The Smc5/6 complex is widely believed to be required for homologous recombination. It is shown that repair defects of Smc5/6 mutants are due to the Nse1-dependent recruitment of dysfunctional complexes to lesions.
Abstract
Of the three structural maintenance of chromosomes (SMC) complexes, Smc5/6 remains the most poorly understood. Genetic studies have shown that Smc5/6 mutants are defective in homologous recombination (HR), and consistent with this, Smc5/6 is enriched at lesions. However, Smc5/6 is essential for viability, but HR is not, and the terminal phenotype of null Smc5/6 mutants is mitotic failure. Here we analyze the function of Nse1, which contains a variant RING domain that is characteristic of ubiquitin ligases. Whereas deletion of this domain causes DNA damage sensitivity and mitotic failure, serine mutations in conserved cysteines do not. However, these mutations suppress the DNA damage sensitivity of Smc5/6 hypomorphs but not that of HR mutants and remarkably decrease the recruitment of Smc5/6 to loci containing lesions marked for HR-mediated repair. Analysis of DNA repair pathways in suppressed double mutants suggests that lesions are channeled into recombination-dependent and error-free postreplication repair. Thus the HR defect in Smc5/6 mutants appears to be due to the presence of dysfunctional complexes at lesions rather than to reflect an absolute requirement for Smc5/6 to complete HR.
INTRODUCTION
The structural maintenance of chromosomes (SMC) complexes are highly conserved and essential mediators of chromosome dynamics. They comprise cohesin, which is essential for sister chromatid cohesion; condensin, which is required for mitotic chromosome condensation; and a third complex known as Smc5/6. The precise function of Smc5/6 has remained somewhat elusive, although most studies have concluded that this complex is a key component of double-stranded DNA break (DSB) repair by homologous recombination (HR; Hirano, 2006; Murray and Carr, 2008). Whether this reflects a direct and specific role in HR or is a consequence of a more fundamental and/or general role in chromosome organization remains to be determined.
The Smc5/6 complex comprises eight subunits. Smc5 and Smc6 are the two large SMC proteins that are related to Smc1 and Smc3 in cohesin and Smc2 and Smc4 in condensin. These proteins have globular N- and C-termini containing Walker A and B ATP-binding domains that are separated by coiled-coil domains that are interrupted by a flexible hinge. By folding at the hinge, the N- and C-termini are paired and bridged by ATP. Protein association studies suggest that Smc5/6 has a similar architecture to cohesin and condensin. Smc5 and Smc6 dimerize at the hinge domains to form a V-shaped structure. The kleisin subunit Nse4 bridges the globular domains of Smc5 and Smc6 and also forms a subcomplex with Nse1 and Nse3 (Sergeant et al., 2005; Palecek et al., 2006). Nse2, an E3 SUMO ligase (Andrews et al., 2005), forms an independent interaction with Smc5 (Duan et al., 2009a, 2009b). Each of these subunits is essential for cell viability. Two additional but less conserved subunits, Nse5 and Nse6, are HEAT repeat proteins that interact with Smc5 and Smc6 and are required for DNA repair but not for cell viability in the fission yeast Schizosaccharomyces pombe (Pebernard et al., 2006).
Because of the essential nature of Smc5/6, most genetic studies of its function have relied on conditional or hypomorphic alleles. In S. pombe, hypomorphs are hypersensitive to a range of DNA-damaging agents. Through a combination of epistasis, pulse-field, and two-dimensional gels and chromatin immunoprecipitation (ChIP) studies, the damage sensitivity has been primarily attributed to a requirement for Smc5/6 late in HR, after joint-molecule formation between paired sister chromatids (Lehmann et al., 1995; Verkade et al., 1999; Ampatzidou et al., 2006; Irmisch et al., 2009). In addition, Smc5/6 also plays a role at stably stalled replication forks, where it is required to recruit the HR initiator Rad52 (Irmisch et al., 2009), and also in the maintenance of DNA damage checkpoint signaling (Harvey et al., 2004). These functions are also likely to contribute to the damage sensitivity of the mutants. By analogy to cohesin, Smc5/6 likely plays a structural role at lesions to enable their repair. However, direct data suggesting what this role might be are lacking.
A paradox arises when settling on HR as the major function for Smc5/6: Smc5/6 is essential for cell viability, but HR is not. Moreover, the terminal phenotype of null mutants is postanaphase mitotic failure (Verkade et al., 1999; Harvey et al., 2004), and although the onset of this defect is accelerated by exogenous DNA damage, mitotic failure does not require it. Furthermore, mitotic failure of Smc5/6 mutants can also be induced when combined with a mutation in the type II topoisomerase, top2-191, and in this case, this is in the absence of DNA damage that is above background levels. Under these conditions, and after replication stress, the mitotic failure is associated with the postanaphase retention of cohesin on chromosome arms, suggesting a tight interplay between these related SMC complexes (Outwin et al., 2009; Tapia-Alveal et al., 2010). In this regard, cohesin is also required for DSB repair, where it has been proposed to facilitate the interaction between sister chromatids for HR (Strom et al., 2004; Unal et al., 2004). However, other mechanisms to facilitate HR are possible, and, of note, mutants of condensin are also defective in DNA repair (Aono et al., 2002). It is likely that these observations serve to highlight the importance of higher-order chromosome structure to enable the engineering required to process lesions, exchange and resolve strands, and recover an intact chromosome.
Smc5/6 is unique in that two of its non-SMC subunits have catalytic activity. Nse2 contains an SP-RING domain and is an E3 SUMO ligase (Andrews et al., 2005). This activity is required for DNA repair but not for cell viability, despite nse2 being an essential gene. Nse1 contains a variant RING (vRING) domain with a C4HC3 organization of zinc-coordinating residues (Fujioka et al., 2002; McDonald et al., 2003; Harvey et al., 2004). Strains with cysteine-to-alanine mutations in the vRING domain are viable but show DNA repair defects. The deletion of the vRING domain is similarly defective in repair and prevents the recruitment or retention of Smc5/6 to nuclear foci induced by DNA damage, a possible explanation for the damage sensitivity of these cells (Pebernard et al., 2008a).
