Abstract
Despite intense, continued interest in global analyses of signaling cascades through mass spectrometry-based studies, the large-scale, systematic production of phosphoproteomics data has been hampered in-part by inefficient fractionation strategies subsequent to phosphopeptide enrichment. Here we explore two novel multidimensional fractionation strategies for analysis of phosphopeptides. In the first technique we utilize aliphatic ion pairing agents to improve retention of phosphopeptides at high pH in the first dimension of a two-dimensional RP-RP. The second approach is based on the addition of strong anion exchange as the second dimension in a three-dimensional reversed phase (RP)-strong anion exchange (SAX)-RP configuration. Both techniques provide for automated, online data acquisition, with the 3-D platform providing the highest performance both in terms of separation peak capacity and the number of unique phosphopeptide sequences identified per μg of cell lysate consumed. Our integrated RP-SAX-RP platform provides several analytical figures of merit, including: (1) orthogonal separation mechanisms in each dimension; (2) high separation peak capacity (3) efficient retention of singly- and multiply-phosphorylated peptides; (4) compatibility with automated, online LC-MS analysis. We demonstrate the reproducibility of RP-SAX-RP and apply it to the analysis of phosphopeptides derived from multiple biological contexts, including an in vitro model of acute myeloid leukemia in addition to primary polyclonal CD8+ T-cells activated in vivo through bacterial infection and then purified from a single mouse.
Reversible phosphorylation plays a central role in the regulation of normal cell physiology. The strong links between aberrant signaling and human disease, along with the potential for specific inhibition of disrupted kinase activity, continue to drive efforts aimed at systematic and large-scale analysis of phosphorylation in cells and tissues. Shortly after introduction of immobilized metal affinity chromatography (IMAC)1 as an enrichment tool prior to mass spectrometry (MS) analysis (1–3), several laboratories demonstrated the feasibility of phosphopeptide identification and quantitation en masse (4–7). In the ensuing years, despite widespread proliferation of improved and innovative (8–14) phosphoproteomics methods, the field struggled with low specificity and poor reproducibility within and across protocols and laboratories. These limitations effectively made dynamic range a secondary issue for the majority of studies. Over the past ca. 5 years, the performance of phosphopeptide enrichment protocols and related methods has stabilized; in fact several groups (15–23) have successfully coupled phosphopeptide enrichment with online or offline fractionation schemes to achieve, in some cases, over 10,000 phosphopeptide identifications. Although these strategies provide for larger phosphosite catalogs, closer inspection reveals that the analytical efficiency, as measured by the number of phosphopeptide identifications per microgram of biological lysate consumed, has remained surprisingly consistent at ≈1–10 phosphopeptides/μg across a wide range of sample types (Table I). One explanation is that the physicochemical properties of phosphopeptides render them less amenable to fractionation by commonly used techniques. For example, although the combination of strong cation exchange (SCX) with reversed phase (RP) has been tremendously successful for fractionation of tryptic peptides generally (24–26), its performance in phosphoproteomics studies has been hampered in-part by poor phosphopeptide binding in the first dimension (SCX) (10, 27) and low separation peak capacity (28). These results are consistent with the modest difference in pKa between the binding sites of typical SCX resins and phosphorylated side chains of serine, threonine, and tyrosine amino acids. These limitations have led to the development of numerous other strategies for fractionation of phosphopeptides including strong anion exchange chromatography (17, 22), continuous pH gradients (29), ERLIC (30, 31), and hydrophilic interaction chromatography (19), all utilizing RP chromatography for the second dimension separation. In this report we describe two approaches for fractionation of phosphopeptides that build upon our recent work (32) in coupling true, nanoflow chromatography with reversed phase-reversed phase (RP-RP) fractionation (28). First we utilize aliphatic-functionalized quarternary amines as ion pairing agents to improve retention of phosphopeptides in the first dimension RP separation performed at high pH. In a second approach, we use the standard ammonium formate RP-RP buffer system (28), and add anion exchange as a second dimension in a 3-D, RP-SAX-RP configuration. The latter provided for the highest performance in terms of separation peak capacity, orthogonality, and the number of unique phosphopeptide sequences identified per μg of cell lysate consumed. We demonstrate the technical reproducibility of RP-SAX-RP at various fractionation depths and input levels. Finally, we use RP-SAX-RP for the quantitative analysis of divergent signaling between two clinically relevant, constitutively active FLT3 mutants in an in vitro model of acute myeloid leukemia, in addition to qualitative identification of phosphopeptides in primary polyclonal CD8+ T-cells activated in vivo through bacterial infection and then purified from a single mouse. Collectively our RP-SAX-RP platform provides significantly improved efficiency (IDs/microgram input) along with multiple analytical figures of merit, including: (1) orthogonal separation mechanisms in each dimension; (2) high separation peak capacity; (3) efficient retention of singly- and multiply phosphorylated pepitdes; (4) compatibility with automated, online LC-MS analysis.
Table I. A survey of recent large scale phosphoproteomics studies. This list is non-comprehensive and some subsets of data were not readily available.
| Reference | Source | Starting material | Phospho peptides | IDs/μg | Phosphorylation sites | Sites/μg |
|---|---|---|---|---|---|---|
| (112) | K562, Hela | 20 mg | 5689 | 0.3 | 5064 | 0.3 |
| (15) | Hela | 12 mg | 68379 | 5.7 | 14265 | 1.2 |
| (113) | Drosoph | 1.5 mg | 10097 | 6.7b | ||
| (8) | K562 | 100 μg | 1100 | 11.0 | ||
| (114) | hESC | 1 mg | 3090 | 3.1 | 3067 | 3.1 |
| (115) | hESC | 10 mg | 51920 | 5.2 | 62642 | 6.3 |
| (96) | HeLa | 10 mg | 24714 | 2.5 | ||
| (69) | 32D | 10 mg | 14700 | 1.5 | ||
| (116) | Jurkat | 10 mg | 11708 | 1.2 | 10665 | 1.1 |
| (117) | hESC | 2 mg | 2548 | 1.3 | ||
| (118) | HeLa | 400 μg | 1940 | 4.9 | ||
| (119) | Hepa1–6 | 10 mg | 5433 | 0.5 | ||
| (120) | Yeast | 10 mg | 74093 | 7.4 | 10656 | 1.1 |
| (121) | Livera | 1 mg | 305 | 0.3 | 274 | 0.3 |
| (122) | Braina | 5 mg | 551 | 0.1 | 466 | 0.1 |
| (123) | Plateletsa | 5 mg | 630 | 0.1 | 674 | 0.1 |
| (110) | Tcellsa | 4 mg | 253 | 0.1 | 281 | 0.1 |
| (124) | Musclea | 3 mg | 879 | 0.3 | ||
| (125) | Braina | 3 mg | 8014 | 2.7 | ||
| (126) | hMSC | 120 μg | 492 | 4.1 | 716 | 6.0 |
| (127) | hMSC PM | 100 μg | 757 | 7.6 | 703 | 7.0 |
| (19) | Hela | 300 μg | 814 | 2.7 | 1004 | 3.3 |
a Denotes analysis of primary tissue.
b Denotes detected features after enrichment.
EXPERIMENTAL PROCEDURES
All multiplierz scripts referenced in the manuscript are freely available on our website at:http://blais.dfci.harvard.edu/index.php?id = 64.
Materials
Magnetic Ni-NTA-agarose was obtained from Qiagen (Valencia, CA). Acetonitrile, EDTA, FeCl3, urea, and ammonium bicarbonate were from Sigma-Aldrich (St. Louis, MO). Trifluoroacetic acid was obtained from Pierce (Rockford, IL). Phosphopeptides EEpSGpSpSEEEAVLQR and LIEDAEpYTAK were synthesized using Fluorenylmethyloxycarbonyl chemistry.
K562 Cell Culture and Preparation of Digested Lysate
K562 cells were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in 5% CO2. Cells (≈5e7) were harvested by centrifugation during log phase. After washing twice with 20 ml phosphate buffered saline, the pellet was lysed with 3 ml of 8 m urea, 100 mm ammonium bicarbonate, and 30 μl each of Sigma-Aldrich phosphatase inhibitor cocktails I and II. Protein concentration was determined using the Bradford Assay (Bio-Rad laboratories, Hercules, CA). Proteins were reduced by adding dithiothreitol to a final concentration of 10 mm and incubating for 30 min at 60 °C, and alkylated with iodoacetamide (final concentration 20 mm) for 30 min in the dark at room temperature. Excess iodoacetamide was quenched by the addition of dithiotreitol to a final concentration of 20 mm. This solution was diluted to a final volume of 12 ml in 0.1 m ammonium bicarbonate. Trypsin (150 μg, 1:50 enzyme:substrate) was added and digestion was performed at 37 °C overnight. The resulting peptide solution was acidified with 10% trifluoroacetic acid (TFA), desalted on a C18 solid phase extraction cartridge and eluted with 25% acetonitrile with 0.1% TFA. Aliquots of eluted peptides were lyophilized by vacuum centrifugation and stored at −80 °C.
Batch Mode Phosphopeptide Enrichment
Magnetic Ni-NTA agarose beads were washed 3× with 400 μl water, and treated with 400 μl of 100 mm EDTA, pH 8.0 for 30 min with end-over-end rotation. EDTA solution was removed, and beads were then washed 3× with 400 μl water, and treated with 600 μl of 10 mm aqueous FeCl3 solution for 30 min with end-over-end rotation. After removing excess metal ions, beads were washed 3× with 400 μl water, and 1× with 400 μl 1:1:1 acetonitrile:methanol:0.01% acetic acid. Tryptic peptides (in 80% MeCN/0.1% TFA; typical peptide concentrations were ≤1 μg/μl; see Supplemental Experimental Procedures for additional details) were then added to the beads, and phosphopeptide capture proceeded for 30 min with end-over-end rotation. After removing the supernatant, beads were washed 3× with 400 μl 80% acetonitrile/0.1% TFA. Phosphopeptides were eluted with 50 μl 1:20 ammonia/water for 30 min, dried to ≈5 μl by vacuum centrifugation, and reconstituted with 20 mm ammonium formate buffer.
Two-dimensional RP-RP and Three-dimensional RP-SAX-RP Systems
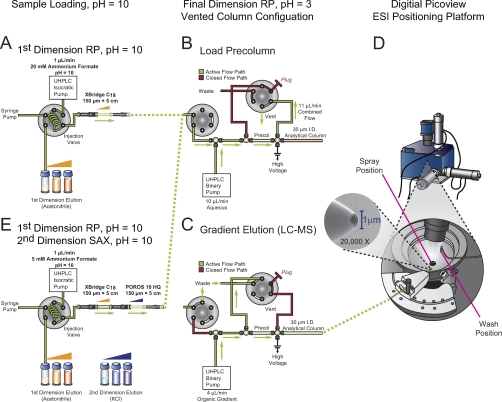
The multidimension fractionation platform (Fig. 1) consisted of a two-pump UPLC system with autosampler (Waters, Milford, MA) and an external valve (Valco, Austin, TX). For RP-RP experiments (Fig. 1A), the first dimension analytical column consisted of a 150 μm I.D. capillary packed with 5 cm of 5 μm C18 (XBridge, Waters). The weak needle wash and isocratic pump delivered various ion pairing agents all at pH 10.0. To transfer peptides between the first and second dimension columns, the trapping and vent valves were positioned to direct flow to the second dimension precolumn (150 μm I.D. capillary packed with 4 cm of POROS10R2, Applied Biosystems, Framingham, MA), the isocratic pump delivered 1 μl/min of pH 10 buffer (with sample loop of 20 μl in-line), and the binary pump delivered 10 μl/min of 0.2 m acetic acid (to dilute organic content and acidify the first dimension column effluent) for 25.5 min. Elution of peptides was accomplished by injection of acetonitrile fractions at pH 10.0 with or without an ion-pairing agent. After trapping, both valve positions were switched, to create a precolumn split and allow for gradient elution (2–30% B in 60 min, A = 0.2 m acetic acid, B = acetonitrile with 0.2 m acetic acid) of peptides to the analytical column (30 μm I.D. capillary packed with 12 cm of 5 μm Monitor C18, Column Engineering, Ontario, CA containing integrated 1 μm emitter tip), and into the mass spectrometer (Fig. 1B) at a flow rate of ≈30 nL/min. Three-dimensional RP-SAX-RP experiments were conducted in a similar fashion, except that an additional anion exchange column (150 μm I.D. capillary packed with 5 cm of POROS10HQ) was connected to the outlet of the first dimension RP column. Elution of peptides trapped on this column was performed using injections of ammonium formate, or potassium chloride at pH 10.0. Experiments were designed to transfer fractions of equivalent peptide complexity to the final dimension column with each injection. Toward this end, organic eluents used to elute peptides from the first to second dimensions typically included a low concentration of salt (70 mm ammonium formate or 10 mm KCl) to simultaneously elute a subset of peptides to the final dimension column in the same trapping event (see Supplemental Experimental Procedures). After completion of a fractionation experiment, columns were regenerated with 10 μl 90% acetonitrile, 10 μl of 1 m KCl in 10% acetic acid, 10 μl of 50% acetonitrile with 500 mm ammonium formation, and 10 μl of 900 mm ammonium formate in 10% acetic acid. In addition, the final dimension column was subjected to a series of three rapid (5 min.) organic gradients.
