Abstract
Low bone mineral density (BMD) is a risk factor of osteoporosis and has strong genetic determination. Genes influencing BMD and fundamental mechanisms leading to osteoporosis have yet to be fully determined. Peripheral blood monocytes (PBM) are potential osteoclast precursors, which could access to bone resorption surfaces and differentiate into osteoclasts to resorb bone. Herein, we attempted to identify osteoporosis susceptibility gene(s) and characterize their function(s), through an initial proteomics discovery study on PBM in vivo, and multiscale validation studies in vivo and in vitro. Utilizing the quantitative proteomics methodology LC-nano-ESI-MSE, we discovered that a novel protein, i.e. ANXA2, was up-regulated twofold in PBM in vivo in Caucasians with extremely low BMD (cases) versus those with extremely high BMD (controls) (n = 28, p < 0.05). ANXA2 gene up-regulation in low BMD subjects was replicated at the mRNA level in PBM in vivo in a second and independent case-control sample (n = 80, p < 0.05). At the DNA level, we found that SNPs in the ANXA2 gene were associated with BMD variation in a 3rd and independent case-control sample (n = 44, p < 0.05), as well as in a random population sample (n = 997, p < 0.05). The above integrative evidence strongly supports the concept that ANXA2 is involved in the pathogenesis of osteoporosis in humans. Through a follow-up cellular functional study, we found that ANXA2 protein significantly promoted monocyte migration across an endothelial barrier in vitro (p < 0.001). Thus, elevated ANXA2 protein expression level, as detected in low BMD subjects, probably stimulates more PBM migration through the blood vessel walls to bone resorption surfaces in vivo, where they differentiate into higher number of osteoclasts and resorb bone at higher rates, thereby decreasing BMD. In conclusion, this study identified a novel osteoporosis susceptibility gene ANXA2, and suggested a novel pathophysiological mechanism, mediated by ANXA2, for osteoporosis in humans.
Osteoporosis is a worldwide public health problem that is most prevalent in the elderly, and is particularly problematic in postmenopausal women. Osteoporosis is characterized by low bone mineral density (BMD)1 and increased risk of osteoporotic fractures. Osteoporotic hip fractures have a high associated morbidity and mortality, and contribute substantially to health care expenditures (1). The annual cost of osteoporosis and fractures in the US elderly alone was estimated at $16 billion in 2002 and $22 billion in 2008 (2), and this is expected to increase substantially over the following decades.
BMD is a quantitative trait determined by multiple factors. Heritability of BMD is estimated to be 0.5–0.9 (3). Extensive genetic and genomic studies, conducted over the past decade, have identified several genes that are associated with BMD variation in humans (4). Collectively, however, these implicated genes explain no more than 10% of BMD variation in any individual human population. So the basis for the majority of variation in BMD that is genetically determined still awaits identification.
Osteoporosis results from excessive bone loss, which is largely because of increased bone resorption by osteoclasts and/or decreased bone formation by osteoblasts. Functional studies, primarily based on mouse models or in vitro cell cultures, have been widely used to establish the relevance of well-known or novel genes to osteoblastogenesis and/or osteoclastogenesis and bone phenotypes (4–8). However, biological relevance established through these approaches may not necessarily be translatable to humans. Consequently, clinical studies with feasible study designs for discovery of genes clinically significant to bone phenotypes/metabolism in vivo, followed by functional exploration and/or validation studies in vivo and in vitro, is highly desirable to identify and characterize the genes and their products for the risk of osteoporosis in humans.
Bone-resorbing osteoclasts are hematopoietic in origin and are derived from cells of monocyte-macrophage lineage (9). In the adult human peripheral skeleton such as the hip bone, the local marrow is not a significant source for generation of osteoclasts; virtually all osteoclasts functioning at these sites are primarily derived from monocytes migrating to bone via the peripheral circulation (10, 11). After migrating from peripheral blood to bone, monocytes differentiate and fuse into immature multinuclear osteoclasts, and are subsequently activated to become mature osteoclasts at sites of bone resorption (9). It is extremely difficult to collect large numbers of osteoclasts in vivo in humans for experimentation; however, peripheral blood monocytes (PBM), which are osteoclast precursors, can be collected in large numbers with relative ease.
Previous studies have identified several cytokines produced by PBM, (e.g. IL-1, IL-6, TNF-α), which are important for osteoclast differentiation, activation, and apoptosis (12–14). In Chinese, differential gene expression in PBM was found to be associated with variations in BMD (15–17). These results demonstrated the utility of employing PBM as a model cell type for studying gene function in relation to risk of osteoporosis in humans. Accordingly, we here propose that in-depth study of PBM in vivo, complemented by in vitro functional studies, has significant potential to identify novel susceptibility genes for osteoporosis and provides new insights into the pathogenesis of osteoporosis in humans.
Proteins are the major executors of gene functions in biological organisms. Changes of physiological conditions are often reflected by alterations in protein expression and/or metabolism (18–20). Shotgun quantitative proteomics, which systematically identifies and quantifies proteins at a proteome-wide scale, has emerged as a novel and powerful methodology for disease-related biomarker discovery (21). However, this approach has yet to be widely applied to bone biology. Dissecting variations of protein expression levels in human primary osteoclast precursors (e.g. PBM), under normal versus diseased conditions, is a potentially fruitful strategy for identifying genes functionally relevant to osteoclastogenesis in vivo in humans.
Utilizing the quantitative proteomics methodology, and a strategy of multidisciplinary and integrative studies, the present work was designed to identify genes important to osteoporosis in vivo in humans, and to preliminarily explore the functional mechanism by which they contribute to the pathophysiology of osteoporosis. Specifically, identification of genes important to osteoporosis was based on studies at three molecule levels (protein, RNA, and DNA), and based on evidence generated from multiple independent study samples.
EXPERIMENTAL PROCEDURES
Human Subjects
This study was approved by appropriate Institutional Review Boards. Signed informed-consent documents were obtained from all study participants before they enrolled in the study.
