Abstract
DNA and histone modifications direct the functional state of chromatin and thereby the readout of the genome. Candidate approaches and histone peptide affinity purification experiments have identified several proteins that bind to chromatin marks. However, the complement of factors that is recruited by individual and combinations of DNA and histone modifications has not yet been defined. Here, we present a strategy based on recombinant, uniformly modified chromatin templates used in affinity purification experiments in conjunction with SILAC-based quantitative mass spectrometry for this purpose. On the prototypic H3K4me3 and H3K9me3 histone modification marks we compare our method with a histone N-terminal peptide affinity purification approach. Our analysis shows that only some factors associate with both, chromatin and peptide matrices but that a surprisingly large number of proteins differ in their association with these templates. Global analysis of the proteins identified implies specific domains mediating recruitment to the chromatin marks. Our proof-of-principle studies show that chromatin templates with defined modification patterns can be used to decipher how the histone code is read and translated.
DNA methylation and histone post-translational modifications (PTM)1 play important roles in regulating chromatin states and thereby the use and readout of the genome. Trimethylation of lyinse 4 (H3K4me3) and lysine 9 (H3K9me3) of histone H3 have, for example, been connected to transcriptional activation and repression, respectively. They therefore present a prototypic pair of antagonistic histone PTMs.
Generally, chromatin marks either influence chromatin packaging directly or via recruitment of specific proteins and multiprotein complexes that mediate downstream effects (1, 2). Candidate approaches of individual factors or using targeted libraries of protein families together with histone tail peptide affinity purification experiments carried out in isolation or on peptide arrays have identified a number of proteins that specifically interact with individual chromatin marks (see for example ref. 3–6). These include factors containing methyl-DNA binding domains as well as chromodomains, plant homeodomain (PHD) fingers, tudor domains, and ankyrin repeats interacting with histone methyl-lysine residues. Further, 14-3-3 proteins interacting with histone phospho-serine residues and bromodomain containing factors binding to histone acetyl-lysine residues have been described (7). In vitro studies have characterized the exact binding specificities of several proteins containing these domains. Also, structural insights are now available for a number of chromatin mark binding complexes (7, 8).
Interestingly, the interactions of individual domains of chromatin modification binding proteins with their cognate marks are rather weak (interaction strength in the micromolar range) (9). Although the study of interactions of individual proteins with DNA methylation or distinct histone PTM marks has been central to our current understanding of chromatin mediated processes, it is emerging that patterns of marks rather than individual modifications direct functional states of chromatin (10, 11). Here, factors containing multiple domains interacting with different chromatin marks have gained high interest (12). Multivalent binding might not only allow for stronger and thereby more discriminatory interaction than single domain binding, but could also direct readout of complex patterns of modifications. Also, multiprotein complexes appear to contain several factors with the same or distinct chromatin mark recognition functionality thereby possibly establishing more stable interaction.
Gaining global insight into the relationship of chromatin modifications and functional states of chromatin ultimately requires isolation and characterization of intact chromatin domains from cells. In absence of such experimental systems in vitro approaches that mimic and incorporate different DNA methylation and histone PTM configurations will likely be extremely useful in defining the complement of factors that targets a given pattern of chromatin marks. Here, DNA and/or histone tail peptide affinity purification experiments can only be of limited value as only individual or shortly spaced combinatorial patterns of modifications can be analyzed (see for example ref. 13).
Nonquantitative mass spectrometry (e.g. MudPIT, ref. 14) analysis of differential affinity purification reactions has been useful in identifying proteins binding a given target (4). However, because these methods do not provide sufficient quantitative information on the proteins recovered in separate experiments in the first place, factors that bind two separate matrices (e.g. sample and control) with different strength will not be necessarily recognized as specific interaction partners of either one. Therefore, different mass spectrometry methods have been introduced that allow identification and sensitive quantification of proteins in matched experiments (15). Especially, isotope labeling by amino acids in cell culture (SILAC) has proven useful in various proteomics based approaches (16). Here, we set out to establish an in vitro system usable for the analysis of complex chromatin modification patterns based on recombinant, uniformly modified chromatin templates in combination with quantitative SILAC-based mass spectrometry analysis. In this manner, we defined the interactome of the H3K4me3 and H3K9me3 chromatin marks. Surprisingly, only some factors were also recruited to corresponding histone N-terminal peptides in parallel experiments. Our results set the stage for using chromatin-based affinity approaches to investigate how the histone code is read and translated on a global scale.
EXPERIMENTAL PROCEDURES
Cell Culture, Labeling, and Nuclear Extract Preparation
HeLa S3 cells were grown in lysine- and arginine-deficient Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal bovine serum (PAA, Pasching, Austria). One cell population was supplemented with normal isotope containing l-lysine and l-arginine (Sigma, Munich, Germany) and another with heavy isotope labeled 13C6-lysine and 13C615N4-arginine (Euriso-Top, Saint-Aubin Cedex, France) generating mass shifts of +6 and +10 Da, respectively. Cells were grown for at least six passages at smaller volumes and then expanded to 2 l in spinner flasks (0.5–1.0 × 106 cells/ml) (16). The cells were then transferred to a 5 l fermenter (Applikon, Schiedam, Netherlands) and grown under standard conditions (2.5–5.0 × 106 cells/ml). Harvested cells were used to prepare nuclear extract according to standard procedures (17).
Peptides
Peptides containing the 20 N-terminal amino acids of histone H3 were synthesized in unmodified and modified form using Fmoc (N-(9-fluorenyl)methoxycarbonyl)-based solid-phase synthesis H3unmodified: ARTKQTARKSTGGKAPRKQL; H3K4me3: ARTK(me3)QTARKSTGGKAPRKQL; H3K9me3: ARTKQTARK(me3)STGGKAPRKQL. Peptides contained a C-terminal non-native lysine biotinylated at the ε-amino group for affinity purification reactions or were transformed to thioacetamidthiophenylesters for native protein ligation (18–20).
Native Protein Ligation
Histone modifications were achieved by native protein ligation using histone H3 (1–20) thioester peptides and recombinant X. laevis histone H3Δ1–20,A21C as described (21). Reactions were carried out in 100 mm potassium phosphate (pH 7.9), 3 m guanidine-HCl, 0.5% v/v benzyl mercaptan, 0.5% v/v thiophenol at 25 °C with vigorous mixing. Crude reaction mixture was diluted 50-fold into SAU-200 buffer (7 m deionized urea, 20 mm sodium acetate (pH 5.2), 1 mm EDTA, 1 mm dithiotreitol, 200 mm NaCl), applied to a 5 ml Hi-Trap SP-Sepharose high performance cation exchange column (GE Healthcare, Munich, Germany), and eluted with a linear NaCl gradient from 200 to 600 mm in 10 column volumes. Protein was dialyzed extensively against 2 mm dithiotreitol at 4 °C, lyophilized and stored at −80 °C. Routinely we set up ligation reactions containing 27 mg histone (2 μmol), 23 mg thioacetamidthiophenylester histone H3 peptide (10 μmol) in 10 ml reaction volume. After purification on average 10 mg ligated histone H3 was obtained (0.6 μmol). Purity and identity of thioester peptides and ligated proteins was confirmed by analytical high-performance liquid chromatography, mass spectrometry, and SDS-PAGE (see supplemental Fig. S1).
Recombinant Chromatin
Recombinant chromatin was prepared essentially as described (22). Briefly, recombinant wild type Xenopus laevis histones were expressed in Escherichia coli and purified as described (23). Assembly of histone octamers containing modified and unmodified histone H3 as well as nucleosome array reconstitution was performed by salt dialysis on biotinylated 12 × 200 × 601 DNA template as described (23, 24). Quality of chromatin reconstitution was monitored by native agarose gel electrophoresis, MNase digest, and analytical ultracentrifugation (see supplemental Fig. S2).