Fig. 1.
Schematic of automated, online nanoflow multidimensional platform for fractionation of phosphopeptides. A, For RP-RP, the autosampler first loads sample and is then used to inject first dimension eluents (acetonitrile, orange). B, C, An additional six-port, two-position valve is used to configure a vented column geometry in the final dimension and provide efficient capture of peptides on the low-pH RP precolumn. B, First dimension fractions are diluted (10:1) and acidified with RP solvent A (0.1 m acetic acid) introduced by an ultra-high pressure binary pump. C, The same binary pump then delivers an organic gradient to elute peptides from the second dimension column for MS/MS analysis. The precolumn effluent split provided a stable column/ESI flow rate of ≈30 nL/min. Active solvent flow paths are highlighted in green. D, A computer-controlled positioning platform (Digital PicoView) automatically moves the emitter tip between “electrospray” and “wash” positions during data acquisition and sample loading, respectively. E, A SAX column is connected to the high pH first dimension to create an online RP-SAX-RP fractionation platform. After loading phosphopeptides, the autosampler is used to inject first- (acetonitrile, orange) or second- (KCl, blue) dimension eluents, respectively.
MS and MS/MS Parameters on the LTQ-Orbitrap XL Mass Spectrometer
The LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific, San Jose, CA) was programmed to operate in data dependent mode, such that the top 10 most abundant precursors in each MS scan were subjected to MS/MS (CAD, electron multiplier detection, collision energy = 35%, isolation width = 2.8 Da, threshold = 20,000). Dynamic exclusion was enabled with a repeat count of 1 and an exclusion duration of 30 s. ESI voltage was 2.2 kV. Additional parameters can be found in Supplemental Experimental Information.
Quantitative Phosphoproteomics Analysis of Acute Myeloid Leukemia (AML) Model System (WT, WT + FL, ITD, D835Y)
Murine BaF3 cells that stably expressed wild-type (WT), or constitutively active mutant forms (ITD and D835Y, respectively) of the receptor tyrosine kinase FLT-3 were cultured as described (33). WT cells were serum starved overnight and were left untreated as a control, or were treated with 50 ng/ml of FLT-3 ligand (FL) for 5 min. Cells were collected, rinsed with cold PBS, and lysed in 8 m urea. Solubilized lysates were diluted to a final urea concentration of 1.8 m with 50 mm ammonium bicarbonate, and protein concentration was measured by BCA assay. Protein lysates (∼2E6 cells corresponding to ∼100 μg protein) were digested with trypsin after reduction with 10 mm dithiotreitol and alkylation with 55 mm iodoacetamide. For each biological condition (WT, WT+FL, ITD, D835Y), 100 μg of tryptic peptides were reconstituted with 30 μl 1 m triethylammonium bicarbonate, 70 μl ethanol, and one tube of iTRAQ reagent (114, WT; 115, WT+FL; 116, D835Y; 117, ITD). Labeled peptides were mixed, dried by vacuum centrifugation, desalted, and reconstituted in 400 μl of 80% MeCN, 0.1% TFA, and phosphopeptides enriched via batch mode enrichment (described above). Phosphopeptides were subjected to a 67 fraction three-dimensional RP-SAX-RP MS/MS analysis using a QSTAR Elite mass spectrometer (AB Sciex, Foster City, CA).
MS and MS/MS Parameters on the QSTAR Elite Mass Spectrometer
MS spectra were acquired for 0.5 s. A minimum threshold of 50 counts was used for MS/MS, with a charge state range of 2+ to 4+ and a multiplier value of 4. Up to 5 precursors could be selected for each cycle, with a 20 s exclusion time for each precursor selected for MS/MS. One broad range collisionally activated dissociation (CAD) (m/z: 140–2000) and narrow range CAD (m/z: 104–125) were acquired for each precursor. A fixed collision energy of 60 eV was used for narrow range CAD scans.
Automated Tip Washing and Positioning
Digital PicoView ESI sources (New Objective, Woburn, MA; model DPV-550 for the Orbitrap XL and DPV-400 for the QSTAR Elite) were used to facilitate positioning of the emitter tip at the orifice of the mass spectrometer during each analysis (Fig. 1D). While loading samples or trapping peptides on the final dimension precolumn, the tip was positioned at a wash station and bathed in a continuous stream of water.
Western Blotting
Proteins were resolved by SDS-PAGE on a 10% acrylamide gel, transferred to nitrocellulose, and detected by enhanced chemiluminescence using a Fugi LAS-4000 imager. Rabbit α-mouse RUNX1 (clone EPR3099) was from Epitomics (Burlingame, CA). Rabbit α-mouse PKCδ phosphoY311 (EP2609Y) was from Abcam (Cambridge, MA). STAT5 phospho Y694 (#9359, C11C5) was from Cell Signaling Technology (Beverly, MA).
Data Processing
Orbitrap data files were directly accessed and converted to .mgf using default parameters of the multiplierz script orbi2mgf.mz. Several scans in the first data file of each experiment were manually summed in XCalibur to derive the mass error of Si(CH3)2O))6. This value was used to re-calibrate precursor masses in .mgf files from each set of experiments using the proportional equation described by Wenger et al. (34) (recalibrated mz = mz ×(445.120025/observed Si(CH3)2O))6 m/z; RECAL Orbi data from mgf.mz). QSTAR files were converted to .mgf using Mascot Daemon (2.2.0, settings are described in Supplemental Experimental Information), and .mgf from a given experiment were re-calibrated using a linear equation derived from fitting experimentally observed masses in a high confidence peptide identification (RECAL QSTAR data from mgf.mz). Files were searched using Mascot version 2.2.1 against a forward-reversed human (38190 forward entries) (35) or mouse (34967 forward entries) (36) NCBI refseq database (both downloaded Nov. 2009) with an appended cRAP (common repository of adventitious proteins(37)) database of 752 entries. Precursor and product ion tolerances were 10 ppm and 0.6 Da, or 100 ppm and 0.1 Da for Orbitrap or QSTAR data, respectively. Search parameters included trypsin specificity, up to two missed cleavages, fixed carbamidomethylation (C, +57 Da), variable deamidation (NQ, +1 Da), oxidation (M, +16 Da), and phosphorylation (STY, +80 Da), and, when applicable, fixed iTRAQ modification (N-term, K +145 Da). After searching, an excel spreadsheet containing the mascot search results was generated using Multiplierz software (Geryon edition 0.7.188) (38). Data were processed to remove reverse database hits and forward hits with false discovery rate (FDR) >1.0%, (forward-reverse_FDR_filter.mz). A list of unique peptides (FDR<1%) from each analysis was generated using the multifile detect feature of multiplierz, and further processed using a script to count the number of unique phosphorylated peptides (39). Unique phosphorylated peptides were defined as unique combinations of sequence and phosphorylation state, i.e. the same sequence phosphorylated at different residues is counted only once, whereas the same sequence appearing with different numbers of phosphorylation sites would be counted according to degree.
Phosphorylation Site Localization
Phosphorylation site localization was evaluated using a multiplierz script that implemented the MD-score method of Savitski et al. (40). Briefly, the MD score of a phosphopeptide is the difference in Mascot score between the highest ranking and the next highest-ranked peptide hit with the same sequence but different assigned phosphorylation site(s). If no such hit exists, the peptide score was used as the MD score. To quantify phosphorylation sites on the protein level, a score threshold was used to filter the peptides at a false localization rate of 1%. This threshold is instrument specific and we employed values recommended by Savistki et al. (40), specifically 11 for Orbitrap data and 10 for QSTAR data.
iTRAQ Reporter Ion Extraction and Processing
For relative quantitation, iTRAQ reporter ion peak intensity values were extracted from narrow range CAD scans (wiff_iTRAQ.mz) and corrected for known isotopic impurities (iTRAQ_software_corrections.mz). A separate LC/MS/MS analysis of the Fe3+-NTA resin supernatant was performed, and signal in each quantitation channel (114, 115, 116, and 117) was summed (across all peptides) and used to derive normalization factors to further correct the phosphopeptide data for small variations in source protein concentration. For peptides with multiple MS/MS scans, the associated iTRAQ reporter signal intensity values were summed prior to calculation of ratios.
Analysis of Reproducibility and Fraction-to-fraction Overlap
Extracted ion chromatograms (XICs) were generated for reproducibly detected peptides from replicate three-dimensional RP-SAX-RP experiments using deisotoped MS scan data. For each peptide, an XIC was generated (across every fraction) using a time window corresponding to the earliest RT - 0.5 min to the latest RT + 0.5 min and a mass tolerance of 0.015 Da. The precursor signal for a peptide was obtained by summing XIC apex intensities across all fractions in an individual experiment. Fraction to fraction overlap for a given peptide was evaluated by expressing the maximum peptide intensity across all fractions as a percentage of the total signal.
Calculation of Peak Capacity
Peak capacity of a given fraction was estimated by dividing the useful peptide elution range (retention time of last peptide identification – retention time of first identification) by the median peptide peak width at the XIC base. Base peak widths were estimated by multiplying half-height XIC peak widths by a factor of 2. The estimated peak capacity of an experiment was obtained by summation of the peak capacities of the individual fractions.
Orthogonality Plots
To visualize the orthogonality of the three-dimensional RP-SAX-RP experiments, we plotted second dimension salt concentration versus first dimension acetonitrile concentration (pH 10.0) and represented the number of phosphopeptides identified in each third dimension LC-MS/MS analysis as circles of proportional diameter. To focus on useful peptide fractionation space, high salt (>700 mm) and organic (>25%) fractions were excluded from analysis. Note that each phosphopeptide identification was considered a data point for the linear regression.
Analysis of Amino Acid Frequencies and Permutation Test
A list of unique phosphopeptide sequences (with no missed cleavages) from phosphoELM (release 8.3; March 2010) in text file format was generated using Phosphopeptides_from_ELM.mz. Acidic amino acid frequencies were evaluated by counting the number of each amino acid and dividing by the total number of residues, and were plotted as log ratios (base 2). To evaluate the significance of amino acid biases between specific analyses, we used a permutation test as described previously.(41) Briefly, peptide sequences in each dataset were combined and then randomly permuted so that the number of peptides in each dataset remained constant. After each permutation, amino acid frequencies and log ratios were recalculated for a total of 10,000 times. One-sided p values were calculated by summing the frequency (probability) of all log ratios equal to or more extreme than the observed log ratios and were doubled to yield two-sided p values. Lastly, two-sided p values were Bonferroni corrected based on the number of amino acids. Frequency of occurrence was considered significant for p ≤ 0.01.
Infection of Mice
C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in specific pathogen-free conditions. All animal procedures were approved by the Center for Animal Resources and Comparative Medicine (ARCM, Harvard Medical School, Boston, MA). Recombinant Listeria monocytogenes expressing chicken ovalbumin (Lm-OVA) (30) was provided by Dr. Michael Starnbach (Harvard Medical School). Bacteria were grown to early exponential phase in brain heart infusion broth at 37 °C for 1 h, spun down by centrifugation at 3600 rpm for 10 min at 4 °C (Allegra 6KR Kneewell Centrifuge, Beckman Coulter, Fullerton, CA) and resuspended in phosphate buffered saline (PBS). Mice were infected at 8 weeks of age by injecting ≈1.6 × 103 colony forming units of live Lm-OVA i.v. into the lateral tail vein. The inoculum dose was confirmed by duplicate dilution plating on streptomycin-containing Tryptic Soy Agar plates. Bacteria and infected animals and tissues were manipulated in Biosafety Level 2 (BL2) biohazard containment cabinets.