All subjects were self-identified as being of European origin. Chronic diseases and conditions that might potentially affect bone mass or bone metabolism were excluded. These diseases and conditions included chronic disorders involving vital organs (heart, lung, liver, kidney, brain), serious metabolic diseases (diabetes, hypo- and hyper-parathyroidism, hyperthyroidism, etc.), other skeletal diseases (Paget's disease, osteogenesis imperfecta, rheumatoid arthritis, etc.), chronic use of drugs affecting bone metabolism (hormone replacement therapy, corticosteroid therapy, anti-convulsant drugs), malnutrition conditions (such as chronic diarrhea, chronic ulcerative colitis, etc.), etc. In addition, subjects taking anti-bone-resorptive or bone anabolic agents/drugs, such as bisphosphonates were also excluded from this study. The purpose of the above exclusion criteria was to minimize the effect of any known environmental and/or therapeutic factors that might potentially influence bone phenotypic variation, thereby increasing statistical power for detecting relevant genetic factors. For Samples 1 and 2 used for PBM gene expression studies, we also excluded diseases or conditions affecting the immune system, such as influenza (within 1 week of recruitment), autoimmune or autoimmune-related diseases such as systemic lupus erythematosus, and immune-deficiency conditions such as AIDS, hematopoietic and lymphoreticular malignancies (e.g. leukemia, lymphoma).
Hip BMD (g/cm2) was measured for study subjects using Hologic 4500 W dual energy x-ray absorptiometry machines (Hologic Inc., Bedford, MA). Hip BMD represents a combined BMD from the femoral neck, trochanter, and interchochanter. The machines were calibrated daily. The coefficient of variation of repeated measurements was 1.87%.
The present work involved four study samples, including three independent case-control samples and one random population sample. Basic characteristics of the four study samples are summarized in Table I. It is worth noting that, according to World Health Organization's diagnostic criteria, all subjects with low BMD in the three case-control samples were diagnosed as having osteopenia (i.e. –2.5 < T score ≤ –1.0) or osteoporosis (i.e. T score ≤ –2.5). Herein, T-score is defined as the number of standard deviations a subject's BMD differs from the average BMD of their gender- and ethnicity-matched young adults.
Table I. Basic characteristics of the study samples. Presented are Mean ± S.E.
| Category | Sample 1 |
Sample 2 |
Sample 3 |
Sample 4 |
|||
|---|---|---|---|---|---|---|---|
| High BMD | Low BMD | High BMD | Low BMD | High BMD | Low BMD | - | |
| Sample Size | 14 | 14 | 40 | 40 | 22 | 22 | 997 |
| Age (yrs) | 68.7 ± 1.1 | 67.7 ± 1.7 | 49.4 ± 1.3 | 50.0 ± 1.3 | 31.8 ± 1.0 | 33.7 ± 0.8 | 50.3 ± 0.6 |
| Height (cm) | 159.50 ± 1.33 | 160.30 ± 2.77 | 163.43 ± 0.98 | 162.94 ± 0.96 | 175.45 ± 2.09 | 167.84 ± 1.67 | 1.71 ± 0.00 |
| Weight (kg) | 81.14 ± 3.41 | 60.79 ± 2.87 | 83.18 ± 2.48 | 62.27 ± 2.17 | 98.45 ± 4.88 | 62.54 ± 1.93 | 80.16 ± 0.56 |
| Hip BMDa (g/cm2) | 2.13 ± 0.22 | −1.39 ± 0.21 | 1.45 ± 0.11 | −1.05 ± 0.07 | 2.53 ± 0.11 | −1.74 ± 0.09 | 0.97 ± 0.00 |
a In the Samples 1–3, hip BMD was presented as Z-score. Z-score is defined as the number of standard deviations a subject's BMD differs from the average BMD of their age-, gender-, and ethnicity- matched population. All the subjects in low BMD groups were diagnosed as having osteopenia or osteoporosis.
Sample 1 for Proteomics Discovery Study
Sample 1 included 28 unrelated postmenopausal Caucasian women, selected from an archived population database and composed of 14 subjects with extremely high hip BMD (Z-score: 2.13 ± 0.22, mean ± S.E.) and 14 subjects with extremely low hip BMD (Z-score: –1.39 ± 0.21, mean ± S.E.). Z-score is defined as the number of standard deviations a subject's BMD differs from the average BMD of their age-, gender-, and ethnicity-matched population.
Sample 2 for mRNA Expression Study
Sample 2 included 80 unrelated Caucasian women, selected from an archived population database. The sample included 40 subjects with extremely high hip BMD (Z-score: 1.45 ± 0.11, mean ± S.E.) and 40 subjects with extremely low hip BMD (Z-score: –1.05 ± 0.07, mean ± S.E.).
Sample 3 for DNA Sequencing Analyses
Sample 3 included 44 unrelated adult Caucasians. The sample included 22 subjects with extremely high hip BMD (Z-score: 2.53 ± 0.11, mean ± S.E.) and 22 subjects with extremely low hip BMD (Z-score: –1.74 ± 0.09, mean ± S.E.).
Sample 4 for Single Nucleotide Polymorphism (SNP) Association Study
Sample 4 included 997 unrelated adult Caucasians.
Peripheral Blood Monocyte (PBM) Isolation in Samples 1 and 2
For Samples 1 and 2, 60 ml peripheral blood was collected from each subject by certificated phlebotomist. EDTA was used as anticoagulant. The fresh blood samples were processed instantly for PBM isolation by experienced technicians. First, peripheral blood mononuclear cells (PBMC) were isolated from whole blood using density gradient centrifugation with Histopaque-1077 (Sigma, H1077–1). Then, PBM were isolated from PBMC using a monocyte negative isolation kit (Dynal Biotech Inc., Lake Success, NY) following the manufacturer's recommendation. The kit contains a highly optimized antibody mix, blocking reagent, and Depletion Dynabeads® to deplete T cells, B cells, and natural killer cells from PBMC, leaving monocytes untouched and free of surface-bound antibody and beads. Per our experience, the purity of PBM isolated using this method was 86% ± 3% (17).
PBM Proteome Profiling and Comparative Analyses in Sample 1
PBM total proteins were extracted using a complete proteome extraction mammalian kit (Calbiochem, San Diego, CA; No. 539779). Protein concentration was measured using Bradford method. Up to 20 μg total protein was precipitated using a protein precipitation kit (Calbiochem, No. 539180). Protein pellets were dissolved in 50 μl 50 mM ammonium bicarbonate with 0.1% RapiGest (Waters, Milford, MA), reduced by 5.0 mM dithiothreitol, alkylated by 15 mM indole acetic acid, and then digested by trypsin (Sigma, T6567). Protein digests were concentrated to ∼20 μl, of which 15 μl was aspirated and brought back to 20 μl, with 0.5% formic acid and 100 fmol yeast alcohol dehydrogenase I digestion standard (Waters, No. 186002328).
