Unexpected redundancy in the nuclear import pathways used by the essential mRNA export factor NXF1 increases progressively from fungi to nematodes and insects to chordates, potentially paralleling the increasing complexity in mRNA export regulation and the evolution of new nuclear functions for NXF1.
Abstract
In human cells, the mRNA export factor NXF1 resides in the nucleoplasm and at nuclear pore complexes. Karyopherin β2 or transportin recognizes a proline–tyrosine nuclear localization signal (PY-NLS) in the N-terminal tail of NXF1 and imports it into the nucleus. Here biochemical and cellular studies to understand the energetic organization of the NXF1 PY-NLS reveal unexpected redundancy in the nuclear import pathways used by NXF1. Human NXF1 can be imported via importin β, karyopherin β2, importin 4, importin 11, and importin α. Two NLS epitopes within the N-terminal tail, an N-terminal basic segment and a C-terminal R-X2-5-P-Y motif, provide the majority of binding energy for all five karyopherins. Mutation of both NLS epitopes abolishes binding to the karyopherins, mislocalized NXF1 to the cytoplasm, and significantly compromised its mRNA export function. The understanding of how different karyopherins recognize human NXF1, the examination of NXF1 sequences from divergent eukaryotes, and the interactions of NXF1 homologues with various karyopherins reveals the evolutionary development of redundant NLSs in NXF1 of higher eukaryotes. Redundancy of nuclear import pathways for NXF1 increases progressively from fungi to nematodes and insects to chordates, potentially paralleling the increasing complexity in mRNA export regulation and the evolution of new nuclear functions for NXF1.
INTRODUCTION
The transport of mRNA from the site of transcription in the nucleus to the site of translation in the cytoplasm is an essential process in eukaryotic gene expression. In human cells, the mRNA export factor NXF1 (also known as TAP) escorts mRNA transcripts out of the nucleus by simultaneously binding mRNA, mRNA adaptor proteins, and phenylalanine–glycine (FG) repeats of the nuclear pore complex (NPC; Stutz and Izaurralde, 2003; Erkmann et al., 2005; Reed and Cheng, 2005; Kohler and Hurt, 2007; Hautbergue et al., 2008; Carmody and Wente, 2009; Kelly and Corbett, 2009). NXF1 is unique among nuclear transport factors, as it is a multidomain protein that bears no structural or mechanistic resemblance to the karyopherin proteins that transport protein cargoes, tRNAs, and microRNAs through the NPC. mRNA export by NXF1 is a process that occurs independent of the GTPase Ran (Gruter et al., 1998).
Human NXF1 (hsNXF1) contains a 110-residue N-terminal tail that precedes four well-characterized globular domains (Figure 1A; Liker et al., 2000; Fribourg et al., 2001; Fribourg and Conti, 2003; Grant et al., 2002; Ho et al., 2002; Senay et al., 2003; Stutz and Izaurralde, 2003). The RNA-binding (RBD) and leucine-rich repeat (LRR) domains bind constitutive transport element (CTE)–containing viral RNAs (Braun et al., 1999). The two domains are also involved in binding cellular mRNAs, likely with the help of adaptor proteins (Bachi et al., 2000; Strasser and Hurt, 2000; Stutz et al., 2000; Huang et al., 2003; Hautbergue et al., 2008). Beyond the two domains that bind RNA are the NTF2-like and UBA domains. The heterodimer of NTF2-like domain with NXT1 and the UBA domain bind FG repeats of nucleoporins to target NXF1 to the NPC for translocation (Santos-Rosa et al., 1998; Katahira et al., 1999; Fribourg et al., 2001; Grant et al., 2002; Senay et al., 2003). The N-terminal tail is the least-well-characterized region of hsNXF1. The tail is predicted to be structurally disordered and contains a 10-residue segment that is critical for targeting hsNXF1 to the nucleus (Bear et al., 1999; Braun et al., 1999; Kang et al., 1999; Katahira et al., 1999; Truant et al., 1999; Bachi et al., 2000). This segment was later identified as part of a proline–tyrosine nuclear localization signal (PY-NLS) that binds the importin karyopherin β2 (Kapβ2 or transportin) (Lee et al., 2006; Imasaki et al., 2007). The N-terminal tail also contributes to interactions with adaptor proteins E1B-AP5, ALY/REF, SR proteins, and NS1, the influenza virus protein, which inhibits mRNA export (Bachi et al., 2000; Stutz et al., 2000; Huang et al., 2003).
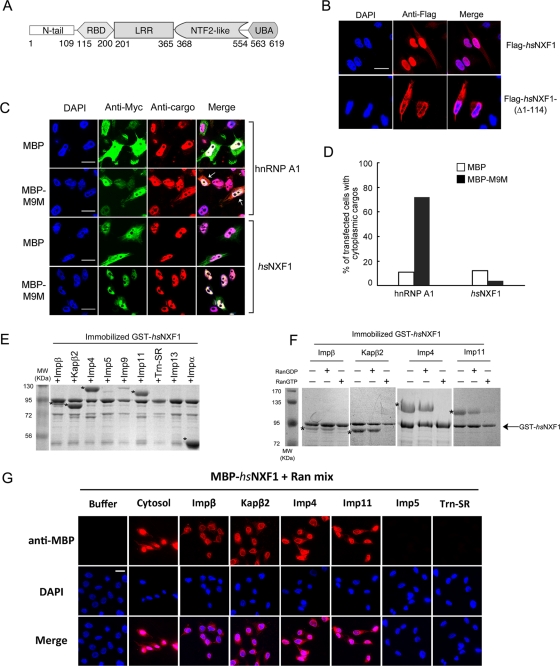
FIGURE 1:
hsNXF1 is imported into the nucleus by multiple karyopherins. (A) The domain organization of hsNXF1. (B) The N-terminal tail of hsNXF1 is necessary for its nuclear localization. Full-length hsNXF1 and deletion mutant hsNXF1-(Δ1-114) were cloned into pFLAG-CMV2 vectors and transfected into HeLa cells. The overexpressed proteins were detected by immunofluorescence using anti-FLAG antibodies. Scale bar, 10 μm. (C) Kapβ2 inhibitor M9M did not alter the subcellular localization of hsNXF1. HeLa cells were transfected with myc-tagged MBP or MBP-M9M, and endogenous Kapβ2 cargoes hnRNP A1 and hsNXF1 were detected by immunofluorescence. Scale bars, 10 μm. (D) Quantitation of C; ∼50 transfected cells from each group were analyzed, and the percentage of cells with significant cytoplasmic cargoes is shown as the bar graph. (E) hsNXF1 interacts with karyopherins Impβ, Kapβ2, Imp4, Imp11, and Impα in pull-down binding assays. Immobilized GST-hsNXF1 was incubated with purified recombinant karyopherins. Bound proteins were visualized by SDS–PAGE and Coomassie staining. (F) Kapβ–hsNXF1 interactions are RanGTP sensitive. Immobilized GST-hsNXF1 was first incubated with karyopherins, washed extensively, and then incubated with buffer, RanGDP, or RanGTP. Bound proteins in E and F were visualized using Coomassie staining. Asterisks in E and F indicate bound Karyopherins. (G) Impβ, Kapβ2, Imp4, and Imp11 are able to import hsNXF1 into HeLa cell nucleus. Nuclear import assays were performed in digitonin-permeabilized HeLa cells with MBP-hsNXF1 in the presence of purified Kapβs or buffer. Samples were fixed and stained with anti-MBP antibody and Alexa 546 anti-mouse secondary antibody, then subjected to immunofluorescence analysis. Scale bar, 10 μm.
