Abstract
Drug-drug interactions (DDIs) with the HIV protease inhibitors (PIs) are complex, paradoxical (e.g., ritonavir/alprazolam), and involve multiple mechanisms. As part of a larger study to better understand these DDIs and to devise a framework for in vitro to in vivo prediction of these DDIs, we determined the inductive effect of ∼2 weeks of administration of two prototypic PIs, nelfinavir (NFV), ritonavir (RTV), and rifampin (RIF; induction positive control) on the cytochrome P450 enzymes CYP1A2, CYP2B6, CYP2C9, and CYP2D6 and the inductive or inductive plus inhibitory effect of NFV, RTV, or RIF on CYP3A and P-glycoprotein in healthy human volunteers. Statistically significant induction of CYP1A2 (2.1-, 2.9-, and 2.2-fold), CYP2B6 (1.8-, 2.4-, and 4-fold), and CYP2C9 (1.3-, 1.8-, and 2.6-fold) was observed after NFV, RTV, or RIF treatment, respectively (as expected, CYP2D6 was not induced). Moreover, we accurately predicted the in vivo induction of these enzymes by quantifying their induction by the PIs in human hepatocytes and by using RIF as an in vitro to in vivo scalar. On the basis of the modest in vivo induction of CYP1A2, CYP2B6, or CYP2C9, the in vivo paradoxical DDIs with the PIs are likely explained by mechanisms other than induction of these enzymes such as induction of other metabolic enzymes, transporters, or both.
Introduction
Clinical use of the anti-HIV protease inhibitors (PIs) is complicated by their unpredictable but profound drug-drug interactions (DDIs). Many of the protease inhibitors, most notably ritonavir (RTV), are known to potently inhibit and/or inactivate CYP3A enzymes (Ernest et al., 2005). For this reason, RTV is almost exclusively used in combination with other PIs to pharmacologically boost their bioavailability by inactivating CYP3A (Cooper et al., 2003). On the basis of in vitro metabolism and the in vivo effect of RTV boosting, CYP3A is believed to be the major clearance mechanism of many of the PIs, including RTV (Kumar et al., 1996; Koudriakova et al., 1998; Unadkat and Wang, 2000). However, on chronic administration, RTV and other PIs are capable of inducing their own clearance despite potent CYP3A inactivation (Hsu et al., 1998). Likewise, amprenavir, another potent CYP3A inactivator (Ernest et al., 2005), has little effect (18% increase) on the clearance of saquinavir, a CYP3A substrate (Unadkat and Wang, 2000). Likewise, after multiple dose administration of RTV, the oral clearance of alprazolam (a CYP3A probe drug) remains unchanged (Norvir product labeling; Abbott, Abbott Park, IL), whereas on acute administration RTV decreases the oral clearance of alprazolam as expected (Greenblatt et al., 2000). Although these observations could be explained by net induction of in vivo CYP3A activity, we have shown this is not the case. In fact, CYP3A activity is reduced by ∼90% after multiple dose RTV treatment, which is accurately predicted from in vitro data using sandwich cultured human hepatocytes (Kirby et al., 2011). Therefore, these data suggest induction of other clearance mechanisms, likely other cytochrome P450 (P450) and/or drug efflux pumps such as P-glycoprotein (P-gp) or multidrug resistance protein 2 (Su et al., 2004; Ye et al., 2010). In agreement with this hypothesis, there are sporadic in vivo reports that RTV appears to induce CYP1A2 (Norvir product labeling; Abbott), CYP2B6, and CYP2C9 (Fichtenbaum and Gerber, 2002; Hughes et al., 2007).
In an attempt to provide a mechanistic framework to accurately predict the multifaceted and seemingly unpredictable DDIs elicited by the PIs, we conducted in vivo studies in healthy volunteers with two prototypic PIs [RTV and nelfinavir (NFV)] and the well known inducer, rifampin (RIF). Our studies were designed to assess the inductive effect of multiple dose treatment (∼14 days) with RTV (400 mg b.i.d.), NFV (1250 mg b.i.d.), or RIF [600 mg once daily (qd)] on the major drug-metabolizing P450 enzymes (CYP1A2, CYP2B6, CYP2C9, CYP2D6, and CYP3A4) and P-gp by administration of probe drug cocktails. We then asked if these DDIs can be predicted from in vitro experiments using human liver microsomes and hepatocytes using the broad spectrum inducer RIF as an in vitro to in vivo scalar for induction of multiple enzymes. We have previously reported and predicted the CYP3A-mediated DDIs from in vitro data (Kirby et al., 2011). In this manuscript, we demonstrate and predict the in vivo induction of CYP1A2, CYP2B6, CYP2C9, and CYP2D6 by RTV, NFV, or RIF. CYP2D6 was included in our study as a negative control because it is not inducible by xenobiotics (Benedetti, 2000). The effect of the PIs on P-gp activity will be the subject of another manuscript because that study also revealed an interesting and clinically significant drug-drug interaction between the CYP2B6 probe drug bupropion (BUP) and digoxin.
Materials and Methods
Study Design.
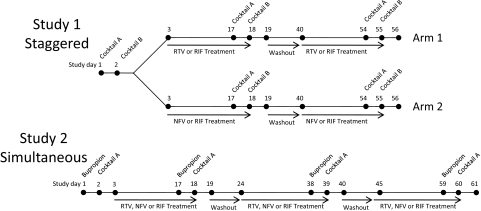
The study design, subject selection criteria, and subject safety monitoring have been described in detail in our previous manuscript focusing on the CYP3A-mediated DDIs with RTV, NFV, and RIF (Kirby et al., 2011). The design of the study with respect to the CYP1A2-, CYP2B6-, CYP2C9-, and CYP2D6-mediated DDIs with RTV, NFV, or RIF is described here (Fig. 1). In brief, two studies were conducted in healthy volunteers. In study 1, the drugs used to measure in vivo CYP1A2 (caffeine, 200 mg p.o.), CYP2C9 (tolbutamide, 500 mg p.o.), and CYP2D6 (dextromethorphan, 30 mg p.o.) activity were administered as part of a P450 phenotyping cocktail (Wang et al., 2001). Study 1 was conducted in two arms (RTV and RIF treatment or NFV and RIF treatment). In study 2, bupropion (150 mg p.o.) was administered to measure in vivo CYP2B6 activity, and all subjects were treated with RTV, NFV, and RIF. The phenotyping drugs were administered before and after ∼14-day treatment with oral RTV (dose escalation to 400 mg b.i.d.), NFV (1250 mg b.i.d.), or RIF (600 mg qd). The order of treatment was randomized in all studies. Our studies used 400 mg b.i.d. RTV because they were initiated before the exclusive use of low-dose RTV (100 mg) as a “booster,” and many of the unpredictable DDIs described above were observed with the higher doses of RTV (≥200 mg b.i.d.). Subjects fasted after midnight before each study session and until 2 h after administration of the phenotyping drugs. The phenotyping drugs were given approximately 12 h after the last dose of RTV, NFV, or RIF to minimize reversible inhibition and thereby more accurately estimate the fold induction of P450 enzymes. Blood and urine samples were collected before and up to 48 h after phenotyping drug administration. Plasma and urine samples were stored at −20°C until analysis.
Fig. 1.
Study design showing administration of phenotyping drugs and the dosing regimen of RTV, NFV, or RIF treatment. Cocktail A: midazolam (2 mg p.o.) and digoxin (0.5 mg p.o.). Cocktail B: midazolam (1 mg i.v.), caffeine (200 mg p.o.), tolbutamide (500 mg p.o.), and dextromethorphan (30 mg p.o.). Bupropion: bupropion (150 mg p.o.). Treatment ∼14 days: ritonavir (RTV, escalating dose to 400 mg b.i.d.), nelfinavir (NFV, 1250 mg b.i.d.), and rifampin (RIF, 600 mg qd).
