Abstract
Transcription factors play a key role in transcription regulation as they recognize and directly bind to defined sites in promoter regions of target genes, and thus modulate differential expression. The overall process is extremely dynamic, as they have to move through the nucleus and transiently bind to chromatin in order to regulate gene transcription. To identify transcription factors that affect glycogen accumulation in Neurospora crassa, we performed a systematic screen of a deletion strains set generated by the Neurospora Knockout Project and available at the Fungal Genetics Stock Center. In a wild-type strain of N. crassa, glycogen content reaches a maximal level at the end of the exponential growth phase, but upon heat stress the glycogen content rapidly drops. The gene encoding glycogen synthase (gsn) is transcriptionally down-regulated when the mycelium is exposed to the same stress condition. We identified 17 deleted strains having glycogen accumulation profiles different from that of the wild-type strain under both normal growth and heat stress conditions. Most of the transcription factors identified were annotated as hypothetical protein, however some of them, such as the PacC, XlnR, and NIT2 proteins, were biochemically well-characterized either in N. crassa or in other fungi. The identification of some of the transcription factors was coincident with the presence of DNA-binding motifs specific for the transcription factors in the gsn 5′-flanking region, and some of these DNA-binding motifs were demonstrated to be functional by Electrophoretic Mobility Shift Assay (EMSA) experiments. Strains knocked-out in these transcription factors presented impairment in the regulation of gsn expression, suggesting that the transcription factors regulate glycogen accumulation by directly regulating gsn gene expression. Five selected mutant strains showed defects in cell cycle progression, and two transcription factors were light-regulated. The results indicate that there are connections linking different cellular processes, such as metabolism control, biological clock, and cell cycle progression.
The fungus Neurospora crassa has been widely used as a model organism for the understanding of fundamental aspects of eukaryotic biology. The knowledge of its genome sequence (1) has allowed the identification of proteins required for gene regulation, such as the transcriptional regulatory proteins. An examination of the classes of transcription factors in the N. crassa genome reveals that the organism carries elements shared by simple and complex metazoan models (2). The availability of a set of deletion strains, each carrying a deletion in a specific ORF encoding a transcription factor, allows the screening for genes linked to a particular phenotype. Here we used this mutant strains set to identify transcription factors that either directly or indirectly regulate glycogen metabolism in N. crassa.
In many organisms, glycogen is a carbon and energy reserve carbohydrate with an intricate metabolism regulation that senses nutrient availability and other environmental conditions. The amount of glycogen found in a particular situation results from the balance between glycogen synthase and glycogen phosphorylase activities. These enzymes regulate, respectively, the synthesis and degradation of this compound and they are both regulated by phosphorylation. Besides reversible changes in their activities, glycogen levels are also correlated with physiological conditions. In addition, other proteins may also be involved in glycogen accumulation because protein activation resulting from different signaling pathways affects glycogen storage (3, 4).
In N. crassa, glycogen content reaches a maximal level at the end of the exponential growth phase. However, under stress conditions, such as heat shock, glycogen content drops rapidly (5, 6). The yeast Saccharomyces cerevisiae accumulates glycogen under heat shock (7), demonstrating that yeast and N. crassa show opposite responses concerning this environmental condition. The glycogen decrease observed in N. crassa might result from the regulation, at transcriptional level, of enzymes involved in the carbohydrate metabolism, as transcription of the gene encoding glycogen synthase (gsn) decreases under heat stress (5, 8). The gsn promoter has one cis-acting STRE DNA motif, which is specifically bound by nuclear proteins activated under heat shock. In S. cerevisiae, STRE is recognized by two transcription factors, the zinc finger proteins Msn2p and Msn4p (Msn2/4p), which mediate the cellular response to multiple stresses and are components of the environmental stress response (9). We have previously combined biochemical techniques and a proteomic approach coupled to mass spectrometry in an attempt to identify N. crassa proteins that are activated upon heat shock and bind to the STRE motif of the gsn promoter (10). Only hypothetical proteins having domains that might be involved in transcription regulation were identified, and none of them had a DNA-binding domain.
To identify transcription factors regulating glycogen metabolism in the fungus N. crassa, we used a mutant strains set with single-gene deletions of known or putative transcription factors to search for mutant strains having glycogen accumulation profiles different from that in the wild-type strain. The mutant strains were analyzed under normal growth temperature (30 °C) and under heat shock stress (45 °C). The results described in this work showed that most of the transcription factors identified have been annotated in the N. crassa database (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html) as hypothetical proteins. However, many are proteins that have been functionally characterized, either in N. crassa or in other fungi. This indicates that glycogen metabolism regulation in eukaryotic cells comprises a complex regulatory network involving metabolic and nutrient sensing, which under certain circumstances could lead to impairment of cellular development.
EXPERIMENTAL PROCEDURES
Neurospora crassa Strains and Growth Conditions
The N. crassa strain FGSC 9718 (mus-51∷ bar mat a), and a set of 147 mutant strains individually knocked-out in genes encoding transcription factors were purchased from the Fungal Genetics Stock Center (FGSC, University of Missouri, Kansas City, Missouri, http://www.fgsc.net) (11). The deletion strains comprise a set of mutants where each open reading frame (ORF)1 has been disrupted from start to stop codon by the insertion of the hph gene (hygromycin B phosphotransferase) as a marker (12). The strains were cultivated in Vogel's minimal medium (13) supplemented with 2% sucrose. After 10 days of culture, conidia were suspended in sterile water and counted. For the heat shock experiments, conidia (107/ml) were first germinated in 60 ml of Vogel's minimal medium supplemented with 2% sucrose, at 30 °C and 250 rpm during 24 h. After this time an aliquot was removed, filtered, frozen in liquid nitrogen and stored at −80 °C until use. The remaining culture was filtered and transferred into fresh Vogel's medium preheated at 45 °C. After 30 min, the mycelia were harvested by filtration, frozen in liquid nitrogen, and stored at −80 °C.
Glycogen and Protein Quantification
Mycelia pads were ground to a fine powder in a prechilled mortar in liquid nitrogen, and extracted into lysis buffer (50 mm Tris-HCl, pH 7.6, 100 mm NaF, 1 mm EDTA, 1 mm PMSF, 0.1 mm tosyl-L-lysine chloromethyl ketone, 1 mm benzamidin, and 1 μg/ml of each pepstatin and aprotinin). Cellular extracts were clarified by centrifugation at 3,000 × g, for 10 min at 4 °C, and the supernatants were used for glycogen and protein quantifications. Glycogen content was measured following the protocol described by Hardy and Roach (14), with slight modifications. Briefly, 100 μl of the crude extract was precipitated with 20% TCA (final concentration). The supernatant was separated after centrifugation (5,000 × g, 10 min, 4 °C), the glycogen was precipitated with 500 μl of 95% cold ethanol, collected by centrifugation, washed twice with 66% ethanol, dried, and digested with α-amylase (10 mg/ml) and amyloglucosidase (30 mg/ml). Free glucose was measured with a glucose oxidase kit, and the glycogen content was normalized to the total protein concentration. Total protein was quantified by the Hartree method (15), using BSA as standard.
