Abstract
The emergence of acquired drug resistance results from multiple compensatory mechanisms acting to prevent cell death. Simultaneous monitoring of proteins involved in drug resistance is a major challenge for both elucidation of the underlying biology and development of candidate biomarkers for assessment of personalized cancer therapy. Here, we have utilized an integrated analytical platform based on SDS-PAGE protein fractionation prior to liquid chromatography coupled to multiple reaction monitoring mass spectrometry, a versatile and powerful tool for targeted quantification of proteins in complex matrices, to evaluate a well-characterized model system of melphalan resistance in multiple myeloma (MM). Quantitative assays were developed to measure protein expression related to signaling events and biological processes relevant to melphalan resistance in multiple myeloma, specifically: nuclear factor-κB subunits, members of the Bcl-2 family of apoptosis-regulating proteins, and Fanconi Anemia DNA repair components. SDS-PAGE protein fractionation prior to liquid chromatography coupled to multiple reaction monitoring methods were developed for quantification of these selected target proteins in amounts of material compatible with direct translation to clinical specimens (i.e. less than 50,000 cells). As proof of principle, both relative and absolute quantification were performed on cell line models of MM to compare protein expression before and after drug treatment in naïve cells and in drug resistant cells; these liquid chromatography-multiple reaction monitoring results are compared with existing literature and Western blots. The initial stage of a systems biology platform for examining drug resistance in MM has been implemented in cell line models and has been translated to MM cells isolated from a patient. The ultimate application of this platform could assist in clinical decision-making for individualized patient treatment. Although these specific assays have been developed to monitor MM, these techniques are expected to have broad applicability in cancer and other types of disease.
Multiple myeloma (MM)1 is an incurable malignancy of plasma cells harbored in the bone marrow, which is clinically characterized by secretion of monoclonal antibodies, calcium dysregulation, anemia, lytic bone lesions, and kidney damage. For five decades, therapeutic regimens for MM have included the alkylating agent, melphalan (p-di-2-chloroethylamino-1-phenylalanine, l-phenylalanine mustard, or l-PAM) (1). Therapy with l-PAM (alone or in combination with novel agents) remains the standard of care for transplant-ineligible patients and is the backbone of high-dose therapy associated with autologous stem cell transplant. Although MM patients initially respond to chemotherapy, treatment failure is inevitable because of the emergence of drug resistance. Because of the importance of detecting acquired drug resistance (ADR) for patient assessment and treatment, numerous mechanisms of ADR have been elucidated for MM in vitro, utilizing cell line models developed by chronic exposure to chemotherapy (2–7). These models were designed to identify potential drug targets to reverse ADR as well as to discover prognostic or chemopredictive biomarkers that could be translated to assessment of MM patients. These isogenic model systems clearly show that drug resistance is derived from multiple factors and that prediction of response can not depend on the measurement of one pathway.
The biology of one such model of ADR, the 8226/LR5 cell line, selected by chronic exposure of RPMI-8226 MM cells to the DNA-damaging agent, melphalan, has been extensively characterized, leading to the identification of causative mechanisms of drug resistance in MM, including activation of NFκB signaling networks, modulated expression of Bcl-2 family members, and enhanced DNA damage repair capacity (2, 5–7). The relationships between these different protein networks have been defined using traditional cancer biology techniques (Fig. 1A).
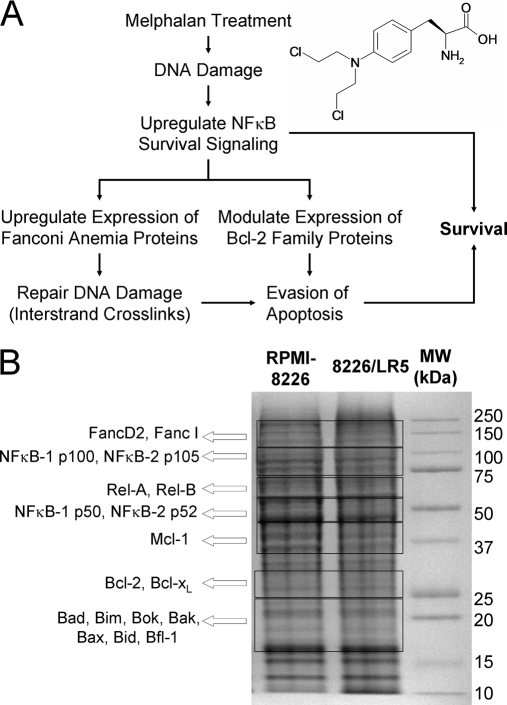
Fig. 1.
GeLC-MRM analysis for examining melphalan resistance. An overview of the response to melphalan (inset at top left) includes NF-κB survival signaling, regulation of apoptosis, and DNA damage repair (A). A signature of resistance in MM is driven by NFκB-1 p50/RelB survival signaling, which causes up-regulation of the anti-apoptotic protein, Bcl-xL, and proteins in the Fanconi Anemia pathway, particularly FANCD2. The use of GeLC-MRM enables assessment of proteins from each of these groups in the same sample. Excision patterns are overlaid on an SDS-PAGE gel visualized with Coomassie Blue (B). The proteins found in each of the seven gel regions are indicated.
The transcription factor, Nuclear Factor-kappa B (NFκB), promotes growth and survival of MM cells; furthermore, mutations that cause constitutive activation of NFκB signaling are frequently detected in later stages of MM progression ((8). NFκB transactivation has also been shown to inhibit apoptosis and to contribute to drug resistance in cancer cells, including MM (9, 10). Moreover, in the melphalan-selected 8226/LR5 myeloma cell line, constitutive activation of NFκB (RelB/p50) signaling regulates expression of apoptotic proteins (particularly Bcl-xL) and DNA damage repair via the Fanconi Anemia (FA) pathway, and confers drug resistance (7). As a result, the inhibition of NFκB is a viable strategy to reverse ADR in MM. Experimental evidence indicates that NFκB subunits may be important prognostic or predictive biomarkers in MM. Therefore, expression measurements of NFκB proteins may be useful in directing and optimizing therapeutic regimens for advancing personalized medicine (7, 11, 12).
Alterations in the apoptotic machinery have also been identified as causative determinants in melphalan resistance (2, 13, 14). Interactions between pro- and anti-apoptotic proteins determine cell fate in response to a host of internal and external stimuli. Expression of these molecules is frequently altered in cancer pathogenesis, and specific changes have been implicated in drug resistance (15, 16). Increased levels of Bcl-xL have been observed in doxorubicin-resistant and melphalan-resistant RPMI-8226-derived cell lines, suggesting that Bcl-xL expression may serve as a biomarker for ADR to DNA-damaging agents (17, 18). Mcl-1 has also been shown to be essential for the survival of human myeloma cells, and overexpression of Mcl-1 protects MM cells from drug-induced apoptosis (19–21). Mcl-1 has been suggested as a high-priority therapeutic target because of its roles in carcinogenesis and chemoresistance (22, 23). Furthermore, the balance between Bim and Mcl-1 expression controls the survival of human myeloma cells (24). Additional family members, including Bfl1/A1 (25,26), Bid (27), and Bim (5) have also been studied to examine the mechanisms determining cell fate in MM. Because of the overlapping function of pro- and anti-apoptotic family members, the measurement of individual proteins will not be sufficient for predicting chemosensitivity across patients. However, comprehensive analysis of apoptotic proteins in the Bcl-2 family may elucidate therapeutic responses and ADR in both model systems and patients. Analysis of these proteins may also reveal candidate biomarkers for disease severity and drug resistance in MM patients.
In models of melphalan resistance, the repair of DNA interstrand crosslinks resulted from increased expression of genes in the DNA-damage response pathway involving Fanconi Anemia (FA) and breast cancer susceptibility proteins (BRCA) (5). The key functional event in this pathway is the monoubiquitination of the repair effectors, FANCD2 and FANCI (28, 29). Activation of this pathway culminates in repair of DNA crosslinks, UV-induced dimers, and double-strand breaks by homologous recombination and translesion synthesis (30). The repair of the interstrand crosslinks contributes to drug resistance in MM and other malignancies (31, 32). Furthermore, enhanced DNA-repair capacity correlates with the development of drug resistance in cell lines (5, 7) as well as with poor patient outcomes (33, 34). Therefore, evaluation of DNA repair pathways could be a viable approach for predicting response and detecting resistance to DNA-damaging agents, including melphalan.
Analysis of these three interrelated aspects of ADR (NFκB survival signaling, regulation of apoptosis, and DNA repair through the FA/BRCA pathway) may give further insight into the underlying biology of ADR and provide a profile for detecting resistance to alkylating agents in patients. To this end, development of quantitative methods to simultaneously evaluate multiple effector pathways will be important for the development of rational personalized therapeutic regimens. To complement antibody-based techniques, multiple reaction monitoring (MRM) mass spectrometry (also called selected reaction monitoring) has emerged as an important method for targeted detection and quantification of proteins in the complex matrices common to biological and clinical samples. The coupling of liquid chromatography and multiple selected reaction monitoring experiments (LC-MRM) was first translated from small molecule analysis to endogenous peptides (35), then to proteotypic peptides from individual target proteins (36) and more recently to the assessment of panels of proteins (37–39). Much of the existing literature focuses on the development and implementation of LC-MRM in biomarker pipelines (40, 41) LC-MRM has been widely applied to quantify biomarkers of human disease by comparing protein expression in samples acquired from patients to those obtained from disease-free controls (42–44). Furthermore, LC-MRM has been proven to be effective in monitoring signaling pathways and networks, which demonstrates additional utility for the elucidation of biological processes (45, 46).
Based on the versatility shown in these prior applications, a method using SDS-PAGE for protein fractionation prior to LC-MRM has been developed for simultaneous quantification of selected protein targets relevant to drug resistance in MM. This choice was based on the prior performance and ease of implementation of SDS-PAGE as a fractionation method prior to liquid chromatography coupled to tandem mass spectrometry peptide sequencing (GeLC-MS/MS), a method that has been extensively used to catalog proteomes (47–51). Although most of these targets would not be detected in GeLC-MS/MS, the use of LC-MRM with absolute quantification (52–55) enables quantification of these proteins by comparison to known amounts of stable-isotope-labeled standard (SIS) peptides. In this article, we report the quantitative analysis of signaling pathways and biological processes related to melphalan-resistance in MM. Specifically, protein expression levels for NFκB subunits, apoptotic proteins, and selected FA proteins have been measured with LC-MRM using the amount of protein that would be extracted from ∼40,000 cultured cells (∼8 μg). This amount of material was used to challenge the sensitivity of the method to prepare for the limits in protein amount imposed by the acquisition of clinical specimens, i.e. bone marrow aspirates. The outcome is the initial stage of a systems biology platform guided by known cancer biology that can be used to monitor drug response and ADR in MM, enabling quantification of multiple candidate biomarkers in the limited number of cells expected from patient specimens.
The utility of the platform is illustrated by comparison of protein expression in two different ADR models based on the RPMI-8226 cell line (which contains a classical translocation of c-Maf and the immunoglobulin heavy chain (IgH) enhancer region (t(14;16)) as well a somatic mutation in the K-Ras oncogene) and the U266 cell line (which is characterized by the translocation between cyclin D1 and IgH (t(11;14)), has wild-type Ras, and is driven by an interleukin-6 autocrine/paracrine stimulatory loop) (13, 56, 57). Both melphalan-resistant cell lines were derived by chronic selection in increasing doses of drug (2, 6, 7). Translation of these assays to protein quantification in isolated MM cells from a bone marrow aspirate demonstrates the feasibility of patient assessment.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) at the highest purity, unless otherwise specified. High performance liquid chromatography (HPLC) solvents (water and acetonitrile) were purchased from Burdick and Jackson (Honeywell, Muskegon, MI). The antibodies against Bcl-2 (554202) and Mcl-1 (559027) were acquired from BD Pharmingen (San Diego, CA). Antibodies directed against Bcl-xL (2764), Bad (9268), Bim (4582), and Bid (2002) were purchased from Cell Signaling Technology (Boston, MA). The antibodies recognizing Bax (SC-493), NFκB1 p50 (SC-8414x), NFκB2 p52 (SC-298), RelA p65 (SC-372) were from Santa Cruz Biotechnology (Santa Cruz, CA).
