Abstract
5-Aminoimidazole-4-carboxyamide-ribonucleoside (AICAR), a prodrug activator of AMP-activated protein kinase (AMPK), increased hepatic expression of cytochrome P450 4a10, 4a14, and 4a31 mRNAs 2-, 3-, and 4-fold, respectively, and liver microsomal lauric acid ω-hydroxylation increased 2.8-fold. Likewise, mRNA levels of the peroxisome proliferator-activated receptor α (PPARα)-responsive genes, Acox1, Acadm, Cpt1a, and Fabp1, were also increased by AICAR treatment. AICAR did not elicit these changes in PPARα null mice. In isolated murine hepatocytes, AICAR and adenosine produced similar effects, and these responses were blocked by the PPARα antagonist [(2S)-2-[[(1Z)-1-methyl-3-oxo-3-[4-(trifluoromethyl)phenyl]-1-propenyl]amino]-3-[4-[2-(5-methyl-2-phenyl-4-oxazolyl)ethoxy]phenyl]propyl]-carbamic acid ethyl ester (GW6471). Inhibition of AMPK using compound C (dorsomorphin or 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine) did not block the induction of the PPARα-responsive genes by AICAR or adenosine, and 6,7-dihydro-4-hydroxy-3-(2′-hydroxy[1,1′-biphenyl]-4-yl)-6-oxo-thieno[2,3-b]pyridine-5-carbonitrile (A-769662), a non-nucleoside, direct activator of AMPK, did not increase expression of PPARα-responsive genes. An inhibitor of adenosine kinase, 5-iodotubercidin, blocked these responses, suggesting that the phosphorylation of AICAR and adenosine to AICAR 5′-monophosphate (ZMP) or AMP, respectively, was required. Concentrations of ZMP and AMP were elevated and ATP levels diminished at 24 h. The PPARα-dependent responses were associated with increased concentrations of oleic acid, a potent PPARα agonist, and diminished levels of oleoyl-CoA. Oleoyl-CoA synthase activity was inhibited by ZMP and AMP with IC50 values of 0.28 and 0.41 mM, respectively. These results suggest that PPARα is activated by increased concentrations of free fatty acids that may arise from impaired fatty acid metabolism caused by altered levels of ATP, AMP, and ZMP after AICAR or adenosine treatment.
Introduction
Members of cytochrome P450 family 4 (Cyp4) contribute to the ω-hydroxylation of fatty acids in liver and kidney (Hsu et al., 2007; Hardwick, 2008). Previous studies performed in our laboratory revealed that human CYP4F2, a prominent fatty acid ω-hydroxylase in liver and kidney, is up-regulated by the pharmacological activation of AMP-activated protein kinase (AMPK) by AICAR, genistein, or resveratrol in human hepatocytes and the human hepatoma-derived HepG2 cell line (Hsu et al., 2011). AMPK is a serine/threonine protein kinase that serves as a sensor of cellular stress and a modulator of lipid and glucose metabolism. AMPK is a heterotrimeric complex that consists of a catalytic subunit (α) and two regulatory subunits (β and γ) (Viollet et al., 2007). Theoretically, AMPK can exist in 12 possible heterotrimeric complexes because multiple isoforms of each subunit have been identified, namely, α1, α2, β1, β2, γ1, γ2, and γ3. Furthermore, it seems that the combination of AMPK subunits can vary according to cell type and tissue (Hardie et al., 2006). The activation of AMPK involves the phosphorylation of threonine-172 within the activation loop of the catalytic domain of the α-subunit by upstream kinases and allosteric activation by AMP, which increases when ATP is depleted during metabolic stress. Once activated, AMPK acts to increase ATP-generating catabolic pathways, including fatty acid oxidation, and decrease ATP-consuming processes such as gluconeogenesis. AMPK is activated under stress conditions such as glucose deprivation, hypoxia, and oxidative stress. Pharmacological activation of AMPK by AICAR requires phosphorylation by adenosine kinase to form ZMP, an AMP mimic. Chemically unrelated activators such as 6,7-dihydro-4-hydroxy-3-(2′-hydroxy[1,1′-biphenyl]-4-yl)-6-oxo-thieno[2,3-b]pyridine-5-carbonitrile (A-769662) can activate AMPK directly by binding to allosteric effector sites. In addition, a number of drugs and drug-like molecules have been shown to activate AMPK indirectly by increasing cellular AMP/ATP ratios and/or stimulating upstream kinases. These compounds include biguanides such as metformin (Zhou et al., 2001) and polyphenols such as resveratrol and genistein (Zang et al., 2006).
Fatty acid oxidation plays an important role in the production of ATP, and for some cell types, fatty acids are the major nutrient for ATP production during fasting. Under normal conditions, mitochondrial and peroxisomal β-oxidation pathways are the primary route of fatty acid catabolism with a relatively minor contribution from the microsomal ω-oxidation pathway (Hsu et al., 2007; Hardwick, 2008). However, during periods of cellular stress, including starvation, the levels of intracellular free fatty acids can increase, resulting in an increased contribution of the ω-hydroxylation pathway to overall fatty acid oxidation. The ω-hydroxylated metabolites can be oxidized by an alcohol dehydrogenase, and the resulting ω-oxo-fatty acid product can be subsequently metabolized by an aldehyde dehydrogenase to a dicarboxylic fatty acid. The dicarboxylic fatty acid product can either enter the β-oxidation pathway or be excreted in the urine. In addition, the ω-oxidation of fatty acids can decrease the accumulation of free fatty acids and reduce the potential for lipotoxicity. AMPK activation increases mitochondrial fatty acid oxidation by inactivating acetyl-CoA carboxylase, which reduces the inhibition of carnitine palmitoyl transferase-1 by malonyl-CoA. In addition, activation of AMPK triggers changes in gene transcription to augment the production of ATP from catabolism of glucose and fatty acids (Cantó and Auwerx, 2010; McGee and Hargreaves, 2010).