Nse1's vRING domain is suggestive of an E3 ubiquitin ligase activity, which has been demonstrated with recombinant proteins. The activity is stimulated by its interaction with MAGE domain subunit Nse3 (Doyle et al., 2010), although there are mixed reports as to the robustness of this activity (Pebernard et al., 2008a), and such an activity for the holocomplex has yet to be demonstrated. Thus, although it is attractive to have both SUMO and ubiquitin ligases in the same complex, it is possible that the vRING domain confers another function to the complex, and notably has been shown to stabilize the interaction of the kleisin Nse4 with Nse3 within the Nse1-3-4 subcomplex (Pebernard et al., 2004, 2008a; Sergeant et al., 2005; Palecek et al., 2006).
Here we report an analysis of Nse1 function in S. pombe. We show that the C-terminal half of Nse1, including the vRING domain, is crucial for mitotic fidelity. In addition, we constructed cysteine-to-serine mutations in the vRING domain of Nse1, which surprisingly do not confer DNA damage sensitivity. Conversely, we show that these mutations actually suppress the repair defects of Smc5/6 mutants, including the SUMO ligase–dead nse2-SA allele (Andrews et al., 2005), which we show to be specifically defective in processing replicative DNA damage. The suppression is accompanied by the channeling of lesions into postreplication repair (PRR) pathways, particularly the error-free branch that functions by template switching. Thus, in this context, recombination occurs independent of Smc5/6 function. Finally, we also show that the suppressing nse1 vRING mutation significantly reduces the recruitment or retention of both wild-type and mutant Smc5/6 complexes to loci containing lesions marked by Rad52 for HR-mediated repair. Thus DNA repair can proceed without enrichment of Smc5/6 at lesions, which suggests that the recruitment of dysfunctional complexes is what confers the repair defects in Smc5/6 mutants.
RESULTS
The C-terminus of Nse1 is required for DNA damage resistance and mitotic fidelity
To learn more regarding the mechanism of function for Smc5/6 mediated by Nse1, we searched for alleles generated by random mutagenesis that conferred both temperature and DNA damage sensitivity. Several were isolated, and in each case the mutants encoded proteins that truncated the C-terminus, including the vRING domain. The strongest allele, nse1-15, was a deletion mutation that resulted in a stop codon after leucine 119 (of 232 residues) and was retained for analysis.
nse1-15 cells showed reduced viability at 25°C, forming colonies that stained with the vital dye phloxine B (Figure 1A). nse1-15 cells were also severely growth inhibited at 36°C and were extremely sensitive at 25°C to agents that induce replicative DNA damage (purine alkylation in methyl methanesulfonate [MMS]) or replication fork arrest (dNTP depletion in hydroxyurea [HU]) (Figure 1A). However, the sensitivity to DNA damage outside of S phase was less severe. Asynchronous S. pombe cultures are predominantly G2 cells and contain only ∼10% S phase cells (Forsburg and Nurse, 1991). UV irradiation of asynchronous nse1-15 cells resulted in only a modest sensitivity compared with other Smc5/6 mutants such as smc6-74, which is damage sensitive throughout the cell cycle (Figure 1B). Conversely, nse1-15 was slightly more sensitive to UV irradiation than smc6-74 when S phase–arrested cells were irradiated (Supplementary Figure S1). 4′,6-Diamidino-2-phenylindole (DAPI) staining of cultures grown at 25°C showed that ∼10% of nse1-15 cells display chromosome segregation defects, which rises to ∼40% of cells after two cell cycles at 36°C (Figure 1C). Thus the C-terminal half of Nse1 is required for DNA damage resistance, particularly in S phase, and for mitotic fidelity, a feature of null mutants of the essential Smc5/6 genes in S. pombe (Verkade et al., 1999; Harvey et al., 2004).
FIGURE 1:
Characterization of nse1-15. (A) Tenfold serial dilutions of wild-type and nse1-15 cells were plated on the indicated medium, and plates were incubated at 25°C for 5 d (Control, MMS, HU) or at 36°C for 4 d. (B) UV survival assays for the indicated strains grown at 25°C. Although somewhat UV sensitive, nse1-15 is not as sensitive as the smc6-74 hypomorph. (C) Microscopy images of control and nse1-15 cells. Liquid cultures were grown at 25 or 36°C for 8 h and fixed. Numbers are percentage of cells showing aberrant mitotic figures (mean ± SD, three counts of 100 cells). Arrows indicate examples of cells with aberrant mitotic figures. Bar, 10 μm.
Point mutations in the vRING domain suppress an S phase–specific repair defect due to loss of Nse2's SUMO ligase activity
Previous work indicated that deletion of the vRING domain (residues 184–219) similarly results in temperature and S phase damage sensitivity. However, unlike nse1-15, deletion of the vRING does not cause mitotic defects, and yet it does confer sensitivity to UV irradiation of asynchronous cultures. The temperature and S phase damage sensitivity was also conferred by cysteine-to-alanine mutations at residues 197 and 199 in the vRING domain, corresponding to cysteines 3 and 4 within the C4HC3 motif (Pebernard et al., 2008a). Independently, we constructed cysteine-to-serine mutations at C199 (position 4, nse1-C199S) and C216 (position 7, nse1-C216S) and found no discernible phenotype. We then crossed these nse1 alleles to nse2-SA, an allele of nse2 with mutations in the SP-RING domain that ablates SUMO ligase activity and confers sensitivity to HU and MMS (Andrews et al., 2005). Remarkably, both nse1-C199S and nse1-C216S substantially suppressed the sensitivity of nse2-SA cells to a range of agents that inflict replicative DNA damage (Figure 2A), including the UV-mimetic 4-nitroquinoline-N-oxide (4-NQO). However, nse2-SA is not significantly sensitive to UV irradiation delivered to asynchronous (primarily G2) cultures (Figure 2B), but, like nse1-15, is UV sensitive in S phase (Supplementary Figure S1). Furthermore, nse2 is an essential gene, and another hypomorph, nse2-1, is sensitive to DNA damage in G2 (McDonald et al., 2003). Therefore the SUMO ligase activity of Nse2 is specifically required for DNA damage resistance in S phase, and the nse2-SA defects are suppressed by Nse1 vRING mutations that do not confer DNA damage sensitivity to wild-type cells.
