There are distinct molecular pathologies for two cystic fibrosis–causing mutations in the first transmembrane span of the CF transmembrane conductance regulator protein. These results have implications for understanding the mechanisms of membrane protein integration and folding and the effects of disease-causing mutants.
Abstract
Many missense mutations in the cystic fibrosis transmembrane conductance regulator protein (CFTR) result in its misfolding, endoplasmic reticulum (ER) accumulation, and, thus, cystic fibrosis. A number of these mutations are located in the predicted CFTR transmembrane (TM) spans and have been projected to alter span integration. However, the boundaries of the spans have not been precisely defined experimentally. In this study, the ER luminal integration profiles of TM1 and TM2 were determined using the ER glycosylation machinery, and the effects of the CF-causing mutations G85E and G91R thereon were assessed. The mutations either destabilize the integrated conformation or alter the TM1 ER integration profile. G85E misfolding is based in TM1 destabilization by glutamic acid and loss of glycine and correlates with the temperature-insensitive ER accumulation of immature full-length CFTR harboring the mutation. By contrast, temperature-dependent misfolding owing to the G91R mutation depends on the introduction of the basic side chain rather than the loss of the glycine. This work demonstrates that CF-causing mutations predicted to have similar effects on CFTR structure actually result in disparate molecular perturbations that underlie ER accumulation and the pathology of CF.
INTRODUCTION
Cystic fibrosis (CF) is a lethal genetic disease caused by a lack of functional cystic fibrosis transmembrane conductance regulator protein (CFTR; Riordan et al., 1989; Cheng et al., 1990). In the cell, the CFTR protein is translated and integrated into the endoplasmic reticulum (ER) membrane. It then traffics through the secretory pathway to the cell surface, where it functions as a chloride channel (Anderson et al., 1991; Bear et al., 1992). More than 90% of CF patients have at least one allele with a deletion of phenylalanine at position 508 (ΔF508) (Kerem et al., 1989). ΔF508 (Cheng et al., 1990) and many other CF mutations (Gregory et al., 1991) result in mutant CFTR that does not properly fold and is retained in the ER by cell protein quality control. Thus loss of function due to accumulation of mutant proteins in the ER is the major molecular pathology leading to CF. CFTR contains five domains: two transmembrane-spanning domains (TMDs), two nucleotide-binding domains (NBDs), and a regulatory region (R). Missense mutations in all domains have been identified that result in mutant CFTR accumulation in the ER (Cystic Fibrosis Mutation Database, www.genet.sickkids.on.ca/app). For the bulk of these mutations, neither the CFTR structural perturbation(s) nor the cellular mechanisms for recognizing the perturbation(s) are well characterized. A detailed understanding of CF-causing mutant effects on the CFTR protein is required to decipher the cellular machinery responsible for recognition and retention of the mutants and for individualizing CF therapeutic strategies.
During translation, each CFTR domains folds and can then associate with previously folded domains to form the final, functional CFTR structure (Du et al., 2005; Kleizen et al., 2005; Thibodeau et al., 2005, 2010; Hoelen et al., 2010). The first domain of CFTR translated is TMD1, which contains six transmembrane (TM) spans. The first TM span of TMD1 is translated near the beginning of CFTR production, when a CF-causing mutation could disrupt local protein secondary structure or intradomain structure. Conversely, a mutation could modestly affect TMD1 domain structure but dramatically alter interdomain interactions and global CFTR folding. The roles of intradomain and interdomain defects in CFTR global misfolding or recognition and retention in the ER have not been clearly elucidated.
Mutations that introduce charge into the hydrophobic interior of a TM span are predicted to disrupt that span in a position-dependent manner (Monne et al., 1998; Partridge et al., 2002). Within TMD1, more than 30 mutations introduce or alter a charged amino acid residue in the predicted TM spans, and many are near TM1 (Therien et al., 2001). The first transmembrane span of a multispanning integral membrane protein is important for targeting, integration into, and ultimate topology within the ER membrane (Hartmann et al., 1989). Therefore TM1 mutant cellular ER accumulation could be caused by disrupted ER targeting, integration, topology formation, or TM span stability. The defect(s) could then augment further CFTR misfolding by disrupting intradomain, interdomain, and/or interprotein interactions. One or more of these misfolding event(s) is ultimately recognized by cellular quality control machinery, resulting in accumulation in the ER. Much of the current knowledge about recognition of aberrant proteins is based on studies of soluble proteins or the extramembrane domains of integral membrane proteins. Understanding the details of the misfolding caused by TM mutations is required for addressing subsequent questions about the identity of the proteins involved in recognizing aberrant transmembrane structures.
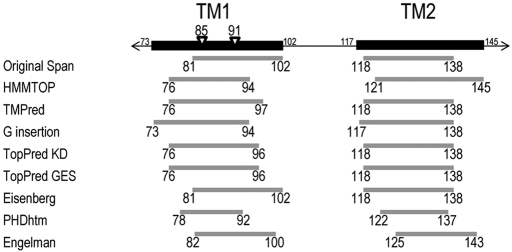
The CF-causing mutants G91R and G85E are in the original predicted TM1 span, residues 81–102 (Riordan et al., 1989). Both mutations were predicted to perturb the TM1 due to introduction of a charged amino acid residue into the putative TM1 span (Xiong et al., 1997). Even though TM1 signal sequence activity in vitro was reduced by both mutations, TM2 appropriately oriented both TMs due to its ability to also act as a signal sequence (Lu et al., 1998). Indeed, TM1 and TM2 have both been implicated in determining CFTR TM span topology (Lu et al., 1998; Chen and Zhang, 1999). The mutant TM1 topologies were indistinguishable from wild-type CFTR, but multidomain constructs containing these mutants have decreased stability in Xenopus laevis oocytes (Xiong et al., 1997). Consistent with reduced stability, full-length G91R CFTR accumulates in the ER, and multiple domains exhibit increased proteolytic susceptibility in mammalian cells (Du and Lukacs, 2009). However, the structural defect(s) that underlie mutant protein destabilization, recognition, and accumulation in the ER remains obscure. The locations of the 85 and 91 positions with respect to various predictions of the TM1 and TM2 spans are shown in Figure 1. These positions are within or peripheral to TM1, depending on the algorithm used.
FIGURE 1:
Predicted TM1 and TM2 spans. Black rectangles including all residues predicted to be in TM1 or TM2 are shown above gray rectangles representing the individual predictions from algorithms listed on the left. The positions of the CF-causing mutants G85E and G91R are indicated by triangles.
The energy associated with the cost of placing a charged residue into a TM span is position dependent (Hessa et al., 2005), and the topology and integration profile of a TM span depend on the placement of positive and negative charge (von Heijne, 1992; Monne et al., 1998). Positioning of a charged residue in a TM span may also alter translocon interactions with that TM span (Pitonzo et al., 2009) or TM span associations with other TM spans (Choma et al., 2000; Zhou et al., 2000, 2001). Therefore precise experimental knowledge of the boundaries of TM1 is required for understanding the role of the glycine at positions 85 and 91 on CFTR folding and of the alterations caused by the disease-associated missense mutations at these positions.
