Abstract
The plasma membrane separates the cell from the external environment and plays an important role in the stress response of the cell. In this study, we compared plasma membrane proteome modifications of yeast cells exposed to mild (0.4 m NaCl) or high (1 m NaCl) salt stress for 10, 30, or 90 min. Plasma membrane-enriched fractions were isolated, purified, and subjected to iTRAQ labeling for quantitative analysis. In total, 88–109 plasma membrane proteins were identified and quantified. The quantitative analysis revealed significant changes in the abundance of several plasma membrane proteins. Mild salt stress caused an increase in abundance of 12 plasma membrane proteins, including known salt-responsive proteins, as well as new targets. Interestingly, 20 plasma membrane proteins, including the P-type H+-ATPase Pma1, ABC transporters, glucose and amino acid transporters, t-SNAREs, and proteins involved in cell wall biogenesis showed a significant and rapid decrease in abundance in response to both 0.4 m and 1 m NaCl. We propose that rapid protein internalization occurs as a response to hyper-osmotic and/or ionic shock, which might affect plasma membrane morphology and ionic homeostasis. This rapid response might help the cell to survive until the transcriptional response takes place.
Exposure of yeast cells to saline stress implies exposure to both specific cation toxicity and osmotic stress (1). Certain ions, such as Na+ or Li+, are toxic for cells because of their ability to inhibit specific metabolic pathways, probably through inhibition of specific targets, as has been shown to be the case for the yeast Hal2 protein and certain RNA-processing enzymes (2, 3). In addition, high salinity results in an imbalance in the membrane potential and thus affects the activity of membrane transporters (4) and disrupts ion homeostasis within cells. The yeast cell response to high salinity has been extensively studied and serves as a model for changes in gene expression in response to external stimuli (5, 6). The response is mediated by several stress-responsive signaling pathways, with the high osmolarity glycerol mitogen-activated protein kinase pathway playing a major role. This pathway is involved in sensing an increase in turgor pressure and transducing the appropriate signals to the gene expression program (1). Moreover, Saccharomyces cerevisiae cells respond to high extracellular NaCl concentrations by increasing both potassium uptake and sodium efflux in order to maintain an appropriate Na+/K+ ratio (5, 7–11).
Transcript expression has been examined as a response of yeast cells to saline stress. The expression of about 7% of the genes in the yeast genome is increased by more than fivefold after a mild and brief saline shock (0.4 m NaCl, 10 min) and most responsive genes show a transient expression pattern, as mRNA levels rapidly decline after 20 min of stress. A similar set of genes shows increased expression in cells subjected to higher saline concentrations (0.8 m NaCl), although, in this case, the response is delayed (5). The transcriptional induction of most genes that are strongly responsive to salt stress is dependent on the presence of the stress-activated mitogen-activated protein kinase Hog1 (12). When cells were incubated with 1 m NaCl, the number of genes showing a more than two fold increase in expression increased over time, being 107 at 10 min, 243 at 30 min, and 354 at 90 min (6). The response after 10 min of salt stress involved transcripts coding for proteins involved in nucleotide and amino acid metabolism, intracellular transport, and protein synthesis, after 30 min of stress the response involved transcripts related to respiration and energy production, and after 90 min of stress the response involved transcripts the response involved cellular detoxification, major facilitator superfamily transporters, and enzymes involved in nitrogen or sulfur metabolism and lipid or fatty acid biosynthesis.
A serious limitation of mRNA-based techniques is the lack of information on post-transcriptional regulation events. The translational response of yeast cells to 1 m NaCl stress consists of strong, but transient, inhibition of protein synthesis (13). Protein abundance can also be altered by post-translational events leading to protein degradation or to modification of subcellular localization. It is therefore not surprising that studies performed on yeast cells treated with NaCl (14) or lithium (15) show a weak correlation between proteomic data and mRNA-based data. So far, the analysis of the proteomic response of yeast cells to salt stress has been restricted to soluble proteins and has used a two-dimensional electrophoresis gel-based approach (16–18); using this method, most proteins showing changes in abundance were found to be involved in various aspects of carbohydrate metabolism, such as the synthesis of the osmo-protectant glycerol, in protein folding, and in protein degradation. Recently, a gel-free global quantitative phosphoproteomic study was performed on S. cerevisiae cells treated with 0.4 m NaCl for 5 or 20 min (14). Many phosphorylation events were identified, showing a very active and dynamic post-translational response to salt stress. Moreover, proteome changes revealed an increase in the abundance of more than 100 proteins after 20 min of salt stress; these included numerous osmotic stress-responsive proteins involved in glycerol production (14).
Although several groups have studied the yeast proteomic response to salt stress, they have focused on the total or soluble protein proteome, rather than the membrane proteome (19). In proteomic studies, membrane proteins are more difficult to analyze than soluble proteins and so usually constitute only a small proportion of data sets. The plasma membrane separates the cell interior from the external medium and plays a very important role in stress sensing, as well as cell defense (1, 20). The first response of the cell to any kind of external stress would be expected to affect plasma membrane protein organization. Plasma membrane proteins are encoded by only about 4% of the S. cerevisiae genome (Organelle Database) and are difficult to detect in global proteomic studies. The number of yeast plasma membrane proteins initially identified was not large. Navarre et al. (21) optimized a S. cerevisiae plasma membrane purification protocol to reduce contamination from other membranes and cytosolic proteins and identified 12 plasma membrane proteins by two-dimensional electrophoresis. Delom et al. (22) used an optimized protocol for plasma membrane preparation based on French press cell rupture and sucrose gradients, resolving the proteins using an ion-exchange chromatographic/lithium dodecyl sulfate-PAGE procedure and were able to identify a total of 90 S. cerevisiae proteins, 25 of which had been previously described as plasma membrane proteins. As regards C. albicans, several studies have used proteomic approaches to identify proteins in plasma membranes (23–25) and 41 integral plasma membrane proteins have been described.
In this study, we used an optimized plasma membrane purification procedure and a quantitative gel-free proteomic approach based on iTRAQ (isobaric Tags for Relative and Absolute Quantitation)1 labeling (26) to monitor changes in the plasma membrane proteome in cells exposed to 0.4 m or 1 m NaCl for 10, 30, or 90 min. Our procedure allowed the identification and quantification of more than 100 plasma membrane proteins. We found that, after mild salt stress, the abundance of 12 plasma membrane proteins was significantly increased and that of 33 decreased. High salt stress caused a significant decrease in abundance of 24 plasma membrane proteins, the abundance of 20 of these also being decreased by mild salt stress.
Our data suggest that salt stress induces a rapid internalization of important plasma membrane proteins as a first response to hyper-osmotic and/or ionic shock. This rapid response might help the cell to survive until the transcriptional response takes place.
EXPERIMENTAL PROCEDURES
Yeast Cultures and Salt Stress
S. cerevisiae strain W303 (MAT α ade2–1, leu2–3,112 his3–11,15 trp1–1 ura3–1 can1–100) was used. Cells were grown at 28 °C to an OD600 of 3.0 in YPD medium (1% yeast extract, 2% peptone, 2% glucose). One culture (control) was harvested and processed for plasma membrane purification, while 0.4 m or 1 m NaCl stress was applied to three cultures for 10, 30, or 90 min by adding a prewarmed (28 °C) solution of NaCl in YPD, then membrane-enriched fractions were prepared from all cultures as described below. A complete control experiment was performed with the same incubation times but without salt addition.
Isolation of Plasma Membrane-enriched Fractions
All steps were performed at 4 °C. Cells were harvested by centrifugation at 5000 rpm for 5 min (ILA.9100, Beckman), washed with 0.5 liters of cold distilled water, and resuspended in 15 ml of cold homogenization medium (250 mm sorbitol, 50 mm imidazole, 1 mm MgCl2, pH 7.5) containing a protease inhibitor mix [1 mm phenylmethylsulfonyl fluoride (PMSF) and 2 μg/ml each of leupeptin, aprotinin, antipain, pepstatin, and chymostatin] per 10 g of cells, and the suspension poured into a 75-ml homogenizer glass flask. Glass beads were added at a ratio of 15 g per 10 g of cells and the suspension homogenized in a Braun MSK homogenizer (Helsungen, Germany) for 3 min at full speed with cooling. Cell debris and unbroken cells were eliminated by two 5 min centrifugations at 3000 rpm and one 5 min centrifugation at 6000 rpm (JA-30.50Ti, Beckman). The supernatant was then centrifuged for 45 min at 14,000 rpm (JA-30.50Ti, Beckman) to pellet crude membranes, which were resuspended in 5 ml of suspension medium (10 mm imidazole, 2 mm MgCl2, pH 7.5, containing the protease inhibitor mix) per 10 g of cells. The suspension containing crude membranes was homogenized and brought to pH 4.8 with 1 m acetic acid and protein aggregates removed by centrifugation at 8000 rpm for 2 min (JA-30.50Ti, Beckman). The supernatant was rapidly neutralized (pH 7.5) with 1 N NaOH and centrifuged for 30 min at 42,000 rpm (70TI, Optima-Beckman) (27), then the pellet was resuspended in 50 mm triethyl ammonium bicarbonate (TEAB), pH 8.0, and stored at 20 °C until use. The final step was membrane stripping to remove peripheral membrane proteins without affecting the integral components. The membranes were incubated in 0.1 m sodium carbonate, 50 mm TEAB, pH 11.5, for 30 min on ice, then centrifuged at 48,000 rpm for 30 min (TLA55, Optima-Beckman) to pellet the stripped membranes, which were resuspended in 50 mm TEAB. The protein concentration was determined using the bicinchoninic acid (Sigma) protein assay.
Preparation of Total Cell Lysates
All steps were performed at 4 °C. Total cell lysates were prepared from 100 ml yeast cultures grown as described above. Control and salt stress cultures were harvested, washed in ice-cold water, and the cell pellets resuspended in lysis buffer (25 mm Tris, 5 mm EDTA, pH 7.5) containing the protease inhibitor mixture and PMSF described above and homogenized using glass beads by vortexing for 8 × 30 s, with cooling on ice for 30 s between each step. The cell lysates were centrifuged at 14,000 rpm for 5 min (Hettich Micro 20) and the supernatants used for immunoblotting.
SDS-PAGE and Immunoblotting
The plasma membranes were mixed with an equal volume of sample buffer (100 mm Tris HCl, pH 6.8, 4 mm EDTA, 4% SDS, 20% glycerol, 0.002% bromphenol blue) containing 1% dithiothreitol and incubated at room temperature for 10 min, then the proteins were resolved by SDS-PAGE and transferred to a PVDF membrane (Millipore, Billerica, MA) using a semidry transfer system (Bio-Rad, Hercules, CA) in 50 mm Tris, 40 mm glycine, 0.0375% SDS, and 20% methanol. The blot was saturated overnight at 4 °C using 3% low fat dried milk in Tris-buffered saline (TBS; 50 mm Tris, 150 mm NaCl, pH 7.6) containing 0.5% Tween 20, then were incubated at room temperature for 1.5 h with rabbit antibodies against plasma membrane H+-ATPase Pma1p (dilution: 1:10 000) or for 2 h with rabbit antibodies against β-1,3-glucanosyltransferase Gas1 (dilution 1: 4000; gift from H. Riezman, Geneva) diluted in TBS, 0.5% Tween 20 containing 0.5% low fat milk. After three washes with TBS containing 0.1% Tween 20, the blot was incubated at room temperature for 1 h with horseradish peroxidase-coupled anti-rabbit IgG antibodies (dilution 1:10000, Chemicon International, Temecula, CA), followed by chemiluminescence detection (Roche Diagnostics, Indianapolis, IN). The signals were captured and quantified using a KODAK 4000R Image Station, driven by KODAK Molecular Imaging Software version 4.0. The rabbit polyclonal antibodies directed against Pma1p have been generated from highly purified plasma membrane Pma1 protein from Schizosaccharomyces pombe (28).