FIGURE 2:
Mutations in Nse1's vRING domain suppress Smc5/6 mutants. (A) Tenfold serial dilutions of the indicated strains were plated onto media containing the following drugs that cause replicative DNA damage: the alkylating agent MMS, the dNTP depleter HU, the topoisomerase I poison camptothecin (CPT), and the UV mimetic 4-NQO. Neither nse1 allele results in damage sensitivity, but both suppress the sensitivity of nse2-SA. (B) UV survival curve for the indicated strains irradiated as asynchronous cultures. Neither of the nse1 alleles nor nse2-SA is sensitive to UV irradiation in G2. (C) Tenfold serial dilutions of the indicated strains were plated onto media containing the indicated concentrations of MMS or HU and incubated for 4 d at 30°C. nse1-C216S suppresses both smc6-74 and smc6-X. (D) UV survival curve of asynchronous cultures in G2 shows that the UV sensitivity of smc6-74 and smc6-X is also suppressed by nse1-C216S.
Thus we conclude that the need for the SUMO ligase activity requires prior function of Nse1 in a vRING-dependent manner. This might include the ubiquitination of a protein(s) by Nse1, an activity that has been shown for Nse1/3 in vitro (Doyle et al., 2010). Furthermore, these data show that Nse2 must have SUMO ligase–independent function(s), possibly via the overall structural integrity of the Smc5/6 complex.
Multiple Smc5/6 hypomorphs are suppressed by nse1-C216S
We next asked whether the suppression of nse2-SA was specific to this mutation and focused on nse1-C216S, as the suppression conferred by this allele was slightly stronger than that by nse1-C199S. We found that nse1-C216S suppressed the MMS, HU, and UV sensitivity of both smc6-74 and smc6-X, almost back to wild-type levels (Figure 2, C and D). Similarly, nse1-C216S also suppressed the sensitivity of nse3-1 and nse4-1 to these agents, although to a lesser extent than the smc6 alleles (Supplementary Figure S2). Thus the suppression seen on the nse2-SA background is neither Nse2- nor S phase–specific, as it suppressed UV sensitivity of asynchronously growing (mostly G2) smc6-74 and smc6-X.
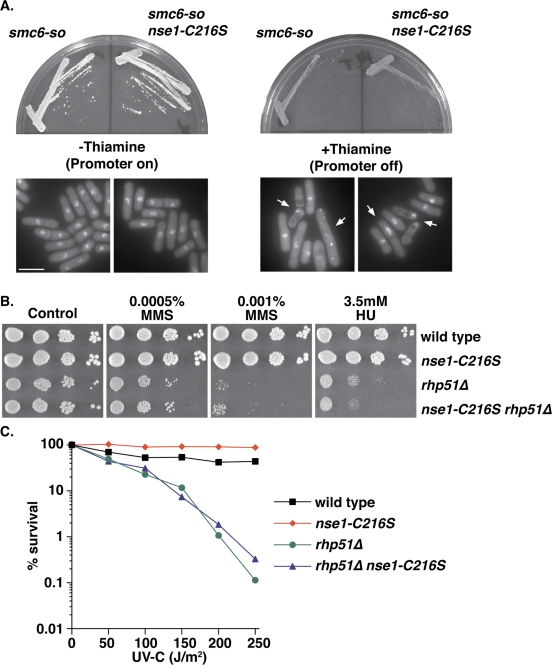
We next asked whether nse1-C216S could suppress null alleles of Smc5/6 genes. To this end, we used smc6-so, a strain in which the sole copy of smc6 is under the control of the thiamine-repressible nmt1 promoter and behaves as wild type in the absence of thiamine but as a null in its presence (Harvey et al., 2004). Clearly, nse1-C216S did not suppress the requirement for Smc6 for cell viability or mitotic fidelity (Figure 3A), and thus the suppression appears to be specific to the repair defects of hypomorphic alleles.
FIGURE 3:
nse1-C216S does not bypass the essential role of the Smc5/6 complex or the requirement for general recombination. (A) smc6-so is a “shut-off” strain, in which the only copy of smc6 is under the control of the thiamine-repressible nmt1 promoter and is lethal in the presence of thiamine. Plates show the indicated strains streaked and incubated for 4 d at 30°C. nse1-C216S does not restore growth in the presence of thiamine. Micrographs show DAPI-stained cells grown in the absence of thiamine or in the presence of thiamine for 32 h. Arrows indicate cells undergoing mitotic failure. Bar, 10 μm. (B) Chronic exposure to MMS or HU shows that rhp51Δ strains are sensitive and that nse1-C216S cannot suppress this sensitivity. Spots are 10-fold serial dilutions of the indicated strains, and plates were incubated respectively at 30°C for 4 d. (C) UV survival curve of asynchronous cultures in G2 show that the UV sensitivity of rhp51Δ is not suppressed by nse1-C216S.