PBM proteomes were profiled using a method of liquid chromatography-nano-electrospray ionization (LC-nano-ESI)-MSE (22), through nanoAcquity ultra performance liquid chromatography coupled with Synapt High Definition Mass Spectrometry (HDMS) (Waters). Proteome data acquisition was controlled by MassLynx 4.1 software (Waters). Briefly, the protein digests (∼500 ng) were injected into a BEH C18 75 μm × 150 mm analytical column (Waters), and separated by solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid) at a flow rate of 0.3 μl/min using a gradient of 2 hours as follows: 3% B initial, 10% B at 1.0 min, 30% B at 75 min, 40% B at 90 min, 95% B at 91 min, 95% B at 95 min, 3% B at 96 min, equilibrate thereafter till 120 min. The eluate was analyzed by HDMS under positive ion V-mode. The following parameters were set for data acquisition: collision energy, 5 volts for MS and ramp 15–40 volts for MSE; scan time, 0.6 s per scan. The HDMS machine was calibrated daily to ensure high accuracy (2.0 ppm for lock mass of m/z 785.8426).
For each PBM proteome digest sample, triplicate LC-nano-ESI-MSE data sets were acquired. Then, the MSE data were processed with ProteinLynx Global Server version 2.3 (Waters) using default parameters. Based on the alternating low- and elevated-energy nature of MSE data, properties of each ion (mass-to-charge ratio, retention time, intensity, etc.) were determined, and a list of all precursor and product ions was produced. Specifically, the ion's intensity was derived from the areas of both the chromatographic and mass spectrometric peaks. The precursor ion intensity threshold was set to be above 1000 counts. Human protein database International Protein Index version 3.56 (153,078 protein entries) was searched by using the following parameters: enzyme specificity, trypsin; number of missed cleavages permitted, 1; fixed modification, Carbamidomethyl C; variable modifications, Acetyl N-TERM, Deamidation N, Deamidation Q, and Oxidation M; mass tolerance for precursor ions, 15 ppm; mass tolerance for product ions, 30 ppm; minimum peptide matches per protein, 1; minimum fragment ion matches per protein, 7; minimum fragment ion matches per peptide, 3; false positive rate, limited to 4% per randomized database searching.
For each sample, only proteins identified at least twice in the triplicate LC-nano-ESI-MSE analyses were reported as truly present. Total ion counts of the three most intense matched peptides were used to quantify each protein. With the standard alcohol dehydrogenase I as references, protein quantification level was exported in femtomol and nanogram. Mean values from triplicate analyses were used to represent protein expression levels in each PBM sample.
Based on PBM proteome profiles in the Sample 1, Kruskal-Wallis tests were used to compare the mean values of protein expression levels to identify proteins differentially expressed between subjects with low BMD versus high BMD.
ANXA2 mRNA Assay in PBM and Comparative Analyses in Sample 2
Total RNA was extracted from PBM using RNeasy Mini kit (Qiagen). ANXA2 mRNA expression levels were determined using Affymetrix HG-U133A GeneChip (R) arrays (17). We used the Robust Multiarray Average (RMA) algorithm (23) to transform the probe-level raw data into mRNA expression data. Among currently available algorithms, the RMA algorithm generates the most reproducible results and shows the highest correlation coefficients with RT-PCR data (24). Based on expression data generated by the RMA algorithm, mRNA differential expression analysis was conducted between subjects with low BMD versus high BMD using a t test.
Genomic DNA Extraction in Samples 3 and 4
Genomic DNA was extracted from 30 ml peripheral blood samples using a DNA isolation kit (Gentra systems, Minneapolis, MN). DNA concentration was assessed by a DU530 UV/VIS spectrophotometer (Beckman Coulter Inc.). DNA quantification was double-checked using PicoGreen® dsDNA Reagent and Kits (Invitrogen, Carlsbad, CA).
ANXA2 Gene Sequencing and Variation Analyses in Sample 3
DNA nucleotide sequences in the chromosome region Chr15: 60619350–60710185, which spans 20-kb upstream and downstream of ANXA2 gene, were analyzed. The sequence information was acquired using the next-generation sequencing method (25) by Complete Genomics Inc. Briefly, the human genome sequencing procedures include DNA library construction, DNA Nano-Balls generation and DNA Nano-Balls array self-assembling, cPAL-based sequencing, imaging, image data analyses including base-calling, DNA Nano-Balls mapping, and sequence assembly (25). Human Genome Build 37.1 was used as a reference to identify sequence variations in each sample. Exact Chi-square tests were conducted to evaluate whether there are significant differences in genotype frequency distribution between subjects with low versus high BMD.
Genetic Association Analyses between ANXA2 and BMD in Sample 4
A total of fifteen SNPs within the ANXA2 gene were covered by, and genotyped with, the Affymetrix Mapping 250k Nsp and 250k Sty arrays at the Vanderbilt Microarray Shared Resource at Vanderbilt University Medical Center, Nashville, TN using the standard protocol recommended by the manufacturer (26). All the SNPs had a genotyping rate of >99%, with minor allele frequencies > 0.15 and p values of Hardy-Weinberg Equilibrium test > 0.001. The PLINK program (27) was used to test whether the SNPs have significant genotypic effects on hip BMD. False discovery rate_BH method was adopted to adjust p values and correct for multiple testing (28).
FASTSNP program (http://fastsnp.ibms.sinica.edu.tw) provides up-to-date information about known and potential functional effect of SNPs (29). For SNPs associated with hip BMD in Sample 3 and Sample 4, we further analyzed their potential functional effects by using the FASTSNP program.