Study Drugs, Chemicals, and Reagents.
All study drugs were supplied via the University of Washington Investigational Drug Services. See Table 1 for provider information for study drugs and drug and metabolite reference standards. Optima-grade water, methanol, and methyl t-butyl ether (MTBE) were purchased from Thermo Fisher Scientific (Waltham, MA). β-Glucuronidase was purchased from MP Biomedicals (Solon, OH). All other chemicals used were reagent grade or higher.
TABLE 1.
Study drugs, reference standard provider, and analytical method information
Study drugs: caffeine, 100-mg tablets (GlaxoSmithKline, Uxbridge, Middlesex, UK); bupropion, 150-mg extended release tablets (Watson Pharmaceuticals, Corona, CA); tolbutamide, 500-mg tablets (Mylan, Pittsburgh, PA); and dextromethorphan, 15-mg Robitussin Cough Gels (Wyeth Consumer Healthcare, New York, NY).
| Reference Standards | Supplier Information | Analytical Method | UPLC/MS/MS Detection Parameters: m/z Transition, Cone Voltage, Collision Energy |
|---|---|---|---|
| Caffeine | Cerilliant (Round Rock, TX) | UPLC/MS/MS and HPLC/UV | 195.1 < 137.9, 15, 20 |
| 1-Methyl uric acid | Sigma-Aldrich (St. Louis, MO) | HPLC/UV | N.A. |
| 1,7-Dimethyl uric acid | Sigma-Aldrich | HPLC/UV | N.A. |
| 1-Methyl xanthine | Sigma-Aldrich | HPLC/UV | N.A. |
| AAMU | Sigma-Aldrich | HPLC/UV | N.A. |
| Bupropion | Cerilliant | UPLC/MS/MS | 240.0 < 184.0, 20, 20 |
| 4-OH-BUP | Cerilliant | UPLC/MS/MS | 256.0 < 238.0, 17, 20 |
| Tolbutamide | Sigma-Aldrich | UPLC/MS/MS | 271.1 < 172.0, 25, 10 |
| 4-OH TOLB | Sigma-Aldrich | UPLC/MS/MS | 287.2 < 188.0, 25, 10 |
| Carboxy tolbutamide | Sigma-Aldrich | UPLC/MS/MS | 301.0 < 201.9, 20, 10 |
| Dextromethorphan | Sigma-Aldrich | UPLC/MS/MS | 272.3 < 215.2, 40, 22 |
| Dextrorphan | Sigma-Aldrich | UPLC/MS/MS | 258.2 < 201.2, 45, 22 |
| 3-Methoxy morphinan | Sigma-Aldrich | UPLC/MS/MS | 258.1 < 201.2, 37, 35 |
| Internal standards for quantification | |||
| d4-Midazolam | Cerilliant | UPLC/MS/MS | 330.0 < 295.2, 40, 27 |
| d4-OH midazolam | Cerilliant | UPLC/MS/MS | 346.0 < 328.2, 40, 22 |
| [13C]Caffeine | Toronto Research Chemicals Inc. (North York, ON, Canada) | UPLC/MS/MS | 198.1 < 139.9, 15, 20 |
| Chlorpropamide | Sigma-Aldrich | UPLC/MS/MS | 277.0 < 174.9, 25, 15 |
| 7-β hydroxy propyl theophylline | Sigma-Aldrich | HPLC/UV | N.A. |
N.A., not applicable.
Drug and Metabolite Analysis.
Caffeine, bupropion, 4-hydroxy bupropion (4-OH BUP), tolbutamide, 4-hydroxy tolbutamide (4-OH TOLB), carboxy tolbutamide, dextromethorphan (DEX), dextrorphan (DOR), 3-methoxy morphinan (3MM), and 5-hydroxy morphinan plasma concentrations were determined by an ultraperformance liquid chromatography (UPLC)/tandem mass spectrometry (MS/MS) method. In brief, 50 μl of an internal standard mixture containing d4-midazolam, d4-hydroxymidazolam, [13C]caffeine, and chlorpropamide was added to plasma samples (1 ml), followed by 100 μl of concentrated ammonium hydroxide and 4 ml of MTBE. Samples were mixed for 30 min, centrifuged for 10 min at 2000g, and then the organic layer was removed. Concentrated HCl (200 μl) was added to the remaining sample, which was again extracted with 4 ml of MTBE. The organic layer was removed, combined with the previous extract, and evaporated to dryness under vacuum. The residue was reconstituted in 100 μl of 50:50 0.1% acetic acid in water/0.1% acetic acid in methanol. Fifteen microliters of this solution was analyzed by a previously described UPLC/MS/MS method (Kirby et al., 2011). See Table 1 for ion collection parameters. A calibration curve and quality control samples were prepared in fetal bovine serum because of difficulty in obtaining caffeine-free plasma from the local blood bank. Urine samples were diluted 1:10 or 1:50 and then pretreated with 1000 U of β-glucuronidase in an acetic acid solution (100 mM, pH 5) overnight (∼16 h) before being extracted using the method described above. A set of calibrators and controls were similarly subjected to the β-glucuronidase treatment for quantification of the urine samples. Precision and accuracy of the controls was less than 20% coefficient of variation and 20% error, respectively.
Stereospecific analysis of the urinary (S,S)-4-OH BUP/(S)-BUP ratio was conducted as described previously (Coles and Kharasch, 2007; Kharasch et al., 2008) with minor modifications including optimized liquid chromatography/MS/MS parameters and dilution of urine samples rather than solid-phase extraction. With these modifications, comparable sensitivity, linearity, reproducibility, and enantiomeric separation were achieved as previously published. Urinary caffeine, paraxanthine, 1-methyl uric acid, 1,7-dimethyl uric acid, 1 methyl xanthine, and 5-acetylamino-6-amino-3-methyluracil (AAMU, a combination of AAMU and 5-acetylamino-6-formylamino-3-methyluracil) concentrations were measured using a previously validated high-performance liquid chromatography (HPLC)/UV method (Nyeki et al., 2001).
Pharmacokinetic Analysis.
Noncompartmental analysis of the plasma concentration-time profiles of caffeine, bupropion, tolbutamide, dextromethorphan, and dextrorphan was performed using WinNonlin Professional version 5.0 (Pharsight, Mountain View, CA). Parameters estimated included area under the plasma concentration-time curve (AUC0-t), where t is the last sample time, AUC0-∞, terminal plasma t1/2, and oral clearance (CLPO, Dose/AUC0-∞). Renal clearance of BUP and 4-OH-BUP were calculated as the ratio of the amount of unconjugated drug excreted in the urine to the plasma AUC over the urine collection interval. The (S,S)-4-OH-BUP/(S)-BUP urinary ratio (UR) was calculated as the molar ratio of total (after deconjugation) (S,S)-4-OH-BUP to (S)-BUP in the 24-h urine. Formation clearances (Clform) of paraxanthine, 4-OH BUP, and 4-OH TOLB were estimated by the ratio of the total amount of metabolite plus downstream metabolites (paraxanthine plus downstream metabolites, 4-OH BUP and 4-OH TOLB, plus carboxy tolbutamide) excreted in the 24-h urine and the AUC0–24 of the parent (caffeine, bupropion, and tolbutamide), respectively. The dextrorphan/dextromethorphan AUC ratio (DOR/DEX) was calculated as the ratio of DOR and DEX plasma AUC0–24. URs for DOR/DEX and 3MM/DEX were calculated by the ratio of total amount of DOR or 3MM, respectively, and the total amount of DEX excreted in the 24-h urine. All subjects with a DOR/DEX UR less than 3.3 before treatment were deemed phenotypic CYP2D6 poor metabolizers and were excluded from further analysis (Schmid et al., 1985). All urinary metabolite data were corrected for equivalent mass of the parent compound.