PY-NLSs are generally 15–30 amino acids long, are basic in character, are found in structurally disordered regions of proteins, and usually contain an N-terminal basic or hydrophobic motif and a C-terminal R-X2-5-P-Y motif (Lee et al., 2006; Cansizoglu et al., 2007; Suel et al., 2008; Suel and Chook, 2009). Kapβ2 binds PY-NLSs with high affinity (Kd ≈ 10–50 nM) to target import cargoes for translocation through the NPC. RanGTP releases PY-NLSs from Kapβ2 in the nucleus. The crystal structure of Kapβ2 bound to a 30-residue fragment of the hsNXF1 PY-NLS showed interactions with only 10 residues immediately surrounding the C-terminal R-x2-5-P-Y motif but not with an N-terminal basic/hydrophobic motif (Imasaki et al., 2007).
Here we report that biochemical and cellular studies to understand the energetic organization of the hsNXF1 PY-NLS unexpectedly revealed that the mRNA export factor is imported into the nucleus via five different karyopherin pathways. hsNXF1 can be imported into the nucleus through the interactions of its N-terminal tail with importin β (Impβ), Kapβ2, Imp4, Imp11, and Impα. Within the N-terminal tail of hsNXF1, an N-terminal basic NLS epitope spanning residues 21–30 is important for binding Impα and for direct interactions with Impβ, Imp4, and Imp11, whereas the R-X2-5-P-Y motif at residues 71–75 is important for Kapβ2 binding. Mutation of both NLS epitopes abolished binding to all five karyopherins, mislocalized hsNXF1 to the cytoplasm, and significantly compromised its functions in gene expression. The understanding of how different karyopherins recognize hsNXF1 and how different karyopherins bind NXF1 proteins from various organisms and the examination of diverse NXF1 sequences revealed the evolutionary development of redundant NLSs in the mRNA export factors. The redundancy of nuclear import pathways for NXF1 increases with the complexity of the eukaryote, suggesting parallel evolution of new nuclear functions for NXF1.
RESULTS
Multiple karyopherins mediate nuclear import of human NXF1
In human cells, hsNXF1 is localized mostly to nucleoplasm and the NPC (Bear et al., 1999; Katahira et al., 1999; Bachi et al., 2000). Despite the ability of hsNXF1 to interact with the NPC through its C-terminal NTF2-like and UBA domains (Santos-Rosa et al., 1998; Katahira et al., 1999; Fribourg et al., 2001; Grant et al., 2002; Senay et al., 2003), a minimal nonclassic NLS spanning residues 61–102 in the N-terminal tail was found to be critical for its nuclear localization through nuclear import by Kapβ2 (Bear et al., 1999; Kang and Cullen, 1999; Katahira et al., 1999; Truant et al., 1999; Bachi et al., 2000). Consistent with these previous findings, we showed that full-length hsNXF1 was localized in the nucleus but that a mutant lacking the N-terminal tail was cytoplasmic (FLAG-hsNXF1(Δ1-114); Figure 1B). Because hsNXF1 is a well-established Kapβ2 cargo (Truant et al., 1999; Bachi et al., 2000; Lee et al., 2006; Imasaki et al., 2007), we expressed the Kapβ2-specific peptide inhibitor M9M (Cansizoglu et al., 2007) in HeLa cells to determine whether Kapβ2 is the main nuclear import factor for hsNXF1. Surprisingly, myc–maltose-binding protein (MBP)-M9M failed to mislocalize hsNXF1 to the cytoplasm even though the inhibitor mislocalized other Kapβ2 cargoes such as hnRNP A1 (Figure 1, C and D), hnRNP M, HIV-1 Rev, and FUS to the cytoplasm (Cansizoglu et al., 2007; Hutten et al., 2009; Dormann et al., 2010). These results suggested that Kapβ2 is not the sole nuclear importer of hsNXF1.
To identify additional nuclear import factors for hsNXF1, we tested its binding to most of the known human import-karyopherins. Immobilized glutathione S-transferase (GST)–hsNXF1 bound recombinant Impβ, Kapβ2, Imp4, Imp11, and Impα with apparent stoichiometric binding, as shown by strong Coomassie-stained bands of the five karyopherins (Figure 1E). hsNXF1 did not bind recombinant Imp5, Imp9, Trn-SR, or Imp13 (Figure 1E). Interactions with Impβ, Kapβ2, Imp4, Imp11, were Ran sensitive, as subsequent incubations with RanGTP released hsNXF1 from the karyopherins (Figure 1F). Impβ, Kapβ2, Imp4, and Imp11 also mediated nuclear import of MBP-hsNXF1 in digitonin-permeabilized HeLa cells (Figure 1G). However, Imp5 and Trn-SR, which did not bind hsNXF1, could not import MBP-hsNXF1 (Figure 1G). Impα was not tested in the nuclear import assays since its effect cannot be distinguished from that of direct hsNXF1–Impβ interactions. Results of the karyopherin-binding and nuclear import assays suggested that in addition to the well-established Kapβ2 pathway, hsNXF1 can be imported into the nucleus through direct interactions with Impβ, Imp4, and Imp11 and via the classic Impα/β pathway.
NLSs for Impβ, Kapβ2, Imp4, Imp11, and Impα reside within the hsNXF1 N-terminal tail
We divided the multidomain hsNXF1 into its modular domains based on available structural information (Liker et al., 2000; Fribourg et al., 2001; Fribourg and Conti, 2003; Grant et al., 2002; Ho et al., 2002; Senay et al., 2003). Immobilized GST fusions of the N-terminal tail (hsNXF1-N; residues 1–109), the RBD (residues 115–200), LRR (residues 201–365), NTF2-like (residues 368–554), and UBA (residues 563–619) domains were tested for binding to Impβ, Kapβ2, Imp4, Imp11, and Impα (Figure 2, A and B, and Supplemental Figure S1). All five karyopherins bound to hsNXF1-N but not to the other domains. Impβ, Kapβ2, Imp4, and Imp11 mediated nuclear import of hsNXF1-N into the nucleus of digitonin-permeabilized HeLa cells (Supplemental Figure S2A). hsNXF1-N also targeted pyruvate kinase to the HeLa cell nuclei (Supplemental Figure S2B), whereas hsNXF1 lacking its N-terminal tail was cytoplasmic (Figure 1B). These results suggested that all the NLSs in hsNXF1 that are recognized by Impβ, Kapβ2, Imp4, Imp11, and Impα are located within its N-terminal tail.
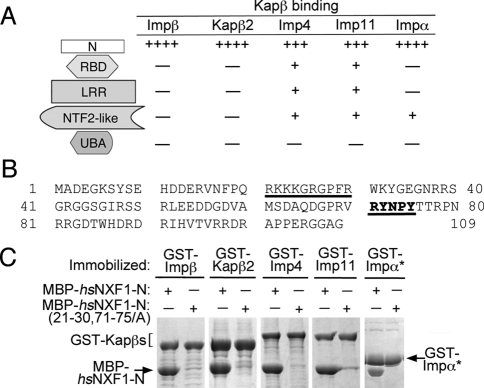
FIGURE 2:
The NLSs of hsNXF1 for Impβ, Kapβ2, Imp4, Imp11, and Impα are all located in the N-terminal tail (hsNXF1-N). (A) Summary of the pull-down binding assays of hsNXF1 domains with Impβ, Kapβ2, Imp4, Imp11, and Impα (data shown in Supplemental Figure S1). The number of plus signs indicates the relative binding strength, and the minus sign indicates no significant binding. (B) The sequence of hsNXF1-N. The two NLS epitopes identified by alanine scanning mutagenesis and ITC (Supplemental Figures S3–S7 and Table 1) are underlined. (C) Alanine mutations at both NLS epitopes of hsNXF1 eliminated binding to Impβ, Kapβ2, Imp4, Imp11, and Impα. Immobilized GST-karyopherins were incubated with MBP-hsNXF1-N or mutant MBP-hsNXF1-N(21-30, 71-75/A). Bound proteins were visualized by SDS–PAGE and Coomassie staining. GST-Impα* refers to Impα without its N-terminal IBB domain (residues 75–529).