Peptide Synthesis and Evaluation
Solid state peptide synthesis (Symphony, Protein Technologies, Tucson, AZ) was used to make stable isotope labeled standards at the 25 micromole scale using standard 9-fluorenylmethoxycarbonyl (FMOC) chemistry; crude peptides are purified by semipreparative reversed-phase HPLC (U3000; Dionex, Sunnyvale, CA) using a C18 reversed-phase column (TP 238, 250 × 10 mm, 10–15 μm particles; Grace Vydac, Deerfield, IL). Twenty-minute gradients were run from 5% to 60% B solvent (A: 2% acetonitrile/0.1% formic acid; B: 90% acetonitrile/0.1% formic acid). Eluted peptides were measured at 204 and 214 nanometer wavelengths and collected with automatic triggering (Foxy Jr, ISCO). Purified peptides were mixed 1:1 with α-cyano-4-hydroxycinnamic acid (10 mg/ml) in 50% aqueous acetonitrile and analyzed with matrix assisted laser desorption ionization mass spectrometry (MALDI MS) to evaluate completeness of synthesis and sequenced with MS/MS (4700, ABSciex, Framingham, MA). Peptide standards were quantified by amino acid analysis using 6N HCl-phenol hydrolysis. Free amino acids were derivatized by FMOC and OPA (to label both primary and secondary amines), separated by reversed-phase HPLC, and detected by fluorescence (AminoQuant, Hewlett Packard).
Synthesis of Isopeptide Standards for Monoubiquitinated FANCD2 and FANCI
To develop standards for the tryptic peptides containing the sites of ubiquitination, the isopeptides (branched peptides with glycyl-glycine moieties connected to the epsilon amino group of a lysine via amide bonds) were synthesized with FMOC-Lys(Boc)-Wang resin or FMOC-Arg(Pbf)-Wang resin at the 25 μmol scale (Tribute, Protein Technologies). Side-chain protection was provided by tBu for Ser and Thr, OtBu for Asp, and Trt for Asn and Gln. The terminal Lys residues were coupled as Boc-Lys(FMOC)-OH. FMOC removal was achieved with piperidine-DMF (1:4) for 30 s with conditional UV monitoring of the dibenzofulvene-piperidine adduct. Coupling was carried out with the protected amino acid monomer, HBTU and NMM (1:1:4) for 2 h. After coupling of the terminal Lys residues, the Fmoc protecting groups on the epsilon nitrogens were removed and sequential coupling of Gly was carried out to achieve the desired isopeptide-resins. The isopeptides were simultaneously deprotected and cleaved from the resins using Reagent B (85% trifluoroacetic acid/5% phenol/5% water/2% triisopropylsilane). The crude products were precipitated using cold diethyl ether and centrifuged before the ether was decanted. Finally, isopeptide pellets were then washed with cold diethyl ether; the isopeptides were then dissolved in 0.1% trifluoroacetic acid (10 ml), frozen, and lyophilized.
Cell Culture
The RPMI-8226 and U266 human MM cell lines were obtained from the American Type Culture Collection (Manassas, VA). The process for development of corresponding melphalan-resistant 8226/LR5 and U266/LR6 cells has been previously described (2). All multiple myeloma cells were cultured in suspension using RPMI 1640 medium (Cellgro; Fisher Scientific) supplemented with 10% heat-inactivated fetal calf serum (Omega Scientific, Tarzana, CA), 100 units/ml penicillin/streptomycin and incubated in a 5% CO2 atmosphere at 37 °C. Melphalan (33 mm stock) was dissolved in ethanol containing 1 m hydrochloric acid. The 8226/LR5 drug resistant cell line was passaged weekly in medium containing 5 μm melphalan; U266/LR6 was similarly maintained in 6 μm melphalan.
Cell Lysis, LC-MRM Sample Preparation, and Western Blotting
Cells (n = 106) were lysed in high salt buffer (50 mm Tris-HCl pH7.4, 0.1% Nonidet P-40, 1 m NaCl, protease inhibitor mixture (Roche, Madison, WI), 25 mm NaF, 2 mm Na3VO4, and 0.1 m Na2HPO4) in order to recover proteins bound to DNA (e.g. active NFκB subunits and Fanconi Anemia proteins. The lysates were sonicated using 1 s pulses and then centrifuged at 18,000 × g for 10 min (Microfuge 22R, Beckman Coulter, Brea, CA). Aliquots of each cell lysate containing 50 μg of protein (corresponding to the extract from 250,000 cells) were denatured by boiling in loading buffer and separated on 4–12% gels (Criterion XT, Bio-Rad, Hercules, CA) for 80 min at 150 V. Proteins were visualized by staining with colloidal Coomassie Brilliant Blue G-250 (Bio-Rad).
Selected regions of each gel lane (as determined in previous immunoblotting experiments and confirmed by LC-MRM screening) were excised (see Fig. 1B). Consistent excision of gel regions was guided by the molecular weight markers and the overall staining pattern of the cell lysate, and not governed by the expected migration of a protein to a single visualized gel band. The proteins were reduced with 2 mm tris(2-carboxyethyl)phosphine (TCEP), alkylated with 20 mm iodoacetamide (IAA), and digested overnight (18–24 h) with trypsin at 37 °C. SIS peptides were added to the samples after in-gel digestion. Then, samples were concentrated to dryness by vacuum centrifugation (Speedvac, Thermo), and resuspended in 30 μl aqueous 2% acetonitrile with 0.1% formic acid prior to mass spectrometry. One sixth of each digest was analyzed by LC-MRM in each technical replicate; individual measurements are made with the amount of protein extracted from ∼40,000 cells (∼8 μg). In lysates prepared from 4.5 × 105 cells and separated by SDS-PAGE as described above, immunoblotting (Western) was performed for qualitative measurements of expression levels for selected proteins using the SuperSignal West Pico Chemiluminescence Kit (Pierce, Rockford, IL).
MM Tumor Cell Selection
As a pilot study for feasibility, an anonymous and de-identified MM patient sample was acquired before treatment and analyzed under a protocol approved by the University of South Florida Institutional Review Board. An aliquot of bone marrow aspirate (10 ml) was drawn for research. For sorting, freshly isolated bone marrow mononuclear cells were stained at a concentration of 107 cells per ml with CD38-FITC, CD138-APC, Kappa-PE or Lambda-PE, CD34-PE-Cy7 (BD Biosciences, San Jose, CA), and CD19-Pacific Blue (Invitrogen, Carlsbad, CA). Samples were acquired and sorted using an Aria (BD, Franklin Lakes, NJ) equipped with 488 nm, 633 nm, and 407 excitation lasers, for isolation of CD138+ (positive for syndecan-1) cells. The purity of the sorted population was greater than 95%. Cell lysis and sample preparation for LC-MRM were the same as described above for cell lines.
LC-MRM Screening and Sample Analysis
Target peptides were selected based on prior LC-MS/MS experiments and using Pinpoint(58) (Thermo, San Jose, CA) or Skyline (59) for in silico digestion of the proteins and selection of peptides and transitions. For LC-MRM screening, peptide candidates were limited to 7–25 amino acids in length; cysteine- and methionine-containing peptides were excluded unless there were few other choices (n < 5). All y fragment ions from y3 to y(n−1) were initially monitored. Consistent co-elution of the transitions and the intensities of the ion signals were used to rank peptide candidates prior to refinement based on the ease of synthesis of SIS peptides. One SIS peptide was developed for quantification, but all peptides detected in these screens were monitored in subsequent biological experiments in order to provide additional confirmation of changes in expression. The endogenous peptides, labeled standards, and selected transitions are provided in Table I.
Table I. Endogenous peptide targets, corresponding internal standards, and selected transitions for monitoring NFκB subunits, Bcl-2 family members, and Fanconi Anemia DNA damage repair proteins. Residues underlined in bold font indicate the sites of incorporation of stable isotope labels or single conservative amino acid replacements. Stable isotope labeled amino acids include proline (13C5, 15N), valine (13C5, 15N), leucine (13C6, 15N), and tyrosine (13C9, 15N). Limits of quantification (LOQ) are estimated using the lower end of the linear range of the calibration curve for each peptide standard. Additional data are available online at http://proteome.moffitt.org/QUAD/66.
| Protein | Peptide (Label) | Transitions | LOQ (fmol) | Additional Peptides |
|---|---|---|---|---|
| NFκB-1, p105 | AGADLSLLDR (A → G) | y5–y8 | 0.1 | 3 |
| NIHLHAHSLVGK | y6–y10 | 5 | ||
| NFκB-1, p50 | NIHLHAHSLVGK | y6–y10 | 5 | |
| NFκB-2, p100 | AGAGAPELLR | y5–y9 | 0.1 | 5 |
| VGADPALLDR | y5–y9 | 0.1 | ||
| DSGEEAAEPSAPSR | b8, y8–y10 | 0.5 | ||
| NFκB-2, p52 | QYAIVFR (Q → N) | y3–y5 | 0.1 | |
| RelA, p65 | GSFSQADVHR | y4–y8 | 0.5 | 6 |
| DLEQAISQR (E → D) | y4–y7 | 0.1 | ||
| RelB | ADFSQADVHR | y3–y7 | 1 | 3 |
| Bcl-2 | FATVVEELFR | y5–y8 | 10 | |
| Bcl-xL | EAGDEFELR | y4–y7 | 0.5 | |
| Mcl-1 | QSLEIISR (E → D) | y3–y7 | 0.1 | 1 |
| Bfl-1 | VLQNVAFSVQK (A → G) | y4–y6, y9 | 5 | |
| Bad | GLGPSPAGDGPSGSGK | y10, y11, y13, y14 | 1 | |
| Bid | IEADSESQEDIIR (D → E) | y5–y11 | 0.1 | 1 |
| Bim | IGDEFNAYYAR | y4, y6, y7, y10 | 0.5 | |
| Bak | QLAIIGDDINR (A → G) | y6–y9 | 1 | |
| Bax | TGALLLQGFIQDR (A → G) | y6–y9 | 2 | |
| Fanc D2 | NSDEINIPR | y3–y7 | 0.1 | 6 |
| Fanc I | TIETSPSLSR | y5–y8 | 0.1 | 7 |
Using a nanoLC system (U3000, Dionex, Sunnyvale, CA or Easy-nLC, Proxeon, Denmark), 5 μl of sample (containing the protein equivalent from 41,700 cells) was loaded onto a trapping column at 5 μl/min (Dionex, 300 μm × 5 mm), washed for 20 min, then eluted onto a C18PepMap100 column (LC Packings/Dionex, Sunnyvale, CA) with 75 μm inner diameter, 3 μm particle size, and 100 Å pore size. Peptides were eluted at a flow rate of 300 nl/min. The solvent system was composed of aqueous 2% acetonitrile with 0.1% formic acid (A) and aqueous 90% acetonitrile with 0.1% formic acid (B).