Isolation of in Vivo-activated Polyclonal Primary CD8+ T Lymphocytes
At the peak of the immune response to Lm-OVA (8 days post infection), mice were sacrificed and the spleens removed in T-cell medium consisting of phenol-red free RPMI 1640/2 mm l-glutamine medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mm Hepes, 1 mm sodium pyruvate, 100 μm nonessential amino acids (Invitrogen, Carlsbad, CA), 50 μm 2-mercaptoethanol and 10% (v/v) fetal bovine serum (Sigma-Aldrich, St. Louis, MO). Splenocytes from each spleen were harvested separately in T-cell medium and filtered through a 40 μm nylon cell strainer (BD Biosciences, San Jose, CA). Splenic CD8+ T cells were enriched by magnetic depletion of non-CD8+ T cells on LD columns (Miltenyi Biotec, Auburn, CA), using biotin-conjugated monoclonal anti-mouse antibodies against B220 (clone RA3–6B2), Gr1 (clone RB6–8C5), CD49b (clone DX5), NK1.1 (clone PK136), CD11b (clone M1/70) (2.5 μg/ml, eBioscience, San Diego, CA), CD4 (clone RM4–5, 0.5 μg/ml, Invitrogen) and TER-119 (clone TER-119, 2.5 μg/ml, BioLegend, San Diego, CA), secondarily labeled with anti-biotin MicroBeads (Miltenyi Biotec), all diluted in ice-cold magnetic-activated cell sorting (MACS) buffer (PBS supplemented with 0.5% bovine serum albumin and 2 mm EDTA). Enriched cells were stained at 4 °C for 30 min using monoclonal PE-conjugated anti-mouse B220 (1 μg/ml, clone RA3–6B2, eBioscience), FITC-conjugated anti-mouse CD8 (5 μg/ml, clone 53–6.7, Biolegend) and APC-conjugated anti-mouse CD44 (1 μg/ml, clone IM7, eBioscience) antibodies diluted in MACS buffer. After washing the stained cells with MACS buffer, activated polyclonal CD8+ T cells, defined by the expression of intermediate and high levels of CD44 (CD44int/high), were isolated in MACS buffer at 4 °C using a BD FACSAria cell sorter (BD Biosciences) installed inside a BL2 biohazard containment cabinet (Immune Disease Institute, Harvard Medical School). CD44int/high CD8+ T cells were identified after gating on live lymphocytes (by plotting forward versus side scatters) and excluding both cell aggregates (by plotting forward scatter pulse width versus pulse area) and B220+ cells, using a BD FACSDiva Software (BD Biosciences). The number of viable cells obtained after sorting was ≈1–3 × 106 cells/spleen, as determined by light microscopy using the Trypan Blue (Sigma-Aldrich) exclusion method. The purity of the sorted cells was >90% viable, >94% CD8+/B220−, and >99% CD44int/high. Collected data was analyzed using FlowJo V7.6.1 Software (Tree Star, Ashland, OR). Isolated CD44int/high CD8+ T cells were lysed as described below.
Preparation of CD8+ T-Cell Lysates
Sorted CD8+ T cells (≈1–3 × 106) were spun down by centrifugation at 1000 rpm for 5 min at 4 °C (Allegra 6KR Kneewell Centrifuge, Beckman Coulter, Fullerton, CA); washed by centrifugation at 8000 rpm for 2 min at 4 °C (Microcentrifuge 5415R, Eppendorf, Westbury, NY) with cold PBS containing 5 mm sodium fluoride, 1 mm sodium orthovanadate (New England Biolabs, Ipswich, MA) and 1× Halt Phosphatase Inhibitor Mixture (Thermo Scientific, Rockford, IL), and lysed with 150 μl of cold 8 m urea in 100 mm ammonium bicarbonate containing phosphatase inhibitors as above. Protein concentrations were determined using the MicroBCA Protein Assay (Thermo Scientific) after diluting an aliquot of the cell lysates with distilled water to <1 M urea. The total amount of protein obtained from the sorted cells was ≈5–10 μg/spleen. Proteins were reduced and alkylated as described above, and then desalted using a self-packed C18 column (360 μm O.D. × 250 μm I.D. with 8 cm of resin). Phosphopeptides were enriched in batch mode as described above and analyzed in a 10 fraction RP-SAX-RP MS/MS experiment using a LTQ-Orbitrap XL (ThermoFisher Scientific, Waltham, MA).
Safety Considerations
Trifluoroacetic acid is corrosive, and should be handled in a fume hood with appropriate protective equipment as described in the manufacturer's material safety data sheet. Recombinant Lm-OVA and infected mice and cells should be handled in a BL2 cabinet.
RESULTS
All native mass spectrometry data and results (mzResults) files, including annotated MS/MS spectra and other metrics as described in the Philadelphia Guidelines are freely accessible online at: pcXKvH75Q+mGbF8do6hCUB0LvzwqP2EjY4l04ccANl7zNxLpR6l8NZ3Vv9sAYlrAQd+24DWWsQgLIiLFz+L4irJPpZMAAAAAAAFkjA==
Multiphosphorylated Peptides Are Poorly Retained on RP at pH 10
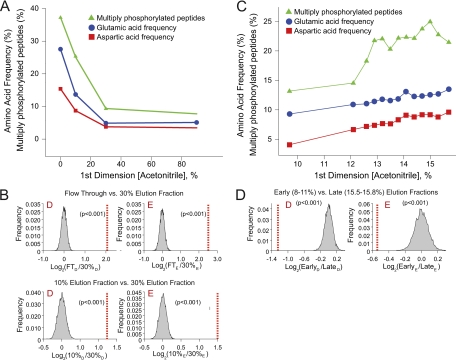
We recently described a true nanoflow, online platform (32) for UHPLC-compatible RP-RP fractionation (28) of tryptic peptides. Fig. 1 (A–D) shows a schematic representation of our two-dimensional nanoflow system and related modifications (Fig. 1E) required for our three-dimensional implementation (see below and Methods). Briefly, our platform is based on a Waters NanoACQUITY UHPLC modified to accept self-packed columns (42) and reconfigured with an additional two-position, six-port valve to enable a precolumn effluent split, in a vented column configuration (43, 44), that allows for gradient elution of peptides in the second dimension at low nanoliter per minute flow rates (Fig. 1B and 1C). A UHPLC binary pump generates the solvent gradient for LC-MS/MS in the usual manner, whereas the second on-board pump is used in isocratic mode to introduce samples or first dimension eluents loaded from autosampler vials. In contrast to previous reports (10, 15–22, 45, 46), our system provides an automated, online platform that leverages the combined advantages of multidimensional fractionation (25, 28) and high efficiency electrospray ionization achieved at ultralow effluent flow rates (42). Given the anecdotal evidence that phosphopeptides are particularly susceptible to losses as a result of sample handling procedures, in addition to the predicted (47) and observed (10, 27) weak retention of phosphopeptides on SCX resins, we reasoned that our RP-RP platform would provide an improved fractionation system for analysis of phosphopeptides. To evaluate the analytical capabilities of this platform, we used Fe-NTA (8) to enrich phosphopeptides from ≈100 μg of human myeloid K562 cell lysate. Peptides were loaded through the NanoAQUITY autosampler, onto the first dimension RP column in pH 10 ammonium formate buffer, and analyzed by RP-RP-MS/MS. Data were acquired for four fractions, including the loading flow-through. In total we identified 1356 unique phosphopeptides (<1% FDR, see supplemental Fig. S1 and supplemental File 1) across all four fractions, corresponding to ∼14 IDs per microgram of cell lysate. Only 73 nonphosphorylated peptides were observed, and less than 12% of all identified peptides were detected in more than one fraction, indicating high selectivity (≈95%) for Fe-NTA based enrichment and minimal fraction-to-fraction overlap, respectively. Interestingly, we noticed that a significant number (306, or ≈23% of the total) of phosphopeptides were detected in the loading flow-through. To explore the physicochemical nature of these peptides, we plotted the percentage of multiply phosphorylated peptides in addition to the frequency of acidic residues, as a function of first dimension acetonitrile concentration (Fig. 2A). We observed that the flow-through fraction contained a higher percentage of multiply-phosphorylated peptides (37%) as compared with that observed across the total of all peptides detected (21%). At pH = 10 each phosphorylated side chain will carry two negative charges (pKa1≈1.2 and pKa2≈6.5) (47, 48) and bind weakly to RP resin, in accordance with solvophobic theory (49); poor retention would be further exacerbated for multiply-phosphorylated peptides, in agreement with data in Fig. 2A. Similarly, acidic residues (D, red and E, blue) occurred with greater frequency in the un-retained and 10% acetonitrile fractions (D+E ≈43% and ≈22%, respectively) as compared with the 30% acetonitrile fraction (≈8%, Fig. 2A), or more generally as compared with all unique phosphopeptide sequences in the phosphoELM database (≈16%) or the human refseq database (22) (≈12%, supplemental Fig. S2). Next, we applied a permutation test (41) to verify the statistical significance of the apparent bias for D (p = <0.001) and E (p = <0.001) residues in phosphopeptides detected in the flow-through and 30% fractions, or similarly between the 10 and 30% acetonitrile fractions (Fig. 2B).
Fig. 2.
Two-dimensional RP-RP analysis of phosphopeptides from K562 proteins. A, Percentages of acidic amino acids and multiply phosphorylated peptides observed in a four-fraction two-dimensional RP-RP analysis. B, Null distributions derived from permutation tests for aspartic and glutamic acid residues. Dashed red line represents the log ratio (base 2) of the observed amino acid frequencies relative to the null distributions. For each permutation test, a p-value < 0.001 indicates a statistically significant bias for a given residue in the flow-through (B, top) or 10% acetonitrile (B, bottom) fractions versus the 30% acetonitrile fraction. C, Percentages of acidic amino acids and multiply phosphorylated peptides observed in a 73 fraction two-dimensional RP-RP analysis with tetrabutylammonium ion as an ion pairing agent. D, Permutation test (as in panel B) indicates that later fractions are enriched for acidic residue-rich peptides.
Based on the above results, we next asked whether alternative organic modifiers could be used to improve phosphopeptide retention on the first dimension column at high pH. In fact, data from previous studies demonstrated that functionalized tertiary and quaternary amines will effectively ion pair with acidic moieties in various biomolecules at high pH to improve retention and separation on RP resins (50–58). Encouragingly several of these compounds have been used in LC-MS applications (50–54, 56). Toward this end, we first prepared a series of synthetic phosphopeptides, including a triply phosphorylated sequence, and then verified that their retention at high pH generally increased in the presence of ion pairing reagents with greater hydrophobicity (supplemental Table S1). Of the compounds tested, we observed that tetrapropylammonium ion provided the strongest and most reproducible retention of phosphopeptides on RP resin at basic pH, even at concentrations as low as 5 mm. Next we again enriched phosphopeptides from human myeloid K562 cell lysate and performed RP-RP-MS/MS in which 73 fractions were generated in 5 mm ammonium formate buffer with 5 mm tetrapropylammonium ion. In this experiment (supplemental Fig. S3), we detected 4647 unique phosphopeptides (<1% FDR, ≈12 ID/μg, see Supplemental File 2) and only 340 nonphosphorylated peptides (selectivity ≈93%). Importantly, no phosphorylated peptides were observed in the flow-through fraction, indicating that the tetrapropylammonium ion provided for improved retention of phosphopeptides derived from complex biological extracts on RP resin under alkaline conditions.
To better elucidate the ion pairing properties of this compound, we repeated the analysis described above and observed a positive correlation between relative acidic content and the acetonitrile concentration required for elution from the first dimension column (Fig. 2C) as well as a statistical enrichment of acidic residues in the late- versus early-eluting fractions (Fig. 2D). These results confirm that the overall acidity of phosphopeptides impedes their binding to RP resin at high pH. Despite the capacity of tetrapropylammonium ion to modulate the hydrophobicity of phosphopeptides, we found that only 41 of the 73 fractions contained an appreciable number (>50) of sequence identifications and that some 52% of phosphopeptides were detected across multiple second dimension fractions. In addition, we observed that ≈18% acetonitrile was sufficient to elute the majority of phosphopeptides from the first dimension column (supplemental Fig. S3). Collectively these data, along with those from previous studies that utilized SCX-based fractionation techniques (10, 15, 27), demonstrate that the acidity of phosphopeptides presents unique challenges with respect to high performance, multidimensional separation strategies for proteomics applications.