Two NLS epitopes contribute differently to interactions with Impβ, Kapβ2, and Impα
hsNXF1 contains a PY-NLS that interacts with Kapβ2 (Lee et al., 2006; Imasaki et al., 2007). Interactions between hsNXF1 residues 68–79, which contain a R-X2-5-P-Y motif, were observed in the crystal structure of Kapβ2 bound to residues 53–82 of hsNXF1 (Imasaki et al., 2007). The absence of electron density for residues 53–67 of hsNXF1 in their structure suggested that a previously predicted hydrophobic motif at 59VAMS62 contributed little to hsNXF1-Kapβ2 interactions and may not be the N-terminal hydrophobic motif of the PY-NLS. We used in vitro pull-down binding assays, isothermal calorimetry (ITC), deletion, and scanning alanine mutagenesis to study the energetic organization of the hsNXF1 PY-NLS. hsNXF1-N bound Kapβ2 with a Kd of 40.5 nM (Table 1 and Supplemental Figure S3; the hsNXF1-N sequence is shown in Figure 2B). The N- and C-terminal truncations mapped residues 1–92 as the smallest hsNXF1 fragment that maintains the high-affinity Kapβ2-binding of hsNXF1-N (Kd of 54 nM; Supplemental Table S1). We used scanning alanine mutagenesis and qualitative pull-down binding assays to identify binding determinants or NLS epitopes in the hsNXF1 PY-NLS (Supplemental Figure S4, A and B). The results suggested binding hotspots at residues 71–75 and 21–30 (Supplemental Figure S4, A and B). We then used ITC to measure the energetic contributions of these potential NLS epitopes. Mutation of residues 71–75 to alanines reduced hsNXF1-N–Kapβ2 affinity by approximately fivefold, whereas mutation of the basic patch spanning hsNXF1 residues 21–30 resulted in approximately threefold affinity reduction (Table 1 and Supplemental Figure S3). These results confirmed that the C-terminal R-X2-5-P-Y motif at 71RYNPY75 as a hotspot for binding Kapβ2 and that hsNXF1 contains a PY-NLS of the basic subclass with its N-terminal basic motif at residues 21–30.
TABLE 1:
Binding affinity of hsNXF1-N proteins for Kapβ2 and Impβ.
| Kapβ | hsNXF1-N | Kda (nM) | ΔH (kcal/mol) | TΔSb (kcal/mol/K) | Kd,mutant/Kd,wild type |
|---|---|---|---|---|---|
| Kapβ2 | Wild type | 40.5 ± 12.6 | −17.21 ± 1.25 | −7.31 ± 1.42 | — |
| 21-30/A | 127.9 ± 19.5 | −13.93 ± 2.65 | −4.68 ± 2.56 | 3.2 | |
| 71-75/A | 215.3.8 ± 61.4 | −12.91 ± 1.12 | −3.98 ± 1.27 | 5.4 | |
| 21-30, 71-75/A | n.d. | n.d. | n.d. | ≫6c | |
| Impβ | Wild type | Kd1 6 ± 5 Kd2 1519 ± 225 | −7.51 ± 0.07 −4.60 ± 0.55 | 3.40 ± 0.58 3.20 ± 0.49 | — |
| 21-30/A | 2234 ± 52 | −2.48 ± 0.52 | 5.09 ± 0.53 | 372d | |
| 71-75/A | Kd1 6 ± 2 | −7.73 ± 0.28 | 3.26 ± 0.48 | 1 (Kd1) | |
| Kd2 2461 ± 723 | −3.28 ± 0.58 | 4.22 ± 0.60 | 2 (Kd2) | ||
| 21-30, 71-75/A | n.d. | n.d. | n.d. | >400d,e |
n.d., not detectable. All experiments were performed three to five times (±SD).
aStoichiometry, 0.9–1.1.
bTΔS = ΔH – ΔG.
cThe lowest measurable Kd of 215 nM in the Kapβ2–hsNXF1-N series was used to estimate Kd,mutant/Kd,wild type.
dRatio taken using Kd1,wild type.
eThe lowest measurable Kd of 2.23 μM in the Impβ–hsNXF1-N series was used to estimate Kd,mutant/Kd,wild type.
hsNXF1-N also binds Impβ with high affinity. The Impβ-binding isotherm of MBP-hsNXF1-N fit a two-site binding model (χ2 ≈ 1.78 × 104), with Kd values of 6 nM and 1.5 μM, respectively (Table 1 and Supplemental Figure S3). N- and C-terminal deletion mutants and alanine scanning mutagenesis of hsNXF1-N suggested that the binding energy for Impβ was likely concentrated in the first 40 residues of hsNXF1, with small contributions from residues 70–109 (Supplemental Figure S5, A–C). MBP-hsNXF1-N(21-30/A) showed no detectable binding by ITC, suggesting that the 21RKKKGRGPFR30 basic patch was indeed essential for interactions with Impβ (Table 1 and Supplemental Figure S3). Mutations of 71RYNPY75 to alanines had no effect on Impβ binding (Table 1 and Supplemental Figure S3).
The 21RKKKGR26 segment of hsNXF1 matches the K-K/R-X-K/R consensus sequence for the monopartite classic NLS (Chelsky et al., 1989; Hodel et al., 2001; Lange et al., 2007; Yang et al., 2010). Furthermore, scanning alanine mutagenesis revealed an Impα-binding hotspot at residues 21–30 (Supplemental Figure S6), suggesting that 21RKKKGR26 might indeed be a bona fide monopartite classic NLS. Scanning alanine mutagenesis of MBP-hsNXF1-N suggested that the interaction between hsNXF1-N and Imp11 is more distributed (Supplemental Figure S7). Similar experiments were unsuccessful with Imp4 due to instability of the immobilized GST-Imp4.
Collectively, the foregoing results showed that interactions of hsNXF1 with Kapβ2, Impβ, and Impα are differentially mediated by two distinct NLS epitopes. The R-X2-5-P-Y motif at residues 71–75 of hsNXF1 is important for Kapβ2 binding, whereas the 21RKKKGRGPFR30 basic patch contributes significantly to interactions with Impβ and Impα.
Mutation of the two NLS epitopes abolished karyopherin binding, mislocalized hsNXF1 in cells, and compromised gene expression
Mutations of the two NLS epitopes 21RKKKGRGPFR30 and 71RYNPY75 to alanines in MBP-hsNXF1-N(21-30,71-75/A) abolished binding to all five karyopherins (Figure 2C). To determine the importance of the NLS epitopes for nuclear import, we transfected pyruvate kinase and enhanced green fluorescent protein (EGFP) fusions of full-length hsNXF1 proteins into HeLa cells (Figure 3A and Supplemental Figure S9). Pyruvate kinase (PK) alone localized to the cytoplasm, whereas PK-hsNXF1 appeared exclusively in the nucleus. Mutations of the individual NLS epitopes in PK-hsNXF1(21-30/A) showed some cytoplasmic NXF1, PK-hsNXF1(71-75/A) showed more cytoplasmic mislocalization, and mutation of both epitopes in PK-hsNXF1(21-30,71-75/A) showed extensive cytoplasmic mislocalization. NLS epitope mutants of hsNXF1-N-PK and EGFP-hsNXF1 showed similar mislocalization patterns as the PK-hsNXF1 mutants in HeLa cells (Supplemental Figures S8 and S9). The 71-75/A mutation had a larger effect than the 21-30/A mutation, suggesting that of the five karyopherins that import hsNXF1, Kapβ2 likely played the most important role.