Peptide and Chromatin Affinity Purifications
Affinity purifications were performed essentially as described using two separate preparations of nuclear extract (22). Each experiment was performed in “forward” (light extract, unmodified chromatin and peptide; heavy extract, modified chromatin and peptide) and “reverse” (light extract, modified chromatin and peptide; heavy extract, unmodified chromatin and peptide) label swap affinity purification. For peptide affinity purifications, 40 μl prewashed streptavidin coated paramagnetic beads (Pierce, Rockford, IL) were saturated with 10 μg biotinylated histone peptide overnight at 4 °C. A 0.5 ml aliquot of precleared HeLa S3 nuclear extract (light or heavy isotope labeled) was incubated with the peptide-bound paramagnetic beads for 4 h while rotating. Beads were washed three times with 1 ml of PD150 buffer (20 mm HEPES pH 7.9, 150 mm NaCl, 0.1% Triton X-100, 5% glycerol). Beads from parallel affinity purification reactions using unmodified and modified peptides were mixed (25) and bound proteins were eluted with LDS sample buffer (Invitrogen, Carlsbad, CA). Chromatin affinity purifications were performed accordingly using 50 μg chromatin and 200 μl paramagnetic beads. To improve SDS-PAGE resolution, eluates of chromatin affinity purification reactions were incubated with 1 kU benzonase (Calbiochem, San Diego, CA) nuclease for 1 h at 37 °C.
Western Blotting
Primary antibodies used were: αH3K4me3 (1:2,000, Abcam, Cambridge, UK), αH3K9me3 (1:1,000, Millipore, Billerica, MA), αFLAG (1:1,000, Sigma, Munich, Germany), and αSMCHD1 (1:1,000, Abcam, Cambridge, UK).
LC-MS/MS
Eluted proteins were separated on 4–12% gradient SDS-PAGE gels (Invitrogen, Carlsbad, CA) and stained with Colloidal Coomassie Blue. Each gel lane was cut into 23 equal gel slices and proteins therein were in-gel digested with trypsin (Promega, Madison, WI) as described (26). Tryptic peptides from each gel slice were extracted and analyzed by nanoflow HPLC (Agilent, Boeblingen, Germany) coupled to nanoelectrospray LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operating in positive ion mode. Peptides were first loaded and desalted onto an in-line trap column (1.5 cm length, 150 μm inner diameter, packed in-house with Reprosil AQ-5 μm/300Å) and then separated on analytical column (15 cm length, 75 μm inner diameter, as trap column) at flow-rate 250 nL/min and linear gradient from 7.5 to 37.5% acetonitrile in 0.1% (v/v) formic acid for 50 min. Data-dependent acquisition of eluting peptides was applied and consisted of one survey scan in Orbitrap (with resolution set to 30,000 at m/z 400 and automatic gain control target at 106) followed by MS/MS of the five most intense precursors in the LTQ using collision-induced decay fragmentation with previously fragmented ions dynamically excluded for 90 s. Each sample was analyzed in three technical replicates.
Data Analysis
Raw MS files from LTQ-Orbitrap XL were analyzed by MaxQuant software (version 1.0.13.13) (27). Peak lists generated by Quant.exe (the first module of MaxQuant) were searched by Mascot search engine (Mascot Daemon 2.2.2; Matrix Science, London, UK) using International Protein Index (IPI) Human protein database (version 3.73, June 2010, containing 89739 entries) supplemented with 179 common contaminants (e.g. keratins, serum albumin) and concatenated with the reverse sequences of all entries. Mascot search parameters were used as follows: Carbamidomethylation of cystein and oxidation of methionine were set as variable modifications, tryptic specificity with no proline restriction and up to two missed cleavages was used. The initial mass tolerance used was 7 ppm and for MS/MS 0.6 Da. Only peptides with minimal length of six amino acids were considered. Peptides were filtered for maximum false discovery rate of 1% in MaxQuant. Only unique and razor peptides with posterior error probability of less than 0.05 and proteins with ratio count of at least three were accepted and used for quantification (27). Results from MaxQuant were analyzed and visualized with R (script details are available in supplemental Experimental Procedures). Proteins showing opposite ratios between forward and reverse label-swap experiments were manually validated using MaxQuant Viewer, marked as potential false positives and included into a separate list that was not used for plotting with R. All proteins are listed in supplemental Tables S1–S4 with accession numbers, number of unique peptides, % sequence coverage, quantification significance, and variability, as reported by MaxQuant.
For enrichment analysis-based hierarchical clustering, the quantified proteins from each experiment were divided into five lists corresponding to enrichment ratio cutoffs of below –4 to –4, –4 to –2, –2 to 2, 2 to 4, and above 4. Proteins from each list were searched for enriched protein domains terms using DAVID (28) without enrichment score cutoff (databases used: UniProt, Sequence Feature, InterPro, PIR Superfamily, PFAM, SMART). The resulting lists were then collated using a Python script (script details are available in supplemental Experimental Procedures; http://www.python.org). All terms being enriched with EASE score from DAVID of better than 0.1 in at least one of the lists were included into a combined list. Hierarchical clustering was done in the R statistical environment using the Euclidean distance function and combined linkage method.
Plasmids
cDNAs encoding mACTL8 (IMAGE ID:IRATp970D1240D), mFANCF (IMAGE ID:IRATp970B10123D), mSPIN1 (IMAGE ID:IRAVp968H0931D), and mADNP (FANTOM3 ID:6330563C07) were obtained from Imagenes. cDNA encoding mZmym3 was amplified from cDNA isolated from NIH3T3 cells (reference sequence GenBank NM_019831.3). cDNAs were cloned into a modified pcDNA3.1 vector fusing the 3′ end to a 2xHA-2XFLAG tag. The following primer pairs were used for PCR amplification. pCDNA-Actl8-HAHAFlFl(BamHI - NotI), fwd: GAGCTCGGATCCATGGCTTCAAGAACCGTTATC, rev: GGGTATGCGGCCGCCATCCTCATATGCTCACCATAC; pCDNA-FancF-HAHAFlFl (BamHI - NotI) fwd: GAGCTCGGATCCATGGAATCCCTTCTGCAGCAC, rev: GGGTATGCGGCCGCTACAGAACTGAGGCCTGCGC; pCDNA-SPIN1-HAHAFlFl(AflII - NotI) fwd: TTTAAACTTAAGCTTATGAAGACCCCATTCGGGAAG, rev: GGGTATGCGGCCGCGGATGTTTTCACCAAATCGTAG; pCDNA-Zmym3-HAHAFlFl(AflII - NotI) fwd: TTTAAACTTAAGATGGACCCCAGTGATTTCCC, rev: GGGTATGCGGCCGCGTCTAGGTCTTCTTCCCCAG; pCDNA-ADNP-HAHAFlFl(BamHI - NotI) fwd: GAATTGGGATCCATGTTCCAACTTCCTGTCAAC, rev: GGGTATGCGGCCGCGGCTTGCTGGCTGCTCAGC. Details of cloning are available upon request.
Cell Culture and Transfection
NIH3T3 and HEK 293T (ATCC) cells were grown at 37 °C in a humidified atmosphere, 5% CO2 using Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, and 1 × penicillin/streptomycin (Invitrogen, Carlsbad, CA). Transfection was performed using JetPEI reagent (Biomol, Hamburg, Germany). Typically 6 μl JetPEI reagent and 3 μg of plasmid were used per coverslip in a 6-well plate (NIH3T3) and 40 μl JetPEI reagent and 20 μg of plasmid were used per 15 cm dish (HEK 293T).