Western Blotting
Confirmation of ANXA2 Protein Differential Expression in Sample 1
Western blotting was conducted using standard procedures. Approximately 50 μg of total protein per sample was loaded. Mouse anti-human ANXA2 monoclonal primary antibody (Abnova, Littleton, CO; No. H00000302-M02) and HRP- conjugated goat anti-mouse IgG secondary antibody (Abnova, No. PAB0096) were used for chemiluminescence detection of ANXA2 using FluorChem FC2 imaging system (Alpha Innotech, San Leandro, CA). HRP-conjugated mouse anti-human beta-actin monoclonal primary antibody (Abcam, Cambridge, MA; No. 8226) was used for detection of beta-actin. Herein, beta-actin served as a loading control for each of the PBM total protein samples. The intensity ratio of ANXA2 band to beta-actin band was used as normalized ANXA2 protein level in each PBM sample. Comparative analyses of the normalized ANXA2 protein levels between subjects with low BMD versus high BMD were conducted using a Student's t test.
Detection of ANXA2 Protein in Human PBM Culture Medium
From a peripheral blood sample donated by a postmenopausal Caucasian woman (>50 yrs old), PBM were isolated using a monocyte isolation kit (Invitrogen, No. 11350D). Freshly isolated PBM (4 × 104 cells) were cultured using RPMI Media 1640 (Invitrogen) for 36 h. The cell culture was collected and centrifuged at 600 × g, and the supernatant was collected and concentrated (2×) using a Speedvac system (Savant Instrument Inc.). ANXA2 protein in the concentrated culture medium was tested by Western blotting using standard procedures. Mouse anti-human ANXA2 monoclonal primary antibody (Abnova, No. H00000302-M01) and HRP-conjugated goat anti-mouse IgG secondary antibody (Sigma, No. A4416) were used.
Monocyte Trans-endothelial Migration Assay
Twenty-four-well flat-bottomed tissue culture plates with trans-well culture inserts were used to establish upper and lower chambers for monocyte migration assays. The inserts had 5-μm pore filters and were coated with 0.2% gelatin (Sigma-Aldrich) overnight before the migration assay. Human umbilical vein endothelial cells (HUVEC, ScienCell, Carlsbad, CA; No. 8000) were cultured using endothelial cell medium (ScienCell, No. 1001) and endothelial cell growth supplement (ScienCell, No. 1052) to achieve 70–80% confluence. For each migration assay, 3 × 104 HUVEC were plated in trans-well culture inserts (Costa, Cambridge, MA) and allowed to grow to confluence for 72 h. Then, SC human monocyte cells (ATCC No. CRL-9855) (5 × 105 cells per well) were seeded on the HUVEC cell layer in 0.1 ml Iscove's modified Dulbecco's medium in the upper chamber. The lower chamber contained 0.6 ml Iscove's Modified Dulbecco's Medium.
Monocyte chemotactic protein 1 (MCP-1) is a known chemoattractant for monocytes (30). Thus, MCP-1 (200 ng/ml) was used as a positive indicator to establish proper conditions for SC monocyte trans-endothelial migration. Fetal bovine serum (FBS) was added to the upper and/or lower chambers at varying concentrations to optimize migration conditions. The effect of ANXA2 on SC trans-endothelial migration was tested under optimal conditions. ANXA2 protein (Origene, Catalogue No. TP305081) was introduced into either the lower or upper chambers at varied concentrations (0 ng/ml, 50 ng/ml, 100 ng/ml, 200 ng/ml, and 400 ng/ml). SC cells were allowed to migrate from the upper to lower chambers for 24 h. After that, SC cells in the lower chamber were counted using a hemocytometer under a microscope. The percentage of SC cells migrating from the upper to the lower chamber was used to represent the monocyte mobility under each experimental condition. Triplicate assays were conducted for each condition. Student's t test was conducted to test whether ANXA2 protein treatment has significant effect on monocyte migration, as compared with the negative control.
RESULTS
ANXA2 Protein was Up-Regulated in PBM In Vivo in Low BMD Subjects
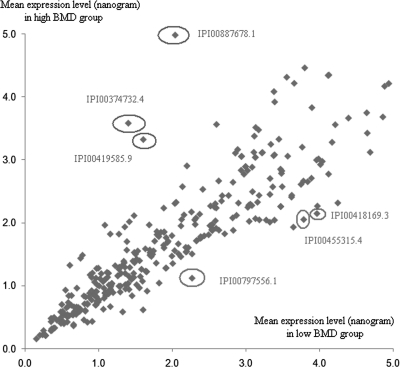
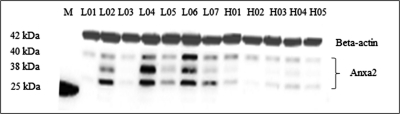
In the first case-control study sample (i.e. Sample 1), a total of 1539 proteins in PBM were identified and quantified. Of these 1539 proteins, 501 were identified in more than 5 PBM samples from both high and low BMD subjects. Then, comparative analyses were conducted to identify differentially expressed proteins among the 501 proteins. Fig. 1 illustrates the protein expression levels for proteins expressed at relatively low levels in low BMD and high BMD subjects. According to Kruskal-Wallis test using the raw data set and the data set normalized by the housekeeping protein beta actin, ANXA2 protein was found to be differentially expressed between low BMD and high BMD subjects (p < 0.05, Table II and supplemental Table S1). Herein, three ANXA2 protein isoforms (IPI00418169.3, IPI00455315.4, and IPI00797556.1) were identified. They were all found to be consistently up-regulated in low BMD subjects compared with high BMD subjects (p < 0.05). As shown in Fig. 2, up-regulation of ANXA2 in PBM in low BMD subjects was verified by Western blotting (p < 0.05, for each of the three bands detected). supplemental Table S2 lists ANXA2 protein sequences identified in a random PBM sample, with sequence coverage of 33.8%, 41.6%, and 39.5% respectively, for IPI00418169.3 (40.4 kDa), IPI00455315.4 (38.6 kDa), and IPI00797556.1 (24.4 kDa).
Fig. 1.
Relative proteome expression in PBM from low versus high BMD subjects. Presented are mean protein expression levels in subjects with high BMD and low BMD, respectively. Approximately 500 ng PBM total proteins were analyzed using LC-nano-ESI-MSE method. Herein, only proteins commonly expressed (in ≥10 PBM samples) and detected at <5.0 ng are presented. ANXA2 protein (IPI00418169.3, IPI00455315.4, and IPI00797556.1) were up-regulated in PBM in low BMD subjects. Representative proteins down-regulated in PBM in low BMD subjects include IPI00419585.9 (Peptidyl-prolyl cis-trans isomerase A, PPIA), IPI00374732.4 (similar to peptidyl-prolyl isomerase A-like, PPIAP19), and IPI00887678.1 (similar to peptidyl-prolyl isomerase A-like, LOC654188). As no evidence of differential expression were found in subsequent Western blot analyses using anti-PPIA antibody (Abnova Catalogue No. H00005478-M01), these proteins and the genes encoding them were not analyzed further in this study.