Although the topology of CFTR has been assessed experimentally (Chang et al., 1994b), the TM span boundaries have not been experimentally determined, and predicted span boundaries for TM1 and TM2 vary significantly (Figure 1). Further complicating TM span prediction, the initial integrated span and the span placement in the final protein structure may not be equivalent for many TM spans (Lu et al., 2000; Kauko et al., 2010). One example of this is TM8 of CFTR, which may have a more extended form during integration that exposes more of the residues to the ER lumen than in the final structure (Carveth et al., 2002). In another instance, TM spans within aquaporin-1 reorient during protein maturation in the ER (Lu et al., 2000). This may make monitoring an integrated span versus a final span complex. In the present work, the TM1 and TM2 ER luminal integration profiles were operationally defined using fitness as a substrate for N-linked core glycosylation. As a membrane protein is integrated into the ER membrane, prior to trafficking through the secretory pathway, core glycosylation of appropriate consensus sequences, NXS/T (X ≠ P), occurs in the ER lumen. This core glycosylation is then modified in the Golgi to produce complex glycosylated proteins (Helenius and Aebi, 2001). For CFTR, two natural glycosylation sites are present that can be used to monitor CFTR integration and cellular trafficking by changes in electrophoretic mobility upon core glycosylation, producing band B at ∼150 kDa, and complex glycosylation, producing a diffuse band C above 170 kDa.
In the ER, the oligosaccharyl transferase (OST) protein complex catalyzes the en bloc covalent attachment of a 14-saccharide unit to the consensus-sequence asparagine residue (Helenius and Aebi, 2001). The OST complex resides in the ER membrane associated with the translocation machinery, and, thus, core glycosylation likely occurs cotranslationally (Nilsson et al., 2003; Chavan and Lennarz, 2006). Access to the active site of OST and efficient core glycosylation require that the asparagine residue is extended into the ER lumen and accessible to OST during its synthesis (Nilsson and von Heijne, 1993), presumably before the domain folds. Generally, the minimum distance from the luminal surface of the ER membrane is accommodated by a minimum of 12 residues (Nilsson and von Heijne, 1993). This distance dependence can be used as a molecular ruler to examine the distance from the integrated profile of a TM span to the active site of OST (Nilsson et al., 1998). This type of analysis has been used in vitro to characterize the effects of single amino acid residues on TM helix integration profiles (Monne et al., 1998; Nilsson et al., 1998) and in cell culture to characterize ER luminal TM span boundaries (Popov et al., 1997; Cheung and Reithmeier, 2005, 2007). N-Linked glycosylation has also been used more generally to identify the ER luminal portions of transmembrane proteins, thus determining protein topology (Chang et al., 1994b; Cheung and Reithmeier, 2007). Recently core glycosylation has been developed as a technique to characterize the TM span properties required for recognition and integration by the translocon (Hessa et al., 2005, 2007; Lundin et al., 2008). With use of this method, it was found that 10 of the 12 CFTR TMs, including TM1 and TM2, could insert independently into the ER membrane (Enquist et al., 2009).
In this study, the span boundaries or integration profiles of TM1 and TM2 were determined and the effects of the CF-causing mutations G85E and G91R assessed. The G91R and G85E mutations were previously predicted to have similar effects on CFTR folding yet are shown here to cause disparate perturbations in the protein. This work further elucidates CFTR membrane-spanning structures and provides mechanistic insight into the molecular pathology of the G85E and G91R CF-causing mutations.
RESULTS
CFTR TM1 and TM2 span predictions
The initial prediction of TM1 and TM2 included residues 81–102 (TM1) and 118–138 (TM2) (Riordan et al., 1989). Since then, additional prediction algorithms to identify span boundaries have been developed. Several of these algorithms were used to predict boundaries for TM1 and TM2 (Figure 1). All methods indicate the presence of a TM span between residues 73 and 102 (TM1) and between residues 117 and 145 (TM2), with the TM boundaries at different positions, depending on the method used. Moreover, when the CF-causing mutations G85E and G91R were analyzed using these algorithms, variable effects of mutations were predicted, including shortening, no affect, no TM, or shifting TM1 boundaries (Supplemental Table S1). Taken together, the absence of relevant biochemical data regarding TM1 and TM2 structural information, the dramatic differences in predicted TM boundaries, and the lack of consensus in the mutant predicted effects highlight the need for an experimental approach.
Natural glycosylation sites in CFTR are not required for its cellular trafficking
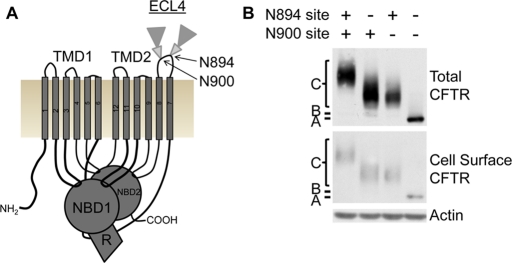
Glycosylation of CFTR occurs at two natural sites between TM7 and TM8 during its cellular trafficking, with core glycosylation occurring in the ER and complex glycosylation occurring subsequent to trafficking out of the ER (Figure 2A). The need of the natural sites for CFTR integration and subsequent trafficking from the ER was tested by their individual or combined removal. The sites, with asparagines at positions 894 and 900, were independently mutated (N to D) to remove the glycosylation consensus sequence. CFTR trafficking to the ER, Golgi, and cell surface of HeLa cells was monitored using glycosylation and cell surface biotinylation. Each natural CFTR site can be core and complex glycosylated independent of the other site and these proteins trafficked to the cell surface (Figure 2B). To verify core and complex glycosylation, samples were treated with specific glycosidases that result in electrophoretic mobility shifts of deglycosylated samples (Supplemental Figure S1).
FIGURE 2:
CFTR natural glycosylation sites are not required for cellular trafficking. (A) Schematic of the five CFTR domains: two transmembrane-spanning domains (TMD1 and 2), two nucleotide-binding domains (NBD1 and 2), and a regulatory region (R). Two natural N-linked glycosylation sites are in extracellular loop 4 (ECL4) between TM7 and TM8, with core glycosylation (small triangle) and complex glycosylation (large triangle) depicted. (B) Cellular trafficking of CFTR containing both or individual or lacking natural glycosylation sites in HeLa cells was monitored by Western blot analysis and verified by glycosidase treatments (Supplemental Figure S1). The positions of CFTR with no glycosylation (band A), core glycosylation (band B), and complex glycosylation (band C) are marked. Top, total cell CFTR; middle, cell surface CFTR identified by biotinylation; actin is a control.
CFTR containing an artificial glycosylation site between TM1 and TM2 is trafficked in the cell
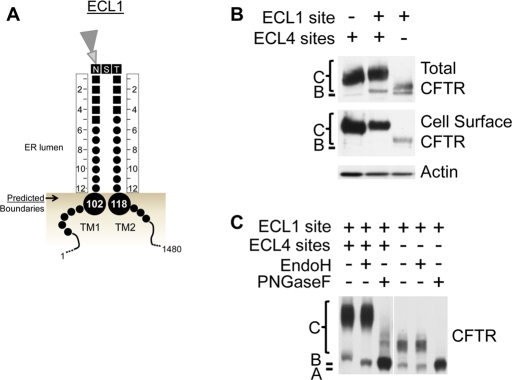
To use glycosylation as a tool to monitor TM1 and TM2 ER integration profiles, the effect of introducing an artificial glycosylation site into extracellular loop 1 (ECL1) between TM1 and TM2 was determined. ER luminal core glycosylation depends on a consensus site (NXS/T) with a minimum extension of a distance of 12 residues N- and C-terminal into the ER lumen or from the ER membrane (Nilsson and von Heijne, 1993). TM1 and TM2 boundary predictions (Figure 1) were used to introduce additional residues both N- and C-terminal to the artificial site to bracket the appropriate distance (Figure 3A). This artificial glycosylation site is introduced between CFTR residues Y109 and D110 and includes five residues added N-terminal and four residues added C-terminal to the NST site, which forms the ECL1 site in this study. In HeLa cells, the ECL1 site is both core and complex glycosylated and trafficked to the cell surface in CFTR containing or lacking the natural ECL4 sites (Figure 3B). Core and complex glycosylation of the ECL1 site containing proteins was verified by electrophoretic mobility shifts after treatment with specific glycosidases (Figure 3C). Mutation of the consensus sequence in the ECL1 site resulted in no glycosylation of the ECL1 loop, confirming glycosylation at the specific site (Supplemental Figure S2). Of importance, the introduced ECL1 site is efficiently core glycosylated with no nonglycosylated band detected, suggesting that this variant is efficiently integrated into the ER membrane (Figure 3, B and C, Figure 4A, and Supplemental Figure S2).
