JMY is a p53 cofactor that was recently shown to nucleate actin assembly. Evidence is presented that the function of JMY in actin nucleation is regulated by an inhibitory activity in cells and that JMY has a previously unrecognized function as a modulator of neuritogenesis.
Abstract
Junction-mediating and regulatory protein (JMY) is a p53 cofactor that was recently shown to nucleate actin assembly by a hybrid mechanism involving tandem actin monomer binding and Arp2/3 complex activation. However, the regulation and function of JMY remain largely uncharacterized. We examined the activity of JMY in vitro and in cells, its subcellular distribution, and its function in fibroblast and neuronal cell lines. We demonstrated that recombinant full-length JMY and its isolated WASP homology 2 domain, connector, and acidic region (WWWCA) have potent actin-nucleating and Arp2/3-activating abilities in vitro. In contrast, the activity of full-length JMY, but not the isolated WWWCA domain, is suppressed in cells. The WWWCA domain is sufficient to promote actin-based bead motility in cytoplasmic extracts, and this activity depends on its ability to activate the Arp2/3 complex. JMY is expressed at high levels in brain tissue, and in various cell lines JMY is predominantly cytoplasmic, with a minor fraction in the nucleus. Of interest, silencing JMY expression in neuronal cells results in a significant enhancement of the ability of these cells to form neurites, suggesting that JMY functions to suppress neurite formation. This function of JMY requires its actin-nucleating activity. These findings highlight a previously unrecognized function for JMY as a modulator of neuritogenesis.
INTRODUCTION
The actin cytoskeleton plays essential roles in basic cellular processes, including migration, adhesion, division (Pollard and Cooper, 2009), and membrane trafficking (Lanzetti, 2007), as well as in specialized processes like neuritogenesis (Kessels et al., 2010). In the case of cell migration, dynamic actin polymerization in protrusive structures such as filopodia and lamellipodia generates the force that drives leading-edge protrusion. Neuronal cells also rely on these membrane protrusions for establishment of polarity, formation of growth cones, and neurite extension during development and differentiation. Understanding how the actin cytoskeleton contributes to these cellular processes requires a complete characterization of the regulators of actin polymerization.
Nucleation is the rate-limiting step in actin polymerization and is accelerated by three classes of nucleators: the Arp2/3 complex and its activators the nucleation-promoting factors (NPFs), formins, and tandem-monomer-binding nucleators (Campellone and Welch, 2010; Firat-Karalar and Welch, 2010). Each has a distinct molecular mechanism of action and mode of regulation. The junction-mediating and regulatory protein (JMY) was recently discovered as a hybrid of the NPF and tandem-monomer-binding nucleator classes (Zuchero et al., 2009). Originally identified as a partner of the transcription coactivator p300, JMY was shown to function in the p53 response by augmenting p53-dependent transcription and apoptosis (Shikama et al., 1999). On DNA damage, JMY is released from Mdm2 (which normally targets it for ubiquitin-dependent degradation) and accumulates in the nucleus (Coutts et al., 2007). These functions are mediated by its unique N-terminal domain and central coiled-coil region. In addition, JMY has a C-terminal WASP homology 2 domain, connector, and acidic region (WWWCA) consisting of three actin-binding WH2 domains (WWW) and Arp2/3-binding connector (C) and acidic (A) regions (Zuchero et al., 2009). The WWW domain of JMY nucleates unbranched actin filaments independent of the Arp2/3 complex by bringing together actin monomers in a manner similar to the tandem-monomer-binding nucleator Spire. In addition, the WWWCA domain activates the Arp2/3 complex to promote the formation of Y-branched actin networks. Thus JMY is unique among known nucleators in combining Arp2/3–dependent and Arp2/3–independent nucleating abilities in the same protein.
How the actin-nucleating activity of JMY is regulated remains undetermined. The N-terminal domains of NPFs from the well-characterized WASP and WAVE families are highly divergent and confer distinct modes of regulation that generally involve modulating the accessibility of the C-terminal WCA domain (Campellone and Welch, 2010). For WASP and N-WASP, the activity of the WCA domain is autoinhibited through intramolecular interactions between it and the upstream domains, and inhibition is relieved by the Rho-family GTPases Cdc42 and Rac, the lipid phosphatidylinositol 4,5-bisphosphate, many SH3-containing proteins, and phosphorylation (Miki et al., 1998; Rohatgi et al., 1999, 2000; Higgs and Pollard, 2000; Kim et al., 2000; Prehoda et al., 2000; Torres and Rosen, 2003; Tomasevic et al., 2007). In contrast, the activity of the WCA domain in WAVEs is trans-inhibited by interacting proteins and activated by Rac, phosphatidylinositol (3,4,5)-triphosphate, IRSp53, and phosphorylation (Eden et al., 2002; Suetsugu et al., 2006; Derivery et al., 2009; Ismail et al., 2009; Lebensohn and Kirschner, 2009). Because the role of full-length JMY in actin assembly has not yet been determined, it is not clear whether JMY is regulated by a WASP-type, a WAVE-type, or another mechanism.
JMY is reported to localize to the nucleus (Coutts et al., 2007; Zuchero et al., 2009) and also to the leading edge in migrating cells. It plays a role in migration by promoting Arp2/3–mediated actin polymerization (Zuchero et al., 2009) and by modulating cell adhesion through down-regulation of cadherin expression and stability (Coutts et al., 2009). In addition, the intrinsic nucleating ability of JMY might be required for its function in the nucleus as a transcriptional coactivator (Coutts et al., 2009). JMY is also important for spindle migration, asymmetric cell division, and cytokinesis in mouse oocytes (Sun et al., 2011). Nevertheless, it remains to be determined whether JMY plays a role in other actin-dependent cellular processes.
Here we investigated the activity of full-length JMY in actin assembly in vitro and in cells and the cellular role of JMY in neurons. We found that full-length JMY and the isolated WWWCA region have comparable actin-nucleating and Arp2/3 complex–activating abilities in vitro but that the ability of full-length JMY to induce actin polymerization in cells is significantly lower. This suggests that the NPF and nucleating activity of full-length JMY is inhibited under native conditions. Moreover, we report a previously unrecognized role for JMY in neurons as a negative regulator of neurite outgrowth during differentiation.
RESULTS
Full-length JMY nucleates actin filaments and activates the Arp2/3 complex in vitro
JMY contains a unique N-terminal domain, three central coiled-coil segments, a polyproline region, and a C-terminal WWWCA domain (Figure 1A). The isolated JMY WWWCA domain was previously shown to both nucleate actin polymerization and promote Arp2/3 complex–mediated actin assembly (Zuchero et al., 2009), but the activity of the full-length JMY protein was not described. To determine whether full-length JMY affects actin assembly in vitro, we purified histidine (His)-tagged full-length recombinant JMY (His-JMY) from insect cells (Figure 1B) and examined its ability to promote actin nucleation in the absence and presence of Arp2/3 complex using the pyrene–actin polymerization assay. His-JMY induced rapid, dose-dependent actin polymerization in the absence of Arp2/3 complex, and actin polymerization was further accelerated in the presence of the Arp2/3 complex (Figure 1C and Supplemental Figure S1A). The Arp2/3–stimulating activity, but not the intrinsic actin nucleation activity, was disrupted by mutating a conserved tryptophan residue (W981) in the A domain (Figure 1C), which was implicated in Arp2/3 complex binding and activation by all NPFs (Marchand et al., 2001). To examine the potential mode of JMY regulation, we next assessed the relative activity of full-length JMY compared with its WWWCA domain. To this end, we purified maltose-binding protein (MBP)–tagged, full-length recombinant JMY (MBP-JMY) and MBP-JMY-WWWCA from insect cells (Figure 1B) and compared their intrinsic nucleating and Arp2/3–activating abilities. At all doses tested, MBP-JMY and MBP-JMY-WWWCA had comparable activities in the absence and presence of the Arp2/3 complex (Figure 1D and Supplemental Figure S1B). This suggests that, under these conditions, recombinant full-length JMY is fully active, similar to recombinant WAVEs, as well as the more recently described NPFs WHAMM and WASH (Campellone et al., 2008; Duleh and Welch, 2010; Jia et al., 2010). It does not appear to be autoinhibited like recombinant N-WASP and WASP (Rohatgi et al., 1999; Tomasevic et al., 2007).