Statistical Analyses.
Because pharmacokinetic (PK) parameters are typically log-normally distributed, statistical analysis was conducted on log-transformed PK parameters. This was done by calculating the geometric mean ratio (GMR) by exponentiation of the average difference of log-transformed PK parameters. If the 90% confidence interval (CI) of this GMR included unity, the treatment was considered to not have significantly altered the PK parameter.
Using historical data of caffeine, bupropion, and tolbutamide pharmacokinetics in healthy volunteers, we conducted an a priori power analysis using plasma AUC as the primary outcome measure. Assuming equal variance between control and treatment groups, our analysis indicated that n = 7 would provide 80% power (α < 0.05) to discern a 100, 40, and 59% change in the plasma AUC of caffeine, bupropion, and tolbutamide, respectively.
In Vitro to In Vivo Prediction of P450 Induction.
In vitro induction of P450 activity and mRNA expression was estimated from our previously published studies in human hepatocytes (Dixit et al., 2007b). In brief, human hepatocytes were treated with increasing concentrations (0–25 μM) of RTV, NFV, or RIF for 72 h (in protein-free media), the cells were harvested, and microsomal P450 activity was evaluated using a validated in vitro phenotyping cocktail (Dixit et al., 2007a). The maximal fold induction (Emax) and concentration resulting in half-maximal induction (EC50) for RTV, NFV, and RIF induction of CYP1A2, CYP2B6, CYP2C9, and CYP2D6 were estimated by fitting eq. 1 (Fahmi et al., 2008) to the data, where I is the total inducer concentration:
In vivo induction of P450 enzymes was predicted using the in vitro derived Emax and EC50 values for each enzyme using eq. 2 (Fahmi et al., 2008), where Iu is the unbound average inducer concentration and d is the in vitro to in vivo scaling factor for induction:
The in vitro to in vivo induction scaling factor for each enzyme was estimated by determining the d value that provided accurate prediction of the in vivo P450 activity GMR (CYP1A2, paraxanthine Clform; CYP2B6, 4-OH BUP Clform; and CYP2C9, 4-OH TOLB Clform) as a result of RIF treatment. These scaling factors were used for the in vitro to in vivo prediction of these P450 enzymes by RTV or NFV using eq. 2.
Results
Subject demographics; treatment periods for RTV, NFV, and RIF; and cocktail administration are described in detail in our previous manuscript addressing the CYP3A-mediated DDIs (Kirby et al., 2011). In brief, 16 healthy volunteers (33 ± 9 years of age, 78 ± 14 kg, and 5 males and 11 females) completed study 1 and 9 volunteers (29 ± 9 years of age, 79 ± 14 kg, and three males and six females) completed study 2.
CYP1A2 (Caffeine).
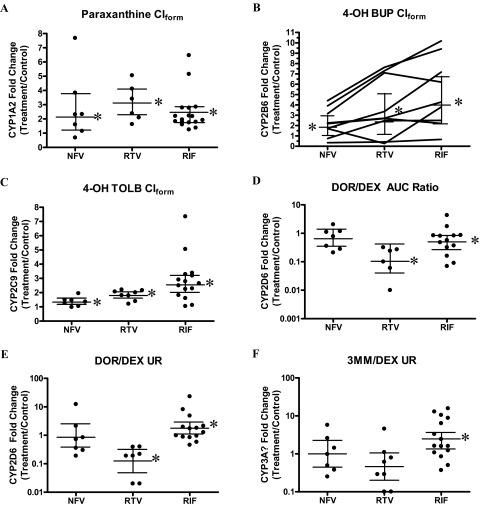
Two subjects in the RTV arm of study 1 were excluded from CYP1A2 analysis because of suspected ingestion of caffeine overnight (24-h plasma caffeine concentrations greater than at 12 h) during the control phase. All treatments resulted in modest induction of in vivo CYP1A2 (Clform GMRs greater than unity), with RTV being the most effective inducer (2.9-fold) and NFV and RIF inducing 2.1- and 2.2-fold, respectively (Fig. 2A; Table 2).
Fig. 2.
Induction of in vivo P450 activity by NFV, RTV, or RIF. A to F show the fold change in in vivo P450 activity for each subject (●) and the GMR with 90% CIs (horizontal bars). A, CYP1A2 activity (paraxanthine Clform) was significantly induced by NFV (2.1-fold), RTV (2.9-fold), or RIF (2.2-fold). B, CYP2B6 activity (4-OH BUP Clform) was variably and significantly induced by NFV (1.8-fold), RTV (2.4-fold), or RIF (4-fold); subjects that showed the greatest induction with RIF also showed the greatest induction with RTV or NFV. C, CYP2C9 activity (4-OH TOLB Clform) was significantly induced by NFV (1.3-fold), RTV (1.8-fold), or RIF (2.6-fold). CYP2D6 activity [DOR/DEX (D) or DOR/DEX UR (E)], as expected, was not significantly induced by NFV or RIF. RTV significantly decreased both CYP2D6 phenotype markers to ∼10% of basal activity. F, the purported CYP3A phenotype marker (3MM/DEX UR) was induced by RIF (2.5-fold), but it was unchanged by NFV or RTV.
TABLE 2.
In vivo CYP1A2, CYP2B6, CYP2C9, or CYP2D6 activity measured by disposition of caffeine, bupropion, tolbutamide, or dextromethorphan, respectively, before and after multiple dose administration of nelfinavir, ritonavir, or rifampin
Bold values are statistically significant (90% CI does not include 1.00).
