The fusion of biological membranes entails a drastic rearrangement of the lipid bilayer. New assays that distinguish fusion from lysis were developed to study an in vitro reconstitution of the yeast vacuolar fusion machinery. These assays revealed that true fusion is accompanied by strongly enhanced membrane permeability to small molecules and by lysis.
Abstract
The fusion of sealed biological membranes joins their enclosed aqueous compartments while mixing their membrane bilayers. Reconstituted fusion reactions are commonly assayed by lipid mixing, which can result from either true fusion or from lysis and its attendant reannealing of membranes. Fusion is also frequently assayed by the mixing of lumenal aqueous compartments, using probes of low molecular weight. With several probes (biotin, methylumbelliferyl-N-acetyl-α-d-neuraminic acid, and dithionite), we find that yeast vacuolar SNAREs (SNAP [Soluble NSF attachment protein] Receptors) increase the permeability of membranes to small molecules and that this permeabilization is enhanced by homotypic fusion and vacuole protein sorting complex (HOPS) and Sec17p/Sec18p, the vacuolar tethering and SNARE chaperone proteins. We now report the development of a novel assay that allows the parallel assessment of lipid mixing, the mixing of intact lumenal compartments, any lysis that occurs, and the membrane permeation of small molecules. Applying this assay to an all-purified reconstituted system consisting of vacuolar lipids, the four vacuolar SNAREs, the SNARE disassembly chaperones Sec17p and Sec18p, the Rab Ypt7p, and the Rab effector/SM protein complex HOPS, we show that true fusion is accompanied by strongly enhanced membrane permeability to small molecules and a measurable rate of lysis.
INTRODUCTION
Subcellular compartmentation is maintained on the exocytic and endocytic pathways by vesicle budding, targeted docking, and fusion. Fusion is constitutive or can be tightly regulated, as for synaptic neurotransmission or insulin secretion. The first step of intracellular membrane fusion is tethering, mediated by Rab/Ypt-family GTPases and by “effector” tethering proteins, which selectively bind to an individual Rab in its GTP-bound form. SNAP (Soluble NSF attachment protein) Receptor (SNARE) proteins of the two tethered membranes then form 4-helical trans-SNARE complexes, in physical and functional association with proteins of the Sec1/Munc18 family and others. Fusion joins the two membrane bilayers into one and mixes their lumenal contents. Dramatic structural rearrangements of the bilayers of apposed membranes are required, yet fusion usually occurs without lysis, which would release lumenal contents into the cytoplasm. Lysis can, however, occur as a side reaction of fusion (Engel and Walter, 2008). Purified and reconstituted neuronal SNAREs will mediate lipid mixing (Weber et al., 1998), reflecting both true fusion (Nickel et al., 1999) and lysis/reannealing (Dennison et al., 2006). v-SNARE proteoliposomes bearing a soluble fluorophore that had docked with t-SNARE–supported bilayer sheets released their lumenal content on the cis- side of the membrane, indicative of lysis rather than true fusion (Wang et al., 2009). These studies have raised the question of how biological membrane fusion occurs without lysis.
We study these processes through the homotypic fusion of yeast vacuoles (Wickner, 2010). Purified vacuoles fuse in a multistep pathway. On incubation with ATP, the vacuolar SNARE complexes are disassembled by Sec17p and Sec18p (Mayer et al., 1996). Vacuoles are tethered by the homotypic fusion and vacuole protein sorting (HOPS) complex (Stroupe et al., 2006; Hickey and Wickner, 2010), a hexameric complex with direct affinity for the vacuolar Rab Ypt7p, for SNAREs, and for phosphoinositides (Stroupe et al., 2006). Tethered vacuoles are drawn together, then the proteins and lipids that are required for fusion form a highly enriched microdomain, the “vertex ring” (Wang et al., 2002), around the tethered and apposed membranes of each vacuole, and the SNAREs pair in trans. Vertex ring enrichment and the ensuing fusion require Ypt7p, the HOPS complex, the four vacuolar SNAREs, the SNARE-disassembly chaperones, Sec17p and Sec18p, and essential vacuolar lipids such as ergosterol, phosphoinositides, phosphatidylethanolamine, and diacylglycerol (Kato and Wickner, 2001; Wang et al., 2003; Fratti et al., 2004; Jun et al., 2004; Mima and Wickner, 2009). Biochemical assays of lumenal compartment mixing and lysis have shown that the proteins that catalyze efficient organelle fusion trigger a substantial release of lumenal calcium (Peters and Mayer, 1998; Merz and Wickner, 2004) but only little lysis (Starai et al., 2007). Up to two-thirds of vacuoles from cells with concomitant overexpression of all four SNAREs undergo lysis during fusion, however (Starai et al., 2007). Vacuole lysis may only be an infrequent side reaction of fusion in wild-type cells, yet the yeast cytosol contains specific and potent peptide inhibitors of vacuolar proteases (Maier et al., 1979), suggesting that lysis might occur at some frequency.
The proteins and lipids that mediate vacuole fusion have been purified and reconstituted into proteoliposomes (Mima et al., 2008; Stroupe et al., 2009). On incubation with HOPS, Sec17p, Sec18p, and ATP, there is rapid and efficient mixing of the lipids between these proteoliposomes, a standard assay of fusion (Struck et al., 1981). We have now studied the topology of this reaction in greater depth, with new assays for lumenal compartment mixing, lipid mixing, and lysis, and find that fusion is accompanied by substantial enhancement of the permeability to small molecules and that lysis occurs at a measurable frequency.
RESULTS
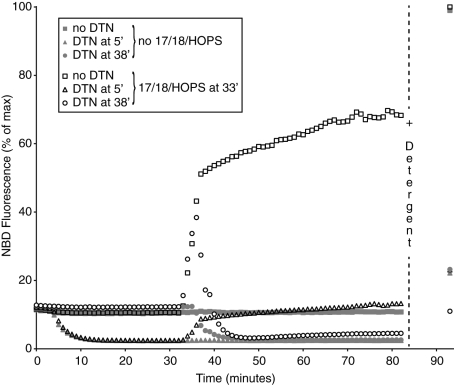
HOPS, Sec17p, and Sec18p cause lipid mixing among proteoliposomes bearing the Rab Ypt7p and the four vacuolar SNAREs (Mima et al., 2008; Stroupe et al., 2009). Proteoliposomes with N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE) and rhodamine-PE, the four vacuolar SNAREs, and Ypt7 were incubated with similar proteoliposomes without fluorophore, and HOPS, Sec17p, and Sec18p were added after 33 min. This addition triggered lipid mixing, causing dequenching of the NBD fluorescence (Figure 1, open squares) to more than half the full dequenching seen when detergent was added at the end of the incubation. If dithionite (DTN) was added at 5 min (triangles), 75% of the NBD-PE was reduced to its nonfluorescent form, as shown after detergent addition or by the greatly reduced dequenching upon Sec17p/Sec18p/HOPS addition at 33 min (open triangles). The percentage of NBD-PE that was accessible to DTN was far more than the percentage expected to be on the outside of sealed and impermeant proteoliposomes. Most strikingly, when the first addition to the proteoliposomes was Sec17p/Sec18p/HOPS at 33 min, and the rapid dequenching had begun, DTN added at 38 min gained access to virtually all the NBD-PE, as judged by the almost complete loss of fluorescence (open circles), even after complete dequenching due to detergent addition at 85 min. This finding also indicates that there was ongoing lysis and/or enhanced membrane permeability to DTN. Specific assays were devised to explore these possibilities.
FIGURE 1:
DTN arrests lipid dequenching when SNARE-bearing liposomes are incubated with Sec17p/Sec18p/HOPS. 4-SNARE RPLs composed of VLM (50 μM donor and 400 μM acceptor) were mixed with ATP/Mg2+, aliquoted into wells, and placed at 27°C. One set of wells received RB150, a second set received 7.5 mM DTN at an early time point (after a 4- to 5-min preincubation at 27°C), and a third set received 7.5 mM DTN at a late time point (3 min after the addition of chaperones or their buffers). Thirty minutes after the first DTN addition (by which time DTN has decayed), Sec17p/Sec18p/HOPS (open symbols) or their respective buffers (filled symbols) were added, and incubations continued for 45 min. Reactions were stopped by the addition of Thesit to 0.1% (wt/vol). Results are expressed as percent of NBD signal remaining after treatment relative to maximal values (i.e., detergent values for reactions with no DTN treatment). Each experiment was carried out in duplicate a minimum of three times and included liposomes prepared on different days. In all cases DTN was added to wells within 1 min of thawing on ice.