Protein Digestion and iTRAQ Labeling
Purified plasma membranes (20 μg of protein) were solubilized in 50 mm TEAB, 0.1% Rapigest (Waters), pH 8.0, by sonication for 5 min in a bath sonicator (Bioruptor, Diagenode), and the proteins reduced by incubation for 1 h at 60 °C with 25 mm tris(2-carboxyethyl)phosphine, then alkylated with 0.26 m methyl-methanethiosulfonate for 10 min at room temperature in the dark. The reduced and alkylated proteins were digested for 16 h at 37 °C using sequencing grade modified trypsin (Promega, Madison, WI) at a protease/protein ratio of 1/20 and the Rapigest lysed by incubating the protein sample in 0.5% trifluoroacetic acid (TFA) for 60 min at 37 °C. After centrifugation of the sample at 54,000 rpm (TLA55, Optima-Beckman) for 45 min at 4 °C, the supernatant was centrifuged for 20 min at 54,000 rpm (TLA55, Optima-Beckman), then the final supernatant was vacuum dried (Speedvac SC 200, Savant) and iTRAQ labeling performed according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). The samples were labeled with tag 114 for the control, tag 115 for the 10 min sample, tag 116 for the 30 min sample, and tag 117 for the 90 min sample.
Reversed Phase Chromatography
Before separation, the samples were dissolved in 0.025% TFA and 5% acetonitrile (ACN), then the labeled peptides were mixed together and 12.9 μg of the mixture desalted using a C18 Pep Map 100 pre-column and subjected to reverse phase chromatography on a C18 PepMap100 (LC Packings) analytical column for 180 min at a flow rate of 300 nl/min using a linear gradient from 8% ACN in water/0.1% TFA to 76% ACN in water/0.085% TFA. The eluted peptides were mixed with α-cyano-4-hydroxycinnamic acid matrix (2 mg/ml in 70% ACN, 0.1% TFA) and spotted directly onto a matrix-assisted laser desorption ionization (MALDI) target using a Probot system (LC Packings Amersham Biosciences).
Mass Spectrometry Analysis
The spotted plate was analyzed on an Applied Biosystems 4800 MALDI time-of-flight (TOF)/TOF Analyzer using a 200-Hz solid state laser operating at 355 nm. MS spectra were obtained using a laser intensity of 3200 and 2000 laser shots per spot in the m/z range of 800 to 4000, whereas MS/MS spectra were obtained by automatic selection of the 12 most intense precursor ions per spot using a laser intensity of 3800 and 2100 laser shots per precursor. Collision-induced dissociation was performed with an energy of 1 kV with air as the collision gas at a pressure of 1 × 106 Torr. Data were collected using Applied Biosystems 4000 Series Explorer™ software. Liquid chromatography-tandem MS (LC-MS/MS) data were processed using ProteinPilot software and the ParagonTM search algorithm (Shilov et al., 2007) (Applied Biosystems/MDS SCIEX/4800 version 2.0). The MS/MS data were used to search the UniProtKB/Swiss-Prot database (276 256 sequences Release 54.0 of 24, July 07 from the website http://www.ebi.ac.uk/FTP/) using the “thorough search” option and a S. cerevisiae taxonomy filter. The “iTRAQ 4plex peptide labeled” sample type and a “biological modification ID focus” were selected in the analysis method. Trypsin was selected as the digestion enzyme, with cysteine alkylation by methyl-methanethiosulfonate as a modification. The results were further processed by the Pro GroupTM Algorithm to determine the minimal set of justifiable identified proteins. Proteins were annotated based on SGD, Gene Bank, and UniProt Databases.
Data were normalized using ProteinPilot. All reported data were based on 99% confidence for protein identification as determined by ProteinPilot (ProtScore ≥ 2.0). Protein identification confidence was expressed as the “Unused Protein Score,” a measurement of the protein identification confidence taking into account peptides from spectra that have not already been “used” by higher scoring proteins.
Quantification of Relative Change
To determine differences in expression of proteins between the salt-treated cells and the control, the average ratio of the identified protein was calculated by ProteinPilot based on the weighted average log ratios of the peptides. Differentially expressed proteins were further analyzed for significant down- or up-regulation, which was calculated by ProteinPilot. A cutoff level of significance of 95% (or p < 0.05) was chosen as a criterion for each individual experiment. Peptides that matched multiple proteins were not included in the quantification by the software. The final ratios were calculated from the average of the ratios obtained in at least two independent experiments in which the change in protein abundance was significant (p < 0.05). In each experiment, bias correction for unequal mixing during the combination of the different labeled samples was performed. This correction is based on the assumption that most proteins do not change in expression. Thus, if samples from each experimental condition are not combined in exactly equal amounts, bias correction fixes this systematic error. The software identifies the median average protein ratio and corrects it to unity, and then applies this factor to all quantification results.
Protease Protection Assay
We performed a modified protease protection assay as described in Davis et al. (29). Cells were grown at 28 °C to an OD600 of 3.0 in YPD medium. One culture (control) was kept at 28 °C for 30 min, while 0.4 m NaCl stress was applied to another culture for 30 min. Cells were collected by centrifugation at 3500 rpm (Hettich Micro 20) for 5 min, washed in cold TE stop buffer (10 mm Tris-HCl pH7, 1 mm EDTA, 20 mm NaN3, 20 mm NaF) and resuspended in TE stop buffer containing 0.5% β-mercaptoethanol. Then samples were incubated for 30 min at 37 °C. Cells were washed once and then resuspended in TE stop buffer containing 0.6 M sorbitol. One-milliliter aliquots of cells were treated with 0, 5, 25, 50, or 100 μg/ml Proteinase K (Roche). Digestion was performed at 30 °C for 30 min. Then 10 mm PMSF was added and total cell lysates were prepared by the addition of 120 μl of lysis buffer (2 m NaOH, 5% β-mercaptoethanol), incubation on ice for 10 min, addition of 120 μl of 100% TCA followed by incubation on ice. The cell lysates were centrifuged at 9700 rpm (Eppendorf 5417C) for 10 min. Pellets were washed once with cold acetone. After a centrifugation at 9700 rpm (Eppendorf 5417C) for 10 min pellets were dried and resuspended in 200 μl of sample buffer (100 mm TrisHCl pH 6.8, 4 mm EDTA, 4% SDS, 20% glycerol, 0.002% bromphenol blue, 1% dithiothreitol). Five min sonication (Bioruptor, Diagenode) and 10 min incubation at 37 °C were used to resuspend the proteins. Twenty microliters were loaded on SDS-PAGE and transferred to a PVDF membrane as described above. Western blot analysis was used to assess susceptibility of protein Gas1p to external protease. As a control of cells integrity during digestion with proteinase K we measured the degradation of the cytosolic protein Cdc48p with rabbit antibodies directed against Cdc48p (dilution 1:2000) (30). Cdc48p was also used as a loading control. The signal of Cdc48p was used for quantification normalization of Gas1p signal.
RESULTS
Yeast Plasma Membrane Purification
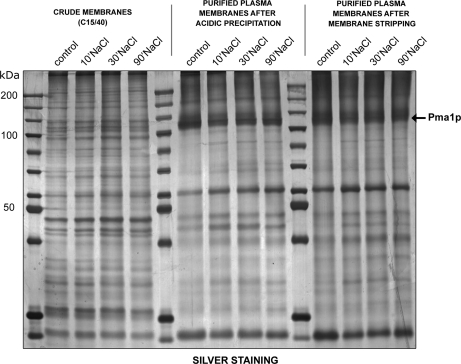
Cultures of wild-type yeast cells were grown in YPD to an OD600 of 3.0. The control culture was harvested and processed for plasma membrane purification, while salt-stress (0.4 m or 1 m NaCl in YPD) was induced in another three cultures for 10, 30, or 90 min, which were then processed for plasma membrane purification (supplemental Fig. S1). In our laboratory, we routinely use a highly reproducible method to purify yeast plasma membranes based on the disruption of intact cells with glass beads, followed by a combination of differential centrifugation and selective acidic precipitation of contaminating organelles, such as mitochondria (27). This procedure results in plasma membranes of high purity, as illustrated by SDS-PAGE (Fig. 1), which shows that the plasma membrane H+-ATPase Pma1p, a typical plasma membrane marker with an apparent molecular mass of ∼100 kDa, is highly enriched in the purified plasma membranes. Finally, the purified plasma membranes are stripped with alkaline sodium carbonate to remove peripheral membrane-associated proteins without affecting integral membrane components or lipid-anchored proteins. The stripping step does not drastically modify the protein pattern on SDS-PAGE gels (Fig. 1), but removes contaminants that could be detected by mass spectrometry. Plasma membrane purification is not affected by salt treatment, as shown by the specific enrichment of Pma1p during the purification process after treatment with 1 m NaCl (Fig. 1) and iTRAQ labeling is therefore a valid method for quantifying changes induced by salt stress.
Fig. 1.
Protein profiles during yeast plasma membrane purification. Five micrograms of proteins were separated on a 10% SDS-PAGE gel and visualized by silver staining. The fractions analyzed were the membrane-enriched fraction after differential centrifugation, purified plasma membranes after acid precipitation, and purified plasma membranes after stripping. Control corresponds to nonstressed cells, while the other cells were treated for the indicated time with 1 m NaCl.
Identification and Quantitative Analysis of Yeast Plasma Membrane Proteins
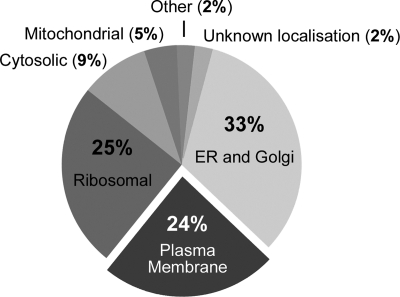
Proteins (20 μg) from each plasma membrane sample were solubilized, reduced, alkylated, digested with trypsin, and the peptides labeled with iTRAQ tags as follows: Control sample, tag 114; 10 min stress, tag 115; 30 min stress, tag 116; and 90 min stress, tag 117. Labeled peptides were mixed and subjected to reverse-phase chromatography. Up to 1080 spots were obtained and subjected to MALDI-TOF/TOF spectrometry. A maximum of 12 precursors per spot were selected for MS/MS analysis. From 452 to 530 proteins were identified in the experiment using 0.4 m NaCl and 358–505 in that using 1 m NaCl, with a confidence of at least 99% (supplemental Table and supplemental Fig. S3). Plasma membrane proteins accounted for 24% of the identified proteins (Fig. 2). The most abundant contaminants were proteins from the early secretory pathway (endoplasmic reticulum and Golgi apparatus) and ribosomal proteins. Of the plasma membrane proteins, 68% were integral membrane proteins and, of these, 67% had more than six transmembrane domains. In total, we identified 109 plasma membrane proteins in the first set of experiments (0.4 m NaCl) and 88 in the second (1 m NaCl) (Table I). Many transporters have been identified and were annotated according to the Transporter Classification database (31).
