Abstract
Information about the physical association of proteins is extensively used for studying cellular processes and disease mechanisms. However, complete experimental mapping of the human interactome will remain prohibitively difficult in the near future.
Here we present a map of predicted human protein interactions that distinguishes functional association from physical binding. Our network classifies more than 5 million protein pairs predicting 94,009 new interactions with high confidence. We experimentally tested a subset of these predictions using yeast two-hybrid analysis and affinity purification followed by quantitative mass spectrometry. Thus we identified 462 new protein-protein interactions and confirmed the predictive power of the network. These independent experiments address potential issues of circular reasoning and are a distinctive feature of this work. Analysis of the physical interactome unravels subnetworks mediating between different functional and physical subunits of the cell. Finally, we demonstrate the utility of the network for the analysis of molecular mechanisms of complex diseases by applying it to genome-wide association studies of neurodegenerative diseases. This analysis provides new evidence implying TOMM40 as a factor involved in Alzheimer's disease. The network provides a high-quality resource for the analysis of genomic data sets and genetic association studies in particular. Our interactome is available via the hPRINT web server at: www.print-db.org.
Accurate high-throughput detection of protein-protein interactions is one of the most challenging tasks in the postgenomic era. Availability of such data has become essential for studying biological pathways, molecular evolution, for assessing protein functions based on functional genetics screens, and for studying molecular mechanisms of diseases (1–3). The size of the human physical interactome is predicted to be between 130,000–600,000 interactions (2, 4, 5). High throughput techniques, such as yeast two-hybrid (Y2H)1 (6, 7) or affinity purification followed by mass spectrometry (8, 9) are being used for the large-scale measurement of protein binding. However, those interactions, together with the protein-protein interactions measured through small-scale experiments (10) only cover 52,000 interactions, i.e. less than 25% of the predicted human interactome (11). Computational prediction of protein interactions can fill this gap until the human interactome has been fully explored using experimental techniques (12). In addition, computational prediction can help guiding experimental screening thereby significantly shortening the time needed until reaching (nearly) complete coverage of an interactome (13).
It is important to distinguish databases assembling data and reporting experimentally tested interactions from others that actually predict previously not reported interactions. We call the second type of interactions ‘de novo' predictions, as these interactions have no experimental evidence through assays directly testing for binding (although there might be indirect experimental evidence, e.g. co-expression or common knock-out phenotypes). The class of databases making such de novo prediction can again be subdivided into two subtypes: those predicting functional interactions (14–16) and others predicting physical association (14, 17–20). A functional interaction typically just indicates membership in a common pathway, whereas physical association refers to direct or indirect binding of proteins in a stable or transient complex. Recent work has underlined the importance of distinguishing the prediction of functional from physical association (19–21). Knowing physical associations is important for elucidating the structure of pathways and for understanding molecular mechanisms underlying high-level phenotypes (1, 4, 11). However, only a few existing databases actually make computational predictions of physical associations of human proteins using heterogeneous types of evidence (18–20).
Here we present an approach that integrates heterogeneous biological data in order to predict and distinguish physical from functional interactions. Applying this framework to human data we were able to predict 94,009 new physical associations with high confidence (probability > 0.7, see Results for more details). We termed this map “human predicted protein interactome” (hPRINT) and validated predictions experimentally based on Y2H and AP-MS analyses. Using these complementary technologies we identified 462 new human protein interactions and we validated the high predictive power of our scoring scheme.
Having established the accuracy of hPRINT, we used this interaction map for studying the physical organization of cellular processes with a specific focus on the molecular causes of neurodegenerative diseases. Our assessment of interactions between gene products that are associated with neurodegenerative diseases reveals that hPRINT can be used for prioritizing candidate genes suggested by genome-wide association studies. Using amyotrophic lateral sclerosis (ALS), Alzheimer's and Parkinson's diseases as examples we demonstrate how hPRINT can assist in the reconstruction of molecular mechanisms linking genes to pathologic phenotypes.
EXPERIMENTAL PROCEDURES
Interaction Prediction
Data Sets
For training and testing, we used data from the Human Protein Reference Database (HPRD) (22), the Comprehensive Resource of Mammalian protein complexes (CORUM) (23), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (24). In order to create a data set of physically interacting genes (PHYSET, 72,450 interactions), we selected only in vivo interactions from HPRD, human interactions from CORUM, and binary and complex interactions defined in human KEGG pathways. In addition, we selected high confidence interactions reported in a previous analysis (25) where each interaction is reported in at least two publications (termed CRGhigh). A data set of functionally related but not physically interacting genes (FUNSET, 412,587 interactions) was extracted from KEGG pathways. FUNSET is composed of gene pairs that are in the same pathway but are not physically interacting. Finally we generated a data set of noninteracting gene pairs (NONSET, 331,596 interactions). NONSET consists of random pairs of genes from distinct KEGG pathways that are not known to interact physically. Hence, NONSET represents interactions that are neither functionally related nor physically binding.
Feature Set
We used 18 features to predict interactions. Five types of evidence are taken from the STRING database (version 8.2): genomic neighborhood, gene fusion, phylogenetic profile, coexpression, and text mining (16). Five additional features are generated using the GoGene tool, which annotates genes based on Gene Ontology (GO) terms and disease annotations using text mining information (including co-occurrence in publications) (26). The features extracted with GoGene are: cellular component, molecular function, biological process, disease, co-occurrence. Next, we used presence of known binding motifs in protein sequences as a predictor for physical binding. This feature (named “domain pairs”) is based on the presence of binding domains predicted by profile Hidden Markov Models (27). Finally, we considered the topology of the STRING interaction network to predict physical interactions. We recalculated the STRING combined score after eliminating the experimental and database features in order to exclude any experimental evidence. Using the resulting STRING interaction scores we extracted seven topological features for each edge of this network: clustering coefficient, minimum spanning tree, extended minimum spanning tree, neighborhood ratio, ratio between shortest path and edge weight, local betweenness, and global betweenness. Detailed descriptions for all features can be found in supplementary material.
Training
We performed a three-class classification, namely physical, functional, and nonrelated. All the PHYSET is used as training data for physical interactions. To avoid a bias toward larger classes, we randomly sampled from FUNSET and NONSET to obtain training sets of approximately even size. A Random Forests with 1000 trees was trained (28). Random Forests generates three probabilities summing up to 1 for each edge: probability of being physical (RFphys), probability of being functional (RFfun), and probability of being nonrelated (NON). This analysis was done using the Random Forests package from R (http://www.r-project.org/).