In the present study, we tested whether activators of AMPK would stimulate the expression of murine Cyp4 genes in vivo to further elucidate the role of AMPK in the regulation of hepatic fatty acid ω-hydroxylation. In doing so, we found that Cyp4a10, Cyp4a14, and Cyp4a31 mRNAs are increased after treatment of mice with AICAR in vivo and in primary cultures of mouse hepatocytes in vitro. Cyp4a10 and Cyp4a14 are established PPARα-responsive genes; however, this had yet to be determined for other mouse Cyp4a genes (Hsu et al., 2007). PPARα is a ligand-activated transcription factor that serves as a biological sensor for intracellular fatty acid levels (Kersten et al., 2000; Pégorier et al., 2004; Desvergne et al., 2006; Lefebvre et al., 2006). Therefore, we tested whether other PPARα-responsive genes are up-regulated by AICAR treatment. To this end, acyl CoA oxidase 1 (Acox1), acyl CoA dehydrogenase, medium chain (Acadm), carnitine palmitoyltransferase 1A (Cpt1a), and fatty acid binding protein (Fabp1) mRNAs were measured after AICAR treatment and compared with vehicle controls. Similar to Cyp4a10, Cyp4a14, and Cyp4a31, these mRNAs were also increased by AICAR treatment. These responses were not seen in PPARα null mice, indicating that the effect depended on PPARα. However, in all cases, the response to AICAR was insensitive to pharmacological inhibition of AMPK activation by compound C (dorsomorphin or 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine). Moreover, A-769662, a mechanistically distinct, direct activator of AMPK (Cool et al., 2006), did not increase the expression of these PPARα target genes. Furthermore, results are presented that demonstrate that the formation of ZMP from AICAR or AMP from adenosine is required for the up-regulation of the PPARα-responsive genes examined. In addition, because PPARα is activated by free fatty acids, the concentrations of free fatty acids in hepatocytes 24 h after treatment with AICAR were measured. Oleic acid and stearic acid were increased significantly after AICAR and adenosine treatment. This elevation was not blocked by compound C; however, this did require conversion of AICAR and adenosine to ZMP and AMP, respectively. Pretreatment of hepatocytes with a PPARα antagonist blocked the AICAR-mediated increase in PPARα-responsive mRNAs, suggesting that ligand-dependent activation of PPARα underlies the increase in these mRNAs after AICAR treatment. Therefore, our studies detailed herein suggest that AICAR and adenosine increase the expression of PPARα target genes by an AMPK-independent mechanism that depends on ZMP or AMP formation, respectively.
Materials and Methods
Animals and Treatments.
All handling of mice was in accordance with protocols that were approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute. Eight-week-old C57BL/6 or 129/Sv mice were injected intraperitoneally with 0.5 ml of a saline solution of AICAR (0.7 mg/g body weight; Toronto Research Chemicals Inc., North York, ON, Canada) or saline alone 2 h after the onset of the dark cycle. Control and treated mice were provided with chow and water ad libitum. Lighting was on a 12-h light/dark cycle. For the time-course studies, mice were sacrificed 6, 12, 24, and 48 h after injection, and livers were harvested. In addition, mice were injected intraperitoneally twice with 24 h between injections, and then sacrificed 24 h after the final injection. Liver microsomes were isolated as described previously (Raucy and Lasker, 1991) for the measurement of lauric acid ω-hydroxylase activity.
Quantitative Real-Time Polymerase Chain Reaction.
Hepatic RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Reverse transcription of total RNA (5 μg) was performed using AffinityScript multiple temperature reverse transcriptase (Agilent Technologies, Santa Clara, CA). The RNA concentration was determined spectrophotomically. The primers used for measurement of Cyp4a12, Cyp4a14, L27, and PPARα mRNA were described previously (Savas et al., 2009). The sequences for all other primers used for qPCR are listed in Table 1. The mouse ribosomal protein L27 served as the housekeeping gene for normalization of mRNA levels. L27 mRNA levels were used for normalization because this gene did not exhibit sensitivity to any of the treatments used in this study. Before performing qPCR, Cyp4a10, Cyp4a14, Cyp4a31, Cyp4f14, and L27 PCR products were generated using the above primers and conventional PCR, and the isolated products were ligated into the EcoRI site of the pCRII-TOPO vector for subcloning (Invitrogen). Sequence determinations confirmed the identity of the cloned insert. Serial dilutions of the resulting plasmids were used to generate standard curves for the measurement of mRNA levels using qPCR. Relative levels of Cyp4f14, Cyp4f15, Cyp4f16, and Cyp4f17 were determined from the standard curves using serial dilutions of the Cyp4f14 plasmid as a template, whereas the L27 plasmid was used as reference standard for qPCR to measure Acox1, Acadm, Cpt1a, Acsl1, Fabp1, and small heterodimer partner (Shp) expression. The cycling reactions were carried out by denaturation for 5 min at 95°C followed by 50 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 30 s.
TABLE 1.
Oligonucleotides used for qPCR
| Target | Accession No. | Coordinates | Orientation | Sequence (5′ to 3′) |
|---|---|---|---|---|
| Cyp4a10 | NM_010011.2 | 45–445 | Forward | GAGTGTCTCTGCTCTAAGCCCA |
| Reverse | AGGCTGGGGTTAGCATCCTCCT | |||
| Cyp4a30b | NM_001100185 | 84–360 | Forward | GTGGTCTCTTTGCTTGGCCTACTG |
| Reverse | CCGTAGCCAATCCAGGGAGCAAAG | |||
| Cyp4a31 | NM_201640 | 165–369 | Forward | AGCAGTTCCCATCACCGCCC |
| Reverse | TGCTGGAACCATGACTGTCCGTTT | |||
| Cyp4a32 | NM_001100181 | 238–557 | Forward | GGGCATGAGCAGTTCAAAGGC |
| Reverse | GAGTCCTGACCAGCCAGCCT | |||
| Cyp4f13 | NM_130882 | 354–685 | Forward | GTGTACCCAATCCTGCGACTCG |
| Reverse | TGCTGTCAGACTCTTGGCAGTTGC | |||
| Cyp4f14 | NM_022434 | 267–533 | Forward | CCCATGGAAGACCCTGCTACTGCT |
| Reverse | CCTTGAGTGCAACAGCGGCTGA | |||
| Cyp4f15 | NM_134127 | 254–487 | Forward | GAGCTTCCTGGCTCTTGGCCCGTG |
| Reverse | GGGCTACCGAAGCTGAGGCATTG | |||
| Cyp4f16 | NM_024442 | 61–259 | Forward | CTGCGGCTAAGTGTGTCTGGGC |
| Reverse | TGGAAAGTCTGGCCCATTTCGGTA | |||
| Cyp4f17 | NM_001101445 | 359–634 | Forward | GCCTGCAGTTGTTGACTGAAAGGA |
| Reverse | CGCTGCCACTTGGCATGCATGATG | |||
| Cyp4f39 | NM_177307 | 405–742 | Forward | GCTGGGTCACATGAGCATGTATCT |
| Reverse | GTCACTGAGCCCTCTGCCAGATGT | |||
| Shp | NM_011850 | 224–438 | Forward | GCACCTGCAGGGAGGCCTTG |
| Reverse | CGGTCTGATGGCTGGGCACC | |||
| Acox1 | NM_015729 | 526–821 | Forward | AAGCCAGCGTTACGAGGT |
| Reverse | CTGTTGAGAATGAACTCTTGG | |||
| Acadm | NM_007382 | 1060–1403 | Forward | ACTCCGGTCGCCGGAACACT |
| Reverse | CCTCCGCCATGGGAATCCGC | |||
| Cpt1a | NM_013495 | 660–983 | Forward | ACGCATGACAGCACTGGCCC |
| Reverse | CCTCCCCAGGGATGCGGGAA | |||
| Fabp1 | NM_017399 | 53–233 | Forward | TGCCACCATGAACTTCTCCGGC |
| Reverse | TCCAGTTCGCACTCCTCCCCC |
Primary Mouse Hepatocytes.