Fig. 2.
Distribution of the proteins in the plasma membrane fraction identified in the LC/MSMS analysis.
Table I. List of identified plasma membrane proteins. Proteins have been identified in three or, if not available, two (*) independent repeats of the experiment.
| N | Systematic name | Protein name | Description | Unused protein scorea |
Peptidesb |
TM/Anchorc | Subcellular Localisationd | Accession number | TCDB numbere | TCDB subfamilye | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.4 M | 1 M | 0.4 M | 1 M | |||||||||
| 1 | YOL130W | Alr1p | Plasma membrane Mg2+ transporter | 4.05–6 | 6.04–8* | 2–3 | 3–4 | 2 | PM | Q08269 | 1.A.35.2 | The CorA Metal Ion Transporter (MIT) Type 2 Family |
| 2 | YBL069W | Ast1p | Protein that interacts with Pma1p | 3.13–6* | x | 1–3 | x | peripheral | PM | P35183 | ||
| 3 | YDR093W | Atc4p/Dnf2p | Aminophospholipid translocase | 4.83–9.7 | 8–8.8* | 2–4 | 4 | 10 | PM | Q12675 | 3.A.3.8 | The Aminophospholipid-ATPase (PLA) Subfamily |
| 4 | YER166W | Atc5p/Dnf1p | Aminophospholipid translocase | x | 2–6* | x | 1–3 | 10 | PM | P32660 | 3.A.3.8 | The Aminophospholipid-ATPase (PLA) Subfamily |
| 5 | YDR384C | Ato3p | Putative ammonium transporter | x | 2* | x | 1 | 6 | PM and MT | Q12359 | 9.B.33.1 | The ATO (ATO) Subfamily |
| 6 | YBR068C | Bap2p | Branched-chain amino acid transporter | 8–10 | 6.92–10 | 4–5 | 3–5 | 12 | PM | P38084 | 2.A.3.10 | The Yeast Amino Acid Transporter (YAT) Subfamily |
| 7 | YDR046C | Bap3p | Branched-chain amino acid transporter | 2.82–5.05 | 4–4.27* | 1–2 | 2 | 12 | PM and MT | P41815 | 2.A.3.10 | The Yeast Amino Acid Transporter (YAT) Subfamily |
| 8 | YER155C | Bem2p | Rho GTPase activating protein (RhoGAP) | 2.08–6.52 | 2–4* | 1–3 | 1–2 | PM and other | P39960 | |||
| 9 | YJL058C | Bit61p | Subunit of TORC2 complex | 2–2.02* | x | 1 | x | peripheral | PM and CS | P47041 | ||
| 10 | YLR229C | Cdc42 | Rho-like GTPase, essential for cell polarity during division | 4.02–4.14 | 4–6 | 2 | 2–3 | anchor | PM | P19073 | ||
| 11 | YNL192W | Chs1p | Chitin synthase I | 11.44–22.16 | 12.1–15.7 | 5–11 | 6–7 | 7 | PM | P08004 | 10.C.2.1 | The Chitin Synthesis Subfamily |
| 12 | YBR023C | Chs3p | Chitin synthase III | 4.0–10 | 2–4.69 | 2–5 | 1–2 | 6 | PM | P29465 | 10.C.2.1 | The Chitin Synthesis Subfamily |
| 13 | YPR124W | Ctr1p | High-affinity copper transporter | 12.85–16.13 | 11.2–16 | 6–8 | 5.–8 | 2 | PM | P49573 | 9.A.11.1 | The Copper Transporter-1 (CTR1–1) Type 1 Subfamily |
| 14 | YKL046C | Dcw1p | Putative mannosidase required for cell wall biosynthesis | 6–8 | 4–8 | 3–4 | 2–4 | anchor | PM | P36091 | ||
| 15 | YMR238W | Dfg5p | Putative mannosidase required for cell wall biosynthesis | 2–5.22* | 2.8–4* | 1–2 | 1–2 | anchor | PM | Q05031 | ||
| 16 | YPL265W | Dip5p | Dicarboxylic amino acid transporter | 4–6.02 | 4–12 | 2–3 | 2–6 | 12 | PM | P53388 | 2.A.3.10 | The Yeast Amino Acid Transporter (YAT) Family |
| 17 | YBR078W | Ecm33p | Protein of unknown function | 12–12.01 | 10.68–12.72 | 6 | 5–6 | anchor | PM and MT | P38248 | ||
| 18 | YDR040C | Ena1p | P-type ATPase sodium pump | 20.32–27.55* | 6.95–8.92* | 10–13 | 3–4 | 10 | PM | P13587 | 3.A.3.9 | The Na+ Efflux-ATPase (ENa) Subfamily |
| 19 | YOL158C | Enb1p | Ferric enterobactin transporter | 2.53–6 | 4–6.01 | 1–3 | 2–3 | 12 | PM, END | Q08299 | 2.A.1.16 | The Siderophore-Iron Transporter (SIT) Subfamily |
| 20 | YDR261C | Exg2p | Exo-1,3-beta-glucanase | 5.52–8.09 | x | 2–4 | x | anchor | PM | P52911 | ||
| 21 | YMR058W | Fet3p | Oxidoreductase required for high-affinity iron uptake | 4.7–8.44 | 2–4.52 | 2–4 | 1–2 | 1 | PM | P38993 | 9.A.10.1 | The Oxidase-dependent Fe2+ Transporter (OFeT-1) Type 1 Subfamily |
| 22 | YMR319C | Fet4p | Low-affinity Fe2+ transporter | 2.01–7.8 | 2.68–8* | 1–3 | 1–4 | 7 | PM | P40988 | 9.A.9.1 | The Low-affinity Fe++ Transporter (FeT-1) Type 1 Subfamily |
| 23 | YDL222C | Fmp45p | Uncharacterized protein of SUR7 family | 6.21–13.56 | x | 3–6 | x | 4 | PM and MT | Q07651 | 9.B.X13 | The Putative Uncharacterized Transport Proteins family X13 |
| 24 | YLL043W | Fps1p | Channel of MIP family, involved in efflux of glycerol | 2* | x | 1 | x | 5 | PM | P23900 | 1.A.8.5 | The Glycerol Efflux Facilitator Subfamily |
| 25 | YLR214W | Fre1p | Ferric and cupric reductase | 18.43–28 | 14.2–21 | 9–14 | 7–10 | 7 | PM | P32791 | 10.F.1.1 | The Iron Reductase Type 1 Subfamily |
| 26 | YOL152W | Fre7p | Putative ferric reductase | 4.92–8* | 2.02–6* | 2–4 | 1–3 | 8 | PM | Q12333 | 10.F.1.2 | The Iron Reductase Type 2 Subfamily |
| 27 | YER145C | Ftr1p | High affinity iron transporter | 8.77–15.8 | x | 4–7 | x | 7 | PM | P40088 | 9.A.10.1 | The Oxidase-dependent Fe2+ Transporter (OFeT-1) Type 1 Subfamily |
| 28 | YBL042C | Fui1p | High affinity uridine transporter | 2.49–6 | 3.2–6 | 1–3 | 1–3 | 10 | PM | P38196 | 2.A.39.3 | The Nucleobase Permease (NCS1–3) Type 3 |
| 29 | YMR307W | Gas1p | 1,3-beta-glucanosyltransferase | 31.76–35.26 | 19.72–36.02 | 15–17 | 9–18 | anchor | PM | P22146 | ||
| 30 | YMR215W | Gas3p | 1,3-beta-glucanosyltransferase | 15.52–19.26 | 9.7–16.32 | 7–9 | 4–8 | anchor | PM | Q03655 | ||
| 31 | YOL030W | Gas5p | 1,3-beta-glucanosyltransferase | 16–18 | 10–12 | 8–9 | 5–6 | anchor | PM | Q08193 | ||
| 32 | YLR342W | Gls1p/Fks1p | Catalytic subunit of 1,3-beta-D-glucan synthase | 97.71–101.38 | 71.87–90.89 | 48–50 | 35–45 | 13 | PM and CS | P38631 | 10.C.2.2 | The Beta-1,3-glucan Synthesis Subfamily |
| 33 | YHR005C | Gpa1p | GTP-binding subunit of the heterotrimeric G protein | 14.82–20.02 | 8.9–10 | 7–10 | 4–5 | anchor | PM | P08539 | ||
| 34 | YER020W | Gpa2p | Nucleotide binding alpha subunit of the heterotrimeric G protein | 8.29–16.98 | 4.42–6 | 4–8 | 2–3 | PM and MT | P10823 | |||
| 35 | YGL084C | Gup1p | Plasma membrane protein involved in remodeling GPI anchors | 3.84–5.97 | 3.7–6* | 1–2 | 1–3 | 10 | PM and ER | P53154 | 10.A.1.4 | The Diacylglycerol O-acyltransferase Subfamily |
| 36 | YGR191W | Hip1p | High-affinity hitidine transporter | x | 2.09–2.32 | x | 1 | 12 | PM | P06775 | 2.A.3.10 | The Yeast Amino Acid Transporter (YAT) Subfamily |
| 37 | YHR094C | Hxt1p | low-affinity glucose transporter | 8.54–12 | 2.75–4.43 | 4–6 | 1–2 | 12 | PM | P32465 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 38 | YMR011W | Hxt2p | high-affinity glucose transporter | 2.22–8.18 | 4–8 | 1–4 | 2–4 | 12 | PM | P23585 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 39 | YDR345C | Hxt3p | low-affinity glucose transporter | 28.16–30.54 | 17.7–26 | 14–15 | 8–13 | 12 | PM | P32466 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 40 | YHR092C | Hxt4p | High-affinity glucose transporter | 22.04–26.04 | 18.1–20 | 11–13 | 9–10 | 12 | PM | P32467 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 41 | YHR096C | Hxt5p | Glucose transporter | 8.31–10.12 | x | 4–5 | x | 12 | PM | P38695 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 42 | YDR343C | Hxt6p | High-affinity glucose transporter | 2* | x | 1 | x | 12 | PM | P39003 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 43 | YDR342C | Hxt7p | Glucose transporter | 38.51–49.98 | 18.62–24.5* | 19–24 | 9–12 | 12 | PM | P39004 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 44 | YBR086C | Ist2p | Increased sodium tolerance protein | 12.34–22.07 | 8–32.8 | 6–11 | 4–16 | 8 | PM | P38250 | 1.A.X1.Y1 | Undetermined Subfamily |
| 45 | YHR135C | KC11/Yck1p | Casein kinase I isoform | 8–30.04 | 10.07–12* | 4–15 | 5–6 | anchor | PM | P23291 | ||
| 46 | YNL154C | KC12/Yck2p | Casein kinase I isoform | 16.02–28.62 | 18.05–22.86 | 8–14 | 9–11 | anchor | PM | P23292 | ||
| 47 | YDR122W | Kin1p | Serine/threonine protein kinase involved in regulation of exocytosis | 2.11–4 | x | 1–2 | x | peripheral | PM | P13185 | ||
| 48 | YLR096W | Kin2p | Serine/threonine protein kinase involved in regulation of exocytosis | 2.01–8.02 | x | 1–4 | x | peripheral | PM | P13186 | ||
| 49 | YNL323W | Lem3p | Alkylphosphocholine resistance protein | 5.52–7.04 | 2.66–3.12* | 2–3 | 1 | 2 | PM and ER | P42838 | ||
| 50 | YPL004C | Lsp1p | Primary component of eisosomes | 6.01–12.01 | 4.89–8 | 3–6 | 2–4 | PM. EIS | Q12230 | |||
| 51 | YNL142W | Mep2p | Ammonium transporter | 2–4* | 2 | 1–2 | 1 | 11 | PM | P41948 | 2.A.49.3 | The Ammonium Transporter (AmT-3) Type 3 Subfamily |