The above Random Forests scores are de novo predictions of interactions because they are not based on any data originating from experimental testing of interactions. In order to integrate prior knowledge of measured interactions we combined the Random Forests scores with experimental lines of evidence using Bayesian integration (implemented in R) as described previously (29). This approach also accounts for correlation between individual lines of evidence.
Evaluation
The different prediction strategies were computationally validated using cross-validation and using independent sets of known interactions. Fivefold cross-validation was performed by randomly sampling training and test sets from the pools of reference interactions. However, cross-validation might overestimate the predictive power of machine learning methods, because it does not take into account systematic differences among independently measured data. Hence, our second strategy hides one data source during the training phase and uses it for testing. Here, we used CRGhigh for independent testing, because it is not commonly used as a training set and so allowing it to be used as an independent test set for comparing all different networks. If a test interaction was reported in another source, it was removed from the training data and only used for testing.
Analysis of Cross Talk between Pathways and Compartments
In order to analyze the cross-talk between pathways we selected all genes annotated for at least one cellular process or environmental information processing pathway in KEGG. We generated a high confidence physical interaction network of these selected genes with interactions having a Random Forests physical interaction score above 0.7. Because many genes are annotated for more than one pathway it is nontrivial to decide if a physical interaction is within or between two pathways. Two different strategies were followed for classifying interactions as “between pathway.” Assume Pg1 and Pg2 are the sets of pathways for which the genes g1 and g2 are annotated. In the first strategy, we call the interaction g1 − g2 ‘between’ if Pg1 ∩ Pg2 = Ø and we added as cross-talk for each pair of pathways in the Cartesian product Pg1 ∩ Pg2. If Pg1 ∩ Pg2 = A ≠ Ø then we treat this as a within interaction and we added contribution as within interaction for each pathway in A. This first approach rests on the assumption that two genes annotated for a common pathway are interacting inside that pathway. However, if genes are also annotated for different pathways the interaction may (in addition) also link those distinct pathways.
Hence, in the second strategy, even if two genes share common pathways, we assumed there is cross-talk between pathways in Pg1 and Pg2. Again we add a contribution as cross-talk for each pair in Pg1 ∩ Pg2. Note, that in contrast to the first strategy, it is possible to have pairs (x, x) in this Cartesian product since Pg1 ∩ Pg2 is not necessarily empty. Such a pair was assumed as within interaction in pathway x. At the end, for each strategy, we generated a N × N matrix showing the cross-talk between N pathways.
We carried out the same analysis for cellular compartments. The only difference is that, instead of KEGG pathways, we used genes that have a cellular localization annotation in the generic version of GO slim (http://www.geneontology.org). Cytoscape was used for drawing the networks (30).
Selecting Genes for Y2H Experiments
Genes potentially related to ALS, Parkinson, Huntington, or Alzheimer diseases were selected using three data sources, Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/omim/, downloaded 28/10/10), KEGG (24), and Genetic Association Database (GAD) (31). From OMIM we selected genes that are known to be related with these diseases; for achieving maximal stringency we only selected genes from OMIM class 3: their mutations were positioned by mapping the wild-type gene and a mutation in that gene created a phenotype that is in association with the disorder. GAD contains results from Genome Wide Association Studies (GWAS) and linkage studies. We selected genes from GAD that show positive association with the diseases. From KEGG we selected all genes participating in the respective disease pathways. The union of all of these genes resulted in 433 nonredundant genes (Entrez Gene IDs).
Functional Enrichment
We calculated functional enrichment (based on GO) of genes interacting with known disease associated genes (OMIM) or candidate genes (GWAS) using Fisher's exact test. The purpose was to show that “linker genes” lying between GWAS and OMIM genes are enriched for specific molecular functions that are different from other genes neighboring OMIM genes. Hence, we did not compute the functional enrichment of linker genes versus the whole genome, but versus other neighbors of OMIM genes. Thus, enrichment of linker genes was computed using as universe not the whole genome but the whole set of OMIM or GWAS gene interactors respectively (supplemental Tables S6, S7). However, using the whole genome as a universe yields similar findings especially in case of the OMIM interactors (supplemental Table S8).
Central Nervous System (CNS) Specificity
CNS specificity for each of the interactions is calculated via applying the Kolmogorow-Smirnov (KS) test. mRNA expression levels in various human tissues were collected from BIOGPS. For each of the 12,056 genes present in the BIOGPS we compared expression in CNS tissues and cell types versus all other tissues using the KS test. Interactions were scored by assigning the lowest p value of the two interacting genes to the edge. This is because of the fact that an interaction is present in a specific tissue only if both partners are expressed, hence it is restricted on the less promiscuous gene.
Experimental Testing of Protein Associations
Y2H
Y2H experiments were performed as described previously (7). In Brief, selected ORFs were transferred into bait (pBTM117c) and prey vectors (pACT4-DM). The L40ccU2 MATa yeast strain was transformed with the bait plasmids and preys were used to transform MATalpha strain L40ccα (32). Bait and prey yeast strains were pair wise ordered in mircotiter plate format according to hPRINT predictions and mated on YPD for 36 h. Diploid yeast were grown on S.D. media supplemented with histidine and uracil for 3 days. Interacting proteins were identified by growth on selective plates (-Leu-Trp-Ura-His) after 6 days. Random noninteracting pairs were tested by mating nonpair wise matching bait and prey plates. Every protein pair was assayed in at least two independent interaction mating experiments.
Cell Line Production
Mouse or human BAC harboring the genes of interest were obtained from the BACPAC Resources Center (http://bacpac.chori.org). The N-terminal NFLAP tagging cassette as well as the C-terminal LAP and DIGtag tagging cassettes were PCR amplified using primers that carry 50 nucleotides of homology to the N- or C terminus, respectively, of each of the target genes. Recombineering and stable transfection of the modified BAC was performed as described (33). Briefly, both, a plasmid carrying two recombinases and the purified tagging cassette, were introduced into the E. coli strain containing the BAC vector using electroporation. Precise incorporation of the tagging cassette was confirmed by PCR and sequencing. Next, the GFP-tagged BACs were isolated from bacteria using the Nucleobond PC100 kit (Macherey-Nagel, Germany).
Subsequently, HeLa Kyoto cells were transfected using Effectene (Qiagen, Dorking, Surrey, UK) and cultivated in selection media containing 400 μg/ml geneticin (G418, Invitrogen, Carlsbad, CA). Finally, HeLa pools stably expressing the tagged transgenes were analyzed by Western blot and immunofluorescence using an anti-GFP antibody (Roche) to verify correct protein size and localization of the tagged transgene. Next, cell pools were subject to analysis using mass spectrometry (8).
