Recent work suggests an important role for the Ran importin system in cilia trafficking. At the onset of ciliogenesis, Ran GTP levels rise markedly at the centrosome. Altering Ran GTP levels by varying RanBP1 expression modulates cilia formation and trafficking.
Abstract
The small GTPase Ran and the importin proteins regulate nucleocytoplasmic transport. New evidence suggests that Ran GTP and the importins are also involved in conveying proteins into cilia. In this study, we find that Ran GTP accumulation at the basal bodies is coordinated with the initiation of ciliogenesis. The Ran-binding protein 1 (RanBP1), which indirectly accelerates Ran GTP → Ran GDP hydrolysis and promotes the dissociation of the Ran/importin complex, also localizes to basal bodies and cilia. To confirm the crucial link between Ran GTP and ciliogenesis, we manipulated the levels of RanBP1 and determined the effects on Ran GTP and primary cilia formation. We discovered that RanBP1 knockdown results in an increased concentration of Ran GTP at basal bodies, leading to ciliogenesis. In contrast, overexpression of RanBP1 antagonizes primary cilia formation. Furthermore, we demonstrate that RanBP1 knockdown disrupts the proper localization of KIF17, a kinesin-2 motor, at the distal tips of primary cilia in Madin–Darby canine kidney cells. Our studies illuminate a new function for Ran GTP in stimulating cilia formation and reinforce the notion that Ran GTP and the importins play key roles in ciliogenesis and ciliary protein transport.
INTRODUCTION
Virtually all mammalian cells display cilia, and these organelles execute essential functions. Ciliary functions include motility (e.g., sweeping motion across respiratory tract epithelium), development of left–right asymmetry during embryogenesis, photosensation (e.g., photoreceptors in the retina), mechanosensation (e.g., fluid flow in the renal tubules), chemosensation (e.g., odorant detection in the olfactory sensory epithelium), and initiation of some signal transduction cascades (e.g., Hh, Wnt, and PDGF; Nonaka et al., 2002; Rosenbaum and Witman, 2002; Huangfu et al., 2003; Haycraft et al., 2005; Schneider et al., 2005; Satir and Christensen, 2007; Lal et al., 2008; Scholey, 2008; Berbari et al., 2009; Gerdes et al., 2009; Johnson and Leroux, 2010). Ciliopathies have been recognized for several years and are often marked by a constellation of clinical features, including retinal degeneration, mental retardation, situs inversus, polydactyly, encephalocele, and cystic disease of the kidney, liver, and pancreas (Badano et al., 2006; Fliegauf et al., 2007). Indeed, polycystic kidney disease and retinitis pigmentosa are two well-described ciliopathies.
Of interest, protein synthesis does not occur in cilia, and recent work has begun to explain how proteins are targeted to this organelle. Specifically, intraflagellar transport (IFT) is now appreciated as essential for assembly and maintenance of almost all flagella and cilia. IFT can function to transport membrane channels and signaling proteins into and along the cilia and flagella (Rosenbaum and Witman, 2002; Follit et al., 2006; Pazour and Bloodgood, 2008; Pedersen and Rosenbaum, 2008). In addition, various ciliary targeting sequences have been identified that appear to direct proteins into the cilia (Geng et al., 2006; Jenkins et al., 2006; Tao et al., 2009; Follit et al., 2010; Hurd et al., 2011). In addition, small G proteins and their regulators, such as Rab6, Rab8, Rab10, Rab11, Rab23, Arf4, Arf6, and Rabin8, were reported to mediate ciliary protein targeting from the Golgi, trans-Golgi network, and ciliary transition zone into cilia and also photoreceptors (Eggenschwiler et al., 2001; Deretic et al., 2005; Nachury et al., 2007; Yoshimura et al., 2007; Babbey et al., 2010; Knoddler et al., 2010). Furthermore, elegant new studies demonstrate that ciliary transition zone proteins regulate the entry and exit of ciliary proteins (Craige et al., 2010; Williams et al., 2011). Nevertheless, our view of ciliary transport is still evolving, and a complete understanding of the mechanisms that target proteins to cilia is elusive.
Recent studies from our group and others suggest that Ran GTP and the importins are involved in targeting proteins to primary cilia. As it is classically understood, the Ran/importin system is a nucleocytoplasmic shuttle (Harel and Forbes, 2004). In interphase cells, the steep gradient of Ran GTP between cytosol (low) and nucleus (high) promotes cargo-importin loading in the cytosol and induces cargo release in the nucleus (Gorlich et al., 2003; Kalab et al., 2006). In the cytosol, importin α binds both cargo protein and importin β, resulting in an importin α/β/cargo heterotrimer that is transferred across the nuclear pore. Once in the nucleus, Ran GTP binds the importins—releasing the nuclear cargo—and Ran GTP/cellular apoptosis susceptibility gene product/importin α and Ran GTP/importin β are separately recycled back to the cytosol. Once in the cytosol, the Ran GTP/importin complexes associate with Ran GTPase-activating protein 1 (RanGAP1) and Ran-binding protein 1 (RanBP1), converting Ran GTP to Ran GDP and releasing the importins (Harel and Forbes, 2004; Pemberton and Paschal, 2005; Yudin and Fainzilber, 2009). The cycle then begins anew. Of interest, knockdown of RanBP1 markedly increases cytosolic Ran GTP and disrupts the critical intracellular Ran GTP gradient (Tedeschi et al., 2007). During cell division, Ran GTP concentrates at the chromosomes and mitotic spindles. At the spindle poles, Ran GTP releases active aster-promoting activities—including TPX2 and NuMa—from the importin α/β heterodimer, and this event subsequently initiates the process of spindle assembly (Clarke and Zhang, 2001; Nachury et al., 2001).
In light of this nuclear transport model, our group and others discovered that some ciliary proteins contain nuclear localization signals (NLS), which mediate a direct interaction between these ciliary proteins and the importins. Of note, the Verhey group found that importin β2 (transportin) binds to a C-terminal NLS-like sequence in KIF17, a kinesin-2 motor that ferries cargo to the ciliary tip. Mutation of the KIF17 NLS-like sequence abrogated its entry into the cilium, indicating that this sequence serves as a ciliary localization signal (CLS) for KIF17 (Dishinger et al., 2010). Similarly, our group demonstrated that importin β2 directs the retinitis pigmentosa protein 2 (RP2) to the primary cilium via an interaction with the M9 domain of RP2 (Hurd et al., 2011). The ciliary proteome reveals that many importins and Ran regulatory proteins reside in ciliary compartments, suggesting that Ran/importin might govern ciliogenesis in addition to controlling the localization of ciliary proteins (Andersen et al., 2003; Avidor-Reiss et al., 2004; Li et al., 2004; Blacque et al., 2005; Efimenko et al., 2005; Keller et al., 2005; Pazour et al., 2005; Stolc et al., 2005; Broadhead et al., 2006; Gherman et al., 2006; Liu et al., 2007).