Table II. Differential expression of ANXA2 protein in sample 1 as Quantified by LC-nano-ESI-MSE. P and Adj. P are p values of Kruskal-Wallis tests using the raw dataset and the datasets normalized by beta actin, respectively. ANXA2 protein expression levels in Sample 1 are presented in supplemental Table I.
| Protein ID | IPI00418169.3 | IPI00455315.4 | IPI00797556.1 |
|---|---|---|---|
| Molecular Weight (kDa) | 40.4 | 38.6 | 24.4 |
| Protein expression level in low BMD subjects (ng, Mean ± S.E.) | 3.97 ± 0.73 | 3.79 ± 0.70 | 2.27 ± 0.43 |
| Protein expression level in high BMD subjects (ng, Mean ± S.E.) | 2.15 ± 0.42 | 2.05 ± 0.40 | 1.12 ± 0.22 |
| Ratio of protein expression level (L:H) | 1.85 | 1.85 | 2.03 |
| p value | 0.03 | 0.03 | 0.01 |
| Adj. p value | 0.04 | 0.04 | 0.03 |
Fig. 2.
Up-regulation of anxa2 protein in subjects in the low versus high BMD group as verified by Western blot. Three bands of ANXA2 protein were imaged on the blot. According to their locations on the blot, the three bands presumably correspond to ANXA2 protein isoforms IPI00418169.3, IPI00455315.4, and IPI00797556.1, as identified by LC-nano-ESI-MSE. Molecular weight of beta-actin is about 41.7 kDa. M: protein marker (Bio-Rad Catalog No. 161–0375) including a protein with molecular weight of about 25 kDa. Proteins were separated on a 10–20% linear gradient polyacrylamide gel (Bio-Rad Catalog No. 161–1124) by electrophoresis. Presented is a representative blot of triplicate experiments.
ANXA2 mRNA was Up-Regulated in PBM In Vivo in Low BMD Subjects
To validate the relevance of ANXA2 gene expression in vivo to BMD variation in humans, as revealed by differential protein expression levels in Sample 1, we further studied ANXA2 mRNA expression levels in an independent case-control study sample (i.e. Sample 2). Consistent with the findings at the protein level, ANXA2 mRNA expression levels, represented by three probes (201590_x_at, 210427_x_at, 213503_x_at), were also up-regulated in the group of low BMD subjects compared with the group of high BMD subjects (p < 0.05).
ANXA2 Gene Sequence Variants were Differentially Distributed in Low Versus High BMD Subjects
In the 3rd case-control study sample (i.e. Sample 3), a total of 202 nucleotide variants in the ANXA2 gene were identified by deep-sequencing. Among these 202 variants, 101 quality-controlled SNPs (minor allele frequency >5% and call rate >80%) were tested for association with hip BMD. We found that three sequence variants (corresponding to SNPs rs12909425, rs11631777, and rs62004988) had nominally significant differences in genotype frequency distribution between low BMD and high BMD subjects (p < 0.05, Table III). FASTSNP program predicted that SNPs rs12909425 and rs62004988 are located within potential binding sites of transcription factors (enhancers). Thus, these two SNPs may have potential effects on ANXA2 transcription.
Table III. Three sequence variations in ANXA2 gene differentially distributed in low vs. high BMD subjects in sample 3. All subjects with low BMD were diagnosed as having osteopenia or osteoporosis. Exact chi-square test's were used to calculate the difference in frequency distribution. BMD: bone mineral density.
| Sequence Variation | Group | Genotype & Frequency | P-Value | ||
|---|---|---|---|---|---|
| rs12909425 | TT | TC | CC | ||
| Low BMD | 3 | 14 | 3 | 0.009 | |
| High BMD | 1 | 8 | 13 | ||
| rs11631777 | GG | GA | AA | ||
| Low BMD | 0 | 1 | 21 | 0.029 | |
| High BMD | 2 | 5 | 15 | ||
| rs62004988 | AA | AG | GG | ||
| Low BMD | 0 | 13 | 9 | 0.041 | |
| High BMD | 2 | 6 | 14 | ||
ANXA2 Gene SNPs were Associated with BMD Variation in a Caucasian Population
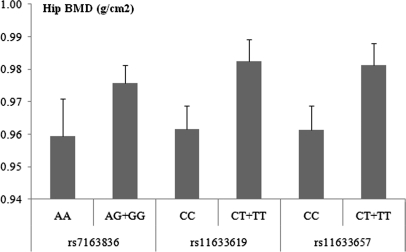
In 997 unrelated Caucasians (i.e. Sample 4), we found that among the total 15 studied SNPs located within the ANXA2 gene and covered by the Affymetrix array, three SNPs (i.e. rs7163836, rs11633619, and rs11633657) were nominally associated with hip BMD, as well as femoral neck BMD (Table IV). As shown in Fig. 3, homozygous carriers of alleles A (minor) at rs7163836, C (major) at rs11633619, and C (major) at rs11633657 have an average decrease in hip BMD of 1.7%, 2.2%, and 2.1%, respectively, compared with heterozygous carriers and non-carriers. After correcting for multiple testing, one of the three SNPs, i.e. rs11633619 remained associated with hip and femoral neck BMD (Table IV). FASTSNP program predicted that of the above three SNPs, rs11633619 is located within a potential binding site of transcription factors (enhancers). Thus, this SNP may have potential effects on ANXA2 transcription.
Table IV. Three SNPs in ANXA2 gene associated with BMD at hip and FN in sample 4. The PLINK program was used to test the association of SNPs with BMD. FN: femoral neck. MAF: minor allele frequency. BMD: bone mineral density.
Fig. 3.
Genotypic effect of three SNPs in ANXA2 gene on hip BMD in sample 4. Presented are Mean and S.E. of hip BMD. Homozygous carriers of allele A (minor) at rs7163836, C (major) at rs11633619, and C (major) at rs11633657 have an average decrease in hip BMD of 1.7%, 2.2%, and 2.1%, respectively, compared with heterozygous carriers and non-carriers.