Affinity Purification, Mass-Spectrometry Protein Identification (AP-MS)
AP-MS was performed according to the recently published QUBIC (Quantitative BAC InteraCtomics) method (8). In short, pulldowns of GFP-tagged, transgenic cell line and of an untransfected control cell line were done in triplicates using monoclonal anti-GFP antibody coupled to μMACS beads (Miltenyi Biotec). Purified proteins were digested in-column and purified peptides were directly subjected to liquid chromatography tandem MS (LC-MS/MS) analysis using a Proxeon EASY-nLC system coupled online to a LTQ-Orbitrap. Raw data was analyzed using the MaxQuant Software (version 1.1.0.28) with label-free protein quantification (34). Significant interactors were determined by a volcano plot-based strategy, combining p values of the standard equal group variance t test with ratios comprised from protein intensities in the pulldowns of the transgenic and the control cell line. MaxQuant settings and significance cut-offs were chosen as described in (8).
RESULTS
Predicting the Human Physical Interactome
For predicting the human protein-protein interactions we developed a novel combined Random Forests/Bayesian learning strategy. First, we integrated information from automated text mining with comparative and functional genomics data, protein domain profiles, and network features resulting in a total of 18 features (Fig. 1, supplemental Table S1). This data was generated in-house (26, 27) and obtained from the STRING database (35). Because we aimed at the de novo prediction of binding experimental data reporting direct evidence for physical protein association was excluded at this step. Experimental binding data was however integrated at a later step for further improving the coverage and accuracy of the interaction map (see Fig. 1A and Experimental Procedures). We generated independent sets of positive reference interactions based on four high-confidence sources (see Experimental Procedures). All subsequent steps were tested independently on these positive reference sets in order to ensure generality of our findings. Random interactions between proteins that were part of the positive reference sets were used as a negative reference set. We employed the Random Forests supervised learning algorithm (28) for integrating the features and predicting interactions. An important feature of our method is the simultaneous classification of three types of protein pairs: physical binding (RFphys), functional association (RFfun), and nonrelated, i.e. pairs of proteins that likely do not interact. These scores reflect the probability for membership in the respective class. RFfun reflects the probability that an interaction is functional but not physical, whereas physical binding (high RFphys) does not preclude functional association. Note that 1 – (RFfun + RFphys) is the probability that the respective protein pair does not interact at all. Using our pipeline we tested more than 5 million protein pairs. hPRINT predicts 94,009 new interactions (RFphys > 0.7) that have no prior experimental evidence in any of the databases that we included. We created a web-interface for hPRINT at www.print-db.org, allowing to search the database and to download the data.
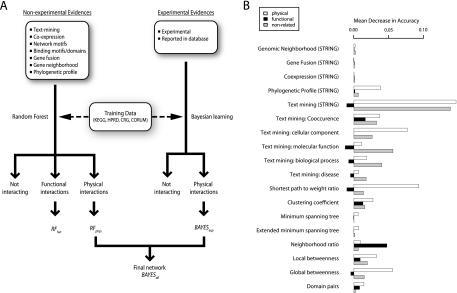
Fig. 1.
A, Workflow and B, feature importance for predicting interactions. In a first step Random Forests physical and functional interactions scores are estimated excluding direct experimental evidence. Features based on experimental evidence are weighted using a Bayesian approach and respective log-likelihood scores are computed. In a second step Random Forests physical (RFphys) and log-likelihood scores from experimental interactions are combined through Bayesian learning in order to give a final score for each interaction. B, Feature importance for Random Forests machine learning expressed as the mean decrease of accuracy when shuffling the respective line of evidence. Some features are predictive but have low feature importance because of low coverage (e.g. Gene Fusion).
Evaluating hPRINT
Based on the positive and negative reference interactions we subjected hPRINT to a range of tests. In addition to cross-validation, we assessed predictions based on test sets obtained from independent sources. This approach ensures that the performance assessment is independent of specificities of the training or test data. First, we compared our approach to other machine learning methods (Fig. 2A and supplemental Figs. S1A, S2A, S3A). Random Forests clearly outperformed all other methods tested, which is consistent with previous studies (21, 36, 37). Next, we compared four published networks and hPRINT in their ability of predicting physical association of human proteins (Fig. 2B–E and supplemental Figs. S1B, S2B, S3B). hPRINT performed consistently better than previous approaches. In order to show that these differences are statistically significant, we performed fivefold cross validation, computing each time the area under the ROC curve (AUC). This provided us with distributions of AUC scores that we compared between hPRINT and STRING (which has the largest overlap with the test set among all competing databases). It turned out that the AUCs of hPRINT are significantly larger than those of STRING (t test, p = 6.7 × 10−07) (supplemental Table S2). In order to underline the importance of distinguishing physical from functional association we also tested if RFfun could predict known physical binding events (Fig. 2B): whereas RFfun is predictive for physical association, it performs much worse than RFphys.
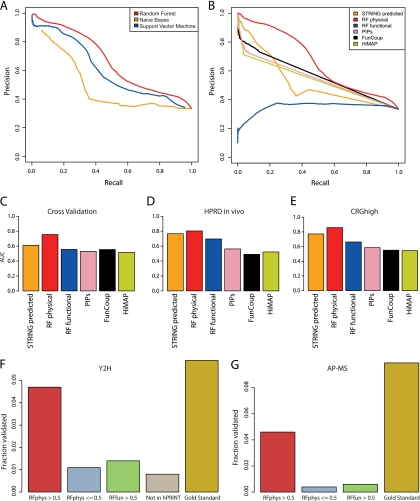
Fig. 2.
