbr NHCs, respectively, originating from two QC cells. The white asterisk marks an adjacent slowly dividing wild-type QC cell.
(M) to (X) Confocal images of clones induced in wild-type BOB;HS:CRE root meristems, analyzed at 3 ([M] to [P]), 5 ([Q] to [T]), and 6 ([U] to [X]) d after 1-h CRE induction. Orange arrowhead points to a wild-type slowly dividing TagRFPER-marked QC cell performing a single division in the 3-d time lapse. Green arrowheads ([P], [T], and [X]) point to a columella stem cell before (P) and after division ([T] and [X]). Blue arrowhead points to an expanding columella daughter cell. Inset in (X) depicts the root analyzed in (M) to (X) stained for starch accumulation as a marker for columella differentiation showing one QC (black arrow) and one columella stem cell layer (blue arrow). Bars = 20 μm.
To investigate the progression of cell division in a wild-type situation, we examined clones induced by HS in a wild-type Col-0 harboring HS:CRE and transformed with an empty BOB construct. Similar to the results described above, we observed that clonally marked fluorescent wild-type QC cells (n = 14) rarely divided, as previously reported (Clowes, 1956; Dolan et al., 1993; Figures 3M to 3X, orange arrowhead). To quantify the effect of RBR removal from QC cells, we counted the number of divisions in RBR-deficient cells (BOB-RBR) and compared it to the number of divisions in wild-type marked clones (BOB). Clones lacking one or two copies of RBR (three NHCs and seven single fluorescence clones) had an average of 1.33 divisions per 24 h compared with wild-type clones (n = 14 clones in 14 roots), which had an average of 0.31 divisions per 24 h (P ≤ 0.002, Mann-Whitney U test). Wild-type clones with a QC inception, although dividing much slower than their rbr counterparts, also invaded columella tissue (Figures 3U, 3W, and 3X, orange arrowhead, n = 14), indicating that division products of the QC are preferentially distributed toward the root cap. The stem cell niche organization was not affected by any of the manipulations or generated clones and maintained single QC and columella stem cell layers (Figure 3X, inset, black and blue arrows, respectively) followed by differentiating starch containing columella cells.
Cell-Autonomous RBR Activity in the Columella Constrains Division and Promotes Differentiation
Phenotypic investigation of rbr NHCs in tissues other than QC revealed that columella stem cells lacking RBR showed a similar excessive cell division phenotype as the QC NHCs. For example, we followed an individual rbr columella stem cell NHC (Figures 4A to 4D) that gave rise to six daughter cells (Figures 4M to 4P, yellow bracket) in a time interval when the adjacent wild-type columella stem cell generated only two daughter cells (Figures 4A to 4P, white asterisks).
Figure 4.
Effects of RBR Deletion on Cell Differentiation.
(A) to (P) Confocal images of clones induced in a WOX5:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root meristem analyzed at 2 ([A] to [D]), 3 ([E] to [H]), 4 ([I] to [L]), and 5 ([M] to [P]) d after CRE-GR activation by dex. Right panels ([D], [H], [L], and [P]) represent schematic illustrations of the relevant clones for each time point in the adjacent panels. The yellow bracket marks an NHC originating in the columella before cell division (D), after one periclinal division ([H], top, blue arrowhead) and after a set of two to three irregular divisions ([L] and [P]). The red bracket marks a columella daughter NHC before (D) and after ([H], bottom, blue arrowhead) oblique division. White asterisks mark wild-type columella stem cell and its daughter dividing slowly in comparison to the columella stem cell NHC (yellow bracket).
(Q) to (S) Confocal images of clones induced in a HS:CRE;rbr-3/rbr-3;BOB-RBR+/+ root tip 3 d after HS reveal more layers of unexpanded columella cells within the rbr NHC (white bracket).
(T) Nomarski image of the root analyzed in (Q) to (S) stained for starch accumulation as a marker for columella differentiation showing additional layers at QC (black arrow) and columella stem cell position (white bracket), indicating failure to initiate the normal columella differentiation program of the latter.