Permeabilization and lysis by proteins that catalyze fusion
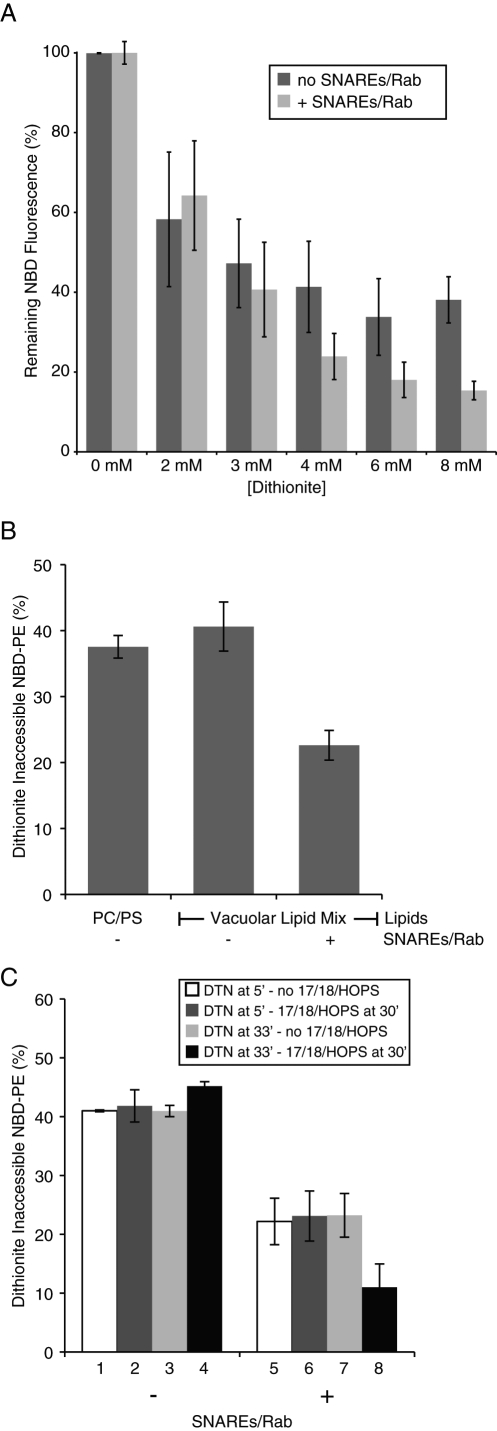
To ensure that these assays used adequate DTN concentrations, vacuolar lipid mix (VLM) liposomes or VLM proteoliposomes bearing Ypt7p and all four SNAREs were incubated with increasing amounts of DTN for 45 min at 27°C, then diluted with detergent to assay the full amount of remaining NBD-PE. Although all the NBD-PE was reduced to its nonfluorescent derivative by concentrations of DTN over 2 mM when the membranes were first dissolved in detergent (Mima et al., 2008; unpublished data), ∼40% of the liposomal NBD-PE remained inaccessible to even high levels of DTN (Figure 2A, dark columns). In contrast, ∼20% of the NBD-PE remained inaccessible to DTN in proteoliposomes bearing the SNAREs and Rab (Figure 2A, light gray columns, and Figure 2B). To compare the roles of SNARE and Rab with the roles of Sec17p/Sec18p/HOPS in creating DTN access to NBD-PE, an experiment was performed as shown in Figure 1, but it used proteoliposomes either with or without SNAREs/Rab. After 85 min, when any added DTN had been oxidatively inactivated, detergent was added, and the remaining fluorescent NBD-PE was measured and expressed as a percentage of that seen in samples that never received DTN. For protein-free liposomes, the addition of Sec17p/Sec18p/HOPS did not increase the access of DTN to NBD-PE (Figure 2C, columns 1–4), whereas SNAREs and Ypt7p enhanced DTN access (compare columns 5–7 with 1–3), which in this case was increased further by the addition of Sec17p/Sec18p/HOPS (Figure 2C, column 8). The presence of all three of these proteins (Sec17p/Sec18p/HOPS) was required for this further increase (unpublished data).
FIGURE 2:
SNAREs and SNARE chaperones enhance DTN access to proteoliposomal lipids. (A) DTN reduces accessible NBD. Vacuole lipid mixture liposomes or 4-SNARE-RPLs (50 μM donors and 400 μM acceptors) were mixed with ATP/Mg2+, aliquoted into wells, and placed at 27°C for 5 min. The indicated concentrations of DTN were added (immediately after DTN thawing), and the reactions were incubated for 45 min. Thesit (0.1% [wt/vol]) was then added to establish the amount of total or remaining NBD fluorescence. Dark columns, protein-free liposomes; light columns, RPLs bearing Rab/SNAREs. Each experiment was conducted a minimum of three times, and two replicates of each condition were done per experiment, using liposomes prepared on different days. (B) The presence of Rab/SNAREs affects permeation of DTN. For each set of RPLs, 50 μM donors and 400 μM acceptors were mixed with ATP/Mg2+, aliquoted into wells, and placed at 27°C for 5 min. Freshly thawed DTN was added to 7.5 mM, and the reactions were further incubated for 45 min. Thesit (0.1% [wt/vol]) was added to end all reactions. Results show the amount of NBD fluorescence that is inaccessible to DTN relative to maximal values for each set (i.e., detergent values of reactions that were incubated without DTN). Experiments were performed in duplicate, a minimum of three times, using liposomes made on different days. (C) Chaperones further enhance lumenal access to DTN in the presence of Rab/SNAREs. Experiments were conducted as in Figure 1, except that liposomes did or did not bear Rab/SNAREs as indicated. Results are presented as the % of NBD fluorescence remaining after treatment with DTN (early or late) in the presence of Sec17p/Sec18p/HOPS or their buffers. White columns correspond to early DTN treatment followed by incubation with buffers; dark gray columns to early DTN treatment followed by incubation with Sec17p/Sec18p/HOPS; light gray columns to late DTN treatment following addition of buffers; and black columns to late DTN treatment following addition of Sec17p/Sec18p/HOPS. In each case, detergent was added after 85 min, when the DTN had become fully inactive, and the value presented is the remaining NBD fluorescence, expressed as percentage of the value from samples never exposed to DTN. Each experiment was carried out in duplicate a minimum of three times and included liposomes prepared on different days (except for protein-free liposomes for which the same set was used throughout).
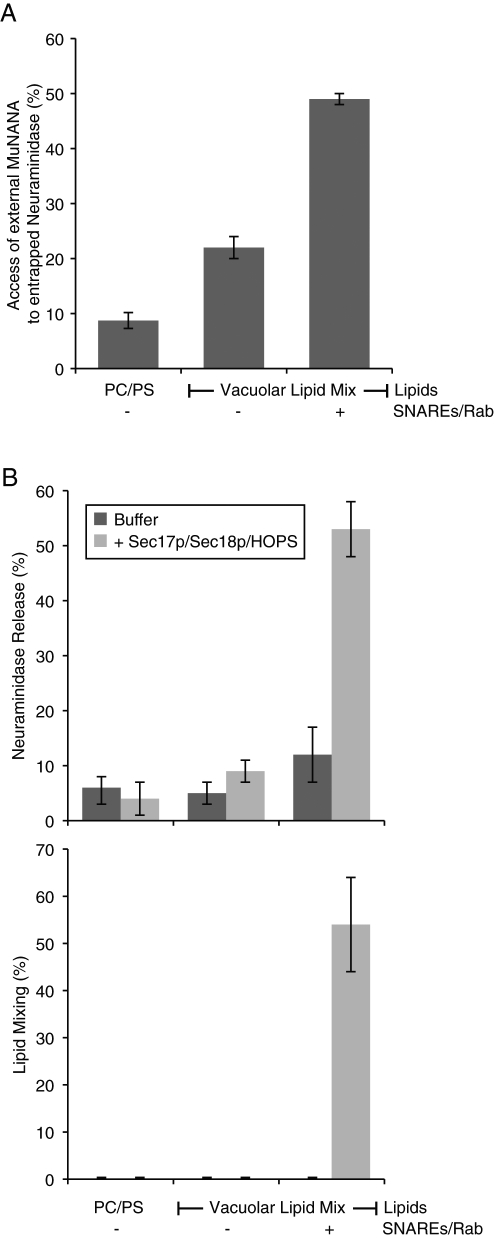
Is this enhanced access of external probe specific to DTN? We prepared proteoliposomes of phosphatidylcholine/phosphatidylserine (PC/PS) or of vacuolar lipid composition without SNAREs/Ypt7p or with these proteins (Figure 3A) with entrapped neuraminidase (a soluble protein of 36 kDa). These were used to compare the hydrolysis of external 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid (MuNANA), in the presence or absence of detergent, to determine the access of MuNANA to lumenal neuraminidase across an intact membrane. Although vacuolar lipids allowed a greater access than PC/PS, there was a strong further enhancement by SNAREs and Rab (Figure 3A), even though there was no Sec17p, Sec18p, or HOPS, and hence no fusion (Mima et al., 2008; Stroupe et al., 2009). These proteoliposomes were assayed for neuraminidase release from sedimentable reconstituted proteoliposomes (RPLs) during incubation in the absence, or presence, of Sec17p/Sec18p/HOPS. Although SNAREs alone led to some lysis (Figure 3B, top panel, dark columns), lysis was dramatically enhanced by Sec17p/Sec18p/HOPS in the presence of SNAREs (gray columns), the very condition that supports lipid mixing (Figure 3B, bottom panel). These assays, however, only measure the yield of lysis, including lysis events that happen after one or more rounds of fusion, and do not permit a direct comparison of the initial rate of lysis to the initial rate of any true fusion.