Three-dimensional Separation Based on RP-SAX-RP Enables Efficient Fractionation and Identification of Phosphopeptides
Based on the results above, we next asked whether addition of anion exchange as a second dimension, in a three-dimension, RP-SAX-RP geometry (Fig. 1E), would provide an effective fractionation platform. In this configuration, phosphopeptides captured by SAX, either from the loading flow-through or via controlled elution from the first dimension RP column at high pH, are subsequently eluted onto the third dimension RP column with increasing concentrations of salt. To test this strategy, we first injected the phosphopeptide standards prepared in ammonium formate buffer (20 mm, pH = 10) onto a tandem set of RP and anion exchange columns. As expected, the triply phosphorylated peptide bound strongly to the SAX column under these conditions and was only observed in the third dimension LC-MS/MS analysis upon elution with 500 mm KCl; less acidic phosphopeptide standards required a combination of acetonitrile and KCl for elution from the first and second dimension columns, respectively (supplemental Table S2). In essence, this system provides for a three-dimensional analysis of phosphopeptides, with two orthogonal dimensions of RP, and a charge based anion exchange separation.
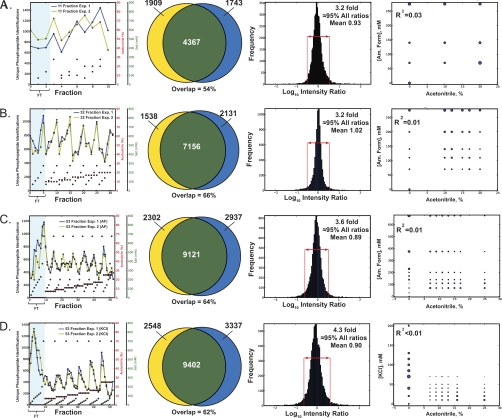
We next sought to establish several analytical performance metrics for RP-SAX-RP fractionation of phosphopeptides derived from a complex biological matrix. Toward this end we prepared multiple aliquots of tryptic peptides, each derived from 100 μg of human myeloid K562 cell lysate. We used batch mode Fe-NTA (8) to enrich phosphopeptides and acquired RP-SAX-RP-MS/MS data at a depth of 11 fractions. This experiment was then repeated, starting with a second aliquot of tryptic peptides, followed by phosphopeptide enrichment, and finally 11 fraction RP-SAX-RP-MS/MS analysis (see supplemental Files 3 and 4). Fig. 3A (left panel) shows an overlay of phosphopeptide identifications from both analyses plotted as a function of fraction number; given the good correlation in total identifications per fraction, we next asked whether individual phosphopeptides eluted across multiple first- and second-dimension fractions or were largely segregated within single, discrete fractions. Using sequence identification as a guide, we observed that ≈78% of all phosphopeptides were constrained within individual fractions. However, the use of peptide ID as a surrogate for “detection” is complicated by the stochastic nature of MS/MS. In an effort to mitigate this effect we next used our recently described multiplierz desktop software environment (38, 59) to generate extracted ion chromatograms (XICs) for phosphopeptides reproducibly identified in both experiments. (see Experimental Methods). Consistent with the results above, this analysis revealed that on average 85% of all ion current associated with a given phosphopeptide was detected in a single fraction. The neighboring Venn diagram (Fig. 3A, second panel) illustrates that we identified 6276 and 6110 unique phosphopeptides, respectively, with 4376 (54%) identified reproducibly across both RP-SAX-RP analyses. Interestingly, we identified 193 tyrosine phosphorylated peptides containing 202 unique sites of tyrosine phosphorylation, including five unique pY sites on BCR-ABL, the primary oncogenic kinase that drives proliferation in K562 cells. To avoid inflation of IDs we counted all occurrences of the same amino acid sequence and phosphorylation state as a single phosphopeptide sequence (i.e. oxidation, deamidation, and the position of the phosphate group(s) are ignored). Next we plotted the corresponding XIC apex intensity ratios for the set of reproducibly detected phosphopeptides as a histogram (third panel), and found that 95% of these ratios fell within ≈threefold of the mean value. To explore the degree of orthogonality across the separation space we represented the number of phosphopeptides identified in each third dimension LC-MS/MS analysis as circles of proportional diameter, and then plotted these as function of both first- and second-dimension eluent concentrations (Fig. 3A, fourth panel). A least squares fit to these data yielded a residual value (R2) of 0.03, indicating a high degree of orthogonality (e.g. poor correlation between first- and second-dimension elution) in the RP-SAX-RP separation.
Fig. 3.
Analytical figures of merit for RP-SAX-RP fractionation of phosphopeptides derived from 100 μg of K562 protein lysate. (A–C) Analysis of technical replicates at a depth of (A) 11, (B) 32, and (C) 53 fractions, respectively. Data are plotted to illustrate (first panel) the reproducible distribution of phosphopeptides across individual fractions (concentrations of acetonitrile and ammonium formate are shown as red and green circles, respectively), overlap in phosphopeptide identifications (second panel), and reproducibility of phosphopeptide intensity ratios (third panel). The number of unique phosphopeptide identifications are represented as circles of proportional diameter (fourth panel) and plotted as a function of first- and second-dimension elution conditions. D, Data plotted as in (A–C) for a 53 fraction RP-SAX-RP analysis performed with KCl as the second dimension eluent.
To further confirm the robustness of our RP-SAX-RP platform we next extended this analysis to fractionation depths of 32 and 53 fractions, respectively (Fig. 3B and 3C; see supplemental Files 5–8). In each case we observed very similar results in terms of the reproducibility of fractionation (first panel), peptide identification (second panel), and peptide intensity ratios (third panel). In addition we found that the RP-SAX-RP separation platform maintained a high degree of orthogonality at each fractionation depth tested (fourth panel). To test whether the use of an alkali salt would significantly affect system performance, we substituted KCl for ammonium formate as the second dimension eluent and repeated the characterization experiment described above (see supplemental Files 9–10). Comparison of the data in Fig. 3C and 3D suggest that ammonium formate and KCl provide comparable data, although as a volatile salt, we suspect that the former may provide more robust performance over time (60). Interestingly, at higher fractionation depths, we noted a concomitant increase in the relative number of phosphopeptides contained in the first dimension flow-through (see shaded region in panel one and corresponding column in Table II). This observation highlights the capacity of our RP-SAX-RP configuration to provide controlled fractionation of phosphopeptides that bind poorly to RP at high pH and would otherwise be difficult to detect comprehensively in a single flow-through fraction. Consistent with the data above (Fig. 2), phosphopeptides detected in the first dimension RP flow-through were enriched in acidic residues as compared with the entire fractionation space (on average, 20% versus 16% overall). Table II lists analytical figures of merit for the RP-SAX-RP analyses described in Fig. 3. As expected, we observed a systematic increase in separation peak capacity as a function of fractionation depth. In addition we achieved a dramatic improvement in efficiency as measured by both the number of phosphopeptide and phosphosite identifications per microgram of input lysate as compared with numerous previous studies (compare Tables I and II, respectively). Collectively, these results indicate that the RP-SAX-RP configuration provides an effective means to fractionate and identify phosphopeptides derived from complex biological matrices and offers multiple analytical figures of merit, including: (1) orthogonal separation mechanisms in each dimension; (2) high separation peak capacity; (3) efficient retention of singly- and multiply-phosphorylated peptides; (4) compatibility with automated, online LC-MS analysis.
Table II. Analytical figures of merit for three-dimensional RP-SAX-RP experiments performed at different fractionation depths. AF, ammonium formate; KCl, potassium chloride; NA, analysis not performed; IDs, identifications; see Experimental Procedures for details regarding calculations. Average Range for Experiment = Retention time (RT) of latest eluting peptide–RT of earliest eluting peptide. % Single Detections = Percentage of phosphopeptides detected in a single, final dimension fraction. Average % Signal in Max Fraction (Overlapping Peptides) = Average maximum signal for reproducibly detected peptides across all fractions expressed as a percentage of total signal. 1st Dim Flow Thru IDs = Number of phosphopeptides not retained on the 1st dimension RP column.
| Experiment | Average Range for Experiment (min) | Estimated Total Peak Capacity | % Single Detections (ID) | Av. % Signal in Max Fraction (Overlapping IDs) | Time (hr) | IDs | μg | IDs/μg | Phos sites | Phos sites/μg | 1st Dim Flow-Thru IDs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 fraction K562–1 | 61.0 | 818 | 76% | 84% | 26.6 | 6276 | 100 | 62.8 | 3806 | 38.1 | 2464 |
| 11 fraction K562–2 | 60.4 | 734 | 79% | 86% | 26.6 | 6110 | 100 | 61.1 | 3708 | 37.1 | 1756 |
| 32 fraction K562–1 | 57.1 | 1907 | 67% | 77% | 77.0 | 8694 | 100 | 86.9 | 5321 | 53.2 | 1995 |
| 32 fraction K562–2 | 57.5 | 1917 | 67% | 78% | 77.0 | 9287 | 100 | 92.9 | 5692 | 56.9 | 2356 |
| 53 fraction K562–1-AF | 55.3 | 3168 | 68% | 83% | 126.5 | 11423 | 100 | 114.2 | 6787 | 67.9 | 3409 |
| 53 fraction K562–2-AF | 54.8 | 3140 | 68% | 83% | 126.5 | 12058 | 100 | 120.6 | 7053 | 70.5 | 4097 |
| 53 fraction K562–1-KCl | 52.0 | 3597 | 74% | 88% | 126.5 | 11950 | 100 | 119.5 | 6871 | 68.7 | 5186 |
| 53 fraction K562–2-KCl | 49.4 | 3513 | 76% | 89% | 126.5 | 12739 | 100 | 127.4 | 7301 | 73.0 | 4912 |
| 10 fraction TCell-1 | 52.7 | 571 | 79% | NA | 26.2 | 2516 | 10 | 251.6 | 1179 | 117.9 | NA |
| 10 fraction TCell-2 | 57.6 | 680 | 79% | NA | 26.2 | 3254 | 10 | 325.4 | 1371 | 137.1 | NA |
| 10 fraction TCell-3 | 56.6 | 607 | 80% | NA | 26.2 | 2575 | 10 | 257.5 | 1099 | 109.9 | NA |
| 10 fraction K562–1 | 51.8 | 573 | 78% | 93% | 26.2 | 4686 | 10 | 468.6 | 2486 | 248.6 | 763 |
| 10 fraction K562–2 | 59.9 | 663 | 79% | 93% | 26.2 | 4703 | 10 | 470.3 | 2521 | 252.1 | 815 |
Quantification of Oncogenic FLT3 Signaling in Acute Myeloid Leukemia
Aberrant signaling from tyrosine kinases is a well established oncogenic event in hematopoietic malignancies (61, 62). For example, mutation of the gene encoding Fms-like tyrosine kinase 3 (FLT3) or overexpression of the corresponding protein product represent the most frequent molecular abnormalities in AML, and occur in some 35 and 90% of patients, respectively. Genetic lesions that lead to either in-frame, tandem internal duplications (ITD) near the juxtamembrane domain or point mutations in the kinase activation loop at D835 (typically D835Y) collectively account for ∼35% of clinical cases. Both mutation classes induce constitutive tyrosine kinase activity and confer IL3-independent growth of hematopoietic cell lines. Interestingly, cumulative evidence to date suggests that ITD and D835Y mutants respond differently to the current suite of targeted therapeutics and that they can activate different downstream targets (63). These observations, along with collective experience to date in the use of therapeutics that target single oncogenes or pathways (64–67), motivate continued efforts to elucidate oncogenic signaling pathways in greater detail. Toward this end, we used iTRAQ stable isotope labels to encode tryptic peptides derived from cells that express wild-type (WT) or mutant FLT3 kinases (supplemental Fig. S4). Phosphopeptides were enriched via Fe-NTA IMAC and analyzed in a 67 fraction, RP-SAX-RP experiment (Fig. 4). A total of 7787 unique phosphopeptides were detected (supplemental File 11), including 182 that were phosphorylated on tyrosine. These data along with those described in Fig. 3 above, demonstrate that RP-SAX-RP provides for high efficiency fractionation and detection of phosphopeptides derived from different cell lines (human K562 and murine BaF3) and on multiple mass spectrometry platforms (Thermo Oribtrap XL and ABI-SCIEX QSTAR Elite). Moreover, the results in Fig. 4 represent a nearly 10-fold increase in the number of phosphorylation sites detected as compared with a recent study from our lab in which we used an offline RP fractionation scheme (five fractions total) to analyze FLT3 signaling in the same cell system, under the same culture conditions (68). The efficiency of our RP-SAX-RP platform is further evidenced by the fact that our data compare favorably in scale (4586 phosphorylation sites) to that in a recent report from Choudhary et al. (14700 unique phosphorylation sites) (69), while consuming some 25-fold less cell lysate (400 μg versus 10 mg).