| 52 | YPR138C | Mep3p | Ammonium transporter | 2.03–4 | 2–2.02* | 1–2 | 1 | 11 | PM | P53390 | 2.A.49.3 | The Ammonium Transporter (AmT-3) Type 3 Subfamily |
| 53 | YLR332W | Mid2p | Protein that acts as a sensor for cell wall integrity signaling | 2 | x | 1 | x | 1 | PM | P36027 | ||
| 54 | YDR033W | Mrh1p | Protein of unknown function | 4.49–16.66 | 4.01–13.77 | 2–8 | 2–6 | 7 | PM and MT | Q12117 | 10.G.1.1 | The Heat Shock Type 1 Subfamily |
| 55 | YPR149W | Nce2p | Non-classical export protein 2 | 7.77–11.4 | 5.4–10.01 | 3–5 | 2–5 | 4 | PM and other | Q12207 | 10.D.1.1 | The Non-classical Secretion Subfamily |
| 56 | YLR138W | Nha1p | Na+/H+ antiporter | 2.45–6.62 | x | 1–3 | x | 12 | PM | Q99271 | 2.A.36.4 | The Monovalent Cation:Proton Antiporter-1 (CPA1–4) Type 4 Subfamily |
| 57 | YIR006C | Pan1p | Protein of actin cytoskeleton-regulatory complex | 4.32–8.28 | x | 2–4 | x | peripheral | PM and CS | P32521 | ||
| 58 | YPL058C | Pdr12p | Plasma membrane ATP binding cassette (ABC) transporter | 23.67–28.32 | 15.55–23.18 | 11–14 | 7–11 | 12 | PM | Q02785 | 3.A.1.205 | The Pleiotropic Drug Resistance (PDR) Subfamily |
| 59 | YDR406W | Pdr15p | Plasma membrane ATP binding cassette (ABC) transporter | 15.25–15.71 | 3.4–4 | 7 | 1–2 | 12 | PM | Q04182 | 3.A.1.205 | The Pleiotropic Drug Resistance (PDR) Subfamily |
| 60 | YOR153W | Pdr5p | Plasma membrane ATP binding cassette (ABC) transporter | 86.47–100.26 | 86.84–89.95 | 43–50 | 43–44 | 12 | PM | P33302 | 3.A.1.205 | The Pleiotropic Drug Resistance (PDR) Subfamily |
| 61 | YOL084W | Phm7p | Protein involved in phosphate metabolism | 13.53–20.06 | x | 6–10 | x | 10 | PM and VAC | Q12252 | 9.B.X2 | The Putative Uncharacterized Transport Proteins family X2 |
| 62 | YGR086C | Pil1p | Primary component of eisosomes | 21.52–32 | 18.28–20.14* | 10–16 | 9–10 | PM,EIS | P53252 | |||
| 63 | YMR008C | Plb1p | Phospholipase B | 16.15–12.85 | 6.74–8.79 | 6–8 | 3–4 | anchor | PM | P39105 | ||
| 64 | YGL008C | Pma1p | Plasma membrane H+−ATPase | 117.45–144.33 | 88–104.69 | 58–72 | 44–52 | 10 | PM | P05030 | 3.A.3.3 | The H+-ATPase (PMA) Subfamily |
| 65 | YOR161C | Pns1p | Protein of unknown function | 6–9.52 | x | 3–4 | x | 10 | PM | Q12412 | 9.B.X1 | The Putative Uncharacterized Transport Proteins family X1 |
| 66 | YDR032C | Pst2p | Protein of unknown function | 8.89–13.72 | 8.01–14* | 4–6 | 4–7 | PM and MT | Q12335 | |||
| 67 | YKR093W | Ptr2p | Integral membrane peptide transporter | 20.61–22.46 | 18.26–22.87 | 10–11 | 9–11 | 11 | PM | P32901 | 2.A.17.2 | The Peptide: H+ Symporter Subfamily |
| 68 | YOR101W | Ras1p | GTP-binding protein | 4–8 | 4–6.04 | 2–4 | 2–3 | anchor | PM | P01119 | ||
| 69 | YNL098C | Ras2p | GTP-binding protein | 16–18.96 | 12–14 | 8–9 | 6–7 | anchor | PM | P01120 | ||
| 70 | YOR301W | Rax1p | Protein involved in bud site selection | 6.03–8.17 | 2.17–6 | 3–4 | 1–3 | 3 | PM | Q08760 | 10.H.1.1 | The COS Subfamily |
| 71 | YPR165W | Rho1p | GTPase of the Rho subfamily of Ras-like proteins | 4.01–8.52 | 5.7–8 | 2–4 | 2–4 | anchor | PM and other | P06780 | ||
| 72 | YNL090W | Rho2p | GTPase of the Rho subfamily of Ras-like proteins | 4.03–5.7 | 4* | 2 | 2 | anchor | PM and other | P06781 | ||
| 73 | YIL118W | Rho3p | GTPase of the Rho subfamily of Ras-like proteins | 8–11.36 | 9.4–14 | 4–5 | 4–7 | anchor | PM and other | Q00245 | ||
| 74 | YNL180C | Rho5p | GTPase of the Rho subfamily of Ras-like proteins | 4.02–4 | 2–4.16* | 2 | 1–2 | anchor | PM and NUC | P53879 | ||
| 75 | YOR049C | Rsb1p | Sphingoid long-chain base transporter | 2–3.95 | x | 1 | x | 7 | PM and ER | Q08417 | 9.B.X20 | The Putative Uncharacterized Transport Proteins family X20 |
| 76 | YMR266W | Rsn1 | Protein of unknown function | x | 4–7.63 | x | 2–3 | 11 | PM | Q03516 | 9.B.X2 | The Putative Uncharacterized Transport Proteins family X2 |
| 77 | YKL051W | Sfk1p | Protein of unknown function | 3.45–10.25 | x | 1–5 | x | 6 | PM | P35735 | 9.B.X1 | The Putative Uncharacterized Transport Proteins family X1 |
| 78 | YER118C | Sho1p | Transmembrane osmosensor | 4.82–5.09 | x | 2 | x | 4 | PM | P40073 | 9.B.X1 | The Putative Uncharacterized Transport Proteins family X1 |
| 79 | YBL007C | Sla1p | Actin cytoskeleton-regulatory complex protein | 2.02–3.15 | x | 1 | x | peripheral | PM and END | P32790 | ||
| 80 | YNL243W | Sla2p | Protein involved in endocytosis | 6.11–12.02 | 2.01–4* | 3–6 | 1–2 | 1 | PM and END | P33338 | ||
| 81 | YOR008C | Slg1p | Cell wall integrity and stress response component | 4* | 2* | 2 | 1 | 1 | PM | P54867 | ||
| 82 | YIL105C | Slm1p | Phosphatidylinositol 4,5-bisphosphate-binding protein | 5.4–7.76* | 2.14–4.42 | 2–3 | 1–2 | anchor | PM | P40485 | ||
| 83 | YIL147C | Sln1p | Histidine kinase osmosensor | 2.01–4.32 | 2* | 1–2 | 1 | 2 | PM | P39928 | ||
| 84 | YGR197C | Sng1p | Nitrosoguanidine resistance protein | 3.71–8.01 | 3.7–6 | 1–4 | 1–3 | 6 | PM | P46950 | 9.B.X21 | The Putative Uncharacterized Transport Proteins family X21 |
| 85 | YDR011W | Snq2p | Plasma membrane ATP binding cassette (ABC) transporter | 48.78–74.43 | 46.07–58.63 | 24–37 | 23–29 | 12 | PM | P32568 | 3.A.1.205 | The Pleiotropic Drug Resistance (PDR) Subfamily |
| 86 | YPL232W | Sso1p | Plasma membrane t-SNARE | 8.45–14.1 | 4.08–10.54 | 4–7 | 2–5 | 1 | PM | P32867 | ||
| 87 | YMR183C | Sso2p | Plasma membrane t-SNARE | 18.01–21.4 | 10–16.1 | 9–10 | 5–8 | 1 | PM | P39926 | ||
| 88 | YLR452C | Sst2p | Protein involved in pheromone response pathway | x | 2.26–2.55* | x | 1 | PM | P11972 | |||
| 89 | YDR536W | Stl1p | Glycerol proton symporter | 8.16–12 | x | 4–6 | x | 12 | PM | P39932 | 2.A.1.1 | The Sugar Porter (SP) Subfamily |
| 90 | YLR305C | Stt4p | Phosphatidylinositol-4-kinase | 22.87–34.02 | 8.73–16.01 | 11–17 | 4–8 | PM and MT | P37297 | |||
| 91 | YML052W | Sur7p | Plasma membrane protein that localizes to MCC patches | 12.03–14 | 6.01–8.64 | 6–7 | 3–4 | 4 | PM | P54003 | 9.B.X13 | The Putative Uncharacterized Transport Proteins family X13 |
| 92 | YBR069C | Tat1p | Amino acid transporter | 4.03–7.87 | 2–11.7 | 2–3 | 1–5 | 12 | PM | P38085 | 2.A.3.10 | The Yeast Amino Acid Transporter (YAT) Subfamily |
| 93 | YOR086C | Tcb1p | Lipid-binding protein | 31.19–41.38 | 9.66–24.46 | 15–20 | 4–12 | 1 - 2 | PM and MT | Q12466 | ||
| 94 | YNL087W | Tcb2p | Lipid-binding protein | 13.28–21.28 | 11.85–20.04 | 6–10 | 5–10 | 1 - 2 | PM and MT | P48231 | ||
| 95 | YML072C | Tcb3p | Lipid-binding protein | 32.17–47.72 | ]21.6–41.57 | 16–23 | 10–20 | 1 - 2 | PM and MT | Q03640 | ||
| 96 | YLR004C | Thi73p | Putative carboxylic acid transporter | 4–4.92 | 4–4.02* | 2 | 2 | 12 | PM and ER | Q07904 | 2.A.1.14 | The Anion: Cation Symporter (ACS) Subfamily |
| 97 | YLR237W | Thi7p | Plasma membrane thiamine transporter | 16.76–19.56 | 17.2–20.4 | 8–9 | 8–10 | 12 | PM | Q05998 | 2.A.39.4 | The Nucleobase Permease (NCS1–4) Type 4 |
| 98 | YGR260W | Tna1p | High affinity nicotinic acid transporter | 2–6 | x | 1–3 | x | 12 | PM and MT | P53322 | 2.A.1.14 | The Anion: Cation Symporter (ACS) Subfamily |
| 99 | YLL028W | Tpo1p | Polyamine transporter | 2–6.2 | 2.01–8* | 1–3 | 1–4 | 12 | PM and VAC | Q07824 | 2.A.1.2 | The Drug: H+ Antiporter-1 (12 Spanner) (DHA1) Subfamily |
| 100 | YGR138C | Tpo2p | Polyamine transporter | 4–5.7* | x | 2 | x | 12 | Pm and VAC | P53283 | 2.A.1.2 | The Drug: H+ Antiporter-1 (12 Spanner) (DHA1) Subfamily |
| 101 | YPR156C | Tpo3p | Polyamine transporter | 6–13.1 | 5.7–8.96 | 3–6 | 2–4 | 12 | PM | Q06451 | 2.A.1.2 | The Drug: H+ Antiporter-1 (12 Spanner) (DHA1) Subfamily |
| 102 | YOR273C | Tpo4p | Polyamine transporter | 15.22–19.4 | 6–12.6 | 7–9 | 3–6 | 12 | PM and VAC | Q12256 | 2.A.1.2 | The Drug: H+ Antiporter-1 (12 Spanner) (DHA1) Subfamily |
| 103 | YCR004C | Ycp4p | Protein of unknown function | 18.44–20.27 | 8–16 | 9–10 | 4–8 | PM and MT | P25349 | |||
| 104 | YLR020C | Yeh2p | Plasma membrane steryl ester hydrolase | 2.36–5.17* | x | 1–2 | x | 1 | PM | Q07950 | ||
| 105 | YLR413W | YLR413w | Protein of unknown function | 16.37–22 | 11.7–16.18 | 8–11 | 5–8 | 4 | PM | Q06689 | 10.A.1.2 | The Fatty Acids Elongation Subfamily |
| 106 | YLR414C | YLR414c | Protein of unknown function | 4.01–6 | 4–7.7* | 2–3 | 2–3 | 4 | PM | Q06991 | 10.A.1.2 | The Fatty Acids Elongation Subfamily |
| 107 | YNL194C | YNL194c | Protein of unknown function | 4.42–9.05 | x | 2–4 | x | 4 | PM | P40169 | 9.B.X13 | The Putative Uncharacterized Transport Proteins family X13 |