FIGURE 3:
Direct assay of lysis and membrane permeability. (A) Access of encapsulated neuraminidase to external MuNANA. For each liposome tested, 50 μM nan+ donors and 400 μM acceptors were mixed with ATP/Mg2+ and 50 μM MuNANA, aliquoted into wells, and prewarmed at 27°C for 5 min. Following this preincubation, either RB150 or Thesit was added to the wells, and the amount of MuNANA hydrolyzed was measured over a 30-min period. The amount of MuNANA hydrolyzed in the absence of detergent is shown as a percentage of the amount that was hydrolyzed when the liposomes were solubilized by detergent. The data presented are the average of a minimum of three experiments (two replicates per treatment, per experiment) that used liposomes made on different days. (B) Encapsulated neuraminidase (36 kDa) is released from liposomes bearing Rab/SNAREs in the presence of Sec17p/Sec18p/HOPS. For each type of liposome, a master mix consisting of 50 μM nan+ donors and 400 μM acceptors mixed with ATP/Mg2+ was aliquoted into six tubes (to measure neuraminidase release, top panel) and six wells (to measure lipid dequenching, bottom panel). The tubes were placed in a 27°C water bath, and the plates were placed in the fluorimeter at 27°C. After a 6-min preincubation, Sec17p/Sec18p/HOPS, their buffers, or Thesit was added (two replicates each; see Materials and Methods), and reactions were incubated for 30 min. After 30 min, Thesit was added to the wells. The contents of the tubes were centrifuged for 30 min, and their supernatants were analyzed for the presence of released neuraminidase by addition of MuNANA. Top, amount of neuraminidase activity in the supernatants of spun reactions (relative to + detergent samples); bottom, extent of lipid dequenching (relative to + detergent final values). Data were averaged from a minimum of four independent experiments (two replicates, per treatment, each experiment) using liposomes made on at least two separate days.
Thus the physiological lipids and proteins that support vacuole fusion also support enhanced membrane permeability, which can extend to lysis and release of contents. Is actual fusion occurring in this reconstituted system as well? Further quantitative understanding required the development of an assay (see next paragraph) that reports the initial events of fusion without lysis, lysis with full release of lumenal macromolecular contents, and permeabilization that permits small molecule passage across otherwise intact membranes.
A new assay of fusion, lysis, permeabilization, and lipid mixing
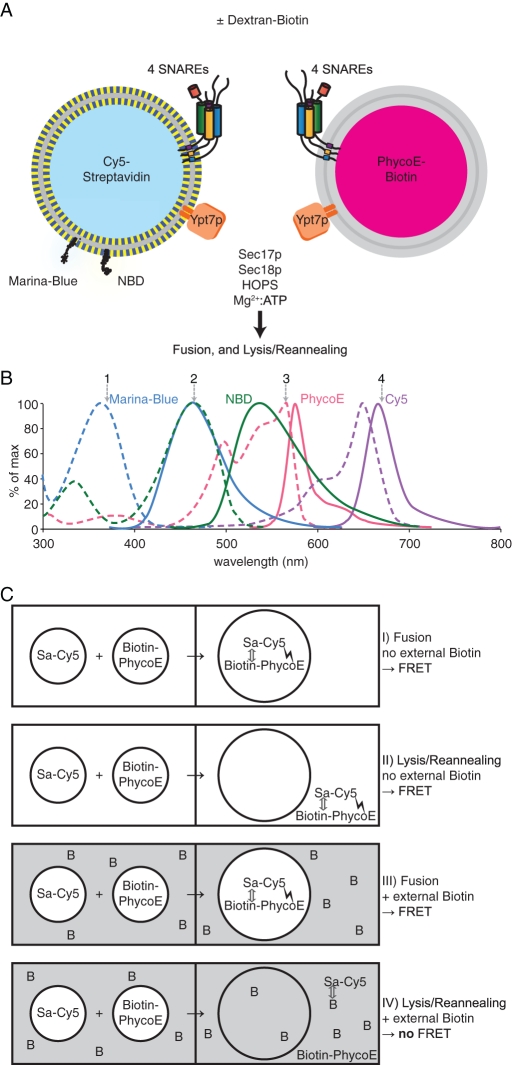
A lipid mix of vacuolar composition was dissolved in detergent-mixed micelles with Ypt7p and the four vacuolar SNAREs. To prepare “donor” proteoliposomes, the detergent solution also contained Marina Blue PE, NBD-PE, and streptavidin, which had been covalently derivatized with the fluorophore Cy5 (Figure 4A, left). To prepare “acceptor” proteoliposomes, the detergent solution bore biotinylated R-phycoerythrin (PhycoE; an exceptionally bright fluorescent protein; Oi et al., 1982), but no fluorescent lipids (Figure 4A, right). Dialysis was performed with a 20 kDa cutoff membrane to remove the detergent to form proteoliposomes, then with a 1000 kDa cutoff membrane to remove unincorporated proteins. The final protein/lipid ratios (Supplemental Figure S2A) and liposome size distributions (average median size of 171 (±13) nm; Supplemental Figure S2B) were comparable to previous characterizations of similar proteoliposomes (Mima et al., 2008). With an average number of entrapped molecules per 100,000 lipid molecules of 3.67 (±0.40) for PhycoE and 4.60 (±1.63) for Cy5-labeled streptavidin (Sa-Cy5), essentially all proteoliposomes contain lumenal markers (∼9 and 11 molecules per average size RPL for PhycoE and Sa-Cy5, respectively; see Supplemental Figure S2C). Marina Blue PE can be excited at 370 nm (Figure 4B, position 1; dashed light blue curve); its emission at 465 nm (position 2; solid light blue curve) is quenched by NBD-PE (dashed green). Dilution of these two lipids upon lipid mixing between donor and acceptor proteoliposomes relieves this quenching and serves as a direct assay of lipid mixing. PhycoE, excited at 565 nm (position 3; dashed pink), emits in a range (solid pink) that excites Cy5 (dashed purple). The emission of Cy5 at 670 nm (position 4; solid purple) is a direct measure of the Förster (or fluorescence) resonance energy transfer (FRET) that ensues when biotinylated PhycoE binds, essentially irreversibly, to Sa-Cy5.
FIGURE 4:
A novel RPL fusion assay that monitors lipid mixing, content mixing, and lysis. (A) Liposomes bearing the four vacuolar SNAREs and Ypt7p and the indicated fluorescent components are incubated with ATP/Mg2+, Sec17p and Sec18p, and HOPS. To monitor lipid mixing, the donor RPLs contain the fluorescently labeled lipids Marina Blue PE and NBD-PE. The Marina Blue fluorescence is quenched by NBD. On fusion of donor RPLs with acceptor RPLs, which do not carry any fluorescent lipids, the relative dilution of the fluorescent lipids results in a decrease in quenching efficiency, and thus an increase in the Marina Blue fluorescence signal intensity. To monitor the mixing of the aqueous compartments within the RPLs, donor RPLs contain Sa-Cy5 , whereas acceptor RPLs contain biotinylated PhycoE. On fusion, the Sa-Cy5 binds the biotin molecules that are coupled to PhycoE. The resulting close proximity of these two fluorescent molecules results in a FRET signal with PhycoE as donor and Cy5 as acceptor. In the presence of excess external dextran-biotin, the binding sites of streptavidin that leaks out of RPLs upon lysis are blocked; thus only true compartment mixing that preserves the integrity of the liposomal membranes increases the PhycoE/Cy5 FRET signal. (B) Fluorescence excitation (dashed lines) and emission (solid lines) spectra of the four involved fluorophores. (C) How the assay distinguishes between fusion and lysis. In the absence of external biotin (I and II), both fusion and lysis increase the PhycoE/Cy5 FRET signal. In the presence of external biotin in excess amounts (III and IV), only true fusion increases the PhycoE/Cy5 FRET signal, as the binding sites of any released streptavidin molecule get blocked by the external dextran-biotin, thus preventing its binding to PhycoE.