(U) to (W) Confocal images of rbr NHC in endodermis tissue (marked by dashed line) induced in a WOX5:CRE-GR;rbr-3/rbr-3;BOB-RBR+/+ root meristem, analyzed at 3 d after CRE-GR activation by dex, reveal an occasional ectopic periclinal division (white arrowhead).
Induced clones are shown in the red (PI + TagRFPER; [A], [E], [I], [M], [Q], and [U]) and cyan (CyPetER; [B], [F], [J], [N], [R], and [V]) channels and in the overlays ([C], [G], [K], [O], [S], and [W]). Bars = 10 μm.
Next, we wanted to determine the differentiation status of the proliferating cells in the rbr null columella clones. However, the combination of confocal imaging and Nomarski optics to visualize starch granules as a marker for differentiation in adjacent cells with and without clones was technically challenging; marked clones identified by confocal microscopy cannot be retraced with certainty using Nomarski optics since the fluorescent marker proteins are no longer visible. Therefore, we took a more general approach using a long (1 h) HS induction in rbr-3/rbr-3;BOB-RBR+/+;HS:CRE seedlings resulting in larger rbr clones and compared the columella of these roots to columella regions in wild-type plants. Induction of broad clones in the columella led to additional layers of unexpanded and undifferentiated columella cells (Figures 4Q to 4T, white bracket; compare with inset in Figure 3X). Lack of starch granules in these proliferating unexpanded cells indicates their failure to initiate the normal columella differentiation program. To quantify the effect of RBR deletion on cell proliferation, we compared the number of cells in columella NHCs versus adjacent regions with a similar area where no clones were visualized. We show that rbr sectors have 2 to 3 times more cells than their adjacent wild-type regions (Table 4; P ≤ 0.04, Wilcoxon sign-rank test). Unlike the observed proliferation in NHC columella stem cells, NHC columella daughter cells resulted in only one to two extra rounds of division (Figures 4A to 4H, red bracket). Nevertheless, wild-type columella daughter cells at the same position do not divide at all (Figures 3P, 3T, and 3X, blue arrowhead) (Dolan et al., 1993). Ultimately these daughter NHCs also expanded (Figures 4H, 4L, and 4P, red bracket) and differentiated (Figure 4T). Null columella daughters committed to differentiation or in expanded starch-containing differentiated columella cells kept growing similar to their wild-type neighboring cells and revealed no phenotypic changes (Figures 4E to 4P, red bracket). We compared the phenotype of rbr columella cells to those of wild-type clones generated by an empty BOB construct. Figures 3M to 3X show a wild-type columella stem cell lineage progression starting with a single cell (Figures 3M, 3O, and 3P, green arrowhead) that follows a typical anticlinal division thereby setting off one stem cell and one daughter cell (Figures 3Q, 3S to 3U, 3W, and 3X, double green arrowheads). The existing columella daughter (Figures 3M, 3O, and 3P, blue arrowhead) did not divide any more (Figures 3Q, 3S to 3U, 3W, and 3X, blue arrowhead). Columella stem cells missing one or two RBR copies performed one division per 24 h (n = 18 clones in seven roots) in comparison to 0.3 divisions per 24 h (n = 13 clones in seven roots) of wild-type (BOB) columella stem cells (P ≤ 0.03, Mann-Whitney U test). Columella daughter cells lacking one or two RBR copies had an average of 0.75 (n = 12 clones in six roots) divisions per day in comparison to RBR wild-type cells that have never displayed any division (n = 7 clones in five roots, P ≤ 0.05, Mann-Whitney U test).
Table 4.