We then asked whether nse1-C216S was a general suppressor of HR defects. All recombination in S. pombe is dependent on the Rad52 homologue encoded by rad22, but rad22Δ cells are poorly viable and rapidly accumulate suppressors (Morishita et al., 2005; Osman et al., 2005). However, cells deleted for the Rad51 homologue, rhp51, are defective for recombination pathways other than single-stranded annealing (Doe et al., 2004), and thus rhp51Δ cells are highly sensitive to DNA damage. nse1-C216S had no effect on the MMS, HU, or UV sensitivity of rhp51Δ cells (Figure 3, B and C), and thus nse1-C216S is not a general suppressor of HR defects. Moreover, nse1-C216S did not modify the damage sensitivity of the cohesin kleisin mutant rad21-K1 (Tatebayashi et al., 1998; Supplementary Figure S3), which is proposed to be HR defective due to a defect in sister chromatid cohesion. Thus nse1-C216S specifically suppresses the HR-mediated repair defects of the Smc5/6 hypomorphs.
nse1-C216S suppresses the DNA damage sensitivity of brc1Δ cells
brc1 encodes a multi-BRCT repeat protein that is required for resistance to replicative DNA-damaging agents but not for resistance to DNA damage outside of S phase. Brc1 is also required for the viability of Smc5/6 hypomorphs and, when overexpressed, suppresses their sensitivity to DNA damage (Verkade et al., 1999; Morishita et al., 2002; Sheedy et al., 2005; Lee et al., 2007; Williams et al., 2010). Brc1 might aid in Smc5/6-dependent repair, represent a parallel pathway of repair, or specifically function to recover from toxic repair intermediates resulting from Smc5/6 dysfunction. To get more insight into the relationship between Brc1 and the Smc5/6 complex, we tested for interactions among smc6-74, nse1-C216S, and brc1Δ. Remarkably, nse1-C216S suppressed the synthetic lethality of brc1Δ smc6-74 cells but did not suppress the sensitivity of brc1Δ smc6-74 cells to MMS and UV irradiation. Moreover, nse1-C216S also suppressed the MMS sensitivity of brc1Δ cells (Figure 4). Because nse1-C216S does not suppress the essential nature of Smc5/6 (Figure 3A), this supports the hypothesis that the damage sensitivity of brc1Δ cells at least in part reflects a requirement for Brc1-dependent repair of intermediate lesions generated by Smc5/6 dysfunction, which in turn is suppressed by nse1-C216S. However, as smc6-74 nse1-C216S brc1Δ cells are significantly more sensitive to both MMS and UV than smc6-74 cells, Brc1 may also act independently of Smc5/6 dysfunction.
FIGURE 4:
nse1-C216S suppresses the MMS sensitivity of brc1Δ and the synthetic lethality of smc6-74 brc1Δ double mutants. (A) Tenfold serial dilutions of the indicated strains were spotted onto plates containing the indicated concentrations of MMS, and plates were incubated at 30°C for 4 d. nse1-C216S suppresses the MMS sensitivity of brc1Δ and suppresses the synthetic lethal interaction of brc1Δ combined with smc6-74. brc1Δ smc6-74 nse1-C216S triple mutants were obtained by tetrad dissection and confirmed by backcrossing. Although nse1-C216S suppressed the synthetic lethality, it cannot suppress the high sensitivity of the triple mutant to MMS. (B) UV survival curve of the indicated strains. Note that brc1Δ cells are not UV sensitive, but the brc1Δ smc6-74 nse1-C216S triple mutant is more sensitive than the smc6-74 parent.
nse1-C216S resistance to DNA damage is via Rad55/57-mediated recombination
We next asked which DNA repair pathways are used under conditions in which nse1-C216S is suppressing the damage sensitivity of smc6-74. Due to the synthetic lethality and growth defects seen in smc6-74 and nucleotide excision repair (NER) mutants (Lee et al., 2007), we were unable to assay the requirement for NER, although neither MMS nor HU induces bulky lesions for NER-based repair, and the doses of UV-C we are using evoke lesions other than pyrimidine dimers and photoproducts, including DNA breaks (Callegari and Kelly, 2006, 2007; Callegari et al., 2010). Base excision repair, a major pathway in the repair of alkylation damage by MMS (Marti et al., 2002), and mismatch repair genes were not required for suppression of smc6-74 by nse1-C216S. Examples of these pathways are shown in Supplementary Figure S4.
Therefore we tested the role of HR in the suppression of smc6-74 by nse1-C216S. We could not test the core HR machinery of Rad51, 52, and 54 (encoded by rhp51, rad22, and rhp54, respectively, in S. pombe) due to epistasis with smc6-74 (Lehmann et al., 1995; Sheedy et al., 2005). However, there are two dimeric complexes of Rad51 paralogues that facilitate the formation and stability of the Rad51 nucleoprotein filament. These are Rad55/57 (encoded by rhp55/57 in S. pombe) and Swi5/Sfr1 (Akamatsu et al., 2007). Remarkably, rhp57Δ nse1-C216S double mutants were significantly more sensitive to MMS and HU than the rhp57Δ parent. Furthermore, rhp57Δ smc6-74 nse1-C216S triple mutants were more sensitive than smc6-74 rhp57Δ cells (Figure 5, A and B). This indicates that although nse1-C216S cells have wild-type sensitivities to HU and MMS, they are significantly reliant on Rhp55/57 for the repair of lesions. Conversely, swi5 was not required for suppression of the MMS or HU sensitivity despite a synthetic increase in sensitivity in swi5Δ smc6-74 double mutants to MMS. Furthermore, as with rhp57Δ, nse1-C216S increased the MMS (but not HU) sensitivity of swi5Δ cells, although, of note, swi5Δ cells are not as MMS (or HU) sensitive as rhp57Δ cells (Figure 5C). We can conclude that there is a strong bias to Rad55/57-dependent repair in nse1-C216S cells and in smc6-74 nse1-C216S double mutants, which is reminiscent of smc6-74 suppression by Brc1 overexpression (Sheedy et al., 2005). This bias might be due to a reduced ability to initiate Swi5/Sfr1-dependent recombination or a channeling of lesions into Rad55/57-dependent repair via the production of a specific intermediate as a result of Smc5/6 dysfunction.