FIGURE 1:
Full-length JMY and its isolated WWWCA domain have comparable actin-nucleating and Arp2/3–activating activities in vitro. (A) Domain organization of JMY and other NPFs are shown. A, acidic domain; B, basic domain; C, connector domain; CC, coiled-coil region; GBD, GTPase-binding domain; N, N-terminal domain; P, polyproline region; PRD, proline-rich domain; SHD, Scar-homology domain; TBR, tubulin-binding region; W, WH2 domain; WAHD1, WASH homology domain 1; WMD, WHAMM membrane interaction domain. (B) Purified His-JMY, His-JMY (W981A), MBP-JMY, and MBP-JMY-WWWCA were resolved by SDS–PAGE and stained with Coomassie blue. (C) Actin (2 μM, 7% pyrene labeled) was polymerized with the indicated concentrations of His-JMY with or without 20 nM Arp2/3 complex. When not indicated, 200 nM His-JMY or His-JMY (W981A) was used. a.u., arbitrary units. (D) Actin was polymerized with the indicated concentrations of MBP-JMY or MBP-JMY-WWWCA with or without 20 nM Arp2/3 complex. (E) Images of actin filaments (2 μM actin) polymerized by 200 nM His-JMY or a GST-N-WASP-WWCA control with or without 10 nM Arp2/3 complex. Arrows highlight examples of branched filaments. Scale bar, 5 μm. (F) Quantification of the percentage of branched filaments for the conditions described in E. Data are the mean ± SEM of three experiments with at least 200 filaments counted in 10 different fields (*p < 0.05; *** p < 0.001).
Because isolation of tightly autoinhibited recombinant WASP required expression in mammalian cells (Tomasevic et al., 2007), we also sought to examine the actin polymerization activity of full-length JMY purified from mammalian cells. We generated a stable 293 human epithelial kidney cell line expressing JMY with an N-terminal localization and affinity purification (LAP) tag (Cheeseman and Desai, 2005) consisting of green fluorescent protein (GFP), a TEV protease cleavage site, and an S peptide (Supplemental Figure S2A). With the use of anti–GFP-antibody affinity chromatography followed by TEV cleavage, S-JMY was purified from the stably transfected cell line but not control untransfected cells (Supplemental Figure S2B). No stoichiometric binding partners copurified with JMY under these conditions, although substoichiometric binding partners may be present. The activity of JMY expressed in mammalian cells was comparable to that of JMY purified from insect cells (Supplemental Figure S2C), suggesting that recombinant JMY is fully active for actin polymerization regardless of the cell type from which it is purified.
To examine the organization of actin filaments nucleated by full-length JMY in the absence and presence of the Arp2/3 complex, we visualized individual filaments in polymerization reactions with His-JMY by fluorescence microscopy (Figure 1, E and F). By itself, His-JMY nucleated unbranched filaments, as described previously for JMY-WWWCA (Zuchero et al., 2009). In the presence of the Arp2/3 complex, His-JMY stimulated the formation of Y-branches, albeit at a lower frequency than the GST-N-WASP-WWCA control. These results are consistent with the ability of full-length JMY to nucleate actin polymerization by itself and activate Arp2/3–mediated branching and polymerization.
JMY-WWWCA promotes actin-based motility by an Arp2/3 complex–dependent mechanism
To examine the relative contributions of JMY's intrinsic nucleating activity and its Arp2/3 complex–stimulating activity to generate actin-based forces in cell cytosol, we next examined the ability of JMY-WWWCA and the Arp2/3 activation–deficient mutant JMY-WWWCA (W981A) to promote actin-driven bead motility in Xenopus laevis egg extracts. In similar experiments, other NPFs promote bead motility by an Arp2/3 complex–dependent mechanism (Yarar et al., 2002). We first purified GST-JMY-WWWCA and GST-JMY-WWWCA (W981A) from Escherichia coli and confirmed that GST-JMY-WWWCA nucleated actin polymerization by itself and activated Arp2/3–mediated actin polymerization, whereas GST-JMY-WWWCA (W981A) only nucleated actin polymerization and was defective in activating the Arp2/3 complex (Figure 2A). When 0.5-μm-diameter beads were coated with JMY-WWWCA and added to cell extracts, they formed actin comet tails (Figure 2B) and underwent motility with a mean rate of 7.7 μm/min (standard error of the mean [SEM] = 0.4 μm/min, n = 20 over three independent experiments; Figure 2C and Supplemental Movie S1). In contrast, beads coated with mutant JMY-WWWCA (W981A) did not undergo motility or form comet tails (Figure 2B). These results demonstrate that JMY-WWWCA is sufficient to polymerize actin and direct actin-based motility in cell cytosol and that motile force requires an ability to activate the Arp2/3 complex.
FIGURE 2:
JMY-WWWCA–coated beads undergo actin-based motility in X. laevis egg extracts. (A) Actin (2 μM, 7% pyrene labeled) was polymerized with 200 nM GST-JMY-WWWCA or GST-JMY-WWWCA (W981A) with or without 20 nM Arp2/3 complex. (B) JMY-WWWCA– and JMY-WWWCA (W981A)–coated beads were incubated in X. laevis egg extracts supplemented with rhodamine–actin, and actin structures were visualized by fluorescence microscopy. Arrows highlight bead clusters. Scale bar, 2.5 μm. (C) Time-lapse images of a rhodamine-labeled actin comet tail (Figure 2B, inset) trailing a moving JMY-WWWCA–coated bead. Images were taken at 10-s intervals. Scale bar, 2.5 μm.
JMY is widely expressed in mammalian tissues and cell lines
To begin to investigate the cellular function of JMY, we determined its expression pattern in mammalian tissues and cell lines. We generated a polyclonal antibody against full-length His-JMY that recognized a major protein of ∼125 kDa, roughly corresponding to the predicted molecular mass of native JMY (111 kDa), in extracts from a wide range of mouse tissues (Figure 3A) and mammalian cell lines (Figure 3B). JMY was expressed in most tissues examined and exhibited particularly high expression levels in brain and testis (Figure 3A). In addition, JMY was expressed in all cultured mammalian cell lines tested, including monkey Cos7 and human HFF fibroblasts, human 293 epithelial cells, human SY5Y, mouse Neuro 2a, and rat PC-12 neuronal cells (Figure 3B). Thus JMY has a wide expression profile in mammalian tissues and cell lines.
FIGURE 3:
JMY is widely expressed in mammalian tissues and cell lines. (A) Extracts from mouse tissues (20 μg/lane) or (B) from SY5Y, PC-12, Neuro 2a, Cos7, HFF, NIH 3T3, 293, and MEF cells were immunoblotted with anti-JMY antibody. Anti-tubulin antibody was used as a loading control.