| 108 | YOL019W | YOL019w | Protein of unknown function | 2.37–10 | 2–6 | 1–5 | 1–3 | 4 | PM and VAC | Q08157 | 9.B.X7 | The Putative Uncharacterized Transport Proteins family X7 |
| 109 | YGR281W | Yor1p | Plasma membrane ATP binding cassette (ABC) transporter | 41.27–54.06 | 28.4–41.5 | 20–27 | 14–20 | 12 | PM | P53049 | 3.A.1.205 | The Pleiotropic Drug Resistance (PDR) Subfamily |
| 110 | YKL126W | Ypk1p | Serine/threonine protein kinase | 4–7.52 | 2.57–4.04* | 2–3 | 1–2 | peripheral | PM and CS | P12688 | ||
| 111 | YGR198W | Ypp1p | Cargo-transport protein involved in endocytosis | 10–12.02 | 2–3.7* | 5–6 | 1 | peripheral | PM and other | P46951 | ||
| 112 | YLR120C | Yps1p | Aspartic protease involved in cell wall biosynthesis | 2–2.92* | x | 1 | x | anchor | PM | P32329 | ||
| 113 | YOL109W | Zeo1p | Peripheral plasma membrane protein | 6–12.04 | 6–10 | 3–6 | 3–5 | peripheral | PM | Q08245 | ||
a The protein identification confidence is expressed as the “Unused Protein Scores,” a measurement of the protein identification confidence taking into account peptides from spectra that have not already been “used” by higher scoring proteins. The two numbers indicate the lowest and highest scores obtained among the three repeats.
| Percent Confidence | ProtScore |
|---|---|
| 99% | 2.0 |
| 95% | 1.3 |
| 90% | 1.0 |
| 66% | 0.47 |
b Number of peptides represents the minimal number of high confidence (99%) peptides used for identification. The two numbers indicate the lowest and highest scores obtained among the three repeats.
c Number of transmembrane domains or the presence of lipid anchor was based on the Yeast Transport Protein Database: http://homes.esat.kuleuven.be/∼sbrohee/ytpdb/index.php/Main_Page and http://www.yeastgenome.org/ (TMHMM transmembrane segments prediction).
d Subcellular localization based on Organell Database: http://organelledb.lsi.umich.edu/, SGD http://www.yeastgenome.org/, and UniProt databases: http://www.uniprot.org/ PM-plasma membrane, VAC-vacuole, CS-cytosol, NUC-nucleus, MT-mitochondrion, ER-endoplasmic reticulum, END-endosome, EIS-eisosome.
e Annotation according to TCDB database (http://www.tcdb.org/).
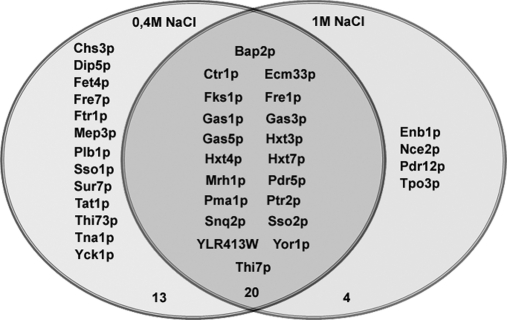
The reporter peaks of the iTRAQ tags in the MS/MS spectra were used for quantification as described in the Materials and Methods. The experiments were repeated three times and the results are the average of three or two significant results. Mild salt stress led to a significant increase in abundance of 12 plasma membrane proteins and a significant decrease in 33 (Table II), whereas high salt stress caused a significant decrease in abundance of 24 plasma membrane proteins and an increase in none (Table III). Twenty plasma membrane proteins showed a decrease in abundance under both conditions, these being the plasma membrane H+-ATPase (Pma1p), three hexose transporters (Hxt3p, Hxt4p, and Hxt7p), three plasma membrane ABC transporters (Pdr5p, Snq2p, and Yor1p), a plasma membrane t-SNARE (Sso2p), one amino acid transporter (Bap2p), a peptide transporter (Ptr2p), the thiamine transporter (Thi7p), four proteins involved in cell wall biogenesis (Gas1p, Gas3p, Gas5p, and Fks1p), two proteins involved in metal ion transport (Fre1p and Ctr1p), and three proteins of unknown function (Mrh1p, Ecm33p, and YLR413W).
Table II. Plasma membrane proteins showing a significant (p < 0,05) change in abundance in response to 0.4 M NaCl treatment. The results are presented as the average ratio in the salt stressed (tag 115 for 10 min, 116 for 30 min and 117 for 90 min) compared to the control (tag 114) sample calculated from three or, if not available, two (*) independent repeats of the experiment. NS: no significant change.
| a) DOWN-REGULATED PROTEINS | |||||
|---|---|---|---|---|---|
| N | Protein name | Description | iTRAQ ratio |
||
| 10 min | 30 min | 90 min | |||
| 1 | Bap2p | Branched-chain amino-acid transporter | 0.73 ± 0.01* | 0.75 ± 0.01* | 0.75 ± 0.02* |
| 2 | Chs3p | Chitin synthase III | 0.69 ± 0.12* | NS | NS |
| 3 | Ctr1p | High-affinity copper transporter | 0.75 ± 0.03* | 0.68 ± 0.06 | 0.66 ± 0.04 |
| 4 | Dip5p | Dicarboxylic amino acid transporter | 0.48 ± 0.16* | NS | NS |
| 5 | Ecm33p | Protein of unknown function | 0.73 ± 0.09 | 0.79 ± 0.04 | 0.83 ± 0.05 |
| 6 | Fet4p | Low-affinity Fe(II) transporter | NS | NS | 0.38 ± 0.11* |
| 7 | Fks1p | Catalytic subunit of 1,3-beta-D-glucan synthase | 0.64 ± 0.07 | 0.76 ± 0.09 | 0.69 ± 0.11 |
| 8 | Fre1p | Ferric and cupric reductase | 0.67 ± 0.09 | 0.61 ± 0.07 | 0.52 ± 0.07 |
| 9 | Fre7p | Putative ferric reductase | NS | 0.60 ± 0.07* | NS |
| 10 | Ftr1p | Plasma membrane Iron transporter | 0.70 ± 0.09 | 0.74 ± 0.11 | 0.63 ± 0.11 |
| 11 | Gas1p | 1,3-beta-glucanosyltransferase | 0.72 ± 0.08 | 0.71 ± 0.05 | 0.75 ± 0.07 |
| 12 | Gas3p | 1,3-beta-glucanosyltransferase | 0.80 ± 0.05* | 0.84 ± 0.05* | 0.76 ± 0.01* |
| 13 | Gas5p | 1,3-beta-glucanosyltransferase | 0.73 ± 0.01* | 0.81 ± 0.07 | NS |
| 14 | Hxt3p | Low-affinity glucose transporter | 0.74 ± 0.02* | 0.69 ± 0.06 | 0.63 ± 0.08 |
| 15 | Hxt4p | High-affinity glucose transporter | 0.8 ± 0.11 | 0.67 ± 0.01 | 0.85 ± 0.05 |
| 16 | Hxt7p | Glucose transporter | NS | 0.79 ± 0.01* | NS |
| 17 | Mep3p | Ammonium transporter | NS | NS | 0.61 ± 0.01* |
| 18 | Mrh1p | Protein of unknown function | 0.73 ± 0.06 | 0.65 ± 0.01 | 0.74 ± 0.15 |
| 19 | Pdr5p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.77 ± 0.09 | 0.76 ± 0.08 | 0.68 ± 0.12 |
| 20 | Plb1p | Lysophospholipase B | 0.75 ± 0.04 | 0.77 ± 0.06 | 0.89 ± 0.04 |
| 21 | Pma1p | Plasma membrane H+-ATPase | 0.69 ± 0.06 | 0.67 ± 0.04 | 0.75 ± 0.08 |
| 22 | Ptr2p | Integral membrane peptide transporter | 0.79 ± 0.02 | 0.59 ± 0.06 | 0.71 ± 0.07 |
| 23 | Snq2p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.84 ± 0.08 | 0.78 ± 0.08 | 0.79 ± 0.04* |
| 24 | Sso1p | Plasma membrane t-SNARE | 0.68 ± 0.03 | 0.67 ± 0.03* | 0.66 ± 0.02* |
| 25 | Sso2p | Plasma membrane t-SNARE | 0.79 ± 0.02 | 0.59 ± 0.06 | 0.71 ± 0.07 |
| 26 | Sur7p | Plasma membrane protein that localizes to MCC patches | 0.80 ± 0.02* | 0.77 ± 0.05 | 0.79 ± 0.07* |
| 27 | Tat1p | Amino acid Transporter | NS | 0.61 ± 0.09 | 0.70 ± 0.03* |
| 28 | Thi73p | Putative carboxylic acid transporter | NS | 0.57 ± 0.04* | 0.72 ± 0.11* |
| 29 | Thi7p | Plasma membrane thiamine transporter | 0.48 ± 0.04 | 0.39 ± 0.03 | 0.54 ± 0.03 |
| 30 | Tna1p | High-affinity nicotinic acid transporter | 0.71 ± 0.03* | NS | NS |
| 31 | Yck1p | Casein kinase I isoform | NS | NS | 0.79 ± 0.1* |
| 32 | YLR413w | Protein of unknown function | 0.52 ± 0.03* | 0.45 ± 0.03 | 0.53 ± 0.06 |
| 33 | Yor1p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.79 ± 0.10 | 0.77 ± 0.04 | 0.82 ± 0.08 |
| b) UP-REGULATED PROTEINS | |||||
|---|---|---|---|---|---|
| N | Protein name | Description | iTRAQ ratio |
||
| 10 min | 30 min | 90 min | |||
| 1 | Fmp45p | Uncharacterized protein of SUR7 family | 2.24 ± 0.09* | 4.37 ± 0.05* | 4.22 ± 0.016* |
| 2 | Hxt1p | Low-affinity glucose transporter | NS | 2.39 ± 0.62 | 2.08 ± 0.49 |
| 3 | Hxt5p | Glucose transporter | NS | 3.52 ± 0.23* | 3.76 ± 0.23 |
| 4 | Ist2p | Increased Sodium Tolerance protein | NS | 1.45 ± 0.22 | 1.52 ± 0.32 |
| 5 | Lsp1p | Primary component of eisosomes | 1.30 ± 0.11* | 1.61 ± 0.05* | 1.43 ± 0.14* |
| 6 | Nce2p | Non-classical export protein 2 | NS | 1.36 ± 0.014* | 1.37 ± 0.11 |
| 7 | Pdr15p | Plasma membrane ATP-binding cassette (ABC) transporter | NS | NS | 1.24 ± 0.09* |
| 8 | Phm7p | protein involved in phosphate metabolism | NS | 2.49 ± 0.38* | 2.14 ± 0.23* |
| 9 | Pil1p | Primary component of eisosomes | NS | 1.73 ± 0.28 | 1.59 ± 0.38* |
| 10 | Pst2p | Protein of unknown function | 1.30 ± 0.21* | NS | 1.33 ± 0.08* |
| 11 | Tcb1p | Lipid-binding protein | NS | 1.35 ± 0.08 | 1.36 ± 0.09 |
| 12 | YNL194c | Protein of unknown function | 1.40 ± 0.16* | 2.41 ± 0.51 | 2.35 ± 0.35 |
Table III. Membrane proteins showing a significant (p < 0.05) change in abundance in response to high 1 M NaCl treatment. The results are presented as the average ratio for the protein in the salt stressed sample (tag 115 for 10 min, 116 for 30 min, and 117 for 90 min) compared to the control sample (tag 114) calculated from three or, if not available, two (*) independent repeats of the experiment. NS: no significant change.