RNA Extraction and Northern Assay
Total RNA was prepared using the LiCl method according to Sokolovsky et al. (16). Total RNA (15 μg) was electrophoresed on a 1.5% agarose-formaldehyde denaturing gel (17), at 65 V during 5 h, and then transferred to neutral nylon membranes (Hybond N, GE HealthCare) in 2 × SSC. The blots were probed with the full-length gsn cDNA (106 to 108 cpm), radiolabeled with [α-32P]-dATP (3,000 μCi/mmol) by random priming (NEBlot kit, Biolabs) in 10 ml of ULTRAhyb hybridization solution (Ambion, Austin, TX), at 42 °C overnight. After hybridization, the blot was washed twice in 2 × SSC, 0.1% SDS for 10 min, and twice in 0.1 × SSC, 0.1% SDS for 15 min, and exposed to an x-ray film.
pacC cDNA Cloning and Production and Purification of the Recombinant Protein
For production of the truncated recombinant PACC transcription factor a 639-bp fragment of the ORF NCU00090 was amplified by PCR from a cDNA plasmid library (pYADE5-Nc) with the oligonucleotides 90-F and 90-R2 (Table I). The underlined sequences correspond to the NdeI and BamHI sites, respectively. The amplified fragment was cloned into the NdeI-BamHI sites of the pET28a vector leading to the plasmid construction pET-ΔPACC for the expression of a truncated protein containing the N-terminal 213 amino acids fused with His-tag. The plasmid construction was confirmed by DNA sequencing. For expression of the PACC recombinant protein, the plasmidial construction was used to transform competent E. coli cells from BL21 (DE3) pLysS strain. Cells were grown at 37 °C in LB medium containing appropriate antibiotics and the recombinant protein was induced by addition of 0.4 mm final concentration of IPTG for 4 h, 37 °C. Cells were harvested by centrifugation, suspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 mm imidazole, 20% v/v glycerol, 0.5 mm PMSF, 25 mm benzamidine and 50 mm NaF) and lysed by sonication. The supernatant was clarified by centrifugation and subjected to affinity chromatography. The recombinant protein was eluted with linear imidazole gradient (20–500 mm) and dialyzed against dialysis buffer (10 mm Tris-HCl pH 7.9, 100 mm KCl, 20% v/v glycerol, 1 mm EDTA, 0.5 mm dithiotreitol). Purified protein was analyzed by SDS-PAGE with Coomassie Brilliant Blue staining (18) and quantified (15).
Table I. Oligonucleotides used in this study.
| Primer | Sequencea | Source | Name | Position (nt) |
|---|---|---|---|---|
| 90-F | 5′-CATATGTCGTCCACACCAGCCCAG-3′ | ORF NCU00090 | – | 1 to 21 |
| 90-R2 | 5′-GGATCCTTACTTGTGAACTGGAGCCTG-3′ | ORF NCU00090 | – | 639 to 622 |
| PacC-F | 5′-GACCCAACAGCCCAACTT-3′ | gsn promoter | pacC probe | −1918 to −1901 |
| GSN-RP3 | 5′-GCAACGAATACTCCCATG-3′ | gsn promoter | pacC probe | −1789 to −1806 |
| GSN-FP4 | 5′-CTGATTGGGAAAGGTCAGA-3′ | gsn promoter | nit2 probe | −1645 to −1626 |
| GSN-RP2 | 5′-CTGTTGACCTGCGTTAAC-3′ | gsn promoter | nit2 probe | −1269 to −1286 |
| XLNR-F2 | 5′-TGAGGGTGAGAAAGTTGC-3′ | gsn promoter | xlnR probe | −2173 to −2156 |
| XLNR-R2 | 5′-TATTCTGCAACGGAACTCC-3′ | gsn promoter | xlnR probe | −2034 to −2053 |
a NdeI and BamHI restriction sites are underlined in the sequences. Positions are according to the ATG start codon of translation. Stop codon inserted in the ORF sequence is shown in bold.
Preparation of Crude Cellular Extract
Mycelium of the wild-type strain grown at 30 °C was used to prepare the cellular extract. About 10 mg of frozen samples were ground to a fine powder under liquid nitrogen in a prechilled mortar, homogenized in a 20 ml of lysis buffer (15 mm HEPES-KOH pH 7.9, 10% w/v glycerol, 500 mm KCl, 5 mm MgCl2, 0.5 mm EDTA, 1 mm dithiotreitol, 0.5 mm PMSF, 10 μg/ml each of antipain and pepstatin A, 25 mm benzamidine, 50 mm NaF) and stirred with glass beads in 8 cycles of 30 s of agitation and 30 s on ice. Crude cellular extract was obtained after centrifugation (3,200 × g, 2 min, 4 °C), dialyzed against buffer D (15 mm HEPES-KOH pH 7.9, 15% w/v glycerol, 100 mm KCl, 1 mm EDTA) at 4 °C for 2 h, and cleared by centrifugation (20,000 × g, 20 min, 4 °C) before loading onto a HiTrap Heparin-Sepharose FF column (GE Healthcare). Total proteins were eluted by using a 0.1–1.5 m KCl linear gradient and the protein fractions were dialyzed against buffer D plus 0.5 mm PMSF, 25 mm benzamidine and 50 mm NaF, frozen in liquid N2 and stored at −80 °C. Total protein was quantified (15).
Electrophoretic Mobility Shift Assay
DNA-protein binding reactions were carried out in 1 × binding buffer (25 mm HEPES-KOH pH 7.9, 20 mm KCl, 10% v/v glycerol, 1 mm dithiotreitol, 0.2 mm EDTA, 0.5 mm PMSF, 12.5 mm benzamidine, 5 μg/ml of each antipain and pepstatin A) containing 2–4 μg poly(dI-dC).(dI-dC) as unspecific competitor, and either 2.0 μg of PACC recombinant protein or 35 μg of crude cellular extract. The radiolabeled DNA probe (∼104 cpm) was added and reactions were incubated during 20 min at room temperature. Free probe was separated from DNA-protein complexes by electrophoresis on a native 4% polyacrilamide gel in 0.5 × Tris borate-EDTA (TBE) buffer (300 V, 10 mA, 10 °C). After electrophoresis, the gel was dried and autoradiographed. For competition assays, an excess of specific DNA competitor was added to the binding reactions 10 min prior the incubation with the radiolabeled probe.
DNA Probes and Competitors for EMSA
To produce the 134-bp nit2, 146-bp pacC, and 139-bp xlnR probes, DNA fragments containing the nit2, pacC, and xlnR cis elements from the gsn promoter, respectively, were amplified from the IV9A-1 plasmid (GenBank#AF417205) using the oligonucleotides described in Table I in the presence of [α-32P]-dATP (3,000 Ci/mmol). The DNA probes were purified on a 2% low-melting point agarose gel. Unlabeled nit2, pacC and xlnR probes, used as specific DNA competitors, were quantified by measuring the absorbance at 260 nm, and added to the binding reaction in 10-fold molar excess.
Flow Cytometry Analysis
Conidia were harvested by centrifugation after growing in Vogel's minimal solid medium with 2% sucrose, and a total of 5 × 104 cells were suspended in 0.5 ml of PI solution (0.1% v/v Triton X-100, 0.1% sodium citrate, 0.9 U/ml RNase A, 50 μg/ml propidium iodide). The cell suspension was kept in the dark for 1.5 h at room temperature. A FACSCaliburTM flow cytometer equipped with CellQUESTTM software (Becton Dickinson, San Juan, CA) was used to analyze cell size using a Forward Scatter detector (FSC-H), and cell complexity using a Side Scatter detector (SSC-H). Propidium iodide incorporation was measured using a fluorescence detector, with an excitation wavelength of 488 nm and an emission wavelength of 585/42 nm. Ten thousand events were evaluated per sample. The experiments were repeated at least three times.
Bioinformatic Tools
Online bioinformatic tools were used to predict the biochemical parameters of the selected transcription factors. The nucleotide sequences of the knocked-out ORFs codifying the transcription factors were identified in the fungus genome at the Broad Institute home page (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html). The polypeptide sequences were compared against the database of sequences available at the National Center for Biotechnology Information (NCBI), using the BlastP tool (http://www.ncbi.nlm.nih.gov/blast/) to identify sequences with known function similar to the search sequence. For the theoretical estimates of the isoelectric point and molecular weight, the ProtPAram tool at the ExPASY server was used (ca.expasy.org). The presence of protein domains was investigated using the N. crassa genome at the FGSC (http://www.fgsc.net/scripts/strainsearchform.asp), SMART (http://smart.embl-heidelberg.de), and Pfam 22.0 (http://pfam.sanger.ac.uk) web sites. The presence of classical Nuclear Localization Signals (cNLS) was determined by PSORTII (http://psort.nibb.ac.jp/form2.html), and the presence of cis DNA elements in the gsn promoter was determined by MatInspector (www.genomatix.de).