Comparison of Cell Number in Wild-Type and rbr Columella Clones
| Plant No. | CNHC | CWT | CNHC/CWT = Division Ratio |
| 1 | 17 | 9 | 1.9 |
| 2 | 7 | 3 | 2.3 |
| 3 | 7 | 2 | 3.5 |
| 4 | 4 | 2 | 2.0 |
| 5 | 10 | 4 | 2.5 |
| 6 | 5 | 2 | 2.5 |
| Averagea | n/r | n/r | 2.45 |
CNHC, number of cells in NHC; CWT, number of cells without recombination in adjacent region equal in area; n/r, not relevant.
Average proliferation ratio in columella and QC prior to differentiation = [Σ(CNHC/CWT)]/n = 2.45 ± 0.57; n = 6. P ≤ 0.04 (Wilcoxon sign-rank test).
Finally, the orientation of the division plane in rbr columella clones was altered in most cases (20 out of 31 cell divisions at all stages of clone expansion [n = 17]; Figures 4E to 4P), suggesting that in this region, RBR is required for establishing a proper division plane. In this context, it is noteworthy that previous experiments revealed a mild effect of reduction in RBR levels on cell division planes of proximal stem cells, contributing to stele, ground tissue, and epidermis (Wildwater et al., 2005). Consistent with this observation, we found that rbr null endodermis cells occasionally (5 out of 14 analyzed endodermis NHCs) showed an extra periclinal division (Figures 4U to 4W, white arrowhead), whereas cortex and epidermis cells in the proximal meristem were not affected by RBR removal. Normally, such a specific reoriented periclinal division takes place only in the ground tissue daughter cell or, at later stages, in the ground tissue stem cell. Divisions in more proximal ground tissue cells are always anticlinal (Dolan et al., 1993).
Our data using null clones corroborate that the meristem region in close proximity to the stem cell niche is most sensitive to manipulation of the RBR pathway. Importantly, for all clones that gave rise to phenotypes within the stem cell niche, we rarely observed any cellular changes in their immediate unmarked neighbors (e.g., white asterisks in Figure 4, n = 10). These data strongly indicate that RBR acts in a cell-autonomous manner in all cell types studied and that its activity level in the stem cell organizer, columella stem cells, and their immediate daughters contribute to the niche-specific features of these cells.
DISCUSSION
The BOB System as a Versatile Tool for Clonal Analysis
Haploid-essential genes required for both male and female gamete development have hitherto been difficult to study. Our new clonal deletion system overcomes current obstacles and generally simplifies sector deletion analysis for all type of mutations. Moreover, clone formation does not rely on prerequisites such as integration of the lox sites into a specific locus or somatic recombination and G2 entrance (Muzumdar et al., 2007; Wang et al., 2007). With the BOB tool, NHCs can be detected in a homozygous complemented background by double fluorescence, allowing efficient propagation of genotypes that can be directly used for analysis. Furthermore, we demonstrate that the CRE-GR fusion allows for tissue-specific induction of recombination. One potential improvement of the BOB system would be the fusion of a destruction box to vYFPNLS to ensure elimination of the protein at least in dividing cells.
There has been a debate on the persistence of the excised circular product from the CRE-mediated recombination (Srivastava and Ow, 2003; Coppoolse et al., 2005). Our results suggest that this fragment does not play a significant role in our clonal deletion system. First, the observed phenotypes in plants harboring deletion clones are very similar to phenotypes observed using silencing methods (Wildwater et al., 2005; A. Cruz-Ramírez, personal communication). Second, we observed a strong reduction of vYFPNLS levels after HS-induced ubiquitous recombination. Third, CRE is activated by inducible systems (HS or GR/dex). Hence, iterative excision and integration, especially when the concentration of the circular fragment is low, are unlikely. Finally, a reintegration of a circular fragment should change the fluorescence signature of the clone (i.e., vYFPNLS would be reactivated and one of the other fluorescent proteins [CyPetER or TagRFPER] would be disconnected from the 35S promoter and no longer transcribed). We have not observed this type of change in clones. Therefore, we conclude that the BOB system generates NHCs, even though the disappearance of the protein under control of the excised gene will not be instantaneous but rather depend on the turnover rate of the specific protein in each case.