Evaluation of hPRINT. A, Precision-Recall curves for comparing Random Forest with naïve Bayesian prediction and Support Vector Machines (SVM). Radial basis factor was used as kernel for training the SVM. B, Precision-Recall curves for comparing Random Forests physical and functional scores with other published networks. The aim of the Random Forests machine learning was the de novo prediction of new interactions; hence, experimental interaction measurements were ignored. In the comparison we also removed experimental evidence from the other data sets. This was also necessary to avoid circular reasoning. Panels (A) and (B) are both based on fivefold cross validation using functional and noninteracting gene pairs (FUNSET and NONSET) together as the negative data set. Supplemental Fig. S1 shows equivalent plots when using other combinations of training and test sets. (C–E) Area Under the ROC Curve (AUC) for (C) fivefold cross validation, (D) using HPRD as an independent test set, E, using CRGhigh as an independent test set. The AUC of RFphys is significantly larger than in all the other cases (supplemental Fig. S2; supplemental Table S3). F, G, Experimental validation using yeast two-hybrid (F) and AP-MS (G). High scoring interactions (RFphys > 0.5) could be confirmed with much higher probability than interactions without any evidence (“Not in hPRINT”), low scoring interactions (RFphys < = 0.5), and interactions predicted to be only functional (RFfun > 0.5). The comparison with the reference set (“Gold Standard”) is a measure of the sensitivity of the assays. Note that in case of AP-MS we cannot define a “Not in hPRINT” set. See Supplementary Material for additional analyses of the experimental testing.
It has recently been proposed that predicted physical interactomes can be used for streamlining the experimental mapping of interactions (13). To test this hypothesis with human proteins and to further corroborate the reliability of hPRINT we conducted experimental testing of predicted interactions using Y2H and AP-MS. For Y2H we selected 433 proteins that are known to be related to at least one of four neurodegenerative diseases (ALS, Parkinson's, Huntington, and Alzheimer's, see Experimental Procedures for details). After removing proteins for which clones were not available in our library or which were autoactive we were left with 281 proteins, giving rise to almost 40,000 possible pairs. Of these we tested 5434 at least twice. These interactions consist of 548 pairs with RFphys scores above 0.5, 3010 had no evidence in hPRINT, and the remaining ones have RFphys scores below 0.5. Also, this set contained 295 interactions from our positive control set, which we used for assessing the sensitivity or retest rate of the assay. Thus, our experimental test set contains various controls all based on the same 281 proteins (i.e. thereby controlling for potential protein set specific biases). We reproducibly detected 81 interactions (54 present in hPRINT), most of which were not reported before. Validation rates are substantially better for high-scoring interactions compared with the negative controls (Fig. 2F). The experimentally validated interactions have significantly higher RFphys scores compared with RFfun (KS test, p = 0.0016) and random interactions (KS test, p = 2.45·10−12). This is also true for cut-off values different than 0.5. supplemental Figs. S4 and S5 show that the predictive power increases as a function of the interaction score. Other databases also performed better than random in predicting the Y2H interactions; however, the predictive power was below that of RFphys (STRING: p = 1.25·10−09, PIP: p = 0.615, HiMAP and FunCoup had too small overlap with the experimentally validated interactions to allow for a quantitative assessment). Hence, using hPRINT we can significantly increase the success rate for interaction screening as compared with random testing of interactions.
Next, we performed AP-MS experiments using 14 proteins with neurological relevance as baits. For these baits hPRINT predicted in total 43 interactions with a RFphys score above 0.5. In case of the AP-MS measurements the set of tested interactions was defined as the set of all predicted interactions with the respective bait protein. Between 1 and 181 proteins were copurified per bait, resulting in a total of 462 interactions (92 present in hPRINT). Again, validation rates are much higher for RFphys than for the negative controls (Fig. 2G), RFphys scores of validated interactions are significantly higher than random (p = 2.2 × 10−7) and higher than RFfun scores (p = 3.05 × 10−7, supplemental Table S3). We also tested how well other databases could predict the experimentally verified interactions. Similar to what we observed with the Y2H test set, the comparison with other databases using the AP-MS test set shows that hPRINT performs best (supplemental Table S3, supplemental Figs. S4 and S6). Benchmarking our predictions against another recently published set of AP-MS measurements (38) yields similar results (supplemental Fig. S7).
Predicted Interactome Covers Many Underexplored Genes
Most existing measurements of protein-protein interactions are biased toward well-studied genes and even high-throughput screens may be biased because of the selection of bait proteins (39, 40). One goal of this study was to at least partly fill this gap by predicting interactions for less well-studied genes. In order to assess the bias toward well-studied genes, genes were grouped based on their citation frequency in PubMed abstracts. Fig. 3 shows the number of interactions as a function of “gene popularity.” Experimentally verified interactions (reported in HPRD, KEGG, CORUM, CRGHigh, and IntAct) are biased toward well-studied genes, whereas in hPRINT this bias is much less pronounced. hPRINT not only predicts new interactions among already well-studied genes for which an abundance of information is already available. Thus, the input data used is less dependent on gene-popularity and our prediction method effectively uses this information. The importance of text mining derived features in our predictions (Fig. 1B) might suggest that our network should be subject to the same bias as experimental data sets. However, our text mining based features are normalized for the number of citations (26), which partly balances the bias against less studied genes. Additionally, our network is utilizing unbiased information such as co-expression or protein sequence, which is available for virtually all gene pairs. In conclusion our network predicts interactions for largely unexplored parts of the genome.
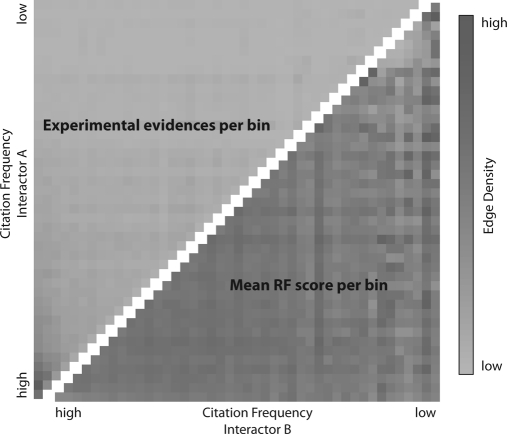
Fig. 3.
Publication Bias. The gene name citation frequency in PubMed abstracts is shown along both axes and genes were grouped in even-sized bins. The color in each grid cell encodes the number of interactions connecting the respective gene products in the two corresponding bins. The upper triangle shows the number of experimentally tested interactions per bin pair; the lower triangle shows the number of predicted interactions in hPRINT (mean RF score per bin). The predicted interactome covers the human genome much more evenly than the known (experimentally tested) interactions, which are heavily biased toward well-studied genes (bottom left corner).