These two proteoliposome preparations are mixed and incubated with ATP, Sec17p, Sec18p, and HOPS, either without an external quencher of the biotin/streptavidin interaction or with a large molar excess of external biotin or dextran-biotin of 10,000 Da or (in standard assays) 70,000 Da (Figure 4A). Projected outcomes of these incubations are shown schematically in Figure 4C. On fusion, in the absence or presence of external dextran-biotin (symbolized as B), biotin-PhycoE binds tightly to Sa-Cy5, bringing the Cy5 and PhycoE fluorophores into close proximity, allowing FRET to occur (Figure 4C, I and III). This assay records only about half of the initial fusion events, as the fusion among proteoliposomes bearing lumenal Sa-Cy5 with each other, or between those bearing only lumenal biotin-PhycoE, does not generate a FRET signal. Lysis in the absence of external dextran-biotin (Figure 4C, II) also allows efficient complex formation and FRET signal. Thus, without external dextran-biotin, the FRET signal is a measure of the lysis events (Figure 4C, II) plus approximately half of the initial fusion events (Figure 4C, I). When a large excess of external dextran-biotin is present, the Sa-Cy5 released by lysis will form a complex with dextran-biotin, blocking the FRET signal (Figure 4C, IV), but true fusion (Figure 4C, III) still yields a FRET signal. Each assay condition is therefore performed with, and without, added external dextran-biotin in separate wells of a 384-well plate in a fluorescence plate reader that automatically reads and records both the lipidic and the proteinaceous fluorescence signals over time. The FRET in the well with dextran-biotin represents approximately half of the fusion events, whereas the FRET in wells without dextran-biotin represents these fusion events plus all lysis.
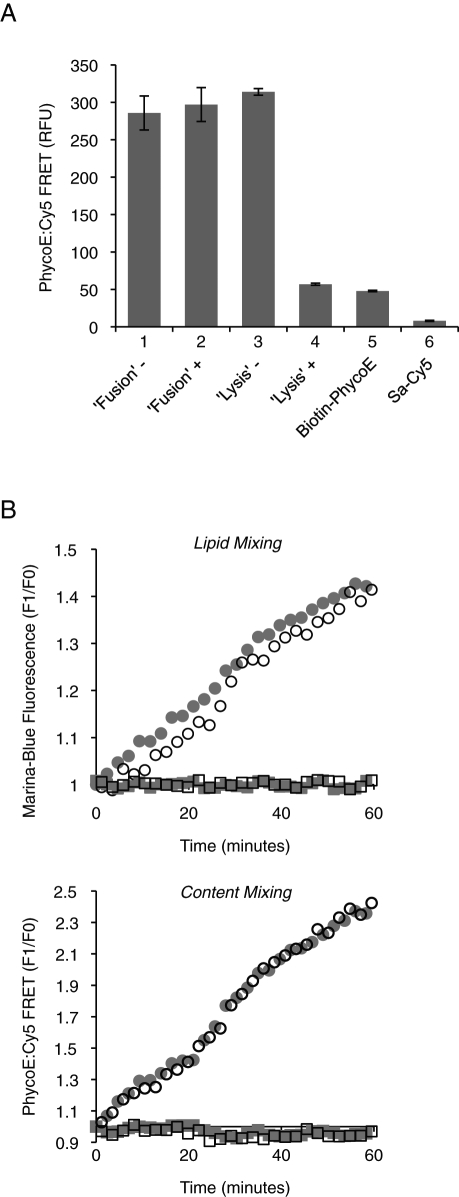
Control experiments have validated this experimental scheme. The same signal was obtained when concentrated biotin-PhycoE was mixed with concentrated Sa-Cy5, as would occur within fused proteoliposomes, then diluted with buffer alone or buffer with excess dextran-biotin (Figure 5A, columns 1 and 2). These findings show that a fusion signal would not be lost if proteoliposomes that had fused in the presence of dextran-biotin underwent subsequent lysis. The FRET signal was also the same when these two fluorescent proteins were diluted before being mixed (column 3), as in lysis. Thus the signals from lysis and from fusion are directly comparable. When the fluorescent proteins were mixed with excess dextran-biotin before being mixed with each other, the ensuing FRET signal (column 4) was reduced to the background level derived from the two separate fluorophores (columns 5 and 6). In addition, the kinetics of lipid mixing are unaffected by the presence or absence of lumenal fluorophores (Figure 5B, top panel), and the kinetics of content mixing are unaffected by the presence or absence of fluorescent lipids (bottom panel).
FIGURE 5:
Assay controls. (A) The intensities of signals resulting from either fusion or lysis are comparable as seen by mimicking these two events. Fusion: 4 μM biotinylated PhycoE was mixed with 8 μM Sa-Cy5, incubated for 30 min at 27°C, and diluted 1:500 (vol/vol) in RB150 buffer with (+) or without (–) dextran-biotin 10,000 (1 μM biotin). Lysis: 8 nM biotinylated PhycoE was mixed with 16 nM Sa-Cy5 in RB150 buffer with (+) or without (–) dextran-biotin 10,000 (1 μM biotin), then incubated for 30 min at 27°C. FRET signals were measured in 384-well plates in a fluorescence plate reader (excitation: 565 nm; emission: 670 nm). The values for 8 nM biotinylated PhycoE alone, or 16 nM Sa-Cy5, are also shown. Data are shown in relative fluorescence units as mean ± SD of n = 3 experiments. (B) FRET between the fluorescently labeled lipids (for lipid mixing) and the entrapped fluorescent probes (for content mixing) do not interfere with each other. Proteoliposomes (250 μM donors and 250 μM acceptors) were mixed in RB150 with Mg2+/ATP and incubated at 27°C with either Sec17p/Sec18p/HOPS (circles) or their respective buffers (squares). Shown is the relative increase in fluorescence for a typical (for at least n = 3 repeats) experiment. Top, lipid mixing of proteoliposomes that carry either only fluorescently labeled lipids (open symbols) or both fluorescently labeled lipids and entrapped fluorescent probes (gray symbols). The maximal extent of fusion is reached at a F1/F0 of 1.65 ± 0.07, as determined by preparing proteoliposomes with half the amount of fluorescent lipids (i.e., 0.75% [mol/mol] of Marina Blue PE and NBD-PE, respectively). Bottom, content mixing of proteoliposomes that carry either only entrapped fluorescent probes (open symbols) or both fluorescently labeled lipids and entrapped fluorescent probes (gray symbols).
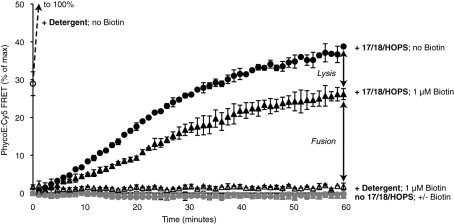
When mixed proteoliposomes were incubated with detergent without dextran-biotin, the FRET signal increased to a maximal value (set to 100%) within minutes (Figure 6, open circle, and Supplemental Figure S3, open diamonds). In contrast, when they were premixed with dextran-biotin before the addition of detergent, there was no development of any FRET signal (open triangles), confirming that the large molar excess of external dextran-biotin was sufficient to block all binding sites of streptavidin. When proteoliposomes were incubated at 27°C with Sec17p, Sec18p, ATP, and HOPS in the presence of external dextran-biotin, there was a steadily increasing PhycoE:Cy5 FRET signal, reaching ∼25% of the maximal (plus detergent, minus biotin) signal after 60 min. As noted earlier in the text, the initial rate of FRET increase will underestimate the initial rate of fusion by approximately twofold, although second and third rounds of fusion may include proteoliposomes that are the product of earlier fusion events between proteoliposomes with like content that are invisible in our assay. Incubations performed in the absence of external dextran-biotin developed additional FRET signal due to lysis. For the first round of fusion or lysis events, where only half the fusion is “visible” to our assay, there are approximately fourfold more fusion than lysis events.