FIGURE 3:
An artificial glycosylation site introduced in ECL1 between TM1 and TM2 is glycosylated and trafficked to the cell surface. (A) Schematic of the predicted TM1 and TM2 ER luminal boundaries (large circles) with the introduced artificial glycosylation site. Residues are shown with CFTR residues as small circles and introduced residues as small squares. Twelve residues are designed to be between the core glycosylation site (NST) and the predicted TM boundaries, which are shown as large circles. (B) Cellular trafficking of CFTR containing the natural ECL4 glycosylation sites and/or the artificial ECL1 site in HeLa cells was analyzed by Western blot analysis. Top, total cell CFTR; middle, cell surface CFTR identified by biotinylation; actin is a control. (C) Core and complex glycosylation of CFTR containing the ECL1 site in the presence or absence of the ECL4 glycosylation sites was verified by digestion with glycosidase selective for core and complex glycosylation (PNGaseF) or core glycosylation (Endo H). The positions of CFTR with no glycosylation (band A), core (band B), and complex glycosylation (band C) are marked.
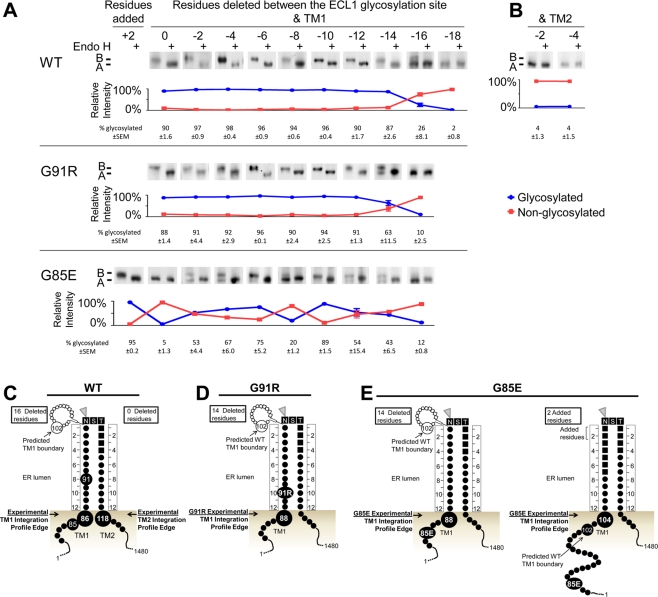
FIGURE 4:
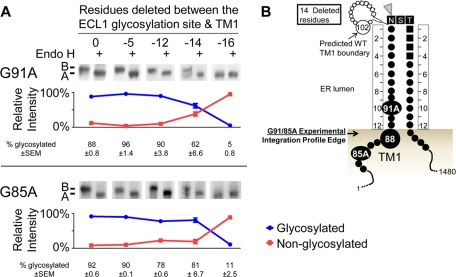
Experimental TM1 ER luminal integration profile edges for WT and CF mutant CFTR. Core glycosylation requires a minimum of 12 residues between the artificial ECL1 site and the ER membrane. The distance between the ECL1 site and TM1 or TM2 was reduced by deleting residues proximal to the site. Core glycosylation analysis of WT, G91R, and G85E CFTR containing the artificial ECL1 site and lacking the ECL4 sites was performed by deletion of residues between the glycosylation site and TM1 (A) or between the glycosylation site and TM2 (B). Constructs were expressed in HeLa cells and analyzed by Western blot. Core glycosylation was detected by its removal with Endo H. Each image shows the core and nonglycosylated CFTR bands. The positions of nonglycosylated (band A) and core glycosylated (band B) CFTR are marked. Below each image, the percentage of glycosylated (blue) and nonglycosylated (red) protein from the untreated sample is shown after quantification, with the average and SEM displayed. Schematics of the experimentally identified ER integration profiles in WT (C) or G91R (D) and G85E (E) mutants.
Experimentally determined TM1 and TM2 ER integration profiles of wild-type CFTR
To examine the TM1 and TM2 ER integration profiles of wild-type (WT) CFTR, an assay was used that employs core glycosylation of the artificial ECL1 site as a molecular ruler in HeLa cells. With use of this assay, the ER integration profile luminal edge for WT CFTR TM1 and TM2, as a basis for comparison to that of the mutant CFTRs, was determined. As described, a 12-residue minimum extension into the ER lumen from the surface of the ER membrane is needed for efficient core glycosylation. Deletion of residues proximal to the site reduces the distance between the ECL1 site and the ER membrane surface and thereby the distance from the integrated TM profile. When the ECL1 site is too close to the ER membrane, it is no longer a good substrate for OST. The deletions required to completely prevent core glycosylation define a location of the integrated span near the luminal edge. Here the integration profile luminal edge is operationally defined as at the next-longest construct that is glycosylated as compared with the nonglycosylated construct. N- or C-terminal residue deletions reduce the distance from the ELC1 site to the integrated TM1 or TM2, respectively. Full-length CFTR constructs containing the ECL1 site were expressed in HeLa cells and ECL1 site core glycosylation monitored. N-Terminal deletion constructs from the ECL1 site were core glycosylated until a 16-residue deletion resulted in partial core glycosylation and an 18-residue deletion was not glycosylated (Figure 4A). With a 16-residue deletion, I86 is 12 residues from the ECL1 site, positioning the TM1 ER luminal integration profile edge near residue 86 (Figure 4C). By contrast, the original TM1 prediction used as a starting point in development of the ECL1 site indicated the TM1 ER luminal boundary was at residue 102 (Figure 3A). The experimentally identified TM1 integration profile is shifted N-terminal to all of the TM1 predictions (Figure 1) with PHDhtm closest, with a boundary of residue 92 predicted.
The predictions are in closer agreement with the experimentally defined TM2 ER luminal integration profile. C-Terminal deletions, including the two-residue deletion, to the ECL1 site tested abolished ECL1 glycosylation (Figure 4B). In the unmodified ECL1 construct, S118 is 12 residues from the ECL1 site, positioning it at the edge of the TM2 ER integration profile (Figure 4C). Thus both the experimentally identified TM2 integration profile edge and the original predicted TM2 boundary are at residue 118 (Figure 3A), as are many of the other predictions (Figure 1).
Determining the ER integration profiles of CF-causing mutants
To assess effects of the CF-causing mutants G91R and G85E on the TM1 ER integration profile, these mutations were analyzed using the ECL1 site core glycosylation assay. The G85E and G91R mutations introduce an ionizable group into or near the predicted TM1 span and might be reasonably expected to alter its ER integration profile (Xiong et al., 1997). If the TM span positioning or integration is altered or destabilized, multiple TM span positions within the ER membrane will be detected as an aberrant or split pattern of core glycosylation (Mingarro et al., 2000). The G91R and G85E mutations in CFTR containing the natural ECL4 sites or the ECL1 site resulted in misfolding and accumulation in the ER (Supplemental Figure S3). G85E reduced CFTR maturation and degradation in constructs containing either the natural ECL4 sites or the artificial ECL1 site (Supplemental Figure S4). Because core glycosylation occurs in the ER prior to complete trafficking or functioning of CFTR, it can be used to characterize TM spans that are integrated into and retained in the ER. Thus, in the same manner as WT ECL1, glycosylation analysis was used to characterize the TM1 integration profiles for the G91R and G85E mutants.