Hepatocytes were isolated from 8-week-old male C57BL/6 or 129/Sv mice using a collagenase/EGTA perfusion method (Lee et al., 2004). Viability was determined by Trypan blue exclusion, and preparations were only used if more than 85% viability was achieved. Hepatocytes were cultured on collagen I-coated six-well plates (BD Biosciences, San Jose, CA) using Williams' medium E containing insulin, transferrin, and selenium (Sigma, St. Louis, MO), 0.1 μM dexamethasone, l-glutamine, penicillin, and streptomycin. The cultures were maintained at 37°C and 5% CO2. After the cultures had been incubated for 24 h, the medium was changed to Williams' medium E containing only l-glutamine, penicillin, and streptomycin. The cultured hepatocytes were then treated with AICAR (500 μM), A-769662 (Tocris Bioscience, Ellisville, MO; 1 or 10 μM), adenosine (10 μM), fenofibrate (100 μM), [[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]acetic acid (WY14643) (Tocris Bioscience; 10 μM), or the equivalent amount of the delivery solvent 4 h after changing the medium. For inhibition studies, the cell cultures were preincubated with compound C (10 μM), 5-iodotubercidin (10 μM), or [(2S)-2-[[(1Z)-1-methyl-3-oxo-3-[4-(trifluoromethyl)phenyl]-1-propenyl]amino]-3-[4-[2-(5-methyl-2-phenyl-4-oxazolyl)ethoxy]phenyl]propyl]-carbamic acid ethyl ester (GW6471) (Tocris Bioscience; 20 μM) for 30 min before the addition of the above-mentioned compounds. After 24 h the cells were harvested for RNA isolation. Stock solutions were prepared in DMSO, and the final concentration of DMSO in the medium was 0.1%.
Lauric Acid Metabolism.
The ω-hydroxylation of lauric acid was measured by using liver microsomes (100 μg) isolated from mice injected with a saline solution of AICAR or saline twice with a 24-h interval in between injections, then mice were sacrificed 24 h after the final injection (48-h treatment). The enzymatic reaction was carried out as described previously (Savas et al., 2009).
Detection of Free Fatty Acids, Fatty Acyl-CoAs, and Nucleotides.
Free fatty acids (oleic acid, stearic acid, linoleic acid, arachidonic acid, palmitic acid, and palmitoleic acid; all standards were purchased from Sigma) were measured in primary hepatocytes isolated from 8-week-old male C57BL/6 mice. Sample extraction and liquid chromatography-mass spectrometry analysis were performed according to the method described by Nomura et al. (2010). Fatty acyl-CoAs were measured according to the protocol described previously (Haynes et al., 2008). Fatty acyl-CoA standards were purchased from Avanti Polar Lipids (Alabaster, AL). Both were analyzed using a Waters (Milford, MA) ACQUITY UPLC System interfaced to an AB SCIEX (Foster City, CA) QTRAP 5500 mass spectrometer. The internal standard used for free fatty acid sample extraction was [13C]oleic acid (Sigma), whereas the internal standard for fatty acyl-CoA extraction was pentadecanoyl CoA. ATP, ADP, AMP, and ZMP were isolated and measured using the method described by Aymerich et al. (2006). A Waters ACQUITY UPLC System equipped with a photo diode array detector was used to measure relative absorbance of the nucleotides at 260 nm using authentic standards for quantitation. ATP, ADP, AMP, and ZMP standards were obtained from Sigma. Concentrations of nucleotides were normalized to cell lysate protein content.
Measurement of Acyl-CoA Synthetase Activity.
Lysates from primary mouse hepatocytes were used to measure total acyl-CoA synthetase activity by determination of the formation of [3H]oleoyl-CoA from [3H]oleic acid (Moravek Biochemicals, Brea, CA) as described previously (Askari et al., 2007). The data are expressed as a rate of formation using 200 μg of lysate protein under initial rate conditions.
Statistical Analysis of Data.
Results are expressed and plotted as mean values ± S.D. Comparisons of control versus treated mice and hepatocytes were performed using an unpaired one-tailed Student's t test (Prism 5 software; GraphPad Software Inc., San Diego, CA); p values <0.05 were considered statistically significant. Nonlinear regressions were performed and subsequent IC50 values were calculated using Prism 5 software.
Results
AICAR Increases Cyp4a10, Cyp4a14, and Cyp4a31 mRNA Expression in Murine Liver.
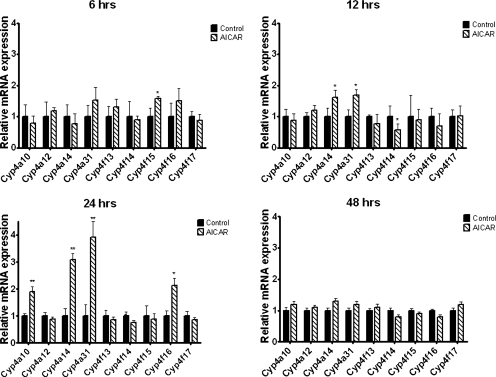
Male and female C57BL/6 mice were injected intraperitoneally with AICAR, and mRNA expression of hepatic Cyp4a and Cyp4f subfamily members was measured for samples collected 6, 12, 24, and 48 h later. Cyp4a10, Cyp4a14, and Cyp4a31 mRNA levels were the most consistently increased, and the maximal fold difference in mRNA abundance was observed at 24 h after injection (approximately 2-, 3-, and 3.4-fold, respectively), and expression returned to basal levels at 48 h after a single injection (Fig. 1). Cyp4f16 mRNA abundance was increased approximately 2-fold, but statistically significant (p < 0.05) elevations of Cyp4f16 were observed only in male mice at the 24-h time point (Fig. 1). Cyp4a29, Cyp4a30b, Cyp4a32, Cyp4f37, Cyp4f39, and Cyp4f40 hepatic mRNA expression levels were near or below the limit of detection. Significant effects of AICAR on the mRNA levels of Cyp4a12, Cyp4f13, Cyp4f14, Cyp4f15, and Cyp4f17 were not evident.
Fig. 1.
Time course for induction of hepatic Cyp4 mRNA after treatment with AICAR. Eight-week-old male C57BL/6 mice were injected intraperitoneally with saline (control) or AICAR (0.7 mg/g) then sacrificed at the indicated times after injection, and mRNA was analyzed using real-time PCR. Data are expressed relative to control (n = 4 mice per time point). Student's t tests were performed to determine significance. *, p < 0.05; **, p < 0.01. Results were similar when the same studies were performed with female mice.
AICAR Treatment Increases the ω-Hydroxylation of a Prototypic Cyp4a Substrate.
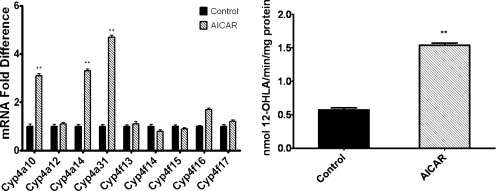
To determine whether the AICAR-mediated increase in mRNA levels of Cyp4a10, Cyp4a14, and Cyp4a31 would result in a subsequent increase in microsomal enzyme activity, assays were performed to measure the metabolism of a Cyp4a substrate, lauric acid, using liver microsomes isolated 24 h after single daily injections of male C57BL/6 mice with AICAR on 2 consecutive days. This treatment resulted in a 2.8-fold increase in the microsomal rate of 12-hydroxylauric acid formation, which corresponded with relative increases in mRNA expression for Cyp4a10 and Cyp4a14 determined for this treatment protocol (Fig. 2).