LC-MRM was performed on TSQ Quantum Ultra and Vantage triple quadrupole mass spectrometers (Thermo, San Jose, CA) spraying from 10 μm electrospray (ESI) tips (New Objective, Woburn, MA) held at 2500 V, with a 200 °C transfer-tube temperature, and 12 V skimmer offset. Acquisition parameters were as follows: Q1 was set to m/z 0.2 in width, Q3 was set to m/z 0.7 in width, and each transition was monitored for 20 milliseconds. Fragmentation was achieved with 1.5 mTorr argon. A minimum of three transitions was detected for each peptide. Quantification was achieved based on the sum of the peak areas for all of the detected transitions using QuanBrowser (Xcalibur, Thermo, San Jose, CA) or Skyline (59). The ratio of peak area of the endogenous peptide to its corresponding SIS peptide was calculated for each peptide in the sample to enable absolute and relative quantification. Three technical replicates (LC-MRM analyses) were performed for each biological sample, and a minimum of three biological replicates have been analyzed for each comparative experiment.
RESULTS AND DISCUSSION
Rationale for Selection of SDS-PAGE Coupled with LC-MRM
Immunoprecipitation and LC-ESI-MS/MS can guide the selection of target peptides and transitions; however, LC-ESI-MRM screening of proteins fractionated by SDS-PAGE has been demonstrated to be effective for developing quantitative assays for signaling proteins and applying them to study cancer biology (45). This method has several advantages, including similarity to Western blot analyses. In addition, the molecular weight separation is useful for reducing the interferences in peptide detection and for separating protein isoforms. Even at the cell numbers (∼40,000 cultured RPMI-8226 cells) used in these experiments, SDS-PAGE separation coupled with LC-MRM screening (GeLC-MRM) is a successful method to elucidate candidate peptide targets from these biologically and clinically relevant proteins. Also, proteins with a wide range of molecular weights can be quantified in a single GeLC-MRM experiment (from <20 kDa to >150 kDa).
Based on prior Western blots, several adjacent bands were initially processed to determine the presence of the protein and enable isolation within a single gel region. By excision of gel regions (as determined by the migration of the molecular weight markers shown in Fig. 1B), these proteins were assessed in parallel from the same aliquot of cells. Furthermore, the gel separation has advantages for isolating isoforms of apoptosis-related proteins and nuclear factor-κB subunits. With NFκB-1 and NFκB-2, the inactive forms (p105 and p100) were separated in-gel from the active subunits (p50 and p52). LC-MRM analysis enables specific quantification of these lower-abundance proteins that are not detected in other types of proteomics experiments. Relative and absolute quantification was achieved by titrating SIS peptides into each sample to produce ion signals similar in intensity to those observed for each endogenous peptide. Because of the possibility of incomplete digestion and peptide recovery from the gel, absolute quantification by LC-MRM establishes a minimum value for protein expression (55). However, well-controlled protein digestion and peptide recoveries are highly reproducible enabling comparisons between different samples.
LC-MRM data provided significantly more information when compared with Western blot analysis, even when using similar or lower amounts of protein. As shown in Fig. 2 and Fig. 3 (with absolute quantification of protein expression listed in Table II), changes in relative protein expression were detected by immunoblotting, whereas both relative and absolute quantification were achieved with LC-MRM. Furthermore, 4.5 × 105 cells were lysed for Western blot analysis (90 μg of protein). For LC-MRM experiments, three technical replicates were performed with a total of 1.25 × 105 cells (∼25 μg of protein). For many of these proteins, even lower amounts of protein could have been used. For example, Bcl-2 and Bcl-xL were detected in less than 104 cells (∼2 μg total protein). The current capacity of LC-MRM enables hundreds to thousands of protein measurements in a single experiment, so a broader array of assays could be developed to quantify the expression of other additional proteins that play a role in drug resistance. The use of GeLC-MRM enables a systems biology approach to the elucidation of mechanism of drug resistance in MM cell lines, with the ultimate goal of biomarker assessment in ex vivo patient specimens. The first step of implementation of this clinical assay platform is illustrated by assessing NFκB, apoptosis-regulating proteins, and selected FA proteins in cell lines and a patient sample.
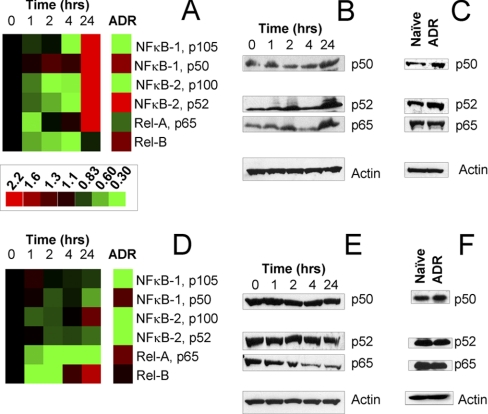
Fig. 2.
Quantification of nuclear factor-κB Survival signaling in naïve, drug treated, and melphalan resistant myeloma cells. NFκB-1, NFκB-2, RelA, and RelB expression were measured with LC-MRM at 0, 1, 2, 4, and 24 h time points after administration of 25 μm melphalan to naïve MM cells growing in culture; protein expression is also quantified in isogenic cell lines with acquired drug resistance (ADR). Ion signals are normalized using spiked SIS peptides; data are plotted relative to the pretreatment control (0 h). Heat maps (A and D) are shown for comparison of naïve MM cells both before and after treatment to cells that have acquired drug resistance; green indicates down-regulation and red shows up-regulation in protein expression. The scale bar applies to both panels. LC-MRM data comparing RPMI-8226 and 8226/LR5 cells (A) are confirmed with Western blotting for NFkB-1 p50, NFkB-2 p52, and Rel-A during drug response (B) and in acquired drug resistance (C). Similar data are shown for naïve U266 and drug-resistant U266/LR6 cells (D–F).
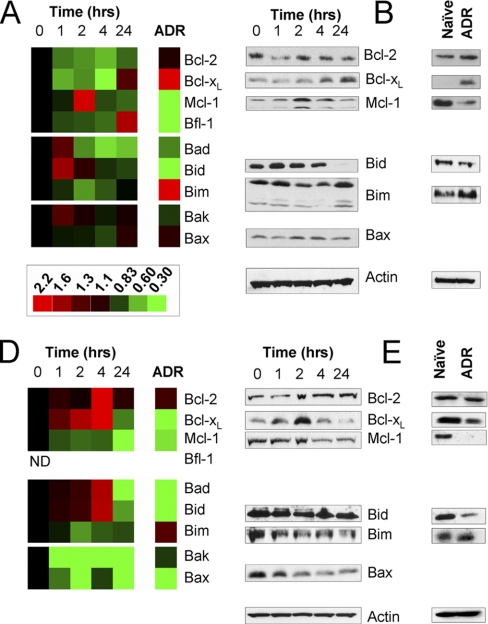
Fig. 3.
Quantification of apoptosis-regulating proteins in the Bcl-2 family in naïve, drug treated, and melphalan resistant myeloma cells. Protein expression was measured with LC-MRM at 0, 1, 2, 4, and 24-hour time points after administration of 25 μm melphalan to naïve MM cells growing in suspension culture; protein expression is also measured in isogenic cell lines with acquired drug resistance (ADR). Ion signals are normalized using spiked SIS peptides; data are plotted relative to the control (0 h). Heat maps (A and D) are shown for comparison of naïve MM cells both before and after treatment to cells that have acquired drug resistance; green indicates down-regulation and red shows up-regulation in protein expression. The scale bar applies to both panels. LC-MRM data comparing RPMI-8226 and 8226/LR5 cells (A) are confirmed with Western blotting for selected apoptotic proteins during drug response (B) and in acquired drug resistance (C). Similar data are shown for naïve U266 and drug-resistant U266/LR6 cells (D–F). ND indicates that the protein was not detected in those cell lines.
Table II. Absolute quantification of selected NFκB survival signaling proteins, Bcl-2 family members, and Fanconi Anemia DNA damage repair proteins in naïve and melphalan resistant multiple myeloma cell lines. The expression level of each protein is reported in attomoles per cell. ND values indicate that the protein was not detected in that cell line. Coefficients of variation for each measurement are given (in %). Student's t-tests were used assess the significance of the differences in protein expression between naïve and drug resistant cell lines.
| Protein | RPMI-8226 |
8226/LR5 |
p value | U266 |
U266/LR6 |
p value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expression (amol/cell) | CV (%) | Expression (amol/cell) | CV (%) | Expression (amol/cell) | CV (%) | Expression (amol/cell) | CV (%) | |||
| NFκB-1 p105 | 0.064 | 14 | 0.027 | 23 | 0.034 | 0.28 | 4.7 | 0.048 | 6.1 | 2.2E-4 |
| NFκB-1 p50 | 0.26 | 18 | 0.41 | 15 | 0.017 | 0.086 | 17 | 0.11 | 14 | 0.11 |
| NFκB-2 p100 | 0.095 | 6.3 | 0.028 | 1.3 | 0.015 | 0.053 | 7.2 | 0.024 | 9.4 | 1.5E-4 |
| NFκB-2 p52 | 0.029 | 20 | 0.076 | 20 | 0.034 | 0.032 | 12 | 0.013 | 7.0 | 0.015 |
| Rel A, p65 | 0.065 | 5.9 | 0.037 | 4.9 | 0.0043 | 0.057 | 16 | 0.079 | 8.9 | 0.027 |
| Rel-B | 0.038 | 0.3 | 0.058 | 0.75 | 0.0069 | 0.050 | 0.3 | 0.056 | 7.8 | 0.32 |
| Bcl-2 | 0.71 | 8.9 | 0.78 | 14 | 0.23 | 0.40 | 0.3 | 0.49 | 2.1 | 0.0044 |
| Bcl-xL | 0.36 | 7.4 | 1.6 | 5.8 | 4.2E-5 | 0.79 | 13 | 0.33 | 3.6 | 8.9E-7 |
| Mcl-1 | 0.79 | 10 | 0.24 | 10 | 3.6E-4 | 0.36 | 7.3 | 0.20 | 10 | 9.6E-4 |
| Bfl-1 | 2.5 | 4.6 | 1.0 | 20 | 2.4E-5 | ND | ND | |||
| Bad | 0.015 | 7.9 | 0.011 | 7.1 | 0.016 | 0.064 | 4.7 | 0.025 | 14 | 0.0021 |
| Bid | 0.34 | 10 | 0.041 | 10 | 5.8E-7 | 0.095 | 2.7 | 0.023 | 6.7 | 2.9E-5 |
| Bim | 0.014 | 17 | 0.043 | 8.0 | 6.9E-4 | 0.053 | 9.1 | 0.069 | 14 | 0.08 |
| Bak | 0.54 | 17 | 0.46 | 2.2 | 0.19 | 0.55 | 6.0 | 0.47 | 9.0 | 0.07 |
| Bax | 3.8 | 19 | 4.4 | 1.5 | 0.33 | 2.3 | 2.6 | 1.1 | 7.9 | 3.0E-4 |
| FANCD2 | 0.062 | 7.4 | 0.12 | 13 | 2.1E-4 | 0.079 | 9.7 | 0.078 | 2.7 | 0.9 |
| FANCI | 0.045 | 19 | 0.078 | 14 | 0.0086 | 0.084 | 4.9 | 0.078 | 4.3 | 0.051 |
LC-MRM Screening and Quantification of Nuclear Factor-κB Subunits
NFκB survival signaling can be monitored using the combination of SDS-PAGE and LC-MRM. The gel separation is also advantageous for isolating the active forms of NFκB-1 and NFκB-2 at 50 kDa and 52 kDa from the inactive forms at 105 kDa and 100 kDa to enable parallel assessment of these proteins from the same sample (see Fig. 1B). From the initial LC-MRM screens, we selected peptides from the C-terminal regions of the full length NFκB-1 and NFκB-2 molecules; additional data was then acquired to select N-terminal peptides in order to monitor the truncated active isoforms in addition to the full length proteins. Assays were developed for NFκB-1 p50 and p105, NFκB-2 p52 and p100, RelA (p65), and RelB. Expression levels in each cell line and coefficients of variation for each measurement are reported in Table II. The statistical significance of the differences between paired naïve and drug-resistant cell lines is reported using p values from Student's t-tests. For comparison of drug-resistant cell lines to their naïve parental cell lines, relative expression values have been plotted in Fig. 2. Selected Western blots are also shown for comparison of protein expression measurements between the two techniques.