Immunofluorescence
Cells were seeded on glass cover slips in 6-well plates. Cover slips were washed two times with phosphate-buffered saline (PBS) and fixed for 10 min in 3% paraformaldehyde in PBS at 37 °C. Cover slips were washed once with PBS and cells were permeabilized for 10 min (0.5% Triton X-100 in PBS). Cells were washed once with 0.2% Triton X-100 in PBS before blocking for 30 min in blocking solution (1 × PBS, 2% BSA, 0.2% Triton X-100 (v/v), 5% normal goat serum (v/v)). Primary antibodies (anti-FLAG M2 (Sigma, Munich, Germany) 1:1,000; anti-H3K9me3; anti-H3K4me3 (both 1:500, Millipore, Billerica, MA)) and fluorescently labeled secondary antibodies (anti-mouse-Alexa555 and anti-rabbit-Alexa488 (Molecular Probes, Eugene, OR) 1:1,000) were applied in blocking solution for 1 h at room temperature. Cover slips were washed two times with 0.2% Triton X-100 in PBS and five times in water. Cover slips were mounted in Mowiol including 50 μg/ml DAPI. Slides were dried overnight at RT and analyzed using a Leica TCS SP5 confocal microscope.
RESULTS AND DISCUSSION
Experimental Set-up
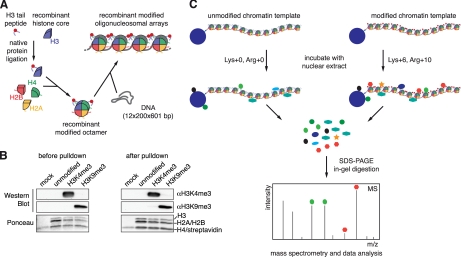
To obtain homogenously modified histone H3 proteins containing H3K4me3 or H3K9me3 we used native protein ligation of synthetic histone H3 (1–20) N-terminal peptides containing these marks and core recombinant histone H3Δ1–20,A21C protein expressed in bacteria (Fig. 1A, supplemental Fig. S1) (21). In conjunction with other core histones H2A, H2B, and H4 octamers were assembled and deposited onto 5′ biotinylated 12 × 200 × 601 DNA template, which contains strong nucleosome positioning potential, using the salt dialysis method (23, 24). Material assembled with unmodified H3, H3K4me3, and H3K9me3 was analyzed by SDS-PAGE and via MNase digest confirming similar histone content and nucleosome density (supplemental Fig. S2). Additional biophysical analysis using analytical ultracentrifugation and atomic force microscopy verified identical hydrodynamic and structural behavior of the different chromatin templates (data not shown).
Fig. 1.
Set-up and workflow of the peptide and chromatin affinity purification system. A, Scheme of reconstitution of recombinant, uniformly modified chromatin using native protein ligation. B, The indicated chromatin templates were analyzed before and after affinity purification reactions using SILAC-labeled nuclear HeLa S3 extract by Western blot using the specified antibodies. Ponceau staining of the membrane served as loading control. C, Workflow of the chromatin affinity purification experiment and its analysis.
Biotinylated, reconstituted oligonuclesomes or synthetic peptides unmodified or containing the H3K4me3 or H3K9me3 modifications were immobilized on magnetic streptavidin beads and incubated with nuclear extract prepared from HeLa S3 cells grown under SILAC conditions. We used normal isotope containing l-lysine and l-arginine for preparing “light” extracts and heavy isotope labeled 13C6-lysine (Lys+6) and 13C615N4-arginine (Arg+10) for generating “heavy” extracts. Extracts were analyzed in trial experiments to verify high synchronicity of “light” and “heavy” material (i.e. both extracts contain the same proteins and in the same amounts but only differ in the labeling of the proteins, supplemental Fig. S3). In forward experiments unmodified chromatin or peptide templates were incubated with “light” extracts, whereas corresponding H3K4me3 or H3K9me3 matrices were incubated with “heavy” extracts. In reverse experiments this order was swapped (label swap experiment).
Western blot analysis of the chromatin templates before and after incubation with extract verified the identity of the modifications (Fig. 1B). Importantly, no loss of H3K4me3 or H3K9me3 after exposure to nuclear extracts was observed. Similarly, mass spectrometric analysis verified identity of the histone H3 N-terminal peptides (data not shown).
Proteins specifically retained on the different matrices were recovered from the beads. Eluates were mixed in 1:1 ratio, run on SDS-PAGE gels and the separated proteins were trypsinized. The tryptic peptides were then analyzed by mass spectrometry and quantitative ratios for heavy and light SILAC pairs were determined (Fig. 1C). To minimize random variation, experiments were repeated twice (biological replicates) for each set-up, i.e. each combination of unmodified versus H3K4me3 or H3K9me3 was run twice in the forward and twice in the reverse direction. Each reaction was analyzed three times (technical replicates) on the mass spectrometer to maximize the probability of faithful identification of all factors in the samples.
H3K4me3 and H3K9me3 Interactomes
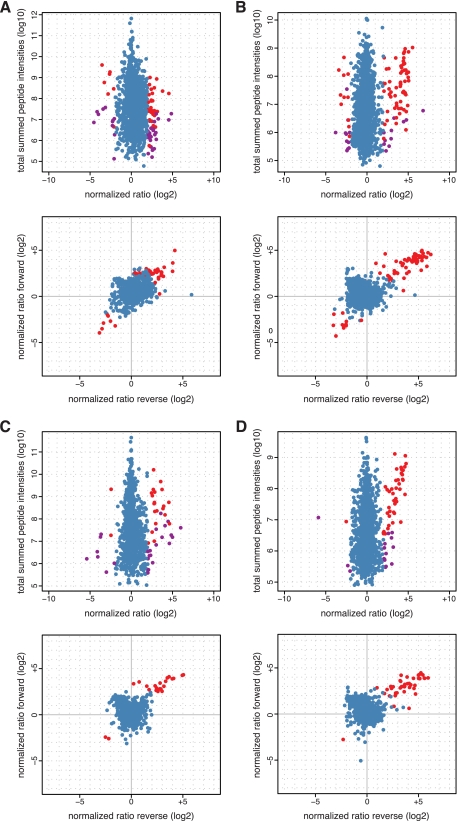
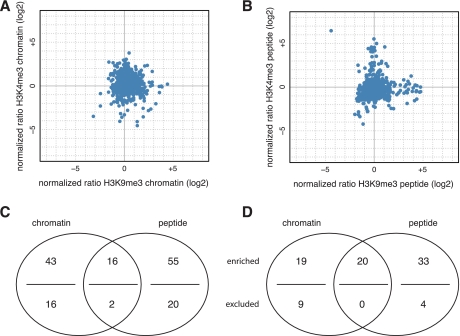
We analyzed a total of ca. 2000 factors in each of the affinity purification experiments out of approximately 5000 proteins present in the Hela S3 nuclear extract (data not shown). These include proteins specifically and unspecifically enriched on the streptavidin beads as well as the chromatin or peptide containing matrices (Fig. 2). By comparing unmodified and modified templates we detected factors enriched (i.e. preferentially binding modified versus unmodified) or excluded (i.e. preferentially binding unmodified versus modified) by the H3K4me3 and H3K9me3 modifications for chromatin as well as peptide-containing matrices to different degrees beyond this background. Applying a relatively stringent threshold of fourfold change, 59 of these were found enriched on H3K4me3 chromatin of which 32 were represented in forward and reverse experimental set-up. Conversely, 18 factors were found excluded from this matrix of which seven were found in both forward and reverse experiments (Table I, Fig. 2A). For the H3K4me3 peptide we found 71 factors enriched with 60 of these represented in forward and reverse experiments; 22 factors were excluded from this matrix with eleven found in forward and reverse experiments (Fig. 2B). For the H3K9me3 chromatin affinity purification we found 39 factors enriched by this modification of which 21 were represented in forward and reverse experiments; nine factors were found excluded from this matrix with two represented in forward and reverse experiments (Table II, Fig. 2C). With the H3K9me3 histone N-terminal tail peptide we found 53 factors enriched with 40 of these found in forward and reverse experiments; four factors were excluded by this modification on the peptide of which one was represented in forward and reverse experiments (Fig. 2D).