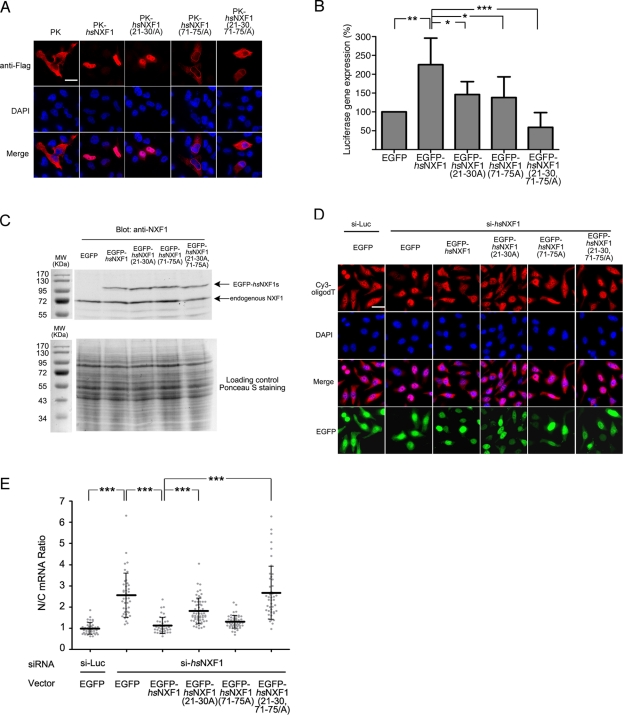
FIGURE 3:
NLS mutations impair nuclear localization of hsNXF1 and its ability to activate luciferase gene expression and export mRNA. (A) FLAG-tagged pyruvate kinase (PK)–hsNXF1 and its NLS mutants were transfected into HeLa cells. Localization of PK fusion proteins were detected by deconvolution microscope. Scale bars, 10 μm. (B) Luciferase reporter gene expression assays of hsNXF1 and its NLS mutants. EGFP-hsNXF1 proteins were cotransfected with pCMV-Luc vector, and the expression levels of the luciferase gene were calculated by normalizing the luciferase signals that were detected by Luciferase Assay System to CellTiter-Glo signals. The results are averages of six independent experiments ± SD. *p < 0.05; **p < 0.01; ***p < 0.001. (C) Expression levels of EGFP-hsNXF1 proteins and endogenous hsNXF1. (D) Wild type and mutant of pEGFP-C1-hsNXF1 vectors were cotransfected with siRNA targeting hsNXF1 for 48 h, and the localization of mRNA was detected by Cy3-oligodT. Scale bar, 10 μm. (E) Quantitation of C; the average nucleus/cytoplasm (N/C) ratios of Cy3 signals in ∼50 transfected cells from each group are shown as middle horizontal bars ± SD. Gray dots, data points. ***p < 0.001.
To determine whether nuclear import of hsNXF1 is critical for mRNA export, we knocked down endogenous NXF1 and examined the rescue effects of wild-type and mutant EGFP-hsNXF1 vectors. Knockdown of NXF1 in HeLa cells by RNA interference caused significant accumulation of mRNA in the nucleus, which indicates that hsNXF1 is essential for mRNA export in mammalian cells (Figure 3, D and E, and Supplemental Figure S10). This defect can be fully rescued by overexpressing wild-type EGFP-hsNXF1 and the EGFP-hsNXF1(71-75/A) mutant (Figure 3, D and E). Rescue by the EGFP-hsNXF1(21-30/A) mutant was partially impaired, and simultaneous mutation of both NLS epitopes in EGFP-hsNXF1(21-30,71-75/A) abolished the rescue ability (Figure 3, D and E). To determine whether nuclear import of hsNXF1 is important for NXF1-mediated gene expression, we examined how overexpressed hsNXF1 and its import mutants affected stimulation of luciferase reporter gene expression. As expected, overexpression of EGFP-hsNXF1 stimulated gene expression (Figure 3B; Gruter et al., 1998; Herold et al., 2000; Braun et al., 2001; Satterly et al., 2007). hsNXF1-mediated stimulation of gene expression was decreased when either the hsNXF1 basic patch (21RKKKGRGPFR30) or its R-X2-5-P-Y motif at residues 71–75 was mutated. Simultaneous mutation of both 21RKKKGRGPFR30 and 71RYNPY75 epitopes lowered gene expression to the level of the EGFP control. These results showed that nuclear import of hsNXF1 is critical for its activity in mRNA export and in mediating gene expression.
Potential NLS epitopes of NXF1 proteins in diverse eukaryotes
Understanding how different karyopherins recognize hsNXF1 was a necessary prerequisite to the identification of potential NLS epitopes in the N-terminal tails of different eukaryotic NXF1s. The sequence of residues 1–200 of hsNXF1 was used to identify NXF1 homologues by BLAST (Altschul et al., 1990). Sequences were available for NXF1 homologues from vertebrates, lancelets, tunicates, echinoderms, nematodes, insects, and fungi. We examined the sequences of NXF1s from fungi (budding yeast, Saccharomyces cerevisiae; fission yeast, Schizosaccharomyces pombe and Schizosaccharomyces japonicus) and animals (nematodes, Caenorhabditis elegans and Caenorhabditis briggsae; insects, Drosophila melanogaster, Drosophila grimshawi, Anopheles gambiae, and Aedes aegypti; and chordates, Homo sapiens, Xenopus tropicalis, Danio rerio, Branchiostoma floridae, and Ciona intestinalis). Although these NXF1 homologues share ∼30% sequence identities and have the same domain organization, their N-terminal tails shared no significant sequence homology (Altschul et al., 1990). In fact, their NXF1-Ns vary considerably in length. For example, the NXF1 homologue in S. cerevisiae Mex67p has a short, 20-residue N-terminal tail, whereas NXF1s of fission yeast, S. pombe (spMex67p) and S. japonicus, contain N-terminal tails that are 40–50 residues long. The N-terminal tails of animal NXF1s are generally longer than 100 residues (Figure 4).
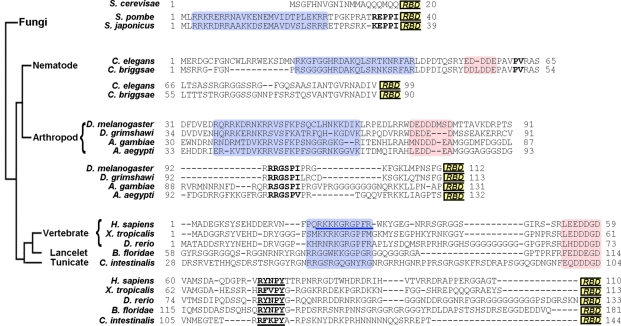
FIGURE 4:
Potential NLS epitopes of NXF1 proteins in diverse eukaryotes. Residues 1–200 of hsNXF1 were used to identify NXF1 homologues by BLAST. Sequences were available for NXF1 homologues from vertebrates, lancelets, tunicates, echinoderms, nematodes, insects, and fungi. Because the NXF1-Ns of divergent organisms shared no significant sequence homology and vary considerably in length, closely related NXF1s from within the groups fission yeast, nematodes, insects, and chordates were aligned by ClustalW. The NXF1-N groups were examined for sequence trends similar to the NLS epitopes in the hsNXF1-N. The four divergent groups show similar organizations of motifs. N-Terminal basic patches are shaded blue, with the N-terminal basic NLS epitope in hsNXF1 underlined in blue; central acidic patches are shaded pink; the C-terminal R/K/P-x2-5-PΦ motifs (Φ is a hydrophobic amino acid) are in bold, with the R-x2-5-P-Y motifs of chordate NXF1s underlined. RBD boxes indicate the beginning of RBD domains.
Instead of generating an alignment of all the very diverse NXF1-Ns, we aligned groups of closely related NXF1s from budding and fission yeasts, nematodes, insects, and chordates (Figure 4; Chenna et al., 2003; Dunn et al., 2008). We examined the NXF1-N groups for sequence/motif trends that are similar to the hsNXF1 NLS epitopes that we characterized. In particular, we looked for basic patches that resembled the basic NLS epitope of hsNXF1-N, segments that resembled the R-X2-5-P-Y epitope of hsNXF1-N, and the seven-residue acidic region that resides between the two NLS epitopes.