Cystic fibrosis–causing mutant G91R shifts the ER integrated profile of TM1
The effect of the G91R mutation on the TM1 ER integrated profile was tested by its introduction into the ECL1 core glycosylation assay. The core glycosylation–derived TM1 ER integration profile places residue 91 in the ER lumen for the wild-type sequence (Figure 4C). By contrast, G91R N-terminal deletion constructs were completely core glycosylated until a 14-residue deletion resulted in partial core glycosylation, and a 16-residue deletion was completely nonglycosylated (Figure 4A), indicating the edge of the mutant TM integration profile shifted by two or three residues. Thus, instead of I86 as in the wild type, L88 is 12 residues from the ECL1 site in the mutant, positioning it at the G91R TM1 ER luminal integration profile edge (Figure 4D).
Cystic fibrosis–causing mutant G85E dramatically alters the TM1 integration profile in the ER membrane
The effect of the G85E mutation on the TM1 ER integration profile was examined by introducing it into the ECL1 core glycosylation assay. The core glycosylation–derived TM1 ER integration profile (Figure 4C) and all prediction algorithms place position 85 within TM1 (Figure 1). In the presence of G85E, the unmodified ELC1 site is not glycosylated (Figure 4A). An additional two residues must be added N-terminal to the ECL1 site to observe its efficient glycosylation (Figure 4A). Unexpectedly, N-terminal deletions of from 0 to 14 residues exhibited an aberrant pattern of core glycosylation, with a mixture of glycosylated and nonglycosylated forms (Figure 4A). This aberrant pattern is in stark contrast to WT TM1 and is consistent with multiple conformations and/or profiles of G85E TM1 in the ER membrane, with the two extreme positions defined by the addition of two residues and the deletion of 14 residues. These positions place residue L88 or R104 at the TM1 ER luminal integration profile edge (Figure 4E). As is the case for G91R, the L88 TM1 integration profile edge is slightly shifted from WT. However, the R104 TM1 integration profile edge is dramatically shifted from WT, indicating a significant disruption in the G85E TM1. Comparison of the experimental integration profile edge to predicted TM1 boundaries reveals that it is surprisingly close to several of the original predicted WT TM1 boundaries (Figure 1) and the Kyte and Doolittle scale–predicted G85E TM1 (Supplemental Table S1).
G85E alters the position of TM1 in the membrane
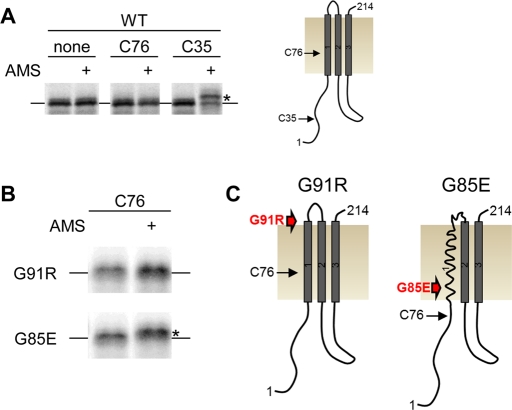
The positioning of G85E TM1 in the membrane as monitored by OST accessibility is altered as compared with WT. To independently validate this result, a cysteine exposure assay was used. In this assay, if a cysteine is within the membrane, it will not react with a membrane-impermeable sulfhydryl reagent, but if it is exposed to aqueous solution, it will react with the reagent. CFTR has a cysteine at position 76, which is within the experimentally derived WT, but not G85E, TM1 integration profile. A construct containing the first three TM spans of CFTR was generated where the naturally occurring cysteines at positions 76 and 128 were mutated to serines. These changes did not measurably alter CFTR maturation as monitored by band C formation (data not shown). A positive control was generated by positioning a nonnative cysteine at residue 35, which is in the cytosol. Constructs of three TMs containing different mutations were generated using in vitro translation in a wheat germ lysate in the presence of canine pancreatic microsomes (Figure 5A). The integrated constructs were probed with the membrane-impermeable reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) to determine whether a cysteine is exposed to solution and, thus, not protected in the membrane. The presence of modification of an exposed cysteine was monitored by gel shift of the reacted construct as compared with unreacted construct. When the three-TM construct contains no cysteine or a cysteine at position 76, no gel shift is seen, indicating that AMS does not have access to this position likely because it is within the membrane (Figure 5A). When the construct contains cysteine at position 35 it is modified by AMS, consistent with the cytosolic location of this position (Figure 5A). The reaction of C76 with AMS in the presence of G91R and G85E was also tested. In WT and G91R, no shift is observed, indicating that position 76 is protected by the membrane (Figure 5, B and C). In contrast, G85E C76 reacts with AMS, indicating that this position is no longer protected by the membrane (Figure 5, B and C). This result is consistent with glycosylation scanning results and an initial positioning of TM1 in the presence of G85E that is more C-terminal than for either WT or G91R. A model for this shift is schematically illustrated in Figure 5.
FIGURE 5:
Positions of TM1 in the ER membrane. The position of a cysteine residue in a CFTR construct containing three TMs (residues 1–214) was examined by a cysteine exposure assay in which a cysteine reaction with the membrane-impermeable reagent AMS only occurs if the cysteine is exposed to the cytosol. Three-TM constructs were generated using an in vitro translation system of wheat germ lysate coupled to column-washed canine pancreatic microsomes, in which the translated protein is radiolabeled with S35-methionine and analyzed by phosphorimage analysis. (A) A CFTR three-TM construct containing no cysteine, C76, or C35 was exposed to AMS and examined for the presence of an interaction by gel shift. Position C35 interacts with AMS and is exposed to the cytosol. A three-TM construct that is nonshifted or shifted is designated by a line or a star, respectively. Right, a schematic of the WT three-TM construct. (B) A CFTR construct containing C76 with G91R or G85E was exposed to AMS and examined for the presence of an interaction by gel shift. G85E has a gel shift, indicating that C76 is exposed to cytosol. (C) Schematics of G91R and G85E mutant CFTR three-TM constructs with the relative positions of C76 and the mutants labeled.
Role of the ionizable side chain in altered G91R and G85E TM1 ER integration profiles
In the G91R and G85E mutants, an ionizable side chain replaces the glycine Cα hydrogen. Glycine has many important roles in the stability of and interaction between transmembrane helices (Curran and Engelman, 2003). To investigate whether the mutant effects on folding and TM boundaries are caused by the loss of glycine or by the introduction of the ionizable group, the 85 and 91 positions were mutated to the neutral residue alanine (G91A and G85A). The alanine mutants were examined for TM1 integration profiles using the ECL1 site core glycosylation assay. In the ECL-site assay, G91A and G85A N-terminal deletion constructs were glycosylated until a 14-residue deletion was partially glycosylated and a 16-residue deletion was not glycosylated (Figure 6A). L88 is at the edge of the TM1 ER integration profile edges for both G91A and G85A (Figure 6B), which are shifted by two or three residues from the WT TM1 ER integration profile edge. This integration profile is the same as the G91R TM1integration profile and one of the two extreme G85E TM1 integration profiles. The two-residue boundary shifts are consistent with the substitution for glycine causing a small but consistent decreased distance between the ECL1 site and the ER membrane. It is striking that the G85A TM1 glycosylation pattern does not indicate multiple profiles for TM1, indicating that the aberrant pattern of G85E TM1 results from introduction of the glutamate side chain (Figure 6A).
FIGURE 6:
Experimental TM1 ER luminal integration profile edges for G91A and G85A mutant CFTR. (A) Core glycosylation analysis of G91A and G85A mutant CFTR containing the artificial ECL1 site by deletion of residues between the glycosylation site and TM1. Constructs were expressed in HeLa cells and analyzed by Western blot. Core glycosylation was detected by its removal with Endo H. Each image shows the core and nonglycosylated CFTR bands. The positions of nonglycosylated (band A) and core glycosylated (band B) CFTR are marked. Below each image, the percentage of glycosylated (blue) and nonglycosylated (red) protein from the untreated sample is shown after quantification, with the average and SEM displayed. (B) A combined schematic of the experimentally identified TM1 ER integration profiles for the G91A and G85A mutants.