The Tissue Specificity of the CRE-GR Constructs
Previous studies indicated that it is impractical to screen for QC and stem cells clones using HS-inducible CRE (Heidstra et al., 2004). Therefore, we designed a series of tissue-specific CRE-GR constructs for induction of deletion clones in favored regions. This approach was proven to be useful since we obtained considerable enrichment of clones in the target tissues. However, the number of specific clones was still lower than expected. For example, WOX5 is expressed in QC cells (Figure 2H, inset), but only 20% of the WOX5 induced clones were found in the QC. One explanation could be that reduction of RBR levels interferes with maintenance and/or specification of niche cell fates such that WOX5 expression expands to neighbor cells; another explanation may be limited movement of cytoplasmic CRE-GR to neighboring cells via plasmodesmata.
Stem Cell Niche–Specific Functions of the RBR Protein
The removal of RBR activity in marked clones suggests a model in which RBR has two roles in the stem cell niche. RBR acts cell-autonomously in the QC as well as in columella stem cells to restrain cell division. In columella stem cells and their daughters, RBR inhibits cell division, while in daughter cells alone it is required for differentiation. Notably, the requirement of RBR to promote columella cell differentiation is most important in close proximity to the stem cell niche: distal displacement of columella cells ultimately imposed differentiation even at low RBR levels.
Why would RBR act in QC cells to suppress cell division? We note that such a role is consistent with the need to maintain the structural organization of QC cells as organizers of the surrounding stem cells. It is interesting that the quiescence of these cells can be relaxed by RBR regulation, and an important question for the future is whether such a regulation could contribute to the postulated function of the QC as a long term reservoir for the columella stem cell population (Kidner et al., 2000; Jiang and Feldman, 2005; Ortega-Martinez et al., 2007).
Why would QC daughter cells, after division, preferentially populate the distal root cap? This feature has also been observed upon ablation of columella stem cells, which triggers division of the proximal QC cell to replace the ablated cell (Xu et al., 2006). While this can be viewed as a passive consequence of stresses and strains within the root tip, there may be a functional advantage for this preference. The distal-most columella cells are the first ones to encounter physical obstacles during root growth and are frequently detached. Perhaps this exposure to physical stress calls for a renewable reservoir for this particular cell type, where QC cells can supply new columella stem cells.
In columella stem cells, RBR levels need to be balanced, low enough to prevent early differentiation (Wildwater et al., 2005) but sufficient to avoid overproliferation (this study). These data not only support previous conclusions from several analyses of RBR function in leaf and pollen development (Desvoyes et al., 2006; Borghi et al., 2010; Chen et al., 2009) but also stress the point that this balance is relevant in stem cell populations and their immediate daughters. This is well illustrated by the induction of periclinal divisions typical for ground tissue stem cells or their daughters by RBR removal: This division in daughter cells is a transient feature defining a particular differentiation state of the cell as it traverses through the meristem; hence, its regulation by RBR can be interpreted as regulation of a step along a differentiation trajectory. However, RBR is not strictly required for progression of terminal differentiation in cells moving away from the niche; differentiation is delayed in the columella region where this can be easily assessed, but cells ultimately differentiate. What causes this difference in competence? Demonstrated interactions between RBR and a set of stem cell–promoting transcription factors, the PLETHORA proteins, whose expression is highest in the stem cell niche, may be responsible for a difference in sensitivity to RBR levels (Galinha et al., 2007). Graded levels of the plant growth regulator auxin may also play a role in this process (Ding and Friml, 2010).
Recent work has implicated mammalian Rb in fate choice and pluripotency of mesenchymal stem cells (Calo et al., 2010), although this work could not identify the domain of action of Rb at cellular resolution. Such studies underpin the need to dissect Rb-mediated functions at a cellular resolution, which may elucidate whether Rb-like proteins have universal roles in stem cells.