Of interest, a fraction of cellular Ran GTP and importin β2 is found at the cilia and centrosomes during interphase and regulates the localization of some ciliary proteins (Keryer et al., 2003; Fan et al., 2007; Dishinger et al., 2010; Hurd et al., 2011). We hypothesize that the concentrated Ran GTP at the centrosomes serves as a docking platform to release ciliary proteins from importins and to allow entry past the newly formed basal bodies into the cilium. By this mechanism, we propose that Ran GTP regulates ciliary protein transport and ciliogenesis.
RESULTS
Ran GTP localizes to cilia and basal bodies
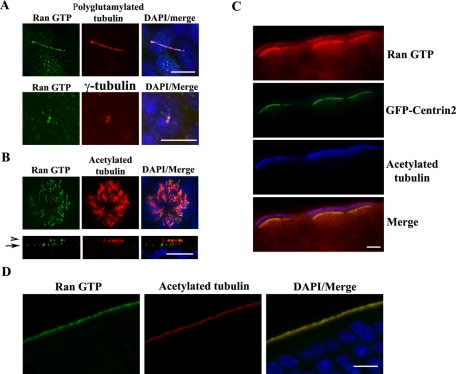
To explore the possible functions of Ran GTP in ciliogenesis, we generated polyclonal antibodies toward Ran GTP using a Ran peptide as previously described by the Macara laboratory (Richards et al., 1995). This antibody detected a single band around 25 kDa in Madin–Darby canine kidney (MDCK), murine inner medullary collecting duct 3 (IMCD3), and human telomerase-immortalized retinal pigment epithelial 1 cells (TERT RPE; Supplemental Figure S1A). Furthermore, this antibody immunoprecipitated Myc-RanQ69L (GTP-locked Ran) but not Myc-RanT24N (GDP-locked Ran) (Supplemental Figure S1B). Using this reagent, we extended our previous studies (Dishinger et al., 2010) and localized Ran GTP in additional mammalian cell types and tissues. We found that Ran GTP localized to primary cilia (Figure 1A, top) and to basal bodies (Figure 1A, bottom) in fully polarized MDCK cells. In differentiated human air tract epithelial cells, Ran GTP localizes to motile cilia and multiple basal bodies (Figure 1B). In green fluorescent protein (GFP)–centrin2 transgenic mouse sections, we determined that Ran GTP primarily localizes to the basal bodies and motile cilia of nasal respiratory epithelium (Figure 1C). We also demonstrated that Ran GTP accumulated at olfactory cilia in GFP-centrin2 mouse sections, similar to our previous findings in wild-type mouse (Dishinger et al., 2010; Figure 1D). We confirmed that Ran GTP antigenic peptides blocked the anti-Ran GTP immunofluorescence signals in TERT RPE cells (Supplemental Figure S1C), thus verifying the specificity of the antibody. These data further confirm the localization of RanGTP in cilia and basal bodies.
FIGURE 1:
Ran GTP localizes to cilia and basal bodies. (A) Ran GTP localizes to primary cilia and basal bodies in MDCK cells. MDCK cells were grown on Transwell filters for 6 d postconfluence, fixed, and stained with anti–Ran GTP (green) and either anti–polyglutamylated tubulin (red; top) or anti-γ-tubulin (red; bottom). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (B) Ran GTP localizes to motile cilia in air tract epithelia. Air tract epithelia were fixed and stained with anti–acetylated tubulin (red) and anti–Ran GTP (green) antibodies. The arrowhead indicates Ran GTP at motile cilia, and the arrow highlights Ran GTP at ciliary basal bodies. (C) Ran GTP localizes to motile cilia and basal bodies in respiratory turbinates. GFP-centrin transgenic mouse respiratory turbinates were stained with anti–Ran GTP (red) and anti–acetylated tubulin (blue). (D) Ran GTP localizes to olfactory sensory neuron cilia. GFP-centrin2 transgenic mouse olfactory epithelial sections were stained with anti-Ran GTP (green) and anti–acetylated tubulin (red). Scale bars, 10 μm.
The accumulation of Ran GTP at the centrosome is linked to ciliogenesis
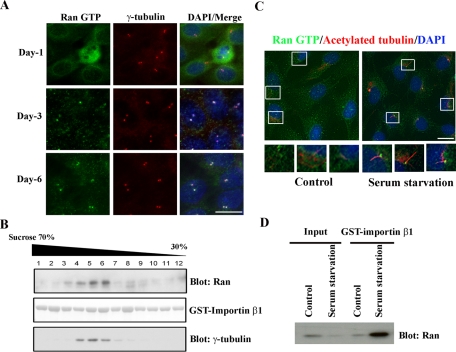
The localization of Ran GTP with cilia and basal bodies prompted us to examine whether Ran GTP fills a functional role in ciliogenesis. First, we assayed the presence of Ran GTP in centrosomes during MDCK polarization, a lengthy process that culminates in cilia formation. We cultured MDCK cells on Transwell filters for various lengths of time and costained with γ-tubulin and Ran GTP antibodies. At day 1, when MDCK cells were not yet polarized, we did not strongly detect Ran GTP at the centrosomes (Figure 2A, top). However, Ran GTP significantly accrued at the centrosomes by days 3 and 6 (Figure 2A, middle and bottom). We also biochemically confirmed the finding that increased levels of Ran GTP were associated with centrosomes by centrosomal fractionation (Figure 2B). We used full-length glutathione S-transferase (GST)–importin β1 to pull down Ran GTP from the isolated centrosomes of fully polarized MDCK cells and probed with a mouse anti-Ran antibody. We verified that Ran GTP is clearly enriched in the centrosomal (γ-tubulin positive) fractions (Figure 2B). In nonpolarized MDCK cells, we weakly detected Ran GTP in the centrosomal fractions (data not shown).