ANXA2 Protein, Secreted by PBM, Promoted Monocyte Trans-endothelial Migration
Western blot analysis demonstrated that ANXA2 protein (∼38 kDa) was present in the conditioned medium of primary human PBM cell culture. Therefore, ANXA2 protein is presumably secreted by human PBM. Subsequently, exogenous ANXA2 protein was introduced to test its effect on monocyte migration. Among our preliminary tests, 0.1% FBS in the upper chamber and 5.0% FBS in the lower chamber are optimal for MCP-1 and ANXA2 to promote SC monocyte migration. Subsequently, under this condition, the effects of ANXA2 on SC trans-endothelial migration were assayed.
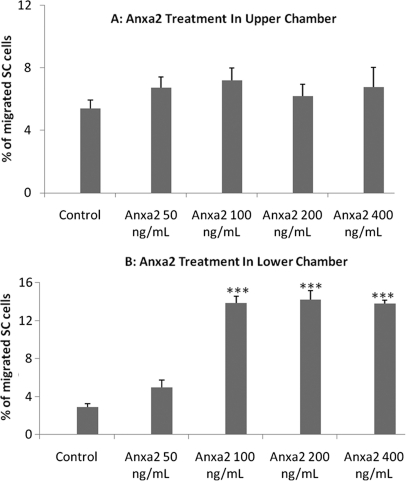
As shown in Fig. 4 (panel A), ANXA2 introduced into the UPPER chamber (50, 100, 200, and 400 ng/ml) moderately promoted SC monocyte trans-endothelial migration, with the strongest effect at the concentration of 100 ng/ml (1.4-fold increase after 24-hour, compared with the negative control), though a significant dose response was not observed. As shown in Fig. 4 (panel B), ANXA2 introduced into the LOWER chamber (100, 200, and 400 ng/ml) strikingly promoted SC monocyte trans-endothelial migration (p < 0.001). 200 ng/ml of ANXA2 produced the strongest effect in promoting the migration (4.9-fold increase after 24-hour, compared with the negative control). Collectively, these assays support that ANXA2 protein plays a significant role in promoting monocyte trans-endothelial migration.
Fig. 4.
Effect of ANXA2 protein on monocyte trans-endothelial migration. Presented for each condition is the SC monocyte migration rate (Mean and S.D.) from triplicate assays. PBM cell line SC cells were seeded onto a layer of HUVEC cells in the upper chamber of the migration setup. Medium in the upper and lower chamber was supplemented with 0.1% and 5.0% FBS, respectively. ANXA2 protein was introduced into either the UPPER or LOWER chambers at varied concentrations (0 ng/ml, 50 ng/ml, 100 ng/ml, 200 ng/ml, and 400 ng/ml). SC cells were allowed to migrate for 24 h. *** p < 0.001, as compared with the negative control without ANXA2 treatment.
DISCUSSION
The present work represents our pioneering effort to identify osteoporosis risk genes and characterize their functions using a novel strategy of multi-disciplinary and integrative studies. We employed state-of-the-art quantitative proteomics methodology to profile in vivo PBM proteomes from cases and controls to discover proteins thus genes that are functionally relevant to osteoporosis in humans. Differentially expressed gene(s) identified were verified by Western blotting. mRNA expression levels for the gene(s) encoding these proteins were subsequently analyzed in an independent study sample. Up-regulation of the ANXA2 gene, at both mRNA and protein expression levels, in PBM in low versus high BMD subjects, strongly supports the functional relevance of ANXA2 to BMD regulation in vivo in humans. To explore further the importance of ANXA2 to bone phenotypes, we studied the relationship between ANXA2 and osteoporosis at the DNA level. In both case-control and population samples, we identified SNPs in the ANXA2 gene that are associated with hip BMD in Caucasians. Collectively, all this evidence supports the concept that ANXA2 is a susceptibility gene for osteoporosis in humans.
Osteoclastogenesis involves monocyte differentiation into osteoclasts. In the adult human peripheral skeleton such as the hip bone, osteoclastogenesis occurs only after PBM have migrated across endothelial surfaces from blood to bone (9). The trans-endothelial migration apparatus utilized in this study, was designed to mimic the endothelial barrier between blood and bone, as PBM's must migrate through capillaries to sites of bone resorption on the surface of bone before they undergo differentiation and form bone-resorbing osteoclasts. As ANXA2 was found to be secreted by human PBM, it is important to understand the role of exogenous ANXA2 protein in monocyte trans-endothelial migration. Our in vitro migration assays suggested that: 1) Extracellular ANXA2, present in the blood of humans, may induce monocyte trans-endothelial migration to bone resorption surfaces; 2) Extracellular ANXA2, present at bone resorption sites in humans, may promote monocyte trans-endothelial migration from the circulation and attract them to bone to differentiate into osteoclasts. Both activities may act synergistically to accelerate more PBM recruitment to bone. Supporting a significant role for ANXA2 protein in regulating monocyte trans-endothelial migration, evidence showed that ANXA2 present on the cell surface of human PBM may contribute to matrix-penetrating activity of monocytes (31, 32).
In addition to the function of ANXA2 in monocyte trans-endothelium migration, evidence from in vitro studies showed that ANXA2 is also important to osteoclast formation and bone resorption. Specifically, in human marrow cultures, ANXA2 promoted osteoclast precursor proliferation and differentiation, and increased osteoclast formation and bone resorption (9, 33–36).
Integrating functional evidence from the present in vivo and in vitro studies and previous in vitro studies, we propose a novel pathophysiological mechanism, regulated by PBM-expressed ANXA2, for osteoporosis in humans. First, increased expression of ANXA2 protein on PBM's, probably via promoting PBM's matrix-penetrating activity, contributes to PBM migration from the blood to sites of bone resorption. Second, ANXA2 protein released after PBM reach bone resorption sites elevates local extracellular ANXA2 concentration. Elevated extracellular ANXA2 concentration at bone resorption sites further attracts PBM's to the sites by enhancing trans-endothelial migration, resulting in an expanded pool of osteoclast precursors at the sites of bone resorption. Meanwhile, ANXA2 also promotes monocyte differentiation, osteoclast formation, and bone resorption. Collectively, the above changes act synergistically to contribute to elevated osteoclastogenesis, resulting in accelerated bone resorption and increased bone loss, and thus potentially, osteopenia and osteoporosis.