METHODS
Growth Conditions
Seeds were fume sterilized in a sealed container with 100 mL bleach supplemented by 3 mL of 37% hydrochloric acid for 2 to 5 h, and then suspended in 0.1% agarose and plated on a growth medium consisting of Murashige and Skoog salts, 1% Suc, 0.8% plant agar, MES, pH 5.8, 50 μg/mL ampicillin, and 1 to 5 μM dex (optional), stratified for 2 d in a 4°C dark room, and grown vertically in long-day conditions (16 h light followed by 8 h of dark). For HS induction, plates with 2 to 3 dpg seedlings were placed in a 37°C incubator for 1 h and analyzed 2 d later.
Microscopy
Seedlings harboring red or cyan clones were preselected under a Leica MZ16F fluorescence stereoscope and further analyzed by confocal microscopy. To excite and collect red, cyan, and yellow fluorescent signals in a Leica SP2 confocal microscope, we performed sequential scanning as follows: the CyPetER and the vYFPNLS were excited together using the 458- and 514-nm laser wavelengths, respectively, and emission was collected at 465 to 506 nm for the CyPetER and 523 to 566 nm for the vYFPNLS. PI, which marks cell walls and dead cells (3 μg/mL, final concentration), and TagRFPER were visualized by exciting at 488 and 543 nm, respectively, and emission was collected at 502 to 522 and 561 to 633 nm. Fluorescence signal intensity was measured using the “quantify” function in the Leica TCS SPII confocal software in a region of interest excluding overexposed areas that contain dead cells.
Although signals from cell walls, dead cells, and TagRFPER marked clones are collected using the same filter settings, they are clearly distinguishable from one another based on the subcellular localization of the fluorescence. PI marked walls of living cells appear as a rectangular outline. Dead cells accumulate PI within the cytoplasm and nucleus and show a high emission intensity coupled with distorted cell shape. TagRFPER clones are characterized by signal from the ER surrounding the circular nucleus.
Cloning
The BOB construct (Figure 1A) consists of a pGreenII backbone (Hellens et al., 2000) with a 35S promoter driving vYFPNLS (Nagai et al., 2002) as a default expressed marker and two incompatible lox variants, lox2272 and loxN, placed in between the 35S and vYFPNLS sequences. Despite our attempt to generate a fast turnoff of the vYFPNLS by a general SV40 nuclear localization signal (NLS; Lassner et al., 1991) as a second indication for genomic deletion of the GOI, vYFPNLS was still visible up to several days in NHCs expressing TagRFPER and CyPetER. Downstream to the 3AT (vYFPNLS terminator), we placed a multicloning site for insertion of the complementing wild-type genomic allele of any GOI, including its native promoter followed by two fluorescent proteins, CyPetER (Nguyen and Daugherty, 2005) and TagRFPER (Merzlyak et al., 2007), with the lox2272 and loxN at their 5′, respectively. To generate the BOB plasmid, four fragments were amplified from the template plasmids, pCB1 (Heidstra et al., 2004), 221e pSCR-H2B-VYFP-3AT, P1R4-DR5:ER-CyPet-nosT, and ER-TagRFP CBR_7 using primer pairs 1 and 2, 3 and 4, 5 and 6, and 7 and 8, respectively (see Supplemental Table 1 online). The PCR products and the binary pGII124 vector, carrying a methotrexate resistance marker, were cut with the appropriate restriction enzymes and simultaneously ligated overnight. Positive colonies were analyzed by restriction and sequencing. To clone the genomic fragment of RBR, including the 2.2-kb 5′ and 1.5-kb 3′ regions in pBOB, we amplified it from Col-0 genomic DNA with primers 9 and 10, digested with NheI and ClaI, and ligated it between the NheI and BstBI sites creating pBOB-RBR. We used primer pairs 13 and 14, 15 and 16, and 17 and 18 (see Supplemental Table 1 online) to amplify Col-0 genomic DNA of WOX5, EN7, and FEZ promoters, respectively, and cloned the PCR products by Gateway (Invitrogen). CRE-GR constructs (Brocard et al., 1998) driven by tissue-specific promoters were generated by three-way Gateway reaction.