Fig. 2.
Repeated doses of AICAR increase the metabolism of lauric acid (a Cyp4a substrate). Liver microsomes were isolated from 8-week-old male C57BL/6 mice that were intraperitoneally injected with saline (control) or AICAR daily for 2 days. Mice were euthanized 24 h after the final injection. Liver microsomes and total mRNA were prepared as described under Materials and Methods. Left, relative mRNA expression was determined as described in the legend to Fig. 1. Right, rates of liver microsomal metabolism of [14C]lauric acid were determined as described under Materials and Methods (n = 4 mice). Student's t tests were performed to determine significance. **, p < 0.01.
Activation of Cyp4a10, Cyp4a14, and Cyp4a31 mRNA Expression by AICAR Is PPARα-Dependent.
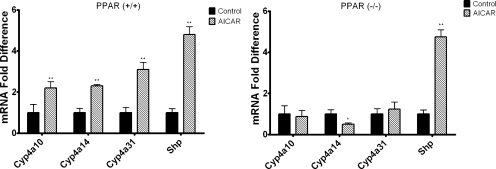
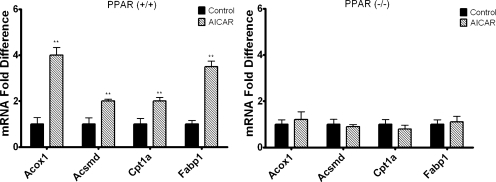
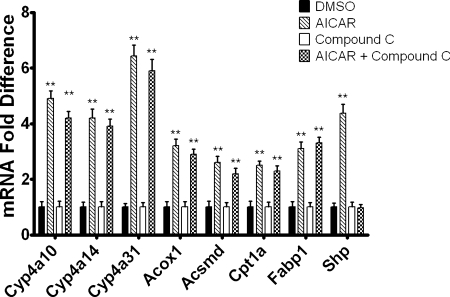
Cyp4a10 and Cyp4a14 are PPARα-responsive genes, and AICAR has been reported to increase the expression of PPARα and PPARα target genes in murine skeletal muscle (Lee et al., 2006). Thus, studies were performed to determine whether the effect of AICAR on Cyp4a10, Cyp4a14, and Cyp4a31 mRNA expression is PPARα-dependent. Wild-type and PPARα null 129/Sv mice were injected intraperitoneally with AICAR or saline and sacrificed 24 h after injection. Cyp4a10, Cyp4a14, and Cyp4a31 mRNA levels were increased 2- to 3-fold in wild-type 129/Sv mice treated with AICAR compared with saline-treated controls (Fig. 3). However, the effect of AICAR on expression of these genes was abrogated in the PPARα null 129/Sv mice (Fig. 3). Cyp4f16 was elevated approximately 1.7-fold in the 129/Sv mice and 1.5-fold in PPARα null mice (data not shown); however, these increases in mRNA expression were not statistically significant. Shp mRNA was measured as a positive control because it has been demonstrated that Shp mRNA expression is increased by AICAR in an AMPK-dependent manner (Kim et al., 2008). Shp mRNA levels were increased by AICAR in both wild-type and PPARα null 129/Sv mice. To determine whether our findings could be extended to other PPARα-responsive genes, the mRNA levels for Acox1, Acsmd, Cpt1a, and Fabp1 were measured 24 h after AICAR treatment. Similar to Cyp4a10, Cyp4a14, and Cyp4a31, mRNA levels for Acox1, Acsmd, Cpt1a, and Fabp1 were increased in AICAR-treated wild-type 129/Sv mice but not in the PPARα null 129/Sv mice (Fig. 4). To confirm that AICAR induces the PPARα-responsive genes at the transcriptional level, hepatocytes isolated from 8-week-old C57BL/6 mice were preincubated with actinomycin D and then treated with AICAR. The presence of actinomycin D inhibited the activation of Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNAs by AICAR (P < 0.01) (data not shown).
Fig. 3.
AICAR-mediated induction of Cyp4a mRNA is PPARα-dependent. Eight-week-old wild-type 129/Sv mice [PPARα(+/+)] (left) or PPARα-null(−/−) (right) mice (129/Sv background) were injected intraperitoneally with AICAR or saline. Livers were harvested 24 h after injection, and mRNA expression was analyzed using qPCR (n = 4). Shp was used as an AMPK-responsive positive control. Student's t tests were performed to determine significance. *, p < 0.05; **, p < 0.01.
Fig. 4.
AICAR-treatment increases the expression of the PPARα-responsive genes Acox1, Acsmd, Cpt1a, and Fabp1 in a PPARα-dependent manner. Eight-week-old wild-type 129/Sv (left) or PPARα-null mice (129Sv background) (right) were injected intraperitoneally with AICAR or saline. Livers were harvested 24 h after injection, and mRNA expression was analyzed using qPCR (n = 4). Student's t tests were performed to determine significance. **, p < 0.01.
Because Cyp4a31 had yet to be established as a PPARα-responsive gene, we measured the expression of Cyp4a31 mRNA in hepatocytes isolated from PPARα null 129/Sv mice or heterozygous littermates that express PPARα treated with either fenofibrate or vehicle control. Cyp4a31 mRNA expression was increased 23-fold after fenofibrate treatment in hepatocytes isolated from mice expressing PPARα, whereas no increase in Cyp4a31 mRNA levels was observed in hepatocytes isolated from PPARα null mice treated with fenofibrate versus the vehicle control (data not shown).
Inhibition of AMPK Does Not Block AICAR-Mediated Increases in Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNAs.
To test whether the effects of AICAR on Cyp4a10, Cyp4a14, Cyp4,a31, and Acox1 mRNA expression depend on the activation of AMPK, primary mouse hepatocytes were pretreated with the AMPK inhibitor compound C (Zhou et al., 2001) and then incubated with AICAR. Shp, an established AMPK-responsive gene (Kim et al., 2008), was not increased by AICAR in the hepatocytes pretreated with compound C. The presence of compound C did not affect, however, the AICAR-mediated increase in Cyp4a10, Cyp4a14, Cyp4a31, Acsmd, Cpt1a, or Fabp1 mRNAs, suggesting that AICAR induces the expression of the corresponding genes by an AMPK-independent mechanism (Fig. 5). For comparison to AICAR-mediated increases in Cyp4a10 (4-fold), Cyp4a14 (4-fold), and Cyp4a31 (6-fold) mRNA levels, treatment of hepatocytes isolated from male C57BL/6 mice treated with the PPARα agonist WY14643 were measured and exhibited 10.6 ± 0.15-, 10.7 ± 0.13-, and 9.0 ± 0.12-fold activation of Cyp4a10, Cyp4a14, and Cyp4a31 mRNA expression, respectively.