RESULTS
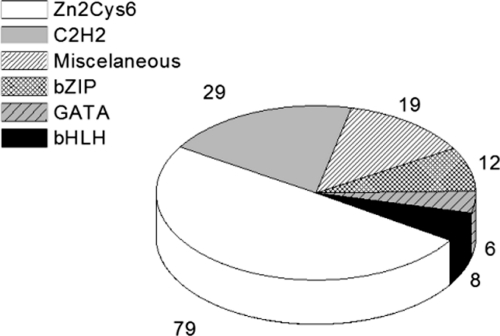
We screened a set of N. crassa mutant strains, each carrying a deletion in a single gene encoding a transcription factor. Fig. 1 shows the transcription factor families to which the proteins belong. Most belong to the Zn2Cys6 fungal binuclear cluster family, which is fungus-specific and the largest class of transcription factors in N. crassa (2). The second largest class of transcription factors analyzed in this work is the C2H2 family, found in both prokaryotic and eukaryotic organisms. A considerable number of miscellaneous factors were screened, including the CAAT-binding transcription factors, and transcription factors carrying the forkhead, homeobox, RING finger, and WD repeat domains. These factors usually play important roles in the cell cycle biology of eukaryotic organisms. A smaller number of transcription factors belonging to the bZIP, GATA, and bHLH families was also screened (2).
Fig. 1.
Families of N. crassa transcription factors. Distribution in percentage of transcription factor families represented by mutant strains analyzed here.
Transcription Factors Controlling Glycogen Accumulation
Previous results from our group have shown that wild-type N. crassa accumulates glycogen at high levels at the end of the exponential growth phase while growing under its regular temperature (30 °C), and that the glycogen content decreases when the mycelium is exposed to heat shock (45 °C) (5, 6). To identify transcription factors regulating glycogen metabolism in the fungus N. crassa, we used a mutant-strain set with single-gene deletions of putative or already known transcription factors to search for mutant strains having glycogen accumulation profiles different from that found in the wild-type strain. The mutant strains were analyzed under normal growth temperature (30 °C) and heat shock stress (45 °C).
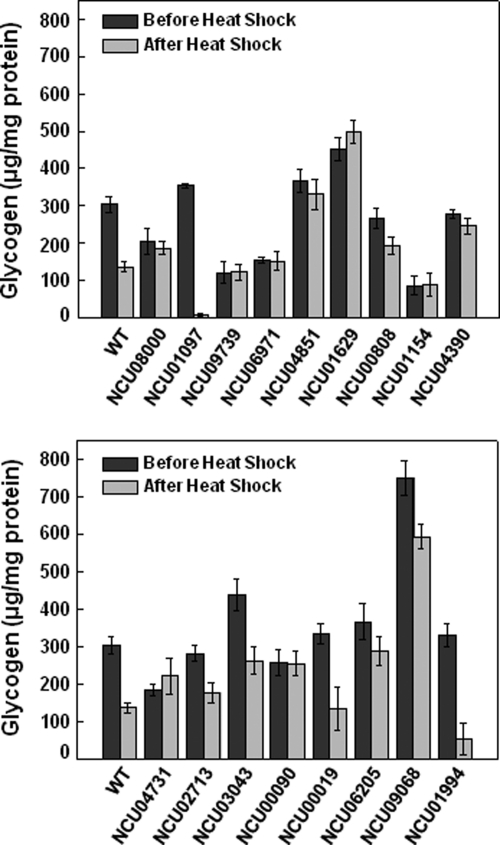
Of the 147 mutant strains analyzed, 17 presented patterns of glycogen accumulation different than the wild-type strain (Fig. 2). Five strains (knocked-out in the ORFs NCU08000, NCU09739, NCU06971, NCU01154, and NCU00090) were selected because they did not show differences in their glycogen content either before or after heat shock. Note that at both temperatures, low levels of glycogen accumulated in the strains with deleted NCU09739, NCU06971, and NCU01154 ORFs. The strains with deleted NCU04851, NCU00808, NCU04390, NCU02713, NCU03043, and NCU06205 ORFs were selected because they showed less pronounced reduction in glycogen levels after heat shock, compared with the wild-type strain. Two mutant strains (mutated in the ORFs NCU01097 and NCU01994) showed extremely large glycogen content reductions after heat shock. Interestingly, the strains mutated in the ORFs NCU01629, and NCU04731 accumulated more glycogen under the heat shock condition than under the normal growth condition. Finally, a hyper-accumulation of glycogen was observed in the NCU01629 and NCU09068 ORFs deleted strains in both temperatures. All of these mutant strains showed impaired control of glycogen accumulation, compared with the wild-type strain, suggesting that the transcription factors missing in the mutant strains might be involved in the regulation of glycogen accumulation.
Fig. 2.
Glycogen accumulation before and after heat shock in selected mutant strains. Glycogen was extracted from mycelia submitted or not to heat shock (transfer from 30 °C to 45 °C), digested with α-amylase (10 mg/ml) and amyloglucosidase (30 mg/ml), and the free glucose was enzymatically determined with a glucose oxidase kit. Results represent the average of at least three independent experiments. WT, FGSC 9718 strain.
The biochemical and molecular characteristics of the selected transcription factors are shown in Table II. Most belong to the Zn2Cys6 zinc finger family, the largest class in N. crassa (2). Four proteins belong to the C2H2 zinc finger family, and two to the GATA zinc finger, a family only found in eukaryotic organisms. The bHLH and bZIP transcription factor families both have one member among the selected proteins, and proteins having either the forkhead domain or the Tup-N-terminal and WD repeats, which belong to the miscellaneous factor family (2), were also selected in our screen. Fifteen selected transcription factors have cNLS, which are amino acid sequences that target cargo proteins into the nucleus, either monopartite or bipartite (19). Two transcription factors (ORFs NCU09739 and NCU06205) do not have amino acid sequences characteristic of cNLS. One transcription factor, annotated as hypothetical protein, might have a nonclassical NLS (ORF NCU09739), and the rco-1 (regulation of conidiation) gene product is the S. cerevisiae Tup1 homolog (ORF NCU06205, described below).
Table II. Classification of the transcription factor family, annotation, biochemistry, and structural characteristics of the proteins selected.