FIGURE 5:
nse1-C216S shows preferential requirement of rhp57 over swi5 for resistance to DNA damage and suppression of smc6-74 under conditions that impede S phase progression. The indicated strains were constructed by tetrad dissection and further confirmed by backcrossing. MMS and HU sensitivity assays were then performed using spots of 10-fold serial dilutions, with the plates incubated at 30°C for 4 d. (A, B) Note that although nse1-C216S is not MMS or HU sensitive, nse1-C216S sensitizes rhp57Δ cells to both MMS and HU, and the deletion of rhp57 abolishes the suppression of smc6-74 by nse1-C216S, with the triple mutant now resembling the nse1-C216S rhp57Δ double mutant. (C) nse1-C216S sensitizes swi5Δ cells to a lesser extent than rhp57Δ, and this is specific to MMS. In the absence of swi5 the MMS suppression of smc6-74 by nse1-C216S can still occur but to a lower extent than in its presence. Note that there is no effect of swi5Δ in HU.
nse1-C216S–mediated suppression of MMS sensitivity is via PRR
The sensitivity of the nse1-C216S rhp57Δ double mutant precluded our ability to determine whether the suppression of smc6-74 uses Rad55/57-dependent HR. Use of HR, however, can evoke different mechanisms, depending on the nature of the lesion. For DNA damage in G2 cells, conventional HR can be used to repair DSBs or lesions that are processed into DSBs as an intermediate of their repair. In S phase, there are two HR-dependent mechanisms. On the collapse of stalled replication forks, HR is required to restart replication. Conversely, polymerase-blocking lesions such as purine alkylation allow the option to bypass the lesion using HR-dependent template switching controlled by PRR (Branzei and Foiani, 2008).
PRR is a two-step process by which lesions can be bypassed during DNA replication but also functions in G2 through a less-defined mechanism (Verkade et al., 2001; Frampton et al., 2006; Szuts et al., 2006; Huang et al., 2009). The initiation of PRR requires monoubiquitination of PCNA on K164 by Rad6/Rad18 (encoded by rhp6 and rhp18, respectively, in S. pombe), which enables the recruitment of bypass polymerases that replicate past a lesion, potentially in a mutagenic manner. The second arm of error-free PRR involves the conversion of monoubiquitinated PCNA to polyubiquitinated molecules with K63 linkages. This is catalyzed by Ubc13/Mms2, and with the aid of Rad5 and the HR machinery, this enables lesion bypass by HR-dependent invasion of the other nascent strand to allow replication off this template (Broomfield et al., 2001; Lee and Myung, 2008). Of importance, template switching is specifically dependent on the Rad55/57 complex (Vanoli et al., 2010).
Rhp18, the initiator of PRR, was clearly required for suppression of smc6-74 by nse1-C216S for both MMS and UV (Figure 6, A and B). Consistent with engagement of PRR, both a ligase-dead rhp18-1 (Lee et al., 2007) mutant and a K164R mutation in PCNA (Frampton et al., 2006) also abolished suppression of MMS sensitivity (Supplementary Figure S5). Rhp18 was not, however, required for suppression in HU (unpublished data). In HU, most replication forks stably arrest and can resume replication once dNTPs are synthesized. Therefore HR is evoked in HU only to restart the small number of stalled replication forks that spontaneously collapse, and there are no lesions to bypass. This phenomenon explains the difference in HU data between rhp57Δ and rhp18Δ, and thus HU was excluded from subsequent epistasis experiments.
FIGURE 6:
nse1-C216S requires the error-free branch of PRR to suppress the MMS DNA damage sensitivity of smc6-74. All strains were constructed by tetrad dissection and confirmed by backcrossing. MMS and UV sensitivity assays were performed as described in the previous figures. (A) rhp18Δ increases the sensitivity of smc6-74 to MMS (A) but not to UV (B), and nse1-C216S cannot suppress this at MMS concentrations of ≥ 0.003% or to all doses of UV. (C) A series of strains lacking ubc13, which encodes the ubiquitin-conjugating enzyme required for error-free PRR, were constructed. These were tested for MMS sensitivity using spots of 10-fold serial dilutions, with plates incubated at 30°C for 4 d. ubc13Δ significantly enhances the sensitivity of smc6-74 to MMS, and nse1-C216S fails to suppress this at MMS concentrations of ≥ 0.003%.
Although nse1-C216S showed an eightfold increase in rates of spontaneous can1 mutagenesis (4.3 × 10−6 vs. 5.6 × 10−7 for wild type), this is significantly lower than other backgrounds that are forced to use this error-prone pathway (Kai and Wang, 2003; Sheedy et al., 2005). Consistent with the requirement for Rad55/57 and the relatively low rates of mutagenesis, we found that Ubc13 was required in a manner identical to Rhp18 (Figure 6C). Therefore, in nse1-C216S smc6-74 strains, a significant proportion of otherwise lethal lesions that are derived from alkylation damage are channeled into error-free PRR, which by its nature requires the initial commitment to error-prone PRR.
Interaction between Nse1 and DNA helicases
Suppression of smc6-74 by Brc1 overexpression depends on PRR, although in this case it is via the error-prone pathway (Sheedy et al., 2005). This suggests that the two modes of suppression, although through different pathways, are each through channeling of lesions into PRR. We therefore asked whether DNA helicases could facilitate the suppression by processing such lesions into the PRR pathway. The RecQ helicase Rqh1 was clearly required for suppression of smc6-74 by nse1-C216S on MMS (Figure 7A). This suggests that this enzyme, presumably with DNA topoisomerase III, can unwind toxic recombination intermediates as part of the suppression process. We then asked whether the helicase activity of Rqh1 was required for the suppression, but found a helicase-dead mutant (rqh1-K547I) to be synthetically lethal with smc6-74, and this lethality was not suppressed by nse1-C216S (Supplementary Figure S6). This suggests that this mutant, Rqh1, is interfering with another enzyme in smc6-74 cells. Indeed, the number of cells produced in the microcolonies over 1 wk was significantly fewer in the triple mutant, showing in this background, as in rhp57Δ cells (Figure 5), that nse1-C216S confers a deleterious effect.