Fig. 2.
Identification of factors recruited by H3K4me3 or H3K9me3 using recombinant, uniformly modified chromatin or histone tail peptide affinity purification and SILAC-MS. A, H3K4me3 chromatin. B, H3K4me3 histone tail peptide. C, H3K9me3 chromatin. D, H3K9me3 histone tail peptide. Upper panels: Scatter plots representing normalized ratios of identified and quantified proteins and total summed peptide intensities. Proteins identified in forward and reverse set-up of the experiment with ratio change above four are colored in red; those only identified in forward or reverse set-up of the experiment but with a ratio change above four are colored in purple. Factors with enrichment or exclusion ratios below four are colored in blue. Lower panels: Scatter plots representing normalized ratios of identified and quantified proteins in both, forward and reverse experiments. Proteins with ratio change above four are shown in red; those with enrichment or exclusion ratios below four are colored in blue. For details on the identified proteins see Tables I and II. In all plots proteins showing opposite ratios between forward and reverse experimental set-up (and therefore potential false positives) are omitted.
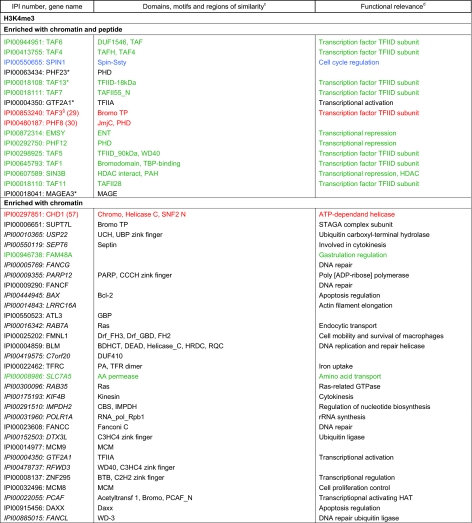
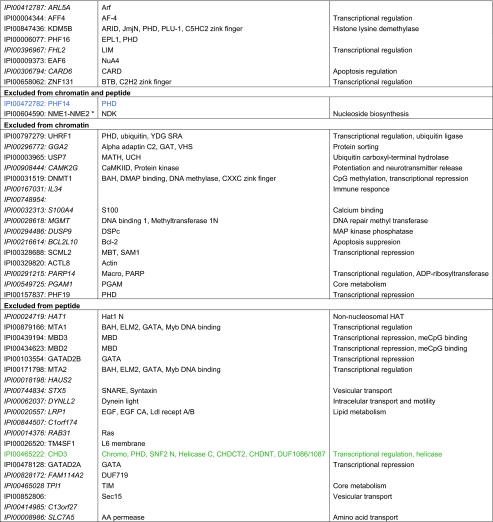
Table I. Factors found enriched or excluded on H3K4me3 chromatin and/or peptide templates from HeLa S3 nuclear extracts. a, b, c, d Factors in italics were identified only in forward or reverse set-up of the experiments. Asterisks mark factors that were identified in forward and reverse set-up of the experiment with either peptide or chromatin templates, but were only found in forward or reverse set-up of the experiment using the other matrix. Factors highlighted in red were verified as direct H3K4me3 binding proteins in independent biochemical experiments using recombinant proteins (please refer to the indicated references for study details). Factors highlighted in blue were also found in a recent SILAC MS study using modified mononucleosomes for affinity purification (46). Factors highlighted in green were also found in a recent SILAC MS study using histone tail peptides for affinity purification (6). Factors highlighted in brown were found overlapping in two recent SILAC MS studies, in histone tail peptide and mononucleosome based experiments (6, 46). Verified interaction factors that were not identified in a recent study using modified mononucleosomes for affinity purification are marked with § (46). Verified interaction factors that were not identified in a recent study using histone tail peptides for affinity purification are marked with # (6).
a Only factors with a four-fold enrichment over the corresponding unmodified templates are given. Table entries are ranked according to the fold enrichment.
b Proteins are identified via the International Protein Index (IPI) number. Only one of in may cases several protein names are listed.
c Protein domains and motifs were derived from Pfam (56).
d Functional relevance according to STRING (54).
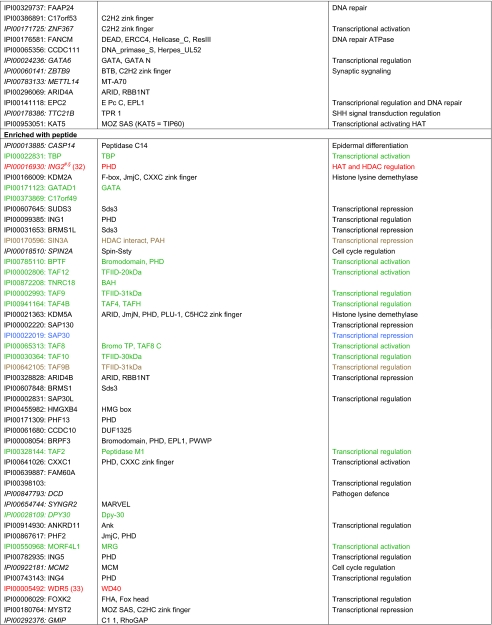
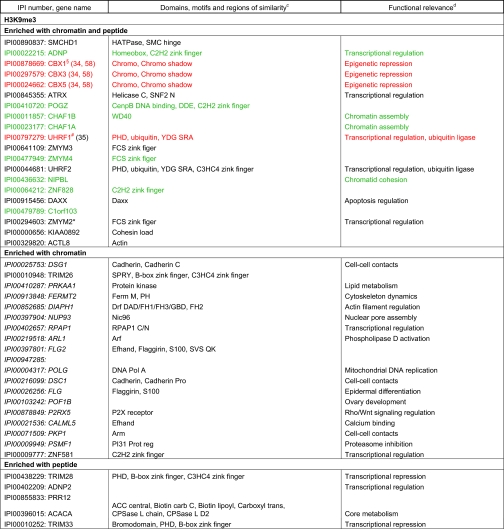
Table II. Factors found enriched or excluded on H3K9me3 chromatin and/or peptide templates from HeLa S3 nuclear extracts. a, b, c, d Factors in italics were identified only in forward or reverse set-up of the experiments. Asterisks mark factors that were identified in forward and reverse set-up of the experiment with either peptide or chromatin templates, but were only found in forward or reverse set-up of the experiment using the other matrix. Factors highlighted in red were verified as direct H3K9me3 binding proteins in independent biochemical experiments using recombinant proteins (please refer to the indicated references for study details). Factors highlighted in blue were also found in a recent SILAC MS study using modified mononucleosomes for affinity purification (46). Factors highlighted in green were also found in a recent SILAC MS study using histone tail peptides for affinity purification (6). Factors highlighted in brown were found overlapping in two recent SILAC MS studies, in histone tail peptide and mononucleosome based experiments (6, 46). Verified interaction factors that were not identified in a recent study using modified mononucleosomes for affinity purification are marked with § (46). Verified interaction factors that were not identified in a recent study using histone tail peptides for affinity purification are marked with # (6).
a Only factors with a four-fold enrichment over the corresponding unmodified templates are given. Table entries are ranked according to the fold enrichment.
b Proteins are identified via the International Protein Index (IPI) number. Only one of in may cases several protein names are listed.
c Protein domains and motifs were derived from Pfam (56).
d Functional relevance according to STRING (54).