The identification of each strain was made according to the FGSC number. Theoretical estimate of physical and chemical characteristics and identification of cNLS were performed according to ProtPAram tools (www.expasy.org/tools/protpar-ref.html) and PsortII (http://psort.nibb.ac.jp/cgi-bin/runpsort.pl), respectively. MW, molecular weight; pI, isoeletric point; cNLS, classical Nuclear Localization Signals; M, monopartite; B, bipartite; NI, not identified.
| FGSC# | ORF | Transcription factor family | Theoretical MW/pI | cNLS | Annotationa | Gene | Reference |
|---|---|---|---|---|---|---|---|
| 11004 | NCU08000 | Zn2Cys6 zinc finger | 105.43/6.78 | 33-PTPKRKK (M) | Cutinase transcription factor 1α | ctf-1α | 23 |
| 600-KRHRR (M) | |||||||
| 11039 | NCU01097 | Zn2Cys6 zinc finger | 90.23/6.34 | 36-KRVKAVTQACHTCRRYK (B) | Hypothetical protein | NI | - |
| 11062 | NCU09739 | Zn2Cys6 zinc finger | 75.09/7.85 | NI | Hypothetical protein | ada-7 | - |
| 11067 | NCU06971 | Zn2Cys6 zinc finger | 103.16/6.44 | 87-PIRRRIS (M) | Transcriptional activator XlnR | xlnR | 28 |
| 128-RKKR (M) | |||||||
| 11089 | NCU04851 | Zn2Cys6 zinc finger | 119.36/7.87 | 268-PKEKRWP (M) | Hypothetical protein | NI | - |
| 221-PKRRNRPAVSCIPCRGRKI (B) | |||||||
| 11102 | NCU01629 | C2H2 zinc finger | 45.91/8.82 | 240-PRPKRQQ (M) | Hypothetical protein | NI | - |
| 11123 | NCU00808 | Zn2Cys6 zinc finger | 66.25/5.79 | 185-PRIKTKK (M) | Hypothetical protein | NI | - |
| 11126 | NCU01154 | GATA zinc finger | 51.01/8.89 | 402-KRKK (M) | Hypothetical protein | sub-1 | - |
| 11134 | NCU04390 | Zn2Cys6 zinc finger | 93.67/6.6 | 89-PQPPRRRKKK (M) | Hypothetical protein | col-22 | - |
| 94-RRKKKPHERDLIDRLKKY (B) | |||||||
| 11139 | NCU04731 | bHLH | 117.54/6.17 | 276-PNSRKRK (M) | Hypothetical protein | sah-2 | 33 |
| 11348 | NCU02713 | C2H2 zinc finger | 31.2/8.99 | 107-KRPR (M) | Hypothetical protein | csp-1 | 30 |
| 11355 | NCU03043 | C2H2 zinc finger | 41.73/ 8.57 | 312-KKHK (M) | Protein FlbC | NI | - |
| 361-RRHKK (M) | |||||||
| 11397 | NCU00090 | C2H2 zinc finger | 67.3/7.19 | 280-PFDARKR (M) | pH response transcription factor pacC/RIM101 | pacC-1 | 37 |
| 285-KRQFDDLNDFFGSVKRR (B) | |||||||
| 11437 | NCU00019 | Forkhead domain | 74.61/7.86 | 33-PSKRRKK (M) | FKH1 protein | NI | - |
| 454- PASSRKRK (M) | |||||||
| 11371 | NCU06205 | Tup-N-Terminal & WD repeats | 66.08/6.42 | NI | Transcriptional repressor | rco-1 | 21 |
| 11392 | NCU09068 | GATA zinc finger | 109.29/8.89 | 287-PIKARKD (M) | Nitrate catabolic enzyme regulatory protein | nit-2 | 39 |
| 325-RKTSIDETSKRNPNRKR (B) | |||||||
| 11342 | NCU01994 | bZIP | 36.01/5.26 | 140-PAQSRRK (M) | Hypothetical protein | NI | - |
| 144- RRKAQNRAAQRAFRERKE (B) |
a Annotation was performed according to Borkovich et al. (2) and the N. crassa database at the FGSC site (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Most of the selected transcription factors have not yet been characterized at the protein level, and were annotated as hypothetical proteins in the N. crassa database (Table II), whereas many of them have been described as being involved with growth and development in the fungus (12). Only a few proteins have functional roles that have already been extensively studied. One is the N. crassa NIT2 protein (ORF NCU09068), a GATA transcription factor that interacts with the Zn2Cys6 NIT4 protein to activate expression of nitrate and nitrite reductases (20). Another is the S. cerevisiae Tup1 ortholog RCO-1 protein (ORF NCU06205), functionally characterized as a regulatory protein that mediates mycelial repression of conidiation gene expression (21). The gene annotated as pacC (ORF NCU00090) is the pacC/RIM101 ortholog, extensively studied in Aspergillus nidulans and S. cerevisiae as encoding a transcription factor involved in pH regulation by activating genes in alkaline conditions and repressing those genes expressed in acidic conditions (22). The ctf-1α gene (ORF NCU08000) product is the well-characterized cutinase transcription factor Ctf1α ortholog, which was described as upregulating genes encoding cutinase enzymes in other filamentous fungi, such as Fusarium solani (23, 24) and F. oxysporum (25). The NCU03043 gene product revealed homology (68% identity) with the FLE1 protein of Podospora anserina (26) and the FlbC protein of different fungi including A. clavatus and A. fumigatus (27). Both FLE1 and FlbC are transcription factors involved in fungi development. Finally, the XlnR ortholog encoded by the ORF NCU06971 is a global transcriptional activator controlling the expression of genes encoding xylanolitic and cellulolytic enzymes, which was first isolated in A. niger (28).
Although most of the transcription factors identified in our screen were annotated as hypothetical proteins, some showed homology with proteins that have already been studied. The ORF NCU02713 deleted strain corresponds to the N. crassa csp-1 (conidial separation-1) mutant previously isolated by Selitrennikoff et al. (29), which showed improper separation of conidia from hyphae. This gene was recently described to encode a light-inducible transcription factor (30). The protein codified by the ORF NCU01994 is the Candida albicans Fcr3 (fluconazole resistance 3) ortholog transcription factor (31). The Fcr3 protein was able to complement a S. cerevisiae mutant strain lacking the transcription factors Pdr1 and Pdr3, which control the expression of several genes involved in Pleiotropic Drug Resistance. The NCU00019 gene product belongs to the forkhead (or Fox, for Forkhead box) transcription factor family, which has been identified in many metazoans as playing important roles in diverse biological processes (32). The ORF NCU04731 encodes a protein having 52% identity with the S. pombe Sre1 transcription factor, a sterol regulatory element binding protein (SREBP) functionally conserved among different fungi. Fungal SREBPs are hypoxic transcription factors required for adaptation to a low oxygen environment (33). Finally, the ORFs NCU01097, NCU09739, NCU04851, NCU01629, NCU00808, NCU01154, and NCU04390 encode hypothetical proteins without any previously described function.
gsn Expression in Selected Mutant Strains
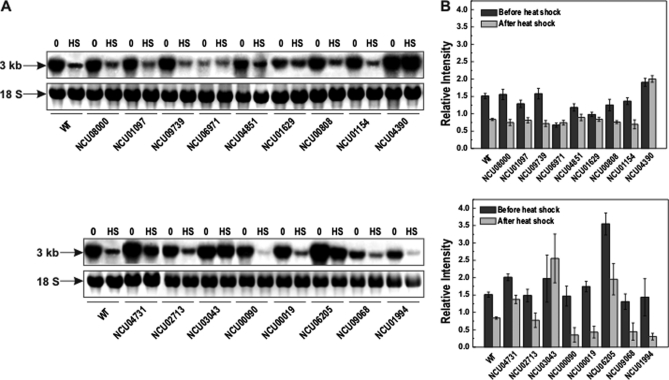
To investigate whether the glycogen accumulation pattern of mutant strains could be related to gsn regulation, gene expression analysis was performed by Northern blot. Previous results from our group have shown a decrease in gsn transcript levels when mycelium of the wild-type N. crassa were transferred from 30 °C to 45 °C (5, 8). In the present study, gsn gene expression in the selected mutant strains was analyzed before and after exposure to 45 °C. Many of the mutant strains presented a gsn transcription profile similar to that observed in the wild-type strain, in which a lower gsn expression was observed after heat shock (Fig. 3). However some mutants showed differences, either because the transcript levels were slightly reduced after heat shock (as seen, for example, for strains with deleted NCU04851, NCU01629, and NCU04731 ORFs), or because the transcript levels were strongly reduced after heat shock (as for strains with deleted NCU00019 and NCU01994 ORFs), as compared with the wild-type strain. Surprisingly, some of the mutant strains showed equal transcript levels both before and after heat shock (strains with NCU06971, NCU04390, and NCU03043 deleted ORFs), indicating loss of gsn gene expression regulation after heat stress. Also, high transcript levels for NCU04390 ORF deleted strain (both before and after heat shock) and NCU06205 ORF deleted strain (before heat shock) were detected.