| DOWN-REGULATED PROTEINS | |||||
|---|---|---|---|---|---|
| N | Protein name | Description | iTRAQ ratio |
||
| 10 min | 30 min | 90 min | |||
| 1 | Bap2p | Branched-chain amino-acid transporter | NS | 0.65 ± 0.032* | 0.65 ± 0.02 |
| 2 | Ctr1p | High-affinity copper transporter | 0.69 ± 0.06 | 0.61 ± 0.12* | 0.72 ± 0.01 |
| 3 | Ecm33p | Protein of unknown function | 0.59 ± 0.03 | 0.60 ± 0.06 | 0.58 ± 0.02 |
| 4 | Enb1p | Ferric entrobacin transporter | 0.67 ± 0.08* | NS | NS |
| 5 | Fks1p | Catalytic subunit of 1,3-beta-D-glucan synthase | NS | 0.81 ± 0.065* | 0.72 ± 0.03* |
| 6 | Fre1p | Ferric and cupric reductase | NS | 0.72 ± 0.02 | 0.74 ± 0.09 |
| 7 | Gas1p | 1,3-beta-glucanosyltransferase | 0.61 ± 0.07 | 0.57 ± 0.03 | 0.58 ± 0.05 |
| 8 | Gas3p | 1,3-beta-glucanosyltransferase | 0.62 ± 0.05* | 0.61 ± 0.01* | 0.67 ± 0.07* |
| 9 | Gas5p | 1,3-beta-glucanosyltransferase | 0.71 ± 10* | NS | NS |
| 10 | Hxt3p | Low-affinity glucose transporter | 0.62 ± 0.09* | 0.59 ± 0.04 | 0.62 ± 0.09 |
| 11 | Hxt4p | High-affinity glucose transporter | 0.77 ± 0.003* | 0.69 ± 0.08 | 0.60 ± 0.06 |
| 12 | Hxt7p | Glucose transporter | NS | 0.54 ± 0.06* | NS |
| 13 | Mrh1p | Protein of unknown function | 0.50 ± 0.09 | 0.47 ± 0.03 | 0.48 ± 0.15 |
| 14 | Nce2p | Non-classical export protein 2 | NS | NS | 0.68 ± 0.05* |
| 15 | Pdr12p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.73 ± 0.08* | 0.73 ± 0.13 | 0.74 ± 0.03 |
| 16 | Pdr5p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.72 ± 0.01 | 0.72 ± 0.05 | 0.70 ± 0.05 |
| 17 | Pma1p | Plasma membrane H+-ATPase | 0.62 ± 0.06 | 0.6 ± 0.06 | 0.58 ± 0.13 |
| 18 | Ptr2p | Integral membrane peptide transporter | 0.75 ± 0.05 | 0.76 ± 0.06 | 0.54 ± 0.06 |
| 19 | Snq2p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.85 ± 0.03* | 0.76 ± 0.03 | 0.79 ± 0.04 |
| 20 | Sso2p | Plasma membrane t-SNARE | 0.68 ± 0.03 | 0.66 ± 0.05 | 0.67 ± 0.03 |
| 21 | Thi7p | Plasma membrane thiamine transporter | 0.7 ± 0.12 | 0.57 ± 0.06* | 0.50 ± 0.10 |
| 22 | Tpo3p | Polyamine transporter | 0.57 ± 0.04* | 0.67 ± 0.2* | 0.64 ± 0.07* |
| 23 | YLR413W | Protein of unknown function | 0.64 ± 0.034* | 0.62 ± 0.07 | 0.57 ± 0.04 |
| 24 | Yor1p | Plasma membrane ATP-binding cassette (ABC) transporter | 0.73 ± 0.04* | 0.69 ± 0.03 | 0.73 ± 0.02 |
Mild salt stress (0.4 m) caused a decrease in 13 additional proteins, of which three are involved in iron uptake (Fre7p, Fet4p, and Ftr1p). In the case of high salt stress (1 m), a decrease in four additional proteins was seen (Enb1p, Nce2p, Pdr12p, and Tpo3p) (Fig. 3). In general, the decrease was rapid (10 min after salt stress); however, the amplitude and rate of the decrease varied, depending on the protein.
Fig. 3.
Proteins showing a decrease in abundance after mild and high salt stress.
Mild salt stress led to an increase in abundance of 12 proteins, including six proteins that have been previously described as salt/osmotic stress-responsive proteins, namely Ist2p (increased sodium tolerance protein), two hexose transporters Hxt1p and Hxt5p (6, 32), Fmp45p (6, 33), Pdr15p (13), and YNL194C (5) (Fig. 4). Interestingly, two of the other proteins are involved in endocytosis, these being two components of eisosomes, Pil1p and Lsp1p (Fig. 4) (34). The other four proteins showing an increase were Phm7p, involved in phosphate metabolism, and three proteins, Nce2p, Pst2p, and Tcb1p, of unknown function.
Fig. 4.
Four up-regulated proteins after mild salt stress.
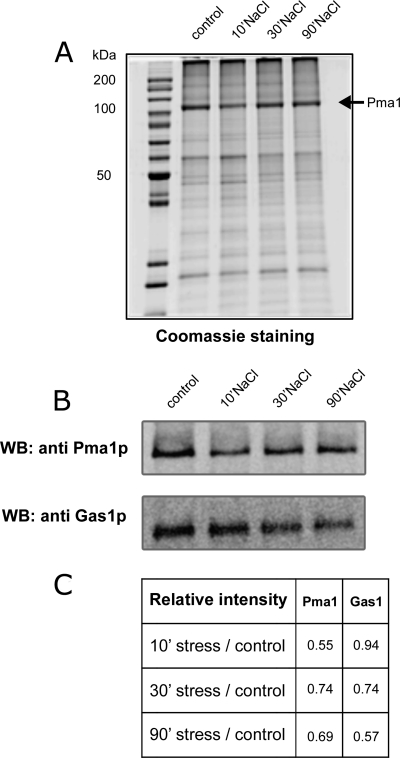
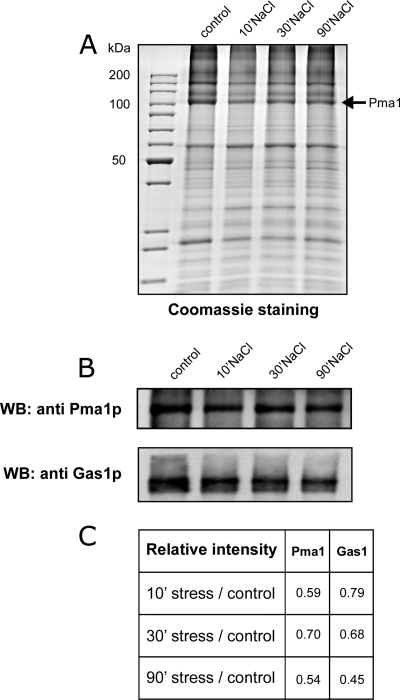
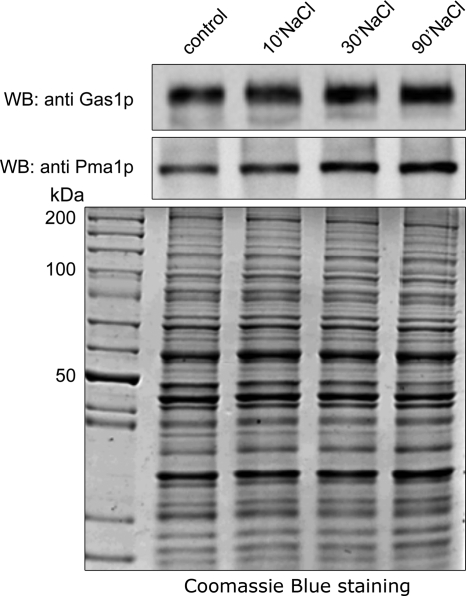
Validation of the quantitative MS results was performed by SDS-PAGE and immunoblotting of purified plasma membranes from 0.4 m (Fig. 5) and 1 m (Fig. 6) NaCl-treated cells with anti-Pma1p and anti-Gas1p antibodies and confirmed the decrease in abundance of both Pma1p and Gas1p (Figs. 5B and 6B). The decrease in abundance of Pma1p was also seen on Coomassie-blue stained SDS-PAGE gels (Figs. 5A and 6A). The decrease of Pma1p was rapid, because it is clearly observed after 10 min of treatment in both conditions. In contrast, the decrease of Gas1p is more progressive (Fig. 5C and 6C). This decrease is because of the salt treatment and not because of cell senescence that could occur during the additional 10, 30, or 90 min incubation. Indeed no modification of abundance was observed for Pma1p or Gas1p when the cells were incubated for 10, 30, or 90 min but without salt treatment (supplemental Fig. S2).