Networks Linking Cellular Processes and Signaling Pathways
Recently it has been noted that viewing signaling pathways as isolated linear chains of reactions may be misleading. Many pathways are in fact interconnected, i.e. signaling pathways are linked to other regulatory or signaling pathways and to basic cellular processes such as endocytosis (41, 42). It is emerging that cells are using highly connected networks to integrate a wide variety of noisy signals, for predicting future conditions in the environment and ultimately for balancing partly conflicting cues to make decisions (43, 44). Having a substantially more comprehensive and less biased map of the human physical interactome allows us to re-examine the degree to which proteins interact within a specific pathway and across pathways. In order to quantify the extent of interpathway connectivity we measured the fraction of interactions bridging different pathways (Fig. 4A, supplemental Fig. S8). Likewise, we quantified the fraction of interactions connecting different cellular compartments (Fig. 4B). Interactions between proteins annotated for different cellular localizations could be either because of binding at interfaces or because of multiple protein localizations. In the latter case, interactions in fact do not “bridge” compartments, but they rather reflect the dynamics of protein (re-)localization. Fig. 4 clearly shows that the fraction of interactions connecting cellular localizations is much larger than the fraction of interactions bridging pathways. Although 50% of the interactions link proteins at different localizations, 29% of the interactions connect proteins annotated for different pathways. This observation reflects the fact that most pathways span several compartments and it shows that the cellular context of proteins is very dynamic. Pathways on the other hand, representing functional subunits of the proteome, are less densely connected between each other. Still, the fact that almost one third of all interactions are inter- rather than intrapathway suggests considerable interconnectedness, emphasizing once more that signal processing and decision making in cells are highly interconnected processes operating at the network level.
Fig. 4.
Fraction of physical interactions connecting (A) pathways and (B) cellular locations. Each pair of pathways or cellular locations is connected by an edge reflecting the fraction of interactions from the smaller of the two groups linking proteins in those groups (“between interactions”). The size of each node shows the number of proteins annotated for that group and the nodes' border thickness is proportional to the fraction of interactions connecting proteins inside each group (“within interactions”). Note that the two panels are at scale, i.e. pathways generally have fewer annotated proteins than cellular locations. Edges with scores below 0.14 are not shown for the sake of simplicity. The full networks have many more edges, e.g. there are physical interactions connecting the Wnt-pathway with cell adhesion.
Using hPRINT for Exploring Genes Associated with Neurodegenerative Diseases
GWAS allow for the unbiased detection of disease modifying genes (45–47). Having identified SNPs in or close to a gene from a large population of individuals it is not always apparent what the molecular mechanisms are linking the causal gene to the disease phenotype (47). Physical protein interaction data has proven to be helpful in similar contexts, but applications to GWAS are still limited (46, 48–52). We reasoned that a network with increased coverage would also be of improved utility for studying GWAS candidate genes.
Here we address the important problem of prioritizing candidate genes identified through GWAS. Our hypothesis was that for a given disease, candidate genes whose products are closer in our network to confirmed causal disease genes are likely to have stronger effects on the disease phenotype, i.e. those genes might be more relevant and easier to replicate. For testing this hypothesis we selected the top ranking genes from AlzGene (53), a database offering a publicly available and regularly updated field-synopsis of published genetic association studies performed on Alzheimer's disease (AD). The overall epidemiological credibility of the top genes is graded as “A” (strong, 19 genes), “B” (moderate, 19 genes), and “C” (weak, 44 genes) (53). Next, we obtained a set of high-confidence disease causing genes from OMIM and quantified the distance between candidate genes from AlzGene and known genes from OMIM (distance was defined as the smallest sum of links connecting the respective proteins in hPRINT). Initially, we performed the analysis using all data, i.e. combining predictive and experimental evidence (using the Bayesian scoring, Fig. 1A). In our network AlzGene candidates are significantly closer to disease genes than random genes (Fig. 5A, supplemental Table S4). Also, genes graded A generally had shorter distances to OMIM genes than genes graded B or C (though this difference was not statistically significant). Next, we tested how important the predictive evidence was for correctly ranking the candidate genes. When using experimental information alone the difference between class B and C genes and randomly selected genes vanished and only class “A” genes were still closer to OMIM genes than expected by chance (supplemental Fig. S9A, supplemental Table S4). Another concern might be that the degree of the nodes that we assessed influenced the results (e.g. if a class A gene has a very high degree this might reduce the distance to all genes in the network). To address this problem we randomly re-wired the network maintaining the degree of each node. Such randomization diminished the differences between the gene classes (supplemental Fig. S9B) showing that the differences seen before are not an artifact caused by high node degrees. These findings suggest that network distance in hPRINT can be used for prioritizing candidate genes from GWA studies and that the predicted interactions add disease relevant information to the network. For prioritizing genes linked to three neurodegenerative diseases, we compiled 75 candidate genes for ALS, Alzheimer's, and Parkinson's disease (54), mapped them onto hPRINT (48 out of 75), and ranked them based on their network distance to known disease genes, respectively (supplemental Table S5). In case of AD the top scoring gene was CLU, which ranked second in AlzGene after ApoE.
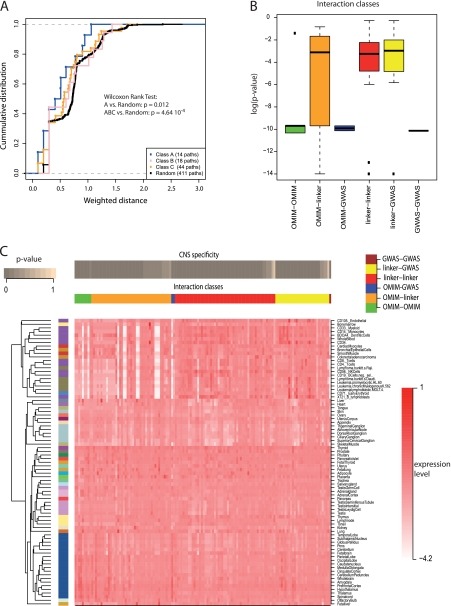
Fig. 5.
Prioritization of Alzheimer's disease candidate genes. A, Empirical cumulative distribution function (ECDF) for the weighted network distance between OMIM genes that are known to cause Alzheimer's and candidate genes with different confidence scores (classes A–C). The distance between class A genes and OMIM genes is generally closer than in case of the other two classes or random genes. The Wilcoxon rank test was used for comparing network distances against distances for random genes. B, Expression specificity in the central nervous system (CNS) quantified as the maximum specificity (minimum p value) of the two interacting genes (see Experimental Procedures for details). Box-plots of CNS specificity are shown for interactions connecting different types of genes (see labels at bottom). CNS specificity is generally higher (low p values) for interactions connecting two disease genes compared with interactions connecting disease genes with other genes. C, Expression specificity of individual interactions across tissues. Each column in the heat map shows the expression of one interaction across all tissues tested. CNS specificity is shown in the top bar. Interactions were grouped in different classes based on the disease classification of the genes (vertical bands, see color code on top). Tissues (rows) were grouped according to the BIOGPS classification (color code on the left). Dendrogram on the left shows clustering of tissues based on expression specificity of the interactions. Resulting groups agree with the BIOGPS classification. Equivalent figures for ALS and Parkinson's are shown in the supplement (supplemental Figs. S10–S13).