Fig. 5.
Increased Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA expression in response to AICAR is not affected by AMPK inhibition. Hepatocytes were pretreated with compound C or DMSO (vehicle control). After 30 min, AICAR was added to the culture medium, and the cells were harvested after 24 h by the addition of TRIzol. Expression was then analyzed using qPCR and normalized to L27 mRNA for triplicate samples. The data are representative of hepatocyte preparations from four individual mice. Student's t test was performed to determine significance. **, p < 0.01.
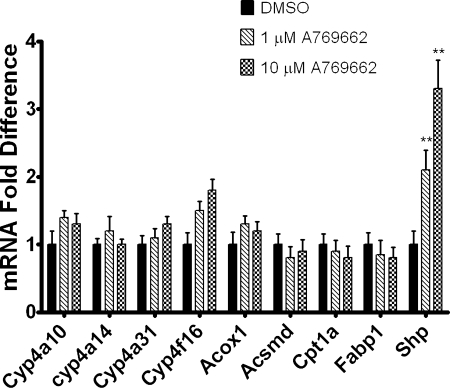
The AMPK Activator A-769662 Does Not Increase Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA Expression.
To further probe our finding that AICAR increases the expression of PPARα-responsive genes in an AMPK-independent manner, the effects of A-769662, a non-nucleoside, direct activator of AMPK that does not depend on AMP/ZMP formation were tested. Shp mRNA expression was increased in hepatocytes treated with A-769662; however, Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNAs were not increased compared with vehicle-treated controls (Fig. 6).
Fig. 6.
The AMPK activator A769662 does not increase Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA expression. Hepatocytes were treated with A-769662 or DMSO (vehicle control). Twenty-four hours after treatment, the cells were harvested by the addition of TRIzol. Expression was then analyzed using qPCR and normalized to L27 mRNA for triplicate samples. The data are representative of hepatocyte preparations from four individual mice. Student's t tests were performed to determine significance. **, p < 0.01.
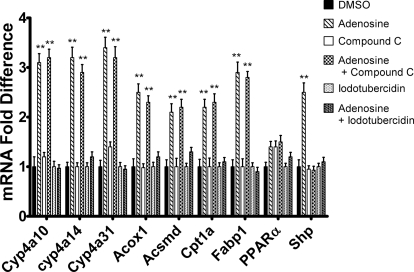
Adenosine Increases Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA Expression.
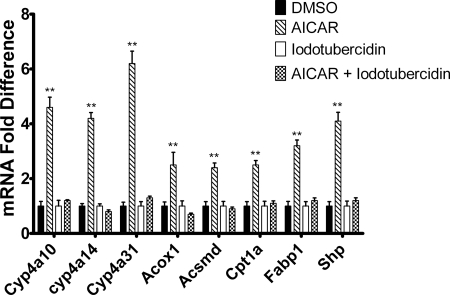
Adenosine has been reported to activate AMPK, and this effect depends on conversion of adenosine to AMP by adenosine kinase (Aymerich et al., 2006). The ability of adenosine to increase Cyp4a10, Cyp4a14, Cyp4a31, and Acox1 mRNA expression in hepatocytes was tested. Adenosine treatment activated expression of the PPARα-responsive mRNAs as well as Shp mRNA (Fig. 7). However, pretreatment with compound C only blocked the increase in Shp mRNA expression, suggesting that activation of AMPK was necessary for activation of Shp expression. Inhibition of adenosine kinase using 5-iodotubercidin abrogated the increase of all mRNAs measured. Although the induction of Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA by adenosine was AMPK-independent, the inhibitor studies suggest that the effect of adenosine depends on the formation of AMP or downstream metabolites. Likewise, AICAR has to be phosphorylated by adenosine kinase to form ZMP, which activates AMPK. We tested therefore whether conversion of AICAR to ZMP is necessary for the activation of the PPARα-responsive genes. When hepatocytes were incubated with 5-iodotubercidin (Henderson et al., 1972), followed by treatment with AICAR 5-iodotubercidin blocked the increase in mRNA expression of Cyp4a10, Cyp4a14, Cyp4a31, Acsmd, Cpt1a, and Fabp1 by AICAR, suggesting that although AICAR-mediated increase in the expression of PPARα-responsive genes is AMPK-independent, it depends on the formation of ZMP or downstream metabolites (Fig. 8). To assess whether either AICAR or adenosine treatment had an effect on adenine nucleotide levels at the 24-h time point, levels of AMP, ADP, and ATP were measured as well as those of ZMP. As shown in Table 2, these treatments elevated levels of AMP 2.3- to 3.5-fold but reduced ATP levels to 44 to 51% of controls at 24 h. In addition, the level of ZMP exceeded that of ATP or AMP at 24 h after AICAR treatment.
Fig. 7.
Adenosine increases Cyp4a10, Cyp4a14, Cyp4a31, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA expression in a AMP-dependent and AMPK-independent manner. Hepatocytes were isolated from 8-week-old male C57BL/6 mice by EGTA/collagenase perfusion. Hepatocytes were pretreated with 5-iodotubercidin (adenosine kinase inhibitor) or DMSO (vehicle control). After 30 min, adenosine (10 μM) was added to the culture medium, and the cells were harvested after 24 h by the addition of TRIzol. Expression was then analyzed using qPCR and normalized to L27 mRNA for triplicate samples. The data are representative of hepatocyte preparations from four individual mice. Student's t tests were performed to determine significance. **, p < 0.01.
Fig. 8.
Conversion of AICAR to ZMP is necessary for the activation of Cyp4a, Acox1, Acsmd, Cpt1a, and Fabp1 mRNA expression. Hepatocytes were isolated from 8-week-old male C57BL/6 mice by EGTA/collagenase perfusion. Hepatocytes were pretreated with 5-iodotubercidin (adenosine kinase inhibitor) or DMSO (vehicle control). After 30 min, AICAR was added to the culture medium, and the cells were harvested after 24 h by the addition of TRIzol. Expression was then analyzed using qPCR and normalized to L27 mRNA for triplicate samples. The data are representative of hepatocyte preparations from four individual mice. Student's t tests were performed to determine significance. **, p < 0.01.
TABLE 2.
Nucleotide concentrations in hepatocytes after treatment with AICAR or adenosine
Data are means ± S.D. (n = 4). To determine significance Student's t tests were performed for AICAR or adenosine versus DMSO (vehicle control, 0.1% final concentration).
| ATP | ADP | AMP | ZMP | |
|---|---|---|---|---|
| Untreated | 1211.4 ± 320.1 | 450.4 ± 20.3 | 230.3 ± 29.7 | N.D. |
| DMSO | 1132.4 ± 120.2 | 371.0 ± 40.0 | 169.8 ± 9.7 | N.D. |
| AICAR | 493.0 ± 70.3* | 299.8 ± 10.2 | 600.4 ± 110.1** | 900.4 ± 49.9** |
| Adenosine | 579.6 ± 60.1* | 249.8 ± 10.3 | 390.3 ± 8.2* | N.D. |
N.D., not detected.