Fig. 3.
gsn gene expression assay by Northern blot before and after heat shock in the selected mutant strains. Mycelia were cultivated at 30 °C for 24 h and then shifted to 45 °C. Samples were collected before (0) and after (HS) temperature shift and total RNA was extracted. Total RNA (15 μg) was separated by electrophoresis in a denaturing formaldehyde gel, transferred to nylon membrane and probed with the [α-32P] radiolabeled full-length gsn cDNA. A, upper panel, gel autoradiography, lower panel, the 18 S rRNA was used as loading control after ethidium bromide staining of the same gels. B, densitometric analysis of the gsn gene expression relative induction (ImageJ software). Results represent the average of at least three independent experiments. WT, FGSC 9718 strain.
An attempt to correlate the gsn expression profile with the glycogen accumulated under both environmental conditions analyzed in this work indicated that in several mutant strains the amount of glycogen correlated with gsn transcript levels (Fig. 3). For example, the NCU06971 ORF deleted strain showed equal amounts of glycogen and the same transcript levels before and after heat shock. In addition, the amount of glycogen and the gsn transcript levels before heat shock were lower than that found in the wild-type strain. A good correlation was also observed for the strains mutated in the ORFs NCU04851, NCU04390, and NCU0199. However, the hyper-accumulation of glycogen observed in the ORF NCU09068 mutant strain may not solely result from gsn expression, as low transcript levels were observed under both environmental conditions. Thus, the transcription factors could act either directly on gene expression or indirectly, by regulating a gene whose product affects gene expression.
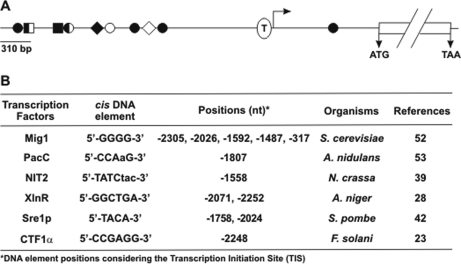
A search for putative transcription factor binding sites on the gsn 5′-flanking region was performed using the MatInspector tool. One putative binding site for the CTFα, PacC, and NIT2 proteins was found at positions –2248, –1803, and –1558 bp, respectively, two putative binding sites for the Sre1 (positions –1758 and –2024) and XlnR (positions –2071 and –2252) orthologue were found, and five sites were identified for the Mig1 orthologue at positions –2305, –2026, –1592, –1487, and –317, relative to the ATG start codon (Fig. 4). The S. cerevisiae Mig1 transcription factor, the Cre1/CreA protein orthologue in filamentous fungi, is a major protein that drives the complex Ssn6-Tup1 by repressing a set of glucose-repressible genes (34). Interestingly, the Tup1 protein is the N. crassa RCO-1 protein (NCU06205) ortholog, which was identified in the present study as a putative transcription factor involved in the regulation of glycogen metabolism.
Fig. 4.
Representation of the gsn gene 5′-flanking region. A, the relative position of the DNA motifs recognized by the transcription factors Mig1 (●), PacC (♦), NIT2 (◊), XlnR (■), Sre1p (○), and CTF1α (□) are indicated. The TATA-box sequence is indicated by T. The gsn ORF is delimited by the ATG start codon and the TAA stop codon. The Transcription Initiation Site (TIS) is represented by an arrow. B, regulatory DNA elements found in the gsn gene 5′-flanking region.
Binding of the Transcription Factors to the gsn Promoter
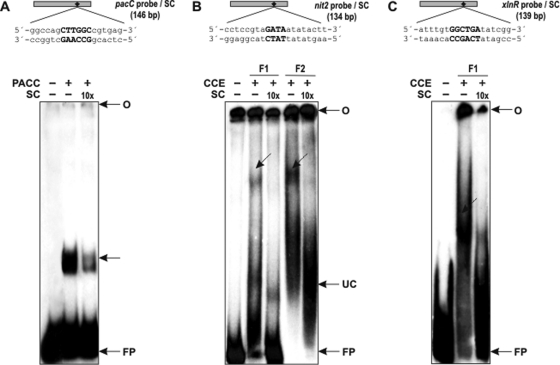
Gel shift analysis was performed to investigate whether some of the selected transcription factors having putative cis motifs in the gsn promoter were able to recognize and bind to DNA fragments containing their respective motifs. As shown in Fig. 5, the recombinant PACC protein bound to the pacC probe and formed a unique and strong DNA-protein complex (Fig. 5A, lane 2), which was reduced in the presence of a 10-fold molar excess of unlabeled pacC probe (Fig. 5A, lane 3). This finding shows that the pacC cis element present in the gsn promoter is an active binding site for the PACC transcription factor. The NIT2 and XLNR putative binding sites were analyzed using crude cellular extract (CCE) prepared from mycelium from the wild-type strain fractionated by affinity chromatography. Two chromatographic fractions were analyzed in binding reactions with the nit2 probe, as shown in Fig. 5B. DNA-protein complexes were observed in the two fractions (Fig. 5B, lanes 2 and 4), which were removed in the presence of 10-fold molar excess of the unlabeled probes (Fig. 5B, lanes 3 and 5). One fraction was used to analyze the xlnR probe and a specific complex was observed (Fig. 5C, lane 2), which was also reduced in the presence of 10-fold molar excess of the unlabeled probe (Fig. 5C, lane 3). Taken together, the results indicate that the transcription factors analyzed were able to bind to their cis elements present in the gsn promoter, suggesting they may have a role in glycogen metabolism regulation.
Fig. 5.
Binding of proteins from N. crassa WT strain to fragments of the gsn promoter. A, upper panel, schematic representation of the pacC probe with the PACC motif (small black diamond) and part of its neighboring sequences. Lower panel, gel shift analysis of PACC motif using 2 μg of the His-ΔPACC recombinant protein. Lane 1, pacC probe, no protein added. The DNA-complexes are indicated by arrows. B and C, upper panels, schematic representation of the nit2 and xlnR probes, respectively, with their motifs (small black diamond) and part of their neighboring sequences. Lower panels, gel shift analysis of NIT2 and XLNR motifs using Heparin-Sepharose chromatographic fractions (F1 and F2). An amount of 35 μg of each fraction obtained by affinity chromatography of crude cellular extract was assayed. Lane 1, nit2 and xlnR probes, no protein added; Lanes 2 and 4, binding in the absence of the specific competitors; Lanes 3 and 5, binding in the presence of the specific competitors. CCE, crude cellular extract; UC, unspecific complex; O, gel origin; SC, specific competitor; FP, free probe.