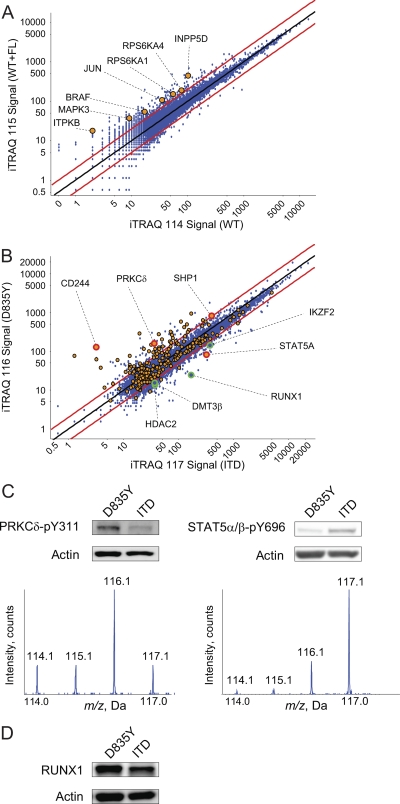
Fig. 4.
RP-SAX-RP analysis of iTRAQ labeled phosphopeptides from an AML model system consisting of wild-type and two oncogenic FLT3 mutants (ITD and D835Y) expressed in murine BaF3 cells. A, Plot of iTRAQ reporter ion intensities (log base 10) for phosphopeptides derived from cells that expressed wild-type FLT3 and were subjected to serum starvation (x axis) or stimulation with FLT3 ligand (y axis), with proteins involved in MAPK signaling (70) highlighted as yellow circles. B, Plot of iTRAQ reporter ion intensities (log base 10) for phosphopeptides derived from cells that stably expressed constitutively active FLT3-ITD (x axis) and -D835Y (y axis) mutants, with phosphotyrosine or phospho-serine/threnine residues displayed as yellow and blue circles, respectively. Selected proteins discussed in the main text are highlighted as large circles with red (pY) and green (pS/pT) outlines, respectively. C, Western blots of PRKCδ-pY311 and STAT5α/β-pY694 from lysates of FLT3-D835Y and -ITD expressing cells and mass spectra indicating iTRAQ reporter signals of the tryptic phosphopeptides spanning these phosphorylation sites (D835Y = 116.1; ITD = 117.1). D, Western blot of total RUNX1 protein from lysates of FLT3-D835Y and -ITD expressing cells.
For the first step in our analysis, we asked what proteins and pathways were activated by stimulation of the WT kinase with FLT3 ligand (FL). We observed regulated (greater than twofold) phosphorylation on members of the MAP kinase pathway (JUN, RPS6KA1, RPS6KA4, MAPK3, and BRAF), as well as proteins involved in lipid (INPP5D and ITPKB) signaling (70) (Fig. 4A, yellow circles). Both of these protein families have been linked to FL-mediated activation of survival and proliferation pathways (71), and serve as a positive control for our approach.
To investigate potential points of divergent signaling, we next compared iTRAQ reporter ion intensities for peptides identified in the context of FLT3-ITD and -D835Y mutants (Fig. 4B). These data revealed that the point mutant was a potent activator of tyrosine signaling (Fig. 4B, yellow circles); of the 244 phosphotyrosine candidate peptides identified (comprising 182 unique sequence-phosphorylation states), 47 exhibited an iTRAQ ratio consistent with increased phosphorylation via D835Y-mediated signaling. In contrast, only seven peptides were preferentially phosphorylated on tyrosine in the context of FLT3-ITD. In agreement with our previous studies (33) we observed that constitutive signaling in the context of D835Y led to tyrosine phosphorylation near the C terminus of the phosphatase SHP1 (Fig. 4B). Data and results from recent studies (72, 73) suggest that these sites may serve as SH-2 domain scaffolds for subsequent assembly of GAB2-SOCS complexes that, in-turn negatively regulate JAK-STAT signaling. In addition, we observed increased phosphorylation on several proteins linked to regulation of signaling in lymphoid cells, including: CD244, PRKCδ, PTK2B, PECAM1, PAG1, PDCD1, CARD11, and PLCG2 (74–81). Relative phosphorylation of PRKCδ and STAT5 in the context of FLT3-ITD and -D835Y was confirmed by Western blot of lysates from biological replicates using site specific antibodies (Fig. 4C). Interestingly we observed the opposite situation for serine and threonine phosphorylation. Of the 602 candidate pS/pT peptide sequences that exhibited significant iTRAQ reporter ion ratios (representing 519 unique sequence/phosphorylation states), 479 of these were preferentially phosphorylated in the context of FLT3-ITD signaling, versus only 123 for point mutant. (Fig. 4B, blue circles). KEGG pathway (70) analysis of ITD-mediated phosphoproteins revealed activation of survival and proliferation pathways (insulin, ERBB, MAPK signaling). In addition, we observed phosphorylation on proteins linked to chromatin remodeling (HDAC2, DMT3β (82)) and transcription (RUNX1, RUNX3, IKZF2). Western blot against total RUNX1 protein (Fig. 4D) confirmed that the increase in phosphorylation observed in the context of FLT3-ITD signaling was not an artifact of protein expression; in fact overall RUNX1 protein levels were higher in cells that expressed the point mutant. DMT3β, RUNX1, and IKZF2 are well-established mediators of hematopoiesis and they are often functionally disrupted in the context of leukemia (83). Moreover, it is now well recognized that oncogenic kinase activity is not sufficient to support complete leukemic transformation (84–86). Of the two FLT3 mutants studied herein, the ITD is strongly associated with poor clinical prognoses (63, 84, 86), and our data are consistent with the hypothesis that FLT3-ITD signaling has a strong propensity, relative to D835Y, to drive epigenetic events that disrupt differentiation and lead to a more aggressive AML phenotype. For example, we identified phosphorylation sites (S77, S78, and S433) on the Ikaros family transcription factor IKZF2 that lie in close proximity to SIN3 binding sites (87). It is tempting to speculate that these sites mediate the interaction of IKZF2 and SIN3, given that the latter is known to bind RUNX1, also observed to be preferentially phosphorylated in the context of ITD signaling, and repress transcription through recruitment of HDACs (88).
As noted above, multiple reports demonstrated that FLT3 mutants activate different downstream targets in vitro, and these observations have been corroborated in vivo with murine bone marrow transplant models. For example, Grundler et al. (89) found that FLT3-ITD induced a myeloproliferative disorder whereas transplantation of cells harboring FLT3-D835Y led to aberrant expansion of the lymphoid compartment in recipient mice. These results correlate well with the clinical manifestations observed for leukemia patients who harbor FLT3-ITD or D835Y mutations; the former is most often observed in the context of myeloid malignancies whereas the latter occurs with high frequency in acute lymphoid leukemia (84). In our study, the biological pathways represented by proteins that were identified as being preferentially phosphorylated by either ITD or D835Y are consistent with both in vivo bone marrow transplant models and human clinical phenotypes. Taken together, these data demonstrate that the increased dynamic range of phosphopeptide identification inherent to RP-SAX-RP fractionation provides for identification and quantification of signaling events related to clinically relevant oncogenic kinase activity in complex biological matrices. The relatively modest quantities of cell lysate required for these data (100 μg lysate per iTRAQ channel) provide further evidence for the high efficiency of our fractionation scheme.
Analysis of Primary CD8+ T-Cells from a Single Mouse
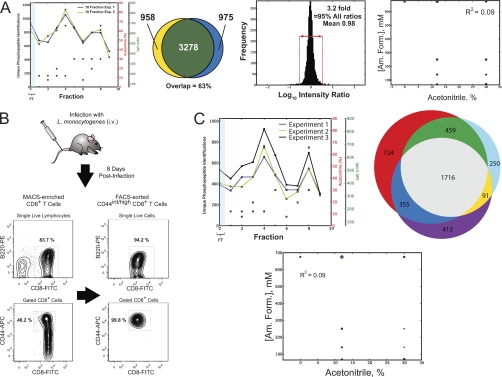
Although cultured cells lines are an invaluable resource for biomedical research, it is well recognized that, as a result of genetic alternations and selective pressures associated with repeated passage under non-physiologic growth conditions, they may not recapitulate the behavior of cells in vivo (90, 91). In principle the use of primary cells will circumvent many of the shortcomings of immortalized cell lines, although in practice this approach is complicated by the frequent mismatch between the availability of biological material and the limited detection and dynamic range provided by a majority of LC-MS platforms. Encouraged by the efficiency gains observed for analysis of cultured cell lines (Figs. 3 and 4), we next asked whether our RP-SAX-RP platform would enable reproducible detection of phosphorylation in rare, primary cell populations. As a first step toward this goal, we revisited the experimental strategy described in Fig. 3, but scaled the input K562 lysate down by an order of magnitude to 10 μg. Technical replicates were processed as described above and analyzed by RP-SAX-RP at a depth of 10 fractions. Again we observed very good agreement for both analyses in terms of the distribution of phosphopeptides across each fraction (Fig. 5A, first panel; see supplemental Files 12–13), overlap in phosphopeptide identifications (second panel), and precursor intensities (third panel). In addition we observed orthogonal behavior for separation of phosphopeptides in the first- and second-dimensions, respectively (Fig. 5A, fourth panel).
Fig. 5.
RP-SAX-RP analysis of phosphopeptides derived from sample-limited scenarios. A, Analysis of technical replicates at a depth of 10 fractions for phosphopeptides derived from 10 μg of K562 protein lysate. Data are plotted to illustrate the (first panel) reproducible distribution of phosphopeptides across individual fractions (concentrations of acetonitrile and ammonium formate are shown as red and green circles, respectively), overlap in phosphopeptide identifications (second panel), and reproducibility of phosphopeptide intensity ratios (third panel). The number of unique phosphopeptide identifications are represented as circles of proportional diameter (fourth panel) and plotted as a function of first- and second-dimension elution conditions. (B–C) RP-SAX-RP analysis of phosphopeptides derived from in vivo activated primary CD8+ T lymphocytes. B, Purity of activated (CD44int/high) CD8+ T cells isolated from a single mouse spleen 8 days after infection with L. monocytogenes-OVA. Contour graphs show the percentages of live B220− (top) and CD44int/high (bottom) CD8+ cells after enrichment of total CD8+ cells by MACS (left panel), and subsequent isolation of B220−/CD44int/high/CD8+ cells by FACS (right panel). Data are representative of three individual mice. (C, left panel) Distribution of unique phosphopeptide identifications across individual fractions for RP-SAX-RP analysis. Each trace (black, yellow, blue) corresponds to data derived from activated T-cells (≈5–10 μg total protein lysate) enriched from a single mouse. (C, right panel) Venn diagram for unique phosphopeptide sequences identified from each mouse. (C, bottom panel) The number of unique phosphopeptide identifications are represented as circles of proportional diameter and plotted as a function of first- and second-dimension elution conditions.
With these results in-hand we next moved on to a sample-limited scenario that would also allow us to test the impact of biological variability on our RP-SAX-RP platform. For this analysis we chose murine primary CD8+ T lymphocytes, which are relatively small (6–15 μm in diameter) specialized cells of the adaptive immune system whose primary function is to destroy host cells infected by viruses or other intracellular pathogens (e.g. bacteria). CD8+ T-cells recognize foreign antigens displayed on the surface of host cells in complex with MHC Class I molecules. Notably, engagement of the T-cell receptor with MHC-antigen complexes drives a diverse array of signaling networks that ultimately results in (1) lysis of infected host cells, (2) growth and clonal expansion of activated T cells, followed some time later by either (3) apoptosis of primed T-cells, or (4) differentiation of activated cells into memory T cells. Although several phosphorylation pathways have been associated with some of these endpoints, the majority of current data is derived from in vitro studies, or through the use of phospho-specific antibodies that collectively target a modest number of phosphoproteins.