The different data sets contain factors that have been shown to directly interact with H3K4me3 or H3K9me3 in independent biochemical experiments, thereby validating our approach. TAF3 (29), PHF8 (30), CHD1 (31), ING2 (32), and WDR5 (33) are known H3K4me3 interacting proteins. The HP1 isoform proteins CBX1, CBX3, CBX5 (34), as well as UHRF1 (35), CDYL1 (36), and MPHOSP8 (37) have been shown to bind H3K9me3. Besides direct interaction partners, the data sets also include proteins that are known to be indirectly recruited to the chromatin marks. For example, TFIID components (e.g. TAF1–2, 4–13) are recruited to H3K4me3 via TAF3 (29). POGZ is bound to H3K9me3 via CBX5 (38). Known or predicted functional interactions between the identified enriched proteins are shown in supplemental Fig. S4. Additional work will be necessary to investigate which of the newly identified components interact directly or indirectly with the chromatin marks.
Overall, the data present a significant expansion of the repertoire of factors that are recruited to H3K4me3 or H3K9me3 chromatin marks. Nevertheless, it is clear that use of other procedures for preparing extracts as well as different cellular sources (cell type, stage of differentiation) will produce distinct data sets. Applying a lower threshold, the overall number of factors identified using standard Hela S3 nuclear extract is much larger (see supplemental Tables S1–S4 for raw data). However, we feel that a fourfold change presents a robust signal and therefore focused for further analysis on this data set. Analysis of the data using values of significance (“B”) as calculated by the MaxQuant software with a cutoff of 0.05 is shown in supplemental Fig. S5 (39).
We ascribe the fact that around 36% of all factors specifically identified in this study reveal high enrichment or exclusion ratios only in forward or reverse experiments but not in both to the high threshold level of fourfold that we set for the analysis. In fact, detailed examination of the proteins only enriched or excluded in the forward or reverse set-up of the experiment showed that these are largely factors that are more difficult to identify (i.e. these have a lower summed peptide total intensity; see purple colored hits in Fig. 2). Low abundance in the extract, small protein size yielding few peptides, and peptide hydrophobicity might contribute to this phenomenon. Also, batch to batch variability of extracts might play a role in factors only identified in forward or reverse experiments. We tried to minimize this possibility by carefully matching the different extracts used (supplemental Fig. S3). Last, false negative identification (i.e. a factor was by chance not detected in forward or reverse experiment) might be a relevant factor in the analysis. Although proteins identified by both forward and reverse experimental set-up present the most stringent hits, proteins that are found in forward or reverse experiments nevertheless result from averages of four independent experiments (two times forward and two times reverse, see above). A number of factors were also represented only in forward or only in reverse set-up of the experiment using either chromatin or peptide templates, but were identified in both directions of the experiment using the other matrix (annotated with asterisk in Tables I and II). We think that these are therefore meaningful candidates and included them in our further analysis.
Fig. 2 also shows that there is general good agreement between proteins identified in forward and reverse experiments. Interestingly, more factors are enriched than excluded by the H3K4me3 and H3K9me3 histone modifications both, in the context of chromatin as well as peptides. While our type of analysis excludes factors that bind histone H3 irrespective of H3K4me3 or H3K9me3 modifications, we do not think that this observation is caused by a technical problem of the method, but rather presents a “true” finding. The available structural data on different histone modification binding proteins identify highly specialized and in many cases narrow binding pockets where multiple interactions mediate recognition and binding of the PTMs (7). This binding mode likely generates a bigger change in free energy than is achieved by a PTM that is added to a stretch of amino acids recognized by a larger protein surface in the unmodified state. Therefore, the attractive mode might generate more discrimination than the repelling mode. Although additional experiments are necessary to test this hypothesis, we nevertheless note that when going from lower to higher eukaryotes the number and type of histone modifications significantly increases. This phenomenon might reflect the fact that the attractive mode indeed generates more robust signal transduction.
Validation of the Approach
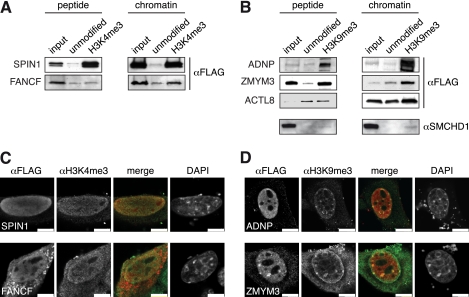
To assess the quality of the new data sets of H3K4me3 and H3K9me3 interacting factors, we choose to independently analyze a set of proteins from each category that had not been analyzed in this context before. We transiently expressed candidate factors in 293 HEK cells and performed peptide or chromatin affinity purification experiments that were analyzed by Western blotting against an engineered FLAG-tag on all factors. As Fig. 3A shows, SPIN1 a factor that is implicated in cell cycle regulation (40) was recovered on the peptide and chromatin H3K4me3 templates, but not on the corresponding unmodified matrices, thereby essentially verifying the results from the SILAC MS analysis. Similarly, we found FANCF, a factor of the Fanconi anemia group (41) specifically enriched on the H3K4me3 chromatin template, but not the corresponding peptide matrix as we had seen in the SILAC MS. Of the H3K9me3 binding factors ADNP, which is a protein containing a neuroactive peptide but that has also nuclear functions (42), and ZMYM3, a Zn-finger protein of unknown function (43) bound specifically to the H3K9me3 peptide and chromatin templates as we had detected in the SILAC MS analysis (Fig. 3B). In contrast, ACTL8, an actin-like protein of unknown function, did not reproduce the findings from the SILAC-MS. We detected the overexpressed protein in comparable amounts bound to the unmodified and H3K9me3 templates. Although this finding might identify a false positive of our screening procedure, we nevertheless point out that ACTL8 contains a PSVLL motif. PxVxL motifs in other proteins have been found to interact with factors of the HP1 type that directly bind H3K9me3 (44). Additional experiments need to find out whether ACTL8 indeed can interact with HP1 and whether overexpression and/or tagging of this factor interfere with its biology. Lastly, we used a specific antiserum against SMCHD1, a factor that might be implicated in the structural maintenance of chromosomes and that was recently found enriched at telomeres (45) to verify that the endogenous, cellular protein is recruited to H3K9me3 (Fig. 3B).
Fig. 3.
Verification of H3K4me3 and H3K9me3 interacting factors. A and B, The indicated proteins containing C-terminal FLAG-tags were transiently expressed in 293T cells. Peptide or chromatin affinity purifications using H3K4me3 (A) or H3K9me3 (B) templates were performed from nuclear extracts and analyzed by Western blot using anti-FLAG antibodies. In case of SMCHD1 extract from untransfected cells was used and the respective affinity purifications were analyzed using anti-SMCHD1 antibodies. Input, 2%. C and D, The indicated proteins containing C-terminal FLAG-tags were transiently expressed in NIH3T3 cells. Immunofluorescence analysis was carried out using anti-FLAG and anti-H3K4me3 (C) or anti-H3K9me3 (D) antibodies. Merged images correspond to the overlay of the two different antibody stainings. DNA was stained with DAPI. Scale bar represents 7.5 μm.
Because the nuclear distribution of H3K4me3 and H3K9me3 are very different, we also analyzed the localization of the new candidate histone methyl-lysine interacting factors in NIH3T3 cells. Here, we found transiently expressed SPIN1 and FANCF diffusely spread in the cell nucleus in a pattern that was reminiscent of H3K4me3 distribution (Fig. 3C). In contrast, transiently expressed ADNP and ZMYM 3 showed enrichment at discrete nuclear foci besides diffuse general localization. As these foci were marked by H3K9me3 and are DNA rich, these represent pericentric heterochromatin. Overall, the results verify five out of six candidates tested as novel interaction partners of H3K4me3 or H3K9me3.