FIGURE 2:
Ran GTP accumulates at basal bodies during primary cilium formation. (A) MDCK cells were seeded on Transwell filters, fixed at the indicated time points, and stained with anti–Ran GTP (green) and anti–γ-tubulin (red). DNA was visualized with DAPI. (B) Ran GTP is enriched in centrosomal fractions. Centrosomes were isolated from fully polarized MDCK cells and fractionated on a discontinuous sucrose gradient. Fractions were washed in 10 mM 1,4-piperazinediethanesulfonic acid, pellets were dissolved in Triton X-100 lysis buffer, and purified samples were subjected to a pull-down assay using full-length GST-importin β1. Proteins bound to GST-importin β1 were eluted, resolved by Bis-Tris PAGE, and immunoblotted with mouse anti-Ran antibody (top). GST-importin β1 was visualized by Ponceau stain (middle). Purified fractions were blotted (i.e., without GST-importin β1 pull down) with anti–γ-tubulin antibody to identify the centrosome-enriched population (bottom). (C) Serum starvation initiates primary cilium formation concomitant with Ran GTP accumulation at basal bodies. TERT RPE cells were cultured on chamberslides with either complete growth media (left) or serum-free media (right) for 48 h. Cells were fixed and stained with anti–acetylated tubulin (red) and anti–Ran GTP (green). Bottom, high-amplification insets of the boxed regions. (D) Serum starvation of TERT RPE cells increases Ran GTP concentration in centrosomes. TERT RPE cells were cultured in complete growth media or serum-free media for 48 h. Then centrosomes were isolated, pulled down using full-length GST-importin β1, and blotted with mouse anti-Ran antibody. Scale bars, 10 μm.
In fibroblasts and some epithelial cells such as TERT RPE and IMCD3, serum starvation initiates primary cilium growth (Schneider et al., 2005; Pugacheva et al., 2007). Usually, primary cilia are fully developed following 24–48 h of serum deprivation. Here, we cultured TERT RPE cells in complete media or serum-free media for 48 h and then stained with anti–acetylated tubulin and anti–Ran GTP to visualize the primary cilia. We did not detect primary cilia or Ran GTP staining in basal bodies when cells were grown in complete media (Figure 2C, left). However, we noted a dramatic enhancement of Ran GTP staining in the basal bodies of primary cilia following serum starvation (Figure 2C, right, and Supplemental Figure S2, B and C). To rule out the possibility that ciliary Ran GTP might contaminate the centrosomal fractions, we did a deciliation time course of TERT RPE cells during nocodazole and cytochalasin B treatment. These agents are commonly used for centrosome isolation procedures. Cilia were completely disassembled after 90 min of treatment with nocodazole and cytochalasin B (Supplemental Figure S2A). In addition, centrosome isolation revealed that Ran GTP levels in the centrosomal fractions were significantly augmented when TERT RPE cells were cultured in serum-free media (Figure 2D). We also confirmed the apparent increase in centrosome-associated Ran GTP in TERT RPE cells after serum starvation by anti-Ran GTP immunoprecipitation (Supplemental Figure S2D). The optimized staining conditions that allow us to visualize centrosomal Ran GTP (cold acetone fixation) yield weak Ran GTP nuclear staining. However, we observed strong nuclear localization of Ran GTP when we used a protocol adapted for nuclear staining: 4% paraformaldehyde fixation followed by 1% SDS permeabilization (Supplemental Figure S1C).
RanBP1 also localizes to cilia and/or basal bodies
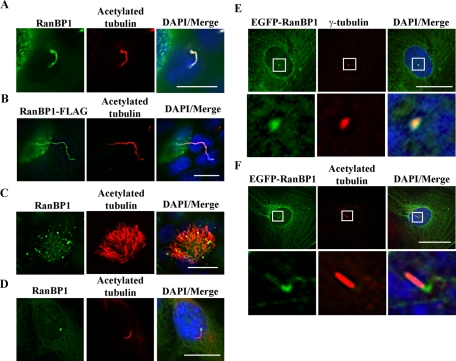
RanBP1 stimulates RanGAP1 to hydrolyze Ran GTP to Ran GDP and also inhibits the activity of the Ran guanine nucleotide exchange factor (GEF) RCC1 (Hayashi et al., 1995; Lounsbury and Macara, 1997). Thus RanBP1 is considered a negative regulator of Ran GTP, and it is predominantly localized to the cytoplasm. Of interest, the cilia/flagella combined proteome database reveals that several importin α and β isoforms, Ran GTPase, and Ran-binding proteins are found in these organelles (Gherman et al., 2006). In addition, a small amount of RanBP1was reported to localize to centrosomes (Di Fiore et al., 2003). We investigated whether RanBP1 might regulate Ran GTP/GDP conversion in cilia and in the basal bodies that initiate ciliogenesis. To address this possibility, we tested the localization of endogenous RanBP1 in IMCD3 cells and air tract epithelial cells. Although RanBP1 distributes throughout the cytoplasm, we also detected RanBP1 costaining along primary cilia with acetylated tubulin in IMCD3 cells (Figure 3A). Similarly, in fully polarized MDCK cells, we moderately expressed inducible RanBP1-FLAG and observed its colocalization with primary cilia upon immunostaining with anti-FLAG and anti–acetylated tubulin antibodies (Figure 3B). In air tract epithelial cells, RanBP1 puncta were distributed along motile cilia (Figure 3C). In TERT RPE cells, we detected endogenous RanBP1 concentrated in the basal bodies of primary cilia following serum starvation (Figure 3D). We also expressed inducible enhanced GFP (EGFP)–RanBP1 in RPE cells and costained with either γ-tubulin or acetylated tubulin antibodies following 48 h of 50 ng/ml doxycycline (Dox) treatment and serum starvation to induce ciliogenesis. As with endogenous RanBP1, we did not observe strong localization of EGFP-RanBP1 in cilia, and we noted that EGFP-RanBP1 is associated with γ-tubulin at centrosomes (Figure 3E) and with the basal bodies of primary cilia (Figure 3F).
FIGURE 3:
RanBP1 localizes to the primary cilia and basal bodies. (A) RanBP1 localizes to primary cilia in IMCD3 cells. IMCD3 cells were cultured on chamberslides for 24 h in complete media and then serum starved for an additional 24 h. Cells were fixed and stained with anti–acetylated tubulin (red) and anti-RanBP1 (green). (B) RanBP1 localizes to primary cilium in MDCK cells. RanBP1-FLAG–inducible MDCK cells were grown on Transwell filters for 5 d and induced with 50 ng/ml DOX for 48 h. Cells were fixed and stained with anti-FLAG (green) and anti–acetylated tubulin (red). (C) RanBP1 localizes to motile cilia in air tract epithelia. Air tract epithelial cells were differentiated on Transwell filters for 3 wk and stained with anti-RanBP1 (green) and anti–acetylated tubulin (red). (D) RanBP1 localizes to the basal body in TERT RPE cells. RPE cells were seeded on chamber slides and cultured with serum-free media for 48 h. Cells were fixed with 4% paraformaldehyde and stained with anti-RanBP1 (green) and anti–acetylated tubulin (red). (E, F) EGFP-RanBP1 localizes to the basal body in TERT RPE cells. Inducible EGFP-RanBP1 RPE cells were seeded on chamberslides and induced with 50 ng/ml DOX for 48 h in serum-free media. Cells were fixed and stained with anti–γ-tubulin (red) in E or anti–acetylated tubulin (red) in F. Bottom, high-amplification insets from the boxed regions. Scale bars, 10 μm.