To study phosphorylation in primary T cells responding to a bacterial challenge in vivo, we first infected mice with Lm-OVA, and then purified activated CD8+ T cells at the peak of the immune response (8 days postinfection) by MACS and fluorescence-activated cell sorting (FACS) (Fig. 5B). Importantly, it has been demonstrated that the majority of activated T cells in mice at this time point are responding to the ongoing infection in a pathogen-specific manner (92). Approximately 1–3 × 106 purified CD44int/high CD8+ T cells (>94% CD8+/B220- and >99% CD44int/high) were obtained from a single spleen (corresponding to ≈5–10 μg of protein), and lysed immediately upon harvest. Phosphopeptides were isolated by Fe-NTA IMAC, and subjected to a 10 fraction RP-SAX-RP analysis. We profiled phosphopeptides in this manner from three individual mice and identified 2516, 3254, and 2575 unique phosphopeptides, respectively (Table II). Across biological triplicates we observed a high degree of reproducibility in terms of the distribution of phosphopeptides across each fraction (Fig. 5C, top left panel, and supplemental Files 14–16) and unique phosphopeptide identifications (Fig. 5C, top right panel). Finally as in our previous analyses (Fig. 3 and Fig. 5A) we again observed orthogonal behavior for separation of phosphopeptides in the first- and second-dimensions, respectively (Fig. 5C, bottom panel).
Among these data we observed phosphorylation sites previously identified in T-cells stimulated in vitro (supplemental Table S3). In addition, we identified phosphorylation on many proteins with known functional roles in T cells, although the specific sites that we detected have not, to the best of our knowledge, been described in activated T cells (supplemental Table S4). Interestingly we also observed phosphorylation on several proteins whose function is largely unexplored in CD8+ T cells. These included regulators of (1) chromatin organization and remodeling, (2) transcription, (3) RNA processing (including pre-mRNA splicing factors), and (4) protein translation. Importantly, the primary CD8+ T lymphocytes used in our analysis were not further manipulated after FACS purification; as a result, the phosphorylation events observed likely reflect physiologic CD8+ T cell responses within the mouse. Collectively, these data provide multiple avenues for future studies designed to decipher the mechanistic role of these, in many cases novel, signaling events in CD8+ T cell activation and fate determination in vivo.
General Recommendations
We conclude with general recommendations and observations that will facilitate continued development and deployment of RP-SAX-RP in other laboratories.
System Capacity
Based on unpublished studies for shotgun analysis of tryptic (e.g. unmodified) peptides, we estimate that the total loading capacity of our first dimension RP column (150 μm I.D. × 5 cm of 5 μm) is currently ≈10 μg. Comparisons of total ion chromatograms from MS analyses of total lysates and phosphopeptides indicate that ≈1% of tryptic peptides are phosphorylated and retained on the Fe-NTA column during enrichment. These data suggest that our current RP-SAX-RP configuration can accommodate phosphopeptides enriched from up to ≈1 mg of biological lysate. Work is ongoing to evaluate the relative performance of larger first dimension columns and to define the point at which unretained peptides from the first dimension begin to exceed the capacity of the second dimension column. We anticipate that careful calibration of these variables will enable robust analysis of phosphopeptides enriched from larger quantities of biological input material.
First and Second Dimension Elution Conditions
We have found that the three-dimensional RP-SAX-RP platform is easily tailored to a variety of sample types and desired fractionation depths. Generally, eluent concentrations in a range of 7 to 25% acetonitrile and 15 to 700 mm ammonium formate represent the useful peptide elution “windows” for first and second dimensions, respectively. Within these limits, the total number of desired fractions can be evenly divided between first and second dimensions, with minimal fraction-to-fraction overlap of peptides. Importantly, our experience to date suggests that these boundary conditions are robust with respect to biological input, obviating the need to run repeated pilot experiments for every sample.
Instrument Time
Total time of analysis is another important consideration when using multidimension fractionation techniques. The analyses described herein required ∼24 h per 10 fractions. For example experiments performed at a depth of 53 fractions required ≈5 days to complete. Work is ongoing to refine third dimension gradient conditions to determine whether total system peak capacity as a function of fractionation depth (supplemental Fig. S5) can be improved in terms of (1) analysis depth (e.g. more fractions) and (2) efficiency (e.g. higher peak capacity per unit of instrument time).
System Reliability
System reliability is particularly important given that a single, multidimension analysis can require several days of instrument time. Consistent with our experience to date using 25 μm I.D. analytical columns with integrated electrospray emitters in a one dimension nanoflow configuration (42), or coupled with high-pH RP in an automated RP-RP configuration (32), our RP-SAX-RP platform has proven to be very robust. The system typically provides uninterrupted analysis of hundreds of fractions, often spanning several months of analysis time, without a major failure. In addition, we have observed that vigorous regeneration of column resins (alternating high salt/high organic injections, for example 1 m KCl with 10% acetic acid, 900 mm ammonium formate with 10% acetic acid, 90% acetonitrile) and use of PEEK coated fused silica for delivering high pH solvents has improved overall column lifetime and system up-time.
DISCUSSION
Results from recent large scale proteomics studies suggest that experimental protocols for phosphopeptide enrichment and analysis have stabilized within individual labs. In fact, related work from our lab (8) and other groups (93, 94) demonstrated that enrichment of phosphopeptides from complex biological matrices can now be performed in an automated fashion. Despite this progress we noted that the yield of phosphopeptides as a function of input material had remained relatively constant across studies from numerous labs (ours included), spanning a wide range of biological sample types and quantities (Table I). Although data that describe an absolute number of phosphopeptides in mammalian systems are not currently available, previous studies have attempted to address relative levels of phosphorylation, and suggest that (1) 30–50% of the proteome is phosphorylated at any given time (95), and (2) a surprising number of proteins are phosphorylated at a stoichiometry of greater than 90% (96). Based on these observations, we asked whether improvements in fractionation could provide a viable avenue for significant increases in the efficiency of phosphopeptide identification. Toward this end we first explored the performance of our recently described (32) implementation of RP-RP fractionation (28) for phosphopeptide analysis. Interestingly, we observed similar behavior for fractionation of phosphopeptides via RP-RP as has been reported for SCX-RP. That is, binding efficiency to the first dimension RP column (ammonium formate, pH = 10) was inversely related to the degree of peptide phosphorylation in particular, as well as overall peptide acidic content. We sought to improve phosphopeptide retention under alkaline conditions through the use of aliphatic-functionalized quaternary amines as ion pairing agents, and found that tetrapropylammonium ion completely abrogated breakthrough of phosphopeptides; moreover, we observed that peptide elution order from the first dimension column was well correlated with both the degree of phosphorylation and relative acidic content (D and E) of phosphopeptides. Despite the successful use of ion pairing chromatography in the analysis of a wide range of molecule classes (50, 54, 97), we observed significant fraction-to-fraction overlap of phosphopeptides in RP-RP fractionation augmented with tetrapropylammonium formate, suggesting that further optimization will be required to utilize these compounds for the analysis of complex biological lysates.
Based on the above results we next placed an anion exchange (SAX) column between the first and (now) third dimension RP columns to ensure efficient capture of phosphopeptides (e.g. those phosphopeptides that were otherwise un-retained on the first dimension column). We found that discrete salt steps facilitated controlled elution of phosphopeptides from the SAX column, providing the basis for an integrated, three-dimensional fractionation platform. Using K562 as a model mammalian system we observed reproducible and orthogonal separation and identification of phosphopeptides across a range of fractionation depths, with efficiencies more than an order of magnitude greater than those previously reported, in some cases exceeding 450 phosphopeptide IDs per μg of cell lysate consumed (Table II). These studies revealed that RP-SAX-RP provides a unique combination of figures of merit as compared with previously reported multidimensional fractionation systems (10, 15–22, 29, 31, 45, 46), including: (1) orthogonal separation mechanisms in each dimension; (2) high separation peak capacity; (3) efficient retention of singly- and multiply phosphorylated peptides; (4) compatibility with automated, online LC-MS analysis. The results reported in Fig. 3 and Table II may represent a lower bound in performance given the improvements in associated proteomics technologies such as specialized algorithms (40, 98, 99) that improve phosphopeptide sequence and site assignment as compared with standard database search algorithms, along with mass spectrometers having higher acquisition rates (100–102) or equipped with complimentary dissociation techniques (103–106). Although the online, capillary format of our RP-SAX-RP platform provides for automated LC-MS/MS acquisition, it is important to note the trade-off in total sample capacity as compared with systems that utilize one or more stages of offline separation. Although these schemes provide for fractionation of large sample quantities, recent reports suggest that sample handling, lyophilization, and nonspecific adsorption of peptides to tube surfaces can reduce yield significantly. For example, Dowell and colleagues reported that online SCX-RP detected twice the number of peptides as compared with an equivalent offline SCX-RP format. Not surprisingly these deleterious effects scale inversely with total input, limiting the performance for offline systems for sample quantities below ≈50–100 μg (107, 108). In addition, it remains to be seen whether efficiencies similar to those demonstrated herein will translate to a wider range of sample types and quantities. Ideally, improved fractionation can directly offset ionization suppression and stochastic undersampling of precursors during MS/MS, both of which compress observed dynamic range. However, the relatively narrow range of efficiencies observed across multiple experiments and cell types listed in Table I could already reflect improved dynamic range from higher performance mass spectrometers or other technological developments. Given that the distribution of phosphoprotein abundances in vivo is not known, the relative impact of improved fractionation, along with other analytical figures of merit in LC-MS/MS (109), will need to be empirically tested across multiple experimental scenarios. Finally, although we have chosen the specific combination of RP and SAX for the first and second dimensions, respectively, it will be interesting to see whether the use of an orthogonal fourth dimension or an alternative separation phase in the current three-dimensional configuration, such as HILIC (19, 45) SCX (10), or ERLIC (30, 31) will provide still further improvements in performance.
Having established several analytical figures of merit for our RP-SAX-RP platform, we next analyzed phosphopeptides in the context of an in vitro model of AML to decipher divergent signaling downstream of FLT3-ITD and -D835Y, two constitutively active kinases commonly associated with leukemogenesis. We found that RP-SAX-RP provided sufficient peak capacity to support accurate iTRAQ-based quantification of discrete signaling events associated with each kinase, and in toto these data recapitulated results from murine bone marrow transplant models and human clinical phenotypes. As a more challenging test case for RP-SAX-RP we analyzed phosphopeptides derived from primary CD8+ T lymphocytes that were activated in vivo, and subsequently isolated from a single mouse. We identified an average of 2782 unique phosphopeptide sequences across biological triplicates, starting with 1–3 × 106 highly enriched T cells (≈5–10 μg of total protein lysate) for each analysis. We observed reproducibility in phosphopeptide identifications comparable to that obtained when analyzing technical replicates of K562 lysate, suggesting that our RP-SAX-RP platform is very robust with respect to variation across biological replicates. These results highlight the analytical capabilities of RP-SAX-RP, and are particularly encouraging in the context of two recent studies, one of which identified 281 phosphorylation sites from 2 mg of biological lysate derived from partially sorted, primary lymphocytes (110), and a second which reported a total of 6248 phosphopeptides across 46 LC-MS/MS analyses of whole mouse spleens (≈2 mg total protein per analysis, or 92 mg total input) (111) in a model designed to study immune response to anthrax exposure. In both studies, the samples were composed of heterogeneous cell populations, each of which will contribute a distinct signaling response to the final ensemble of phosphopeptides measured. The first case included a diverse repertoire of lymphocytes normally found in blood, including: subtypes of B cells, CD4+ T cells, CD8+ T cells, NK cells, and NK-T cells. Interestingly in the latter study, the authors noted an inability to analyze phosphorylation in mouse primary T cells, and hence used phosphoprofiling of whole spleens as a surrogate for host immune response to anthrax exposure. However, analysis of whole spleen encompasses all the aforementioned B, T, NK, and NK-T lymphocytes, in addition to myeloid (dendritic, macrophage, and neutrophil), erythroid, as well as cells of endothelial and stromal origin. In contrast, our RP-SAX-RP platform facilitates analysis of highly enriched, antigen specific CD8+ T cells responding to an ongoing bacterial infection. Accordingly, our data are largely free of confounding effects from contaminating cells and should therefore provide direct insights into the pathways regulating CD8+ T-cell responses in clinically relevant contexts (e.g. infection, vaccine development, etc.). Collectively our data and results demonstrate that RP-SAX-RP provides for reproducible, high efficiency phosphopeptide fractionation and will be particularly valuable for applications in which availability of biological materials is limited.
Acknowledgments
We thank Dr. Natasha Barteneva (Flow and Imaging Cytometry Core, Immune Disease Institute, Harvard Medical School) for sorting the primary CD8+ T cells. The authors thank Keith Fadgen, Jim Murphy, and Geoff Gerhardt of Waters Corp. for valuable discussions and technical assistance.