Comparisons of expression of NFκB proteins in naïve RPMI-8226 and melphalan-resistant 8226/LR5 cell lines are shown in Fig. 2A with corresponding Western blots in Fig. 2C. In 8226/LR5 cells, the signal for inactive NFκB-1 p105 was significantly lower: 0.43 of the value in RPMI-8226 cells. However, the signal for active NFκB-1 p50 increased by a factor of 1.6, indicating that the stores of inactive NFκB-1 were being converted to the active form to promote cell survival. Similar trends were observed for NFκB-2; the inactive form at 100 kDa significantly decreased by a factor of 0.29 in 8226/LR5 as compared with RPMI-8226, whereas the active form at 52 kDa increased by a factor of 2.6. These data show a concerted shift, which enables activation of NFκB-1/2 signaling. RelB, the NFκB transactivating subunit linked to acquired melphalan resistance, increases 1.5-fold in drug resistant cells. RelA (p65), an additional NFκB binding partner, decreased by a factor of 0.57 in 8226/LR5 as compared with RPMI-8226 cells. The change in RelA was not detected by Western blotting.
Based on the absolute quantification of NFκB subunits in RPMI-8226 cells (Table II), total expression of NFκB1 was up-regulated by ∼35% and more of the protein population was converted into the p50 isoform, which increases from 80% of NFκB-1 expression in RPMI-8226 to 94% in 8226/LR5. Total expression of NFκB-2 was not significantly changed in this model of melphalan resistance, but more of the protein population was converted into the p52 isoform, which increased from 23% in RPMI-8226 to 73% in 8226/LR5. Increases in expression were noted for RelB and p50, which mediate resistance to melphalan (7).
Comparisons of the expression levels of NFκB proteins in U266 and U266/LR6 cell lines are shown in Fig. 2D, with corresponding Western blots in Fig. 2F. In comparing relative protein expression levels in U266 and U266/LR6, significant decreases in expression were noted for p105 (0.17), p100 (0.16), and p52 (0.43), whereas RelA significantly increased (1.4). No significant change in expression was noted for RelB or p50. The total expression of NFκB-1 and NFkB-2 both decreased by a factor of 0.43 in U266/LR6 as compared with U266. Expression of p50 shifted from 23% of total NFκB-1 to 70% in drug-resistant cells, but the percentage of NFκB-2 expressed as p52 was unchanged. Because RelB was expressed at approximately half of the level of p50, it could be considered to be the limiting factor for RelB/p50-mediated transcriptional activity. When comparing the data from this set of cell lines to those derived from RPMI-8226, the expression levels of RelB were similar in U266 naïve cells and 8226/LR5 melphalan resistant cells (Table II), suggesting that up-regulation of RelB may not be necessary to achieve drug resistance via NFκB survival signaling.
NFκB LC-MRM to Examine Initial Responses to Acute Melphalan Treatment in Naïve MM Cells
In addition to the steady-state measurements of differences between naïve and drug resistant cells, the initial response of drug sensitive RPMI-8226 cells to melphalan was also explored using time-course measurements of protein expression. Melphalan was administered to the cells at the half-maximal inhibitory concentration (IC50) determined by cell viability assays to be 25 μm. The initial response to the stress of DNA damage is likely to be a critical step in the development of drug resistance, and it may provide clues to the mechanisms that cells employ for survival. NFκB signaling proteins were measured in cells prior to the administration of melphalan in the media (time 0 control) and in aliquots of the same suspension culture at 1, 2, 4, and 24 h. Heat maps of relative expression are shown in Fig. 2A for RPMI-8226 and Fig. 2D for U266 with Western blots for comparison in Fig. 2B and Fig. 2E, respectively. Clear trends were observed for both inactive and active forms of the NFκB-1 and NFκB-2 that show initial decreases; however, the levels not only rebounded but showed significant increases by the 24-hour time point. Peptide signals for NFκB-1 p105 and p50 increased by factors of 2.9 ± 0.42 and 3.9 ± 0.29 after 24 h of exposure to melphalan. Similarly, peptide signals for NFκB-2 p100 and p52 increased by factors of 2.2 ± 0.14 and 4.9 ± 0.27 over the 24 h period after drug administration. These data indicate that the rates of synthesis of NFκB isoforms are increasing, along with accelerated conversion from the inactive forms to the active forms. RelA expression also initially decreased, but at 24 h, the peptide ion signal increased by a factor of 2.8 ± 0.28. RelB did not significantly change; the value for relative expression at 24 h is 0.93 of control. Most changes in NFκB survival signaling observed after acute treatment of naïve cells (at their IC50) were consistent with the steady-state differences observed when comparing RPMI-8226 and 8226/LR5 cells based on the selection pressure of chronic drug exposure (see Fig. 2A).
In U266 cells, relative expression of both NFkB-1 isoforms are not significantly different after acute treatment at the IC50 (25 μm). NFκB-2 p52 (0.74 ± 0.045) and RelA (0.34 ± 0.015) are significantly reduced, whereas p100 (1.4 ± 0.052) and RelB (1.8 ± 0.18) are significantly increased.
LC-MRM Screening and Quantification of Bcl-2 Family Apoptotic Proteins
Within this set of lower molecular-weight proteins, multiple screens often yielded only a single candidate peptide for quantitative assay development; however, these individual peptides can be consistently monitored. For the first set of experiments, the most abundant isoform of each apoptotic protein was selected for analysis. Expression of selected anti-apoptotic and pro-apoptotic proteins were quantified in the cell line models with absolute quantification reported in Table II and relative expression plotted as heat maps shown in Fig. 3. Western blots are shown for selected anti-apoptotic proteins to verify the LC-MRM results. Data are reported for Bcl-2, Bcl-xL, Mcl-1, Bfl-1, Bad, Bid, Bim, Bak, and Bax. Additional assays were developed for Bok, BNIP3, Bik, Hrk, and Bcl-w, but they could not be detected in these experiments because of low expression levels in these cell lines.
Six proteins were differentially expressed between the RPMI-8226 and melphalan resistant 8226/LR5 cell lines (p < 0.05); heat maps are shown in Fig. 3A with Western blots for comparison in Fig. 3C. These changes in drug resistant cells are consistent with previously reported data; however, previously undetected differences were also found. Significant changes were observed for peptides used to monitor the pro-apoptotic proteins, Bad, Bid, and Bim. Decreased expression of Bad (0.73) and Bid (0.12) were noted. In contrast, expression of Bim increased by a factor of 3.0. Up-regulation of Bim mRNA and protein expression (measured by Western blotting) has been shown to contribute to the development of melphalan-resistance in MM cells (5). Signals corresponding to peptides representing the anti-apoptotic proteins, Bcl-xL, Mcl-1, and Bfl-1, were also significantly different between the two cell types. Of the anti-apoptotic proteins, Bcl-xL increases in drug resistant cells by a factor of 4.4. Conversely, Mcl-1 and Bfl-1 expression decreased in drug resistant 8226/LR5 cells to 0.30 and 0.40 of their respective expression levels detected in RPMI-8226 cells.
A comparison of U266 and U266/LR6 showed significant differences between these two cell lines associated with melphalan-resistance. In addition, U266-derived cell lines regulated apoptosis in different ways than RPMI-8226-derived cell lines. As an example, Bfl-1 was not detected in either U266 or U266/LR6. Six apoptotic proteins were differentially expressed in this pair of naïve and drug-resistant cell lines (p < 0.05). Heat maps are shown in Fig. 3D with Western blots for comparison in Fig. 3F. In U266/LR6, Bcl-2 increased (1.2-fold), whereas both Bcl-xL and Mcl-1 decreased to 0.42 and 0.56 of their expression level in U266. The decrease in Bcl-xL was particularly intriguing, because U266 survival is known to be driven by high expression of that anti-apoptotic protein (13, 56, 57). Bad (0.39), Bid (0.24), and Bax (0.48) also significantly decreased after development of ADR. Common trends were noted between RPMI-8226- and U266-derived cell lines for Bid and Mcl-1; Bim was up-regulated in both drug-resistant cell lines when compared with the naïve parental cells, but the difference between Bim expression levels in U266/LR6 and U266 was not significant (p = 0.08).
Modulation of Apoptotic Proteins in Response to Acute Drug Treatment in Naïve Cells
Heat maps of protein expression in RPMI-8226 cells before and after acute treatment with melphalan (at their IC50: 25 μm) are shown in Fig. 3A with the corresponding Western blots in Fig. 3B. The expression of Bcl-2 did not change significantly over the course of this experiment. One hour after treatment, Bcl-xL decreased by a factor of 0.62 and started to rebound 4 h after treatment. At 24 h after treatment, expression of Bcl-xL increased to 1.30 ± 0.09 times the level in control samples. Mcl-1 increased by a factor of 1.95 ± 0.41 at 2 h, and decreased to 0.75 ± 0.06 after 24 h of treatment. Bfl-1 increased by a factor of 1.74 ± 0.76 after 24 h. Bid increased by a factor of 1.64 at 1 h, and decreased to 0.83 ± 0.17 at 24 h after treatment; the attenuation of Bid expression may also play a role in the initial survival mechanism. Bim expression decreased by a factor of 0.65 ± 0.04 at 2 h, and recovered to 0.99 ± 0.03 at 24 h. The transient increase in Mcl-1 expression and simultaneous decrease in Bim expression may contribute to the initial mechanism of cell survival. The expression of Bak and Bax did not change with the drug treatment. In these treatment samples, CV values were generally higher than in the steady-state measurements; however, the data indicate clear trends in the expression of the apoptotic proteins.
Changes in expression of the apoptosis-regulating proteins showed trends that would be consistent with the steady-state differences between the RPMI-8226 and the 8226/LR5 drug-resistant cell lines, if the trends in Bcl-xL, Bid, Bim, and Mcl-1 expression continued. However, the observation of protein expression at longer time points could not be measured because of the increase in apoptosis, which produced a large fraction of dead cells (the IC50 was determined by cell viability measurements at 72 h).
Data for the U266 treatment response are shown in Fig. 3D with corresponding the Western blots in Fig. 3E. Transient increases in Bcl-2 and Bcl-xL expression were noted with peaks at 4 h after treatment (3.6-fold and 2.1-fold higher expression, respectively). However, at 24 h, Bcl-2 expression was not significantly different from control, but Bcl-xL expression decreased to 0.71 ± 0.08 of the pretreatment control value. Mcl-1 expression decreased throughout this time course; expression levels at 24 h after treatment were reduced to 0.41 ± 0.05 of control levels. Bad and Bid expression peaked at 4 h (1.7-fold and 1.8-fold increases, respectively), but significant decreased at 24 h to 0.49 ± 0.08 and 0.65 ± 0.04, when compared with controls. The relative expression of Bim decreased to 0.66 at 2 h, but expression at 24 h after treatment is similar to control levels. Bak and Bax were expressed at lower levels at each time point after treatment; their expression levels were 0.27 ± 0.01 and 0.48 ± 0.05, respectively, at 24 h post-treatment. Drug response of U266 cells appears to be driven by up-regulation of Bcl-2 and Bcl-xL and reduction in Bad, Bid, and Bax.