Interactions between karyopherins and the N-terminal tails of chordate NXF1s
NXF1s of the five chordates that we examined (H. sapiens, X. tropicalis, D. rerio, B. floridae, and C. intestinalis) shared basic patches homologous to the 21RKKKGRGPFR30 basic patch of hsNXF1, acidic regions that aligned with 53LEEDDGD59 of hsNXF1, and the R-X2-5-P-Y motifs (Figure 4). In fact, the R-X2-5-P-Y motifs of all five chordate NXF1s matched the R-Y/F-X-P-Y consensus that is characteristic of energetically strong R-X2-5-P-Y motifs (Suel et al., 2008; Suel and Chook, 2009). Pull-down binding assays with recombinant NXF1-Ns showed that Kapβ2 bound human, X. tropicalis, and D. rerio NXF1-Ns (hsNXF1-N, xtNXF1-N, and drNXF1-N, respectively) and their R-X2-5-P-Y motifs were critical for the interactions (Table 1 and Figure 5, A–C). Impβ bound strongly to hsNXF1-N but more weakly to X. tropicalis and D. rerio NXF1-Ns (Table 1 and Figure 5, A and B). Imp4 bound hsNXF1-N but not X. tropicalis and D. rerio NXF1-Ns, and all three vertebrate NXF1-Ns bound Imp11 (Figure 5A).
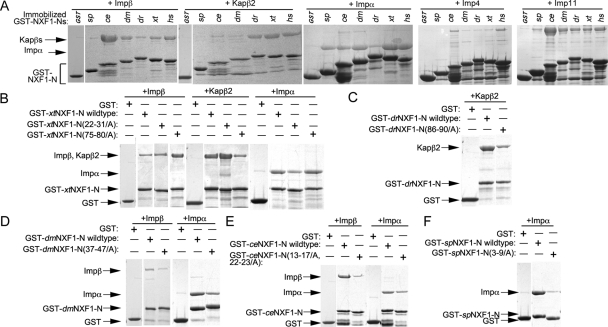
FIGURE 5:
Interactions of NXF1-Ns from different organisms with karyopherins. (A) Immobilized GST-NXF1-Ns of S. pombe, C. elegans, D. melanogaster, D. rerio, X. tropicalis, and H. sapiens were incubated with purified recombinant Impβ, Kapβ2, Impα, Imp4, or Imp11. Bound proteins were visualized by SDS–PAGE and Coomassie staining. Mutations within the N-terminal basic patches and C-terminal R-x2-5-P-Y motifs of NXF1-Ns from X. tropicalis (B), D. rerio (C), D. melanogaster (D), C. elegans (E), and S. pombe (F) were tested for interactions with Impβ, Kapβ2, or Impα. Bound proteins in A–F were visualized using Coomassie staining.
The monopartite classic NLS in 21RKKKGR26 of hsNXF1 is preserved in 22KKRKGR27 of xtNXF1, but no monopartite or bipartite classic NLSs are evident in the N-terminal tails of D. rerio, B. floridae, or C. intestinalis NXF1s (Figure 4). These observations were supported by pull-down binding assays that showed binding of Impα to the basic patch of the human and X. tropicalis NXF1-Ns but not to that of the D. rerio NXF1-N (Figure 5, A and B). These results suggested that many chordate NXF1s are likely Kapβ2 and Imp11 cargoes, and some are also imported into the nucleus by Imp4 and through direct Impβ binding and/or by the classic Impα/β pathway.
Interactions between karyopherins and insect and nematode NXF1s
Nematode (C. elegans and C. briggsae) and insect (D. melanogaster, D. grimshawi, A. gambiae, and A. aegypti) NXF1s appear to all have N-terminal basic patches followed by small acidic regions but not R-X2-5-P-Y motifs (Figure 4). Instead, insect NXF1s have PY-like R-X3-P-I/V motifs C-terminal of their acidic regions that could potentially bind Kapβ2. The two nematodes have PAVPV segments that showed poor resemblance to the R-X2-5-P-Y motif (Suel et al., 2008). Pull-down binding assays showed that neither C. elegans nor Drosophila NXF1-Ns (ceNXF1-N and dmNXF1-N, respectively) bound Kapβ2 (Figure 5A), suggesting that the PY-like R-X3-P-I/V motifs in insect NXF1s are poor substitutes for the R-X2-5-P-Y motif of PY-NLSs. In contrast, the basic patches in NXF1-Ns from both C. elegans and Drosophila contribute to direct interactions with Impβ (Figure 5, A, D, and E). Imp4 bound ceNXF1-N but not dmNXF1-N, and Imp11 bound to both ceNXF1-N and dmNXF1-N (Figure 5A).
Although there are no obvious monopartite classic NLSs in the C. elegans and Drosophila NXF1-Ns, the sequences of their basic patches matched the bipartite classic NLS consensus sequence of K/R-K/R-X10–12-K/R3/5, where K/R3/5 represents three lysine or arginine residues out of five consecutive amino acids (Figure 4; Dingwall and Laskey, 1991). These observations were supported by pull-down binding assays that showed binding of Impα to C. elegans and Drosophila NXF1-Ns (Figure 5A). Mutations of residues in the basic patches of both NXF1s decreased Impα binding (Figure 5, D and E). Collectively, these results showed that the classic Impα/β and direct Impβ pathways rather than the Kapβ2 pathway likely mediate nuclear import of NXF1 in nematodes and insects.
Interactions between karyopherins and the N-terminal tails of S. pombe NXF1
The shorter N-terminal tails of fission yeast (S. pombe and S. japonicus) Mex67p contain N-terminal basic patches but no acidic regions or R-X2-5-P-Y motifs (Figure 4). R/K-X2-P-I segments at the C-terminal end of the tails most closely resembled the R-X2-5-P-Y motif of a PY-NLS. The basic patches appeared to contain bipartite classic NLSs (Figure 4). Pull-down binding assays showed that Impα binds to the S. pombe Mex67p N-terminal tail or spNXF1-N, binds very weakly to Imp11, and does not bind Impβ, Kapβ2, or Imp4 (Figure 5, A and F). These results suggested that nuclear import of NXF1 in S. pombe is most likely to be mediated by the classic Impα/β pathway.
Summary of karyopherin binding to NXF1-Ns from diverse organisms
The trend of NXF1-Ns binding to human karyopherins is conserved in binding assays using S. cerevisiae Kap95p, Kap60p, and Kap104p (Supplemental Figure S11). Binding analysis of diverse NXF1-Ns showed that the numbers of redundant NLSs in NXF1s and the karyopherins that mediate their nuclear localization increase progressively from fungi to nematodes and insects to chordates (Table 2). The S. cerevisiae NXF1 contains neither N-terminal tail nor NLS. The S. pombe NXF1 appeared to use the classic Impα/β pathway. Nematodes and insects used the classic Impα/β and direct Impβ and Imp11 pathways, and chordates used three to five different nuclear import pathways to target their NXF1s to the nucleus.
TABLE 2:
Summary of interactions between karyopherins and the N-terminal tails of NXF1s from diverse eukaryotes.
| NXF1-Na | Kapβ2 | Impβ | Imp4 | Imp11 | Impα |
|---|---|---|---|---|---|
| Human | ++++ | ++++ | +++ | +++ | ++++ |
| X. tropicalis | ++++ | +++ | − | +++ | ++++ |
| D. rerio | ++++ | + | − | +++ | + |
| D. melanogaster | − | +++ | − | +++ | +++ |
| C. elegans | − | +++++ | ++ | +++++ | +++ |
| S. pombe | − | − | − | + | +++ |
aBinding data are shown in Figure 5A.
DISCUSSION
hsNXF1 is a well-established nuclear import cargo of Kapβ2 (Truant et al., 1999; Bachi et al., 2000; Lee et al., 2006; Imasaki et al., 2007). The karyopherin binds a PY-NLS in the N-terminal tail of hsNXF1 (Lee et al., 2006). Through extensive mutagenesis, qualitative and quantitative binding assays, we showed that the PY-NLS of hsNXF1 spans residues 1–92, binds Kapβ2 with a KD of 40 nM, and is a member of the basic and not the previously predicted hydrophobic subclass of PY-NLSs. We identified binding determinants or NLS epitopes in two distinct segments of hsNXF1 that correspond to an N-terminal basic epitope at residues 21–30 and the R-X2-5-P-Y motif at residues 71–75. The latter is a marginal hotspot whereby mutation of the entire five-residue motif decreased Kapβ2 binding by fivefold, whereas mutation of the former decreased affinity by threefold. The basic/hydrophobic and R-X2-5-P-Y epitopes of previously identified PY-NLSs are connected by 3- to 11-residue-long linkers (Lee et al., 2006). The unusually long 40-residue PY-NLS linker in hsNXF1 significantly extends previous limits for linker length without compromising high-affinity interactions with Kapβ2.