The concordance between AlzGene and our network-based analysis is interesting in two respects: AlzGene is also based on an automated ranking of candidate genes. But instead of using network information it ranks genes based on their reproducibility across several genetic linkage and association studies. Hence, we achieve agreement based on complementary data. This implicates first, that our network analysis might be particularly useful for traits with smaller numbers of independent association studies that could be used to confirm candidate genes. And second, the correlation between molecular interactions and reproducibility in association studies suggests that effect size might be a function of molecular proximity to established disease genes.
Linker Genes are Enriched for Common Functions
In order to further corroborate the relevance of genes identified through the network analysis and to obtain first hints toward molecular mechanisms we analyzed the genes and interactions connecting candidate GWAS genes to known disease genes (i.e. genes from OMIM). For each candidate gene we identified its closest known disease causing gene and selected all “linker genes” lying between these two genes in hPRINT. These linker genes are particularly interesting, because they are typically not known to affect disease phenotypes, but they may be important for understanding the disease mechanisms. These linker genes could not easily have been identified without the network information.
We assessed the relevance and consistency of linker genes by measuring the functional enrichment among them based on Gene Ontology (GO) terms. Interestingly, linker genes of all three diseases are enriched for related cellular processes (supplemental Tables S6 and S7). Apoptosis (programmed cell death) and cytoskeleton rearrangements and cargo transportation are two terms that appear frequently among linker genes in all three diseases. These functions are clearly connected to the etiology of neurodegenerative diseases (55), further underlining the potential role of linker genes in the establishment of disease phenotypes.
Interactions Connecting Disease Genes Are CNS Specific
We then calculated the central nervous system (CNS) specificity for disease genes, linker genes, and their interactions based on expression data from BIOGPS (56, 57). The CNS specificity score of interactions is based on the simple notion that both proteins constituting an interaction must be expressed in a given tissue or cell type. Hence, CNS specificity of an interaction is high when a given pair of proteins is expressed in the CNS (see Experimental Procedures).
We noticed that interactions between disease genes (either GWAS or OMIM) are more CNS specific than interactions involving linker genes. (Figs. 5B, 5C, and supplemental Fig. S10–S13). Hence, genes with CNS specific interactions connecting to known disease genes are more likely to be of higher relevance. Supplemental Table S5 lists the top candidate disease genes interacting with known disease genes in a CNS specific manner. Based on this ranking CLU is again predicted to be one of the top candidates for Alzheimer's disease. Also Translocase of outer mitochondrial membrane 40 homolog (TOMM40) ranked highly as an AD candidate gene based on both the shortest path and CNS specificity scores (supplemental Table S5). There has been a debate whether mutations in TOMM40 are actually related with higher risk in developing Alzheimer's disease (58) or whether the correlation of TOMM40 with Alzheimer's is because of linkage disequilibrium (59, 60). More recent work suggests that TOMM40 is indeed involved in AD etiology (61) and our findings support this view.
We also noticed that interactions derived by applying the shortest path algorithm, though they are not all CNS specific, cluster various tissues and especially CNS successfully (Fig. 5C). This observation implies that the physiological differences between tissues are not because of a large fraction of tissue specific proteins (62). Rather, tissue specificity seems to be achieved through activation of a specific set of interactions or protein complexes (Fig. 5C and supplemental Figs. S12, S13).
DISCUSSION
hPRINT uses a combination of Random Forests and Bayesian learning approaches in order to integrate various types of evidence for predicting physical protein interactions and integrating those predictions with known information. This unique combination of machine learning methods, the emphasis on distinguishing physical binding from functional association, the coverage of the human genome, and the extensive experimental testing of our predictions set hPRINT apart from existing resources.
The specific design of our prediction pipeline combines the following goals: (1) it makes robust predictions even in the complete absence of experimental binding evidence; (2) because using Random Forests it allows for nonlinear interactions between the features; (3) final interaction scores also include published experimental evidence. Other designs would have failed to meet at least one of these criteria. For example, including experimental evidence in the first step (and thus dropping the second Bayesian learning step) would potentially have led to circular reasoning. An additional more subtle disadvantage is that in that case Random Forests would have given strong preference to experimental evidence because it almost perfectly predicts binding in the training set. Thus, other types of evidence that are needed for actual predictions would not have been trained correctly for situations when experimental evidence is absent. Our two-step procedure circumvents both of these problems.
When assembling the reference interactions that we used for training and testing we have tried to avoid circular reasoning as much as possible especially by excluding experimental evidence. However, complete independence from all the information we used for predicting interactions is not possible (e.g. in the case of text mining). Essentially all published predicted networks suffer from this limitation. We addressed this problem in two different ways: first, we removed all text mining-based features and second, we conducted independent experimental testing. Supplemental Fig. S14 shows that the quality of the predictions does not drop when removing text mining-based features, even though, of course, the density of the network is reduced. This analysis confirms that the quality measures shown in Fig. 2 are not biased in favor of the predictions because of potential circular reasoning when using text mining. Independent experimental testing should address all possible biases—even undetected ones. We evaluated hPRINT and the other databases based on three new experimental data sets: one was very recently published and not available for the training of hPRINT (38), and two novel screens were performed in the framework of this project and are reported here. Using the two complementary experimental methods, Y2H and AP-MS, we demonstrated the predictive power of hPRINT and we confirmed the importance of distinguishing physical and functional gene associations. In addition, in our experiments we tested the performance of hPRINT and other databases using large scale screening setups. We set out to experimentally test hPRINT predictions with standardized experimental setups rather than testing our method using literature-derived gold standards.