As described above for comparison of the protein expression during the response to melphalan in RPMI-8226 cells to steady-state expression in drug-resistant 8226/LR5 cells, trends observed for U266 cells' treatment response also share similarities with steady-state expression in U266/LR6. Decreases in Bcl-xL and Mcl-1 as well as Bad, Bid, and Bax at 24 h after treatment were consistent with lower expression levels in U266/LR6.
Comparison of Expression of the Groups of Anti-Apoptotic Proteins, Pro-Apoptotic Initiators and Sensitizers, and Pro-Apoptotic Effectors
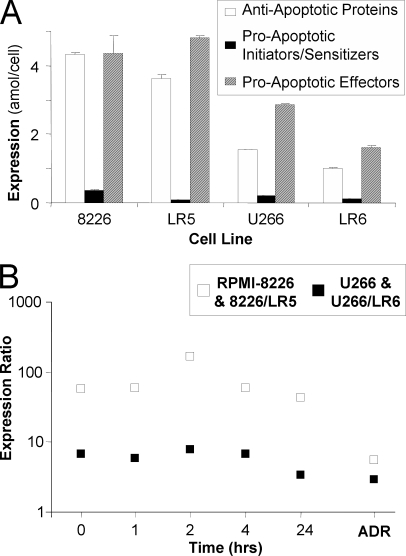
In addition to examining the individual expression values of the apoptosis-regulating proteins, their behavior as a group was also explored using the quantitative data reported in Table II. Total protein expression was calculated for anti-apoptotic proteins (Bcl-2, Bcl-xL, Mcl-1, and Bfl-1), pro-apoptotic initiators and sensitizers (Bad, Bid, and Bim), and pro-apoptotic effectors (Bak and Bax) in order to examine the balance of signaling for programmed cell death versus survival signaling in naïve cells, both before and after treatment, and in cells with ADR. Summed expression data are plotted in bar-graphs in Fig. 4A. Errors were calculated using pooled variance. In RPMI-8226, the expression levels of anti-apoptotic proteins (4.32 ± 0.08 amol/cell) were similar to expression levels for pro-apoptotic effectors (4.37 ± 0.52 amol/cell); whereas pro-apoptotic initiators and sensitizers were expressed at less than 10% of the levels of the other two groups (0.37 ± 0.02 amol/cell). In 8226/LR5 cells, pro-apoptotic effectors were expressed at higher levels (4.82 ± 0.05 amol/cell) and anti-apoptotic protein expression decreased (3.64 ± 0.12 amol/cell), when compared with RPMI-8226. Pro-apoptotic initiators and sensitizers are expressed a 3.9-fold lower levels in 8226/LR5 (0.096 ± 0.003 amol/cell) than in RPMI-8226.
Fig. 4.
Examining the balance of pro-apoptotic and anti-apoptotic protein expression. In each cell line, the sum of the expression levels of anti-apoptotic proteins (white bars), pro-apoptotic initiators and sensitizers (black bars), and pro-apoptotic effectors (diagonally striped bars) are compared (A). In addition, the ratio of Mcl-1 to Bim is plotted (B) to illustrate the potential of LC-MRM to explore relationships between individual apoptotic proteins. Data are plotted on a log scale for drug response in RPMI-8226 (white) and U266 (black); ratios calculated for protein expression in 8226/LR5 (white) and U266/LR6 (black) are also shown for comparison.
In the comparison of U266 and U266/LR6, both similarities and differences in the regulation of apoptosis can be noted. In U266 cells, expression of anti-apoptotic proteins (1.55 ± 0.01 amol/cell) was lower than pro-apoptotic effectors (2.86 ± 0.05 amol/cell) and initiators and sensitizers were again expressed at significantly lower levels (0.21 ± 0.003 amol/cell) than the other two groups. In drug-resistant U266/LR6 cells, the expression levels of all three groups are lower than in U266 cells. Anti-apoptotic proteins were expressed at a total level of 1.02 ± 0.01 amol/cell; pro-apoptotic effector proteins were expressed at a total level of 1.61 ± 0.07 amol/cell, whereas initiators and sensitizers were expressed at a total level of 0.12 ± 0.005 amol/cell.
Signaling through Mcl-1/Bim
The interaction between Mcl-1 and Bim has been described as a point of signal integration for deciding between survival and cell death (24). To examine this signaling in greater detail, the expression ratio of Mcl-1 to Bim was plotted for naïve cells, before and after treatment, and for cells with ADR in Fig. 4B. In RPMI-8226 cells, the ratio is 56. Two hours after treatment, the ratio increased to 170, but decreased to 43 at 24 h after treatment. In drug-resistant 8226/LR5 cells, Mcl-1 expression decreased and Bim expression increased, so that the ratio value was only 5.6. In U266 cells, the Mcl-1/Bim expression ratio was 6.8. When compared with RPMI-8226, minimal changes were observed in the ratio in U266 cells after acute treatment with melphalan. A maximum ratio value of 7.7 was observed at 2 h with a decrease to 3.4 at 24 h. U266/LR6 cells express Mcl-1 at 2.9-fold higher levels than Bim. In both sets of cell lines, the drug-resistant cells have lower Mcl-1/Bim expression ratios than the naïve parental cell lines.
Isoform Analysis of Differentially Expressed Apoptotic Proteins
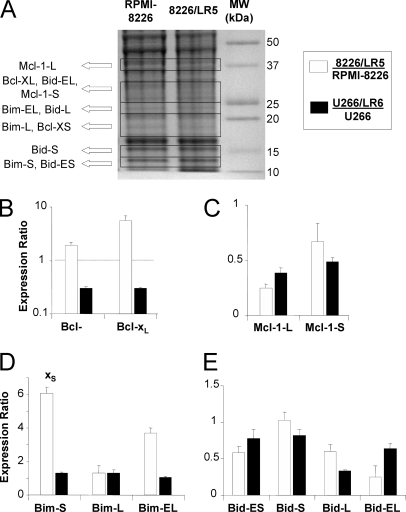
Based on these initial results, comprehensive isoform analysis was performed for Bcl-2 family members with differential expression in naïve and drug-resistant cells: Bcl-xL, Mcl-1, Bid, and Bim for more comprehensive analysis of their isoforms (Bad and Bfl-1 were excluded because they have only one known isoform). LC-MRM can distinguish protein isoforms by detecting and quantifying peptide sequences specific to each isoform, even when the proteins have high sequence homology. Gel separation also plays a useful role in distinguishing protein isoforms. Two Bcl-x, two Mcl-1, four Bid, and three Bim isoforms were monitored in a single experiment by using LC-MRM as shown in Fig. 5. These experiments were specifically designed to detect isoforms and not truncation products. The truncated forms are unstable (e.g. cleaved Bim(60) and are expected to be a very low percentage of the total protein population. Bcl-xL has been reported to be resistant to caspase cleavage both in vitro and in intact cells (61). Bid and Mcl-1 can be cleaved to produce active truncated Bid (tBid) and pro-apoptotic truncated Mcl-1. The lower molecular weight isoforms of the truncated proteins, Mcl-1 and Bid, either did not co-migrate in the gel with the lower molecular weight isoforms or they were measured with isoform-specific peptides that are not contained in the truncated forms of the longer isoform expected in the same region of the gel. Gel-excision patterns of these isoforms are shown in Fig. 5A. Bar-graphs show the average and standard deviation of the expression ratio obtained by comparing drug-resistant cells to naïve parental cells for each protein isoform. Significance was determined by Student's t-tests (p < 0.05).
Fig. 5.
GeLC-MRM Examination of isoforms of Bcl-2 family proteins modulated in melphalan resistance. After observing changes in the expression of the most abundant isoforms of Bcl-xL, Mcl-1, Bid, and Bim, the expression levels of the additional isoforms were investigated in the same cell line models. The gel-excision map is shown (A) with expression levels for Bcl-x (B), Mcl-1 (C), Bim (D), and Bid (E) isoforms. Data are presented as ratios between protein expression in naïve and drug-resistant cells with 8226/LR5 versus RPMI-8226 plotted in white and U266/LR6 versus U266 in black.
Bcl-xL is monitored in the gel region containing proteins from 25–30 kDa; Bcl-xS is monitored in the gel between 17 and 20 kDa. As shown in Fig. 4B, the peptide ion signals for Bcl-xS and Bcl-xL increased by factors of 1.9 and 5.6, respectively, in 8226/LR5 cells when compared with RPMI-8226 cells. In U266/LR6, the expression of Bcl-xL and Bcl-xS is lower, at 0.30 and 0.39, of the respective levels in U266 cells. In both cases, the expression of both Bcl-x isoforms have similar trends in melphalan resistance.
Mcl-1-L was detected in the band between 35 and 39 kDa, and Mcl-1-S was detected between 25 and 30 kDa. Because MCL-1-S shares 85% sequence homology with Mcl-1-L, assays could be developed for two peptides (SWFGISNK and WHQECAAGFCR), which are specific to Mcl-1-S, and one peptide (NHETAFQGMLR), which is specific to Mcl-1-L. Mcl-1-L is reported to be cleaved by caspases C-terminal to Asp127 and Asp157 (61). The molecular weight of truncated Mcl-1-L, containing residues 128–350, is 24.6 kDa, and it migrated to the same band that contains Mcl-1-S. Cleavage C-terminal to Asp157 would produce a lower molecular weight protein that would migrate farther down the gel. To avoid interference from truncated Mcl-1-L, an Mcl-1-S-specific peptide, WHQECAAGFCR, was used for quantification. The shorter isoform, Mcl-1-S, decreased to 0.67 of the amount in the RPMI-8226 cells, and the peptide representing Mcl-1-L, decreased to 0.22 of the amount in the RPMI-8226 cells (Fig. 4C). In U266/LR6 cells, Mcl-1-S and Mcl-1-L expression levels also decreased to 0.49 and 0.39 of the levels detected in the U266 parental cell line. In both models of drug resistance, all Mcl-1 isoforms were significantly down-regulated in ADR.
Bim isoforms were detected in the following gel regions: 20–25 kDa (Bim-EL), 14–16 kDa (Bim-L), and 11–14 kDa (Bim-S). Bim-EL expression increased by a factor of 3.7 and Bim-S increased 6.1-fold in 8226/LR5 when compared with RPMI-8226 cells (Fig. 4D). Bim-L was not significantly different between the two cell lines. In U266/LR6 versus U266, the expression levels of Bim-S and Bim-L are increased 1.3-fold, but Bim-EL was not significantly different. In these models of melphalan-resistance, changes in the expression of Bim isoforms were unique to each cell line.
The following regions were excised for monitoring Bid isoforms: 25–30 kDa (Bid-EL), 20–25 kDa (Bid-L), 14–16 kDa (Bid-S), and 11–14 kDa (Bid-ES). The same assay, monitoring the VASHTPSLLR peptide, was used to quantify Bid-EL, Bid-L, and Bid-ES after SDS-PAGE separation. The sequences of Bid-L and Bid-ES are shorter than Bid-EL, but have complete sequence overlap with segments of Bid-EL. On the other hand, Bid-S shares only 54% sequence homology with Bid-EL, so an isoform-specific peptide (TSSGILPGTSPR) was used to distinguish Bid-S from other Bid isoforms. Pro-apoptotic cleavage of Bid (to form tBid) was reported to occur C-terminal to Asp59 by caspase 8 (62), C-terminal to Ser67 by lysosomal protease (63), C-terminal to Gly70 by Calpain (64), and C-terminal to Asp75 by Granzyme B (65). Regardless of the cleavage site, tBid migrates in the gel in the region from 11–15 kDa, where Bid-S and Bid-ES are monitored. Bid-S was quantified with the isoform specific peptide, TSSGILPGTSPR, which is not contained in tBid. However, tBid would be detected by monitoring VASHTPSLLR peptide to detect Bid-ES. The initial measurement of Bid-L was repeated with measurements for Bid-EL, Bid-S, and Bid-ES. The relative amounts of peptide signals corresponding to Bid-EL (0.24) and Bid-ES (0.58) decreased in a manner similar to Bid-L (0.60), but no significant change was detected for the Bid-S signal (Fig. 4E). Comparison of Bid isoforms in U266/LR6 cells indicated down-regulation of expression levels of Bid-ES (0.78), Bid-S (0.82), Bid-L (0.33), and Bid-EL (0.63) when compared with U266 cells. In both models of ADR, the expression levels of most of the Bid isoforms were decreased.