Flow Cytrometric Analysis
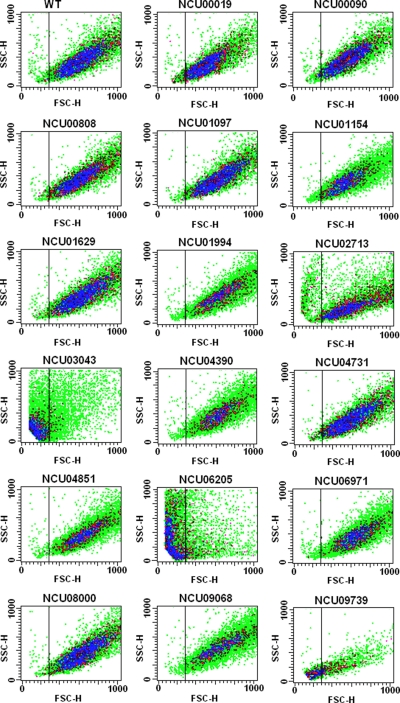
Flow cytometric analysis was used to investigate the cell size, cell complexity, and level of propidium iodide (PI) incorporation of the selected mutant strains. PI is a red fluorescent compound that binds to DNA and can be used to evaluate DNA content in individualized cells by flow cytometry. The intensity of fluorescence is proportional to the quantity of DNA available to PI intercalation. The DNA content in a cell can vary as a function of the cell cycle phase, therefore the comparative intensity of fluorescence is G0/G1 < S < G2/M. Apoptotic cells carry condensed and fragmented DNA promoting a lower fluorescence than cells at G0/G1. This analysis was performed for 10,000 cells from each mutant strain, and compared with the wild-type strain. Fig. 6 shows the results obtained from the Forward Scatter and Side Scatter detectors, which reveal cellular size and complexity, respectively. From all strains analyzed, the NCU02713, NCU03043, NCU06205, and NCU09739 ORFs deleted strains presented altered light scattering profiles, compared with the wild-type strain. The NCU02713 ORF deleted strain showed a discrete increase of cells that was smaller and slightly less complex. The NCU03043 and NCU09739 ORFs deleted strains presented expressive increase of smaller size cells and decreased complexity, whereas the NCU06205 ORF deleted strain showed a distinct increase of smaller cells together with some decrease in complexity. It is important to mention that the parameter of complexity (i.e. shape of the nucleus, the amount and type of cytoplasmic granules and membrane roughness) can be affected by cell size.
Fig. 6.
Morphological analysis of the mutant strains by flow cytometry. Analysis of cell size (FSC-H) and cell complexity (SSC-H) was performed using 10,000 events. Results from one of at least three independent experiments are shown.
Fig. 7 shows the results obtained from mutants that showed changes on fluorescence profile by PI treatment. All mutants with defective morphology presented some alteration in PI incorporation, as revealed by overlapping the mutants and wild-type strain profiles (right panels). The areas M1 and M2 represent cells at transition phases G0/G1 and G2/M of the cell cycle, respectively. The NCU02713 ORF deleted strain presented a high PI content for cells at the G2/M transition phase, indicating that this strain can carry some cellular impairment associated with the G2/M phase. The NCU03043 and NCU06205 ORFs deleted strains showed similar profiles, with a high content of cells between the M1 and M2 areas. This area represents cells at the S phase, indicating that these strains can carry impairments associated with the S phase or related to the S/G2 transition phase. The NCU09739 ORF deleted strain presented a high content of cells prior to the M1 area. Here, low PI incorporation might be a consequence of either loss of DNA content because of an irregular cell cycle, or DNA fragmentation associated to apoptosis. It is noteworthy that the mutant strains shown in Fig. 7 display visible and severe growth defects, such as low conidiation and reduction in both the extension and production of aerial hyphae, compared with the wild-type strain, indicating that the proteins are required for normal vegetative growth and development.
Fig. 7.
Analysis of propidium iodide (PI) incorporation by the selected mutant strains using flow cytometry. PI fluorescence (FL2-H) was measured for 10,000 events. The M1 and M2 areas reveal cells at the G0/G1 and G2/M phases of the cell cycle, respectively. The area between M1 and M2 is related to cells at the S phase of the cell cycle. The sub-M1 area is related to cells with low PI incorporation. Results from one of at least three independent experiments are shown.
DISCUSSION
The release of the complete N. crassa genome (1) and the availability of a mutant strains collection with each strain deleted in a single gene have allowed us to perform a screening aimed to investigate specific aspects of the fungus cell biology. In this type of screen, the phenotype analyzed corresponds to a loss of function, so that conclusions can be drawn concerning the involvement of a particular gene product in the phenotype scored as emphasized in a similar screen in S. cerevisiae (35). In this work, we detected alterations in glycogen accumulation in N. crassa strains deleted in transcription factors, to identify not yet previously described transcriptional regulators of glycogen metabolism. Transcription factors often control gene transcription through binding to specific DNA-binding sites, which can either promote (activate) or repress (inhibit) the recruitment of the transcription initiation machinery. To fully understand a gene function, it is helpful to understand the regulatory network context in which the gene participates, and that includes identification of the transcription factors involved in its regulation.
The fungus N. crassa has been widely used as a model organism, as it exhibits both asexual development and sexual differentiation. Following the availability of its genome sequencing, it has emerged as a suitable model organism for higher eukaryote studies because of its multicellularity and the high number of genes without orthologues available in public databases. It is typically a haploid organism, undergoing only a very transient diploid stage immediately prior to meiosis. In addition, the existence of gene-silencing mechanisms, such as the Repeat Induced Point Mutation, that eliminate duplicated sequences (36), makes it an advantageous organism in the type of screen performed in the present work, which is based on mutant strains having single gene deletion.
Herein we used a quantitative assay to measure glycogen accumulation in a set of strains with deleted transcription factors. Our results demonstrate that transcription factors belonging to different families regulate glycogen metabolism during vegetative growth and also under a stress condition such as heat shock. It should be noted that the amount of glycogen that is accumulated results from the balance between glycogen synthase and glycogen phosphorylase activities. These enzymes are regulated in an opposite way; phosphorylation activates glycogen phosphorylase and inhibits glycogen synthase. In N. crassa glycogen synthase activity is inhibited under heat shock (45 °C) whereas glycogen phosphorylase activity is activated under the same condition (5, 6). Many of the transcription factors identified in our screen have been functionally characterized in different organisms, and some of them may play a role in the control of glycogen metabolism. This hypothesis is reinforced by the existence of DNA-binding sites specific for the transcription factors found in the gsn gene 5′-flanking region.
The transcription factor PacC responds to changes in extracellular pH by activating specific alkaline genes and repressing specific acid genes (37). Here, we demonstrated that this transcription factor binds to gsn promoter. The transcription factor XlnR is described as a transcriptional activator controlling the expression of genes encoding xylanolytic and cellulolytic enzymes in filamentous fungi (28). Our results indicate the latter transcription factor may also control the expression of genes encoding enzymes required for the metabolism of other carbon sources, such as the enzymes involved in glycogen metabolism, thus up-regulation of gsn expression results in glycogen accumulation. Interestingly, it has been shown that the XlnR-induced expression of genes encoding xylanolytic enzymes is modulated by the carbon catabolite repressor Cre1/CreA transcription factor in Aspergillus (38). The Cre1/CreA fungal protein is the S. cerevisiae Mig1 transcription factor orthologue that has five putative binding sites in the promoter gsn, suggesting that this transcription factor is a protein that deserves further investigation concerning the regulation of glycogen metabolism.
Another transcription factor identified in our screen is the N. crassa NIT2 protein (AreA in A. nidulans), a member of the GATA factors family, already characterized in N. crassa as a positive regulator of genes encoding enzymes for nitrogen source catabolism under nitrogen limiting conditions (39). Based on our results, we can suggest that this transcription factor acts as a repressor of carbon metabolism, because the knocked-out strain showed loss of glycogen accumulation regulation, despite having low gsn gene expression as compared with the wild-type strain. A link between carbon and nitrogen regulation was reported by Lockington et al. (40), who described the effect of both carbon and nitrogen sources on the amount of cellulases secreted in A. nidulans. Although the result was preliminary, the authors suggested the existence of a link in the regulation of the carbon and nitrogen utilization pathways in filamentous fungi.