Initially, it might be surprising that the Y2H experiments identified only 81 interactions among 5434 tested protein pairs. However in such an approach it is very important to test a large number of noninteracting pairs as well as the predicted interactions, because we anticipated an extremely low success rate in the negative and random control set (5). Therefore, by design only half of the tested interactions had any prediction score in our database and only 548 had an RFphys score above 0.5. Because in vivo interactions often depend on specific cellular conditions (e.g. presence of co-factors) we do not expect that all predicted interactions can be verified using these standardized high throughput assays. In fact, our validation rate compares well to the retest rate for the positive reference set (Fig. 2C, 2D), indicating that the low sensitivity of the experimental techniques accounts for the relatively low number of interactions found rather than the false positive rate of the hPRINT predictions. Hence, these experiments do not serve to provide validation of individual interactions, but they provide very good support for a quality assessment of the hPRINT predictions and other databases in a quantitative and unbiased way. The main finding from these experiments is that the recovery rate of predictions from other databases (supplemental Table S3) or using RFfun (Fig. 2F, 2G) is significantly lower than from RFphys predications. Even though some of the experimentally observed binding events might not constitute true in vivo interactions and some of the interactions found in the negative sets might be actually true interactions the overall statistics would not change significantly–especially the relative differences between the networks would not change. This notion is supported by the statistical significance of the performance differences, which also reflects robustness against noise in the measurements.
The superior performance of hPRINT compared with previous attempts in predicting protein-protein binding is explained by four facts. First, the Random Forests machine learning method is more flexible than competing methods and it makes significantly fewer assumptions about the nature of the predictors and their relationships to each other. Second, the complete exclusion of experimental binding evidence in the training phase is important for robust de novo prediction of protein binding. Third, we are using additional features such as the network features that have not been used in combination before. Fourth, the distinction of functional and physical interactions in the machine learning turned out to be very important. Though being intuitive, this distinction has not always been made in the past. That is not to say that predicting functional relationships is useless (52). Rather, they reflect different aspects of the system and explicitly distinguishing those aids subsequent analyses built on top of the network.
Our analysis of disease association data shows that dense networks like hPRINT might improve candidate gene prioritization and assist in inferring molecular mechanisms. For example the fact that several linker genes are known to be disease related even though that information was not used in our analysis demonstrates the utility of network-based methods for identifying relevant genes. In that respect, this study represents a proof of principle.
By integrating known with high-confidence predicted interactions we almost double the currently known physical interactome. We anticipate that this resource will be instrumental for directing future screening of interactions and for conducting systems-level analysis of cellular processes. In particular, hPRINT will be valuable for studying disease mechanisms and for short listing candidate genes identified on a genetic basis such as GWAS.
Acknowledgments
We thank Anne Tuukkanen (Technische Universität Dresden, Germany) for help with the analysis of domain motives.
Footnotes
* This work was supported by the Klaus Tschira Foundation, European Community's Seventh Framework Programme (PhenOxiGEn FP7-223539, Ponte FP7-247945, SyBoSS FP7-242129), German BMBF (NGFNp NeuroNet-TP3 01GS08171 (to U.S.), DiGtoP 01GS0859), German BMWi (GeneCloud), and the Max Planck Society.
 This article contains supplemental Figs. S1 to S14 and Tables S1 to S8.
This article contains supplemental Figs. S1 to S14 and Tables S1 to S8.
1 The abbreviations used are:
- ALS
- amyotrophic lateral sclerosis
- APMS
- affinity purification-mass spectrometry
- CNS
- central nervous system
- CORUM
- Comprehensive Resource of Mammalian protein complexes
- GAD
- Genetic Association Database
- GWAS
- genome-wide association studies
- HPRD
- Human Protein Reference Database
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- OMIM
- Online Mendelian Inheritance in Man
- QUBIC
- Quantitative BAC InteraCtomics
- STRING
- Search Tool for the Retrieval of Interacting Genes
- Y2H
- yeast two-hybrid.
REFERENCES
- 1. Beyer A., Bandyopadhyay S., Ideker T. (2007) Integrating physical and genetic maps: from genomes to interaction networks. Nat. Rev. Genet. 8, 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stumpf M. P., Thorne T., de Silva E., Stewart R., An H. J., Lappe M., Wiuf C. (2008) Estimating the size of the human interactome. Proc. Natl. Acad. Sci. U.S.A. 105, 6959–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gunsalus K. C., Ge H., Schetter A. J., Goldberg D. S., Han J. D., Hao T., Berriz G. F., Bertin N., Huang J., Chuang L. S., Li N., Mani R., Hyman A. A., Sönnichsen B., Echeverri C. J., Roth F. P., Vidal M., Piano F. (2005) Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436, 861–865 [DOI] [PubMed] [Google Scholar]
- 4. Bork P., Jensen L. J., von Mering C., Ramani A. K., Lee I., Marcotte E. M. (2004) Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 14, 292–299 [DOI] [PubMed] [Google Scholar]
- 5. Venkatesan K., Rual J. F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K. I., Yildirim M. A., Simonis N., Heinzmann K., Gebreab F., Sahalie J. M., Cevik S., Simon C., de Smet A. S., Dann E., Smolyar A., Vinayagam A., Yu H., Szeto D., Borick H., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Cusick M. E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabási A. L., Vidal M. (2009) An empirical framework for binary interactome mapping. Nat. Methods 6, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 7. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 8. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramirez F., Schlicker A., Assenov Y., Lengauer T., Albrecht M. (2007) Computational analysis of human protein interaction networks. Proteomics 7, 2541–2552 [DOI] [PubMed] [Google Scholar]
- 11. Stelzl U., Wanker E. E. (2006) The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 10, 551–558 [DOI] [PubMed] [Google Scholar]
- 12. Pitre S., Alamgir M., Green J. R., Dumontier M., Dehne F., Golshani A. (2008) Computational methods for predicting protein-protein interactions. Adv. Biochem. Eng. Biotechnol. 110, 247–267 [DOI] [PubMed] [Google Scholar]
- 13. Schwartz A. S., Yu J., Gardenour K. R., Finley R. L., Jr, Ideker T. (2009) Cost-effective strategies for completing the interactome. Nat. Methods 6, 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDermott J., Guerquin M., Frazier Z., Chang A. N., Samudrala R. (2005) BIOVERSE: enhancements to the framework for structural, functional and contextual modeling of proteins and proteomes. Nucleic Acids Res. 33, W324–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhodes D. R., Tomlins S. A., Varambally S., Mahavisno V., Barrette T., Kalyana-Sundaram S., Ghosh D., Pandey A., Chinnaiyan A. M. (2005) Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 23, 951–959 [DOI] [PubMed] [Google Scholar]
- 16. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8: a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown K. R., Jurisica I. (2005) Online predicted human interaction database. Bioinformatics 21, 2076–2082 [DOI] [PubMed] [Google Scholar]