Quantification of Selected Fanconi Anemia Proteins
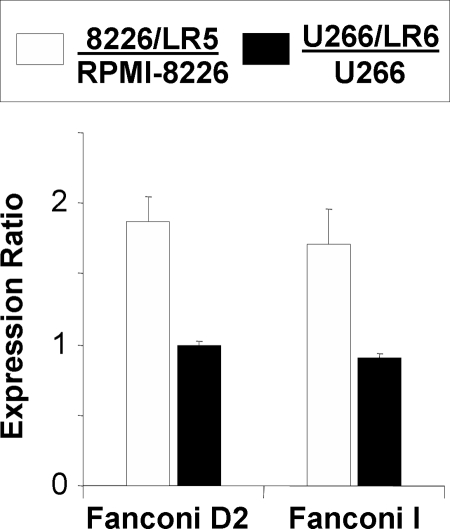
Based on their central role in the FA pathway, the DNA-damage repair effectors, FANCD2 and FANCI were quantified in each cell line. These measurements correspond to total expression of FANCD2 and FANCI, because the gel regions encompass both the unmodified protein and the mono-ubiquitinated active form. Expression levels and coefficients of variation for each measurement are reported in Table II; the statistical significance is reported using p values from Student's t-tests. For comparison of drug-resistant cell lines to their naïve parental cell lines, expression ratios were plotted in Fig. 6. Significant increases in the expression of FANCD2 (1.9-fold) and FANCI (1.7-fold) were observed in 8226/LR5 when compared with RPMI-8226 cells. These LC-MRM measurements indicate the same trends in expression that were previously noted using other methods, including Western blot analysis (protein expression) gene expression profiling (mRNA), and reverse transcriptase-polymerase chain reaction (RT-PCR; mRNA) (5–7). Increases in FANCD2 expression in drug-resistant cells have been noted in Western blots (6). RT-PCR experiments measured an increase in gene expression of 1.11 ± 0.2 for FANCD2 in 8226/LR5 compared with RPMI-8226, but the gene expression of FANCI has not been evaluated. Significant increases in FANCD2 and FANCI mRNA levels have been previously demonstrated in drug-selected U266 cells. However, LC-MRM did not observe similar increases in protein expression in U266/LR6 cells relative to U266 cells (FANCD2 (0.99) and FANCI (0.93)).
Fig. 6.
Expression of Fanconi anemia DNA damage repair effectors in naïve and drug resistant MM cell lines. Expression ratios are plotted for FANCD2 and FANCI comparing 8226/LR5 versus RPMI-8226 (white) and U266/LR6 versus U266 (black).
In addition to performing relative quantification to assess the differences between naïve RPMI-8226 and 8226/LR5 drug-resistant cells, the absolute expression of these FA proteins was also quantified using these LC-MRM assays. The expression values measured for FANCD2 and FANCI were 62 ± 4.6 and 45 ± 8.6 zeptomoles per RPMI-8226 cell and 79 ± 7.7 and 84 ± 4.1 zeptomoles per U266 cell. These similar expression levels are consistent with these proteins functioning as a heterodimer in the initiation of DNA repair. In addition, the absolute quantification data indicates that the levels of expression of FANCD2 and FANCI are comparable in drug-resistant 8226/LR5 and naïve U266 cells, indicating that increases in protein expression may not be necessary to provide sufficient amounts of DNA repair for overcoming the damage induced by chronic melphalan exposure in U266/LR6 cells.
Western blots can also indicate relative levels of ubiquitination in addition to changes in protein expression. Unfortunately, precise excision of these two bands was not possible, so SDS-PAGE was not used to isolate the active (ubiquitinated) protein for measurement of protein expression in the active and inactive forms. Evaluation of the ubiquitination sites on FANCD2 and FANCI using LC-MRM could not be achieved in the amounts of material described here, which contained ∼2.5 fmol of each protein, because the limits of quantification were estimated at 10 fmol for the glycyl-glycyl-lysine-containing peptide from FANCD2 and 20 fmol for the peptide from FANCI based on the calibration curves for their synthetic isopeptide standards. In both cases, the sequences also indicate potential challenges with peptide-based monitoring because of the selection of trypsin for proteolysis. The mono-ubiquitinated peptide and flanking amino acids for FANCD2 are R.KQLSSTVFK.YK; the N-terminal lysine residue is the monoubiquitination site. For FANCI, the lysine is also at the N terminus of the peptide, R.KAMFANQLDAR.K. In both cases, tryptic cleavage may be affected and partially blocked by the branched peptide. Additional variability in cleavage near the C terminus may also increase the number of peptides that would need to be monitored for accurate quantification. Measurement of these post-translational modifications would be possible with more material, but applying more lysate would likely exceed the capacity of both the gel and LC column. An assay for quantification of the mono-ubiquitination of FANCD2 and FANCI would most likely require an immunoprecipitation strategy for protein enrichment.
Translation to CD138+ Cells Isolated from MM Patient Bone Marrow Aspirate
MM tumor cells were isolated from the bone marrow aspirate of a patient that had not received any prior treatment. MM tumor cells from patients generally yield significantly lower amounts of protein than cell line models. As an artifact of the cell culture, MM cell lines increase in size and total protein content, when compared with primary tumor cells acquired from patients. Therefore, we increased the number of cells that were analyzed to 500,000. After lysis of 0.5 × 106 cells (determined by counting flow cytometry events), the amount of protein extracted was 8 μg (which is 1/12 the amount recovered from the same number of cultured myeloma cells: lysis of 500,000 RPMI-8226 cells produces 100 μg of total protein). For LC-MRM, these samples were analyzed in triplicate, injecting 1/3 of the total sample each time (equivalent to ∼1.7 × 105 cells). Quantification of the proteins involved in NFκB signaling, regulation of apoptosis, and FA DNA-damage repair is reported in Table III. Overall, ten out of 17 proteins could be monitored; however, best results were obtained for translation of the assays for the Bcl-2 family proteins and the poorest results for translation of NFκB assays. To increase the success rate for assay translation, higher numbers of cells will need to be acquired from the patient. In this experiment, the bone marrow aspirate volume was 5 ml. Additional material can be readily obtained; the volume of the draw can be increased to as much as 80 ml, but the increased burden on the patient must be taken into account. The limitation would be more critical in evaluating patients that had responded well to therapy or to patients with minimal residual disease, which would produce low-purity samples that yielded only small populations of MM cells. However, this technique still indicated an improvement over approaches based on Western blotting, as this number of cells would currently yield only a single Western blot analysis.
Table III. Protein expression in CD138+ cells isolated from the bone marrow aspirates of a multiple myeloma patient. The amounts in attomoles per cell and coefficients of variation of the measurements (in %) are listed for each detected protein.
| Protein | Expression (amol/cell) | CV (%) |
|---|---|---|
| NFκB-2, p100 | 0.013 | 9.0 |
| Bcl-2 | 0.12 | 4.1 |
| Bcl-xL | 0.0082 | 13 |
| Bcl-xS | 0.012 | 15 |
| Bad | 0.015 | 7.5 |
| Bid | 0.0068 | 9.5 |
| Bim | 0.11 | 6.2 |
| Bak | 0.26 | 5.4 |
| Bax | 0.25 | 9.0 |
| Fanc I | 0.0014 | 9.0 |
CONCLUSIONS
A GeLC-MRM based platform for assessment of protein expression related to NFκB survival signaling, apoptosis, and the Fanconi Anemia DNA damage-response has been developed using models for ADR to melphalan in two MM cell lines (RPMI-8226 and U266), which have different underlying oncogenic biology. Both relative and absolute quantification were performed using SIS peptides, which we have made available to the scientific community on a cost-recovery basis (online at http://proteome.moffitt.org/QUAD/) (66). This set of quantitative assays provides an example of known cancer biology guiding experimental design for LC-MRM, leading to increased knowledge of acquired drug resistance and providing additional techniques for studying cancer biology in patient specimens.
LC-MRM protein quantification enables the determination of both relative expression levels in naïve and drug resistant cells, as well as calculation of the protein copy number per cell. In the analysis of NFκB signaling, the up-regulation of RelB/p50 expression in the ADR of 8226/LR5 was notable. In the naïve RPMI-8226 cells' response to acute treatment with melphalan, concerted up-regulation was observed for NFκB-1 and NFκB-2 isoforms. Both of these findings are consistent with previously demonstrated survival signaling associated with drug resistance (7) though the measurement of lower molecular weight active subunits with LC-MRM must be used as surrogate for activation (rather than a direct measurement through electrophoretic mobility shift assays). Based on the contrast of NFκB responses in RPMI-8226 and U266, NFκB signaling through RelB/p50 may play the same role in the response to acute melphalan treatment in the U266 model system and maintenance of drug resistance under chronic exposure conditions, but changes in NFκB signaling are clearly not driven by increases in protein expression. Regulation may occur through a different mechanism (e.g. phosphorylation by IκKs or by modulation of IκB inhibitor expression). However, the consistent increase of the fraction of NFκB-1 expressed as the p50 isoform in both drug-resistant cell lines indicate that the ratio of p50 to total NFκB-1 could be a candidate biomarker for melphalan-resistance.
In monitoring apoptosis-regulating proteins in these cell line models, the critical importance of evaluating the expression of the Bcl-2 family of proteins in the same sample was revealed, because numerous changes are occurring simultaneously to promote cell survival during drug treatment and in ADR. The redundancy of the effects of these molecules necessitates a comprehensive monitoring approach. Increases in expression of any anti-apoptotic protein can promote survival, whereas increases in expression of pro-apoptotic proteins will tip the balance to initiate cell death. In RPMI-8226, apoptosis in response to melphalan seems to be regulated by different anti-apoptotic proteins at different time points. In sequential order, Mcl-1, Bfl-1, and Bcl-xL are all involved in the response to melphalan. In 8226/LR5 cells, Mcl-1 was down-regulated and Bim was up-regulated, suggesting attempts to initiate apoptosis in melphalan-treated cells that must be overcome by anti-apoptotic signals. Bcl-xL up-regulation is likely the dominant survival signal in 8226/LR5 melphalan-resistant cells, which could explain their lower sensitivity to apoptosis-inducing stimuli from melphalan treatment. In U266 cells, both Bcl-2 and Bcl-xL appear to be involved with survival after acute treatment with melphalan, but the survival of U266/LR6 melphalan-resistant cells is driven primarily by decreases in the expression of anti-apoptotic proteins. In both model systems, consistent trends are noted for Bid and Mcl-1 (which decrease in ADR) as well as Bim (which increases in ADR). Bcl-xL also emerges as a candidate biomarker for ADR. However, understanding the regulation of apoptosis is likely to require measurement of the entire Bcl-2 family. Thus, focusing on a single Bcl-2 family member or a single pair of anti- and pro-apoptotic proteins is unlikely to result in successful evaluation of a disease as heterogeneous as MM. To that end, we propose that a panel of predictive or prognostic assays based on the expression of apoptosis-regulating proteins will need to encompass the entire Bcl-2 family of proteins and that LC-MRM is the preferred method for this approach.