Footnotes
* This work was supported by the Brigham & Women's Hospital, the Dana-Farber Cancer Institute, and the National Institutes of Health, NHGRI (P50HG004233), and NINDS (P01NS047572).
 This article contains supplemental Figs. S1 to S5, Tables S1 to S4, and Files.
This article contains supplemental Figs. S1 to S5, Tables S1 to S4, and Files.
1 The abbreviations used are:
- IMAC
- immobilized metal affinity chromatography
- AML
- Acute Myeloid Leukemia
- CAD
- collisionally activated dissociation
- ESI
- electrospray ionization
- FDR
- False Discovery Rate
- FL
- FLT3 ligand
- FLT3
- FMS-like tyrosine kinase 3
- LC/MS
- liquid chromatography/mass spectrometry
- Lm-OVA
- Recombinant Listeria monocytogenes expressing chicken ovalbumin
- ITD
- internal tandem duplication
- MS/MS
- mass spectrometry/mass spectrometry or tandem mass spectrometry
- NTA
- nitrilotetraacetic acid
- RP-RP
- reversed phase-reversed phase
- RP-SAX-RP
- reversed phase-strong anion exchange-reversed phase
- RT
- retention time
- SCX
- strong cation exchange
- TFA
- trifluoroacetic acid
- UPLC
- ultra-high pressure liquid chromatography
- WT
- wild type.
REFERENCES
- 1. Nuwaysir L., Stults J. (1993) Electrospray ionization mass spectrometry of phosphopeptides isolated by on-line immobilized metal-ion affinity chromatography. J. Am. Soc. Mass Spectrom. 4, 662–669 [DOI] [PubMed] [Google Scholar]
- 2. Posewitz M. C., Tempst P. (1999) Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 71, 2883–2892 [DOI] [PubMed] [Google Scholar]
- 3. Watts J. D., Affolter M., Krebs D. L., Wange R. L., Samelson L. E., Aebersold R. (1994) Identification by electrospray ionization mass spectrometry of the sites of tyrosine phosphorylation induced in activated Jurkat T cells on the protein tyrosine kinase ZAP-70. J. Biol. Chem. 269, 29520–29529 [PubMed] [Google Scholar]
- 4. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 5. Zhou H., Watts J. D., Aebersold R. (2001) A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 19, 375–378 [DOI] [PubMed] [Google Scholar]
- 6. Oda Y., Nagasu T., Chait B. T. (2001) Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 19, 379–382 [DOI] [PubMed] [Google Scholar]
- 7. Ficarro S., Chertihin O., Westbrook V. A., White F., Jayes F., Kalab P., Marto J. A., Shabanowitz J., Herr J. C., Hunt D. F., Visconti P. E. (2003) Phosphoproteome analysis of capacitated human sperm. Evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J. Biol. Chem. 278, 11579–11589 [DOI] [PubMed] [Google Scholar]
- 8. Ficarro S. B., Adelmant G., Tomar M. N., Zhang Y., Cheng V. J., Marto J. A. (2009) Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal. Chem. 81, 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai C. F., Wang Y. T., Chen Y. R., Lai C. Y., Lin P. Y., Pan K. T., Chen J. Y., Khoo K. H., Chen Y. J. (2008) Immobilized metal affinity chromatography revisited: pH/acid control toward high selectivity in phosphoproteomics. J. Proteome Res. 7, 4058–4069 [DOI] [PubMed] [Google Scholar]
- 10. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 12. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 13. Annan R. S., Huddleston M. J., Verma R., Deshaies R. J., Carr S. A. (2001) A multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal. Chem. 73, 393–404 [DOI] [PubMed] [Google Scholar]
- 14. Leitner A. (2010) Phosphopeptide enrichment using metal oxide affinity chromatography. Trac-Trends Anal. Chem. 29, 177–185 [Google Scholar]
- 15. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song C., Ye M., Han G., Jiang X., Wang F., Yu Z., Chen R., Zou H. (2010) Reversed-Phase-Reversed-Phase Liquid Chromatography Approach with High Orthogonality for Multidimensional Separation of Phosphopeptides. Anal. Chem. 82, 53–56 [DOI] [PubMed] [Google Scholar]
- 17. Han G., Ye M., Zhou H., Jiang X., Feng S., Jiang X., Tian R., Wan D., Zou H., Gu J. (2008) Large-scale phosphoproteome analysis of human liver tissue by enrichment and fractionation of phosphopeptides with strong anion exchange chromatography. Proteomics 8, 1346–1361 [DOI] [PubMed] [Google Scholar]
- 18. Pinkse M. W., Mohammed S., Gouw J. W., van Breukelen B., Vos H. R., Heck A. J. (2007) Highly Robust, Automated, and Sensitive Online TiO 2-Based Phosphoproteomics Applied To Study Endogenous Phosphorylation in Drosophila melanogaster. J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 19. McNulty D. E., Annan R. S. (2008) Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics 7, 971–980 [DOI] [PubMed] [Google Scholar]
- 20. Lemeer S., Pinkse M. W., Mohammed S., van Breukelen B., den Hertog J., Slijper M., Heck A. J. (2008) Online Automated in Vivo Zebrafish Phosphoproteomics: From Large-Scale Analysis Down to a Single Embryo. J. Proteome Res. 7, 1555–1564 [DOI] [PubMed] [Google Scholar]
- 21. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 22. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 23. Zhou H., Low T. Y., Hennrich M. L., van den Toorn H., Schwend T., Zou H., Mohammed S., Heck A. J. R. (2011) Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics, M110.006452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd. (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676 [DOI] [PubMed] [Google Scholar]
- 25. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242 [DOI] [PubMed] [Google Scholar]
- 26. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 27. Ballif B. A., Villén J., Beausoleil S. A., Schwartz D., Gygi S. P. (2004) Phosphoproteomic Analysis of the Developing Mouse Brain. Mol. Cell. Proteomics 3, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 28. Gilar M., Olivova P., Daly A. E., Gebler J. C. (2005) Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J. Separation Sci. 28, 1694–1703 [DOI] [PubMed] [Google Scholar]
- 29. Dai J., Wang L. S., Wu Y. B., Sheng Q. H., Wu J. R., Shieh C. H., Zeng R. (2008) Fully automatic separation and identification of phosphopeptides by continuous pH-gradient anion exchange online coupled with reversed-phase liquid chromatography mass spectrometry. J. Proteome Res. 8, 133–141 [DOI] [PubMed] [Google Scholar]
- 30. Alpert A. J. (2008) Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal. Chem. 80, 62–76 [DOI] [PubMed] [Google Scholar]
- 31. Gan C. S., Guo T., Zhang H., Lim S. K., Sze S. K. (2008) A Comparative Study of Electrostatic Repulsion-Hydrophilic Interaction Chromatography (ERLIC) versus SCX-IMAC-Based Methods for Phosphopeptide Isolation/Enrichment. J. Proteome Res. 7, 4869–4877 [DOI] [PubMed] [Google Scholar]
- 32. Zhou F., Cardoza J. D., Ficarro S. B., Adelmant G. O., Lazaro J.-B., Marto J. A. (2010) Online Nanoflow RP-RP-MS Reveals Dynamics of Multicomponent Ku Complex in Response to DNA Damage. J. Proteome Res. 9, 6242–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Askenazi M., Jiang J., Luckey C. J., Griffin J. D., Marto J. A. (2009) A robust error model for iTRAQ quantification reveals divergent signaling between oncogenic FLT3 mutants in acute myeloid leukemia. Mol. Cell. Proteomics, M900452-MCP900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wenger C. D., McAlister G. C., Xia Q., Coon J. J. (2010) Sub-part-per-million precursor and product mass accuracy for high-throughput proteomics on an electron transfer dissociation-enabled orbitrap mass spectrometer. Mol. Cell. Proteomics 9, 754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot/human.protein.faa.gz.
- 36. ftp://ftp.ncbi.nih.gov/refseq/M_musculus/mRNA_Prot/mouse.protein.faa.gz.
- 37. Craig R., Cortens J. C., Fenyo D., Beavis R. C. (2006) Using Annotated Peptide Mass Spectrum Libraries for Protein Identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 38. Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webber J. T., Askenazi M., Marto J. A. (2011) mzResults: An interactive viewer for interrogation and distribution of proteomics results. Mol. Cell. Proteomics 10, M110.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the mascot delta score. Mol. Cell. Proteomics 10, M110.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ficarro S. B., Parikh J. R., Blank N. C., Marto J. A. (2008) Niobium(V) oxide (Nb2O5): Application to phosphoproteomics. Anal. Chem. 80, 4606–4613 [DOI] [PubMed] [Google Scholar]
- 42. Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved Electrospray Ionization Efficiency Compensates for Diminished Chromatographic Resolution and Enables Proteomics Analysis of Tyrosine Signaling in Embryonic Stem Cells. Anal. Chem. 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 43. Licklider L. J., Thoreen C. C., Peng J., Gygi S. P. (2002) Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal. Chem. 74, 3076–3083 [DOI] [PubMed] [Google Scholar]
- 44. Meiring H. D., van der Heeft E., ten Hove G. J., de Jong A. (2002) Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J. Separation Sci. 25, 557–568 [Google Scholar]
- 45. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. (2008) A Multidimensional Chromatography Technology for In-depth Phosphoproteome Analysis. Mol. Cell. Proteomics 7, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 47. Gauci S., van Breukelen B., Lemeer S. M., Krijgsveld J., Heck A. J. (2008) A versatile peptide pI calculator for phosphorylated and N-terminal acetylated peptides experimentally tested using peptide isoelectric focusing. Proteomics 8, 4898–4906 [DOI] [PubMed] [Google Scholar]
- 48. Halligan B. D., Ruotti V., Jin W., Laffoon S., Twigger S. N., Dratz E. A. (2004) ProMoST (Protein Modification Screening Tool): a web-based tool for mapping protein modifications on two-dimensional gels. Nucleic Acids Res. 32, W638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vailaya A., Horváth C. (1998) Retention in reversed-phase chromatography: partition or adsorption? J. Chromatogr. A 829, 1–27 [DOI] [PubMed] [Google Scholar]
- 50. Cheng C. F., Tsang C. W. (1999) Separation of N-nitrosoamino acids by C18 reversed-phase ion-pair high-performance liquid chromatography and compatible detection by electrospray ionization mass spectrometry. J. Chromatogr. A 849, 389–402 [DOI] [PubMed] [Google Scholar]
- 51. Coulier L., Bas R., Jespersen S., Verheij E., van der Werf M. J., Hankemeier T. (2006) Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 78, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 52. Doneanu C. E., Chen W., Gebler J. C. (2009) Analysis of oligosaccharides derived from heparin by ion-pair reversed-phase chromatography/mass spectrometry. Anal. Chem. 81, 3485–3499 [DOI] [PubMed] [Google Scholar]
- 53. Holcapek M., Jandera P., Zderadicka P. (2001) High performance liquid chromatography-mass spectrometric analysis of sulphonated dyes and intermediates. J. Chromatogr. A 926, 175–186 [DOI] [PubMed] [Google Scholar]
- 54. Holcapek M., Volná K., Jandera P., Kolárová L., Lemr K., Exner M., Církva A. (2004) Effects of ion-pairing reagents on the electrospray signal suppression of sulphonated dyes and intermediates. J. Mass Spectrom. 39, 43–50 [DOI] [PubMed] [Google Scholar]
- 55. Mao S. J., Hou S. X., Liang Z., Bi Y. Q., Wu Y., Li H., Jin H. (2006) Ion-pair reversed-phase HPLC: assay validation of sodium tanshinone IIA sulfonate in mouse plasma. J. Chromatogr. B 831, 163–168 [DOI] [PubMed] [Google Scholar]
- 56. Premstaller A., Oberacher H., Huber C. G. (2000) High-performance liquid chromatography-electrospray ionization mass spectrometry of single- and double-stranded nucleic acids using monolithic capillary columns. Anal. Chem. 72, 4386–4393 [DOI] [PubMed] [Google Scholar]
- 57. Waichigo M. M., Danielson N. D. (2006) Ethylammonium formate as an organic solvent replacement for ion-pair reversed-phase liquid chromatography. J. Chromatogr. Sci. 44, 607–614 [DOI] [PubMed] [Google Scholar]
- 58. Wybraniec S. (2006) Effect of tetraalkylammonium salts on retention of betacyanins and decarboxylated betacyanins in ion-pair reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1127, 70–75 [DOI] [PubMed] [Google Scholar]
- 59. Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolters D. A., Washburn M. P., Yates J. R., 3rd (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 61. Scheijen B., Griffin J. D. (2002) Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene 21, 3314–3333 [DOI] [PubMed] [Google Scholar]