JMY localizes to both nucleus and cytosol
Given that JMY has been reported to localize to both the nucleus and cytosol (or leading edge; Coutts et al., 2007, 2009; Zuchero et al., 2009), we next assessed its relative distribution between these two compartments by fractionation of monkey Cos7 and mouse Neuro 2a cell lysates into nuclear and cytosolic fractions. The nuclear marker protein NonO and the cytosolic marker protein tubulin fractionated with their respective compartments with little cross-contamination, confirming the fidelity of the procedure (Figure 4, A and B). In each cell line, endogenous JMY was found in both cytosolic and nuclear fractions but was enriched in the cytosol (mean ± SEM of cytosolic JMY is 81% ± 1% [Cos7], 83% ± 1% [Neuro 2a]; Figure 4, A and B). Further fractionation of the cytosolic extracts into membrane-associated and soluble components demonstrated that JMY did not associate with membranes (unpublished data), in contrast to other NPFs, including WHAMM, the closest vertebrate homologue of JMY in sequence (Campellone et al., 2008).
FIGURE 4:
JMY localizes to both the nucleus and the cytosol. (A, B) Nuclear and cytoplasmic fractions from (A) Cos7 or (B) Neuro 2a cells treated with or without retinoic acid (RA) for 72 h were resolved by SDS–PAGE and immunoblotted with antibodies to JMY, NonO (a nuclear marker), and tubulin (a cytosolic marker). Relative amounts of JMY in the nucleus and cytosol were determined by densitometry and are represented as the mean ± SEM of three experiments. (C) Images of GFP-JMY in Cos7 cells stained for actin filaments with Alexa 568–phalloidin. JMY localizes to both the nucleus and the cytosol, and is sometimes enriched in membrane ruffles (top). The chosen images represent extremes of GFP-JMY nuclear accumulation (top, 16% GFP-JMY intensity in the nucleus; bottom, 41% GFP-JMY intensity in the nucleus). Scale bars, 5 μm. (D) Scatter plot of the percentage of GFP-JMY intensity in the nucleus and cytosol of GFP-JMY–transfected Cos7 cells. Data are from three experiments with 50 cells quantified per experiment.
To further assess the subcellular distribution of JMY, we transfected Cos7 cells with an N-terminally enhanced GFP–tagged variant of JMY (GFP-JMY) and observed its localization. Consistent with the results of fractionation studies, GFP-JMY was visualized in both the nucleus and cytosol (Figure 4C). In some instances, cytosolic GFP-JMY appeared to be concentrated with actin filaments at the cell periphery in membrane ruffles, similar to what has been reported for endogenous JMY at the leading edge of mouse melanoma cells and human neutrophil cells (Zuchero et al., 2009). In agreement with the fractionation data, quantification of the total intensity of GFP-JMY fluorescence in the nucleus versus the cytosol revealed that 19% ± 1% was in the nucleus and 81% ± 1% was in the cytosol (mean ± SEM; Figure 4, C and D). Thus our results demonstrate that most of the JMY in cells is in the cytosol, supporting the notion that JMY carries out actin-dependent functions in this compartment.
Full-length JMY is less active than JMY-WWWCA in promoting ectopic actin assembly in cells
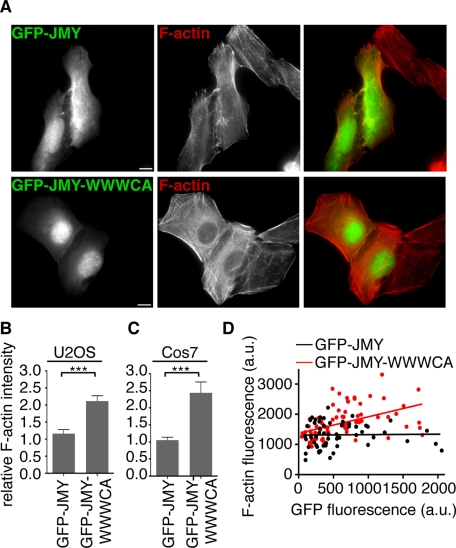
To investigate the mode of regulation of JMY in cells, we assessed the effect of GFP-JMY expression on F-actin abundance. For other NPFs, the effect of protein overexpression on the overall quantity of F-actin in cells is correlated with their mechanism of regulation. For WASP and N-WASP, which are regulated by autoinhibition (Miki et al., 1998; Kim et al., 2000; Prehoda et al., 2000), overexpression of the full-length protein does not lead to an overall increase in cellular F-actin (Campellone et al., 2008). In contrast, for WAVEs, which are regulated by trans-inhibition (Eden et al., 2002; Derivery et al., 2009; Ismail et al., 2009; Lebensohn and Kirschner, 2009), overexpression of the full-length protein leads to a global increase in cellular F-actin (Campellone et al., 2008). To address whether JMY may be regulated by autoinhibition or trans-regulation in cells, we compared the effects of expressing full-length GFP-JMY or truncated GFP-JMY-WWWCA on F-actin content by quantifying the ratio of fluorescent phalloidin staining intensity in transfected compared with nearby nontransfected cells. Of interest, U2OS and Cos7 cells transfected with GFP-JMY-WWWCA had a significant increase in F-actin content relative to nearby nontransfected control cells, whereas F-actin content in cells transfected with GFP-JMY was indistinguishable from that in untransfected controls (Figure 5, B and C). Nevertheless, over the population of cells, GFP-JMY and GFP-JMY-WWWCA were expressed at similar levels (in U2OS extracts [GFP-JMY] = 131 nM, [GFP-JMY-WWWCA] = 151 nM; in Cos7 extracts [GFP-JMY] = 63 nM, [GFP-JMY-WWWCA] = 76 nM). When we quantified and correlated the level of fluorescence of GFP-JMY-WWWCA or GFP-JMY with the intensity of actin fluorescence in individual U2OS cells, there was a positive correlation for GFP-JMY-WWWCA (p < 0.001, r2 = 0.22) but no correlation for full-length GFP-JMY (p = 0.9, r2 = 0.0003; Figure 5D). These observations suggest that JMY-WWWCA is constitutively active, whereas full-length JMY is inhibited in its ability to polymerize actin in cells.
FIGURE 5:
Expression of JMY-WWWCA causes a global increase in cellular F-actin content, but expression of full-length JMY does not. (A) Top, GFP-JMY (green in merge on right) and F-actin (stained with Alexa 568–phalloidin; red in merge on right) in transfected and nontransfected U2OS cells. Bottom, GFP-JMY-WWWCA and F-actin in transfected and nontransfected U2OS cells. Scale bars, 5 μm. (B, C) Quantification of the ratio of F-actin staining intensity in transfected/nontransfected (B) U2OS or (C) Cos7 cells. Data are the mean ± SEM of three experiments with 25 cells examined per sample (***p < 0.001). (D) Graph of GFP-JMY or GFP-JMY-WWWCA fluorescence intensity vs. Alexa 568–phalloidin fluorescence intensities plotted for individual transfected U2OS cells. Data are from three experiments with 25 cells per experiment. Lines represent the best fit for each condition (linear regression analysis: for GFP-JMY-WWWCA, positive linear correlation with a slope of 0.5 ± 0.1, r2 = 0.22, p < 0.0001; for GFP-JMY, no correlation, r2 = 0.0003, p = 0.9).