Down-regulation of RanBP1 promotes ciliogenesis, and overexpression of RanBP1 disrupts primary cilia formation
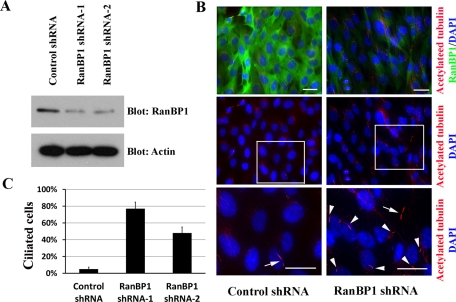
We discovered that Ran GTP and its negative regulator RanBP1 localize to cilia and basal bodies, and we noted that Ran GTP accumulation at basal bodies coincides with ciliogenesis. Therefore we manipulated RanBP1 levels in order to assess the functional roles of RanBP1 in cilia formation. We designed two short hairpin RNA (shRNA) targeting constructs and created stable RanBP1-knockdown TERT RPE cell lines. We confirmed that both cell lines exhibit significantly reduced RanBP1 protein levels (Figure 4A). To evaluate the effects of reduced RanBP1 levels on primary cilia, we cultured RanBP1-knockdown and control cells in complete growth media for 48 h and stained with anti–acetylated tubulin and anti-RanBP1. As expected, we did not detect primary cilia formation in control cells, and we noted the presence of mitotic cells displaying a cytokinetic bridge (Figure 4B, left, middle and bottom, arrow). Of interest, we observed prominent primary cilia in RanBP1 knockdown cells (Figure 4B, right, middle and bottom, arrowheads) in culture conditions promoting cell cycle progression (Figure 4B, right, arrow). Enhanced ciliogenesis in the RanBP1 knockdown cells is quantified in Figure 4C.
FIGURE 4:
RanBP1 knockdown initiates primary cilium formation. (A) TERT RPE cells were individually infected with two RanBP1 shRNAs and one control shRNA (luciferase shRNA), and stable pools were selected. Cell lysates were resolved by Bis-Tris PAGE and immunoblotted with the antibodies indicated. (B) TERT RPE cells stably expressing RanBP1 shRNAs or control shRNA were grown on chamberslides in complete DME/F12 growth media for 48 h. Top, samples were fixed and stained with anti–acetylated tubulin (red) and anti-RanBP1 antibodies (green). Middle, the acetylated tubulin staining individually. Bottom, high-amplification inset of the outlined region. Nuclei were visualized by DAPI staining. Arrows, cytokinetic bridges of mitotic cells; arrowheads, primary cilia. (C) Quantification of the experiments performed in B: 100 cells from each of the three cell lines were counted. The bar graph represents the mean of three individual experiments ± SD. Scale bars, 10 μm.
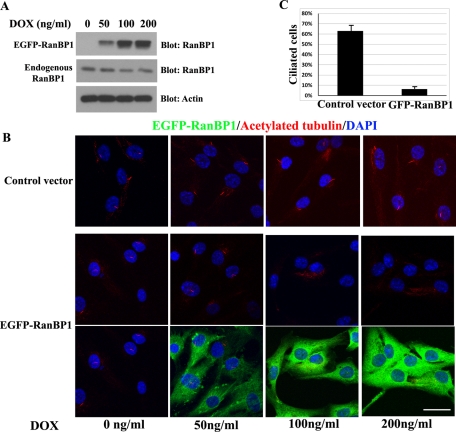
Because reduction of RanBP1 promotes ciliogenesis, we reasoned that overexpression of RanBP1 might impair primary cilium formation. To this end, we further characterized the TERT RPE cell lines stably expressing inducible EGFP-RanBP1. We induced EGFP-RanBP1 expression using various doses of Dox coupled with 24 h of serum starvation to maximize ciliary growth. EGFP-RanBP1 expression increased with higher doses of Dox. We observed that 50 ng/ml Dox induced a 1.5-fold increase in EGFP-RanBP1 compared with endogenous RanBP1, whereas 100 ng/ml or 200 ng/ml Dox induced approximate threefold to fourfold enhancement in EGFP-RanBP1 relative to endogenous RanBP1 (Figure 5A). EGFP-RanBP1–inducible cells treated with low levels of Dox (50 ng/ml) and all control cells (0–200 ng/ml Dox) were still able to form primary cilia; however, inducible EGFP-RanBP1 cells exposed to higher doses of Dox (100 and 200 ng/ml) lose almost all cilia formation following serum starvation (Figure 5B). To exclude the possibility that EGFP expression abrogates cilia formation, we made control EGFP TERT RPE cells. Similar levels of EGFP expression alone do not disrupt ciliogenesis (Supplemental Figure S4, A and B). Thus we validated the finding that RanBP1 antagonizes cilia formation, and these results are quantified in Figure 5C (control vs. EGFP-RANBP1 cells, 200 ng/ml Dox). It has been reported that RanBP1 knockdown or overexpression can alter cell growth, as well as the cell cycle, and this could alter ciliogenesis (Battistoni et al., 1997; Guarguaglini et al., 1997, 2000; Di Fiore et al., 2003; Tedeschi et al., 2007). However, the manipulations of RanBP1 used in this study had only minor effects on the cell cycle (Supplemental Figures S3 and S4, C and D). Knockdown of RanBP1 did reduce cells in S and G2+M (Supplemental Figure S3), but this did not seem sufficient to explain the large increase in ciliated cells. Overexpression of RanBP1 had only minor effects in reducing S phase (Supplemental Figure S4, C and D) yet markedly perturbed ciliogenesis.
FIGURE 5:
Expression of EGFP-RanBP1 disrupts primary cilium formation. (A) TERT RPE cells were infected with an inducible EGFP-RanBP1 retroviral construct, and a stable pool was selected. After growing the cells on chamberslides or 6-cm culture dishes for 24 h, the cells were induced in serum-free media with 50–200 ng/ml Dox for the next 24 h. Cells were harvested, and 20-μg protein samples were electrophoresed on a Bis-Tris PAGE and blotted with RanBP1 and actin antibodies. (B) Cells were fixed and stained with anti–acetylated tubulin. (C) Quantification of the experiments depicted in B: 100 cells from each cell line were counted (EGFP-RanBP1 and control cells treated with 200 ng/ml Dox). Results represent the mean of three individual experiments ± SD. Scale bars, 10 μm.