Interestingly, only SPIN1 and ADNP, but neither FANCF, ZMYM3, nor SMCHD1 were found in recent histone tail peptide (6) or mononucleosome (46) H3K4me3 and H3K9me3 affinity purification experiments. Although there is clearly some overlap with the data sets of the peptide based studies (see Tables I and II) the differences of the mononucleosome- and our oligonucleosme-based studies are striking. As the mononucleosome-based data were already analyzed with a very low twofold cutoff of enrichment, these findings might indicate that multiple nucleosomes in the form of arrays provide different and potentially better landing platforms for factors binding to chromatin marks. Although we do not know the basis for the discrepancies in the separate studies, we note that both previously published experiments (6, 46) performed fewer biological replicates, analyzed the data with far lower enrichment cutoffs and did not provide data on the quality of the SILAC extracts used. Additional work is required to resolve potential technical differences in the different studies and to assess the overall quality of the data sets provided.
Comparison of H3K4me3 and H3K9me3 Interactomes
There is generally no correlation of the factors identified with the H3K4me3 and the H3K9me3 histone PTMs, which reflects the different biological contexts that these modifications have been implicated in (Fig. 4A). Only a few proteins were identified in both, the H3K4me3 and H3K9me3 data sets (e.g. DAXX), but then only with chromatin or peptide templates (DAXX was identified only on chromatin with H3K4me3, but on chromatin and peptide templates with H3K9me3). These might represent factors that have a general affinity for tri-methylated lysine residues irrespective of sequence context. Promiscuous binding to (mono- and di-) methyl-lysine marks in different histone sequence environments has for example been observed for proteins containing malignant brain tumor domains (47).
Fig. 4.
Overall evaluation of chromatin and histone tail peptide H3K4me3 and H3K9me3 affinity purifications. Comparison between H3K4me3 and H3K9me3 interactomes in the context of chromatin (A) and peptide (B) affinity purification reactions. Only proteins identified with both modifications are plotted. Chromatin and peptide affinity purifications identify distinct as well as overlapping sets of proteins recruited to H3K4me3 (C) and H3K9me3 (D). Venn diagrams show the number of factors identified with each approach with a fourfold cutoff, both enriched with the specific modification (above line) and excluded by the modification thereby preferentially binding to the unmodified template (below line).
Several proteins (e.g. DNMT1, UHRF1) show enrichment with one but exclusion with the other modification. Although these are only represented in either the peptide or chromatin data sets using the stringent fourfold cutoff (e.g. DNMT1 was found enriched not only on the H3K9me3 peptide, but also the H3K9me3 chromatin, but with a 3.7 ratio), such factors could be of high biological interest as they might reflect proteins that mediate binary switches between different totally exclusive chromatin states. On a global level, binary chromatin domains have been described for the activating H3K4me3 and repressing H3K27me3 histone PTMs, where regions that contain both chromatin marks in the pluripotent cell state resolve during differentiation into either of the two states (48).
Interestingly, we find far more factors excluded by H3K4me3 chromatin and peptide templates compared with the H3K9me3 PTM (see Fig. 2). This might indicate that generally more factors can bind to the unmodified very N-terminal region of H3 compared with the H3K9 context. We wonder whether this phenomenon reflects the fact that in higher eukaryotes the default chromatin state is repressive (49). The list of factors excluded by H3K4me3 indeed contains some proteins, which according to gene ontology are implicated in transcriptional repression. Comparison of the interactomes of additional activating and repressive chromatin marks might provide further insights into this behavior.
Comparison of Chromatin and Peptide Affinity Purifications
The total numbers of factors identified by the chromatin or peptide affinity purifications for the H3K4me3 and the H3K9me3 PTMs are quite similar (Fig. 4B). However, the degree of overlap between the proteins recruited by the corresponding chromatin and peptide templates is rather limited. A far larger number of factors are identified only by the peptide-based approach or only by the chromatin-based approach. In light of the fact that both, H3K4me3 and H3K9me3 modifications are very distal from the H3 core domain (the H3 tail region encompasses the first 36 amino acids, ref. 50) and are thereby at the periphery of the nucleosome this finding is rather surprising. A priori one might expect to identify the same factors with both approaches and to potentially recruit and recover more proteins on the chromatin templates. After all, additional interaction surfaces (e.g. DNA, other histone tail and core regions) are available in this native setting. By and large, both approaches might be highly complementary. Future studies will need to tell whether the candidates identified with either approach are more functionally relevant than the other. However, the context of patterns of chromatin modifications especially in the context of DNA methylation as well as in a trans-histone setting can certainly only be investigated by the chromatin-based approach.
For each histone PTM we identified slightly more factors with the peptide templates compared with the chromatin templates. We think this might be a reflection of the potentially higher substitution rate of the peptides on the magnetic streptavidin beads. For immobilization of chromatin representing the same amount of free H3 tails far more beads are needed. Likely, there is steric exclusion on the streptavidin tetramer as well as on the bead surface by the chromatin complex. Also, binding of factors to histone PTMs in the context of chromatin might be subject to repulsive forces by DNA as well as steric hindrance thereby fine-tuning the binding of proteins or multiprotein complexes. Last, we do not know whether the histone tails are fully available on the surface of the nucleosomes in general or under the experimental buffer conditions used (51).
In the case of peptide affinity purifications, a stronger tendency of detecting a protein with one modification as not enriched (1:1 ratio of unmodified to modified templates) and in the other modification as enriched can be seen compared with the chromatin affinity purifications (see Fig. 4A, clouds along the axes). Obviously, the chromatin environment presents the more stringent and more discriminating binding surface.
Although the data sets of the H3K9me3 peptide and chromatin affinity purifications as well as the H3K4me3 peptide affinity purifications show a similar overall distribution with relative narrow and focused clustering of the background (blue dots in Fig. 2) as well as distinct highly enriched or excluded factors (red dots in Fig. 2), the results of the H3K4me3 chromatin affinity purification appear different. Here, the background shows an overall wider distribution and only very few factors with high change ratios were identified. Because the results of the H3K4me3 peptide affinity purification are different, this distinction cannot be caused by the histone PTM itself, but must be a consequence of embedding this methyl-lysine mark in a chromatin context. Although we have so far not detected any difference in the biochemical and biophysical behavior of the unmodified, H3K4me3 or H3K9me3 recombinant chromatin preparations, the accessibility of the histone H3 tail might be limited by lysine 4 tri-methylation. This might be a nucleosome restricted effect or be caused by overall chromatin conformational properties (i.e. compaction status). Indeed, single histone modifications affecting the overall behavior of recombinant chromatin templates have been described (52).