FIGURE 7:
nse1-C216S requires Rqh1 and Fml1 for smc6-74's DNA damage sensitivity suppression and requires Fml1 for partial resistance to DNA damage. The strains were constructed by tetrad dissection and confirmed by backcrossing. These were tested for MMS sensitivity using spots of 10-fold serial dilutions, with plates incubated at 30°C for 4 d. (A) rqh1Δ enhances the MMS sensitivity of smc6-74, and this is not suppressed by nse1-C216S. (B) Sensitivity of both of the fml1Δ smc6-74 and fml1Δ nse1-C216S double mutants is enhanced over that of the most-sensitive single mutant. In the triple mutant fml1Δ smc6-74 nse1-C216S, nse1-C216S is not able to suppress the enhanced MMS sensitivity of fml1Δ smc6-74 cells to MMS concentration of ≥ 0.003%. Therefore Fml1 is required for nse1-C216S suppression of smc6-74's MMS sensitivity.
Another helicase/translocase, Fml1, has also been implicated in processing of damaged replication forks and is perhaps required to reverse replication forks (Sun et al., 2008). fml1Δ has been reported to suppress Smc5/6 mutants in S. pombe (Sun et al., 2008) and in Saccharomyces cerevisiae (Chen et al., 2009; Choi et al., 2010; Chavez et al., 2011). We found fml1Δ smc6-74 cells to be extremely sensitive to MMS, but this was subject to high-frequency spontaneous suppression (Figure 7B). This reversion may explain the previously reported suppression of the HU sensitivity of smc6-X by fml1Δ. However, two groups showed that deletion of the fml1 homologue in S. cerevisiae, MPH1, suppresses the HU and MMS sensitivity of Smc5/6 mutants (Chen et al., 2009; Choi et al., 2010; Chavez et al., 2011), which is a clear difference between the yeasts. Using strains that were confirmed to be free of additional mutations by backcrossing, we observed that fml1 was required for the suppression of smc6-74 by nse1-C216S on 0.003% MMS and greater (Figure 7B), and expression of a helicase-dead mutant lacking the DEAH motif failed to rescue this phenotype (Supplementary Figure S6). Surprisingly, fml1Δ nse1-C216S double mutants were more sensitive to MMS than the fml1Δ parent, and together with the extreme sensitivity of fml1Δ smc6-74, this suggests that Smc5/6-dependent repair of alkylation damage is independent of repair mediated by Fml1 function.
Clearly, the suppression of MMS sensitivity is in large part via channeling into the PRR pathway and involves these helicases, presumably through their helicase activities, which suggests that this represents an Smc5/6-independent response that still uses the HR machinery.
nse1-C216S decreases the recruitment/retention of Smc5/6 at lesion-containing loci
The localization of Smc5/6 is dynamically regulated. On the basis of hemagglutinin (HA)-tagged Nse4 and ChIP or ChIP-on-chip, Smc5/6 was observed to be distributed throughout the S. pombe genome but to be enriched at tRNA genes in cycling cells, at centromeric loci in HU, and at telomeric loci in MMS (Pebernard et al., 2008b). Smc5/6 was also enriched at stalled and collapsed replication forks in S. pombe (Irmisch et al., 2009) and at DSBs in S. cerevisiae (Lindroos et al., 2006) and in humans (Potts et al., 2006). We used ChIP to assay the effect of nse1-C216S on this localization under conditions of replication stalling (HU), alkylation damage (MMS), and replication fork collapse (HU plus caffeine, the latter of which inhibits the S phase checkpoint; Wang et al., 1999) as a potential explanation for our observed suppression. Primer sets for quantitative PCR analysis (Supplementary Table 1) were chosen on the basis that they localize to stalled forks in HU (at ade6, enrichment is less marked), as well as collapsed replication forks when the intra–S phase checkpoint is inhibited (Pebernard et al., 2008b; Irmisch et al., 2009; Zaratiegui et al., 2011). This included centromere-proximal tRNA genes, which are know to be potent pause sites for DNA replication (Deshpande and Newlon, 1996; Szilard et al., 2010; Zaratiegui et al., 2011). Finally, MMS should induce global alkylation of purines that in S phase are processed or bypassed by HR-dependent mechanisms. We tested this assertion by assaying the recruitment of the recombination initiator Rad52 to these loci using HU plus caffeine as a positive control, and indeed all primer sets were enriched for Rad52, indicating the presence of lesions at these loci that are primed for Rad52-dependent repair (Supplementary Figure S7).
Similar to the reported data in HU, we observed enrichment of Smc5/6 at centromeric loci, both in the outer (OTR) and inner (tRNALeu and tRNAMet) repeats. However, we observed that in MMS-treated cells, Smc5/6 was enriched at all loci tested (Figure 8). Moreover, enrichment at the subtelomeric repeats (STE1) was only prominent in MMS- and not HU-treated cells. In an nse1-C216S background, baseline levels of Smc5/6 were similar to those of wild type, except at the telomeres, where levels were reduced by ∼50%. Of importance, little or no enrichment above background in either HU or MMS was observed (Figure 8), although expression of Smc5/6 subunits was not affected (unpublished data), and the cells are perfectly viable and have wild-type sensitivities to DNA-damaging agents (Figure 2). Therefore enrichment of Smc5/6 to loci under conditions of replication stress and DNA damage cannot be essential for the repair of genomic lesions caused by these agents.