RanBP1 knockdown enhances Ran GTP in centrosomes
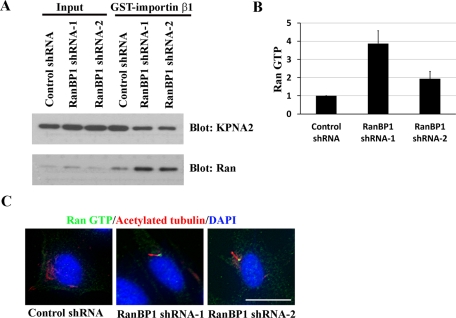
Previous studies demonstrated that RanBP1 knockdown increases cytosolic Ran GTP (Tedeschi et al., 2007). In the present investigation, we found that RanBP1 knockdown stimulates ciliogenesis independent of serum starvation. We inferred that RanBP1 suppression may drive ciliary growth by increasing Ran GTP concentrations at the basal bodies. To test this notion, we isolated centrosomes from RanBP1-knockdown and control cells and subjected the purified fractions to a GST-importin β1 pull-down experiment. Indeed, the RanBP1-knockdown cells demonstrate increased centrosome-associated Ran GTP levels (Figure 6A). KPNA2 (importin α1) served as a positive GST-importin β1 pull-down control. Quantification of Ran GTP levels in control and RanBP1-knockdown cells is depicted in Figure 6B. Next we corroborated the presence of increased Ran GTP at basal bodies in situ. We cultured control and RanBP1-knockdown cells in complete growth media for 48 h and costained with Ran GTP and acetylated tubulin or γ-tubulin antibodies. Indeed, we identified markedly enhanced Ran GTP staining at the basal bodies of primary cilia in RanBP1-knockdown cells. In sharp contrast, we detected neither primary cilia nor significant levels of Ran GTP at the centrosomes in control knockdown cells (Figure 6C and Supplemental Figure S5A). Quantification of Ran GTP staining in control and RanBP1 knockdown cells is displayed in Supplemental Figure S5B.
FIGURE 6:
RanBP1 knockdown increases Ran GTP at centrosomes and initiates primary cilia formation. (A) RanBP1 knockdown increases Ran GTP at centrosomes. Control or RanBP1 knockdown TERT RPE cells were cultured in complete growth media for 48 h. Centrosome fractions were isolated and pulled down using full-length GST-importin β1. Proteins bound to GST-importin β1 were eluted and immunoblotted with the indicated antibodies. (B) Quantification of Ran GTP immunoblots (GE Healthcare Typhoon scanner) from the experiment shown in A. (C) Ran GTP is enriched at the basal bodies of primary cilia in RanBP1-knockdown cells. Control or RanBP1-knockdown TERT RPE cells were grown on chamberslides in complete media for 48 h. Cells were fixed and stained with anti–acetylated tubulin (red) and rabbit anti–Ran GTP (green). Scale bars, 10 μm.
RanBP1 knockdown disrupts localization of KIF17 to the distal tips of primary cilia
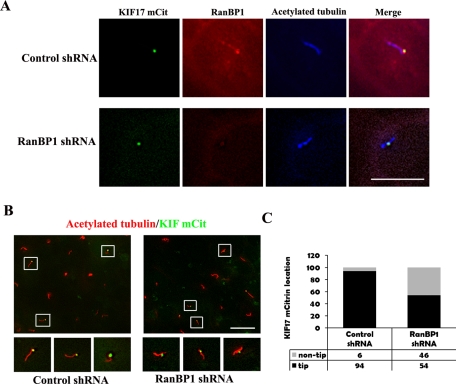
Recently we reported that Ran GTP and importin β2 regulate ciliary entry of KIF17, a kinesin-2 motor. It was demonstrated that expression of RanG19V (GTP-locked Ran) abolished ciliary entry of KIF17. Because Ran GTP is significantly increased in the basal bodies of RanBP1-knockdown cells, we next analyzed the fate of KIF17 in these cells. RanBP1-knockdown IMCD3 cells were transiently transfected with KIF17-mCit, cultured in serum-free media for 24 h, fixed, and stained with anti–acetylated tubulin and anti-RanBP1. Consistent with previous reports, we found KIF17-mCit localized to the tips of primary cilia in control cells (Figure 7A, top). In addition, we noted that RanBP1 colocalizes with KIF17-mCit at the tips of primary cilia (Figure 7A, top). However, KIF17-mCit localization at ciliary tips was perturbed in RanBP1-knockdown cells, and KIF17-mCit was observed in a midway position along the body of the cilium (Figure 7A, bottom). The global disruption of proper ciliary tip KIF17-mCit localization among a field of RanBP1 knockdown cells (as compared with controls) is shown in Figure 7B. Quantification of the immunofluorescence data confirms that virtually all of the ciliary tips contain KIF17-mCit in control cells; however, nearly half of the ciliary tips are missing KIF17-mCit in RanBP1-knockdown cells (Figure 7C).
FIGURE 7:
RanBP1 knockdown disrupts proper KIF17-mCit localization at the tips of primary cilia. (A) IMCD3 cells stably expressing control shRNA (top) and RanBP1 shRNA (bottom) were seeded on chamberslides and grown in complete media for 24 h. Next, they were transiently transfected with KIF17-mCit and switched to serum-free media for an additional 24 h. Cells were fixed and stained with anti-RanBP1 (red) and anti–acetylated tubulin antibodies (blue). KIF17-mCit is labeled in green. (B) Control shRNA (left) and RanBP1-knockdown cells (right) transiently expressing KIF17-mCit were prepared as described in A. Cells were stained with anti–acetylated tubulin (red), and KIF17-mCit is shown in green. Bottom, high-amplification insets of the boxed regions. (C) The graph represents the quantification of experiments depicted in B: 100 KIF17-mCit positive cilia were counted. Black bars indicate KIF17-mCit localized to ciliary tips, gray bars indicate KIF17-mCit not localized to ciliary tips. Scale bars, 10 μm.
DISCUSSION
Studies from our group and others suggest that the importins, traditionally thought of as nucleocytoplasmic shuttles, may function as ciliary transporters as well. Our earlier work demonstrated that a small transmembrane protein Crb3b (also called Crumbs3-CLPI) binds to importin β1. In addition, Crb3b colocalizes with importin β1 in the primary cilia, which suggests that importin β1 may target Crb3b to this organelle (Fan et al., 2007). In a separate report, we showed that importin β2 (also called transportin) directly interacts with RP2. Of note, knockdown of importin β2 blocked the ciliary localization of both endogenous RP2 and EGFP-RP2, implicating importin β2 as the ciliary targeting protein for RP2 (Hurd et al., 2011). This is in agreement with concurrent studies from our group that identified two conserved NLS sequences encoded in KIF17, a kinesin-2 motor. It was discovered that the C-terminal NLS sequence serves as an authentic CLS. Mutation of the CLS barred KIF17 ciliary entry, and this observation led to the elucidation of a mechanism in which importin β2 binds the CLS of KIF17 and regulates its ciliary localization in a Ran GTP–dependent manner (Dishinger et al., 2010). In collaboration with the Verhey laboratory, we also found that Ran GTP was enriched in the primary cilia of NIH 3T3 cells (Dishinger et al., 2010).