Role of the ionizable side chain in trafficking of G91R and G85E
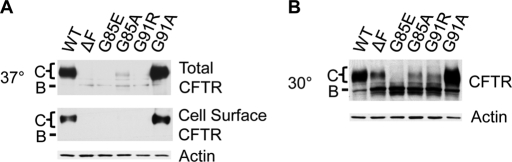
The data from the glycosylation assay demonstrate that the G85E mutant splits the integration profile of TM1, whereas the G91R, G85A, and G91A mutants do not. To examine how these mutants affect the cellular trafficking of CFTR, trafficking at physiologic (37°C) and reduced temperatures (30°C) was monitored. TM1-mutant CFTRs with natural glycosylation sites were expressed in HeLa cells, with CFTR cellular trafficking observed by glycosylation and cell surface biotinylation (Figure 7A). G91A is both core and complex glycosylated and traffics to the cell surface, indicating that introduction of arginine rather than loss of glycine causes G91R ER accumulation. Consistent with this, the G91A mutant has unaltered topology and WT-like degradation in X. laevis oocytes (Xiong et al., 1997). However, G85A was not assessed in the Xenopus system. In stark contrast to G91A, G85A accumulates in the ER, suggesting that both introduction of charge and loss of glycine at position 85 contribute to G85E ER accumulation.
FIGURE 7:
Cellular trafficking of CFTR constructs containing the natural glycosylation sites. HeLa cell trafficking of WT, ΔF508, G85E, G85A, G91R, and G91A mutant CFTR at 37°C was analyzed by Western blot analysis (A). Top, total cell CFTR; middle, cell surface CFTR identified by biotinylation; actin is a control. Low-temperature trafficking rescue at 30°C was performed and analyzed by Western blot analysis (B). The ΔF508 control is retained in the ER at 37°C and partially traffics from the ER at 30°C. The positions of core (band B) and complex (band C) glycosylated CFTR are marked.
The ΔF508 mutation exhibits a temperature-sensitive trafficking from the ER, in which it is retained at 37°C, but it partially traffics from the ER at lower temperatures (Denning et al., 1992). To test the effect of temperature on the TM1 mutants, they were grown at 30°C with ΔF508 monitored as a control (Figure 7B). At both temperatures, G91A traffics like WT CFTR. At the lower temperature, both G91R and G85A mutants partially traffic from the ER and are thus temperature sensitive, similar to ΔF508. It is striking that the G85E mutant exhibited temperature-insensitive ER accumulation. This observation cannot be accounted for by lower protein expression of G85E, as band B is not measurably altered. Of importance, G85E temperature-insensitive ER accumulation correlates with the G85E-perturbed TM1 integration profile with an edge at R104.
DISCUSSION
Many disease-causing mutations in CFTR are predicted to introduce ionizable side chains into or near its hydrophobic TM spans (Cheung and Deber, 2008). Prediction and testing of these mutant consequences are impeded by the lack of detailed CFTR structural data and difficulties in producing the full-length protein. Thus much present work relies on structural models derived from homologues and predictions of domain and transmembrane span boundaries. The predicted boundaries for TM1 and TM2 vary widely, depending on the algorithm used; therefore experimentally determined TM span boundaries are needed to accurately characterize the native and CF-causing mutant TM spans. This study determined the TM1 and TM2 ER luminal integration profile edges and CF-causing mutant G91R and G85E effects on TM1, using the mammalian ER luminal core glycosylation machinery.
Before identification of these boundaries, the two natural glycosylation sites in ECL4 were removed. The natural glycosylation sites were not required for cellular trafficking (Figure 2), consistent with their previously described nonessential roles for trafficking from the ER and CFTR chloride channel function (Howard et al., 1995; Chang et al., 2008; Glozman et al., 2009). Recently these sites have been found to influence the efficiency of CFTR productive protein folding and early secretory trafficking (Glozman et al., 2009) and cell surface retention and turnover in post-ER cellular compartments (Chang et al., 2008; Glozman et al., 2009). Although these effects likely alter overall CFTR levels in the cell, the effects on ER biogenesis do not contribute measurably to the integration process monitored in this study.
An artificial glycosylation site was introduced into ECL1 in CFTR devoid of natural glycosylation sites. This construct was specifically designed to include 12 residues between the predicted TM spans and the consensus sequence, unlike other artificial ECL1 glycosylation sites previously used to monitor cellular localization (Chang et al., 1994a; Cui et al., 2007). The studied ECL1 site here can be core and complex glycosylated, indicating that this region is tolerant of the glycosylation manipulation. However, the total protein quantity of the construct is decreased, as are the levels as measured by pulse chase (Supplemental Figure S4), consistent with previous work that the natural ECL4 sites influence CFTR turnover. We also cannot rule out an altered channel function. Yet steady-state band B is not measurably decreased, and, thus, the slight reduction in total CFTR may result from altered turnover during later steps in CFTR trafficking as observed for other nonnatively glycosylated CFTR forms (Chang et al., 2008; Glozman et al., 2009).
The TM span topology and membrane boundaries for membrane spanning proteins have been studied using the ER core glycosylation machinery both in vitro and in cell culture (Cheung and Reithmeier, 2005, 2007). In one such study, the major disease-causing mutation in the anion exchanger 1 (AE1) resulted in incorrect positioning of a TM span (Cheung and Reithmeier, 2005). In the work presented here, CF-causing mutant effects on TM1 were identified by core glycosylation in mammalian cells. In both cases, this experimental approach provides insight into disease-causing TM span structural perturbations in the mammalian cell that are otherwise difficult to detect. The greatest challenge to core glycosylation analysis is monitoring the small produced electrophoretic differences, which were overcome here by a combination of glycosidase treatments and high-resolution electrophoretic separation.
The core glycosylation machinery is associated with the ER translocon, and core glycosylation likely occurs cotranslationally (Chavan and Lennarz, 2006). Yet the timing of the reaction with respect to TM span integration and translation of other parts of the protein has not been determined in detail. Furthermore, in TM span predictions based on translocon integration energy, the selected span can vary from the identified crystallographic TM span (Kauko et al., 2010). Hence some TM spans may be able to shift and reposition during protein translation and folding and as interacting TM spans are formed (Kauko et al., 2010). Core glycosylation may also be reflecting other TM span interactions during integration. For instance, CFTR TM8 forms an interaction with the translocon machinery during integration (Pitonzo et al., 2009). Ionic interactions between TM spans within membranes have also been found to be important for interaction with other TMs or molecules (Choma et al., 2000; Zhou et al., 2000, 2001), which may be reflected in the pattern of core glycosylation and position of the TM span. It is therefore reasonable to assume that core glycosylation may be indicative of a TM span ER integration profile that occurs prior to the final folded protein structure. A tilting or shift of TM1 subsequent to the glycosylation modification would therefore not be reflected, or perhaps allowed, in the glycosylation-based analysis. If so, the technique allows identification of a cotranslational TM span position.