| Control |
Nelfinavir |
Ritonavir |
Rifampin |
|||||
|---|---|---|---|---|---|---|---|---|
| Average ± S.D. | Average ± S.D. | GMR (90% CI) | Average ± S.D. | GMR (90% CI) | Average ± S.D. | GMR (90% CI) | ||
| CYP1A2 (Caffeine) n = CON 14, NFV 7, RTV 6, RIF 14 | AUC0-∞ (h · μg/ml) | 56.7 ± 46.6 | 30.8 ± 12.9 | 0.49 (0.29–0.84) | 13.90 ± 6.8 | 0.35 (0.26–0.45) | 22.6 ± 7.3 | 0.47 (0.38–0.58) |
| Clearance (ml/min) | 85.4 ± 45.0 | 131 ± 69.0 | 2.03 (1.20–3.45) | 285 ± 128 | 2.90 (2.22–3.80) | 168 ± 73.0 | 2.14 (1.73–2.66) | |
| Clform (ml/min) | 64.3 ± 44.5 | 108 ± 71.2 | 2.13 (1.20–3.81) | 211 ± 88.8 | 2.89 (2.02–4.12) | 132 ± 76.4 | 2.23 (1.79–2.78) | |
| CYP2B6 (Bupropion) n = 9 | AUC0-∞ (h · ng/ml) | 1786 ± 1543 | 1648 ± 1117 | 1.10 (0.75–1.61) | 1201 ± 775 | 0.98 (0.57–1.71) | 962 ± 761 | 0.60 (0.50–0.73) |
| Clearance (l/min) | 5.66 ± 8.80 | 5.49 ± 11.2 | 0.91 (0.62–1.34) | 2.82 ± 1.39 | 1.01 (0.58–1.77) | 8.97 ± 15.7 | 1.66 (1.36–2.02) | |
| Clform (ml/min) | 192 ± 347 | 279 ± 590 | 1.80 (1.08–3.00) | 174 ± 149 | 2.40 (1.13–5.09) | 619 ± 1310 | 3.98 (2.33–6.80) | |
| (S,S)-OH-BUP/(S)-BUP UR | 5.7 ± 5.3 | 9.0 ± 7.5 | 1.77 (1.18–1.84) | 17.7 ± 16.1 | 3.30 (1.97–5.53) | 34.4 ± 34.3 | 4.84 (2.88–8.12) | |
| CYP2C9 (Tolbutamide) n = CON 16, NFV 7, RTV 8, RIF 15 | AUC0-∞ (h · μg/ml) | 596 ± 203 | 455 ± 229 | 0.68 (0.54–0.85) | 295 ± 106 | 0.50 (0.43–0.58) | 216 ± 101 | 0.34 (0.28–0.42) |
| Clearance (ml/min) | 15.7 ± 6.0 | 21.6 ± 8.3 | 1.47 (1.18–1.84) | 33.2 ± 16.7 | 2.00 (1.73–2.31) | 48.3 ± 25.4 | 2.94 (2.39–3.62) | |
| Clform (ml/min) | 6.92 ± 3.59 | 9.80 ± 3.86 | 1.34 (1.13–1.58) | 13.4 ± 9.61 | 1.80 (1.56–2.08) | 19.5 ± 11.1 | 2.55 (2.02–3.22) | |
| CYP2D6 (Dextromethorphan) n = CON 15, NFV 7, RTV 7, RIF 14 | AUC0-∞ (h · ng/ml) | 39.4 ± 56.0 | 46.3 ± 58.4 | 0.87 (0.41–1.82) | 65.1 ± 67.0 | 4.54 (2.35–8.77) | 10.3 ± 19.0 | 0.27 (0.17–0.42) |
| Clearance (ml/min) | 97.1 ± 124 | 102 ± 157 | 1.16 (0.55–2.42) | 19.8 ± 26.3 | 0.22 (0.11–0.43) | 285 ± 292 | 3.73 (2.38–5.86) | |
| DOR/DEX | 3.78 ± 3.99 | 1.17 ± 1.30 | 0.64 (0.35–1.20) | 0.94 ± 1.44 | 0.11 (0.04–0.31) | 1.97 ± 2.36 | 0.50 (0.28–0.88) | |
| DOR/DEX UR Ratio | 306 ± 313 | 375 ± 650 | 0.85 (0.30–2.41) | 37.1 ± 34.1 | 0.13 (0.05–0.32) | 493 ± 407 | 1.77 (1.07–2.94) | |
| CYP3A? (Dextromethorphan) n = CON 15, NFV 7, RTV 8, RIF 15 | 3MM/DEX UR Ratio | 0.72 ± 0.75 | 0.76 ± 1.00 | 1.00 (0.44–2.26) | 0.25 ± 0.19 | 0.46 (0.20–1.09) | 1.49 ± 1.35 | 2.48 (1.28–4.80) |
GMR (treatment/control); CON, control.
CYP2B6 (Bupropion).
All three treatments—nelfinavir, ritonavir, and rifampin—significantly induced the CYP2B6 marker 4-OH BUP Clform by 1.8-, 2.4-, and 4-fold, respectively (Fig. 2B; Table 2). The magnitude of CYP2B6 induction by NFV, RTV, and RIF was highly variable, and in general those subjects that showed the greatest degree of CYP2B6 induction by RIF also had the greatest degree of induction by RTV or NFV. Because the racemic 4-OH-BUP Clform is a combination of the formation rate-limited (S,S)-4-OH BUP and the elimination rate-limited (R,R)-4-OH BUP metabolites, we evaluated the stereospecific 0- to 24-h urinary ratio of the formation rate-limited metabolite (S,S)-4-OH-BUP [(S,S)-4-OH BUP/(S)-BUP UR; Table 2] to confirm the magnitude of CYP2B6 induction observed after RTV, NFV, or RIF treatment as assessed by the racemic 4-OH BUP Clform. Both of these markers showed comparable increases in CYP2B6 activity, with the (S,S)-4-OH-BUP/(S)-BUP UR being a slightly more sensitive measure for RTV and RIF treatments. Rifampin was the only treatment that significantly altered the bupropion clearance (66% increase) and AUC (40% decrease), which is likely due to RIF inducing the non-CYP2B6 clearance of bupropion.
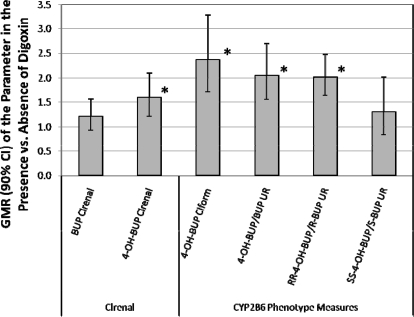
When calculating the 4-OH BUP Clform, we used only the 24-h urine because we observed an unexpected mutual DDI between BUP and the P-glycoprotein probe, digoxin, during the 24- to 48-h period after bupropion administration (see Fig. 1 for study design). This interaction was observed when comparing BUP urinary excretion during the 24- to 48-h period (when digoxin was present) with 0 to 24 h (when it was absent; Fig. 3). When digoxin was present, BUP renal clearance (Clrenal) did not change, but the racemic 4-OH-BUP Clrenal, 4-OH-BUP Clform, racemic 4-OH-BUP/BUP UR, and (R,R)-4-OH-BUP/(R)-BUP UR were significantly increased. It is interesting to note that there was no significant effect of digoxin on the (S,S)-4-OH-BUP/(S)-BUP UR. The dramatic effect of BUP or OH-BUP on digoxin pharmacokinetics will be described in our future manuscript.
Fig. 3.
The effect of an unexpected DDI between the P-glycoprotein probe drug, digoxin, on bupropion pharmacokinetics. Relevant pharmacokinetic parameters of BUP and 4-OH-BUP, in the presence (24–48 h) and absence of digoxin (0–24 h), were compared as GMR and 90% CI. Digoxin caused a significant increase (*, 90% CI does not include unity) in the 4-OH-BUP Clrenal (1.6-fold), racemic 4-OH-BUP Clform (2.4-fold), racemic 4-OH-BUP/BUP UR (2-fold), and (R,R)-4-OH-BUP/(R)-BUP UR (2-fold), but not the (S,S)-4-OH-BUP/(S)-BUP UR.
CYP2C9 (Tolbutamide).
All treatments resulted in a statistically significant induction of in vivo CYP2C9 activity measured by tolbutamide AUC, plasma clearance, or 4-OH TOLB Clform (Table 2; Fig. 2C). 4-OH TOLB Clform is a good measure of CYP2C9 activity because literature data indicate very little contribution of other enzymes (CYP2C19 and CYP2C8) to this pathway (Komatsu et al., 2000). In vivo CYP2C9 activity was increased 2.6-, 1.8-, and 1.3-fold by RIF, RTV, and NFV, respectively. Induction of CYP2C9 was more variable with RIF (range 1–7.4-fold) compared with RTV (range 1.2–2.2-fold) or NFV (range 0.98–1.95-fold).
CYP2D6 (Dextromethorphan).
One subject with a DOR/DEX UR ≤ 3.3 before treatment was deemed a CYP2D6 poor metabolizer and therefore removed from further analysis of CYP2D6 activity. Treatment with NFV did not significantly alter any measured pharmacokinetic parameters of DEX (Table 2; Fig. 2, D–F, note the log y-scale). Ritonavir significantly increased the DEX AUC (4.5-fold) and decreased the clearance of DEX (GMR 0.22). Two commonly used CYP2D6 markers, the DOR/DEX and the DOR/DEX UR, were significantly decreased by RTV (GMR 0.11 and 0.13., respectively). Two subjects in the RTV arm had plasma DOR concentrations that were below the limit of quantification, therefore, for the comparison of DOR/DEX, the number of subjects was six. Rifampin significantly decreased the DEX AUC (GMR 0.27) and increased DEX clearance (3.7-fold). Rifampin treatment significantly affected the CYP2D6 markers (DOR/DEX and DOR/DEX UR), but in an opposing fashion (GMR 0.5 and 1.8, respectively). The DOR/DEX ratio was decreased, implying inhibition of CYP2D6, whereas the DOR/DEX UR was increased, implying induction of CYP2D6. This discrepancy is likely the result of the effect of RIF on the non-CYP2D6 clearance of DEX and DOR (CYP3A and glucuronidation).