Surprisingly, inhibition of Kapβ2 by the M9M peptide inhibitor did not mislocalize endogenous hsNXF1 in HeLa cells, suggesting that Kapβ2 is not its sole nuclear import factor. We showed that the N-terminal tail of hsNXF1 contains multiple redundant and overlapping NLSs that are recognized by Kapβ2, Impβ, Imp4, Imp11, and Impα. The five karyopherins differentially bind the same two NLS epitopes that are recognized by Kapβ2. The basic patch at residues 21–30 is used in interactions with all five karyopherins, whereas the R-X2-5-P-Y motif at residues 71–75 is used only for binding Kapβ2. The overlapping nature of the NLSs suggests that a single molecule of hsNXF1 likely binds only one karyopherin molecule at a time. Mutations of both NLS epitopes greatly diminished nuclear localization of hsNXF1, failed to rescue the nuclear accumulation of mRNA in hsNXF1 knockdown cells, and perturbed NXF1-mediated gene expression as observed by the significant decrease in reporter gene expression.
Our biochemical and biophysical characterization of the hsNXF1 NLS epitopes that bind Kapβ2, Impβ, and Impα allowed extension of these studies to other eukaryotes. The N-terminal tails of NXF1s from fission yeasts, nematodes, insects, and chordates share similar sequence/motif organizations even though they are very diverse in sequence and length. The N-terminal tails of nematode, insect, and chordate NXF1s contain N-terminal basic patches of 10–30 residues, followed by acidic patches of approximately six to eight residues and C-terminal R/K/P-X2-5-P-Φ motifs. The N-terminal tails of two fission yeast NXF1s show similar trends but lack the central acidic patches. No basic, acidic patches or R/K/P-X2-5-P-Φ motifs are present in the N-terminal tail of S. cerevisiae. The N-terminal basic patches of the NXF1s are reminiscent of the N-terminal basic NLS epitope of hsNXF1, whereas their C-terminal R/K/P-X2-5-P-Φ motifs resemble the R-X2-5-P-Y motif of the hsNXF1 PY-NLS. Functions of the central acidic patches are not known.
Individual karyopherins are highly conserved in eukaryotes, both in their sequences and cargo recognition (Enenkel et al., 1995; Lange et al., 2008; Suel et al., 2008; Marfori et al., 2011). The diverse NXF1 N-terminal tails bound similarly to human and S. cerevisiae karyopherins, suggesting that karyopherin specificities for their NLSs are conserved from human to yeast. We found that the number of karyopherins that can mediate nuclear import of NXF1s increased steadily from fungi to nematodes and insects to chordates. Mex67p of S. cerevisiae has no NLS and is known to be localized not to the nucleoplasm but to NPCs (Segref et al., 1997; Katahira et al., 1999). NXF1s from S. pombe, C. elegans, Drosophila, and human are known to be nuclear, and their N-terminal tails are important for their nuclear localization (Bear et al., 1999; Katahira et al., 1999; Bachi et al., 2000; Tan et al., 2000; Yoon et al., 2000; Herold et al., 2001; Wilkie et al., 2001). Mex67p of S. pombe bound mostly Impα, whereas the karyopherin repertoires for C. elegans and D. melanogaster NXF1s were expanded to include Impα, Imp11, and direct interactions with Impβ. The complexity of nuclear import is further increased in chordates, with the use of at least four karyopherins: Impβ, Kapβ2, Imp11, and Impα.
The NLS epitopes recognized by Impβ and Impα are all located within the N-terminal basic patches of the NXF1 proteins, whereas Kapβ2 recognized the R-X2-5-P-Y motifs in chordate (H. sapiens, X. tropicalis, and D. rerio) NXF1s. Of interest, the slightly divergent R/K-X2-P-I, P-X2-P-V, and R-X2-3-P-I/V motifs in S. pombe, C. elegans, and D. melanogaster, respectively, were unsuitable for Kapβ2 binding. Therefore it appears that strong R-X2-5-P-Y motifs evolved only in chordates to expand nuclear import to Kapβ2. The motif, in combination with the more primitive basic patch, produced functional basic PY-NLSs in the NXF1s of these higher eukaryotes, resulting in a total of three to five different nuclear import pathways that target NXF1s to the nuclei of human cells.
It is puzzling that the means of transporting NXF1 into the nucleus are different from S. cerevisiae to humans even though its mRNA export function is conserved. What are the advantages of increased complexity in NXF1 nuclear import or increased redundancy of NXF1 nuclear import pathways in higher eukaryotes? The simplistic suggestion that redundant nuclear import pathways are necessary to ensure correct localization of NXF1 to the nucleus for the crucial process of mRNA export is rather unsatisfactory, given that S. cerevisiae Mex67p has no NLSs and does not need to be localized to the cell nucleus at all. It is more likely that redundant NLSs in NXF1s are important to regulate mRNA export and its coupling to the upstream and downstream gene expression processes of transcription, splicing, and/or translation.
NXF1 binds mRNAs weakly, but the interaction is significantly enhanced by adaptor proteins REF and SR proteins (Hautbergue et al., 2008). In higher eukaryotes, adaptor proteins couple mRNA export to upstream processes of capping and splicing (Izaurralde et al., 1995; Zhou et al., 2000; Masuda et al., 2005; Cheng et al., 2006). Interactions with mRNA and adaptor proteins were mapped to hsNXF1 residues 61–118 and 1–362, respectively (Bachi et al., 2000; Stutz et al., 2000; Huang et al., 2003), thus overlapping significantly with karyopherin binding. In the nucleus, the termination of NXF1 import is likely coupled to its interactions with mRNA, adaptor proteins, and upstream processes of capping and splicing. In the cytoplasm, the karyopherins that import NXF1 may contribute to its release from adaptor proteins and mRNA prior to translation. Furthermore, differential binding of Kapβ2, Impβ, Imp4, Imp11, and Impα to the N- and C-terminal NLS epitopes of hsNXF1 may affect its interactions with various subsets of adaptor proteins, thus providing a means of regulating assembly and disassembly of diverse populations of mRNA export complexes.
Finally, the striking difference in nuclear localization of NXF1 in higher eukaryotes but not in S. cerevisiae may reflect new and still-undetermined functions of NXF1 in the nucleus of higher eukaryotes. The increasing complexity of NXF1 nuclear import in higher eukaryotes may be correlated with similar complexity in nuclear functions of NXF1. The architecture of modular NLS epitopes within the flexible and structurally disordered N-terminal tail of NXF1 may have allowed significant evolvability to form multiple NLSs (Suel et al., 2008). This in turn could have provided a path for NXF1 to switch from using one karyopherin to another and ultimately from one cellular process to another.