FIGURE 6:
Fusion is accompanied by a certain degree of lysis. RPLs (250 μM donors and 250 μM acceptors) were mixed in RB150 with Mg2+/ATP and incubated at 27°C in the presence (triangles) or absence (circles) of external dextran-biotin 70,000 (1 μM biotin) with Sec17p/Sec18p/HOPS (black symbols), their respective buffers (light gray symbols), or detergent (open symbols). The means of the resulting PhycoE/Cy5 FRET signals of a triplicate assay are displayed with SD, as percentages of the maximal values obtained by incubation with detergent in the absence of external biotin. The mean of the ratio of signals for conditions with or without biotin with four pairs of parallel, but independently prepared, 4-SNARE RPLs incubated with Sec17p/Sec18p/HOPS is 1.43 ± 0.064 (SD).
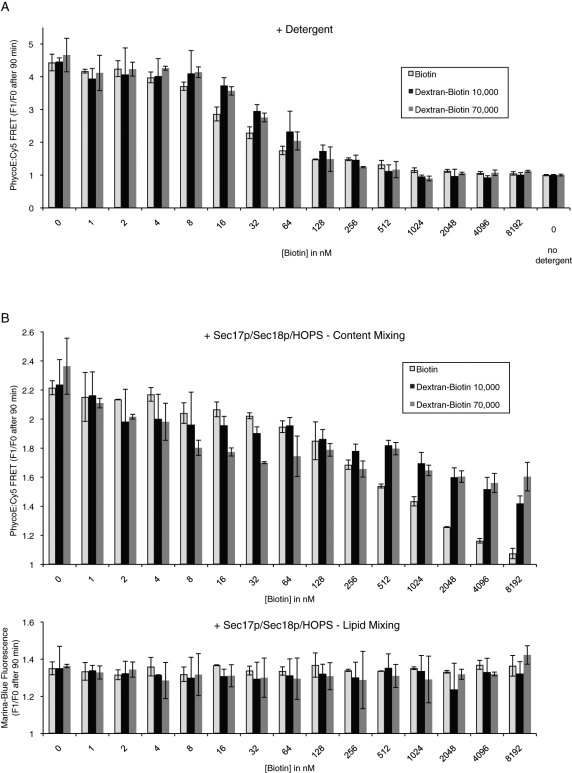
To ensure that appropriate levels of external quencher were used in these assays, we titrated biotin, dextran-biotin 10,000, or dextran-biotin 70,000 into standard fusion incubations. In the presence of detergent, where quencher had full access to the Sa-Cy5, ∼0.5 μM biotin was needed to fully inhibit the FRET signal to the background level seen without detergent or Sec17p/Sec18p/HOPS (Figure 7A, right), a background that is largely due to the PhycoE emission at the wavelength of Cy5 detection. We therefore used concentrations of dextran-biotin 70,000 equaling 1 μM biotin in our standard assay conditions.
FIGURE 7:
Reactions with increasing concentrations of external biotin reveal a fusion-dependent membrane permeabilization. (A) RPLs (250 μM donors and 250 μM acceptors) were mixed in RB150 with increasing amounts of biotin (biotin [light gray columns], dextran-biotin 10,000 [black columns], or dextran-biotin 70,000 [dark gray columns]) and lysed by addition of Thesit to 0.1% (wt/vol). As control, one sample did not receive any detergent to establish the level of background signal. Shown is the relative increase of PhycoE/Cy5 FRET signal after 90 min as the average of n = 3 independent experiments ± SD values. (B) RPLs (250 μM donors and 250 μM acceptors) were mixed in RB150 with Mg2+/ATP and increasing amounts of biotin (biotin [light gray columns], dextran-biotin 10,000 [black columns], or dextran-biotin 70,000 [dark gray columns]) and incubated with Sec17p/Sec18p/HOPS at 27°C for 90 min. The relative increase of the PhycoE/Cy5 FRET signal after 90 min is displayed as the average of n = 3 independent experiments ± SD values.
Fusion factors permit biotin permeation
Under standard assay conditions, the development of a FRET signal between lumenal proteins occurs in a dextran-biotin–inaccessible compartment, as it is fully quenched by the 1 μM external dextran-biotin when given full access through detergent addition (Figure 6). We therefore tested whether biotin itself could permeate into the proteoliposomes. Incubations of the proteoliposomes with HOPS, Sec17p, and Sec18p developed a FRET signal that was largely inaccessible to 512 nM to 8192 nM dextran-biotin (compare Figure 7B, top panel, to 7A). The formation of the lumenal fluorophore FRET complex was quenched, however, when biotin itself was added over this concentration range (Figure 7B, top panel, open columns). The increasing concentrations of biotin or dextran-biotin did not impact the RPL dynamics themselves, as the lipid dequenching signals were stable over this concentration range (Figure 7B, bottom panel). Because the FRET complex that formed in the presence of dextran-biotin was shielded by the membrane bilayer, biotin itself was crossing an intact bilayer to block FRET complex development.
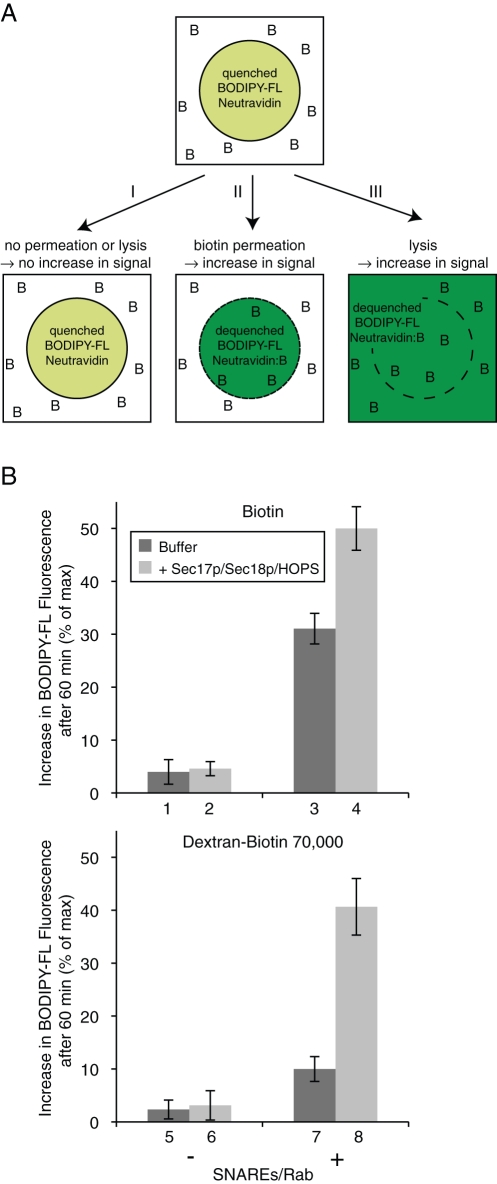
To directly assay the permeation of biotin (Figure 8A), we exploited the fluorescence dequenching of BODIPY-FL–NeutrAvidin when it binds biotin (Emans et al., 1995; Hama et al., 2007). Liposomes bearing lumenal BODIPY-FL–NeutrAvidin as their sole fluorophore were prepared with, or without, vacuolar SNAREs and Ypt7p, then incubated with either Sec17p/Sec18p/HOPS or their buffers in the presence of added biotin or dextran-biotin 70,000 (Figure 8B, top and bottom panel, respectively). There was little permeation of either biotin or biotin-dextran into protein-free liposomes (columns 1 and 5), even when Sec17p/Sec18p/HOPS were present (columns 2 and 6). The presence of the vacuolar SNAREs and Rab caused a measurable increase in access to dextran-biotin 70,000 (column 7) but a substantially greater increase in access to external biotin (column 3), indicating directly that these proteins cause enhanced membrane permeability. As residual amounts of detergent can affect membrane permeability in some systems (Ueno, 1987; Rigaud et al., 1998), we assessed the levels of remaining detergent in our liposomes. We found a final detergent/lipid molar ratio of ∼1:400 and no significant difference between protein-free and SNARE/Rab-bearing liposomes (Supplemental Figure S1). Residual detergent alone is therefore unlikely to cause the observed permeability. When incubations also contained Sec17p/Sec18p/HOPS, dextran-biotin 70,000 gained access to the BODIPY-FL–NeutrAvidin (column 8), indicative of lysis. It is important to note that this assay measures the cumulative permeation or lysis during a 60 min incubation, not just the initial event of fusion or lysis as for the assay described in Figure 4 and used in Figures 5–7. Much of the lysis seen in Figure 8 may be from proteoliposomes that had undergone prior fusion events. In fact, the cumulative release of content during fusion reactions is substantially higher than the amount of lysis measured when recording primary events only (compare Supplemental Figure S4, A and B).