JMY does not play a crucial role in the migration of mouse embryonic fibroblast cells
Previous studies suggested that JMY plays a role in cell migration in U2OS human osteosarcoma cells (Coutts et al., 2009; Zuchero et al., 2009). To investigate whether JMY functions in cell migration in other cell types, we examined the effect of silencing JMY by RNA interference (RNAi) in mouse embryonic fibroblasts (MEFs) and NIH 3T3 fibroblasts. These cells were transfected with control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) small interfering RNA (siRNA) or JMY siRNA, and depletion of JMY was confirmed by immunoblotting (Figure 6A and Supplemental Figure S3A). To measure cell migration, confluent cell monolayers were scratch-wounded, and the percentage of wound closure was monitored over 4.5 h for MEFs and 12 h for NIH 3T3 cells by phase contrast microscopy. JMY-silenced cells and control cells had a statistically similar percentage of wound closure after 4.5 h for MEF cells (Figure 6, B and C; mean ± SEM of 76% ± 5% closure for JMY silenced cells, 82% ± 5% for controls, p = 0.45) and 12 h for NIH 3T3 cells (Supplemental Figure S3, B and C; mean ± SEM of 62% ± 3% closure for JMY-silenced cells, 61% ± 3% for controls, p = 0.96). Because previous studies showed that the NPF WAVE2 localizes to lamellipodia and is crucial for cell motility (Suetsugu et al., 2003; Yamazaki et al., 2003; Yan et al., 2003), we also tested whether the failure to observe a migration defect in JMY-silenced cells might be due to functional redundancy between JMY and WAVE2. We performed similar wound-healing assays in matched WAVE2−/− MEF cells (Yan et al., 2003) also silenced for JMY expression as described earlier (Figure 6A). Although WAVE2−/− cells were defective in wound closure compared with WAVE+/+ controls, silencing of JMY expression did not cause a more pronounced migration defect (Figure 6, C and D; 53% ± 4% closure for WAVE2−/− vs. 51% ± 3% for WAVE2−/− and JMY silenced, p = 0.68). Thus we were unable to detect a role for JMY during the motility of MEF and NIH 3T3 cells, implying that JMY may not be a universal regulator of cell migration.
FIGURE 6:
JMY silencing does not affect cell migration in mouse embryonic fibroblasts. (A) Extracts from WAVE2+/+ and WAVE2−/− cells transfected with GAPDH or JMY siRNAs for 72 h were resolved by SDS–PAGE and immunoblotted with anti-JMY and anti-tubulin antibodies. (B) Time course (0–4.5 h) of wound closure in WAVE2+/+ and WAVE2−/− cells 72 h after transfection with either GAPDH or JMY siRNA. Scale bars, 10 μm. (C) Quantification of the percentage of wound closure 4.5 h after wound initiation. Data represent the mean ± SEM of the remaining wound area in three experiments (***p < 0.001).
JMY negatively influences neurite outgrowth upon neuronal cell differentiation in an actin-dependent manner
Because we did not observe an effect of JMY silencing on the migration of MEF cells, and because JMY is highly expressed in brain relative to other tissues, we hypothesized that it might have a tissue-specific function in neuronal cells. We therefore investigated a role for JMY in neuronal differentiation and neurite formation using Neuro 2a mouse neuroblastoma cells, a well-studied cell line that can be induced to differentiate and undergo neuritogenesis by retinoic acid (RA) treatment (Wu et al., 1998). We first examined whether JMY protein levels changed during RA-induced differentiation. Of interest, we observed a strong down-regulation of JMY protein levels over the first 48 h of RA treatment compared with the tubulin loading control (Figure 7, A and B), suggesting that JMY expression is suppressed during differentiation. Biochemical fractionation of Neuro 2a cell lysates into nuclear and cytosolic fractions did not reveal any change in the relative distribution of JMY between the two compartments upon RA-induced differentiation (Figure 4B), suggesting that the two pools of JMY are down-regulated proportionally.
FIGURE 7:
JMY negatively regulates neurite outgrowth in Neuro 2a cells in an actin-dependent manner. (A) Extracts from Neuro 2a cells that were induced to differentiate with RA and harvested at the indicated time points were resolved by SDS–PAGE and blotted with anti-JMY antibodies and with anti-tubulin antibodies as a loading control. (B) Quantification of JMY expression levels relative to tubulin by densitometry. Data are the mean ± SEM in three experiments. (C) Extracts from Neuro 2a cells treated with control GAPDH siRNA or each of two independent JMY siRNAs for 96 h were resolved by SDS–PAGE and immunoblotted with anti-JMY antibodies and anti-tubulin antibodies. (D) Quantification of the percentage of cells bearing neurites in Neuro 2a cells transfected with control GAPDH siRNA and two independent JMY siRNAs. Data are the mean ± SEM of three experiments with 500 cells quantified for each (*p < 0.05; **p < 0.01). (E) F-Actin stained with Alexa 488–phalloidin in control and JMY-silenced Neuro 2a cells. Arrows highlight cells bearing neurites. Scale bar, 10 μm. (F) Extracts from Neuro 2a cells treated with control GAPDH or JMY siRNAs, then transfected with LAP, LAP-JMY*–, LAP-JMY (W981A)*–, LAP-JMYΔWWWCA–, and LAP-JMY-WWWCA*–expressing plasmids and induced with retinoic acid for 48 h, were resolved by SDS–PAGE and immunoblotted with anti-JMY antibodies and anti-tubulin antibodies. (G) Quantification of the percentage of cells bearing neurites in Neuro 2a cells treated with control GAPDH or JMY siRNAs, then transfected with LAP–, LAP-JMY*–, LAP-JMY (W981A)*–, LAP-JMYΔWWWCA–, and LAP-JMY-WWWCA*–expressing plasmids and induced with retinoic acid for 48 h. Data are the mean ± SEM of three experiments with 500 cells quantified for each (**p < 0.01; ***p < 0.001).
To investigate whether JMY is functionally important during neuronal differentiation, we examined the effect of JMY silencing by RNAi on RA-induced differentiation, neurite outgrowth, and neuronal morphogenesis. Neuro 2a cells were transfected either with control GAPDH siRNA or with one of two independent JMY siRNAs and induced to differentiate by RA treatment. Depletion of JMY was confirmed by immunoblotting (Figure 7C). We first investigated the ability of cells to form neurites upon JMY depletion. Surprisingly, we observed a statistically significant, twofold increase in the number of cells bearing neurites when JMY was silenced (Figure 7, D and E). To determine whether the effect on neurite outgrowth was a specific result of JMY silencing, we transfected cells with an siRNA-resistant version of LAP-JMY (LAP-JMY*), which was expressed in siRNA-treated cells as verified by immunoblotting (Figure 7F). Expression of LAP-JMY*, but not LAP alone, rescued the effect on neurite outgrowth, confirming that the previous phenotype is a specific result of JMY silencing (Figure 7, F and G). We also investigated the effect of JMY depletion on neuronal morphogenesis and F-actin content but did not observe a significant effect on a variety of neurite characteristics, including mean neurite length, mean number of neurites per cell, mean number of neurite branchpoints per unit neurite length, or mean F-actin content of neuronal cells (Supplemental Figure S4). Collectively these results indicate that JMY functions as a negative modulator of neurite outgrowth and that this function may be restricted to the early stages of neuritogenesis.
Finally, we assessed whether the inhibitory effect of JMY on neurite formation requires its actin polymerization activity. To this end, we generated an siRNA-resistant, actin-polymerization defective mutant of JMY missing the WWWCA domain (LAP-JMYΔWWWCA) and tested its ability to rescue the phenotype caused by JMY silencing. Depletion of endogenous JMY and ectopic expression of siRNA-resistant LAP-JMYΔWWWCA were confirmed by immunoblotting (Figure 7F). In contrast with LAP-JMY*, expression of LAP-JMYΔWWWCA in JMY-silenced cells did not rescue the neurite outgrowth phenotype (Figure 7F). Expression of siRNA-resistant LAP-JMY-WWWCA* also did not rescue the neurite outgrowth phenotype caused by JMY silencing, suggesting that simply increasing the F-actin content of differentiated Neuro 2a cells is not sufficient for rescue and that other domains of JMY play an important role. To examine the relative contributions of JMY's intrinsic nucleating and Arp2/3 complex–stimulating activities to neurite formation, we next tested the ability of an siRNA-resistant Arp2/3 activation–deficient mutant, LAP-JMY (W981A)*, to rescue the phenotype caused by JMY silencing. Expression of LAP-JMY (W981A)* partially rescued the neurite outgrowth phenotype, suggesting that JMY's function in inhibiting neurite formation requires its ability to act as an actin assembly protein and that both its intrinsic nucleating and Arp2/3 complex–stimulating activities contribute to this process.