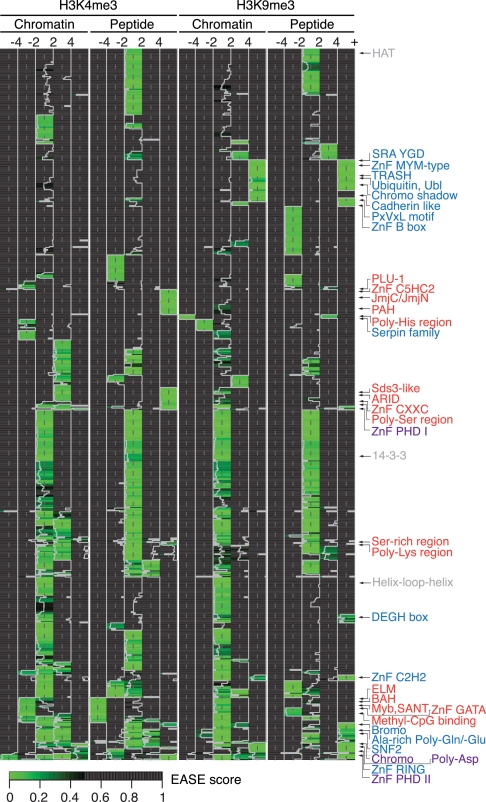
Distinct Protein Domains Mediate Recruitment to H3K4me3 and/or H3K9me3
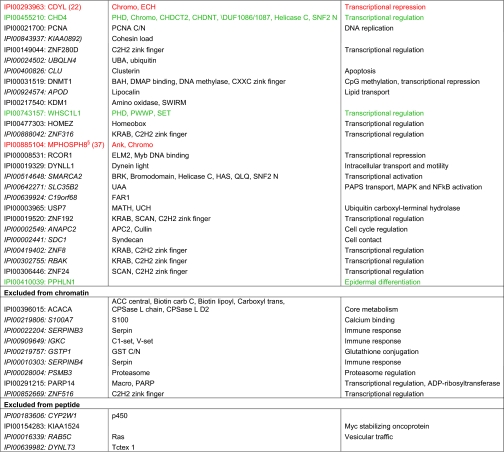
To gain further insight into the recruitment of factors to histone methyl-lysine marks we performed clustering analysis focusing on protein domains (Fig. 5). Prominent chromatin associated protein regions and motifs are highly enriched on the histone methyl-lysine marks. These include several domains directly implicated in methyl-lysine recognition: chromo, PHD, SRA. Also, domains that provide binding interfaces for recruitment of additional proteins are visible e.g. chromoshadow. Other motifs like PxVxL, which has been shown to bind to the chromoshadow domain of HP1 proteins (44, 53), are present in factors presumably indirectly recruited via binding to factors directly interacting with the histone methyl-lysine marks. Common background proteins (ribosomal proteins, tubulins) are present within the middle cutoff group, as are some domains specific for chromatin (helix-loop-helix), peptide (HAT) or both 14-3-3, regardless of the modification used.
Fig. 5.
Domain enrichment in affinity purification reactions comparing H3K4me3 and H3K9me3 PTMs using chromatin and peptide templates. Heatmap representing protein functional and structural domains and motifs enriched within each of the five indicated ratio cutoff groups (databases used: UniProt Sequence Feature, InterPro, PIR Superfamily, PFAM, SMART). Categories enriched or excluded (with a ratio of at least four) by H3K4me3 (red), H3K9me3 (blue) or both (purple) are annotated.
Although we detect domains that have been implicated in the general context of chromatin (e.g. bromodomains interacting with histone acetylation marks, SNF2 present in nucleosome remodeling factors), DNA (e.g. Zn fingers of various type, PLU-1, ARID, Myb, SANT) or methylated DNA (e.g. methyl-CpG binding domain), these are novel in the context of histone methyl-lysine marks. It will have to be seen which of these domains are direct histone methyl-lysine mark binding regions and which mediate additional protein-protein interactions in an indirect mode of recruitment. Also, domains that make contact to DNA or other histone regions of the nucleosome might stabilize weaker interactions with sensitivity to and discrimination of the histone PTMs. Analysis for functional interactions using STRING indeed indicates connection groups between several of the identified factors (supplemental Fig. S4) (54). For example, recruitment of the TFIID complex components has been described via TAF3 binding H3K4me3 (29). Another prominent interaction cluster here is between proteins from the Fanconi anemia group, implicated in the DNA damage response (41). Several of the enriched protein domains might function in the translation of the histone PTMs. This might involve enzymatic activities (e.g. JmjC/JmjN domains in histone demethylases) as well as regions directly impacting onto chromatin structure.
Interestingly, some protein regions are enriched on both, H3K4me3 and H3K9me3 PTMs (e.g. ZnF, PHD, chromo) while being structural and functional parts of different proteins. Obviously, here the domains have evolved to recognize (tri-) methylated lysine residues in different sequence context, but with high discrimination against the unmodified state exemplifying the multiple use of successful and efficient protein folds and motifs.
CONCLUSIONS
Combination of affinity purification experiments of synthetic peptides representing different histone PTMs either in isolation or in the context of chromatin and quantitative mass spectrometric analysis using SILAC provides a comprehensive set of factors potentially mediating the readout and translation of these marks. Our results show that chromatin affinity purification largely defines a set of factors distinct from what can be found with modified histone tail peptides. We think this might be a general finding as comparison of similar analysis recently performed on histone tail peptides (6) or mononucleosomes (46) even show less overlap of factors recruited to different methyl-lysine marks (see also Tables I and II). The chromatin templates provide the advantage of offering additional binding interfaces such as DNA, histone core, other histones and/or other chromatin marks. Thereby, they potentially enable recruitment of multiprotein machineries making several independent contacts to chromatin. Future experiments have to show whether the chromatin data sets indeed contain functionally more relevant factors. In any case, our combined data sets of H3K4me3 and H3K9me3 chromatin and peptide affinity purifications paves the way for additional studies investigating the exact role of the identified proteins in recognition of these methyl-lysine marks as well as in directing chromatin structure and function. Here, combination with array-based technologies that allow screening of direct interactions provides an orthogonal technology (3, 5). Despite characterization of direct interactions with the chromatin marks, emphasis needs to be put onto multiprotein assemblies and interplay of factors that are recruited indirectly. Our global analysis of the protein domains enriched on the chromatin marks as well as the biological connection of factors based on STRING analysis provides a starting point.
Multiple factors might translate individual chromatin marks in different functional settings. In this context, interplay with other histone modifications as well as DNA methylation could manifest an important regulatory mechanism, which has been proposed in the histone code hypothesis (10). Indeed, it is emerging that combinations of chromatin marks mark different domains of chromatin. For example, pericentromeric heterochromatin in higher eukaryotes is enriched in H3 lysine 9 trimethylation (H3K9me3), H4 lysine 20 trimethylation (H4K20me3), H3 lysine 27 monomethylation (H3K27me1) as well as DNA methylation. Linkage to H3 arginine 2 dimethylation (H3R2me2) and H4 arginine 3 dimethylation (H4R3me2) has also been described (55). The use of uniformly modified, recombinant chromatin templates containing different combinations and patterns of chromatin marks for affinity purification experiments will provide an excellent starting point to define the complement of factors that mediate the functional status of such chromatin regions, whose biochemistry has been elusive for a long time.
Acknowledgments
We thank Uwe Plessmann and Winfried Lendeckel for technical support as well as Christian Kelstrup and the MaxQuant summer school 2009 for introduction to R. We are grateful to Matthias Selbach and Fabian Hosp for helpful discussions on enrichment clustering. Ilian Atanassov provided the exhaustive list of HeLa nuclear extract proteins.
Footnotes
* This work was supported by the Max Planck Society (H.U., W.F.), the NoE “The Epigenome” (W.F.) and the Minna-James-Heineman Foundation through a Minerva grant (W.F.). M.N. is supported by a fellowship from the Ph.D. program “Molecular Biology”—International Max Planck Research School at the Georg August University Göttingen.
 This article contains supplemental Figs. S1 to S5, Tables S1 to S4 and Procedures.
This article contains supplemental Figs. S1 to S5, Tables S1 to S4 and Procedures.