FIGURE 8:
Permeation of biotin across membranes depends on the presence of SNAREs. (A) Scheme of the biotin permeation assay. Liposomes with entrapped BODIPY-FL–NeutrAvidin are incubated in the presence of biotin (as free or dextran-bound biotin; depicted as B). As long as neither permeation nor lysis occurs, the BODIPY-FL–NeutrAvidin remains in its quenched state (I). When biotin gets access to the lumenal compartment due to membrane permeation (II), or when the identity of inside and outside compartments gets lost due to lysis (III), the binding of biotin to NeutrAvidin dequenches the BODIPY-FL fluorescence signal. (B) BODIPY-FL–labeled NeutrAvidin–containing liposomes (500 μM lipid), which were either protein free (columns 1, 2, 5, and 6) or bore SNARE and Rab proteins (columns 3, 4, 7, and 8), were mixed in RB150 with Mg2+/ATP and incubated for 60 min at 27°C in the presence of biotin (top panel) or dextran-biotin 70,000 (bottom panel) with either Sec17p/Sec18p/HOPS (light gray columns) or their respective buffers (dark gray columns). The increase of BODIPY-FL fluorescence after 60 min relative to the maximal increase obtained by detergent addition is displayed as the average of n = 3 independent experiments ± SD values.
DISCUSSION
In an earlier study (Mima et al., 2008), it was reported that concentrated DTN solutions gradually lost their potency over the course of 30 min at 27°C due to reaction with dissolved molecular oxygen. The dequenching (lipid-mixing) rates were seen to be equivalent to those of proteoliposomes that had been pretreated with DTN either 30 min before Sec17p/Sec18p/HOPS addition, allowing the DTN to become inactive, or 5 min before Sec17p/Sec18p/HOPS addition, when the DTN was assumed to still be active. This finding suggested that fusion was not accompanied by any lysis. Molecular oxygen, however, is present only at 0.2 mM in water (Murray and Riley, 1970), and thus the oxidation of high concentrations of DTN is limited by the diffusion of oxygen into the solution. The rate of oxidative destruction of dilute DTN under our fusion reaction conditions, at neutral pH in 20 μl in a low volume 384-well plate with a high surface-to-volume ratio, is more rapid. When proteoliposomes received fresh DTN shortly after the addition of HOPS, Sec17p, and Sec18p (Figure 1, open circles), virtually all the NBD-PE fluorescence was destroyed, due to some combination of lysis (Figure 3B) and enhanced permeability of SNARE-proteoliposomes to small molecules. Current assays do not report the relative rates of fusion and lysis; we therefore developed assays to specifically assess the rates of 1) initial fusion or lysis, and 2) membrane permeation. These assays used four distinct macromolecules (neuraminidase, streptavidin, PhycoE, and dextran-biotin 10,000 or 70,000) as well as three small molecules (DTN, MuNANA, and biotin), and showed with all these probes that the very proteins that catalyze fusion—SNAREs, SNARE chaperones, Rab, and Rab effector complex—promote both membrane permeability to small molecules and lysis in addition to true fusion.
The most common assay currently used to study the fusion of reconstituted proteoliposomes measures lipid mixing between proteoliposomes bearing fluorescent lipids and nonfluorescent proteoliposomes. This assay is rapid and quantitative but does not distinguish fusion events in which membranes remain sealed from the events of lysis and reannealing or hemifusion. Other assays (Nickel et al., 1999; Ohya et al., 2009) measure lumenal compartment mixing events in which the membranes remain sealed, but do not report the relative frequency of lysis. In addition, assays of release of lumenal proteins (e.g., Figure 3B) do not measure the number of successful fusion events that may have preceded lysis and do not assess the relative rates of fusion and lysis. Assays that rely on the entrapment of small molecules (Dennison et al., 2006) have shown that small solutes are poorly retained in SNARE proteoliposomes. This behavior may not be limited to reconstituted systems, as trans-SNARE complex formation between purified vacuoles induces a striking flux of calcium from the vacuolar lumen (Merz and Wickner, 2004), and the identity or existence of any specific calcium channel has proven elusive (W.Wickner, personal communication). Additional assays are needed that allow for monitoring the fate of the lumenal compartments (direct comparison of fusion and lysis events), while measuring changes that happen to the lipid bilayer (to observe hemifusion events, e.g.) and recording the permeability of those membranes that remain intact.
We now present one such assay, using RPLs made by dialysis with commercially available fluorescence reporters. There is clear spectral separation of the fluorophores used to measure lipid mixing and lumenal compartment fates (Figure 4B), and control experiments show that neither the lipidic nor lumenal reporters interfere with proteoliposome dynamics (Figure 5B) and that the measurements of fusion and lysis are directly comparable (Figure 5A). This assay is focused on measurement of the initial round of fusion or lysis events; there is no change to the FRET signal from two initially fused proteoliposomes if they subsequently lyse, nor is there any additional FRET signal from the fusion of proteoliposomes that had previously lysed. In the presence of >1 μM dextran-biotin, Sa-Cy5/biotin-PhycoE complex forms inside fused, sealed proteoliposomes (as shown by the absence of FRET complex when the permeability barrier is breached by detergent), yet the formation of this complex is gradually blocked by increasing amounts of underivatized biotin (Figure 7). Thus membranes that are undergoing fusion have enhanced permeability to a small molecule such as biotin while remaining sealed to macromolecules. This permeation is in some proportion to the concentration of biotin over a range in which full permeability (as when detergent is present) would allow a complete blockage of FRET complex formation; however, we do not yet have a quantitative assessment of the increase in permeability coefficient for this compound, or others, across the membrane.
Our studies resolve two salient issues in the field of membrane fusion. The reconstituted vacuolar fusion machinery is shown here to directly cause both true fusion and lysis in a directly measurable ratio. An earlier study with neuronal SNARE proteoliposomes bearing entrapped, complementary oligonucleotides (Nickel et al., 1999) showed some lysis occurring at 37°C that was unrelated to the presence of SNAREs. We do not see this particular background lysis (Supplemental Figure S3), but we used vacuolar, not neuronal, SNAREs, used different lipid composition and buffer conditions, and incubated at 27°C rather than 37°C. We also present direct evidence for the permeability of intact fusing liposomal membranes to small molecules. The capacity of biotin to block the formation of the biotin-PhycoE/Sa-Cy5 FRET complex in the lumenal, dextran-biotin–inaccessible space is in accord with our findings with MuNANA and DTN. For vacuoles freshly isolated from yeast, <5% undergo lytic events that release a lumenal macromolecule such as green fluorescent protein during fusion (Starai et al., 2007), whereas permeability to the small lumenal ion calcium increases strikingly (Peters and Mayer, 1998; Merz and Wickner, 2004). It will require substantial further study to determine whether other SNARE-mediated events, such as “kiss and run” exocytosis of small molecules from membrane vesicles (Rizzoli and Jahn, 2007), may be related to this same membrane permeabilization.
Several limitations remain in the current assays of membrane fusion events. Our proteoliposomes are only one-tenth the diameter of vacuoles and, like almost all current reconstituted proteoliposomes, lack their physiological protein and lipid asymmetries. It will be particularly interesting to see the impact of such, more physiological, conditions, as recent studies (Zick and Wickner, in preparation) have shown that the propensity of docked and fusion-activated membranes to undergo lysis at a certain frequency is inherent to the current reconstituted system, and was so far seen to be largely unaltered over a wide range of concentrations of soluble components and integral membrane constituents. Although our current assays allow concurrent measurement of true fusion, lysis, lipid mixing, and membrane permeability, it will also be important to be able to monitor trans-SNARE complex assembly and lipid/protein microdomain formation. Our current assays have the potential to reveal conditions that support lipid mixing without content mixing or lysis, indicative of hemifusion, but we have not yet observed this intermediate state.
MATERIALS AND METHODS
Protein isolation
The SNARE proteins were essentially isolated as previously described (Mima et al., 2008). Vti1p and Nyv1p were gel-filtered into RB150/ß-OG (20 mM HEPES, pH 7.4, 150 mM NaCl, 10% glycerol [vol/vol], 1% [wt/vol] ß-octyl glucoside) after purification using Sephacryl S-200 HR (GE Healthcare Biosciences, Pittsburgh, PA). A complete detergent exchange was confirmed by determining the 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) concentrations in elution fractions (Urbani and Warne, 2005); only fractions with no residual amounts of CHAPS were pooled and used in the reconstitution experiments. Ypt7p (Hickey et al., 2009), Sec17p (Schwartz and Merz, 2009), His6-Sec18p (Haas and Wickner, 1996), and HOPS (Hickey and Wickner, 2010) were isolated as previously described.