DISCUSSION
JMY was recently discovered as a unique actin assembly protein that nucleates actin both as a tandem-monomer-binding nucleator and an NPF (Zuchero et al., 2009) and also functions as a p300 coactivator in the p53 response (Shikama et al., 1999; Coutts et al., 2009). Here we show that full-length JMY is fully active as an actin nucleator and NPF in vitro but that its activity is suppressed in cells compared with its isolated WWWCA domain, suggesting that full-length native JMY is regulated by an inhibitory mechanism. Moreover, we uncover a new function for JMY in neurite outgrowth during neuronal differentiation.
Our results demonstrate that purified recombinant full-length JMY is a potent actin nucleator and Arp2/3–activating NPF and has comparable activity to its WWWCA domain in vitro. Moreover, recombinant JMY purified from stably transfected mammalian cells is as active as JMY purified from insect cells. These data do not provide direct support for regulation of JMY by autoinhibition. Moreover, we did not find support for trans-regulation of JMY by interacting partners based on the observations that JMY overexpressed in mammalian cells does not copurify with any stoichiometric binding partners and does not fractionate with high–molecular weight complexes in brain extracts like the WAVEs (Eden et al., 2002; Gautreau et al., 2004; Suetsugu et al., 2006; unpublished data). Thus our analysis of the activity of recombinant JMY is inconclusive with regard to understanding the mechanism of JMY regulation. This is perhaps not surprising, as the biochemical activity of recombinant versions of other NPFs exhibit variability depending on differences in experimental conditions. Purified recombinant WASP and N-WASP, which are known to be regulated by an autoinhibitory mechanism (Kim et al., 2000; Rohatgi et al., 2000), exhibit variable activity in vitro, ranging from constitutively active to modestly active (Rohatgi et al., 1999; Yarar et al., 1999; Tomasevic et al., 2007). In contrast, native WASP and N-WASP purified from tissue sources are fully autoinhibited (Higgs and Pollard, 2000; Ho et al., 2004). Similarly, recombinant WAVE complex, which is regulated by trans-inhibition, exhibits activity ranging from full to inactive, depending on the expression and purification methods (Innocenti et al., 2004; Suetsugu et al., 2006; Derivery et al., 2009; Ismail et al., 2009; Chen et al., 2010), whereas native WAVE complex is fully inhibited (Eden et al., 2002; Lebensohn and Kirschner, 2009). Thus the activity of NPFs in vitro depends on the source of protein and conditions for purification, with the native protein presumably exhibiting a more natural mode of regulation. Understanding how JMY is regulated may require purification of the native protein from tissue sources to determine whether it is autoinhibited or copurifies with interacting partners that participate in trans-regulation.
In contrast to the comparable actin-polymerizing activity of full-length JMY relative to its WWWCA domain in vitro, cells expressing full-length JMY show no increase in F-actin content compared with the higher F-actin levels in cells expressing JMY-WWWCA. This observation suggests that full-length JMY is inhibited in its ability to polymerize actin relative to the WWWCA domain in cells. Cells expressing full-length N-WASP also do not exhibit an increase in F-actin content, whereas cells expressing full-length WAVE2 show a significant increase (Campellone et al., 2008). By analogy, this suggests that JMY might be regulated by autoinhibition like WASP/N-WASP. In addition to autoinhibition, it is also possible that JMY is regulated by other mechanisms such as posttranslational modification. For example, WASP (Cory et al., 2003; Torres and Rosen, 2003), WAVE1 (Kim et al., 2006), and WAVE2 (Lebensohn and Kirschner, 2009) are regulated by phosphorylation. Further investigation of autoinhibition and posttranslational modification as possible regulatory mechanisms of JMY is required.
A distinguishing feature of JMY among actin nucleators is its dual localization to both the cytosol and nucleus. In contrast to previous studies that identified the nucleus as the primary location of JMY (Coutts et al., 2007; Zuchero et al., 2009), the results of our cell fractionation studies show that ∼80% of JMY is located in the cytosol. Of interest, the fraction of nuclear JMY ranges from ∼10 to 40% in individual cells, suggesting that JMY may shuttle in and out of the nucleus depending on the physiological state of the cell. Although previous studies demonstrated that JMY accumulates in the nucleus upon DNA damage (Coutts et al., 2007, 2009) and relocates from the nucleus to the leading edge in migrating cells (Zuchero et al., 2009), neither of these triggers was activated in the conditions we used for fractionation and localization experiments. We also did not observe redistribution of JMY between the nucleus and cytosol upon RA-induced differentiation of Neuro 2a cells. Clearly, more work is needed to determine the mechanisms that regulate JMY distribution between nuclear and cytosolic compartments.
The cytosolic pool of JMY does not localize to specific cellular structures, other than a small proportion that colocalizes with actin in membrane ruffles. Despite its localization to ruffles, we did not observe a defect in cell migration in JMY-depleted MEF and NIH 3T3 cells, regardless of whether MEF cells expressed WAVE2, an NPF that is important for lamellipodia formation and cell migration (Yamazaki et al., 2003). This observation contrasts with recent studies showing that JMY plays a role in cell migration by activating Arp2/3–mediated actin polymerization (Zuchero et al., 2009) and by down-regulating cadherin expression and stability (Coutts et al., 2009). It is noteworthy that we used MEF and NIH 3T3 cells to examine the function of JMY in wound-healing assays, whereas other groups used U2OS epithelial cells. Thus a role for JMY in migration does not appear to be broadly distributed across all cell types.
In addition to examining its role in fibroblast cell migration, we also tested the function of JMY in neurons because it is highly expressed in brain relative to other tissues. The actin cytoskeleton plays important roles in various stages of neuronal development, including neurite outgrowth and differentiation, axon path finding, and dendritic spine formation and maintenance (Kessels et al., 2010). Although the molecular machinery that regulates actin polymerization in neurons is not fully characterized, different actin nucleators, including the Arp2/3 complex (Korobova and Svitkina, 2008) and its NPFs N-WASP (Banzai et al., 2000; Pinyol et al., 2007; Wegner et al., 2008), WAVE1 (Kim et al., 2006), and WAVE2 (Ito et al., 2010), as well as the tandem actin-monomer-binding nucleator Cordon-bleu (Ahuja et al., 2007), were shown to positively regulate stages of neuronal development through their role in growth cone formation and dynamics. In contrast, we found that JMY is a negative regulator of neurite outgrowth because its depletion in Neuro 2a cells results in a significant increase in the number of cells that form neurites. Our observation that JMY protein levels are significantly down-regulated upon RA-induced differentiation and neurite outgrowth is also consistent with such an inhibitory function. Given that JMY is an actin-nucleating protein, we further investigated whether its function in modulating neurite outgrowth involves its actin polymerization activity and what relative contributions its intrinsic nucleating and Arp2/3-stimulating activities make to this process. Rescue experiments with full-length JMY, a mutant that cannot polymerize actin, and a mutant that cannot activate the Arp2/3 complex, show that negative regulation of neurite outgrowth requires JMY's actin-nucleating activity and that both intrinsic and Arp2/3 complex–dependent activities participate. Rescue experiments with the WWWCA domain of JMY further reveal that the ability of this constitutively active domain to cause a global increase in cellular F-actin is not sufficient for JMY's function during neurite outgrowth, and that the upstream N-terminal and coiled-coil domains are also required, likely for enabling proper cellular localization and regulation. One key challenge in the future will be to understand the molecular mechanism of JMY's inhibitory role in neurite outgrowth through its effect on actin polymerization.