Genetic Background and Crossing Scheme
pBOB and pBOB-RBR were transformed to Col-0 plants carrying a HS:CRE construct by the floral dip method (Clough and Bent, 1998) and tested for vYFPNLS expression before and for CyPetER or TagRFPER expression 24 h after 20 to 60 min HS. To select for a single BOB-RBR insertion in Col-0 HS:CRE background, we used DNA gel blotting. DNA from T1 plants was digested with XbaI and hybridized with an RBR-specific probe generated by PCR using primers 11 and 12 on Col-0 DNA. A single insertion line was used as a female gametophyte donor to cross with rbr-3/+ plants (GABI_170G02) that produce escape rbr-3 male gametophytes. Sulfadiazin-resistant (encoded by the GABI T-DNA insertion in the RBR gene) seedlings expressing vYFPNLS were genotyped for the rbr-3 allele, analyzed for ploidy by FACS, and selfed twice. rbr-3/rbr-3;BOB-RBR+/+ plants were selected and verified again for single insertion by second DNA gel blot assay. Plant DNA was digested with HaeIII and probed with a 545-bp NcoI/BglII vector-specific fragment isolated from pGII124. Single-insertion BOB-RBR plants were subsequently transformed with constructs of CRE-GR driven by a tissue-specific promoter.
FACS Sample Preparation
Two to three inflorescences or three to four leaves (without petioles and main vein) were chopped with a fine double-edge razor blade and suspended in 500 μL of cold nuclear isolation buffer (Galbraith et al., 1983). This crude extract was filtered through a 60-μm mesh, stained with 10 μL PI (5 mg/mL), and treated with 5 μL RNaseA (100 μg/mL) for 10 min. These nuclei were analyzed for ploidy by a BD influx cell sorter.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: RBR (locus AT3G12280) and BOB (GenBank JF927991).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The BOB System Does Not Affect Growth.
Supplemental Figure 2. Selection for Single Insertion Diploid rbr-3/rbr-3; BOB-RBR Plants.
Supplemental Figure 3. Reduction of vYFPNLS Fluorescence as a Result of Clone Formation.
Supplemental Figure 4. Genomic Deletion and RNAi Silencing of RBR Display Similar Phenotypes.
Supplemental Table 1. Primer List.
Acknowledgments
CRE-GR sequence in the second Gateway box was a gift from Pierre Chambon (Institute for Genetics and Cellular and Molecular Biology, Strasbourg, France). We are grateful to Amal J. Johnston (Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany), Wilhelm Gruissem (Department of Biology, Eidgenössische Technische Hochschule, Zurich, Switzerland), and Ari Pekka Mähönen (Institute of Biotechnology, University of Helsinki, Finland) for materials, to Jean Livet (Département de biologie du développement, l'Institut de la Vision, Paris, France) and Gabino Sanchez Perez (Bioinformatics group, Utrecht University, The Netherlands) for discussions, to Frits Kindt (Department of Biology, Utrecht University, The Netherlands) for assisting with image processing, and to Ger Arkesteijn (The Faculty of Veterinary Medicine, Utrecht University, The Netherlands) for performing FACS analysis. G.W. was supported by The Netherlands Organization for Scientific Research grant 2005/03618/ALW and by an European Research Council-Advanced investigator grant to B.S. R.H. was supported by The Netherlands Organization for Scientific Research Horizon grant 050-71-054.
AUTHOR CONTRIBUTIONS
G.W. and B.S. designed the research. G.W. performed the research. G.W., R.H., and B.S. wrote the article.