MATERIALS AND METHODS
Plasmids
GST fusion constructs were generated by inserting PCR fragments corresponding to the regions of the genes of interest into pGEXTEV vectors (modified pGEX4T3 [GE Healthcare, UK] with TEV site; Chook and Blobel, 1999). The GST fusion constructs include full-length human Impβ, Kapβ2, Imp4, Imp5, Imp9, Imp11, Trn-SR, Imp13, and RanBP1; mouse Impα2-ARM (residues 75–496); full-length human NXF1 or hsNXF1; hsNXF1 fragments hsNXF1-N (residues 1–109), RBD (residues 115–200), LRR (residues 201–365), NTF2-like (residues 368–554), and UBA (residues 563–619), and hsNXF1(1–40), hsNXF1(40-80), hsNXF1(30-80), hsNXF1(1-80), hsNXF1(70-109), and hsNXF1(80-109); and N-terminal tails of X. tropicalis NXF1 (residues 1–115), D. rerio NXF1 (residues 1–136), D. melanogaster NXF1 (residues 1–109), C. elegans NXF1 (residues 1–87), and S. pombe Mex67p (residues 1–31). Synthetic oligonucleotides corresponding to residues 1–87 from C. elegans and residues 1-31 from S. pombe were annealed and inserted into the pGEXTEV vector. MBP fusion constructs of hsNXF1 (full length and fragments) were subcloned from pGEXTEV-hsNXF1 constructs into pMALTEV (modified pMAL [New England BioLabs, Ipswich, MA] with TEV site; Chook et al., 2002) vectors. Mouse Impα2 without the IBB domain (Impα2-ΔIBB, residues 75–529) was cloned into pET21a vector (EMD Biosciences, San Diego, CA). p10, Ran (Chook et al., 2002) were cloned into pET9c vector. Mammalian expressing vectors pEGFP-c1-NXF1 and pCMV-Luc were kindly provided by E. Izaurralde (Max Planck Institute, Tubingen, Germany) and D. Levy (New York University, New York, NY), respectively. The Kapβ2 pathway inhibitor vector pCS2-MT-MBP-M9M and the control vector pCS2-MT-MBP were described previously (Cansizoglu et al., 2007). Full-length hsNXF1 and deletion mutant Δ1-114 were amplified by PCR and cloned into pFLAG-CMV2 vector. Human PK tag was inserted before hsNXF1 gene in pFLAG-CMV2 vectors. NXF1 mutations were made by site-directed mutagenesis using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), and all constructs were sequenced before use.
Recombinant protein preparation
All recombinant proteins were expressed in BL21 (DE3) Escherichia coli cells by induction with 0.5 mM isopropyl-β-d-thiogalactoside overnight at 25°C. For pull-down binding assays, bacteria expressing the GST fusion proteins were lysed by sonication and centrifuged. The supernatants were incubated with glutathione (GSH) Sepharose (GE Healthcare, UK), followed by extensive washes with transfer buffer (TB; 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.3, 110 mM KOAc, 2 mM dithiothreitol [DTT], 2 mM MgOAc, 1 mM ethylene glycol tetraacetic acid) with 20% glycerol. Immobilized GST fusion proteins in TB with 40% glycerol were stored at −20°C. Bacteria expressing GST fusions of Impβ, Kapβ2, Imp4, Imp5, Imp9, Imp11, Trn-SR, and Imp13 were lysed using cell homogenizer EmulsiFlex-C5 (Avestin, Ottawa, Canada) and the proteins purified by GSH affinity chromatography. GST-Imp4 and GST-Imp11 were used for nuclear import assays. For all other experiments, the GST-Kapβs were cleaved with TEV protease and further purified by ion-exchange (HiTrap Q; GE Healthcare, UK) and gel filtration (Superdex 200; GE Healthcare, UK) chromatography. Mouse Impα2-ARM (residues 75–529) and RanBP1 were purified in similar ways (Chook et al., 2002).
To purify MBP fusion proteins, bacterial lysates were incubated with amylose beads (New England BioLabs, Ipswich, MA) and the fusion proteins eluted with 20 mM HEPES, pH 7.5, 50 mM NaCl, 2 mM EDTA, 2 mM DTT, 10% glycerol, and 10 mM maltose. For binding assays with the hsNXF1-N alanine scanning mutants, MBP–hsNXF1-N proteins were concentrated and dialyzed against TB containing 20% glycerol before use. For all other experiments, MBP fusion proteins were further purified by ion-exchange chromatography.
Human Ran and mouse Impα2-ΔIBB were expressed as histidine (His)-tagged proteins and purified by affinity and ion-exchange chromatography (Chook et al., 2002; Dong et al., 2009a, 2009b). For the RanGTP-mediated dissociation assays, recombinant Ran was loaded with either the GTP or GDP (control) before use, as previously described (Suel et al., 2008; Suel and Chook, 2009). His6-NTF2 was purified by affinity chromatography followed by gel filtration (Chook et al., 2002).
In vitro pull-down binding assays
In vitro pull-down binding assays were performed by incubating immobilized GST-fusion proteins with potential binding partners in TB buffer containing 20% glycerol at 4°C for 30 min, followed by extensive washing with the same buffer. Bound proteins were visualized using SDS–PAGE and Coomassie blue staining. Approximately 5 μg of immobilized GST-NXF1 proteins or fragments was incubated with ∼20 μg of purified karyopherins. About half of the bound proteins was loaded for gel analysis. Approximately 10–20 μg of immobilized GST-karyopherins was incubated with ∼20 μg of MBP-NXF1 fragments, and ∼25% of bound proteins was loaded for gel analysis.
RanGTP-mediated dissociation assay
To load Ran with GDP or GTP, ∼8 mg/ml recombinant human Ran was incubated with 4 mM of either GDP or GTP in the presence of 20 mM EDTA and 40 mM DTT for 30 min at room temperature, followed by adding 80 mM MgOAc at 4°C for 15 min. Approximately 5 μg of immobilized GST-NXF1 was first incubated with ∼20 μg of Kapβs for 30 min at 4°C, followed by extensive washing with TB buffer with 20% glycerol. A second incubation was done with 80 μg of RanGDP or RanGTP. After extensive washing with TB buffer with 20% glycerol, half of the bound proteins were separated by SDS–PAGE and visualized with Coomassie blue staining.
Cell culture, transfection, and fluorescence microscopy
HeLa Tet-on cells (Clontech, Mountain View, CA) and 293T cells were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA) at 37°C in a humidified atmosphere containing 5% CO2 in air. Transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After 16 h of transfection, HeLa Tet-on cells were subjected to standard immunostaining procedures as described previously (Cansizoglu et al., 2007) with goat anti–myc-fluorescein isothiocyanate (Bethyl Laboratories, Montgomery, TX), mouse monoclonal antibody 4C2 (a gift from M. Matunis, Johns Hopkins University, Baltimore, MD), goat anti–mouse-Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA), and mouse anti-NXF1 monoclonal antibody 53H8 (Sigma-Aldrich, St. Louis, MO). Cells were stained with 4,6-diamidino-2-phenylindole (DAPI) and then mounted onto slides for imaging. Cells transfected with EGFP fusion proteins were directly stained with DAPI for imaging after fixation and permeabilization. Cells were examined in a DeltaVision RT Deconvolution microscope (Applied Precision, Issaquah, WA) using a 60× oil objective lens. Images were acquired by SoftWoRx software (Applied Precision, Issaquah, WA) and processed with ImageJ software (National Institutes of Health, Bethesda, MD).
Nuclear import assays
HeLa Tet-on cells were grown to 50% confluency on coverslips, washed in cold TB buffer, and permeabilized with 35 mg/ml digitonin on ice for 5 min. Permeabilized cells were incubated with import reaction mixture (5 μM MBP-hsNXF1, 3 μM Ran, 0.3 μM RanBP1, 0.3 μM NTF2, 1 mM GTP, 8 mM magnesium acetate, with or without 5 μM of the individual recombinant karyopherins) for 30 min at room temperature, followed by washing and fixing. The MBP proteins were detected by immunofluorescence using mouse anti-MBP monoclonal antibody.
Isothermal titration calorimetry
Binding affinities of MBP-hsNXF1-N proteins to Impβ and Kapβ2 were quantitated using ITC as described previously (Cansizoglu et al., 2007; Suel et al., 2008). ITC experiments were performed with a MicroCal Omega VP-ITC calorimeter (MicroCal, Northampton, MA). Proteins were dialyzed against buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, and 2 mM β-mercaptoethanol. From 100 to 300 μM MBP-hsNXF1-N proteins was titrated into a sample cell containing 10–20 μM recombinant Impβ or Kapβ2. Most ITC experiments were performed at 20°C with 35 rounds of 8-μl injections. Data were plotted and analyzed using MicroCal Origin software, version 7.0.