To the best of our knowledge, this is the first study in which ANXA2 has been identified in primary human PBM and implicated significant in variations in bone phenotypes in humans. Supported by evidence from three levels of genetic information flow (i.e. protein < = RNA < = DNA), and supported by functional evidence from both in vivo and in vitro studies, our findings provide novel insights into the pathogenesis of osteoporosis in humans. The findings attest that comparative expression proteomics methodology opens a new way to identify gene(s) for complex diseases (herein osteoporosis). The approach is particularly significant given that genes are identified through potentially functional proteins that are associated with phenotypic and clinical differences in humans. The novel multidisciplinary and integrative study strategy, utilized in this study, should also be applicable to other complex diseases in humans. This strategy has substantial potential to help identify genes associated with these diseases, characterize their function, and elucidate the pathophysiological mechanisms underlying the development of these diseases.
Footnotes
* This study was benefited and/or partially supported by grants from NIH (P50AR055081, R21AG27110, R01AR057049, R01AR050496, R01AG026564, and R03TW008221), Franklin D. Dickson/Missouri Endowment and Edward G. Schlieder Endowment. This study was also benefited by the grant support from Natural Science Foundation of China (NSFC) (30600364, 30771222, and 30900810), NSFC-Canadian Institutes of Health Research (CIHR) Joint Health Research Initiative Proposal (30811120436), NSFC/Research Grants Council (RGC) of Hong Kong Joint Research Scheme (30731160618), Shanghai Leading Academic Discipline Project (S30501), and startup fund from Shanghai University of Science and Technology. YWL and YZ were partially supported by R01DC008603 and R01DC008603-S1.
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
All authors state that they have no conflicts of interest.
1 The abbreviations used are:
- BMD
- bone mineral density
- PBM
- peripheral blood monocyte
- PBMC
- peripheral blood mononuclear cells
- HDMS
- high definition mass spectrometry
- RMA
- robust multiarray average
- HUVEC
- human umbilical vein endothelial cells
- MCP-1
- monocyte chemotactic protein 1
- FBS
- fetal bovine serum
- PPIA
- peptidyl-prolyl cis-trans isomerase A.
REFERENCES
- 1. Harvey N., Dennison E., Cooper C. (2010) Osteoporosis: impact on health and economics. Nat. Rev. Rheumatol. 6, 99–105 [DOI] [PubMed] [Google Scholar]
- 2. Blume S. W., Curtis J. R. (2011) Medical costs of osteoporosis in the elderly Medicare population. Osteoporos. Int. 22, 1835–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Recker R. R., Deng H. W. (2002) Role of genetics in osteoporosis. Endocrine 17, 55–66 [DOI] [PubMed] [Google Scholar]
- 4. Xu X. H., Dong S. S., Guo Y., Yang T. L., Lei S. F., Papasian C. J., Zhao M., Deng H. W. (2010) Molecular genetic studies of gene identification for osteoporosis: the 2009 update. Endocr. Rev. 31, 447–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galli C., Zella L. A., Fretz J. A., Fu Q., Pike J. W., Weinstein R. S., Manolagas S. C., O'Brien C. A. (2008) Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology 149, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wan Y., Chong L. W., Evans R. M. (2007) PPAR-gamma regulates osteoclastogenesis in mice. Nat. Med. 13, 1496–1503 [DOI] [PubMed] [Google Scholar]
- 7. Sjögren K., Lagerquist M., Moverare-Skrtic S., Andersson N., Windahl S. H., Swanson C., Mohan S., Poutanen M., Ohlsson C. (2009) Elevated aromatase expression in osteoblasts leads to increased bone mass without systemic adverse effects. J. Bone Miner. Res. 24, 1263–1270 [DOI] [PubMed] [Google Scholar]
- 8. Nakanishi R., Akiyama H., Kimura H., Otsuki B., Shimizu M., Tsuboyama T., Nakamura T. (2008) Osteoblast-targeted expression of Sfrp4 in mice results in low bone mass. J. Bone Miner. Res. 23, 271–277 [DOI] [PubMed] [Google Scholar]
- 9. Roodman G. D. (2006) Regulation of osteoclast differentiation. Ann. N.Y. Acad. Sci. 1068, 100–109 [DOI] [PubMed] [Google Scholar]
- 10. Parfitt A. M. (2001) Skeletal heterogeneity and the purposes of bone remodelling: implications for the understanding of osteoporosis. In: Marcus R., Zfeldman D., Kelsey J. eds. Osteoporosis. San Diego: Academic Press, 433–444 [Google Scholar]
- 11. Parfitt A. M., Mundy G. R., Roodman G. D., Hughes D. E., Boyce B. F. (1996) A new model for the regulation of bone resorption, with particular reference to the effects of bisphosphonates. J. Bone Miner. Res. 11, 150–159 [DOI] [PubMed] [Google Scholar]
- 12. Pacifici R. (1996) Estrogen, cytokines, and pathogenesis of postmenopausal osteoporosis. J. Bone Miner. Res. 11, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 13. Cohen-Solal M. E., Boitte F., Bernard-Poenaru O., Denne M. A., Graulet A. M., Brazier M., De Vernejoul M. C. (1998) Increased bone resorbing activity of peripheral monocyte culture supernatants in elderly women. J. Clin. Endocrinol. Metab. 83, 1687–1690 [DOI] [PubMed] [Google Scholar]
- 14. Cohen-Solal M. E., Graulet A. M., Denne M. A., Gueris J., Baylink D., de Vernejoul M. C. (1993) Peripheral monocyte culture supernatants of menopausal women can induce bone resorption: involvement of cytokines. J. Clin. Endocrinol. Metab. 77, 1648–1653 [DOI] [PubMed] [Google Scholar]