One possibility is that JMY might function as a downstream effector of Rac, Rho, or Cdc42, which have been shown to regulate cytoskeletal dynamics in neurons (Govek et al., 2005). Rac and Cdc42 function in lamellipodia and filopodia formation in the growth cone (Kozma et al., 1997) and promote neurite formation (Sarner et al., 2000; Aoki et al., 2004). On the other hand, activation of Rho inhibits neurite formation and induces retraction by antagonizing Rac activity and promoting the polymerization of a thick layer of cortical actin filaments that may interfere with the cytoskeletal remodeling needed for neurite formation (Yamaguchi et al., 2001). Consistent with this, toxin-mediated inactivation of Rho (Jalink et al., 1994) or expression of dominant-negative Rho (Sebok et al., 1999) induced neurite formation in rat adrenal medulla PC12 and mouse neuroblastoma N1E-115 cells. Given the similar effects of JMY and Rho inhibition on neurite formation, it seems reasonable to speculate that JMY might be an effector protein of Rho that functions in the assembly of the cortical actin cytoskeleton in neurons. A balance between antagonistic activities of different Rho GTPases is suggested to regulate neuritogenesis (Leeuwen et al., 1997), and JMY might participate in establishing this balance.
A second mechanism by which JMY may modulate neurite outgrowth is via a role as a transcriptional regulator. Retinoic acid serves as a master regulator of gene expression during neuronal differentiation by binding to nuclear RA receptors and modulating expression of genes that participate in neuronal differentiation (Clagett-Dame et al., 2006). Because JMY functions in transcriptional regulation by binding to p300 (Shikama et al., 1999), a coactivator for diverse transcription factors, it might exert its inhibitory function by modulating RA-mediated gene expression. Future work will distinguish between the roles of JMY as a transcription cofactor or actin assembly protein in neurons.
Although our study sheds light on the cellular regulation and function of JMY, numerous questions remain. One concerns the nature of JMY regulation in cells. Actin-nucleating proteins are regulated by small GTPases, protein–protein interactions, and/or posttranslational modifications, and it is likely that JMY is also regulated by similar mechanisms. A second question concerns why JMY combines two very different cellular functions—transcriptional regulation and actin assembly—in one protein. Given the emerging role for nuclear actin in transcription regulation (Skarp and Vartiainen, 2010), it is tempting to speculate that JMY acts as a molecular link between the actin cytoskeleton and the transcription machinery. It will be important to examine transcription- and actin-related functions of JMY in cellular processes, including neuritogenesis, to understand whether they work together or independently. Answering these questions will contribute to our understanding of the cellular function of JMY and the general mechanisms that regulate actin dynamics in the cytosol and nucleus.
MATERIALS AND METHODS
Plasmids
Plasmids are listed in Supplemental Table S1. The JMY coding sequence was amplified by PCR from full-length mouse JMY cDNA (accession BC090835; Open Biosystems, Thermo Biosystems, Huntsville, AL) using primers listed in Supplemental Table S2. The JMY-W981A point mutation and four silent point mutations in the RNAi-resistant LAP-JMY*, LAP-JMY (W981A)*, and LAP-JMY-WWWCA* were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) in accordance with the manufacturer's instructions using primers listed in Supplemental Table S2. The DNA sequence of each construct was verified.
Protein expression and purification
His-JMY, His-JMY (W981A), MBP-JMY, MBP-JMY-WWWCA, and rArp2/3 complex were expressed and purified using the baculovirus expression system. Sf9 and Hi5 insect cells were cultured at 28°C in Grace's medium supplemented with 2% fetal bovine serum (FBS; JR Scientific, Woodland, CA), penicillin, and streptomycin. Bacmids and baculoviruses were generated in E. coli and Sf9 cells using the Bac-to-Bac system according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Recombinant proteins were then expressed by infecting Hi5 cells for 72 h at 28°C with the appropriate baculovirus strains. Hi5 cells were pelleted, lysed by freeze–thaw in nickel-nitriloacetic acid (Ni-NTA) lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and protease inhibitors [10 μg/ml each of aprotinin, leupeptin, pepstatin, and 1 mM phenylmethylsulfonyl fluoride]) for the purification of His-JMY and His-JMY (W981A) or MBP-binding lysis buffer (20 mM Tris, pH 7.6, 250 mM NaCl, 100 mM KCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, 5% glycerol, and protease inhibitors) for the purification of MBP-JMY and MBP-JMY-WWWCA and clarified by centrifugation. His-JMY and His-JMY (W981A) were purified from the resulting supernatant by Ni-NTA chromatography (Qiagen, Valencia, CA), followed by anion-exchange chromatography on a HiTrap QP column (GE Healthcare, Piscataway, NJ) and finally gel filtration chromatography on a Superdex 200 column (GE Healthcare) into gel filtration buffer (20 mM 3-(N-morpholino)propanesulfonic acid, pH 7.0, 100 mM KCl, 2 mM MgCl2, 5 mM ethylene glycol tetraacetic acid [EGTA], 1 mM EDTA, 0.5 mM DTT, 0.2 mM ATP, 10% glycerol, and protease inhibitors). MBP-JMY and MBP-JMY-WWWCA were purified from the resulting supernatant by amylose affinity chromatography (New England BioLabs, Ipswich, MA), followed by gel filtration chromatography into gel filtration buffer on a Superdex 200 and Superdex 75 column, respectively. rArp2/3 complex was purified as described previously (Goley et al., 2004).
GST-JMY-WWWCA, GST-JMY-WWWCA (W981A), and GST-N-WASP-WWCA were expressed in E. coli BL21-Rosetta cells grown to OD600 = 0.6 and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 4 h at 37°C. Following expression, bacteria were pelleted and lysed by freeze–thaw and sonication with 1 mg/ml lysozyme in glutathione S-transferase (GST)–binding lysis buffer (1× phosphate-buffered saline [PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.46 mM KH2PO4], pH 7.4, 250 mM KCl, and protease inhibitors). The proteins were purified by affinity chromatography on glutathione–Sepharose (GE Healthcare), followed by gel filtration chromatography on a Superdex 75 column (GE Healthcare) into gel filtration buffer.
For affinity purification of recombinant JMY from mammalian cells, JMY was tagged with an N-terminal LAP tag (LAP-JMY) consisting of GFP, a TEV protease cleavage site, and the S peptide portion of RNAse (Cheeseman and Desai, 2005). The 293 cells were transfected with LAP-JMY, and stable cell lines that express LAP-JMY were selected in 1 mg/ml G418. LAP-based purification was performed as described previously (Cheeseman and Desai, 2005) with the following modifications. Briefly, cells were lysed in LAP lysis buffer (75 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 300 mM KCl, 15% glycerol, 0.4% NP-40, and protease inhibitors), and LAP-JMY was first isolated from the cell extracts using protein A–Sepharose beads (GE Healthcare) coated with anti-GFP antibodies. Bound protein was eluted by cleaving with 0.06 mg/ml TEV protease in TEV cleavage buffer (50 mM HEPES, pH 7.5, 1 mM EGTA, 1 mM MgCl2, 300 mM KCl, 10% glycerol, 0.05% NP-40, and 1 mM DTT). The supernatant after TEV cleavage contained S-JMY along with other binding proteins.
Antibody generation and immunoblotting
Polyclonal anti-JMY and anti-GFP antibodies were generated by immunizing rabbits (Covance, Berkeley, CA) with full-length His-JMY and His-GFP, respectively. Antibodies were affinity-purified using standard procedures (Harlow and Lane, 1988). Anti-JMY was used for immunoblotting at 1:4000. Other primary antibodies used for immunoblotting were anti-JMY (M-300; 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), anti–tubulin E7 (1:5000, developed by M. Klymkowsky at the University of Colorado, Boulder, CO, and maintained by the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA), and anti-NonO (1:4000; BD Biosciences, San Diego, CA). Horseradish peroxidase–conjugated secondary antibodies were from GE Healthcare. For immunoblotting, extracts were resolved by SDS–PAGE and transferred onto nitrocellulose membranes that were then probed with primary and secondary antibodies and visualized by chemiluminescence (GE Healthcare). For determining tissue-specific expression of JMY, a mouse tissue blot (Imgenex, San Diego, CA) was used.