, P < 0.05;
, P < 0.01.
Adenosine and AICAR Alter the Abundance of Free Fatty Acids and Their CoA Esters.
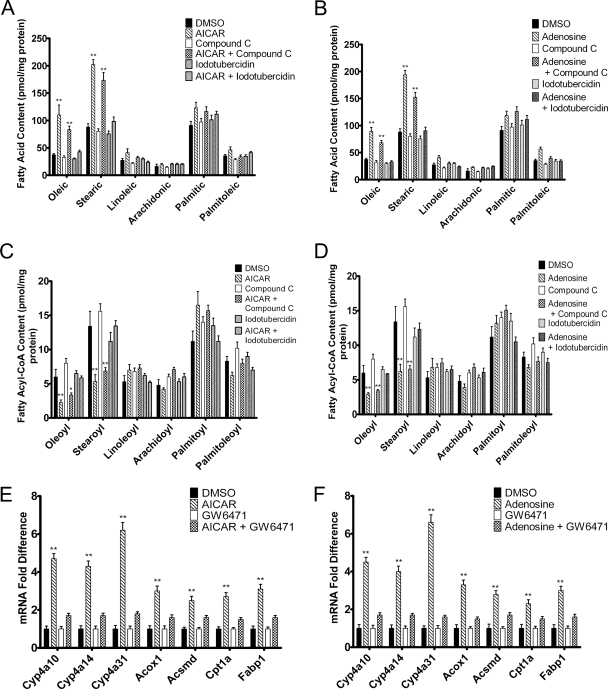
To investigate the mechanism by which AICAR and adenosine increase the expression of PPARα-responsive genes independently of AMPK, studies were performed to determine whether these compounds induced changes in the concentrations of free fatty acids. Nonesterified fatty acids were demonstrated previously to activate PPARα transcriptional activity (Forman et al., 1997; Kliewer et al., 1997). Treatment of primary human hepatocytes with AICAR and adenosine resulted in an increase in the abundance of oleic acid by 2.7- and 2.4-fold, respectively, compared with vehicle-treated samples (Fig. 9, A and B). Stearic acid was also significantly increased by 2.4- and 2.2-fold after AICAR and adenosine treatment, respectively. Although linoleic acid, arachidonic acid, palmitic acid, and palmitoleic acid showed a trend toward being increased after AICAR and adenosine treatment, these increases did not reach statistical significance. Preincubation of hepatocytes with 5-iodotubercidin, but not compound C, blocked this increase in the concentration of free fatty acids measured (Fig. 9, A and B). Although treatments with compound C decreased the concentrations of the free fatty acids measured, this small effect was not significant. To further test a role for ligand-dependent activation of PPARα, hepatocytes were treated with AICAR and adenosine in the presence of the PPARα antagonist ligand GW6471. The use of this antagonist blocked the up-regulation of PPARα-responsive mRNAs by AICAR and adenosine (Fig. 9, E and F). Oleic acid is an abundant high-affinity agonist for PPARα, exhibiting a 2 nM dissociation constant (Hostetler et al., 2005). Thus, the elevation of oleic acid concentrations could contribute to the activation of PPARα observed in these studies.
Fig. 9.
Ligand activation of PPARα is required for AICAR- and adenosine-stimulated increased mRNA expression of PPARα-responsive genes. A to D, hepatocytes were treated with AICAR (A and C) or adenosine (B and D) in the presence and absence of compound C or 5-iodotubercin, and free fatty acids and fatty acyl-CoAs were isolated from hepatocytes followed by quantitation using ultra performance liquid chromatography- tandem mass spectrometry as described under Materials and Methods. Free fatty acid and fatty acyl-CoA concentrations were normalized to total protein concentration of cells that were lysed for analysis. E and F, for the experiments using the PPARα antagonist GW6471 hepatocytes were preincubated with this compound or DMSO (vehicle control) followed by treatment with AICAR (E) or adenosine (F) 30 min later. The hepatocytes were harvested after 24 h by the addition of TRIzol, and the mRNA levels were measured using qPCR and normalized to L27 mRNA for triplicate samples. Hepatocytes were isolated from four individual mice, and Student's t tests were performed to determine significance. **, p < 0.01.
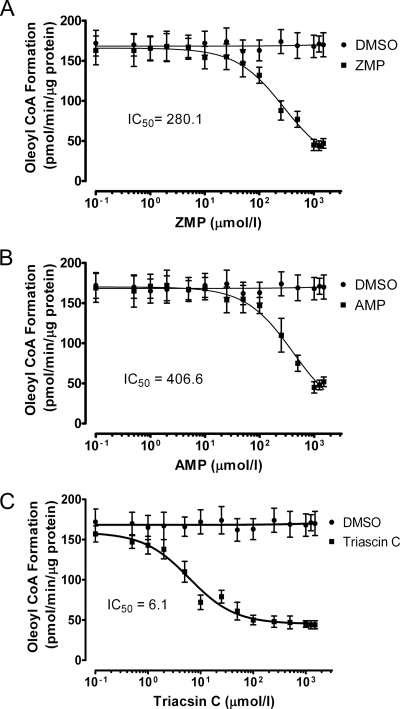
Because in vitro studies indicate that both saturated and unsaturated acyl-CoAs are high-affinity PPARα ligands that can increase coactivator recruitment (Hostetler et al., 2005), the relative abundance of fatty acyl-CoAs was measured by using mass spectrometry. In contrast to the observed elevation of oleic acid and stearic acid, levels of oleoyl-CoA and stearoyl-CoA were decreased by >50% after treatment with AICAR or adenosine. Furthermore, this effect was 5-iodotubercidin-sensitive, indicating that the formation of ZMP or AMP was necessary (Fig. 9, C and D). This suggested that the observed elevation of the two fatty acids might reflect a reduction in the rate of esterification to CoA catalyzed by ATP-dependent long-chain fatty acyl-CoA synthetases (ACSLs), which might reflect diminished levels of ATP. Moreover, AMP has been demonstrated to inhibit long-chain fatty acyl-CoA synthesis (Pande and Mead, 1968; Alexandre et al., 1969). To determine whether ZMP inhibits the formation of oleoyl-CoA from oleic acid, its effects on oleoyl-CoA formation in liver lysates were examined (Fig. 10). ZMP was observed to inhibit the oleoyl-CoA synthesis with an IC50 value of 280 μM, which is less than the concentration of AICAR used to treat hepatocytes. Likewise, AMP inhibited the reaction with an apparent IC50 of 406 μM, which is within the range of AMP concentrations observed after adenosine treatments at the concentration used to treat hepatocytes to activate AMPK (Aymerich et al., 2006). As a positive control, the established ACSL inhibitor triascin C exhibited an IC50 value of 6.1 μM with a maximum inhibition of approximately 70%, which is consistent with published values (Igal et al., 1997; Askari et al., 2007).
Fig. 10.