Liposome preparation
Proteoliposomes for NBD/rhodamine lipid dequenching assays were prepared as described (Mima et al., 2008) with modifications. The cardiolipin in lipid mixes was replaced by 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). Following solvent evaporation under a stream of nitrogen, lipid films were further dried under vacuum for 3 h, then hydrated on ice in RB150/Mg2+/ß-OG (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 10% glycerol [vol/vol], 1% [wt/vol] ß-octyl glucoside) for 1 h with intermittent vortexing. SNARE proteins and Ypt7p were added, and the solution was nutated for 2 h at 4°C. The molar ratios of protein/lipid were 1:1000 for SNAREs and 1:2000 for Ypt7p. For protein-free liposomes, equivalent amounts of the respective buffers were added instead. The detergent solutions were dialyzed in 20 kDa cutoff dialysis cassettes three times (3 h, 3 h, and overnight) against a 1000-fold volume of RB150/Mg2+, then floated on a four-step Histodenz (Sigma-Aldrich, St. Louis, MO) gradient (40%, 30%, 20%, and RB150/Mg2+). The RPLs harvested at the topmost interface were dialyzed 1:2000 against RB150/Mg2+ for 3 h, and lipid phosphate was determined as described (Chen et al., 1956). For donor liposomes that contained neuraminidase (nan+), 20 mg of purified protein was added per 1 ml of RPL preparation after lipid film hydration. For PC/PS liposomes, the lipid mixes are POPC (62.5% or 65.5% for donor or acceptor liposomes, respectively), POPS (34.5%), and NBD-PE and Rh-PE (1.5% each for donors).
Proteoliposomes for the simultaneous analysis of lipid mixing and content mixing were prepared with the following modifications. The Rh-PE in donor liposomes was replaced by Marina Blue PE (Invitrogen, Carlsbad, CA). Lipid mixes with 35 mM ß-OG were dried under vacuum in a centrifugal evaporator (SpeedVac SC100; Savant Instruments, now Fisher Scientific, Waltham, MA) for 3 h, then rehydrated for 30 min at 23°C in 5× RB150/Mg2+ (100 mM HEPES, pH 7.4, 750 mM NaCl, 5 mM MgCl2, 50% glycerol [vol/vol]) with intermittent vortexing. Lipid/detergent solutions were twice treated in a bath sonicator for 5 min to assure complete dispersion. Proteins were added at molar protein/lipid ratios of 1:1000 for SNAREs and 1:10,000 for Ypt7p; for donor RPLs, Sa-Cy5 (KPL, Gaithersburg, MD) was added at 8 μM; for acceptor RPLs, Biotin-XX-R-Phycoerythrin (Invitrogen) was added at 4 μM. Solutions were dialyzed in 20 kDa cutoff dialysis cassettes three times (3 h, 3 h, and overnight) against a 1000-fold volume of RB150/Mg2+ to form RPLs, then dialyzed in 1000 kDa tubing (SpectrumLabs, Rancho Dominguez, CA) three times (at least 12 h each) against 1000-fold volumes of RB150/Mg2+ to remove nonentrapped probes. Final concentrations were determined by lipid phosphate assays. RPLs were stored at 4°C until use.
DTN stock solutions
To minimize variability, sodium DTN (Sigma-Aldrich, St. Louis, MO) was dissolved (100 mM in ice-cold TrisCl buffer, pH 10; degassed for 20–30 min under vacuum) the day it was received from the vendor. The solution was immediately aliquoted into eight-tube PCR strips, snap-frozen in liquid nitrogen, and stored at –80°C. For experiments using a range of concentrations, individual PCR strips were prepared, such that addition of 1.5 μl of DTN stock solution would yield the desired final concentration of DTN per well. Less than 5 min elapsed from the time the DTN was opened until the stocks were frozen.
DTN sensitivity in the absence of chaperones
For each set of liposomes, donors (50 μM lipids) and acceptors (400 μM lipids) were premixed in RB150/Mg2+/ATP (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM ATP, 1.2 mM MgCl2), aliquoted into 384-well plates, and incubated at 27°C for 5 min. An equal volume of ice-cold RB150 or freshly thawed 7.5 mM DTN was added per well, and reactions were incubated for 45 min at 27°C. Thesit (Sigma-Aldrich, St. Louis, MO) was added to all wells to a final concentration of 0.1% (wt/vol) to dequench the remaining NBD fluorescence, which was then measured in a fluorescence plate reader (excitation: 460 nm; emission: 538 nm; cutoff: 515 nm).
DTN sensitivity in the presence of chaperones
Liposomes were premixed in RB150/Mg2+/ATP and prewarmed for 5 min as described earlier in this section. Each set of liposomes was aliquoted into 12 wells of a 384-well plate. Four wells received DTN (to 7.5 mM) immediately after it had thawed on ice (“early DTN”), four other wells received ice-cold RB150 at the same time. The remaining four wells (i.e., “late DTN”) received DTN from another freshly thawed portion 3 min after the addition of the chaperones. Thirty minutes after the early addition of DTN, 6 of the 12 wells received Sec17p (324 nM), Sec18p (1 μM), and HOPS (40 nM); the other six received the corresponding buffers. Reactions were incubated for 45 min at 27°C after the addition of chaperones or their buffers. Thesit was then added to 0.1% (wt/vol) to all wells to terminate the reaction. The NBD fluorescence was measured throughout in a fluorescence plate reader (excitation: 460 nm; emission: 538 nm; cutoff: 515 nm).
Neuraminidase purification
Neuraminidase purification (Hoyer et al., 1991) was modified as follows. A freshly grown colony of BL21 carrying plasmid pSX61, which encodes the NanH gene from Salmonella typhimurium under its own promotor (a generous gift from New England BioLabs, Ipswich, MA), was inoculated in 100 ml of Lysogeny Broth plus ampicillin (LB/Amp), grown for 7 h at 37°C and then diluted 1:1500 into two 3 l portions of LB/Amp. Cultures grew overnight with vigorous shaking at 37°C. Cells were collected by low-speed centrifugation; suspended in 1/20th volume of 10 mM TrisCl, pH 7.6; again collected by centrifugation; resuspended in 10 mM TrisCl, pH 7.6 (35 ml total volume); and frozen dropwise in liquid nitrogen. The cell suspension was thawed on ice and passed three times through a French pressure cell (940 psi). Cell debris was removed by centrifugation (50,000 rpm, 60Ti rotor, 1 h), and the supernatant was loaded onto a 1.5 cm × 9 cm column of DE52 (Whatman, Piscataway, NJ) that had been equilibrated with 10 mM TrisCl, pH 7.6. Unbound material was passed through a Centricon 100K filter device (Millipore, Billerica, MA), then concentrated using a Centricon 30K filter device. Purity (at >95%) was confirmed by SDS–PAGE and Colloidal Coomassie staining.
Neuraminidase assay in the absence of chaperones
For each set of liposomes, nan+ donors (50 μM lipid) and acceptors (400 μM lipid) were mixed in RB150/Mg2+/ATP and 50 mM MuNANA, aliquoted into 384-well plates, and allowed to prewarm for 5 min at 27°C in a fluorescence plate reader. Either RB150 or Thesit (to a final concentration of 0.1% [wt/vol]) was added, and the hydrolysis of MuNANA (Gold Biotechnology, St. Louis, MO) by neuraminidase was monitored for 30 min (excitation: 365 nm; emission: 460 nm; cutoff: 445 nm).