Fig. 5.
Purified plasma membrane analysis after 0. 4 m salt treatment. Ten micrograms of purified plasma membrane proteins (control or treated with 0.4 m NaCl for 10, 30, or 90 min) were separated on a 10% SDS-PAGE gel and visualized by Coomassie Blue staining (A) or immunoblotting using anti-Pma1p and anti-Gas1p antibodies (B). C, This table shows the treated/untreated ratio for Pma1 and Gas1 based on the immunodetection. Similar results were obtained in three independent experiments.
Fig. 6.
Purified plasma membrane analysis after 1 m salt treatment. Ten micrograms of purified plasma membrane proteins (control or treated with 1 m NaCl for 10, 30, or 90 min) were separated on a 10% SDS-PAGE gel and visualized by Coomassie Blue staining (A) or immunoblotting using anti-Pma1p and anti-Gas1p antibodies (B). C,This table shows the treated/untreated ratio for Pma1 and Gas1 based on the immunodetection. Similar results were obtained in three independent experiments.
The rapid decrease in abundance of salt-responsive proteins could be because of endocytosis. Internalization of plasma membrane proteins can lead to either their degradation in the vacuole or their storage in internal vesicles. Quantitative analysis of plasma membrane proteins does not provide any information about the fate of the proteins after internalization. To distinguish between the degradation and storage of internalized proteins, we analyzed the Pma1p and Gas1p content of total cell extracts after salt stress by immunoblotting and found that neither Pma1p nor Gas1p was degraded (Fig. 7). This suggests that these proteins are rapidly internalized following salt stress and stored inside the cell before they are recycled to the plasma membrane or degraded in the vacuole.
Fig. 7.
Immunoblotting for Gas1p and Pma1p in total extracts from control cells and cells treated with 1 m NaCl. Ten micrograms of proteins in a total cell extract of non-stressed and stressed cells (1 m NaCl) were separated on SDS-PAGE and transferred to a PVDF membrane and immunoblotting performed using anti-Gas1p or anti-Pma1p antibodies (top panels). The Coomassie Blue-stained SDS-PAGE gel is shown below as a loading control.
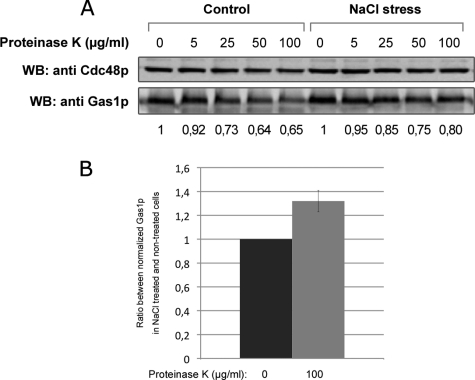
To confirm this hypothesis, we performed a protease protection assay to measure the susceptibility of a plasma membrane protein to be degraded by the addition of a protease in the external medium (29). We decided to follow the degradation of Gas1p because this protein faces the external medium and therefore should be rapidly degraded by external proteases. In addition Gas1p is one of the proteins that responded to salt treatment. Internalization of Gas1p after salt stress should protect the protein against degradation. Salt-treated and nontreated cells have been incubated with increasing amounts of Proteinase K for 30 min and Gas1p signal has been detected from total extract by immunoblotting (Fig. 8). In nontreated cells, we observed the degradation of Gas1p after Proteinase K treatment. This degradation increases with the concentration of protease used. Moreover the protection of the cytosolic protein Cdc48p shows that the degradation occurs well on the surface of the cells and is not because of cell lysis. In salt-treated cells, we observed that a fraction of the signal corresponding to Gas1p is protected against degradation (Fig. 8A). Quantification was performed from three repeats on cells treated with 0 or 100 μg/ml of Proteinase K. Gas1p signal was first normalized with Cdc48p signal and then the ratio between the nontreated and treated cells with protease was calculated. This analysis shows that 30% of Gas1p was protected against degradation in cells treated with NaCl (Fig. 8B). This confirms that Gas1p is internalized after the stress and accumulates inside the cell without being degraded.
Fig. 8.
Protease protection assay on nontreated and salt-treated yeast cells. Yeast cells were grown on YPD and treated (NaCl stress) or not (control) with 0.4 m NaCl for 30 min. Then the cells were incubated with indicated amount of Proteinase K for 30 min. After protease treatment cells were washed, protease inhibitor was added and total extract was prepared. Gas1p was detected by immunodetection (A). For quantification, Gas1p signal was first normalized with Cdc48p signal used as cell integrity and loading control. Numbers represent the decrease of Gas1p signal compared with the conditions with no protease. B, Ratios between normalized numbers obtained in (A) from salt-stressed and control cells at 0 and 100 μg/ml Proteinase K were calculated from three independent experiments. Error bar shows standard deviation.
DISCUSSION
In this study, we used a gel-free proteomic approach for the quantitative profiling of yeast plasma membrane proteins isolated after salt treatment of the cells.
Using a well-established procedure to purify yeast plasma membrane proteins and a gel-free approach to focus on integral membrane proteins, we were able to identify a very high number of yeast plasma membrane proteins compared with previous studies (21, 22). In our study, the purification process coupled to nanoLC-MALDI-TOF/TOF analysis allowed the identification of 113 plasma membrane proteins, of which 68% were integral plasma membrane proteins and 32% were lipid-anchored (18%) or tightly associated with the plasma membrane (14%). Based on two databases (Organelle DB (http://organelledb.lsi.umich.edu) and Saccharomyces genome database (http://www.yeastgenome.org)), we estimate the number of annotated plasma membrane proteins around 240. This means that we could identify almost 50% of the putative plasma membrane proteins. It is likely that the percentage of identified proteins among the proteins really present in the plasma membrane is higher because all genes are not expressed all the time in the cell.
Several proteins exhibited significant changes in abundance after salt treatment. Interestingly 20 plasma membrane proteins showed a reduction in abundance after both 0.4 m and 1 m NaCl treatment. In some cases, the changes in protein levels were moderate, but repeated independent experiments showed a significant decrease in abundance of particular proteins. One of these was the plasma membrane H+-ATPase Pma1p. Pma1p drives secondary active transport and contributes to intracellular pH regulation (35). A decrease in the activity (36) or amount (37) of Pma1p because of mutation results in a reduction in the membrane potential and a concomitant decrease in the uptake of toxic sodium cations. Although Pma1p is a very stable protein, it can be rapidly internalized under certain circumstances, such as perturbation of the lipid composition of the plasma membrane (38) or in the case of some mutations, which provoke its rapid endocytosis (39–40). In previous studies, quantification of mRNAs showed that Pma1p mRNA levels were not modified following 1 m NaCl stress (6, 13). Our data suggest that salt stress modifies the global organization of the plasma membrane, resulting in rapid internalization of Pma1p. Decreased levels of Pma1p in the plasma membrane after salt stress could be very favorable for cells exposed to toxic sodium ions. Reduced amino acid uptake is a general phenomenon observed in yeast cells grown in the presence of salt (4) and one reason for this might be a reduced level of amino acid transporters in the plasma membrane. Here, we observed a decrease in abundance of two transporters, Bap2p, a high-affinity transporter involved in the uptake of leucine, isoleucine, and valine, and Ptr2p, a di- and tripeptide transporter. This result is consistent with a reduced amino acid uptake. Moreover, the decreased uptake might also result from the reduced levels of Pma1p in the plasma membrane, resulting in a weaker proton gradient and less energy for amino acid uptake. Previous data show that, using 1 m NaCl stress, transcription of the ABC transporter PDR5 is induced (6), but its translation is inhibited (13). The long-term effect of this complex regulation has not been analyzed. However, we observe a rapid decrease in Pdr5p and two other ABC transporters, Snq2p and Yor1p, following salt stress. However, another ABC transporter, Pdr15p, showed an increase after 0.4 m NaCl treatment. The role of ABC transporters in salt stress has not been established, but it seems that these proteins are important in this process.
Sodium is not actively imported into yeast cells. The pathways for sodium uptake in yeast probably involve illicit pathways through various plasma membrane transporters, which, together, cause a cation leak. Many yeast transporters involved in the uptake of hexoses, amino acids, ammonium, or cations possess additional unspecific cation transport activity (10). In theory, down-regulation of these transporters could counteract unspecific Na+ uptake. We observed a decrease in abundance of three hexose transporters (Hxt3p, Hxt4p, and Hxt7p), an amino acid transporter (Bap2p), a peptide transporter (Ptr2p), and two proteins involved in metal ion transport (Fre1p and Ctr1p). Previous global mRNA studies (6) did not show any decrease in transcript levels for these proteins; in fact, in some cases, e.g. hexose transporters and Bap2 permease, they showed an increase in transcript levels upon salt stress. However, we can postulate that the first response of the cell to salt stress is a rapid internalization of the major transporters of the plasma membrane, which could attenuate the effect of the stress, and that this is followed by the transcriptional response. We assume that the rapid internalization of plasma membrane proteins is controlled by endocytosis in response to hyperosmotic and/or ionic shock, which might affect the ionic homeostasis of the cell. Immunoblotting for Pma1p and Gas1p in total cell lysates showed similar levels in salt-stressed and control cells. Moreover, the protease protection assay shows that Gas1p is partially protected from external proteases after salt-treatment. The same experiment shows that Gas1p is not rapidly degraded by endogenous proteases after its internalization. This suggests that, after endocytosis, important plasma membrane proteins accumulate inside the cell, probably in recycling endosomes. We speculate that this internal pool of proteins could be recycled to the plasma membrane after stress release or after adaptation of the cells to the stress. Alternatively, the internalized proteins could be degraded later if necessary.
This rapid response could help the cell to survive before the transcriptional response takes place. Currently, we do not know whether the observed effect occurs as a consequence of a regulated process. Two facts support this hypothesis: (1) we observed a very rapid (10 min of stress) decrease in protein levels that involved only a subset of plasma membrane proteins and (2) previous studies have shown no transcriptional effect for these genes (6). On the other hand, salt-stressed cells encounter combined stress because of sodium toxicity and changes in water potential (1). Osmotic shock can cause morphological deformation of the yeast cell surface (41) which could induce membrane internalization. Dupont and coworkers (42) investigated the effect of osmotic changes on the plasma membrane and showed that hyperosmotic shock induced by high concentrations of glycerol leads to morphologic changes in the plasma membrane, such as the formation of plasma membrane invaginations, followed by membrane internalization. It is possible that the high salt concentration might have influenced plasma membrane morphology, thus inducing internalization. The mechanism responsible for the observed changes remains to be investigated.