Luciferase reporter gene assay and Western blotting
The experiments were performed according to Chakraborty et al. (2006). Briefly, 293T cells grown on 30-mm six-well plates were cotransfected with pCMV-Luc (2 μg) and either wild-type or mutant pEGFP-C1-hsNXF1 (2 μg) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After 12 h of transfection, cells were lysed, and luciferase activities of each sample were measured using Luciferase Assay Reagent (Promega, Madison, WI) in triplicate. CellTiter-Glo assays were performed similarly with CellTiter-Glo reagent (Promega, Madison, WI) according to manufacturer's instructions. Averages of the luciferase signals (SLuc) were divided by the average of CellTiter-Glo signals (SCell) to reduce the difference of cell numbers between samples. The ratio SLuc/SCell was normalized to that of EGFP control (100%) and represented as percentage in the bar graph.
Approximately 25 μg of proteins from cell lysates was loaded onto 12% SDS–PAGE gels and transferred to polyvinylidene fluoride membrane, followed by sequential incubation with mouse anti-NXF1 monoclonal antibody 53H8 (Sigma-Aldrich, St. Louis, MO) and horseradish peroxidase–labeled goat anti-mouse antibody (GE Healthcare, UK). The signals were detected using an Amersham ECL Western Blotting System (GE Healthcare, UK).
Oligo(dT) in situ hybridization
HeLa Tet-on cells (Clontech, Mountain View, CA) were plated on coverslips in 24-well plates 16 h before transfection. The rescue constructs of either wild-type or mutant pEGFP-C1-hsNXF1 containing silent mutations at siRNA targeting region were cotransfected with siRNA targeting hsNXF1 (Dharmacon, Lafayette, CO) into the cells using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instruction. After 48 h, the cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min and permeabilized with 0.2% Triton X-100. The plates were washed in PBS, and oligo(dT) in situ hybridization was performed as described (Chakraborty et al., 2006), except that the cells were incubated at 42°C overnight in a humidifying chamber with a Cy3-labeled oligo(dT) probe (Invitrogen, Carlsbad, CA) at a final concentration of 10 μg/ml. After hybridization, the coverslips were washed twice with 2× SSC (1× SSC contains 0.15 M sodium chloride and 0.015 M sodium citrate, pH 7) and mounted onto slides with ProLong Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA). Images were acquired by SoftWoRx software (Applied Precision, Issaquah, WA) and processed with Image J software (National Institutes of Health, Bethesda, MD).
Sequence alignment
Multiple sequence alignments were performed using ClustalW (Chenna et al., 2003) with manual adjustment. Uniprot accession numbers for the NXF1 or Mex67p sequences are Q9Y8G3 (S. pombe), B6JXN8 (S. japonicas), Q9XVS7 (C. elegans), A8WY32 (C. briggsae), Q9U1H9 (D. melanogaster), B4JKG4 (D. grimshawi), Q7QK79 (A. gambiae), Q17MK6 (A. aegypti), Q9UBU9 (H. sapiens), Q28C94 (X. tropicalis), and Q5CZT0 (D. rerio). GenBank accession numbers are XP_002589241 (B. floridae) and XP_002129680 (C. intestinalis).
Supplementary Material
Acknowledgments
We thank M. Matunis E. Izaurralde, and D. Levy for gifts of antibody and vectors, Zhu Wang for cDNA of C. elegans NXF1, Tao Yue cDNA for D. melanogaster NXF1, and Daniel Verduzco for RNAs of D. rerio NXF1. We thank Alex D'Brot for karyopherin constructs and proteins and Nick Grishin, Rama Ranganathan, and Mike Rosen for discussions. We acknowledge the assistance of the UT Southwestern Live Cell Imaging Facility, a Shared Resource of the Harold C. Simmons Cancer Center, supported in part by a National Cancer Institute Cancer Center Support Grant, 1P30 CA142543-01. This work was funded by National Institutes of Health Grants R01-GM069909 (Y.M.C.) and R01-GM07159, R01-AI079110, and R01-AI089539 (B.M.A.F.), the Welch Foundation (I-1532; Y.M.C.), a Leukemia and Lymphoma Society Scholar Award (Y.M.C.), the UT Southwestern Endowed Scholars Program (Y.M.C.), and the American Heart Association (09PRE2150104; Z.C.Z.).
Abbreviations used:
- BLAST
basic local alignment search tool
- GST
glutathione S-transferase
- Imp
importin
- ITC
isothermal calorimetry
- Kap
karyopherin
- MBP
maltose-binding protein
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- NXF1
nuclear export factor 1
- PK
pyruvate kinase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0222) on September 30, 2011.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachi A, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J, Tan W, Zolotukhin AS, Tabernero C, Hudson EA, Felber BK. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- Braun IC, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Satterly N, Fontoura BM. Nuclear export assays for poly(A) RNAs. Methods. 2006;39:363–369. doi: 10.1016/j.ymeth.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Chook YM, Jung A, Rosen MK, Blobel G. Uncoupling Kapbeta2 substrate dissociation and ran binding. Biochemistry. 2002;41:6955–6966. doi: 10.1021/bi012122p. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009a;16:558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009b;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Sanchez R, Treichel N, Marzluff WF, Kutay U. Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA. 2005;11:45–58. doi: 10.1261/rna.7189205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Conti E. Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15. EMBO Rep. 2003;4:699–703. doi: 10.1038/sj.embor.embor883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RP, Hurt E, Neuhaus D, Stewart M. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat Struct Biol. 2002;9:247–251. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DN, Coburn GA, Kang Y, Cullen BR, Georgiadis MM. The crystal structure and mutational analysis of a novel RNA-binding domain found in the human Tap nuclear mRNA export factor. Proc Natl Acad Sci USA. 2002;99:1888–1893. doi: 10.1073/pnas.042698599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Hutten S, Walde S, Spillner C, Hauber J, Kehlenbach RH. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J Cell Sci. 2009;122:1100–1110. doi: 10.1242/jcs.040154. [DOI] [PubMed] [Google Scholar]
- Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, Yamada M, Sato M. Structural basis for substrate recognition and dissociation by human transportin 1. Mol Cell. 2007;28:57–67. doi: 10.1016/j.molcel.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Kang Y, Bogerd HP, Yang J, Cullen BR. Analysis of the RNA binding specificity of the human tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Corbett AH. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic. 2009;10:1199–1208. doi: 10.1111/j.1600-0854.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Devine SE, Corbett AH. A PY-NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA-binding protein Hrp1. J Biol Chem. 2008;283:12926–12934. doi: 10.1074/jbc.M800898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liker E, Fernandez E, Izaurralde E, Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M, Kobe B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci USA. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senay C, Ferrari P, Rocher C, Rieger KJ, Winter J, Platel D, Bourne Y. The Mtr2-Mex67 NTF2-like domain complex. Structural insights into a dual role of Mtr2 for yeast nuclear export. J Biol Chem. 2003;278:48395–48403. doi: 10.1074/jbc.M308275200. [DOI] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- Suel KE, Chook YM. Kap104p imports the PY-NLS-containing transcription factor Tfg2p into the nucleus. J Biol Chem. 2009;284:15416–15424. doi: 10.1074/jbc.M809384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel KE, Gu H, Chook YM. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008;6:e137. doi: 10.1371/journal.pbio.0060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Zolotukhin AS, Bear J, Patenaude DJ, Felber BK. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Kang Y, Cullen BR. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J Biol Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Zimyanin V, Kirby R, Korey C, Francis-Lang H, Van Vactor D, Davis I. Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA. 2001;7:1781–1792. [PMC free article] [PubMed] [Google Scholar]
- Yang SN, Takeda AA, Fontes MR, Harris JM, Jans DA, Kobe B. Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J Biol Chem. 2010;285:19935–19946. doi: 10.1074/jbc.M109.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Love DC, Guhathakurta A, Hanover JA, Dhar R. Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export. Mol Cell Biol. 2000;20:8767–8782. doi: 10.1128/mcb.20.23.8767-8782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.