Arp2/3–complex activity assays
Rabbit skeletal muscle actin and pyrene-labeled actin were prepared and pyrene actin polymerization assays were performed essentially as described previously (Goley et al., 2004). Pyrene actin and unlabeled actin were mixed in G-buffer (5 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, and 0.2 mM DTT) to generate a 3 μM G-actin solution with 7% pyrene–actin. Fourteen μl of reaction mixture containing proteins to be tested in the assay was mixed with 6 μl of 10× initiation buffer (20 mM MgCl2, 10 mM EGTA, and 5 mM ATP), and this mixture was added to 40 μl of G-actin mix to start polymerization. Fluorescence was detected at 20-s intervals at 365-nm excitation and 407-nm emission wavelengths using a microplate reader (Infinite F200 Pro; Tecan, Männedorf, Switzerland) for experiments with MBP-JMY and MBP-JMY-WWWCA or a spectrofluorimeter (Fluorolog-3; Horiba Jobin Yvon, Edison, NJ) for all other experiments. Magellan, version 7.0, software for the microplate reader and DataMax, version 2.2.12B, software for the spectrofluorimeter were used for data collection. Polymerization reactions were allowed to reach steady state, and curves were normalized for differences in steady-state fluorescence.
For actin branching experiments, 3 μM actin mix was polymerized under the same conditions as those described for the pyrene–actin polymerization assays. Samples were collected as soon as the polymerization reactions reached steady state and immediately stabilized with rhodamine–-phalloidin (Molecular Probes, Invitrogen). Stabilized filaments were diluted, applied to poly-l-lysine–coated coverslips (Sigma-Aldrich, St. Louis, MO), and imaged using an BX51 microscope (Olympus, Center Valley, PA) equipped with a 60× objective and an Orca-ER camera (Hamamatsu, Hamamatsu, Japan), using μManager software (Edelstein et al., 2010). For each condition, branched and unbranched filaments in 10 different images were manually counted using ImageJ software (National Institutes of Health, Bethesda, MD) in three separate experiments and the percentage branching was calculated. A filament with more than one branch was counted as one. Image analysis and contrast adjustment were done using Photoshop (Adobe, San Jose, CA).
Bead motility assays
For bead motility experiments, polystyrene microspheres (0.5 μm, nonfunctionalized; Polysciences, Warrington, PA) were incubated on ice with 1 μM GST-JMY-WWWCA or GST-JMY-WWWCA (W981A) for 1 h and then blocked with 5 mg/ml bovine serum albumin (BSA) for 15 min. Beads were washed and resuspended in CSF-XB (10 mM HEPES, pH 7.7, 2 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, 5 mM EGTA, and 50 mM sucrose) and kept at 4°C. Beads (1 μl) were incubated with 1 μl of actin (2 μM, 20% rhodamine labeled) and 8 μl of X. laevis egg extract (Maresca and Heald, 2006), and 2 μl of this reaction was squashed between a microscope slide and 22-mm-square glass coverslips, sealed, and observed after 15–30 min using fluorescence microscopy as described later. To determine rates of movement, fluorescence images were recorded every 10 s, and rates of movement were determined by averaging the distance in a 1- to 3-min time interval using ImageJ software.
Cell fractionation
For nuclear and cytosolic fractionation, cells were washed with PBS and lysed in cytosolic lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, and protease inhibitors) for 15 min on ice. The homogenate was then centrifuged at 3000 × g for 3 min to sediment nuclei, and the pellet was used to make the nuclear fraction. The supernatant was resedimented at 3000 × g for 5 min, and the resulting supernatant was the cytosolic fraction. The nuclear pellet was washed with cytosolic lysis buffer, resuspended in nuclear lysis buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 10% glycerol, and protease inhibitors), sonicated on ice, and sedimented at 15,000 × g for 5 min, and the resulting supernatant was the nuclear fraction. Nuclear and cytosolic extract volumes were equalized before taking samples for SDS–PAGE to ensure that equal amounts of each fraction were loaded on the gel. Equal volumes of each fraction were resolved by SDS–PAGE and blotted with anti-JMY antibodies, and the intensity of JMY bands in each fraction was determined by densitometry using Photoshop. Relative JMY levels in the nuclear and cytoplasmic fractions were calculated after determining the ratio of nuclear and cytosolic JMY band intensities.
Mammalian cell culture, transfection, and RNA interference
Cos7, U2OS, 293, HFF, NIH 3T3, WAVE2+/+, and WAVE2−/− MEFs (Yan et al., 2003) and SY5Y and Neuro 2a cells were cultured at 37°C in 5% CO2 in DMEM (Invitrogen) supplemented with 10% FBS (JR Scientific), penicillin, and streptomycin. PC12 cells were cultured at 37°C in 5% CO2 in RPMI 1640 (Invitrogen) supplemented with 10% FBS, penicillin, and streptomycin. For localization of recombinant tagged proteins, mammalian cells were transfected with the indicated DNA constructs using Lipofectamine LTX (Invitrogen) for Neuro 2a cells and Lipofectamine 2000 (Invitrogen) for other cell lines, in accordance with the manufacturer's instructions. At 24 h posttransfection, cells were seeded onto glass coverslips for subsequent immunofluorescence experiments. To silence JMY expression by RNAi, Neuro 2a cells were transfected independently with 20 nM each of two different siRNAs (JMY siRNA#1:sc-35725, Santa Cruz Biotechnology, and JMY siRNA#2, ID 75606, Ambion, Invitrogen) using RNAiMAX (Invitrogen). For rescue experiments, Neuro2a cells were transfected with the expression plasmids 24 h posttransfection with the siRNAs. For differentiation experiments, Neuro 2a cells at 24 h posttransfection were seeded onto glass coverslips coated with 1 mg/ml poly-d-lysine (Sigma-Aldrich) at a density of 104 cells/cm2.
For wound-healing experiments, WAVE2+/+, WAVE2−/−, and NIH 3T3 cells were plated on glass-bottom plates (MatTek, Ashland, MA) that were marked to guide the localization of wounds. Cells were transfected with 20 nM control GAPDH siRNA or JMY siRNA#1 once for MEF cells on day 1 and twice for NIH 3T3 cells on days 1 and 2 and were grown to confluency over 72 h. One day before wounding, cells were serum starved in DMEM with 0.5% FBS for 16 h. After serum starvation, three parallel wounds were made by scratching the cell monolayer with a 27-G syringe. Cells were washed with PBS to remove detached cells and replaced with fresh complete medium. Phase contrast images of eight different wounds per sample were captured using an Olympus IX71 microscope with a 20× (0.75 numerical aperture [NA]) PlanApo phase objective every 1.5 h over a period of 4.5 h for MEF cells and every 3 h over a period of 12 h for NIH 3T3 cells (image acquisition described later). The wound area at each time point was determined using ImageJ (Abramoff et al., 2004), and the percentage of wound closure was defined as the difference between the wound area at 0 h (defined as 100) and the remaining wound area at 4.5 h.
Immunofluorescence microscopy, image acquisition, and microscopic quantification
For immunofluorescence experiments, cells were washed with PBS, fixed in PBS with 2.5% paraformaldehyde for 30 min at 37°C, and permeabilized with 0.1% Triton X-100 for 5 min. They were then blocked with PBS with 1% BSA and 1% FBS and treated with 4 U/ml Alexa Fluor 488–phalloidin or Alexa Fluor 568–phalloidin (Invitrogen) in PBS with 1% BSA. Coverslips were mounted on glass slides using ProLong Gold antifade reagent (Invitrogen).