The 3MM/DEX UR has been proposed as a marker of in vivo CYP3A activity. This parameter was significantly affected only by treatment with RIF (GMR 2.48) (Table 2; Fig. 2F), which is in contrast to our previously reported data in which RTV, NFV, and RIF all significantly altered CYP3A activity as measured by intravenous and oral midazolam clearance (Kirby et al., 2011).
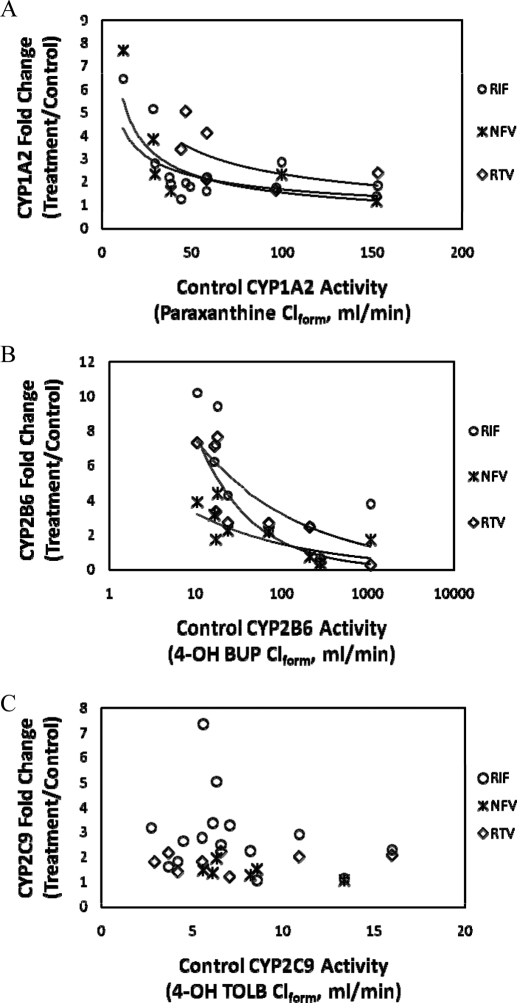
Correlation of Basal P450 Activity and Observed Fold Induction.
Previous reports have shown that the magnitude of in vivo induction of CYP3A is correlated with CYP3A activity before treatment (Gorski et al., 2003). Therefore, we examined if this was true for the in vivo activity of CYP1A2, CYP2B6, or CYP2C9 after RIF, NFV, or RTV treatment (Fig. 4). There was a modest to strong nonlinear inverse correlation of activity before treatment and fold induction of CYP1A2 (R2 = 0.44, 0.70, and 0.39) or CYP2B6 (R2 = 0.47, 0.45, and 0.82) activity by RIF, NFV, or RTV, respectively. In contrast, no such correlation was observed for CYP2C9 activity (R2 < 0.05 for all treatments).
Fig. 4.
Correlation analysis of observed in vivo induction of CYP1A2 (A), CYP2B6 (B), and CYP2C9 (C) after treatment with RIF, NFV, or RTV relative to control P450 activity (4-OH BUP, paraxanthine, or 4-OH TOLB Clform, respectively). In vivo CYP2B6 (A) and CYP1A2 (B) but not CYP2C9 (C) fold induction by RTV, NFV, or RIF treatment was inversely and nonlinearly correlated with basal activity. Those subjects with the highest basal CYP2B6 or CYP1A2 activity tended to show only modest induction of in vivo activity of these enzymes after treatment with the inducers. Note the log scale for control CYP2B6 activity, whereas CYP1A2 and CYP2C9 are displayed on a linear scale. Regression analysis results for CYP1A2 (RIF, y = 12.7x−0.44 R2 = 0.44; NFV, y = 24.1x−0.6 R2 = 0.70; and RTV, y = 29.9x−0.55 R2 = 0.39) and CYP2B6 (RIF, y = 17.4x−0.37 R2 = 0.47; NFV, y = 7.1x−0.34 R2 = 0.45; and RTV, y = 37.3x−0.68 R2 = 0.83).
In Vitro to In Vivo Prediction of P450 Induction.
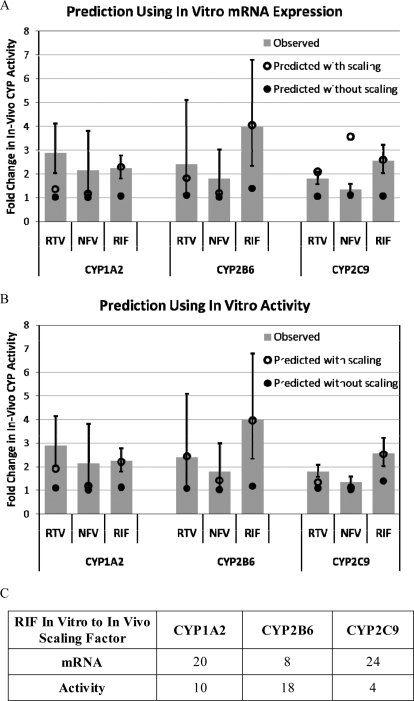
The estimated parameters (Emax and EC50) for RTV, NFV, or RIF induction of CYP1A2, CYP2B6, and CYP2C9 enzyme activity and mRNA expression in human hepatocytes (Table 3) were obtained by re-analysis of our previously published data (Dixit et al., 2007b). The in vitro to in vivo induction scaling factors for RIF were found to be 20, 8, and 24 (mRNA expression) or 10, 18, and 4 (activity) for CYP1A2, CYP2B6, and CYP2C9, respectively (Fig. 5C). These scaling factors scale the fold induction of the P450 mRNA expression or activity measured in human hepatocytes with the fold induction of the P450 activity observed in vivo assuming the average unbound concentration of the inducer is the driving force for induction. Using these scaling factors, four of six induction interactions with RTV or NFV were accurately predicted (within the observed 90% CI) using mRNA expression (Fig. 5A) and five of six using enzyme activity (Fig. 5B).
TABLE 3.
Parameters describing the in vitro induction of CYP1A2, CYP2B6, and CYP2C9 mRNA expression and activity in plated human hepatocytes by rifampin, ritonavir, or nelfinavir
Estimated value (percentage of coefficient of variation of the estimate) in four pooled hepatocyte lots.
| P450 | Parameter | Rifampin |
Ritonavir |
Nelfinavir |
|||
|---|---|---|---|---|---|---|---|
| mRNA | Activity | mRNA | Activity | mRNA | Activity | ||
| CYP1A2 | Emax | 0.160 ± 0.034* | 1.7 (36) | 0.145 ± 0.032* | 1.1 (22) | 0.134 ± 0.034* | 1.6 (30) |
| EC50 (μM) | 5.4 (107) | 1.3 (111) | 5.4 (90) | ||||
| CYP2B6 | Emax | 10.9 (43) | 9.4 (31) | 7.0 (16) | 3.7 (29) | 4.3 (45) | 3.0 (33) |
| EC50 (μM) | 11.1 (94) | 22.6 (55) | 8.0 (41) | 5.5 (87) | 11.0 (102) | 7.9 (84) | |
| CYP2C9 | Emax | 0.165 ± 0.015* | 3.0 (34) | 2.9 (52) | 1.4 (35) | 2.0 (28) | 2.8 (43) |
| EC50 (μM) | 2.7 (133) | 7.5 (135) | 1.9 (157) | 1.0 (153) | 6.4 (119) | ||
Estimates of EC50 were above the highest tested concentration (25 μM); therefore, the slope of the line with intercept forced to 0 was used for extrapolation purposes. Reported value is mean slope ± S.D.