In this study, we expand upon the existing paradigm and explore the functional role of Ran GTP in the centrosome/basal body and its relationship to ciliogenesis. To this end, we tested the Ran GTP distribution in several cell lines and tissues that display different types of cilia. We found that Ran GTP is remarkably enriched in both cilia and the basal bodies. We recognized a tight association between cell culture conditions that favor cilia formation—for instance, polarization of MDCK cells and serum starvation of TERT RPE cells—and a marked accumulation of Ran GTP at the centrosomes. In MDCK cells, ciliogenesis begins when the cells are fully polarized, and this process usually takes 5–7 d to complete. In fibroblasts and some epithelial cells, serum-free culture initiates primary cilia formation within 24–48 h. These findings prompted us to propose that up-regulation of Ran GTP may be a crucial effector for ciliogenesis.
If Ran GTP is indeed a required factor in ciliogenesis, then we imagined that adjusting intracellular Ran GTP levels should yield profound effects on cilia formation. Fortuitously, earlier work showed that Ran GTP regulatory proteins reside in the centrosomes, among them the well-described RanBP1 (Di Fiore et al., 2003). RanBP1 is not a catalytic partner that regulates GTP/GDP turnover of Ran. Rather, RanBP1 binds to RanGAP1, and RanGAP1 promotes the hydrolysis of Ran GTP to Ran GDP (Bischoff et al., 1995; Kuhlmann et al., 1997; Lounsbury and Macara, 1997). In addition, RanBP1 inhibits the activity of RCC1, a Ran GEF, and thereby suppresses the Ran GDP to Ran GTP conversion (Hayashi et al., 1995). In short, the presence of RanBP1 promotes the formation of Ran GDP.
Thus we focused our attention on RanBP1 as a molecular tool to regulate Ran GTP. We demonstrated that endogenous and epitope-tagged RanBP1 localizes to cilia and/or basal bodies in several ciliated cell lines. Next, we reduced RanBP1 levels by shRNA-knockdown techniques and evaluated its effects on cilia formation and intracellular Ran GTP in TERT RPE cells. We found that RanBP1-knockdown RPE cells robustly initiated primary cilia formation independent of serum starvation, normally a required condition for ciliogenesis. We noted that Ran GTP is simultaneously increased in the centrosomes/basal bodies of RanBP1 RPE–knockdown cells. These experiments provide additional support for the concept that Ran GTP enrichment at the centrosome promotes cilia formation.
It is known that knockdown of RanBP1 can have small effects on the progression of the cell cycle and induce apoptosis (Tedeschi et al., 2007). In our system, we saw a slight, 4–6% decrease of the mitotic index in RanBP1-knockdown TERT RPE cells by fluorescence-activated cell sorting (FACS) analysis (Supplemental Figure S3, A and B); however, this could not explain the nearly 20- to 30-fold increase we saw in ciliogenesis. Similarly, in IMCD3 cells RanBP1 knockdown resulted in specific defects in cilia trafficking, independent of ciliogenesis. In addition, overexpression of RanBP1 has been reported to impair S phase entry and the mitotic cycle and to induce multipolar spindles (Battistoni et al., 1997; Guarguaglini et al., 1997, 2000; Di Fiore et al., 2003). We did not detect any spindle or centrosome cohesion defects in cells overexpressing EGFP-RanBP1 in TERT RPE cells. Our FACS results showed that overexpression of EGFP-RanBP1 did not block cell cycle exit from G1 to G0 after serum starvation (Supplemental Figure S4, C and D). In the report by Di Fiore et al. (2003), the authors observed an approximately fourfold increase of overexpressed RanBP1 in their experiment, but only the cells with the highest levels showed mitotic centrosomal abnormalities. This suggests that the loss of cilia formation in the presence of EGFP-RanBP1 is not a secondary effect of cell cycle defects. Thus our findings suggest that ciliogenesis correlates best with levels of Ran GTP in the cytoplasm and centrosome and not with cell cycle effects.
Although we do find Ran GTP and RanBP1 concentrated at the ciliary tips in our study, we do not yet understand their function in this specialized location. It is interesting to note that the specific placement of importin β and RanBP1 mRNAs within the neuronal axon results in their localized protein synthesis when the axon is injured. Subsequently, importin α and the dynein motor are released from their association with Ran GTP. The proteins reshuffle to form an importin α/importin β/dynein cassette, and this complex transports specific cargoes to the cell body to signal axon injury (Lai et al., 2008; Yudin et al., 2008). It is tempting to speculate that the loss of KIF17-mCit from ciliary tips in RanBP1-knockdown cells may be explained by a universal Ran/importin/RanBP1–regulated anterograde/retrograde transport system in polarized cellular extensions such as axons or cilia.
Because Ran GTP accrues in the basal bodies, we attempted to identify the specific Ran GEF responsible for the GDP/GTP turnover in this organelle. The Ran GEF RCC1 exists exclusively in the nuclei, and we did not detect RCC1 in the centrosome by immunocytochemical or immunoblot analysis (S. Fan and B. L. Margolis, unpublished results). Recently, it was reported that RanBP10 might act as a cytosolic Ran GEF (Schulze et al., 2008). However, we did not observe any effects on ciliogenesis when we overexpressed RanBP10 in RPE cells (S. Fan and B. L. Margolis, unpublished results). There is also the possibility that Ran GTP is created in the nucleus and then enters the cytoplasm to concentrate at the basal body.