FIGURE 8:
nse1-C216S decreases recruitment of the Smc5/6 complex to chromatin. Liquid cultures of wild-type and nse1-C216S strains expressing an endogenously HA-tagged nse4 were left untreated or either treated for 4 h at 30°C with 10 mM HU or for 5.5 h with 0.005% MMS. HU treatment in these genetic backgrounds causes forks to stall, and MMS treatment causes fork collapse and DNA damage. Recruitment of HA-Nse4 to previously described loci as major sites of recruitment for wild-type Smc5/6 complex was tested by anti-HA ChIP. The graph shows the fold of enrichment (mean ± SE, n ≥ 3) of the HA-tagged Nse4 normalized to an untagged control at different loci. nse1-C216S cells failed to enrich Nse4-HA to loci in HU- and MMS-treated cells and showed a ∼50% reduction in Nse4-HA levels at telomeres (STE1).
nse1-C216S suppresses an enhanced recruitment of Nse2-SA mutant complexes to lesion-containing loci
We next asked whether nse1-C216S had a similar effect on mutant Smc5/6 complexes, and to this end we used nse2-SA, which, when combined with the HA-tagged nse4 allele, had no significant growth defects compared with the parental strains. Other Smc5/6 mutants, such as smc6-74 and smc6-X, which are more strongly suppressed by nse1-C216S, had a significant growth defect on the nse4-HA background, as did other combinations of hypomorphs and epitope-tagged alleles. These growth defects were associated with chromosome aberrations and therefore cannot be used for ChIP experiments. Nse2-SA complexes interacted with chromosomes and were actually more enriched than wild-type complexes upon HU or MMS treatment (Figure 9A). At the telomeres (STE1), Smc5/6 enrichment was specifically more evident in MMS and not seen in HU, which is consistent with findings for the wild-type complex (Figure 8; Pebernard et al., 2008b). In the origins tested (ars2004, 3005), enrichment in HU was considerably greater in nse2-SA than in wild-type cells.
FIGURE 9:
Mutant complexes are recruited to chromatin upon HU or MMS treatment, and this is reduced in an nse1-C216S background. (A) Liquid cultures of wild-type, nse2-SA, and nse2-SA nse1-C216S strains expressing an endogenously HA-tagged nse4 were left untreated or treated for 4 h at 30°C with 10 mM HU or for 5.5 h with 0.005% MMS. The graph shows the fold of enrichment (mean ± SE, n = 6) of the HA-tagged Nse4 normalized to an untagged control at different loci. In nse2-SA, the recruitment to all loci upon HU or MMS treatment is greater than seen in wild-type cells. In the presence of nse1-C216S, the recruitment of mutant complexes is reduced either to wild-type or lower than wild-type levels in HU-treated cells at all loci tested. The same is the case at centromeres (OTR), telomeres (STE1, tRNALeu), and ars3005 in MMS-treated cells. (B) Wild-type and nse2-SA cells were treated with HU as described in A, and after 4 h cells were washed and released into fresh media. Samples were taken every 30 min to measure the time of residence and fold enrichment at ars2004, ars3005, and OTR in nse2-SA (red) and wild-type (blue) cells. HA-Nse4 is enriched at these loci in nse2-SA and persists at higher levels upon recovery from HU arrest.
We repeated the ChIP analysis in a time course of HU release, as cells recover more synchronously from HU than from MMS. At ars2004 and 3005 and at the centromeres (OTR), more HA-tagged Nse4 was associated with chromatin in nse2-SA cells at the block point. Although it decreased over time upon release, higher levels than wild type persisted upon reentry into the cell cycle (Figure 9B). Of note, HU treatment of nse2-SA cells is associated with chromosome segregation defects (Andrews et al., 2005), and these are also suppressed by nse1-C216S, as evidenced by the viability of these strains in HU (Figure 2).
In HU, where replication forks stall, the increased recruitment of Smc5/6 seen in nse2-SA cells was reduced to wild-type HU-treated levels by nse1-C216S at most loci but was back to untreated levels at replication origins (ars2004, 3005; Figure 9A). In MMS-treated cells, there was also an overall trend of reduced enrichment of Smc5/6 at lesion-containing loci in nse2-SA nse1-C216S cells, although the largest reduction was seen at tRNALeu and the subtelomeric repeats (Ste1), which corresponds to the regions of Smc5/6 enrichment at a global genomic level (Pebernard et al., 2008b). Because nse1-C216S suppresses the sensitivity of nse2-SA to both HU and MMS (Figure 2), this suggests that the higher level of Nse2-SA complexes at tRNA genes and telomeres has a greater pathological effect in MMS than in HU.
To corroborate these findings, we also assayed Smc5/6 localization under conditions of catastrophic global replication fork collapse (acute HU treatment followed by caffeine treatment; Wang et al., 1999; Figure 10). Like Rad52 (Supplementary Figure S7), Smc5/6 was enriched at all loci in wild-type cells, most notably at the telomeres. In nse1-C216S cells, there was modest enrichment at all loci, which contrasts to HU alone, in which there was no enrichment (Figure 8). For nse2-SA cells, enhanced enrichment was again observed, and this was to a greater degree at the centromeres (OTR), tRNA genes, and telomeres. This pattern was reminiscent of a combination of HU and MMS data (replication stalling and DNA damage). In double mutants, recruitment at all loci was reduced but not to the same magnitude as seen in HU-treated cultures (Figure 10A). Of importance, although nse1-C216S shows wild-type sensitivity to chronic HU exposure and suppresses the sensitivity of nse2-SA to HU, nse1-C216S showed approximately threefold lower survival after the acute HU plus caffeine treatment and did not suppress the sensitivity of nse2-SA to this regimen (Figure 10B).
FIGURE 10:
nse1-C216S reduces the recruitment of wild-type and nse2-SA mutant complexes to chromatin under conditions of replisome collapse. (A) ChIP assays using HA-tagged Nse4 expressed in the indicated backgrounds, using cells that were either not treated or arrested in HU prior to treatment with caffeine. Data are mean ± SEM, n = 3–5. (B) Survival assays for the cells treated in A, normalized to HU-alone-treated wild-type cells. Data are mean ± SD, n = 3.
Therefore the suppression of nse2-SA, and presumably all Smc5/6 mutants, that is conferred by nse1-C216S is associated with a reduced recruitment or retention to lesion-containing loci. Furthermore, there is a correlation between the magnitude of reduced mutant complexes at these loci and survival of the genotoxic stress.