In this study, core glycosylation was used to identify the WT TM1 and TM2 ER luminal integration profile edges, which are near but not overlapping with the previously predicted spans. The experimentally determined TM2 integration profile is most consistent with the predictions (Figure 1). Of interest, it is more hydrophobic than TM1 (Wigley et al., 1998), an important parameter for TM span prediction by most methods. The TM1 ER integration profile is significantly N-terminal to the predicted span boundaries, placing C-terminal residues previously predicted to reside in TM1 within the ER lumen. This positioning is supported by the exposure of cysteine at position 76 to solution, whereby 76 resides in the membrane in many predictions (Figure 1). The exposed, more–C-terminal residues may form a structural extension, an amphipathic helix or reentrant loop, interact with parts of CFTR or cellular proteins, or shift into the membrane during a later folding step. On the basis of the criteria for span repositioning (Kauko et al., 2010), which uses transmembrane insertion efficiency predictions (Hessa et al., 2007), the TM1 span is within the range for potential repositioning (data not shown). The only experimentally determined CFTR TM1 integration profile is that identified here, and other studies in full-length protein will be required to validate the final span position.
Thus CF mutations, such as G91R, in the C-terminal region may be within or exposed to the ER lumen during translation and integration. Consistent with this is the modest shift in the G91R-CFTR TM1 ER integration profile. The data here and in previous reports indicate that G91R, but not G91A, disrupts CFTR trafficking in the cell and has significant effects on the stability and assembly of full-length CFTR (Xiong et al., 1997; Younger et al., 2006; Rosser et al., 2008; Du and Lukacs, 2009). Arginine introduction likely propagates throughout the structure of CFTR, much like ΔF508-mediated alterations in NBD1 and ICL4 (Thibodeau et al., 2005; 2010; Hoelen et al., 2010), are most evident in the proteolytic susceptibility of NBD2 (Du et al., 2005).
The core glycosylation experiments demonstrate that G85E is within TM1 and causes at least two TM1 positions with distinct ER integration profiles. There are several alternate integration profiles within the monitored G85E constructs, for which multiple potential explanations exist. One is that G85E samples distinct conformations during integration, and specific conformational distributions between the different constructs are being monitored. The mutation G85E might also lead to different interactions between the TM span and the translocation machinery, similar to interactions mediated by an acidic residue within TM8 of CFTR (Pitonzo et al., 2009). Polar residues have also been shown to drive associations between TM spans (Choma et al., 2000; Zhou et al., 2000, 2001); therefore G85E could result in altered interactions between TM1 and other TM spans that are reflected in the glycosylation pattern. This would indicate that the perturbed pattern for G85E could be due to more than a simple alteration of the 12-residue rule and that perturbations associated with introduction of an acidic residue may be occurring. However, positioning of the G85E TM1 in the membrane monitored by cysteine exposure was found to be more C-terminal in a three-TM span construct that lacked the other nine CFTR TM spans. Thus the simplest interpretation is that the integration profile defect is consistent with a distinct placement or destabilization of G85E TM1 in the membrane. Of interest, the most C-terminal extreme G85E TM1 boundary is within several residues of the original predicted WT TM1 span boundary (Figure 1) and a boundary predicted by TopPred KD (Supplemental Table S1).
Because core glycosylation is likely cotranslational, these defects occur at an early step prior to formation of later domain and multidomain structures. Thus G85E destabilization is an early-folding defect potentially recognizable in the ER before translation is complete. Furthermore, the G85A nonpolar mutation does not perturb the TM1 integration profile and continues to cause CFTR ER accumulation. Consequently, G85E misfolding results from both introduction of an ionizable group and glycine loss, which respectively correlate with temperature-insensitive and temperature-sensitive accumulation in the ER. The specific missteps caused by the two mutants, which have different molecular pathologies, could be recognized individually, in combination, or as a consequent common misfolding domain or multidomain event. In this regard, a Derlin-1–containing complex mediates the retrotranslocation and ER-associated degradation of misfolded proteins (Lilley and Ploegh, 2004; Ye et al., 2004). This complex has been implicated in the recognition and removal of improperly folded CFTR, including G85E mutant CFTR (Sun et al., 2006; Younger et al., 2006; Wang et al., 2008). Experiments designed to test the role of Derlin-1 in the recognition of the G85A mutant would be a reasonable future step toward distinguishing between these two models.
Previous work demonstrated that the G85E and G91R mutations also disrupt later steps in CFTR folding, particularly interdomain interactions, which were proposed to underlie mutant recognition by ER quality control machinery (Xiong et al., 1997). The results presented here demonstrate that G85E dramatically alters the integration profile of TM1. Such an alteration would occur at the earliest steps of translation and integration and could be recognized as a very early misfolding event by ER quality control machinery. The G91R mutant was predicted to have a similar effect on CFTR (Xiong et al., 1997). Yet the experimental evidence presented here distinguishes G91R from G85E with respect to perturbations from the ionizable side chain, the role of glycine, and temperature sensitivity. Small-molecule compounds that correct the cellular processing of ΔF508 were identified through a high-throughput screen (Pedemonte et al., 2005). These compounds likely impart their effects through improved mutant CFTR folding at the ER and stability at the cell surface, with one of the most effective compounds being corrector compound 4 (Pedemonte et al., 2005). It is striking that the corrector compound 4 exhibited mutant-specific effects, partially rescuing the G91R but not G85E CFTR (Grove et al., 2009). This suggests that the molecular pathologies identified in these studies may have significance for determining CF mutants that can benefit from specific treatments to rescue defective CFTR. The detailed mechanistic study of these disease-causing mutants is therefore important to augment the fundamental understanding of membrane protein misfolding and relevant for CF therapeutics.
MATERIALS AND METHODS
Plasmids and DNA techniques
An expression plasmid of full-length, wild-type CFTR (pCMV-CFTR-pBQ6.2) was a gift from J. Rommens (Hospital for Sick Children, Toronto, Canada) and was mutagenized using standard protocols for site-directed mutagenesis (Sambrook et al., 1989). Site-directed mutagenesis was performed by PCR techniques using PfuUltra High-Fidelity DNA Polymerase (Stratagene, Santa Clara, CA). All mutations were confirmed by DNA sequencing. The sense primers are listed in Supplemental Table S2. A glycosylated sequence, NEFDQNSTGQGF, was introduced between CFTR residues Y109 and D110. Residues immediately proximal to the consensus sequence, NST, were removed by site-directed mutagenesis on the CFTR construct containing the artificial ECL1 site. The mutations G91R, G91A, G85E, and G85A were introduced into CFTR constructs containing the natural, artificial, and deletion mutants on the artificial site.
Cell culture and transfection
HeLa Tet-On cells (Clontech, Mountain View, CA), referred to as HeLa cells, were routinely maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Gemini Bio-Products, West Sacramento, CA), 50 μg/ml penicillin, and 50 U/ml streptomycin using standard culture techniques. Plasmids were transfected using Lipofectamine 2000 reagent (Invitrogen) and expressed for 16–24 h. Cells were washed with phosphate-buffered saline (PBS, pH 7.4), lysed in RIPA buffer (20 mM Tris, pH 7.6, 150 mM NaCl, 0.1% SDS, 1% IGEPAL, 0.5% deoxycholic acid, 1 mg EDTA-free protease inhibitor tablet [Roche, Indianapolis, IN]) at 4°C for 1 h, and centrifuged at 13,000 × g. Sample buffer (60 mM Tris, pH 6.8, 5% glycerol, 2% SDS, bromophenol blue, 280 mM β-mercaptoethanol) was added to supernatant and incubated at 37°C for 20 min.
Temperature-sensitive trafficking of CFTR mutants was performed as follows. HeLa Tet-On cells were transfected as described, expressed for 16 h at 37°C, and then moved to 30°C for 16–24 h. Lysis and protein analysis were performed as described.
Western blot analysis
Cell lysate proteins were separated by electrophoresis on 6% (wt/vol) polyacrylamide gels using a Tris-glycine buffering system and transferred to polyvinylidene fluoride Immobilon membranes (Millipore, Billerica, MA). Western blot analysis was performed using primary CFTR antibody M3A7 (Millipore) or 596 (University of North Carolina School of Medicine, Chapel Hill, NC), actin antibody (Millipore), and secondary antibody peroxidase-AffiniPure goat anti–mouse immunoglobulin G (IgG; Jackson ImmunoResearch Laboratories, West Grove, PA) and developed with ELC Plus detection reagent (GE Healthcare, Piscataway, NJ) and film.