Although most transcription factors here identified belong to the zinc finger family, at least one member of the bHLH, bZIP, forkhead, and WD repeat proteins was also identified. Interestingly, the bHLH transcription factor identified in this work (NCU04731) has, at the C-terminal region, a domain found in ER membrane-bound transcription factors called SREBP. The SREBPs can be distinguished from other bHLH proteins by the presence of a tyrosine instead of an arginine residue in their basic regions (41). The first SREBP fungal orthologue was identified in S. pombe (42), and more recently it was characterized in other fungal species, such as Candida albicans (43), A. fumigatus (44), and Cryptococcus neoformans (45). The ability to respond to sterol is conserved between mammalian and fungal SREBPs, however the fungal proteins are hypoxic transcription factors required for growth by regulating genes under low-oxygen conditions (33). The knocked-out N. crassa strain for this transcription factor showed impaired glycogen accumulation compared with the wild-type strain, which did not correlate with gsn gene expression. Considering that glycogen accumulates under anaerobic conditions, a potential involvement of this transcription factor in this metabolic process cannot be ruled out and deserves further investigation.
A very interesting transcription factor identified in our screen is the RCO-1, characterized in N. crassa as a regulatory protein that mediates mycelial repression of conidiation gene expression (21). It is a homologue of S. cerevisiae Tup1, a multidomain protein that mediates transcriptional repression of genes concerned with a variety of processes. In S. cerevisiae, Tup1 and Ssn6 proteins comprise a protein complex that is required for repression of several apparently unrelated genes, including glucose-repressible genes. They need to be physically associated in order to be recruited to promoters by trans-acting DNA-binding proteins (34). Our results indicate that this transcription factor could also control glycogen metabolism as a repressor of gsn gene expression in order to favor free glucose inside the cell. Recent results have shown that RCO-1 participates in photoadaptation in N. crassa by repressing gene transcription after a long exposure to light (46), showing that a mutation that affects the conidiation process also presents clock effects (47).
As previously mentioned, the csp-1 gene product identified in our screen is also a light-inducible transcription factor (30), and a strain mutated in this gene developed superficially normal-looking conidia that failed to completely separate and remained tightly linked (29). The fact that both proteins (RCO-1 and CSP-1) play roles in conidiation and in circadian rhythms, and that the strains mutated in these proteins showed improper glycogen accumulation, led us to speculate on the existence of a connection between circadian clocks and glycogen metabolism, similar to what was described for trehalose in N. crassa (48). In the latter case, the gene encoding trehalose synthase is a clock-controlled gene (ccg-9), thus connecting the requirement for trehalose in clock regulation. In Drosophila, Zheng and Sehgal (49) demonstrated that the AKT and TOR-S6K pathways, which are the major regulators of nutrient metabolism, cell growth, and senescence, affect the brain circadian clock that drives behavioral rhythms. Another recent example of a link between metabolism and circadian rhythms was described by Doi et al. (50), providing direct evidence of the action of the circadian clock in the regulation of mammalian glycogen metabolism. The authors demonstrated that the CLOCK transcription factor regulates the circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2 (glycogen synthase 2), which is the rate-limiting enzyme of glycogenesis in the liver.
The fact that deletion of some of the transcription factors affects cell cycle progression, as demonstrated by flow cytometry assay, is especially interesting. New cell cycle transcription factors have been discovered, which constitute an important tool for studies concerning the regulation of cell cycle transcription. Using a systematic screen to reveal new S. cerevisiae cell cycle transcription factors, White et al. (51) identified a series of transcription factors having functional roles in different biological processes, including glucose and lipid metabolism. Although the results obtained in our screen did not directly show a role for the transcription factors we identified, they constitute a valuable group of candidate proteins acting as regulators in glycogen metabolism control. Our results open new opportunities for investigating key issues concerning glycogen metabolism regulation, such as how glycogen metabolism could be connected to cell-cycle regulation, biological clock, and other aspects of cellular metabolism. A better understanding of such connections will bring insights into the importance of the energy balance in biological processes.
Acknowledgments
We thank Dr. Rui Curi from Instituto de Ciências Biomédicas, USP, São Paulo, Brazil, for the flow cytometry assays.
Footnotes
* This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) by grants to M. C. Bertolini and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). R. D. G. and F. B. C. are graduate fellows supported by CAPES and FAPESP, respectively, F. Z. F. is a post-doctoral fellow from FAPESP, and M. C. B. is a CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) fellow.
1 The abbreviations used are:
- ORF
- open reading frame
- SREBP
- sterol regulatory element binding protein
- STRE
- Stress Response Element
- PMSF
- phenylmethylsulfonyl fluoride.
REFERENCES
- 1. Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L. J., Smirnov S., Purcell S., Rehman B., Elkins T., Engels R., Wang S., Nielsen C. B., Butler J., Endrizzi M., Qui D., Ianakiev P., Bell-Pedersen D., Nelson M. A., Werner-Washburne M., Selitrennikoff C. P., Kinsey J. A., Braun E. L., Zelter A., Schulte U., Kothe G. O., Jedd G., Mewes W., Staben C., Marcotte E., Greenberg D., Roy A., Foley K., Naylor J., Stange-Thomann N., Barrett R., Gnerre S., Kamal M., Kamvysselis M., Mauceli E., Bielke C., Rudd S., Frishman D., Krystofova S., Rasmussen C., Metzenberg R. L., Perkins D. D., Kroken S., Cogoni C., Macino G., Catcheside D., Li W., Pratt R. J., Osmani S. A., DeSouza C. P., Glass L., Orbach M. J., Berglund J. A., Voelker R., Yarden O., Plamann M., Seiler S., Dunlap J., Radford A., Aramayo R., Natvig D. O., Alex L. A., Mannhaupt G., Ebbole D. J., Freitag M., Paulsen I., Sachs M. S., Lander E. S., Nusbaum C., Birren B. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868 [DOI] [PubMed] [Google Scholar]
- 2. Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Plamann M., Goodrich-Tanrikulu M., Schulte U., Mannhaupt G., Nargang F. E., Radford A., Selitrennikoff C., Galagan J. E., Dunlap J. C., Loros J. J., Catcheside D., Inoue H., Aramayo R., Polymenis M., Selker E. U., Sachs M. S., Marzluf G. A., Paulsen I., Davis R., Ebbole D. J., Zelter A., Kalkman E. R., O'Rourke R., Bowring F., Yeadon J., Ishii C., Suzuki K., Sakai W., Pratt R. (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hardy T. A., Huang D., Roach P. J. (1994) Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in. Saccharomyces cerevisiae. J. Biol. Chem. 269, 27907–27913 [PubMed] [Google Scholar]
- 4. Freitas F. Z., de Paula R. M., Barbosa L. C. B., Terenzi H. F., Bertolini M. C. (2010) cAMP signaling pathway controls glycogen metabolism in Neurospora crassa by regulating the glycogen synthase gene expression and phosphorylation. Fungal Genet. Biol. 47, 43–52 [DOI] [PubMed] [Google Scholar]
- 5. de Paula R., de Pinho C. A., Terenzi H. F., Bertolini M. C. (2002) Molecular and biochemical characterization of the Neurospora crassa glycogen synthase encoded by the gsn cDNA. Mol. Genet. Genomics 267, 241–253 [DOI] [PubMed] [Google Scholar]
- 6. Noventa-Jordão M. A., de Lourdes M., Polizeli T. M., Bonini B. M., Jorge J. A., Terenzi H. F. (1996) Effects of temperature shifts on the activities of Neurospora crassa glycogen synthase, glycogen phosphorylase and trehalose-6-phosphate synthase. FEBS Lett. 378, 32–36 [DOI] [PubMed] [Google Scholar]
- 7. Parrou J. L., Teste M. A., François J. (1997) Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143, 1891–1900 [DOI] [PubMed] [Google Scholar]
- 8. Freitas F. Z., Bertolini M. C. (2004) Genomic organization of the Neurospora crassa gsn gene. Possible involvement of the STRE and HSE elements in the modulation of gene transcription during heat shock. Mol. Genet. Genomics 272, 550–561 [DOI] [PubMed] [Google Scholar]