ZMP inhibits oleoyl-CoA formation by hepatocyte lysates. Lysates (200 μg of total protein) from primary mouse hepatocytes were incubated in the presence of [3H]oleic acid, ATP, CoA, and ZMP (A), AMP (B), or triascin C (C) for 20 min at 37°C. The resulting [3H]oleoyl-CoA was isolated, and radioactivity was measured by using a scintillation counter. The results represent experiments that were performed using hepatocytes from four separate isolations, and the incubations were carried out in duplicate.
Discussion
Results presented here indicate that treatment of mice with AICAR induces the expression of mRNAs for Cyp4a10, Cyp4a14, and Cyp4a31 2- to 4-fold at 24 h after injection. A commensurate elevation of microsomal lauric acid ω-hydroxylase activity was observed after twice-daily injections of AICAR. Consistent with this finding, other PPARα target genes, Acox1, Acsmd, Cpt1a, and Fabp1, were also elevated after AICAR treatment in a PPARα-dependent manner. These responses were not observed in PPARα null mice. AICAR was used in these studies as a prodrug to activate AMPK after conversion to ZMP by adenosine kinase. These studies monitored the expression of Shp as a positive control for activation of AMPK, and the stimulation of Shp expression by AICAR was not affected by the absence of PPARα in the null animals.
Primary cultures of mouse hepatocytes were used to examine whether the effect of AICAR depended on AMPK. AICAR treatment of hepatocytes elicited similar effects as observed in vivo for the PPARα target genes and Shp. The 4- to 6-fold responses to AICAR observed for Cyp4a10, Cyp4a14, and Cyp4a31 are approximately 50% of the 10-fold response observed for the synthetic PPARα agonist, Wy14,643, and the responses to AICAR were abrogated by the PPARα antagonist GW6471. In contrast to Shp, the PPARα-dependent responses to AICAR were not significantly diminished by compound C, an AMPK inhibitor, or elicited by a non-nucleoside, direct-acting AMPK activator, A769662, suggesting that the response did not depend on AMPK activation. Nevertheless, the adenosine kinase inhibitor 5-iodotubercidin inhibited the PPARα-dependent responses, and the PPARα-mediated responses depended on conversion of AICAR to ZMP. Moreover, adenosine that is converted to AMP upon import by adenosine kinase produced a similar stimulation of PPARα activity, which was also inhibited by 5-iodotubercidin and insensitive to compound C. Direct measurements of nucleotide levels indicated that concentrations of AMP were elevated 2- to 3-fold, and concentrations of ATP were reduced by half 24 h after AICAR or adenosine treatment. In addition, the concentration of ZMP was elevated to levels exceeding that of ATP or AMP at 24 h after AICAR treatment. These results suggested that elevation of AMP or ZMP led to the activation of PPARα.
PPARs are ligand-activated transcription factors that are activated by the binding of fatty acids and related endogenous activators to these receptors (Kersten et al., 2000; Pégorier et al., 2004; Desvergne et al., 2006; Lefebvre et al., 2006). For this reason, a highly sensitive ultra performance liquid chromatography-tandem mass spectrometry approach was used to characterize the effects of AICAR or adenosine on the concentrations of the most abundant long-chain fatty acids. Both AICAR and adenosine elicited a >2-fold increase in the concentrations of oleic acid and stearic acid at 24 h after treatment, but suppressed levels of their corresponding CoA esters. These effects were inhibited by 5-iodotubericidin but not by compound C. Because oleic acid is a prominent, high-affinity agonist for PPARα, these results suggest increased concentrations of oleic acid could contribute to the observed activation of PPARα. This does not preclude additional mechanisms for activation of PPARα that might arise from AMPK-independent effects of AMP or ZMP on signal transduction pathways that in turn might modulate the phosphorylation state of PPARα or modulate the activity of coactivators (Kersten et al., 2000; Pégorier et al., 2004; Desvergne et al., 2006; Lefebvre et al., 2006).
The elevation of nonesterified oleic acid and stearic acid and diminished levels of oleoyl-CoA and stearoyl-CoA suggested that elevated concentrations of ZMP or AMP coupled with reduced levels of ATP might lead to partial inhibition of ACSLs. Inhibition of ACSLs by triascin C has been shown previously to activate PPARα (Forman et al., 1997), and it is likely that this reflects elevation of nonesterified fatty acids that can activate PPARα of which oleic acid is a potent example. Although AMP has been reported previously to inhibit ACSLs (Pande and Mead, 1968; Alexandre et al., 1969), the effects of ZMP were unknown. The studies reported here indicate that ZMP inhibited oleoyl-CoA synthase activity in liver lysates with an IC50 of 0.28 mM. Likewise, AMP was observed to inhibit oleoyl-CoA synthesis with an IC50 of 0.40 mM. Because basal concentrations of AMP are reported to be approximately 0.26 mM in hepatocytes (Beis and Newsholme, 1975; Noma, 2005), a significant inhibitory effect would be predicted for the 2- to 3-fold elevation of AMP concentrations described in this article. Our results also indicate that concentrations of AMP and ZMP are elevated at the 24-h time point and associated with reduced levels of ATP consistent with other reports (Guigas et al., 2007; Jacobs et al., 2007). The diminished concentrations of ATP observed after AICAR or adenosine treatment may also have a negative impact on ACSL activity. ACSLs use ATP to generate a bound fatty acyl AMP intermediate (Soupene and Kuypers, 2008; Ellis et al., 2010). CoA subsequently reacts with the bound acyl AMP to form acyl CoA by displacing AMP. As a result, ACSLs produce AMP as well as consume ATP, and thus, contribute directly to an elevated AMP/ATP. Moreover, another ATP is used for the phosphorylation of AMP to form ADP that can serve as substrate for ATP formation by glycolysis or fatty acid oxidation. Thioesterases may further exacerbate this effect by hydrolyzing the acyl-CoA, creating a futile cycle. The diminished concentrations of ATP observed after AICAR or adenosine treatment may also have a negative impact on the β-oxidation of fatty acids or incorporation of fatty acids into glycerolipids because acyl-CoAs are required for these pathways. Thus, a reduction in ACSL activity could increase concentrations of free fatty acids and diminish their utilization. In this regard, it is interesting that several ACSLs are PPARα target genes (Soupene and Kuypers, 2008; Ellis et al., 2010), which provides a mechanism to enhance ACSL activity in response to increased concentrations of free fatty acids.
Acknowledgments
We thank Uzen Savas and Mei Hsu for helpful advice throughout this research and the preparation of this article.
This work was supported by the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant R01-HD004445]. N.B. was the recipient of fellowships from the United Negro College Fund/Merck Science Initiative and the Joint University of San Diego/Scripps Research Institute Training for Future Faculty Program supported by the Fletcher Jones Foundation.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.184242.