Glycosylation analysis
HeLa Tet-On cells were transfected and lysed as described. A total of 500 U of glycosidase, PNGaseF, or endoglycosidase H (Endo H) was added to 40 μl of lysis supernatant and incubated at 37° for 2 h. Samples were analyzed by electrophoresis and Western blotting as described.
High-resolution SDS–PAGE analysis of full-length CFTR core glycosylation samples was performed as described, with the following differences. Samples were analyzed on a 13-cm separating gel (6% acrylamide, 375 mM Tris, pH 8.8, 0.1% SDS) with 1.5-cm stacking gel (4% acrylamide, 125 mM Tris, pH 6.8, 0.1% SDS) at constant milliamperes (20–30 mA) until the 150-kDa molecular weight marker was 3–4 cm from the gel bottom. Mock-treated and glycosidase-treated samples were loaded next to each other for comparison.
Cell surface biotinylation
HeLa Tet-On cells were transfected and CFTR expressed as described. Cells were washed with PBS and exposed to membrane-impermeable EZ-Link Sulfo-NHS Biotinylation Reagent (Thermo Scientific, Waltham, MA) for 30 min at 4°C. The reagent was quenched by three washes with 200 mM glycine and 25 mM Tris, pH 8. Cells were washed with PBS and lysed in RIPA buffer as described. Lysate supernatant was incubated with ImmunoPure Immobilized Streptavidin (Thermo Scientific) at 4°C for 1 h. Beads were collected and washed vigorously three times with RIPA buffer. The biotinylated samples were eluted from beads with sample buffer and analyzed by SDS–PAGE and Western blot analysis as described.
In vitro translation and cysteine accessibility assay
mRNAs for in vitro translation experiments were transcribed in vitro using SP6 RNA polymerase as previously described (Woolhead et al., 2004). Nascent chains of WT and mutated CFTR containing residues 1–214 were prepared by in vitro translation of truncated mRNAs. A wheat germ lysate cell-free in vitro translation system was used for creating translational intermediates as previously described (Do et al., 1996; Liao et al., 1997). Wheat germ lysate was prepared as described (Erickson and Blobel, 1983) with an additional final centrifugation step (55,000 rpm, 9 min, 4°C, TLS55 rotor; Beckman Coulter, Brea, CA) to clear lysate. Column-washed canine pancreatic microsomes were added to reactions with 8 equivalent units/25 μl reaction. Translations were carried out for 60 min and terminated with RNaseA on ice for 10 min.
Samples were split into two 12.5-μl samples, and buffer without or with AMS was added to a final of 5 mM AMS. The reaction was allowed to proceed at 25°C in the dark for 15 min. The reaction was stopped by addition of 20 mM dithiothreitol. Microsomes were sedimented through a 100-μl sucrose cushion (0.5 M sucrose, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 40 mM KOAc, 5 mM Mg(OAc)2) in a TLA 100 rotor (Beckman Optima TL centrifuge) at 100,000 rpm for 10 min at 4°C. The pellets were suspended in SDS sample buffer and examined after high-resolution SDS–PAGE on a 13-cm separating 14% polyacrylamide gel by phosphorimage analysis. 14C-Labeled molecular weight protein markers (GE Healthcare) were loaded on the gel.
Pulse-chase experiment and analysis
HEK293 cells (American Type Culture Collection, Manassas, VA) were maintained in complete media containing DMEM (Invitrogen) supplemented with 10% fetal calf serum (Gemini Bio-Products), 50 μg/ml penicillin, and 50 U/ml streptomycin using standard culture techniques. CFTR constructs were transfected at 350,000 cells/ml in suspension using polyetheylenimine (PEI Polysciences, Warrington, PA) transfection reagent, plated in six-well format, and expressed for 48 h prior to pulse-chase analysis. Cells were washed twice with starvation media (DMEM without methionine/cysteine; Invitrogen) and then incubated in starvation media for 30 min at 37°C, 5% CO2. Cells were pulse labeled by removing starvation media and adding 300 μl of pulse media (DMEM with EasyTag EXPRESS35S Protein Labeling Mix at 350 μCi/ml; PerkinElmer, Waltham, MA) and incubating for 30 min at 37°C, 5% CO2. The pulse media was removed, and cells were washed twice with complete media, incubated in 2 ml of complete media at 37°C, 5% CO2, and collected at the indicated times. At 0, 1, 4, 8, 24, and 48 h time points, cells were washed twice with ice-cold PBS and lysed in RIPA buffer at 4°C for 1 h with rocking. Lysates were cleared by centrifugation at 13,000 × g for 10 min at 4°C and supernatant collected.
Immunoprecipitation of 350 μl of cleared lysate was performed by addition of 2.5 μg of M3A7 antibody (Millipore) and 25 μl of bed volume of protein G–agarose (Roche) and incubated at 4°C overnight in a roller drum. Beads were collected by centrifugation and washed twice with RIPA, and proteins were released with 50 μl of sample buffer. Samples were analyzed by 7% glycine SDS–PAGE and transferred to nitrocellulose (Millipore) membranes. Radiolabeled protein samples were analyzed by phosphorimage analysis on a Typhoon 9410 Variable Mode Imager (GE Healthcare). Quantitation was performed using ImageQuant5.1 (GE Healthcare). The data presented in Supplemental Figure S4 are the average of four experiments, with error bars indicating the SE of the mean. Western blot analysis was performed of whole-cell lysate and immunoprecipitations using primary antibody 596 and secondary peroxidase-AffiniPure goat anti–mouse IgG and developed using Amersham ELCPlus Western blotting detection reagent.
TM span predictions
The full-length CFTR sequence was used to identify predicted TM spans within the TM1 and TM2 regions by previous reports or online prediction algorithms. The original predicted span (Riordan et al., 1989) and Eisenberg and Engelman predictions (Wigley et al., 1998) were previously reported. Online predictions used are HMMTOP, version 2.0 (www.enzim.hu/hmmtop/index.html; Hoelen et al., 2010; Kanelis et al., 2010), TMPred (www.ch.embnet.org/software/TMPRED_form.html; Pitonzo et al., 2009), ΔG insertion (http://dgpred.cbr.su.se/index.php?p =home; Hessa et al., 2007), TopPred KD (Kyte and Doolittle) or Goldman, Engelman, and Steitz scales (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred; von Heijne, 1992; Claros and von Heijne, 1994), and PHDhtm using refined PHDRhtm TM span (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_phd.html; Rost and Sander, 1993, 1994; Combet et al., 2000).
Supplementary Material
Acknowledgments
We thank members of the Thomas lab for helpful suggestions and constructive criticism. This work was supported by grants from the National Institutes of Health (DK49835 and DE12309) and the Cystic Fibrosis Foundation (05XX0) to P.J.T. and National Institutes of Health Ruth Kirschstein predoctoral fellowship F30 (DK087186) to A.E.P.
Abbreviations used:
- CFTR
cystic fibrosis transmembrane conductance regulator
- ECL
extracellular loop
- TM
transmembrane span
- TMD
transmembrane-spanning domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0396) on October 12, 2011.