- 62. Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 63. Knapper S. (2007) FLT3 inhibition in acute myeloid leukaemia. Br. J. Haematol. 138, 687–699 [DOI] [PubMed] [Google Scholar]
- 64. Traxler P. (2003) Tyrosine kinases as targets in cancer therapy - successes and failures. Expert Opinion Ther. Targets 7, 215–234 [DOI] [PubMed] [Google Scholar]
- 65. Quintás-Cardama A., Cortes J. (2008) Therapeutic Options Against BCR-ABL1 T315I-Positive Chronic Myelogenous Leukemia. Clin. Cancer Res. 14, 4392–4399 [DOI] [PubMed] [Google Scholar]
- 66. Snead J. L., O'Hare T., Eide C. A., Deininger M. W. (2008) New strategies for the first-line treatment of chronic myeloid leukemia: Can resistance be avoided? Clin. Lymph. Myeloma 8, S107–S117 [DOI] [PubMed] [Google Scholar]
- 67. Chu S. H., Small D. (2009) Mechanisms of resistance to FLT3 inhibitors. Drug Resistance Updates 12, 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Y., Ficarro S. B., Li S., Marto J. A. (2009) Optimized orbitrap HCD for quantitative analysis of phosphopeptides. J. Am. Soc. Mass Spectrom. 20, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 69. Choudhary C., Olsen J. V., Brandts C., Cox J., Reddy P. N., Böhmer F. D., Gerke V., Schmidt-Arras D. E., Berdel W. E., Müller-Tidow C., Mann M., Serve H. (2009) Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell 36, 326–339 [DOI] [PubMed] [Google Scholar]
- 70. Kanehisa M., Goto S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Masson K., Rönnstrand L. (2009) Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell. Signal. 21, 1717–1726 [DOI] [PubMed] [Google Scholar]
- 72. Klingmüller U., Lorenz U., Cantley L. C., Neel B. G., Lodish H. F. (1995) Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell 80, 729–738 [DOI] [PubMed] [Google Scholar]
- 73. Minoo P., Zadeh M. M., Rottapel R., Lebrun J. J., Ali S. (2004) A novel SHP-1/Grb2-dependent mechanism of negative regulation of cytokine-receptor signaling: contribution of SHP-1 C-terminal tyrosines in cytokine signaling. Blood 103, 1398–1407 [DOI] [PubMed] [Google Scholar]
- 74. Mathew S. O., Rao K. K., Kim J. R., Bambard N. D., Mathew P. A. (2009) Functional role of human NK cell receptor 2B4 (CD244) isoforms. Eur. J. Immunol. 39, 1632–1641 [DOI] [PubMed] [Google Scholar]
- 75. Miyamoto A., Nakayama K., Imaki H., Hirose S., Yi J., Abe M., Tsukiyama T., Nagahama H., Ohno S., Hatakeyama S., Nakayama K. I. (2002) Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cδ. Nature 416, 865–869 [DOI] [PubMed] [Google Scholar]
- 76. Brdicka T., Pavlistová D., Leo A., Bruyns E., Korinek V., Angelisová P., Scherer J., Shevchenko A., Shevchenko A., Hilgert I., Cerný J., Drbal K., Kuramitsu Y., Kornacker B., Horejsí V., Schraven B. (2000) Phosphoprotein associated with glycosphingolipid-enriched microdomains (Pag), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J. Exp. Med. 191, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Newman P. J., Newman D. K. (2003) Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler. Thromb. Vasc. Biol. 23, 953–964 [DOI] [PubMed] [Google Scholar]
- 78. Qian D., Lev S., van Oers N. S., Dikic I., Schlessinger J., Weiss A. (1997) Tyrosine phosphorylation of Pyk2 is selectively regulated by Fyn during TCR signaling. J. Exp. Med. 185, 1253–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guo B., Kato R. M., Garcia-Lloret M., Wahl M. I., Rawlings D. J. (2000) Engagement of the Human Pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity 13, 243–253 [DOI] [PubMed] [Google Scholar]
- 80. Matsumoto R., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Wang D., Lin X. (2005) Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-[kappa]B activation. Immunity 23, 575–585 [DOI] [PubMed] [Google Scholar]
- 81. Parry R. V., Chemnitz J. M., Frauwirth K. A., Lanfranco A. R., Braunstein I., Kobayashi S. V., Linsley P. S., Thompson C. B., Riley J. L. (2005) CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 25, 9543–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mizuno S., Chijiwa T., Okamura T., Akashi K., Fukumaki Y., Niho Y., Sasaki H. (2001) Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97, 1172–1179 [DOI] [PubMed] [Google Scholar]
- 83. Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H. (1996) The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 16, 3967–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gilliland D. G., Griffin J. D. (2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532–1542 [DOI] [PubMed] [Google Scholar]
- 85. Knudson A. G. (2001) TIMELINE: Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 1, 157–162 [DOI] [PubMed] [Google Scholar]
- 86. Small D. (2006) FLT3 mutations: biology and treatment. Hematology/the Education Program of the American Society of Hematology. Am. Soc.Hematol. 178–184 [DOI] [PubMed] [Google Scholar]
- 87. Koipally J., Georgopoulos K. (2002) A molecular dissection of the repression circuitry of Ikaros. J. Biol. Chem. 277, 27697–27705 [DOI] [PubMed] [Google Scholar]
- 88. Imai Y., Kurokawa M., Yamaguchi Y., Izutsu K., Nitta E., Mitani K., Satake M., Noda T., Ito Y., Hirai H. (2004) The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol. Cell. Biol. 24, 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Grundler R., Miething C., Thiede C., Peschel C., Duyster J. (2005) FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood 105, 4792–4799 [DOI] [PubMed] [Google Scholar]
- 90. Nolan G. P. (2007) What's wrong with drug screening today. Nat. Chem. Biol. 3, 187–191 [DOI] [PubMed] [Google Scholar]
- 91. Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Murali-Krishna K., Altman J. D., Suresh M., Sourdive D. J., Zajac A. J., Miller J. D., Slansky J., Ahmed R. (1998) Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8, 177–187 [DOI] [PubMed] [Google Scholar]
- 93. Ficarro S. B., Salomon A. R., Brill L. M., Mason D. E., Stettler-Gill M., Brock A., Peters E. C. (2005) Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 19, 57–71 [DOI] [PubMed] [Google Scholar]
- 94. Nilsson C. L., Dillon R., Devakumar A., Shi S. D., Greig M., Rogers J. C., Krastins B., Rosenblatt M., Kilmer G., Major M., Kaboord B. J., Sarracino D., Rezai T., Prakash A., Lopez M., Ji Y., Priebe W., Lang F. F., Colman H., Conrad C. A. (2009) Quantitative phosphoproteomic analysis of the STAT3/IL-6/HIF1α signaling network: An initial study in GSC11 glioblastoma stem cells. J. Proteome Res. 9, 430–443 [DOI] [PubMed] [Google Scholar]
- 95. Hunter T. (1998) The Croonian Lecture 1997. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Phil. Trans. R. Soc. Lond. B 353, 583–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 [DOI] [PubMed] [Google Scholar]
- 97. Harrison C. R., Sader J. A., Lucy C. A. (2006) Sulfonium and phosphonium, new ion-pairing agents with unique selectivity towards polarizable anions. J. Chromatogr. A 1113, 123–129 [DOI] [PubMed] [Google Scholar]
- 98. Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 99. Payne S. H., Yau M., Smolka M. B., Tanner S., Zhou H. L., Bafna V. (2008) Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J. Proteome Res.7, 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Taylor L., Zheng Y., Zhang C., Lock C., Tate S. A., Pawson T. (2010) Identification of species biomarkers of Listeria bacteria by high resolution TOF-MS. Proceedings of the 58th ASMS Conference on Mass Spectrometry and Allied Topics [Google Scholar]
- 102. Nagaraj N., D'Souza R. C. J., Cox J., Olsen J. V., Mann M. (2010) Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 [DOI] [PubMed] [Google Scholar]
- 103. Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 105. Ulintz P. J., Bodenmiller B., Andrews P. C., Aebersold R., Nesvizhskii A. I. (2008) Investigating MS2/MS3 matching statistics: a model for coupling consecutive stage mass spectrometry data for increased peptide identification confidence. Mol. Cell. Proteomics: MCP 7, 71–87 [DOI] [PubMed] [Google Scholar]
- 106. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dowell J. A., Frost D. C., Zhang J., Li L. (2008) Comparison of two-dimensional fractionation techniques for shotgun proteomics. Anal. Chem. 80, 6715–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Slebos R. J., Brock J. W., Winters N. F., Stuart S. R., Martinez M. A., Li M., Chambers M. C., Zimmerman L. J., Ham A. J., Tabb D. L., Liebler D. C. (2008) Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 7, 5286–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rudnick P. A., Clauser K. R., Kilpatrick L. E., Tchekhovskoi D. V., Neta P., Blonder N., Billheimer D. D., Blackman R. K., Bunk D. M., Cardasis H. L., Ham A.-J. L., Jaffe J. D., Kinsinger C. R., Mesri M., Neubert T. A., Schilling B., Tabb D. L., Tegeler T. J., Vega-Montoto L., Variyath A. M., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Carr S. A., Fisher S. J., Gibson B. W., Paulovich A. G., Regnier F. E., Rodriguez H., Spiegelman C., Tempst P., Liebler D. C., Stein S. E. (2010) Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Mol. Cell. Proteomics 9, 225–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Carrascal M., Ovelleiro D., Casas V., Gay M., Abian J. (2008) Phosphorylation analysis of primary human t lymphocytes using sequential imac and titanium oxide enrichment. J. Proteome Res. 7, 5167–5176 [DOI] [PubMed] [Google Scholar]
- 111. Manes N. P., Dong L., Zhou W., Du X., Reghu N., Kool A. C., Choi D., Bailey C. L., Petricoin E. F., 3rd, Liotta L. A., Popov S. G. (2011) Discovery of mouse spleen signaling responses to anthrax using label-free quantitative phosphoproteomics via mass spectrometry. Mol. Cell. Proteomics 10, M110 000927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pan C., Olsen J. V., Daub H., Mann M. (2009) Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics 8, 2796–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bodenmiller B., Mueller L. N., Mueller M., Domon B., Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nature Methods 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 114. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 115. Swaney D. L., Wenger C. D., Thomson J. A., Coon J. J. (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 106, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46 [DOI] [PubMed] [Google Scholar]
- 117. Brill L. M., Xiong W., Lee K. B., Ficarro S. B., Crain A., Xu Y., Terskikh A., Snyder E. Y., Ding S. (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Malik R., Lenobel R., Santamaria A., Ries A., Nigg E. A., Körner R. (2009) Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8, 4553–4563 [DOI] [PubMed] [Google Scholar]
- 119. Pan C., Gnad F., Olsen J. V., Mann M. (2008) Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics 8, 4534–4546 [DOI] [PubMed] [Google Scholar]
- 120. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Han G., Ye M., Zhou H., Jiang X., Feng S., Jiang X., Tian R., Wan D., Zou H., Gu J. (2008) Large-scale phosphoproteome analysis of human liver tissue by enrichment and fractionation of phosphopeptides with strong anion exchange chromatography. Proteomics 8, 1346–1361 [DOI] [PubMed] [Google Scholar]
- 122. Xia Q., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., Peng J. (2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zahedi R. P., Lewandrowski U., Wiesner J., Wortelkamp S., Moebius J., Schütz C., Walter U., Gambaryan S., Sickmann A. (2008) Phosphoproteome of resting human platelets. J. Proteome Res. 7, 526–534 [DOI] [PubMed] [Google Scholar]
- 124. Højlund K., Bowen B. P., Hwang H., Flynn C. R., Madireddy L., Geetha T., Langlais P., Meyer C., Mandarino L. J., Yi Z. P. (2009) In vivo Phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J. Proteome Res. 8, 4954–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wiśniewski J. R., Nagaraj N., Zougman A., Gnad F., Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9, 3280–3289 [DOI] [PubMed] [Google Scholar]
- 126. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 127. Thingholm T. E., Larsen M. R., Ingrell C. R., Kassem M., Jensen O. N. (2008) TiO2-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J. Proteome Res. 7, 3304–3313 [DOI] [PubMed] [Google Scholar]