Images were acquired using 20× (0.75 NA), 60× (1.40 NA), or 100× (1.35 NA) PlanApo objective lenses on an Olympus IX71 microscope equipped with a Coolsnap HQ camera (Photometrics, Tucson, CA). Images were captured using MetaMorph (Universal Imaging, Molecular Devices, Sunnyvale, CA) or μManager (Edelstein et al., 2010) software and converted to 8-bit tiff files, and brightness/contrast levels were adjusted using Photoshop. Quantification was done in a blinded manner by scoring cells that were chosen randomly. Statistical significance and p values were assessed by analysis of variance and Student's t tests using Prism software (GraphPad Software, La Jolla, CA).
For quantification of total GFP-JMY levels in the cytosol and nucleus of cells expressing GFP-JMY, images of transfected cells were acquired as described earlier. Nuclear and cytosolic boundaries were outlined, and the GFP-JMY fluorescence intensity in each area was measured using Photoshop. Relative GFP-JMY levels in the nucleus and cytosol were calculated by dividing the fluorescence intensity of each area by the total fluorescence intensity of the cell.
For quantification of relative F-actin levels in cells expressing GFP-JMY and GFP-JMY-WWWCA, images of transfected and nontransfected cells in the same field were acquired as described earlier. Cell boundaries were outlined, and the mean intensity of Alexa Fluor 568–phalloidin fluorescence was measured using Photoshop. F-Actin fluorescence intensities from transfected cells were then normalized relative to that of nearby nontransfected cells that had a similar overall area. For images with more than one nontransfected cell, the average fluorescence intensity of these cells was used as the reference intensity for normalization. For correlation and linear regression analysis between GFP and actin fluorescence intensities in transfected cells, images were acquired using identical illumination conditions, mean background intensities were subtracted from the GFP and Alexa Fluor 568–phalloidin fluorescence intensities, and actin fluorescence values were then plotted as a function of GFP values in arbitrary units. For quantification of mean F-actin intensities in differentiated Neuro 2a cells that were transfected with control GAPDH siRNA or one of the two independent JMY siRNAs, images were acquired using identical illumination conditions, then mean background intensities were subtracted from mean Alexa Fluor 568–phalloidin fluorescence intensities, and average phalloidin intensities were plotted.
Neurite initiation and morphological quantification
At 24 h posttransfection with plasmids expressing JMY or siRNA to silence its expression, Neuro 2a cells were induced to differentiate with 20 μM RA (Sigma-Aldrich) in DMEM with 2% FBS for 48 and 72 h. The cells were quantified for the following parameters: percentage of cells with neurites, defined as an outgrowth with a length more than twice the diameter of the cell body; average neurite length, defined as the distance from cell body to the distal tip, measured using the NeuronJ plugin (Meijering et al., 2004) for ImageJ (Abramoff et al., 2004); average number of neurites per cell; and mean number of neurite branchpoints per unit neurite length, for which branchpoints are defined as secondary neurites of any length arising from the primary neurite.
Supplementary Material
Acknowledgments
We thank Ken Campellone, Steve Duleh, and Taro Ohkawa for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health/National Institute of General Medical Sciences (GM059609) and the University of California Cancer Research Coordinating Committee to M.D.W. and a predoctoral fellowship from the American Heart Association (0715035Y) to E.N.F.
Abbreviations used:
- FBS
fetal bovine serum
- JMY
junction-mediating and regulatory protein
- NPF
nucleation-promoting factor
- RA
retinoic acid
- WWWCA
WASP homology 2 domain, connector, and acidic region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0585) on September 30, 2011.
REFERENCES
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with Image. J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Nakamura T, Matsuda M. Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J Biol Chem. 2004;279:713–719. doi: 10.1074/jbc.M306382200. [DOI] [PubMed] [Google Scholar]
- Banzai Y, Miki H, Yamaguchi H, Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in neurite extension in PC12 cells and rat hippocampal primary culture cells. J Biol Chem. 2000;275:11987–11992. doi: 10.1074/jbc.275.16.11987. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clagett-Dame M, McNeill EM, Muley PD. Role of all-trans retinoic acid in neurite outgrowth and axonal elongation. J Neurobiol. 2006;66:739–756. doi: 10.1002/neu.20241. [DOI] [PubMed] [Google Scholar]
- Cory GO, Cramer R, Blanchoin L, Ridley AJ. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol Cell. 2003;11:1229–1239. doi: 10.1016/s1097-2765(03)00172-2. [DOI] [PubMed] [Google Scholar]
- Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2007;8:84–90. doi: 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci USA. 2009;106:19872–19877. doi: 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009;66:777–790. doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2010;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Antibodies: A Laboratory Manual. [Google Scholar]
- Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Morishita R, Shinoda T, Iwamoto I, Sudo K, Okamoto K, Nagata K. Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol Psychiatry. 2010;15:976–986. doi: 10.1038/mp.2010.69. [DOI] [PubMed] [Google Scholar]
- Jalink K, van Corven EJ, Hengeveld T, Morii N, Narumiya S, Moolenaar WH. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J Cell Biol. 1994;126:801–810. doi: 10.1083/jcb.126.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Schwintzer L, Schlobinski D, Qualmann B. Controlling actin cytoskeletal organization and dynamics during neuronal morphogenesis. Eur J Cell Biol. 2010;90:926–933. doi: 10.1016/j.ejcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Kim Y, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L. Actin in membrane trafficking. Curr Opin Cell Biol. 2007;19:453–458. doi: 10.1016/j.ceb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwen FN, Kain HE, Kammen RA, Michiels F, Kranenburg OW, Collard JG. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol. 1997;139:797–807. doi: 10.1083/jcb.139.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling M, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Sarner S, Kozma R, Ahmed S, Lim L. Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells. Mol Cell Biol. 2000;20:158–172. doi: 10.1128/mcb.20.1.158-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebok A, Nusser N, Debreceni B, Guo Z, Santos MF, Szeberenyi J, Tigyi G. Different roles for RhoA during neurite initiation, elongation, and regeneration in PC12 cells. J Neurochem. 1999;73:949–960. doi: 10.1046/j.1471-4159.1999.0730949.x. [DOI] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- Skarp KP, Vartiainen MK. Actin on DNA—an ancient and dynamic relationship. Cytoskeleton. 2010;67:487–495. doi: 10.1002/cm.20464. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Sun SC, Sun QY, Kim NH. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Mol Hum Reprod. 2011;17:296–304. doi: 10.1093/molehr/gar006. [DOI] [PubMed] [Google Scholar]
- Tomasevic N, Jia Z, Russell A, Fujii T, Hartman JJ, Clancy S, Wang M, Beraud C, Wood KW, Sakowicz R. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry. 2007;46:3494–3502. doi: 10.1021/bi062152y. [DOI] [PubMed] [Google Scholar]
- Torres E, Rosen MK. Contingent phosphorylation/dephosphorylation provides a mechanism of molecular memory in WASP. Mol Cell. 2003;11:1215–1227. doi: 10.1016/s1097-2765(03)00139-4. [DOI] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Fang Y, Lu ZH, Ledeen RW. Induction of axon-like and dendrite-like processes in neuroblastoma cells. J Neurocytol. 1998;27:1–14. doi: 10.1023/a:1006910001869. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Katoh H, Yasui H, Mori K, Negishi M. RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J Biol Chem. 2001;276:18977–18983. doi: 10.1074/jbc.M100254200. [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yan C, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, D'Alessio JA, Jeng RL, Welch MD. Motility determinants in WASP family proteins. Mol Biol Cell. 2002;13:4045–4059. doi: 10.1091/mbc.E02-05-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.