In summary, our studies suggest that the accumulation of Ran GTP at the centrosome might function as a local transit center: releasing ciliary cargoes from importin α/β complexes, allowing these cargo proteins to reshuffle binding partners, enabling the cargoes to dock with ciliary elements such as IFT complexes, and ultimately initiating and maintaining ciliogenesis. In RPE and similar nonpolarized cells such as fibroblasts, interfering with the formation of Ran GTP by RanBP1 overexpression was sufficient to block ciliogenesis. In contrast, increasing the level of Ran GTP by RanBP1 knockdown appears to promote ciliogenesis in RPE cells. In a polarized cell such as IMCD3, modulating Ran GTP levels did not appear to affect ciliogenesis but did affect trafficking of the Kif17 motor. Thus careful control of Ran GTP levels may be necessary for optimal protein trafficking into and within the cilia. Perturbation of trafficking induced by lowering Ran GTP levels may be sufficient to prevent ciliogenesis, especially in nonpolarized cells. However, other explanations are possible for Ran-mediated modulation of ciliogenesis. For example, in spindle formation, the Ran importin system releases spindle assembly factors such as HURP and TPX2 that promote formation of the spindles (Kalab and Heald, 2008). In a similar manner Ran and importins could regulate ciliary microtubular dynamics during cilia formation in addition to having effects on ciliary transport. Regardless of mechanism, it is clear from our studies that there is a coordinated regulation of Ran GTP levels at the centrosome and cilia that is essential for proper cilia formation and function.
MATERIALS AND METHODS
Constructs
Human RanBP1 was amplified by PCR from a HeLa cDNA library and subcloned into pRetroX-Tight-Pur vector using BamHI and EcoRI. Later, a FLAG or EGFP tag was inserted at the N-terminus of RanBP1. Myc-tagged Ran was generated by PCR from a human expressed sequence tag (EST; American Type Culture Collection, Manassas, Virginia) and then subcloned into pcDNA3.1 Zeo(+) vector (Invitrogen, Carlsbad, CA) using BamHI and XhoI. Myc-RanQ69L and Myc-RanT24N were generated by PCR mutagenesis. GST-importin β1 was generated by PCR from a full-length EST (mouse importin β1; Open Biosystems, Huntsville, AL) and cloned into pGEX-4T3 (GE Healthcare, Piscataway, NJ) using BamHI. KIF17-mCitrine (mCit) was developed by the Verhey laboratory (Dishinger et al., 2010).
Cell culture
HEK 293T and MDCK II cells were grown in DMEM/high-glucose medium; IMCD3 and TERT RPE cells were grown in DMEM/F12 medium. All culture media were supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, and 2 mM l-glutamine.
Human primary airway epithelial cells were isolated from the tracheobronchial segments of donor lungs obtained at the time of double-lung transplantation and were cultured on collagen-coated plates using bronchial epithelial growth media (BEGM) (Cambrex Bioscience, Walkersville, MD). To differentiate cultures into mucociliary epithelium, passage 1 cells were seeded on collagen-coated Transwell filters and submerged in BEGM until the cells were confluent. Cells were then shifted to an air–liquid interface and maintained in a 1:1 mixture of BEGM and DMEM for 3 wk (Sajjan et al., 2004; Schneider et al., 2005).
To generate stably expressing Myc-RanQ69L and Myc-RanT24N MDCK cell lines, we transfected cells with the constructs described earlier using FuGENE 6 (Roche, Indianapolis, IN), and then selected cells with 200 μg/ml zeocin 48 h later.
To create inducible RanBP1-FLAG, inducible EGFP-RanBP1, and RanBP1-knockdown MDCK or TERT RPE cells, HEK 293T cells were transiently transfected with pGAG/POL and pVSVG plus either pRetroX-Tight-Pur (for inducible expression) or pSIREN-RetroQ (for shRNA expression) constructs using FuGENE6. Retroviral supernatants were collected, filtered, mixed with an equal volume of fresh medium plus 4 μg/ml polybrene (Clontech, Mountain View, CA), and then added to MDCK or TERT RPE cells. Stably expressing pools were selected using 5 μg/ml puromycin. To induce RanBP1 expression, 50–200 ng/ml Dox was added for 24 h in most experiments. For serum starvation, cells were cultured in their typical media minus FBS.
Antibodies
Rabbit anti–Ran GTP antibody was raised to a peptide corresponding to residues 196–207 of human Ran conjugated to keyhole limpet hemocyanin (Cocalico Biologicals, Reamstown, PA). Mouse anti–acetylated tubulin, anti–polyglutamylated tubulin, anti–γ-tubulin, anti-KPNA2, and rabbit anti-FLAG and anti-actin were purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti-Ran was purchased from BD Biosciences (San Diego, CA). Rabbit anti-RanBP1 and rabbit anti-Myc were from Bethyl Laboratories (Montgomery, TX), and mouse anti-Myc (clone 4A6) was from Millipore (Billerica, MA). Goat anti-mouse and goat anti-rabbit secondary antibodies for immunofluorescence were from Molecular Probes (Invitrogen). Horseradish peroxidase (HRP)–conjugated antibodies for immunoblots were purchased from GE Healthcare.
Protein knockdown
Human RanBP1 sequences GGGCAAAACTGTTCCGATTTG and GGGCCATCCGCCTCCTCATGC were selected as RanBP1 shRNA targeting sequences for TERT RPE cells. Mouse RanBP1 sequences GTGCAAAGCTGTTCCGGTTTG and GGACCATCCGCCTTCTTATGA were chosen for RanBP1 knockdown in IMCD3 cells. These sequences were ligated into pSIREN RetroQ (Clontech). A luciferase shRNA construct served as the control.
Immunofluorescence and confocal microscopy
We performed immunofluorescence as described previously. In brief, cells were fixed with 4% paraformaldehyde, followed by 0.25% Triton X-100 permeabilization. For ciliary staining, 0.05% Triton X-100 was used to maintain delicate ciliary architecture. For γ-tubulin staining, we used cold acetone to fix the cells for 4 min. We blocked the samples in 3% goat serum/phosphate-buffered saline (PBS) for 30 min. Primary antibody dilutions were as follows: 1:500 anti–Ran GTP, 1:500 anti–RanBP1, 1:2000 anti–acetylated tubulin and anti–polyglutamylated tubulin, 1:1000 anti–γ-tubulin, and 1:1000 anti-FLAG. Secondary antibodies were diluted 1:2000. Paraformaldehyde-fixed mouse respiratory turbinate and olfactory sections were permeabilized with 0.1% Trion X-100 for 20 min before blocking and staining as described earlier. Most images were obtained using an inverted epifluorescence microscope (Eclipse TE2000U; Nikon, Melville, NY). In some cases, images were acquired using a Meta Laser Scanning Confocal Microscope (LSM 510; Carl Zeiss MicroImaging, Thornwood, NY).