- 18. Lefebvre C., Rajbhandari P., Alvarez M. J., Bandaru P., Lim W. K., Sato M., Wang K., Sumazin P., Kustagi M., Bisikirska B. C., Basso K., Beltrao P., Krogan N., Gautier J., Dalla-Favera R., Califano A. (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDowall M. D., Scott M. S., Barton G. J. (2009) PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 37, D651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexeyenko A., Sonnhammer E. L. (2009) Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 19, 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi Y., Bar-Joseph Z., Klein-Seetharaman J. (2006) Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins 63, 490–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keshava Prasad T. S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Harrys Kishore C. J., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., Pandey A. (2009) Human Protein Reference Database–2009 update. Nucleic Acids Res. 37, D767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruepp A., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Stransky M., Waegele B., Schmidt T., Doudieu O. N., Stümpflen V., Mewes H. W. (2008) CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 36, D646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bossi A., Lehner B. (2009) Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plake C., Royer L., Winnenburg R., Hakenberg J., Schroeder M. (2009) GoGene: gene annotation in the fast lane. Nucleic Acids Res. 37, W300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henschel A., Winter C., Kim W. K., Schroeder M. (2007) Using structural motif descriptors for sequence-based binding site prediction. BMC Bioinformatics 8, (Suppl 4) S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breiman L. (2004) Random Forests. Machine Learning 45, 5–32 [Google Scholar]
- 29. Elefsinioti A., Ackermann M., Beyer A. (2009) Accounting for redundancy when integrating gene interaction databases. PloS One 4, e7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker K. G., Barnes K. C., Bright T. J., Wang S. A. (2004) The genetic association database. Nat. Genet. 36, 431–432 [DOI] [PubMed] [Google Scholar]
- 32. Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K. S., Knoblich M., Haenig C., Herbst M., Suopanki J., Scherzinger E., Abraham C., Bauer B., Hasenbank R., Fritzsche A., Ludewig A. H., Büssow K., Buessow K., Coleman S. H., Gutekunst C. A., Landwehrmeyer B. G., Lehrach H., Wanker E. E. (2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15, 853–865 [DOI] [PubMed] [Google Scholar]
- 33. Poser I., Sarov M., Hutchins J. R., Hériché J. K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A. W., Pelletier L., Kittler R., Hua S., Naumann R., Augsburg M., Sykora M. M., Hofemeister H., Zhang Y., Nasmyth K., White K. P., Dietzel S., Mechtler K., Durbin R., Stewart A. F., Peters J. M., Buchholz F., Hyman A. A. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Snel B., Lehmann G., Bork P., Huynen M. A. (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin N., Wu B., Jansen R., Gerstein M., Zhao H. (2004) Information assessment on predicting protein-protein interactions. BMC Bioinformatics 5, 154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi Y., Dhiman H. K., Bhola N., Budyak I., Kar S., Man D., Dutta A., Tirupula K., Carr B. I., Grandis J., Bar-Joseph Z., Klein-Seetharaman J. (2009) Systematic prediction of human membrane receptor interactions. Proteomics 9, 5243–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Heriche J. K., Sykora M., Augsburg M. M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. (2010) Systematic Analysis of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iossifov I., Rodriguez-Esteban R., Mayzus I., Millen K., Rzhetsky A. (2009) Looking at cerebellar malformations through text-mined interactomes of mice and humans. PLoS Comput Biol 5, e1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cokol M., Iossifov I., Weinreb C., Rzhetsky A. (2005) Emergent behavior of growing knowledge about molecular interactions. Nat. Biotechnol. 23, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 41. Sorkin A., von Zastrow M. (2009) Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kestler H. A., Kühl M. (2008) From individual Wnt pathways towards a Wnt signalling network. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perkins T. J., Swain P. S. (2009) Strategies for cellular decision-making. Mol. Syst. Biol. 5, 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hyduke D. R., Palsson B. Ø. (2010) Towards genome-scale signalling-network reconstructions. Nat. Rev. Genet. 11, 297–307 [DOI] [PubMed] [Google Scholar]
- 45. Frazer K. A., Murray S. S., Schork N. J., Topol E. J. (2009) Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251 [DOI] [PubMed] [Google Scholar]
- 46. Wu X., Jiang R., Zhang M. Q., Li S. (2008) Network-based global inference of human disease genes. Mol. Syst. Biol. 4, 189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cordell H. J. (2009) Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lage K., Karlberg E. O., Størling Z. M., Olason P. I., Pedersen A. G., Rigina O., Hinsby A. M., Tümer Z., Pociot F., Tommerup N., Moreau Y., Brunak S. (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 [DOI] [PubMed] [Google Scholar]
- 49. Bergholdt R., Storling Z. M., Lage K., Karlberg E. O., Olason P. I., Aalund M., Nerup J., Brunak S., Workman C. T., Pociot F. (2007) Integrative analysis for finding genes and networks involved in diabetes and other complex diseases. Genome Biol. 8, R253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Linghu B., Snitkin E. S., Hu Z., Xia Y., Delisi C. (2009) Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol. 10, R91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thomas D. (2010) Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu. Rev. Public Health 31, 21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee I., Lehner B., Vavouri T., Shin J., Fraser A. G., Marcotte E. M. (2010) Predicting genetic modifier loci using functional gene networks. Genome Res. 20, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bertram L., McQueen M. B., Mullin K., Blacker D., Tanzi R. E. (2007) Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 39, 17–23 [DOI] [PubMed] [Google Scholar]
- 54. Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S., Manolio T. A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 106, 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grünblatt E. (2008) Commonalities in the genetics of Alzheimer's disease and Parkinson's disease. Expert Rev. Neurother. 8, 1865–1877 [DOI] [PubMed] [Google Scholar]
- 56. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Devi L., Prabhu B. M., Galati D. F., Avadhani N. G., Anandatheerthavarada H. K. (2006) Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 26, 9057–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu C. E., Seltman H., Peskind E. R., Galloway N., Zhou P. X., Rosenthal E., Wijsman E. M., Tsuang D. W., Devlin B., Schellenberg G. D. (2007) Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics 89, 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bu G. (2009) Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 10, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roses A. D., Lutz M. W., Amrine-Madsen H., Saunders A. M., Crenshaw D. G., Sundseth S. S., Huentelman M. J., Welsh-Bohmer K. A., Reiman E. M. (2010) A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 10, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pontén F., Gry M., Fagerberg L., Lundberg E., Asplund A., Berglund L., Oksvold P., Björling E., Hober S., Kampf C., Navani S., Nilsson P., Ottosson J., Persson A., Wernérus H., Wester K., Uhlén M. (2009) A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5, 337 [DOI] [PMC free article] [PubMed] [Google Scholar]