1 The abbreviations used are:
- PTM
- post translational modification
- SILAC
- stable isotope labeling by amino acids in cell culture
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K9me3
- histone H3 lysine 9 trimethylation
- PHD
- plant homeodomain
- MudPIT
- multidimensional protein identification technology
- TAF
- TATA box binding protein (TBP)-associated factor
- PHF8
- PHD finger protein 8
- CHD1
- chromodomain-helicase-DNA-binding protein 1
- ING2
- inhibitor of growth family member 2
- WDR5
- WD repeat domain 5
- HP1
- heterochromatin protein 1
- CBX
- chromobox homolog
- UHRF1
- ubiquitin-like with PHD and ring finger domains 1
- CDYL
- chromodomain protein Y-like
- MPHOSP8
- M-phase phosphoprotein 8
- SPIN1
- spindling 1
- FANCF
- Fanconi anemia, complementation group F
- ADNP
- activity-dependent neuroprotector homeobox
- ZMYM3
- zinc finger MYM-type 3
- ACTL8
- actin-like 8
- SMCHD1
- structural maintenance of chromosomes flexible hinge domain containing 1
- TFIID
- TATA binding protein
- POGZ
- pogo transposable element with ZnF domain
- ZnF
- zinc finger
- DAXX
- death-domain associated protein
- DNMT1
- DNA (cytosine-5-)-methyltransferase 1
- H3K27me3
- histone H3 lysine 27 trimethylation
- SRA
- SET and RING associated
- HAT
- histone acetyl transferase
- SNF2
- sucrose non fermentable 2
- ARID
- AT-rich interaction domain
- Myb
- myeloblastosis oncogene like
- SANT
- switching-defective protein 3 (Swi3) adaptor 2 (Ada2) nuclear receptor co-repressor (N-CoR) transcription factor (TF)IIIB
- H4K20me3
- histone H4 lysine 20 trimethylation
- H3K27me1
- histone H3 lysine 27 monomethylation
- H3R2me2
- histone H3 arginine 2 dimethylation
- H4R3me2
- histone H4 arginine 3 dimethylation.
REFERENCES
- 1. Fischle W., Wang Y., Allis C. D. (2003) Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 2. Campos E. I., Reinberg D. (2009) Histones: annotating chromatin. Annu. Rev. Genet 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 3. Bua D. J., Kuo A. J., Cheung P., Liu C. L., Migliori V., Espejo A., Casadio F., Bassi C., Amati B., Bedford M. T., Guccione E., Gozani O. (2009) Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One 4, e6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan D. W., Wang Y., Wu M., Wong J., Qin J., Zhao Y. (2009) Unbiased proteomic screen for binding proteins to modified lysines on histone H3. Proteomics 9, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu H., Galka M., Iberg A., Wang Z., Li L., Voss C., Jiang X., Lajoie G., Huang Z., Bedford M. T., Li S. S. (2010) Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J. Proteome Res. 9, 5827–5836 [DOI] [PubMed] [Google Scholar]
- 6. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 7. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams-Cioaba M. A., Min J. (2009) Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 87, 93–105 [DOI] [PubMed] [Google Scholar]
- 9. Fischle W., Wang Y., Allis C. D. (2003) Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 [DOI] [PubMed] [Google Scholar]
- 10. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 11. Sims R. J., 3rd, Reinberg D. (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 9, 815–820 [DOI] [PubMed] [Google Scholar]
- 12. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 14. Liu H., Lin D., Yates J. R., 3rd (2002) Multidimensional separations for protein/peptide analysis in the post-genomic era. BioTechniques 32, 898,, 900, 902 passim [DOI] [PubMed] [Google Scholar]
- 15. Wilm M. (2009) Quantitative proteomics in biological research. Proteomics 9, 4590–4605 [DOI] [PubMed] [Google Scholar]
- 16. Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 17. Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. (1983) Eukaryotic gene transcription with purified components. Methods Enzymol. 101, 582–598 [DOI] [PubMed] [Google Scholar]
- 18. Biancalana S., Hudson D., Songster M. F., Thompson S. A. (2001) Fmoc chemistry compatible thio-ligation assembly of proteins. Lett. Peptide Sci. 7, 291–297 [Google Scholar]
- 19. Futaki S., Sogawa K., Maruyama J., Asahara T., Niwa M., Hojo H. (1997) Preparation of peptide thioesters using Fmoc-solid-phase peptide synthesis and its application to the construction of a template-assembled synthetic protein (TASP). Tetrahedron Letters 38, 6237–6240 [Google Scholar]
- 20. von Eggelkraut-Gottanka R., Klose A., Beck-Sickinger A. G., Beyermann M. (2003) Peptide (alpha)thioester formation using standard Fmoc-chemistry. Biopolymers 71, 352–353 [Google Scholar]
- 21. Shogren-Knaak M. A., Peterson C. L. (2004) Creating designer histones by native chemical ligation. Methods Enzymol. 375, 62–76 [DOI] [PubMed] [Google Scholar]
- 22. Franz H., Mosch K., Soeroes S., Urlaub H., Fischle W. (2009) Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J. Biol. Chem. 284, 35049–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luger K., Rechsteiner T. J., Richmond T. J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 24. Huynh V. A., Robinson P. J., Rhodes D. (2005) A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 345, 957–968 [DOI] [PubMed] [Google Scholar]
- 25. Schulze W. X., Mann M. (2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 26. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 29. Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., Timmers H. T. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 30. Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. (2010) PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct. Mol. Biol. 17, 445–450 [DOI] [PubMed] [Google Scholar]
- 31. Flanagan J. F., Mi L. Z., Chruszcz M., Cymborowski M., Clines K. L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. (2005) Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185 [DOI] [PubMed] [Google Scholar]
- 32. Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 34. Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 [DOI] [PubMed] [Google Scholar]
- 35. Karagianni P., Amazit L., Qin J., Wong J. (2008) ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell Biol. 28, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fischle W., Franz H., Jacobs S. A., Allis C. D., Khorasanizadeh S. (2008) Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 283, 19626–19635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kokura K., Sun L., Bedford M. T., Fang J. (2010) Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 29, 3673–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nozawa R. S., Nagao K., Masuda H. T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. (2010) Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12, 719–727 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 40. Zhang K. M., Wang Y. F., Huo R., Bi Y., Lin M., Sha J. H., Zhou Z. M. (2008) Characterization of Spindlin1 isoform2 in mouse testis. Asian J. Androl 10, 741–748 [DOI] [PubMed] [Google Scholar]
- 41. D'Andrea A. D. (2010) Susceptibility pathways in Fanconi's anemia and breast cancer. N. Engl. J. Med. 362, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mandel S., Rechavi G., Gozes I. (2007) Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev. Biol. 303, 814–824 [DOI] [PubMed] [Google Scholar]
- 43. Beever C., Lai B. P., Baldry S. E., Peñaherrera M. S., Jiang R., Robinson W. P., Brown C. J. (2003) Methylation of ZNF261 as an assay for determining X chromosome inactivation patterns. Am. J. Med. Genet. A 120A, 439–441 [DOI] [PubMed] [Google Scholar]
- 44. Smothers J. F., Henikoff S. (2000) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10, 27–30 [DOI] [PubMed] [Google Scholar]
- 45. Déjardin J., Kingston R. E. (2009) Purification of proteins associated with specific genomic Loci. Cell 136, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bartke T., Vermeulen M., Xhemalce B., Robson S. C., Mann M., Kouzarides T. (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bonasio R., Lecona E., Reinberg D. (2010) MBT domain proteins in development and disease. Semin Cell Dev. Biol. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 49. Allis C. D., Jenuwein T., Reinberg D. (eds.), Caparros, M. L. (assoc. ed.) (2006) Epigenetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 50. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 51. Woodcock C. L., Ghosh R. P. (2010) Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2, a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 53. Thiru A., Nietlispach D., Mott H. R., Okuwaki M., Lyon D., Nielsen P. R., Hirshberg M., Verreault A., Murzina N. V., Laue E. D. (2004) Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosenfeld J. A., Wang Z., Schones D. E., Zhao K., DeSalle R., Zhang M. Q. (2009) Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics 10, 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cochrane G. R., Galperin M. Y. (2010) The 2010 Nucleic Acids Research Database Issue and online Database Collection: a community of data resources. Nucleic Acids Res. 38, D1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sims R. J., 3rd, Chen C. F., Santos-Rosa H., Kouzarides T., Patel S. S., Reinberg D. (2005) Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280, 41789–41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 [DOI] [PubMed] [Google Scholar]