Immunoprecipitation and Western blotting
MDCK and TERT RPE cells were lysed in Triton X-100 lysis buffer (1% Triton X-100, 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol). After centrifugation, supernatants were collected. For anti–Ran GTP immunoprecipitation, 2 μl of rabbit anti–Ran GTP serum was added to cleared lysates and incubated overnight at 4°C. Then 40 μl of 50% protein A–Sepharose was added and incubated for another 2 h at 4°C. Immune complexes were washed three times with HNTG buffer (20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol) and once with PBS. Samples were eluted in LDS (lithium dodecyl sulfate) sample buffer and electrophoresed on 10% Bis-Tris Novex gels (Invitrogen). After transferring proteins to polyvinylidene fluoride, membranes were blocked with 5% bovine serum albumin–Tris-buffered saline (TBS) and incubated with primary antibodies for 1–4 h in blocking buffer. HRP goat anti-mouse or HRP goat anti-rabbit secondary antibodies were diluted in 5% nonfat milk–TBS and incubated with the blots for 1 h. For GST pull-down experiments, 20 μl of a 50% slurry containing full-length GST-importin β1 was added to the centrosomal extracts and incubated for 2 h at 4°C. The GST-importin β1 beads were washed, eluted, and electrophoresed on Bis-Tris gels as described earlier.
Centrosome fractionation
Centrosomes were isolated from fully polarized MDCK cells as described previously (Hurd et al., 2011). To isolate centrosomes from TERT RPE cells, cells were grown in either regular culture media or serum-free media for 48 h at intermediate cell density. Cells were incubated in 5 μg/ml nocodazole and 5 μg/ml cytochalasin B at 37°C for 90 min, washed, lysed, and centrifuged as described previously (Hurd et al., 2011). Centrosomes were extracted with Triton X-100 lysis buffer. GST-importin β1 pull-down experiments and anti–Ran GTP immunoprecipitations were performed as described earlier.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DK84725 (B.L.M.), DC00011 (J.C.M), and DC009606 (J.R.M.). Some of the images were acquired using the Zeiss Confocal Microscope at the Morphology and Image Analysis Core of the University of Michigan. Fluorescence-activated cell sorting analyses were performed by the Flow Cytometry Core at the University of Michigan.
Abbreviations used:
- CLS
ciliary localization signal
- Dox
doxycycline
- GEF
guanine nucleotide exchange factor
- IFT
intraflagellar transport
- IMCD3
inner medullary collecting duct 3
- MDCK
Madin–Darby canine kidney
- NLS
nuclear localization signal
- RanBP1
Ran-binding protein 1
- RanGAP
Ran GTPase-activating protein
- RP2
retinitis pigmentosa protein 2
- TERT RPE
human telomerase-immortalized retinal pigment epithelial
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0267) on October 12, 2011.
REFERENCES
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Babbey CM, Bacallao RL, Dunn KW. Rab10 associates with primary cilia and exocyst complex in renal epithelial cells. Am J Physiol Renal Physiol. 2010;299:F495–F506. doi: 10.1152/ajprenal.00198.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Battistoni A, Guarguaglini G, Degrassi F, Pittoggi C, Palena A, Di Matteo G, Pisano C, Cundari E, Lavia P. Deregulated expression of the RanBP1 gene alters cell cycle progression in murine fibroblasts. J Cell Sci. 1997;110:2345–2357. doi: 10.1242/jcs.110.19.2345. [DOI] [PubMed] [Google Scholar]
- Berbari NF, O'Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, et al. Functional genomics of the cilium, a sensory organelle. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Broadhead R, et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle. Trends Cell Biol. 2001;11:366–371. doi: 10.1016/s0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc Natl Acad Sci USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Ciciarello M, Mangiacasale R, Palena A, Tassin AM, Cundari E, Lavia P. Mammalian RanBP1 regulates centrosome cohesion during mitosis. J Cell Sci. 2003;116:3399–3411. doi: 10.1242/jcs.00624. [DOI] [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol. 2007;178:387–398. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Follit JA, Li L, Vucica Y, Pazour GJ. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol. 2010;188:21–28. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Okuhara D, Yu Z, Tian X, Cai Y, Shibazaki S, Somlo S. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Battistoni A, Pittoggi C, Di Matteo G, Di Fiore B, Lavia P. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem J. 1997;325:277–286. doi: 10.1042/bj3250277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G, Renzi L, D'Ottavio F, Di Fiore B, Casenghi M, Cundari E, Lavia P. Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 2000;11:455–465. [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Yokoyama N, Seki T, Azuma Y, Ohba T, Nishimoto T. RanBP1, a Ras-like nuclear G protein binding to Ran/TC4, inhibits RCC1 via Ran/TC4. Mol Gen Genet. 1995;247:661–669. doi: 10.1007/BF00290397. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Margolis BL. Localization of retinitis pigmentosa 2 to cilia is regulated by importin beta2. J Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, Hurd TW, Zhang L, McEwen DP, Brown RL, Margolis B, Verhey KJ, Martens JR. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Leroux MR. cAMP and cGMP signaling: sensory systems with prokaryotic roots adopted by eukaryotic cilia. Trends Cell Biol. 2010;20:435–444. doi: 10.1016/j.tcb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR 3rd, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M, Tassin AM. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol Cell. 2003;14:4260–4271. doi: 10.1091/mbc.E02-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoddler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann J, Macara I, Wittinghofer A. Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch'ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci USA. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal M, Song X, Pluznick JL, Di Giovanni V, Merrick DM, Rosenblum ND, Chauvet V, Gottardi CJ, Pei Y, Caplan MJ. Polycystin-1 C-terminal tail associates with beta-catenin and inhibits canonical Wnt signaling. Hum Mol Genet. 2008;17:3105–3117. doi: 10.1093/hmg/ddn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr Top Dev Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Macara IG. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J Biol Chem. 1995;270:14405–14411. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Sajjan U, Keshavjee S, Forstner J. Responses of well-differentiated airway epithelial cell cultures from healthy donors and patients with cystic fibrosis to Burkholderia cenocepacia infection. Infect Immun. 2004;72:4188–4199. doi: 10.1128/IAI.72.7.4188-4199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport motors in cilia: moving along the cell's antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze H, Dose M, Korpal M, Meyer I, Italiano JE Jr, Shivdasani RA. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules. J Biol Chem. 2008;283:14109–14119. doi: 10.1074/jbc.M709397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao B, Bu S, Yang Z, Siroky B, Kappes JC, Kispert A, Guay-Woodford LM. Cystin localizes to primary cilia via membrane microdomains and a targeting motif. J Am Soc Nephrol. 2009;20:2570–2580. doi: 10.1681/ASN.2009020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Ciciarello M, Mangiacasale R, Roscioli E, Rensen WM, Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Fainzilber M. Ran on tracks—cytoplasmic roles for a nuclear regulator. J Cell Sci. 2009;122:587–593. doi: 10.1242/jcs.015289. [DOI] [PubMed] [Google Scholar]
- Yudin D, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.