- Cyp4
- cytochrome P450 family 4
- AMPK
- AMP-activated protein kinase
- PPARα
- peroxisomal proliferator-activated receptor α
- Acox1
- acyl CoA oxidase 1
- Acadm
- acyl CoA dehydrogenase, medium chain
- Cpt1a
- carnitine palmitoyltransferase 1A
- Fabp1
- fatty acid binding protein
- AICAR
- 5-aminoimidazole-4-carboxyamide-ribonucleoside
- ZMP
- AICAR 5′-monophosphate
- Shp
- small heterodimer partner
- DMSO
- dimethyl sulfoxide
- qPCR
- quantitative real-time polymerase chain reaction
- ACSL
- ATP-dependent long-chain fatty acyl-CoA synthetase
- A-769662
- 6,7-dihydro-4-hydroxy-3-(2′-hydroxy[1,1′-biphenyl]-4-yl)-6-oxo-thieno[2,3-b]pyridine-5-carbonitrile
- compound C
- dorsomorphin or 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine
- WY14643
- [[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]acetic acid
- GW6471
- [(2S)-2-[[(1Z)-1-methyl-3-oxo-3-[4-(trifluoromethyl)phenyl]-1-propenyl]amino]-3-[4-[2-(5-methyl-2-phenyl-4-oxazolyl)ethoxy]phenyl]propyl]-carbamic acid ethyl ester.
Authorship Contributions
Participated in research design: Bumpus and Johnson.
Conducted experiments: Bumpus.
Performed data analysis: Bumpus and Johnson.
Wrote or contributed to the writing of the manuscript: Bumpus and Johnson.
References
- Alexandre A, Rossi CR, Sartorelli L, Siliprandi N. (1969) The action of atractyloside and AMP on long-chain fatty acid oxidation and on the ATP-dependent fatty acid thiokinase. FEBS Lett 3:279–282 [DOI] [PubMed] [Google Scholar]
- Askari B, Kanter JE, Sherrid AM, Golej DL, Bender AT, Liu J, Hsueh WA, Beavo JA, Coleman RA, Bornfeldt KE. (2007) Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-γ-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes 56:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich I, Foufelle F, Ferré P, Casado FJ, Pastor-Anglada M. (2006) Extracellular adenosine activates AMP-dependent protein kinase (AMPK). J Cell Sci 119:1612–1621 [DOI] [PubMed] [Google Scholar]
- Beis I, Newsholme EA. (1975) The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem J 152:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. (2010) AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci 67:3407–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. (2006) Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab 3:403–416 [DOI] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W. (2006) Transcriptional regulation of metabolism. Physiol Rev 86:465–514 [DOI] [PubMed] [Google Scholar]
- Ellis JM, Frahm JL, Li LO, Coleman RA. (2010) Acyl-coenzyme A synthetases in metabolic control. Curr Opin Lipidol 21:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. (1997) Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci U S A 94:4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigas B, Taleux N, Foretz M, Detaille D, Andreelli F, Viollet B, Hue L. (2007) AMP-activated protein kinase-independent inhibition of hepatic mitochondrial oxidative phosphorylation by AICA riboside. Biochem J 404:499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Hawley SA, Scott JW. (2006) AMP-activated protein kinase–development of the energy sensor concept. J Physiol 574:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JP. (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol 75:2263–2275 [DOI] [PubMed] [Google Scholar]
- Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill AH., Jr (2008) Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res 49:1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JF, Paterson AR, Caldwell IC, Paul B, Chan MC, Lau KF. (1972) Inhibitors of nucleoside and nucleotide metabolism. Cancer Chemother Rep 2 3:71–85 [PubMed] [Google Scholar]
- Hostetler HA, Petrescu AD, Kier AB, Schroeder F. (2005) Peroxisome proliferator-activated receptor α interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem 280:18667–18682 [DOI] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Griffin KJ, Johnson EF. (2007) Human cytochrome p450 family 4 enzymes: function, genetic variation and regulation. Drug Metab Rev 39:515–538 [DOI] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Lasker JM, Johnson EF. (2011) Genistein, resveratrol, and 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside induce cytochrome P450 4F2 expression through an AMP-activated protein kinase-dependent pathway. J Pharmacol Exp Ther 337:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igal RA, Wang P, Coleman RA. (1997) Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem J 324:529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs RL, Lingrell S, Dyck JR, Vance DE. (2007) Inhibition of hepatic phosphatidylcholine synthesis by 5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside is independent of AMP-activated protein kinase activation. J Biol Chem 282:4516–4523 [DOI] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W. (2000) Roles of PPARs in health and disease. Nature 405:421–424 [DOI] [PubMed] [Google Scholar]
- Kim YD, Park KG, Lee YS, Park YY, Kim DK, Nedumaran B, Jang WG, Cho WJ, Ha J, Lee IK, et al. (2008) Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase-dependent regulation of the orphan nuclear receptor SHP. Diabetes 57:306–314 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, et al. (1997) Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci U S A 94:4318–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Peng H, Gelbart T, Beutler E. (2004) The IL-6- and lipopolysaccharide-induced transcription of hepcidin in HFE-, transferrin receptor 2-, and β2-microglobulin-deficient hepatocytes. Proc Natl Acad Sci U S A 101:9263–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Kim M, Park HS, Kim HS, Jeon MJ, Oh KS, Koh EH, Won JC, Kim MS, Oh GT, et al. (2006) AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARα and PGC-1. Biochem Biophys Res Commun 340:291–295 [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Chinetti G, Fruchart JC, Staels B. (2006) Sorting out the roles of PPARα in energy metabolism and vascular homeostasis. J Clin Invest 116:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M. (2010) AMPK-mediated regulation of transcription in skeletal muscle. Clin Sci (Lond) 118:507–518 [DOI] [PubMed] [Google Scholar]
- Noma T. (2005) Dynamics of nucleotide metabolism as a supporter of life phenomena. J Med Invest 52:127–136 [DOI] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. (2010) Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140:49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande SV, Mead JF. (1968) Long chain fatty acid activation in subcellular preparations from rat liver. J Biol Chem 243:352–361 [PubMed] [Google Scholar]
- Pégorier JP, Le May C, Girard J. (2004) Control of gene expression by fatty acids. J Nutr 134:2444S–2449S [DOI] [PubMed] [Google Scholar]
- Raucy JL, Lasker JM. (1991) Isolation of P450 enzymes from human liver. Methods Enzymol 206:577–587 [DOI] [PubMed] [Google Scholar]
- Savas U, Machemer DE, Hsu MH, Gaynor P, Lasker JM, Tukey RH, Johnson EF. (2009) Opposing roles of peroxisome proliferator-activated receptor α and growth hormone in the regulation of CYP4A11 expression in a transgenic mouse model. J Biol Chem 284:16541–16552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA. (2008) Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 233:507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet B, Mounier R, Leclerc J, Yazigi A, Foretz M, Andreelli F. (2007) Targeting AMP-activated protein kinase as a novel therapeutic approach for the treatment of metabolic disorders. Diabetes Metab 33:395–402 [DOI] [PubMed] [Google Scholar]
- Zang M, Xu S, Maitland-Toolan KA, Zuccollo A, Hou X, Jiang B, Wierzbicki M, Verbeuren TJ, Cohen RA. (2006) Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes 55:2180–2191 [DOI] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108:1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]