Absolute quantification by LC-MRM also enables analysis of the balance of anti- and pro-apoptotic signals both as a whole and for specific pairs of proteins (e.g. Mcl-1/Bim). Although the Bcl-2 family proteins interact with one another by forming heterodimers, the levels of anti-apoptotic proteins could be lower than those of pro-apoptotic proteins, because cell death effectors (i.e. Bax) can be in inactive conformations, requiring activation by protein-protein interaction. In the four cell lines studied here, pro-apoptotic initiators and sensitizers are expressed at significantly lower levels than anti-apoptotic proteins and pro-apoptotic effectors. Drug-resistant cell lines exhibit consistent decreases in initiators and sensitizers, when compared with naïve cell lines, indicating that potential for induction of apoptosis may be reduced by decreasing their expression. However, the initiators and sensitizers were not expressed at a lower level than anti-apoptotic proteins and pro-apoptotic effectors in the patient sample. Further experiments will be required to determine if the reduced expression of initiators and sensitizers is an artifact of cell culture or if it has clinical significance. The overall success in translation of the assays for the apoptotic proteins to the patient sample bodes well for their implementation in evaluating therapeutic responses and detecting drug resistance in patients.
Analysis of the Fanconi Anemia components, FANCD2 and FANCI, indicated up-regulation in 8226/LR5 cells when compared with RPMI-8226 cells, as previously noted by quantification of mRNA and Western blotting (5–7). Although mRNA and protein levels may not correlate well (67) the up-regulation of mRNA and protein was noted in response to melphalan in RPMI-8226-derived cell lines. On the other hand, FANCD2 and FANCI protein expression by LC-MRM was noted to be similar between U266/LR6 and U266 in contrast to what would have been predicted by the previous analysis of mRNA expression and the phenotypic similarity to the RPMI-8226-derived cell lines (6, 7). However, absolute quantification of these two proteins did indicate a consistent expression level between the two drug-resistant cell lines (8226/LR5 and U266/LR6); this amount of FANCD2 and FANCI may be sufficient for effecting DNA damage repair during chronic exposure to melphalan. Furthermore, because these FA proteins act as part of a complex pathway, the expression of other family members may also have bearing on drug resistance. Therefore, continued efforts are underway to develop and confirm LC-MRM tools to quantify all of the proteins in the FA pathway. Importantly, our data to date demonstrate a highly quantitative assessment of the DNA-repair machinery with significant translational applications.
Insight into these mechanisms is of great importance for the future development of therapeutic regimens for MM. In order to learn more about the biology of drug resistance and to gain greater insight from each patient sample, these sets of assays are currently being expanded. For NFκB, additional assays can be developed for IκK and IκB proteins, as well as proteins that are transcriptionally regulated by NFκB, similar to the previous LC-MRM analysis of the β-catenin signaling network in colon cancer (45). These assays will be useful to further characterize the activation of this signaling pathway and downstream protein expression. For the apoptosis-regulating proteins, additional assays can be developed for other Bcl-2 family proteins, including NOXA and PUMA, as well as other downstream proteins, like intact and cleaved caspases, that influence apoptotic signaling. In addition, LC-MRM could be used to examine selected post-translational modifications or the technique can be combined with co-immunoprecipitation to quantify the dimerization of Bcl-2 family proteins under different treatment conditions to further examine the balance of pro- and anti-apoptotic signaling. For DNA damage response, additional assays could be developed to measure expression of other FA proteins, ATM, ATR, Chk1, and components of the MRE complex. Additional information could also be gained by examining other proteins involved in MM pathogenesis(68–70) to evaluate the biological background of each tumor in parallel with the determinants of drug resistance. These assays could also be expanded to detect mutations in specific proteins (71).
Finally, the use of these assays provides the ability to systematically examine the reversal of ADR (72, 73) or to evaluate the effects of combination therapy. When guided by known cancer biology, the development of LC-MRM assays to quantify protein targets relevant to each type of treatment will enable systems-level analysis of drug response and the mechanisms of ADR. This information could help define the molecular basis for personalized cancer treatment. The next step in the application of this current biomarker panel will focus on monitoring patients during the course of therapy in order to detect ADR and to enable the selection of appropriate therapeutic regimens for each MM patient, which is of paramount importance for improving treatment outcomes. The current GeLC-MRM platform analyzing seven bands is limited in throughput to one sample per day. Implementation in clinical trials with low to moderate sample size (10 < n <100) will not be challenging in terms of the available instrument time. Because treatment decisions are typically made on a biweekly or monthly basis, results reported a few days after the office visit will not decrease in the level of patient care. The use of these assays in broader patient populations would require strategies to reduce the analysis time in order to increase sample throughput and reduce costs. Because there was little interference detected in LC-MRM for the lower molecular weight proteins, short gradients or direct infusion can be considered to increase sample throughput.
Acknowledgments
Amino acid analysis was performed by Virginia Johnson and Lawrence Dangott at the Texas A&M University Protein Chemistry Laboratory. We thank Thermo for the placement of a Vantage triple quadrupole mass spectrometer in our facility for demonstration purposes, and the Bankhead-Coley Cancer Research program of the Florida Department of Health for purchase of this instrument (09-BE04-SIG). We would like to thank the reviewers for their suggestions and Amy Koomen for editing the manuscript.
Footnotes
* This work and the Moffitt Proteomics Facility are supported by the US Army Medical Research and Materiel Command under Award No. DAMD17-02-2-0051 for a National Functional Genomics Center, the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant, and the Moffitt Foundation. This work was also supported by the NCI through RO1 CA077859 (to WSD) and R21 CA152345 (to LP). The TSQ Quantum Ultra triple quadrupole mass spectrometer was purchased with a grant from the Bankhead-Coley Cancer Research program of the Florida Dept. of Health (06BS-02-9614).
1 The abbreviations used are:
- MM
- multiple myeloma
- LC-MRM
- liquid chromatography coupled to multiple reaction monitoring mass spectrometry
- NFκB
- nuclear factor-κB
- ADR
- acquired drug resistance
- FA
- Fanconi Anemia
- BRCA
- breast cancer susceptibility proteins
- GeLC-MRM
- SDS-PAGE protein fractionation prior to liquid chromatography coupled to multiple reaction monitoring
- SIS
- stable-isotope-labeled standard
- IC50
- half-maximal inhibitory concentration.
REFERENCES
- 1. Kyle R. A., Rajkumar S. V. (2009) Treatment of multiple myeloma: a comprehensive review. Clin. Lymphoma Myeloma 9, 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bellamy W. T., Dalton W. S., Gleason M. C., Grogan T. M., Trent J. M. (1991) Development and characterization of a melphalan-resistant human multiple myeloma cell line. Cancer Res. 51, 995–1002 [PubMed] [Google Scholar]
- 3. Hazlehurst L. A., Foley N. E., Gleason-Guzman M. C., Hacker M. P., Cress A. E., Greenberger L. W., De Jong M. C., Dalton W. S. (1999) Multiple mechanisms confer drug resistance to mitoxantrone in the human 8226 myeloma cell line. Cancer Res. 59, 1021–1028 [PubMed] [Google Scholar]
- 4. Shain K. H., Dalton W. S. (2001) Cell adhesion is a key determinant in de novo multidrug resistance (MDR): new targets for the prevention of acquired MDR. Mol. Cancer Therapeutics 1, 69–78 [PubMed] [Google Scholar]
- 5. Hazlehurst L. A., Enkemann S. A., Beam C. A., Argilagos R. F., Painter J., Shain K. H., Saporta S., Boulware D., Moscinski L., Alsina M., Dalton W. S. (2003) Genotypic and phenotypic comparisons of de-novo and acquired melphalan resistance in an isogenic multiple myeloma cell line model. Cancer Res. 63, 7900–7906 [PubMed] [Google Scholar]
- 6. Chen Q., Van der Sluis P. C., Boulware D., Hazlehurst L. A., Dalton W. S. (2005) The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood 106, 698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yarde D. N., Oliveira V., Mathews L., Wang X., Villagra A., Boulware A., Shain K. H., Hazlehurst L. A., Alsina M., Chen D. T., Beg A. A., Dalton W. S. (2009) Targeting the Fanconi anemia/BRCA pathway circumvents drug resistance in multiple myeloma. Cancer Res. 69, 9367–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keats J. J., Fonseca R., Chesi M., Schop R., Baker A., Chng W. J., Van Wier S., Tiedemann R., Shi C. X., Sebag M., Braggio E., Henry T., Zhu Y. X., Fogle H., Price-Troska T., Ahmann G., Mancini C., Brents L. A., Kumar S., Greipp P., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M., Valdez R., Trent J., Stewart A. K., Carpten J., Bergsagel P. L. (2007) Promiscuous mutations activate the non-canonical NF-kB pathway in multiple myeloma. Cancer Cell 12, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yinjun L., Jie J., Yungui W. (2005) Triptolide inhibits transcription factor NF-kappaB and induces apoptosis of multiple myeloma cells. Leukemia Res. 29, 99–105 [DOI] [PubMed] [Google Scholar]
- 10. Yang H. H., Ma M. H., Vescio R. A., Berenson J. R. (2003) Overcoming drug resistance in multiple myeloma: the emergence of therapeutic approaches to induce apoptosis. J. Clin. Oncol. 21, 4239–4247 [DOI] [PubMed] [Google Scholar]
- 11. San Miguel J. F., Schlag R., Khuageva N. K., Dimopoulos M. A., Shpilberg O., Kropff M., Spicka I., Petrucci M. T., Palumbo A., Samoilova O. S., Dmoszynska A., Abdulkadyrov K. M., Schots R., Jiang B., Mateos M. V., Anderson K. C., Esseltine D. L., Liu K., Cakana A., van de Velde H., Richardson P. G. (2008) Bortezomib plus Melphalan and Prednisone for Initial Treatment of Multiple Myeloma. New Engl. J. Med. 359, 906–917 [DOI] [PubMed] [Google Scholar]
- 12. Reeder C. B., Reece D. E., Kukreti V., Chen C., Trudel S., Laumann K., Hentz J., Pirooz N. A., Piza J. G., Tiedemann R., Mikhael J. R., Bergsagel P. L., Leis J. F., Fonseca R., Stewart A. K. (2010) Once- versus twice-weekly bortezomib induction therapy with CyBorD in newly diagnosed multiple myeloma. Blood 115, 3416–3417 [DOI] [PubMed] [Google Scholar]
- 13. Catlett-Falcone R., Landowski T. H., Oshiro M. M., Turkson J., Levitzki A., Savino R., Ciliberto G., Moscinski L., Fernández-Luna J. L., Nuñez G., Dalton W. S., Jove R. (1999) Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10, 105–115 [DOI] [PubMed] [Google Scholar]