- 15. Deng F. Y., Liu Y. Z., Li L. M., Jiang C., Wu S., Chen Y., Jiang H., Yang F., Xiong J. X., Xiao P., Xiao S. M., Tan L. J., Sun X., Zhu X. Z., Liu M. Y., Lei S. F., Chen X. D., Xie J. Y., Xiao G. G., Liang S. P., Deng H. W. (2008) Proteomic analysis of circulating monocytes in Chinese premenopausal females with extremely discordant bone mineral density. Proteomics 8, 4259–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farber C. R. (2010) Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J. Bone Miner. Res. 25, 2359–2367 [DOI] [PubMed] [Google Scholar]
- 17. Liu Y. Z., Dvornyk V., Lu Y., Shen H., Lappe J. M., Recker R. R., Deng H. W. (2005) A novel pathophysiological mechanism for osteoporosis suggested by an in vivo gene expression study of circulating monocytes. J. Biol. Chem. 280, 29011–29016 [DOI] [PubMed] [Google Scholar]
- 18. Kesisis G., Kontovinis L. F., Gennatas K., Kortsaris A. H. (2010) Biological markers in breast cancer prognosis and treatment. J. Buon. 15, 447–454 [PubMed] [Google Scholar]
- 19. Kuo Y. B., Chan C. C., Chang C. A., Fan C. W., Hung R. P., Hung Y. S., Chen K. T., Yu J. S., Chang Y. S., Chan E. C. (2011) Identification of Phospholipid Scramblase 1 as a biomarker and it's prognostic value for Colorectal Cancer. Mol. Med. 17, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Jong M. C., Pramana J., van der Wal J. E., Lacko M., Peutz-Kootstra C. J., de Jong J. M., Takes R. P., Kaanders J. H., van der Laan B. F., Wachters J., Jansen J. C., Rasch C. R., van Velthuysen M. L., Grènman R. A., Hoebers F. J., Schuuring E., van den Brekel M. W., Begg A. C. (2010) CD44 Expression Predicts Local Recurrence After Radiotherapy In Larynx Cancer. Clin. Cancer Res. 16, 5329–5338 [DOI] [PubMed] [Google Scholar]
- 21. Zhi W., Purohit S., Carey C., Wang M., She J. X. (2010) Proteomic technologies for the discovery of type 1 diabetes biomarkers. J. Diabetes Sci. Technol. 4, 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 23. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 24. Millenaar F. F., Okyere J., May S. T., van Zanten M., Voesenek L. A., Peeters A. J. (2006) How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics 7, 137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drmanac R., Sparks A. B., Callow M. J., Halpern A. L., Burns N. L., Kermani B. G., Carnevali P., Nazarenko I., Nilsen G. B., Yeung G., Dahl F., Fernandez A., Staker B., Pant K. P., Baccash J., Borcherding A. P., Brownley A., Cedeno R., Chen L., Chernikoff D., Cheung A., Chirita R., Curson B., Ebert J. C., Hacker C. R., Hartlage R., Hauser B., Huang S., Jiang Y., Karpinchyk V., Koenig M., Kong C., Landers T., Le C., Liu J., McBride C. E., Morenzoni M., Morey R. E., Mutch K., Perazich H., Perry K., Peters B. A., Peterson J., Pethiyagoda C. L., Pothuraju K., Richter C., Rosenbaum A. M., Roy S., Shafto J., Sharanhovich U., Shannon K. W., Sheppy C. G., Sun M., Thakuria J. V., Tran A., Vu D., Zaranek A. W., Wu X., Drmanac S., Oliphant A. R., Banyai W. C., Martin B., Ballinger D. G., Church G. M., Reid C. A. (2010) Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 [DOI] [PubMed] [Google Scholar]
- 26. Lei S. F., Tan L. J., Liu X. G., Wang L., Yan H., Guo Y. F., Liu Y. Z., Xiong D. H., Li J., Yang T. L., Chen X. D., Guo Y., Deng F. Y., Zhang Y. P., Zhu X. Z., Levy S., Papasian C. J., Hamilton J. J., Recker R. R., Deng H. W. (2009) Genome-wide association study identifies two novel loci containing FLNB and SBF2 genes underlying stature variation. Hum. Mol. Genet. 18, 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., Sham P. C. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. 57, 289–300 [Google Scholar]
- 29. Yuan H. Y., Chiou J. J., Tseng W. H., Liu C. H., Liu C. K., Lin Y. J., Wang H. H., Yao A., Chen Y. T., Hsu C. N. (2006) FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 34, W635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson E. A., Yoshimura T., Leonard E. J., Tanaka S., Griffin P. R., Shabanowitz J., Hunt D. F., Appella E. (1989) Complete amino acid sequence of a human monocyte chemoattractant, a putative mediator of cellular immune reactions. Proc. Natl. Acad. Sci. U. S. A. 86, 1850–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brownstein C., Deora A. B., Jacovina A. T., Weintraub R., Gertler M., Khan K. M., Falcone D. J., Hajjar K. A. (2004) Annexin II mediates plasminogen-dependent matrix invasion by human monocytes: enhanced expression by macrophages. Blood 103, 317–324 [DOI] [PubMed] [Google Scholar]
- 32. Falcone D. J., Borth W., Khan K. M., Hajjar K. A. (2001) Plasminogen-mediated matrix invasion and degradation by macrophages is dependent on surface expression of annexin II. Blood 97, 777–784 [DOI] [PubMed] [Google Scholar]
- 33. Li F., Chung H., Reddy S. V., Lu G., Kurihara N., Zhao A. Z., Roodman G. D. (2005) Annexin II stimulates RANKL expression through MAPK. J. Bone Miner. Res. 20, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 34. Menaa C., Devlin R. D., Reddy S. V., Gazitt Y., Choi S. J., Roodman G. D. (1999) Annexin II increases osteoclast formation by stimulating the proliferation of osteoclast precursors in human marrow cultures. J. Clin. Invest. 103, 1605–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu G., Maeda H., Reddy S. V., Kurihara N., Leach R., Anderson J. L., Roodman G. D. (2006) Cloning and characterization of the annexin II receptor on human marrow stromal cells. J. Biol. Chem. 281, 30542–30550 [DOI] [PubMed] [Google Scholar]
- 36. Takahashi S., Reddy S. V., Chirgwin J. M., Devlin R., Haipek C., Anderson J., Roodman G. D. (1994) Cloning and identification of annexin II as an autocrine/paracrine factor that increases osteoclast formation and bone resorption. J. Biol. Chem. 269, 28696–28701 [PubMed] [Google Scholar]