REFERENCES
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- Bear CE, Li CH, Kartner N, Bridges RJ, Jensen TJ, Ramjeesingh M, Riordan JR. Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR) Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- Carveth K, Buck T, Anthony V, Skach WR. Cooperativity and flexibility of cystic fibrosis transmembrane conductance regulator transmembrane segments participate in membrane localization of a charged residue. J Biol Chem. 2002;277:39507–39514. doi: 10.1074/jbc.M205759200. [DOI] [PubMed] [Google Scholar]
- Chang XB, Hou YX, Jensen TJ, Riordan JR. Mapping of cystic fibrosis transmembrane conductance regulator membrane topology by glycosylation site insertion. J Biol Chem. 1994a;269:18572–18575. [PubMed] [Google Scholar]
- Chang XB, Hou YX, Jensen TJ, Riordan JR. Mapping of cystic fibrosis transmembrane conductance regulator membrane topology by glycosylation site insertion. J Biol Chem. 1994b;269:18572–18575. [PubMed] [Google Scholar]
- Chang XB, Mengos A, Hou YX, Cui L, Jensen TJ, Aleksandrov A, Riordan JR, Gentzsch M. Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator. J Cell Sci. 2008;121:2814–2823. doi: 10.1242/jcs.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan M, Lennarz W. The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci. 2006;31:17–20. doi: 10.1016/j.tibs.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Chen M, Zhang JT. Topogenesis of cystic fibrosis transmembrane conductance regulator (CFTR): regulation by the amino terminal transmembrane sequences. Biochemistry. 1999;38:5471–5477. doi: 10.1021/bi982153t. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Cheung JC, Deber CM. Misfolding of the cystic fibrosis transmembrane conductance regulator and disease. Biochemistry. 2008;47:1465–1473. doi: 10.1021/bi702209s. [DOI] [PubMed] [Google Scholar]
- Cheung JC, Reithmeier RA. Membrane integration and topology of the first transmembrane segment in normal and Southeast Asian ovalocytosis human erythrocyte anion exchanger 1. Mol Membr Biol. 2005;22:203–214. doi: 10.1080/09687860500093115. [DOI] [PubMed] [Google Scholar]
- Cheung JC, Reithmeier RA. Scanning N-glycosylation mutagenesis of membrane proteins. Methods. 2007;41:451–459. doi: 10.1016/j.ymeth.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Du K, Lukacs GL. Cooperative assembly and misfolding of CFTR domains in vivo. Mol Biol Cell. 2009;20:1903–1915. doi: 10.1091/mbc.E08-09-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Enquist K, Fransson M, Boekel C, Bengtsson I, Geiger K, Lang L, Pettersson A, Johansson S, von Heijne G, Nilsson I. Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J Mol Biol. 2009;387:1153–1164. doi: 10.1016/j.jmb.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- Glozman R, Okiyoneda T, Mulvihill CM, Rini JM, Barriere H, Lukacs GL. N-Glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. J Cell Biol. 2009;184:847–862. doi: 10.1083/jcb.200808124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RJ, Rich DP, Cheng SH, Souza DW, Paul S, Manavalan P, Anderson MP, Welsh MJ, Smith AE. Maturation and function of cystic fibrosis transmembrane conductance regulator variants bearing mutations in putative nucleotide-binding domains 1 and 2. Mol Cell Biol. 1991;11:3886–3893. doi: 10.1128/mcb.11.8.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Rosser MF, Ren HY, Naren AP, Cyr DM. Mechanisms for rescue of correctable folding defects in CFTRDelta F508. Mol Biol Cell. 2009;20:4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hoelen H, Kleizen B, Schmidt A, Richardson J, Charitou P, Thomas PJ, Braakman I. The primary folding defect and rescue of DeltaF508 CFTR emerge during translation of the mutant domain. PLoS One. 2010;5:e15458. doi: 10.1371/journal.pone.0015458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, DuVall MD, Devor DC, Dong JY, Henze K, Frizzell RA. Epitope tagging permits cell surface detection of functional CFTR. Am J Physiol. 1995;269:C1565–C1576. doi: 10.1152/ajpcell.1995.269.6.C1565. [DOI] [PubMed] [Google Scholar]
- Kanelis V, Hudson RP, Thibodeau PH, Thomas PJ, Forman-Kay JD. NMR evidence for differential phosphorylation-dependent interactions in WT and DeltaF508 CFTR. EMBO J. 2010;29:263–277. doi: 10.1038/emboj.2009.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauko A, Hedin LE, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane alpha-helices during membrane protein folding. J Mol Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xiong X, Helm A, Kimani K, Bragin A, Skach WR. Co- and posttranslational translocation mechanisms direct cystic fibrosis transmembrane conductance regulator N terminus transmembrane assembly. J Biol Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- Lundin C, Kim H, Nilsson I, White SH, von Heijne G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc Natl Acad Sci USA. 2008;105:15702–15707. doi: 10.1073/pnas.0804842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingarro I, Nilsson I, Whitley P, von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne M, Nilsson I, Johansson M, Elmhed N, von Heijne G. Positively and negatively charged residues have different effects on the position in the membrane of a model transmembrane helix. J Mol Biol. 1998;284:1177–1183. doi: 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, Saaf A, Whitley P, Gafvelin G, Waller C, von Heijne G. Proline-induced disruption of a transmembrane alpha-helix in its natural environment. J Mol Biol. 1998;284:1165–1175. doi: 10.1006/jmbi.1998.2217. [DOI] [PubMed] [Google Scholar]
- Nilsson IM, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- Partridge AW, Therien AG, Deber CM. Polar mutations in membrane proteins as a biophysical basis for disease. Biopolymers. 2002;66:350–358. doi: 10.1002/bip.10313. [DOI] [PubMed] [Google Scholar]
- Pedemonte N, Lukacs GL, Du K, Caci E, Zegarra-Moran O, Galietta LJ, Verkman AS. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitonzo D, Yang Z, Matsumura Y, Johnson AE, Skach WR. Sequence-specific retention and regulated integration of a nascent membrane protein by the endoplasmic reticulum Sec61 translocon. Mol Biol Cell. 2009;20:685–698. doi: 10.1091/mbc.E08-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov M, Tam LY, Li J, Reithmeier RA. Mapping the ends of transmembrane segments in a polytopic membrane proteinScanning N-glycosylation mutagenesis of extracytosolic loops in the anion exchanger, band 3. J Biol Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rosser MF, Grove DE, Chen L, Cyr DM. Assembly and misassembly of cystic fibrosis transmembrane conductance regulator: folding defects caused by deletion of F508 occur before and after the calnexin-dependent association of membrane spanning domain (MSD) 1 and MSD2. Mol Biol Cell. 2008;19:4570–4579. doi: 10.1091/mbc.E08-04-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem. 2006;281:36856–36863. doi: 10.1074/jbc.M607085200. [DOI] [PubMed] [Google Scholar]
- Therien AG, Grant FE, Deber CM. Interhelical hydrogen bonds in the CFTR membrane domain. Nat Struct Biol. 2001;8:597–601. doi: 10.1038/89631. [DOI] [PubMed] [Google Scholar]
- Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, et al. The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem. 2010;285:35825–35835. doi: 10.1074/jbc.M110.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure predictionHydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Wang B, Heath-Engel H, Zhang D, Nguyen N, Thomas DY, Hanrahan JW, Shore GC. BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRDeltaF508 via the derlin-1 complex. Cell. 2008;133:1080–1092. doi: 10.1016/j.cell.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Vijayakumar S, Jones JD, Slaughter C, Thomas PJ. Transmembrane domain of cystic fibrosis transmembrane conductance regulator: design, characterization, and secondary structure of synthetic peptides m1-m6. Biochemistry. 1998;37:844–853. doi: 10.1021/bi972293n. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Xiong X, Bragin A, Widdicombe JH, Cohn J, Skach WR. Structural cues involved in endoplasmic reticulum degradation of G85E and G91R mutant cystic fibrosis transmembrane conductance regulator. J Clin Invest. 1997;100:1079–1088. doi: 10.1172/JCI119618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.