References
- Adamski N.M., Anastasiou E., Eriksson S., O’Neill C.M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Futterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard J., Feil R., Chambon P., Metzger D. (1998). A chimeric Cre recombinase inducible by synthetic, but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 26: 4086–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., Lees J.A. (2010). Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hafidh S., Poh S.H., Twell D., Berger F. (2009). Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc. Natl. Acad. Sci. USA 106: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clowes F.A.L. (1956). Nucleic acids in root apical meristems of Zea. New Phytol. 55: 29–34 [Google Scholar]
- Coppoolse E.R., de Vroomen M.J., van Gennip F., Hersmus B.J., van Haaren M.J. (2005). Size does matter: cre-mediated somatic deletion efficiency depends on the distance between the target lox-sites. Plant Mol. Biol. 58: 687–698 [DOI] [PubMed] [Google Scholar]
- Desvoyes B., Ramirez-Parra E., Xie Q., Chua N.H., Gutierrez C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 107: 22344–22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Ebel C., Mariconti L., Gruissem W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hoess R.H., Ziese M., Sternberg N. (1982). P1 site-specific recombination: Nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. USA 79: 3398–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jiang K., Feldman L.J. (2005). Regulation of root apical meristem development. Annu. Rev. Cell Dev. Biol. 21: 485–509 [DOI] [PubMed] [Google Scholar]
- Johnston A.J., Kirioukhova O., Barrell P.J., Rutten T., Moore J.M., Baskar R., Grossniklaus U., Gruissem W. (2010). Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet. 6: e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C., Sundaresan V., Roberts K., Dolan L. (2000). Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211: 191–199 [DOI] [PubMed] [Google Scholar]
- Lassner M.W., Jones A., Daubert S., Comai L. (1991). Targeting of T7 RNA polymerase to tobacco nuclei mediated by an SV40 nuclear location signal. Plant Mol. Biol. 17: 229–234 [DOI] [PubMed] [Google Scholar]
- Livet J., Weissman T.A., Kang H., Draft R.W., Lu J., Bennis R.A., Sanes J.R., Lichtman J.W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62 [DOI] [PubMed] [Google Scholar]
- McLeod M., Craft S., Broach J.R. (1986). Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Mol. Cell. Biol. 6: 3357–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak E.M., Goedhart J., Shcherbo D., Bulina M.E., Shcheglov A.S., Fradkov A.F., Gaintzeva A., Lukyanov K.A., Lukyanov S., Gadella T.W., Chudakov D.M. (2007). Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 4: 555–557 [DOI] [PubMed] [Google Scholar]
- Metzger D., Clifford J., Chiba H., Chambon P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA 92: 6991–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Luo L., Zong H. (2007). Modeling sporadic loss of heterozygosity in mice by using mosaic analysis with double markers (MADM). Proc. Natl. Acad. Sci. USA 104: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nguyen A.W., Daugherty P.S. (2005). Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23: 355–360 [DOI] [PubMed] [Google Scholar]
- Ortega-Martinez O., Pernas M., Carol R.J., Dolan L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V., Ow D.W. (2003). Rare instances of Cre-mediated deletion product maintained in transgenic wheat. Plant Mol. Biol. 52: 661–668 [DOI] [PubMed] [Google Scholar]
- Wang W., Warren M., Bradley A. (2007). Induced mitotic recombination of p53 in vivo. Proc. Natl. Acad. Sci. USA 104: 4501–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Willemsen V., Bauch M., Bennett T., Campilho A., Wolkenfelt H., Xu J., Haseloff J., Scheres B. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell. 15: 913–922 [DOI] [PubMed] [Google Scholar]
- Winston W.M., Molodowitch C., Hunter C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459 [DOI] [PubMed] [Google Scholar]
- Wyrzykowska J., Schorderet M., Pien S., Gruissem W., Fleming A.J. (2006). Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Hofhuis H., Heidstra R., Sauer M., Friml J., Scheres B. (2006). A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Yoo B.C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y.M., Lough T.J., Lucas W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]