- 9. Martinez-Pastor M., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., Estruch F. (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 10. Freitas F. Z., Chapeaurouge A., Perales J., Bertolini M. C. (2008) A systematic approach to identify STRE-binding proteins of the gsn glycogen synthase gene promoter in Neurospora crassa. Proteomics 8, 2052–2061 [DOI] [PubMed] [Google Scholar]
- 11. McCluskey K. (2003) The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52, 245–262 [DOI] [PubMed] [Google Scholar]
- 12. Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., Litvinkova L., Weiss R. L., Borkovich K. A., Dunlap J. C. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vogel H. J. (1956) A convenient growth medium for Neurospora crassa (medium N). Microbiol. Genet. Bull. 13, 42–43 [Google Scholar]
- 14. Hardy T. A., Roach P. J. (1993) Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J. Biol. Chem. 268, 23799–23805 [PubMed] [Google Scholar]
- 15. Hartree E. F. (1972) Determination of protein: A modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 48, 422–427 [DOI] [PubMed] [Google Scholar]
- 16. Sokolovsky V., Kaldenhoff R., Ricci M., Russo V. E. A. (1995) Fast and reliable mini-prep RNA extraction from Neurospora crassa. Fungal Genet. Newslett. 37, 41–43 [Google Scholar]
- 17. Sambrook J., Russell D. W. (2001) Molecular Cloning. A Laboratory Mannual (3rd ed.) Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY [Google Scholar]
- 18. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 19. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng B., Marzluf G. A. (1998) Interaction between major nitrogen regulatory protein NIT2 and pathway-specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa. Mol. Cell. Biol. 18, 3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamashiro C. T., Ebbole D. J., Lee B. U., Brown R. E., Bourland C., Madi L., Yanofsky C. (1996) Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peñalva M. A., Arst H. N., Jr. (2002) Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66, 426–446, Table of Contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li D., Kolattukudy P. E. (1997) Cloning of cutinase transcription factor 1, a transactivating protein containing Cys6Zn2 binuclear cluster DNA-binding motif. J. Biol. Chem. 272, 12462–12467 [DOI] [PubMed] [Google Scholar]
- 24. Li D., Sirakova T., Rogers L., Ettinger W. F., Kolattukudy P. E. (2002) Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 277, 7905–7912 [DOI] [PubMed] [Google Scholar]
- 25. Rocha A. L., Di Pietro A., Ruiz-Roldán C., Roncero M. I. (2008) Ctf1, a transcriptional activator of cutinase and lipase genes in Fusarium oxysporum is dispensable for virulence. Mol. Plant Pathol. 9, 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coppin E. (2002) The fle1 gene encoding a C2H2 zinc finger protein co-ordinates male and female sexual differentiation in Podospora anserina. Mol. Microbiol. 43, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 27. Adams T. H., Wieser J. K., Yu J. H. (1998) Asexual sporulation in. Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Peij N. N., Gielkens M. M., de Vries R. P., Visser J., de Graaff L. H. (1998) The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64, 3615–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Selitrennikoff C. P., Nelson R. E., Siegel R. W. (1974) Phase-specific genes for macroconidiation in Neurospora crassa. Genetics 78, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lambreghts R., Shi M., Belden W. J., Decaprio D., Park D., Henn M. R., Galagan J. E., Bastürkmen M., Birren B. W., Sachs M. S., Dunlap J. C., Loros J. J. (2009) A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181, 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang X., Talibi D., Weber S., Poisson G., Raymond M. (2001) Functional isolation of the Candida albicans FCR3 gene encoding a bZip transcription factor homologous to Saccharomyces cerevisiae Yap3p. Yeast 18, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 32. Wang M., Wang Q., Zhao H., Zhang X., Pan Y. (2009) Evolutionary selection pressure of forkhead domain and functional divergence. Gene 432, 19–25 [DOI] [PubMed] [Google Scholar]
- 33. Bien C. M., Espenshade P. J. (2010) Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 9, 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith R. L., Johnson A. D. (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325–330 [DOI] [PubMed] [Google Scholar]
- 35. Wilson W. A., Wang Z., Roach P. J. (2002) Systematic identification of the genes affecting glycogen storage in the yeast. Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 232–242 [DOI] [PubMed] [Google Scholar]
- 36. Dunlap J. C., Borkovich K. A., Henn M. R., Turner G. E., Sachs M. S., Glass N. L., McCluskey K., Plamann M., Galagan J. E., Birren B. W., Weiss R. L., Townsend J. P., Loros J. J., Nelson M. A., Lambreghts R., Colot H. V., Park G., Collopy P., Ringelberg C., Crew C., Litvinkova L., DeCaprio D., Hood H. M., Curilla S., Shi M., Crawford M., Koerhsen M., Montgomery P., Larson L., Pearson M., Kasuga T., Tian C., Baştürkmen M., Altamirano L., Xu J. (2007) Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57, 49–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tilburn J., Sarkar S., Widdick D. A., Espeso E. A., Orejas M., Mungroo J., Peñalva M. A., Arst H. N., Jr. (1995) The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamayo E. N., Villanueva A., Hasper A. A., de Graaff L. H., Ramón D., Orejas M. (2008) CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 45, 984–993 [DOI] [PubMed] [Google Scholar]
- 39. Fu Y. H., Marzluf G. A. (1990) nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol. Cell Biol. 10, 1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lockington R. A., Rodbourn L., Barnett S., Carter C. J., Kelly J. M. (2002) Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet. Biol. 37, 190–196 [DOI] [PubMed] [Google Scholar]
- 41. Párraga A., Bellsolell L., Ferré-D'Amaré A. R., Burley S. K. (1998) Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 Å resolution. Structure 15, 661–672 [DOI] [PubMed] [Google Scholar]
- 42. Hughes A. L., Todd B. L., Espenshade P. J. (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120, 831–842 [DOI] [PubMed] [Google Scholar]
- 43. Lane S., Zhou S., Pan T., Dai Q., Liu H. (2001) The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21, 6418–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., Barbuch R., Bard M., Lawrence C. B., Cramer R. A., Jr. (2008) A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4, e1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bien C. M., Chang Y. C., Nes W. D., Kwon-Chung K. J., Espenshade P. J. (2009) Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol. Microbiol. 74, 672–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olmedo M., Navarro-Sampedro L., Ruger-Herreros C., Kim S. R., Jeong B. K., Lee B. U., Corrochano L. M. (2010) A role in the regulation of transcription by light for RCO-1 and RCM-1, the Neurospora homologs of the yeast Tup1-Ssn6 repressor. Fungal Genet. Biol. 47, 939–952 [DOI] [PubMed] [Google Scholar]
- 47. Brody S., Oelhafen K., Schneider K., Perrino S., Goetz A., Wang C., English C. (2010) Circadian rhythms in Neurospora crassa: Downstream effectors. Fungal Genet. Biol. 47, 159–168 [DOI] [PubMed] [Google Scholar]
- 48. Shinohara M. L., Correa A., Bell-Pedersen D., Dunlap J. C., Loros J. J. (2002) Neurospora clock-controlled gene (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot. Cell 1, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng X., Sehgal A. (2010) AKT and TOR signaling set the pace of the circadian pacemaker. Curr. Biol. 20, 1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Doi R., Oishi K., Ishida N. (2010) CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J. Biol. Chem. 285, 22114–22121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White M. A., Riles L., Cohen B. A. (2009) A systematic screen for transcriptional regulators of the yeast cell cycle. Genetics 181, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lundin M., Nehlin J. O., Ronne H. (1994) Importance of a flanking AT-rich region in target site recognition by the CG box-binding zinc finger protein MIG1. Mol. Cell. Biol. 14, 1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arst H. N., Peñalva M. A. (2003) pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 19, 224–231 [DOI] [PubMed] [Google Scholar]