Fig. 5.
In vitro to in vivo prediction of P450 induction using in vitro mRNA expression (A) or enzymatic activity (B). The GMR and 90% CI of the observed in vivo induction of CYP1A2, CYP2B6, and CYP2C9 are shown in gray bars and error bars, respectively. Prediction of the in vivo P450 induction without using RIF in vitro to in vivo scaling (○) generally resulted in minimal to no predicted in vivo induction. In contrast, incorporating the RIF in vitro to in vivo induction scaling factor (●) generally resulted in predicted in vivo P450 induction within the 90% CI of that observed. The RIF scaling factors vary across enzymes and whether mRNA expression or enzymatic activity is used for scaling (C).
Discussion
All treatments resulted in 2- to 3-fold induction of CYP1A2 activity (caffeine ClPO or paraxanthine Clform). There was excellent correlation between the fold change in caffeine CLPO and paraxanthine Clform (R2 = 0.90, slope = 1.01). Thus, caffeine CLPO is an adequate marker of change in CYP1A2 activity in vivo. Induction of CYP1A2 by the PIs and RIF is somewhat surprising because RIF has not been shown (or is not known) to be an aryl hydrocarbon receptor ligand and RTV and NFV are low-affinity aryl hydrocarbon receptor ligands (EC50 > 20 μM) (Frötschl et al., 1998). Previous studies showed that RIF (500–600 mg qd) induces in vivo CYP1A2 ∼20 to 30% (Branch et al., 2000; Backman et al., 2006; Kanebratt et al., 2008) using a single-point caffeine/paraxanthine plasma concentration ratio that is confounded by increased clearance of paraxanthine (mediated by multiple enzymes, including CYP1A2) (Lelo et al., 1989). Our results indicate greater induction of CYP1A2 by RIF (120%) using paraxanthine Clform. Therefore, we speculate that in vivo induction of CYP1A2 by the PIs and rifampin may be a result of nuclear receptor cross-talk (Pascussi et al., 2008) or insensitivity of the in vitro nuclear receptor activation experiments to mimic the in vivo response.
All treatments (NFV, RTV, and RIF) significantly induced CYP2B6 activity measured by 4-OH BUP Clform (1.8-, 2.4-, and 4-fold, respectively). In human liver microsomes, CYP2B6 contributes ∼93% to the formation of 4-OH BUP, with a minor contribution from CYP2C19 (Chen et al., 2010). Thus, CYP2C19 contribution to our CYP2B6 measure is expected to be minimal. A previous study with RTV (400 mg b.i.d.) increased 4-OH BUP Clform (2.1-fold) and decreased BUP AUC 33% (Kharasch et al., 2008). RTV has been shown to cause a dose-dependent decrease in BUP AUC (66 and 22% for 600 and 100 mg b.i.d., respectively) (Park et al., 2010). The reason we did not observe a statistically significant decrease in BUP AUC is unclear. CYP2B6 expression is known to vary as much as 200-fold (Wang and Tompkins, 2008), perhaps a result of genetic polymorphisms, variable expression of CYP2B6, the nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR), and/or environmental factors (Mo et al., 2009). Induction of CYP2B6 is mediated by PXR and CAR (Faucette et al., 2006). NFV, RTV, and RIF are known PXR ligands (Dussault et al., 2001). RTV and NFV are not ligands of the CAR splice variant (CAR3) (Gupta et al., 2008), whereas RIF is not believed to be a CAR ligand (Faucette et al., 2006). This implies that CYP2B6 induction by RTV, NFV, or RIF is PXR mediated. One subject in our study (with the second-highest basal 4-OH BUP Clform) showed no increase in CYP2B6 activity after NFV, RTV, or RIF treatment, but this subject showed the expected response for PXR-mediated CYP3A induction after RIF treatment (Kirby et al., 2011). These data reiterate that induction of CYP2B6 is quite variable.
All treatments (NFV, RTV, and RIF) modestly induced CYP2C9 activity with GMRs of 1.3, 1.8, and 2.6, respectively. Consistent with this observation, RTV decreases the anticoagulant effect of warfarin and acenocoumarol (Hughes et al., 2007; Bonora et al., 2008). CYP2C9 expression is regulated, at least in part, by PXR (Sahi et al., 2009). A correlation analysis of fold induction of CYP1A2, CYP2C9, CYP2B6, and CYP3A showed no significant correlations (R2 ≤ 0.1; data not shown), implying minimal coregulation of these enzymes or an insufficient dynamic range to discern a correlation.
NFV had no significant effect on DEX pharmacokinetics. On the other hand, ritonavir significantly increased the DEX AUC (4.5-fold), decreased the clearance (0.22-fold), and significantly decreased the DOR/DEX and DOR/DEX UR GMRs (∼0.10), implying potent inhibition of CYP2D6. On the basis of the unbound plasma concentration of RTV at the time of DEX dosing (∼0.03 μM) and the modest reversible inhibition Ki (∼4 μM) (von Moltke et al., 1998), reversible inhibition of CYP2D6 by RTV should be minimal. Therefore, we speculate that RTV is either a mechanism-based inactivator of CYP2D6 or a long-lasting metabolite of RTV is a more potent CYP2D6 inhibitor. RIF decreased the AUC and increased the clearance of DEX, likely by induction of CYP3A or conjugation of DEX because CYP2D6 is not inducible by xenobiotics (Benedetti, 2000). However, the DOR/DEX decreased (GMR = 0.5), implying inhibition of CYP2D6, but the DOR/DEX UR increased (GMR = 1.77), implying induction of CYP2D6. These conflicting data suggest that these purported measures of CYP2D6 activity may not be reliable under induced conditions.
Our data show RTV and NFV are in vivo inducers of P450 enzymes other than CYP3A. Therefore, we asked the question, “Can in vivo induction of P450 enzymes be predicted from in vitro hepatocyte experiments and can these data be used to explain some of the unpredictable DDIs with the PIs?” To answer this question, we used our previously published human hepatocyte RIF induction data (Dixit et al., 2007b) (mRNA expression and activity) and in vivo induction of P450 activity (this study) to estimate an in vitro to in vivo induction scaling factor for RIF, an approach similar to that previously used for CYP3A induction (Kirby et al., 2011). Using the induction scaling factors for mRNA or activity, four of six and five of six DDIs were well predicted (prediction fell within the 90% CI of the observed DDI), respectively. In contrast, when the induction scaling factor was not used, the greatest predicted fold induction was 1.4 (RIF − CYP2C9), which was below the observed 90% CI (2.02–3.22). For all of the other DDIs, the prediction without scaling was no induction (≤20%). The difference between scaling factors for mRNA and activity may result from several factors, including differing degrees of mRNA translation into active protein in vitro and/or different shape of the in vitro fold induction versus inducer concentration profile (Emax and EC50).