Mild salt stress led to up-regulation of several plasma membrane proteins that have been previously described as salt or osmotic stress-responsive proteins, these being Ist2p (increased sodium tolerance protein), Pdr15p (13), the hexose transporters Hxt1p and Hxt5p (6, 32), and two proteins belonging to the Sur7-family, Fmp45p (33) and YNL194C (5). Interestingly, among the up-regulated proteins, we found proteins involved in endocytosis, namely Pil1p and Lsp1p, two components of yeast eisosomes. Eisosomes are immobile protein assemblies involved in endocytosis (34) and their up-regulation may support our hypothesis of protein internalization upon mild salt stress. Other up-regulated proteins were a phosphate metabolism protein, Phm7p, and three proteins of unknown function, Nce2p, Pst2p, and Tcb1p. Curiously, we did not find a change in levels of the salt-responsive sodium pump Ena1p in our analysis. The protein was identified in two repetitions of the 0.4 m NaCl stress and one of the 1 m NaCl stress experiment; however, a significant change in abundance was only observed once, which was not sufficient to be retained according to our criterion that a significant difference should be detected in at least two independent repeats. One explanation is that the strain W303 used in this study has four ENA genes coding for highly similar proteins (>97% similarity) (43) and not all of them are up-regulated upon salt stress. The peptides that were used for identification and quantification of the ENA proteins in our study are all common to all four ENA proteins and it was therefore not possible to distinguish ENA1 from the other ENA proteins. The up-regulation of ENA1 could thus be masked by the presence of the other ENA isoforms, which are constitutively expressed (43, 44).
Finally, it should be noted that we did not observe any increase in plasma membrane protein levels after 1 m NaCl treatment and this could be because of a high concentration of salt causing a delayed transcriptional response compared with mild stress (5) and thus to a delayed increase in protein levels.
In conclusion, proteomics based on the chemical tagging of highly purified yeast plasma membranes is a powerful tool that allows the monitoring of the plasma membrane proteome under different conditions. This approach avoids the limitations of traditional gel-based techniques and allows the identification and quantification of a high number of hydrophobic proteins. Quantitative analysis of the plasma membrane proteome of yeast cells treated with mild and severe salt stress revealed numerous significant changes in protein levels. The low salt concentration causes up-regulation of several plasma membrane proteins, including already known stress-responsive proteins and new targets. Interestingly, both salt treatments (0.4 m and 1 m) led to the down-regulation of particular plasma membrane proteins. We postulate that salt stress can cause enhanced endocytosis of membrane proteins in response to hyper-osmotic and/or ionic shock.
Acknowledgments
We thank Howard Riezman for providing the anti-Gas1p antibodies, Michel Ghislain for anti-Cdc48p antibodies, André Goffeau and all members of our group for support and the critical reading of the manuscript. AS and JFH are Research Fellows at the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (Belgium).
Footnotes
* This work was funded by the Interuniversity Attraction Poles Program-Belgian Science Policy, the Communauté française de Belgique-Actions de Recherches Concertées (grant #ARC-0510-329), the Région wallonne-DGTRE, and the FRS-FNRS.
 This article contains supplemental Figs. S1 to S3.
This article contains supplemental Figs. S1 to S3.
1 The abbreviations used are:
- iTRAQ
- isobaric Tags for Relative and Absolute Quantitation
- TEAB
- triethyl ammonium bicarbonate
- TFA
- trifluoroacetic acid
- ACN
- acetonitrile
- PMSF
- phenylmethylsulfonyl fluoride.
REFERENCES
- 1. Hohmann S. (2002) Osmotic stress signaling and osmoadaptation in Yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murguia J. R., Bellés J. M., Serrano R. (1995) A Salt-Sensitive 3′(2′),5′-Bisphosphate Nucleotidase Involved in Sulfate Activation. Science 267, 232–234 [DOI] [PubMed] [Google Scholar]
- 3. Murguia J. R., Bellés J. M., Serrano R. (1996) The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 271, 29029–29033 [DOI] [PubMed] [Google Scholar]
- 4. Norbeck J., Blomberg A. (1998) Amino acid uptake is strongly affected during exponential growth of Saccharomyces cerevisiae in 0.7 M NaCl medium. Fems. Microbiol. Lett. 158, 121–126 [DOI] [PubMed] [Google Scholar]
- 5. Posas F., Chambers J. R., Heyman J. A., Hoeffler J. P., de Nadal E., Ariño J. (2000) The transcriptional response of yeast to saline stress. J. Biol. Chem. 275, 17249–17255 [DOI] [PubMed] [Google Scholar]
- 6. Yale J., Bohnert H. J. (2001) Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276, 15996–16007 [DOI] [PubMed] [Google Scholar]
- 7. Attfield P. V. (1997) Stress tolerance: The key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15, 1351–1357 [DOI] [PubMed] [Google Scholar]
- 8. Rios G., Ferrando A., Serrano R. (1997) Mechanisms of salt tolerance conferred by overexpression of the HAL1 gene in Saccharomyces cerevisiae. Yeast 13, 515–528 [DOI] [PubMed] [Google Scholar]
- 9. Mulet J. M., Leube M. P., Kron S. J., Rios G., Fink G. R., Serrano R. (1999) A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol. Cell. Biol. 19, 3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sychrová H. (2004) Yeast as a model organism to study transport and homeostasis of alkali metal cations. Physiol. Res. 53, S91–S98 [PubMed] [Google Scholar]
- 11. Arino J., Ramos J., Sychrovñ H. (2010) Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74, 95–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito H., Tatebayashi K. (2004) Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 136, 267–272 [DOI] [PubMed] [Google Scholar]
- 13. Melamed D., Pnueli L., Arava Y. (2008) Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. Rna 14, 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soufi B., Kelstrup C. D., Stoehr G., Fröhlich F., Walther T. C., Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 15. Bro C., Regenberg B., Lagniel G., Labarre J., Montero-Lomelí M., Nielsen J. (2003) Transcriptional, proteomic, and metabolic responses to lithium in galactose-grown yeast cells. J. Biol. Chem. 278, 32141–32149 [DOI] [PubMed] [Google Scholar]
- 16. Norbeck J., Blomberg A. (1996) Protein expression during exponential growth in 0.7 M NaCl medium of Saccharomyces cerevisiae. Fems. Microbiol. Lett. 137, 1–8 [DOI] [PubMed] [Google Scholar]
- 17. Gori K., Hébraud M., Chambon C., Mortensen H. D., Arneborg N., Jespersen L. (2007) Proteomic changes in Debaryomyces hansenii upon exposure to NaCl stress. Fems. Yeast Res. 7, 293–303 [DOI] [PubMed] [Google Scholar]
- 18. Yin Z. K., Stead D., Walker J., Selway L., Smith D. A., Brown A. J. P., Quinn J. (2009) A proteomic analysis of the salt, cadmium and peroxide stress responses in Candida albicans and the role of the Hog1 stress-activated MAPK in regulating the stress-induced proteome. Proteomics 9, 4686–4703 [DOI] [PubMed] [Google Scholar]
- 19. Szopinska A., Morsomme P. (2010) Quantitative proteomic approaches and their application in the study of yeast stress responses. OMICS 14, 639–649 [DOI] [PubMed] [Google Scholar]
- 20. Hohmann S., Mager H. (2003) Yeast stress response, Springer-Verlag Berlin, Heidelberg [Google Scholar]
- 21. Navarre C., Degand H., Bennett K. L., Crawford J. S., Mortz E., Boutry M. (2002) Subproteomics: Identification of plasma membrane proteins from the yeast Saccharomyces cerevisiae. Proteomics 2, 1706–1714 [DOI] [PubMed] [Google Scholar]
- 22. Delom F., Szponarski W., Sommerer N., Boyer J. C., Bruneau J. M., Rossignol M., Gibrat R. (2006) The plasma membrane proteome of Saccharomyces cerevisiae and its response to the antifungal calcofluor. Proteomics 6, 3029–3039 [DOI] [PubMed] [Google Scholar]
- 23. Insenser M., Nombela C., Molero G., Gil C. (2006) Proteomic analysis of detergent-resistant membranes from Candida albicans. Proteomics 6, S74–81 [DOI] [PubMed] [Google Scholar]
- 24. Alvarez F. J., Douglas L. M., Konopka J. B. (2007) Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell 6, 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cabezón V., Llama-Palacios A., Nombela C., Monteoliva L., Gil C. (2009) Analysis of Candida albicans plasma membrane proteome. Proteomics 9, 4770–4786 [DOI] [PubMed] [Google Scholar]
- 26. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 27. Goffeau A., Dufour J. P. (1988) Plasma membrane ATPase from the yeast Saccharomyces cerevisiae. Methods Enzymol. 157, 528–533 [DOI] [PubMed] [Google Scholar]
- 28. Dufour J. P., Goffeau A. (1978) Solubilization by lysolecithin and purification of the plasma membrane ATPase of the yeast Schizosaccharomyces pombe. J. Biol. Chem. 253, 7026–7032 [PubMed] [Google Scholar]
- 29. Davis N. G., Horecka J. L., Sprague G. F., Jr. (1993) Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Decottignies A., Evain A., Ghislain M. (2004) Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast 21, 127–139 [DOI] [PubMed] [Google Scholar]
- 31. Saier M. H., Jr., Tran C. V., Barabote R. D. (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirayama T., Maeda T., Saito H., Shinozaki K. (1995) Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol. Gen. Genet. 249, 127–138 [DOI] [PubMed] [Google Scholar]
- 33. Runner V. M., Brewster J. L. (2003) A genetic screen for yeast genes induced by sustained osmotic stress. Yeast 20, 913–920 [DOI] [PubMed] [Google Scholar]
- 34. Walther T. C., Brickner J. H., Aguilar P. S., Bernales S., Pantoja C., Walter P. (2006) Eisosomes mark static sites of endocytosis. Nature 439, 998–1003 [DOI] [PubMed] [Google Scholar]
- 35. Lefebvre B., Boutry M., Morsomme P. (2003) The yeast and plant plasma membrane H+ pump ATPase: divergent regulation for the same function. Prog. Nucleic Acid Res. Mol. Biol. 74, 203–237 [DOI] [PubMed] [Google Scholar]
- 36. Perlin D. S., Brown C. L., Haber J. E. (1988) Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 263, 18118–18122 [PubMed] [Google Scholar]
- 37. Vallejo C. G., Serrano R. (1989) Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast 5, 307–319 [DOI] [PubMed] [Google Scholar]
- 38. Wang Q., Chang A. (2002) Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci. U.S.A. 99, 12853–12858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gong X., Chang A. (2001) A mutant plasma membrane ATPase, Pma1–10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. U.S.A. 98, 9104–9109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferreira T., Mason A. B., Pypaert M., Allen K. E., Slayman C. W. (2002) Quality control in the yeast secretory pathway: a misfolded PMA1 H+-ATPase reveals two checkpoints. J. Biol. Chem. 277, 21027–21040 [DOI] [PubMed] [Google Scholar]
- 41. Adya A. K., Canetta E., Walker G. M. (2006) Atomic force microscopic study of the influence of physical stresses on Saccharomyces cerevisiae and Schizosaccharomyces pombe. Fems. Yeast Res. 6, 120–128 [DOI] [PubMed] [Google Scholar]
- 42. Dupont S., Beney L., Ritt J. F., Lherminier J., Gervais P. (2010) Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochim. Biophys. Acta. 1798, 975–985 [DOI] [PubMed] [Google Scholar]
- 43. Garciadeblas B., Rubio F., Quintero F. J., Bañuelos M. A., Haro R., Rodriguez-Navarro A. (1993) Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 236, 363–368 [DOI] [PubMed] [Google Scholar]
- 44. Wieland J., Nitsche A. M., Strayle J., Steiner H., Rudolph H. K. (1995) The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 14, 3870–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]