Neuraminidase assay in the presence of chaperones
For each set of liposomes, a master mix equivalent to 25 individual reactions (nan+ donors [50 μM lipid] and acceptors [400 μM lipid] mixed with ATP/Mg2+) was aliquoted into six 1.5 ml tubes (three reactions per tube, two replicates per treatment) to measure the amount of neuraminidase that is released during a reaction. The remainder was aliquoted into a 384-well plate to measure lipid dequenching of the samples. The tubes were placed in a 27°C water bath and prewarmed for 6 min. The 384-well plate with the remaining reactions was placed in the fluorescence plate reader and also prewarmed at 27°C for 6 min. Following each preincubation, chaperones (Sec17p [324 nM], Sec18p [1 μM], and HOPS [40 nM]), their buffers, or Thesit (to a final concentration of 0.1% [wt/vol]) was added to the tubes (only chaperones or their buffers were added to the reactions in the 384-well plate). All reactions were incubated at 27°C for another 30 min. After 30 min, reactions in the fluorimeter received Thesit (to 0.1% [wt/vol]) to measure maximal lipid dequenching, and the reactions in the tubes were placed on ice for 5 min, received an equal volume of RB150 (60 μl), and were transferred to an ultracentrifuge tube and centrifuged at 68,000 rpm in a TLA100 rotor (Beckman Coulter, Indianapolis, IN) at 4°C. A 90 μl portion of each supernatant was transferred into a 96-well plate. After a 5 min prewarming period, all wells received MuNANA to 50 mM, and the hydrolysis of MuNANA by neuraminidase was followed for 30 min (wavelengths as stated earlier in the text).
Simultaneous lipid mixing and content mixing assays
To simultaneously measure lipid mixing and content mixing, donor and acceptor RPLs (250 μM lipid each) were mixed on ice with Sec17p (32.5 nM), Sec18p (0.9 μM), and HOPS (40 nM), or their respective buffers, Mg2+ (1 mM), and ATP (1 mM). Reactions were incubated in 384-well plates at 27°C in a fluorescence plate reader for 60–90 min. Fluorescence signals were recorded for Marina Blue (excitation: 370 nm; emission: 465 nm; cutoff: 420 nm) and PhycoE:Cy5 FRET (excitation: 565 nm; emission: 670 nm; cutoff: 630 nm). Maximal values were determined by addition of Thesit to 0.1% (wt/vol). Biotin was added to the reaction mixtures where indicated as biotin (Thermo Fisher Scientific, Rockford, IL), dextran-biotin MW 10,000 (2.5 mol biotin per mol dextran; Invitrogen), or dextran-biotin MW 70,000 (3 mol biotin per mol dextran; Invitrogen).
Biotin permeation assay
To monitor the permeation of biotin across membranes, liposomes, which either do or do not bear SNARE and Rab proteins, were prepared by dialysis as described earlier in the text, with 10 μM BODIPY-FL–labeled NeutrAvidin. For the labeling, 200 μl of NeutrAvidin (10 mg/ml in 0.2 M NaHCO3; Thermo Fisher Scientific) was mixed with 5 μl of BODIPY-FL succinimidyl ester (10 mg/ml in dimethylformamide; Invitrogen) and incubated at room temperature for 1 h. The labeled protein was purified with Sephadex G-50, and the degree of labeling was determined according to the manufacturer's instructions as 2.18 BODIPY-FL molecules per NeutrAvidin tetramer.
To measure biotin permeation, these RPLs (500 μM lipid) were mixed on ice in RB150 (containing 1 mM ATP and 1 mM MgCl2) with Sec17p (32.5 nM), Sec18p (0.9 μM), and HOPS (40 nM), or their respective buffers, and 20 μM biotin (as free biotin or dextran-biotin 70,000). Reactions were incubated in 384-well plates at 27°C in a fluorescence plate reader for 60 min, and the increase in BODIPY-FL fluorescence due to binding of biotin to the labeled NeutrAvidin (Emans et al., 1995; Hama et al., 2007) was recorded (excitation: 480 nm; emission: 512 nm; cutoff: 495 nm). Maximal values were determined by the addition of Thesit to 0.1% (wt/vol).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM23377-35. We are especially grateful to Bill Wickner for his guidance, support, and purifying proteins. Many thanks go to Charles Barlowe for collaboration and to Mark Savellano for guidance with dynamic light scattering. We thank Amy Orr, Holly Jakubowski, and Deborah Mayka for expert assistance. Michael Zick was supported by a research fellowship (ZI 1339/1-1) from the DFG (German Research Foundation).
Abbreviations used:
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DTN
dithionite
- FRET
Förster (or fluorescence) resonance energy transfer
- HOPS
homotypic fusion and vacuole protein sorting complex
- LB/Amp
Lysogeny Broth plus ampicillin
- MuNANA
4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid
- NBD-PE
N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine
- PC
phosphatidylcholine
- PhycoE
R-phycoerythrin
- PS
phosphatidylserine
- RPL
reconstituted proteoliposome
- Sa-Cy5
Cy5-labeled streptavidin
- SNARE
SNAP (Soluble NSF attachment protein) Receptor
- VLM
vacuolar lipid mix
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0680) on October 5, 2011.
REFERENCES
- Chen PS, Toribara TY, Warner H. Microdetermination of phosphorus. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emans N, Biwersi J, Verkman AS. Imaging of endosome fusion in BHK fibroblasts based on a novel fluorimetric avidin-biotin binding assay. Biophys J. 1995;69:716–728. doi: 10.1016/S0006-3495(95)79947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Walter P. Membrane lysis during biological membrane fusion: collateral damage by misregulated fusion machines. J Cell Biol. 2008;183:181–186. doi: 10.1083/jcb.200805182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Wickner W. Homotypic vacuole fusion requires Sec17p (yeast alpha-SNAP) and Sec18p (yeast NSF) EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- Hama Y, Urano Y, Koyama Y, Choyke PL, Kobayashi H. Activatable fluorescent molecular imaging of peritoneal metastases following pretargeting with a biotinylated monoclonal antibody. Cancer Res. 2007;67:3809–3817. doi: 10.1158/0008-5472.CAN-06-3794. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125. doi: 10.1074/jbc.M109.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Roggentin P, Schauer R, Vimr ER. Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl alpha 2-3 linkages. J Biochem. 1991;110:462–467. doi: 10.1093/oxfordjournals.jbchem.a123603. [DOI] [PubMed] [Google Scholar]
- Jun Y, Fratti RA, Wickner W. Diacylglycerol and its formation by phospholipase C regulate Rab- and SNARE-dependent yeast vacuole fusion. J Biol Chem. 2004;279:53186–53195. doi: 10.1074/jbc.M411363200. [DOI] [PubMed] [Google Scholar]
- Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 2001;20:4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier K, Muller H, Holzer H. Purification and molecular characterization of two inhibitors of yeast proteinase B. J Biol Chem. 1979;254:8491–8497. [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Merz AJ, Wickner WT. Trans-SNARE interactions elicit Ca2+ efflux from the yeast vacuole lumen. J Cell Biol. 2004;164:195–206. doi: 10.1083/jcb.200310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima J, Wickner W. Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc Natl Acad Sci USA. 2009;106:16191–16196. doi: 10.1073/pnas.0908694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CN, Riley JP. The solubility of gases in distilled water and sea water–III. Argon. Deep Sea Res Oceanographic Abstr. 1970;17:203–209. [Google Scholar]
- Nickel W, Weber T, McNew JA, Parlati F, Sollner TH, Rothman JE. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc Natl Acad Sci USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, Drechsel D, Kalaidzidis Y, Zerial M. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- Oi VT, Glazer AN, Stryer L. Fluorescent phycobiliprotein conjugates for analyses of cells and molecules. J Cell Biol. 1982;93:981–986. doi: 10.1083/jcb.93.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Mayer A. Ca2+/calmodulin signals the completion of docking and triggers a late step of vacuole fusion. Nature. 1998;396:575–580. doi: 10.1038/25133. [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Levy D, Mosser G, Lambert O. Detergent removal by non-polar polystyrene beads. Eur Biophys J. 1998;27:305–319. [Google Scholar]
- Rizzoli SO, Jahn R. Kiss-and-run, collapse and “readily retrievable” vesicles. Traffic. 2007;8:1137–1144. doi: 10.1111/j.1600-0854.2007.00614.x. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol. 2009;185:535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C, Hickey CM, Mima J, Burfeind AS, Wickner W. Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci USA. 2009;106:17626–17633. doi: 10.1073/pnas.0903801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochem. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- Ueno M. Asymmetric disposition of detergents within vesicle bilayer and its effect on ion permeation through the membrane. Biochim Biophys Acta. 1987;904:140–144. doi: 10.1016/0005-2736(87)90095-2. [DOI] [PubMed] [Google Scholar]
- Urbani A, Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Wang L, Merz AJ, Collins KM, Wickner W. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J Cell Biol. 2003;160:365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Seeley ES, Wickner W, Merz AJ. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Wang T, Smith EA, Chapman ER, Weisshaar JC. Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1–5 ms resolution. Biophys J. 2009;96:4122–4131. doi: 10.1016/j.bpj.2009.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.