The induction scaling values being greater than 1 and variable across enzymes implies that the in vitro sensitivity of P450 induction in human hepatocytes is substantially less than that observed in vivo and a “one size fits all” approach to induction scaling across P450 enzymes is unacceptable. For example, the in vitro induction Emax of CYP1A2 was less than 2-fold for all three inducers whereas greater than 2-fold-induction of CYP1A2 activity was observed in vivo for all three inducers. Many factors may contribute to these differences, including 1) different inducer exposure profiles in vitro and in vivo; 2) different expression profiles of the nuclear receptors in vitro and in vivo; 3) an intrinsic insensitivity of the in vitro hepatocytes as a result of being in an artificial environment lacking potentially important paracrine factors; and/or 4) a lack of three dimensional architecture in the in vitro hepatocytes (our previous experiments were conducted in plated rather than sandwich cultured hepatocytes), which may alter the uptake or efflux transport and thereby affect hepatic accumulation of the inducer. Our analysis illustrates the need for in vitro experimental design modifications to mitigate the insensitivity of in vitro hepatocytes relative to in vivo, potentially alleviating the need for scaling factors and/or the variability of scaling factors across enzymes.
In summary, we have shown that multiple dose treatment of RTV (400 mg b.i.d.) or NFV (1250 mg b.i.d.) significantly but modestly induces CYP1A2, CYP2B6, and CYP2C9 activity. Thus, coadministration of this higher dose RTV or NFV with narrow therapeutic index drugs predominantly cleared by CYP1A2, CYP2B6, or CYP2C9 may require dose adjustment to maintain efficacy. Because RTV is now almost exclusively administered at low doses (100–200 mg), we used our scaled in vitro to in vivo prediction method to predicted the fold induction of CYP1A2, CYP2B6, or CYP2C9 by low-dose RTV (100–200 mg b.i.d., with average plasma concentrations ranging from 1 to 2 μM). We predicted ≤30% induction of CYP1A2, CYP2B6, or CYP2C9 at this low RTV dose, a magnitude of induction unlikely to necessitate dose adjustment. However, these predictions only account for induction by RTV and not for the coadministered PIs. In addition, our data show that the magnitude of induction of CYP1A2, CYP2B6, and CYP2C9 can be predicted using in vitro human hepatocytes if an in vitro to in vivo induction scaling factor that is based on RIF is implemented. Additional analyses of in vitro and in vivo data are needed to see if this approach can be applied to predict in vivo P450 induction by other inducers and if other scalars provide better prediction. However, our results do not explain the more perplexing DDIs observed with the PIs, such as the lack of RTV-alprazolam interaction after chronic RTV administration or autoinduction of RTV clearance. The modest induction of CYP1A2 or CYP2C9 by RTV (or for that matter CYP2B6) is unlikely to contribute significantly to the clearance of alprazolam (contribution of CYP1A2, CYP2C9, or CYP2B6 is <1%) (Gorski et al., 1999) or the autoinduction of RTV. Thus, induction of other clearance mechanisms of alprazolam or other unknown mechanisms must contribute to this paradoxical DDI. Clearly, additional studies are needed to determine how the PIs or alprazolam are cleared despite net inactivation of CYP3A and only modest induction of other minor contributing P450 enzymes.
Acknowledgments
We thank Eric Helgeson and Christine Hoffer for clinical study coordination.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM032165]; the National Institutes of Health National Institute on Drug Abuse [Grants K24-DA00417, R01-DA14211]; the National Institutes of Health National Center for Research Resources [Grant M01-RR00037] through the Clinical Research Center Facility at the University of Washington; the National Institutes of Health National Institute of General Medical Sciences [Grant GM07550] (Pharmacological Sciences training grant; to B.K.); a Simcyp sponsored fellowship (to B.K.); and an Achievement Rewards for College Scientists Foundation fellowship (to B.K.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/dmd.111.038646.
- PI
- anti-HIV protease inhibitor
- DDI
- drug-drug interaction
- RTV
- ritonavir
- P-gp
- P-glycoprotein
- RIF
- rifampin
- NFV
- nelfinavir
- P450
- cytochrome P450
- qd
- once daily
- MTBE
- methyl t-butyl ether
- 4-OH BUP
- 4-hydroxy bupropion
- 4-OH TOLB
- 4-hydroxy tolbutamide
- DEX
- dextromethorphan
- DOR
- dextrorphan
- 3MM
- 3-methoxy morphinan
- UPLC
- ultraperformance liquid chromatography
- MS/MS
- tandem mass spectrometry
- BUP
- bupropion
- HPLC
- high-performance liquid chromatography
- AAMU
- 5-acetylamino-6-amino-3-methyluracil
- AUC
- area under the plasma concentration-time curve
- UR
- urinary ratio
- PK
- pharmacokinetic
- GMR
- geometric mean ratio
- CI
- confidence interval
- PXR
- pregnane X receptor
- CAR
- constitutive androstane receptor
- DOR/DOX
- DOR/DOX AUC ratio
- CLPO
- oral clearance
- Clform
- formation clearance
- Clrenal
- renal clearance.
Authorship Contributions
Participated in research design: Collier, Desai, Dixit, Kharasch, Kirby, Thummel, and Unadkat.
Conducted experiments: Dixit, Kirby, and Whittington.
Contributed new reagents or analytic tools: Whittington.
Performed data analysis: Dixit, Kirby, and Whittington.
Wrote or contributed to the writing of the manuscript: Collier, Desai, Dixit, Kharasch, Kirby, Thummel, Unadkat, and Whittington.
References
- Backman JT, Granfors MT, Neuvonen PJ. (2006) Rifampicin is only a weak inducer of CYP1A2-mediated presystemic and systemic metabolism: studies with tizanidine and caffeine. Eur J Clin Pharmacol 62:451–461 [DOI] [PubMed] [Google Scholar]
- Benedetti MS. (2000) Enzyme induction and inhibition by new antiepileptic drugs: a review of human studies. Fundam Clin Pharmacol 14:301–319 [DOI] [PubMed] [Google Scholar]
- Bonora S, Lanzafame M, D'Avolio A, Trentini L, Lattuada E, Concia E, Di Perri G. (2008) Drug interactions between warfarin and efavirenz or lopinavir-ritonavir in clinical treatment. Clin Infect Dis 46:146–147 [DOI] [PubMed] [Google Scholar]
- Branch RA, Adedoyin A, Frye RF, Wilson JW, Romkes M. (2000) In vivo modulation of CYP enzymes by quinidine and rifampin. Clin Pharmacol Ther 68:401–411 [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu HF, Liu L, Nguyen K, Jones EB, Fretland AJ. (2010) The in vitro metabolism of bupropion revisited: concentration dependent involvement of cytochrome P450 2C19. Xenobiotica 40:536–546 [DOI] [PubMed] [Google Scholar]
- Coles R, Kharasch ED. (2007) Stereoselective analysis of bupropion and hydroxybupropion in human plasma and urine by LC/MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci 857:67–75 [DOI] [PubMed] [Google Scholar]
- Cooper CL, van Heeswijk RP, Gallicano K, Cameron DW. (2003) A review of low-dose ritonavir in protease inhibitor combination therapy. Clin Infect Dis 36:1585–1592 [DOI] [PubMed] [Google Scholar]
- Dixit V, Hariparsad N, Desai P, Unadkat JD. (2007a) In vitro LC-MS cocktail assays to simultaneously determine human cytochrome P450 activities. Biopharm Drug Dispos 28:257–262 [DOI] [PubMed] [Google Scholar]