- 14. Campone M., Vavasseur F., Le Cabellec M. T., Meflah K., Vallette F. M., Oliver L. (2001) Induction of chemoresistance in HL-60 cells concomitantly causes a resistance to apoptosis and the synthesis of P-glycoprotein. Leukemia 15, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 15. Yip K. W., Reed J. C. (2008) Bcl-2 family proteins and cancer. Oncogene 27, 6398–6406 [DOI] [PubMed] [Google Scholar]
- 16. Kaufmann S. H., Vaux D. L. (2003) Alterations in the apoptotic machinery and their potential role in anticancer drug resistance. Oncogene 22, 7414–7430 [DOI] [PubMed] [Google Scholar]
- 17. Tu Y., Renner S., Xu F., Fleishman A., Taylor J., Weisz J., Vescio R., Rettig M., Berenson J., Krajewski S., Reed J. C., Lichtenstein A. (1998) BCL-X expression in multiple myeloma: possible indicator of chemoresistance. Cancer Res. 58, 256–262 [PubMed] [Google Scholar]
- 18. van de Donk N. W., Kamphuis M. M., van Dijk M., Borst H. P., Bloem A. C., Lokhorst H. M. (2003) Chemosensitization of myeloma plasma cells by an antisense-mediated downregulation of Bcl-2 protein. Leukemia 17, 211–219 [DOI] [PubMed] [Google Scholar]
- 19. Wuillème-Toumi S., Robillard N., Gomez P., Moreau P., Le Gouill S., Avet-Loiseau H., Harousseau J. L., Amiot M., Bataille R. (2005) Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia 19, 1248–1252 [DOI] [PubMed] [Google Scholar]
- 20. Gojo I., Zhang B., Fenton R. G. (2002) The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin. Cancer Res. 8, 3527–3538 [PubMed] [Google Scholar]
- 21. Le Gouill S., Podar K., Amiot M., Hideshima T., Chauhan D., Ishitsuka K., Kumar S., Raje N., Richardson P. G., Harousseau J. L., Anderson K. C. (2004) VEGF induces MCL-1 upregulation and protects multiple myeloma cells against apoptosis. Blood 104, 2886–2892 [DOI] [PubMed] [Google Scholar]
- 22. Akgul C. (2009) Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell. Mol. Life Sci. 66, 1326–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warr M. R., Shore G. C. (2008) Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr. Mol. Med. 8, 138–147 [DOI] [PubMed] [Google Scholar]
- 24. Gomez-Bougie P., Bataille R., Amiot M. (2004) The imbalance between Bim and Mcl-1 expression controls the survival of human myeloma cells. Eur. J. Immunol. 34, 3156–3164 [DOI] [PubMed] [Google Scholar]
- 25. Simmons M. J., Fan G., Zong W. X., Degenhardt K., White E., Gélinas C. (2008) Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene 27, 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Werner A. B., de Vries E., Tait S. W., Bontjer I., Borst J. (2002) Bcl-2 Family Member Bfl-1/A1 Sequesters Truncated Bid to Inhibit Its Collaboration with Pro-apoptotic Bak or Bax. J. Biol. Chem. 277, 22781–22788 [DOI] [PubMed] [Google Scholar]
- 27. Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 28. D'Andrea A. D. (2003) The Fanconi Anemia/BRCA signaling pathway: disruption in cisplatin-sensitive ovarian cancers. Cell Cycle 2, 290–292 [PubMed] [Google Scholar]
- 29. Wang W. (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 8, 735–748 [DOI] [PubMed] [Google Scholar]
- 30. Thompson L. H., Hinz J. M. (2009) Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat. Res. 668, 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spanswick V. J., Craddock C., Sekhar M., Mahendra P., Shankaranarayana P., Hughes R. G., Hochhauser D., Hartley J. A. (2002) Repair of DNA interstrand crosslinks as a mechanism of clinical resistance to melphalan in multiple myeloma. Blood 100, 224–229 [DOI] [PubMed] [Google Scholar]
- 32. Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A. (2008) DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 8, 193–204 [DOI] [PubMed] [Google Scholar]
- 33. Dimopoulos M. A., Souliotis V. L., Anagnostopoulos A., Bamia C., Pouli A., Baltadakis I., Terpos E., Kyrtopoulos S. A., Sfikakis P. P. (2007) Melphalan-induced DNA damage in vitro as a predictor for clinical outcome in multiple myeloma. Haematologica 92, 1505–1512 [DOI] [PubMed] [Google Scholar]
- 34. Chen C. C., Taniguchi T., D'Andrea A. (2007) The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J. Mol. Med. 85, 497–505 [DOI] [PubMed] [Google Scholar]
- 35. Kusmierz J. J., Sumrada R., Desiderio D. M. (1990) Fast atom bombardment mass spectrometric quantitative analysis of methionine-enkephalin in human pituitary tissues. Anal. Chem. 62, 2395–2400 [DOI] [PubMed] [Google Scholar]
- 36. Barr J. R., Maggio V. L., Patterson D. G., Jr., Cooper G. R., Henderson L. O., Turner W. E., Smith S. J., Hannon W. H., Needham L. L., Sampson E. J. (1996) Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin. Chem. 42, 1676–1682 [PubMed] [Google Scholar]
- 37. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 38. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 41. Whiteaker J. R., Zhang H., Zhao L., Wang P., Kelly-Spratt K. S., Ivey R. G., Piening B. D., Feng L. C., Kasarda E., Gurley K. E., Eng J. K., Chodosh L. A., Kemp C. J., McIntosh M. W., Paulovich A. G. (2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 6, 3962–3975 [DOI] [PubMed] [Google Scholar]
- 42. Kirsch S., Widart J., Louette J., Focant J. F., De Pauw E. (2007) Development of an absolute quantification method targeting growth hormone biomarkers using liquid chromatography coupled to isotope dilution mass spectrometry. J. Chromatogr. A. 1153, 300–306 [DOI] [PubMed] [Google Scholar]
- 43. Kuhn E., Wu J., Karl J., Liao H., Zolg W., Guild B. (2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 44. Barnidge D. R., Goodmanson M. K., Klee G. G., Muddiman D. C. (2004) Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-MS/MS using protein cleavage and isotope dilution mass spectrometry. J. Proteome Res. 3, 644–652 [DOI] [PubMed] [Google Scholar]
- 45. Chen Y., Gruidl M., Remily-Wood E., Liu R. Z., Eschrich S., Lloyd M., Nasir A., Bui M. M., Huang E., Yeatman T., Koomen J. M. (2010) Quantification of beta-catenin signaling components in colon cancer cell lines, tissue sections, and microdissected tumor cells using reaction monitoring mass spectrometry. J. Proteome Res. 9, 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fang Y., Robinson D. P., Foster L. J. (2010) Quantitative analysis of proteome coverage and recovery rates for upstream fractionation methods in proteomics. J. Proteome Res. 9, 1902–1912 [DOI] [PubMed] [Google Scholar]
- 48. Piersma S. R., Fiedler U., Span S., Lingnau A., Pham T. V., Hoffmann S., Kubbutat M. H., Jiménez C. R. (2010) Workflow comparison for label-free, quantitative secretome proteomics for cancer biomarker discovery: method evaluation, differential analysis, and verification in serum. J. Proteome Res. 9, 1913–1922 [DOI] [PubMed] [Google Scholar]
- 49. Wang H., Chang-Wong T., Tang H. Y., Speicher D. W. (2010) Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J. Proteome Res. 9, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schirle M., Heurtier M., Kuster B. (2003) Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Biol. 2, 1297–1305 [DOI] [PubMed] [Google Scholar]
- 52. Barnidge D. R., Dratz E. A., Martin T., Bonilla L. E., Moran L. B., Lindall A. (2003) Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal. Chem. 75, 445–451 [DOI] [PubMed] [Google Scholar]
- 53. Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
- 54. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Havlis J., Shevchenko A. (2004) Absolute quantification of proteins in solutions and in polyacrylamide gels by mass spectrometry. Anal. Chem. 79, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 56. Alas S., Bonavida B. (2003) Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin's lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin. Cancer Res. 9, 316–326 [PubMed] [Google Scholar]
- 57. Oshiro M. M., Landowski T. H., Catlett-Falcone R., Hazlehurst L. A., Huang M., Jove R., Dalton W. S. (2001) Inhibition of JAK kinase activity enhances Fas-mediated apoptosis but reduces cytotoxic activity of topoisomerase II inhibitors in U266 myeloma cells. Clin. Cancer Res. 7, 4262–4271 [PubMed] [Google Scholar]
- 58. Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., Strasser A. (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 61. Michels J., O'Neill J. W., Dallman C. L., Mouzakiti A., Habens F., Brimmell M., Zhang K. Y., Craig R. W., Marcusson E. G., Johnson P. W., Packham G. (2004) Mcl-1 is required for Akata6 B-lymphoma cell survival and is converted to a cell death molecule by efficient caspase-mediated cleavage. Oncogene 23, 4818–4827 [DOI] [PubMed] [Google Scholar]
- 62. Hengartner M. O. (2000) The biochemistry of apoptosis. Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 63. Stoka V., Turk B., Schendel S. L., Kim T. H., Cirman T., Snipas S. J., Ellerby L. M., Bredesen D., Freeze H., Abrahamson M., Bromme D., Krajewski S., Reed J. C., Yin X. M., Turk V., Salvesen G. S. (2001) Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 276, 3149–3157 [DOI] [PubMed] [Google Scholar]
- 64. Mandic A., Viktorsson K., Strandberg L., Heiden T., Hansson J., Linder S., Shoshan M. C. (2002) Calpain-mediated bid cleavage and calpain-independent Bak modulation: two separate pathways in cisplatin-induced apoptosis. Mol. Cell. Biol. 22, 3003–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heibein J. A., Gopsing I. S., Barry M., Pinkoski M. J., Shore G. C., Green D. R., Bleackley R. C. (2000) Granzyme B–mediated cytochrome C release is regulated by the Bcl-2 family members Bid and Bax. J. Exp. Med. 192, 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Remily-Wood E. R., Liu R. Z., Xiang Y., Chen Y., Thomas C. E., Rajyaguru N., Kaufman L. M., Ochoa J. E., Hazlehurst L., Pinilla-Ibarz J., Lancet J., Zhang G., Haura E., Shibata D., Yeatman T., Smalley K. S. M., Dalton W. S., Huang E., Scott E., Bloom G., Eschrich S. A., Koomen J. M. (2011) A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clin. Appl. 7, doi: 10.1002/prca.201000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Greenbaum D., Colangelo C., Williams K., Gerstein M. (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Anguiano A., Tuchman S. A., Acharya C., Salter K., Gasparetto C., Zhan F., Dhodapkar M., Nevins J., Barlogie B., Shaughnessy J.D., Jr., Potti A. (2009) Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J. Clin. Oncol. 27, 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Claudio J. O., Zhan F., Zhuang L., Khaja R., Zhu Y. X., Sivananthan K., Trudel S., Masih-Khan E., Fonseca R., Bergsagel P. L., Scherer S. W., Shaughnessy J., Stewart A. K. (2007) Expression and mutation status of candidate kinases in multiple myeloma. Leukemia 21, 1124–1127 [DOI] [PubMed] [Google Scholar]
- 70. Bommert K., Bargou R. C., Stühmer T. (2006) Signalling and survival pathways in multiple myeloma. Eur. J. Cancer 42, 1574–1580 [DOI] [PubMed] [Google Scholar]
- 71. Dumontet C., Landi S., Reiman T., Perry T., Plesa A., Bellini I., Barale R., Pilarski L. M., Troncy J., Tavtigian S., Gemignani F. (2010) Genetic polymorphisms associated with outcome in multiple myeloma patients receiving high-dose melphalan. Bone Marrow Transplant. 45, 1316–1324 [DOI] [PubMed] [Google Scholar]
- 72. Xiao H., Xiao Q., Zhang K., Zuo X., Shrestha U. K. (2010) Reversal of multidrug resistance by curcumin through FA/BRCA pathway in multiple myeloma cell line MOLP-2/R. Ann. Hematol. 89, 399–404 [DOI] [PubMed] [Google Scholar]
- 73. Landais I., Hiddingh S., McCarroll M., Yang C., Sun A., Turker M. S., Snyder J. P., Hoatlin M. E. (2009) Monoketone analogs of curcumin, a new class of Fanconi anemia pathway inhibitors. Mol. Cancer 8, 133 [DOI] [PMC free article] [PubMed] [Google Scholar]