- Dixit V, Hariparsad N, Li F, Desai P, Thummel KE, Unadkat JD. (2007b) Cytochrome P450 enzymes and transporters induced by anti-human immunodeficiency virus protease inhibitors in human hepatocytes: implications for predicting clinical drug interactions. Drug Metab Dispos 35:1853–1859 [DOI] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Wang EH, Synold TW, Forman BM. (2001) Peptide mimetic HIV protease inhibitors are ligands for the orphan receptor SXR. J Biol Chem 276:33309–33312 [DOI] [PubMed] [Google Scholar]
- Ernest CS, 2nd, Hall SD, Jones DR. (2005) Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J Pharmacol Exp Ther 312:583–591 [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Maurer TS, Kish M, Cardenas E, Boldt S, Nettleton D. (2008) A combined model for predicting CYP3A4 clinical net drug-drug interaction based on CYP3A4 inhibition, inactivation, and induction determined in vitro. Drug Metab Dispos 36:1698–1708 [DOI] [PubMed] [Google Scholar]
- Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. (2006) Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 317:1200–1209 [DOI] [PubMed] [Google Scholar]
- Fichtenbaum CJ, Gerber JG. (2002) Interactions between antiretroviral drugs and drugs used for the therapy of the metabolic complications encountered during HIV infection. Clin Pharmacokinet 41:1195–1211 [DOI] [PubMed] [Google Scholar]
- Frötschl R, Chichmanov L, Kleeberg U, Hildebrandt AG, Roots I, Brockmöller J. (1998) Prediction of aryl hydrocarbon receptor-mediated enzyme induction of drugs and chemicals by mRNA quantification. Chem Res Toxicol 11:1447–1452 [DOI] [PubMed] [Google Scholar]
- Gorski JC, Jones DR, Hamman MA, Wrighton SA, Hall SD. (1999) Biotransformation of alprazolam by members of the human cytochrome P4503A subfamily. Xenobiotica 29:931–944 [DOI] [PubMed] [Google Scholar]
- Gorski JC, Vannaprasaht S, Hamman MA, Ambrosius WT, Bruce MA, Haehner-Daniels B, Hall SD. (2003) The effect of age, sex, and rifampin administration on intestinal and hepatic cytochrome P450 3A activity. Clin Pharmacol Ther 74:275–287 [DOI] [PubMed] [Google Scholar]
- Greenblatt DJ, von Moltke LL, Harmatz JS, Durol AL, Daily JP, Graf JA, Mertzanis P, Hoffman JL, Shader RI. (2000) Alprazolam-ritonavir interaction: implications for product labeling. Clin Pharmacol Ther 67:335–341 [DOI] [PubMed] [Google Scholar]
- Gupta A, Mugundu GM, Desai PB, Thummel KE, Unadkat JD. (2008) Intestinal human colon adenocarcinoma cell line LS180 is an excellent model to study pregnane X receptor, but not constitutive androstane receptor, mediated CYP3A4 and multidrug resistance transporter 1 induction: studies with anti-human immunodeficiency virus protease inhibitors. Drug Metab Dispos 36:1172–1180 [DOI] [PubMed] [Google Scholar]
- Hsu A, Granneman GR, Bertz RJ. (1998) Ritonavir. Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin Pharmacokinet 35:275–291 [DOI] [PubMed] [Google Scholar]
- Hughes CA, Freitas A, Miedzinski LJ. (2007) Interaction between lopinavir/ritonavir and warfarin. CMAJ 177:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanebratt KP, Diczfalusy U, Bäckström T, Sparve E, Bredberg E, Böttiger Y, Andersson TB, Bertilsson L. (2008) Cytochrome P450 induction by rifampicin in healthy subjects: determination using the Karolinska cocktail and the endogenous CYP3A4 marker 4beta-hydroxycholesterol. Clin Pharmacol Ther 84:589–594 [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Mitchell D, Coles R. (2008) Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol 48:464–474 [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Mitchell D, Coles R, Blanco R. (2008) Rapid clinical induction of hepatic cytochrome P4502B6 activity by ritonavir. Antimicrob Agents Chemother 52:1663–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby BJ, Collier AC, Kharasch ED, Whittington D, Thummel KE, Unadkat JD. (2011) Complex drug interactions of HIV protease inhibitors 1: inactivation, induction, and inhibition of cytochrome P450 3A by ritonavir or nelfinavir. Drug Metab Dispos 39:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Ito K, Nakajima Y, Kanamitsu S, Imaoka S, Funae Y, Green CE, Tyson CA, Shimada N, Sugiyama Y. (2000) Prediction of in vivo drug-drug interactions between tolbutamide and various sulfonamides in humans based on in vitro experiments. Drug Metab Dispos 28:475–481 [PubMed] [Google Scholar]
- Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N. (1998) Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos 26:552–561 [PubMed] [Google Scholar]
- Kumar GN, Rodrigues AD, Buko AM, Denissen JF. (1996) Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J Pharmacol Exp Ther 277:423–431 [PubMed] [Google Scholar]
- Lelo A, Kjellen G, Birkett DJ, Miners JO. (1989) Paraxanthine metabolism in humans: determination of metabolic partial clearances and effects of allopurinol and cimetidine. J Pharmacol Exp Ther 248:315–319 [PubMed] [Google Scholar]
- Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. (2009) Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab 10:730–753 [DOI] [PubMed] [Google Scholar]
- Nyéki A, Biollaz J, Kesselring UW, Décosterd LA. (2001) Extractionless method for the simultaneous high-performance liquid chromatographic determination of urinary caffeine metabolites for N-acetyltransferase 2, cytochrome P450 1A2 and xanthine oxidase activity assessment. J Chromatogr B Biomed Sci Appl 755:73–84 [DOI] [PubMed] [Google Scholar]
- Park J, Vousden M, Brittain C, McConn DJ, Iavarone L, Ascher J, Sutherland SM, Muir KT. (2010) Dose-related reduction in bupropion plasma concentrations by ritonavir. J Clin Pharmacol 50:1180–1187 [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Duret C, Daujat-Chavanieu M, Vilarem MJ, Maurel P. (2008) The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu Rev Pharmacol Toxicol 48:1–32 [DOI] [PubMed] [Google Scholar]
- Sahi J, Shord SS, Lindley C, Ferguson S, LeCluyse EL. (2009) Regulation of cytochrome P450 2C9 expression in primary cultures of human hepatocytes. J Biochem Mol Toxicol 23:43–58 [DOI] [PubMed] [Google Scholar]
- Schmid B, Bircher J, Preisig R, Küpfer A. (1985) Polymorphic dextromethorphan metabolism: co-segregation of oxidative O-demethylation with debrisoquin hydroxylation. Clin Pharmacol Ther 38:618–624 [DOI] [PubMed] [Google Scholar]
- Su Y, Zhang X, Sinko PJ. (2004) Human organic anion-transporting polypeptide OATP-A (SLC21A3) acts in concert with P-glycoprotein and multidrug resistance protein 2 in the vectorial transport of Saquinavir in Hep G2 cells. Mol Pharm 1:49–56 [DOI] [PubMed] [Google Scholar]
- Unadkat JD, Wang Y. (2000) Protease inhibitors, in Metabolic Drug Interactions (Levy RH, Thummel KE, Trager WF, Hansten PD, Eichelbaum M. eds) pp 647–652, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- von Moltke LL, Greenblatt DJ, Duan SX, Daily JP, Harmatz JS, Shader RI. (1998) Inhibition of desipramine hydroxylation (cytochrome P450–2D6) in vitro by quinidine and by viral protease inhibitors: relation to drug interactions in vivo. J Pharm Sci 87:1184–1189 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gorski JC, Hamman MA, Huang SM, Lesko LJ, Hall SD. (2001) The effects of St John's wort (Hypericum perforatum) on human cytochrome P450 activity. Clin Pharmacol Ther 70:317–326 [PubMed] [Google Scholar]
- Ye ZW, Camus S, Augustijns P, Annaert P. (2010) Interaction of eight HIV protease inhibitors with the canalicular efflux transporter ABCC2 (MRP2) in sandwich-cultured rat and human hepatocytes. Biopharm Drug Dispos 31:178–188 [DOI] [PubMed] [Google Scholar]