DISCUSSION
The notion that Smc5/6 is required for HR is based on the repair defects of hypomorphic yeast strains and from RNA interference experiments in mammalian cells (Murray and Carr, 2008). Most of the hypomorphic mutants were selected as DNA damage–sensitive allele and so naturally confer a defect in repair. However, the terminal mitotic lethality of null mutants (Verkade et al., 1999; Harvey et al., 2004) must evoke that Smc5/6 plays a more fundamental role in chromosomal organization.
The phenotypes of nse1-15 highlight the importance of the C-terminal domain in mitotic fidelity. However, the viability of nse1-15 at 25°C indicates that Smc5/6 function is only partially impaired. The vRING domain has been shown to stabilize the Nse1-3-4 subcomplex (Sergeant et al., 2005; Pebernard et al., 2008a). However, nse1-C199S and nse1-C216S are mutations predicted to disrupt zinc-coordinating residues within the RING domain, and yet these strains are viable at all temperatures and have wild-type sensitivities to DNA damage. The cysteine-to-alanine and RING deletion mutations (Pebernard et al., 2008a) may be more structurally disruptive to the Nse1-3-4 subcomplex. In either case, the phenotypes may result from defective ubiquitination by Nse1 (Doyle et al., 2010). However, we and others (Pebernard et al., 2008a) have not been able to show this activity for the endogenous complex derived from both S. pombe and human cells. Thus other or additional functions for this domain, including mediating chromatin interactions, should not be discounted.
Given the strong HR defects in smc6 mutants, the suppression by nse1-C216S is remarkable. This extends to all tested hypomorphs to varying degrees and was neither gene nor allele specific. We also found that Nse2's SUMO ligase activity is only required for replicative DNA damage responses, but only in the presence of wild-type Nse1. The only non-Smc5/6 mutant that was also suppressed by nse1-C216S was brc1Δ. This suggests that nse1-C216S prevents the formation of chromosomal lesions that require Brc1-dependent resolution and is consistent with the synthetic lethality between brc1Δ and all Smc5/6 hypomorphs that are also suppressed by nse1-C216S.
The suppression of smc6-74 by nse1-C216S is significantly via HR-dependent mechanisms of the PRR pathway. For DNA damage in G2, this is the repair of DSBs, including lesions that can be converted to DSBs as a repair intermediate. Despite its name, the PRR pathway is functional in G2 and required for DNA repair out of the context of replication (Verkade et al., 2001; Frampton et al., 2006; Szuts et al., 2006; Huang et al., 2009). For HU treatment, HR-dependent mechanisms can promote the restart of spontaneously collapsed replication forks (Branzei and Foiani, 2008). For alkylation damage by MMS, HR promotes the channeling of lesions into the error-free branch of PRR for damage tolerance, although this does not repair the alkylated base. This is specifically dependent on the Rad55/57 complex, which has been implicated in recombination-dependent template switching (Vanoli et al., 2010), and thus this must be able to occur without the aid of wild-type Smc5/6 function. Intermediates of alkylation damage repair by excision pathways can generate substrates for recombinational repair independent of lesion bypass (Memisoglu and Samson, 2000; Sugimoto et al., 2005). Consistent with this, nse1-C216S rhp57Δ double mutants are more sensitive than either parent, and nse1-C216S cells show increased induced mutagenesis. Because nse1-C216S mutants do not enhance the MMS sensitivity of PRR-defective rhp18Δ cells, the enhanced sensitivity in nse1-C216S rhp57Δ cells must reflect a requirement for Rad55/57-dependent recombination in nse1-C216S cells that is independent of lesion bypass by PRR. This may be due to an inability to initiate Swi5/Sfr1-dependent recombination, or perhaps Smc5/6 dysfunction generates an intermediate that is not recognized by Swi5/Sfr1. Clearly, there is much to be learned about the specificity of these Rad51 paralogues. Furthermore, PRR-dependent damage resistance also functions in G2 without the need for the replisome to collide with a lesion (Frampton et al., 2006; Daigaku et al., 2010; Karras and Jentsch, 2010), and clearly nse1-C216S can suppress G2 HR defects in the smc6 mutants that depend on PRR. Thus not all recombination depends on Smc5/6 function.
In many genetic backgrounds, we observed a dose-dependent suppression of smc6-74's MMS sensitivity both by nse1-C216S and by Brc1 overexpression. Dose dependence is also seen when assaying which genes are required for the suppression (Sheedy et al., 2005; Lee et al., 2007). Excision pathways can remove lesions before they interfere with replication. However, at higher concentrations, the probability of the replisome colliding with an unrepaired lesion is higher, and so the dose dependence reflects the relative need of the recombination-dependent events and is a likely explanation for our observed threshold effect. Because there are many pathways to repair and tolerate lesions, epistasis experiments uncover requirements for particular genes in terms of suppression above threshold doses, some more significantly than others, but do not imply an absolute requirement for one pathway or another in response to a genotoxin.
MMS-treated strains carrying an Nse1 RING deletion fail to form microscopic Nse4-GFP foci (Pebernard et al., 2008a). However, from our ChIP data, we conclude that the suppression by nse1-C216S correlates with a reduced recruitment (or retention) of Smc5/6 to loci containing lesions following DNA damage. This implies that the damage sensitivity of the RING deletion strains, including nse1-15, stems from a defect other than Smc5/6 recruitment to lesions. Of note, the sensitivity of nse2-SA to HU (±caffeine) and MMS correlates with increased levels of Smc5/6 at lesion-containing loci, and this increased signal persists after HU removal. This increased occupancy by Smc5/6 may interfere with DNA repair and/or chromosome segregation, both of which are defective in HU-treated nse2-SA cells (Andrews et al., 2005). Furthermore, this suggests that the SUMO ligase activity of Nse2 may be important in the dynamics of the spatiotemporal organization of Smc5/6 and that this is important to return the complex to a state that exists in unchallenged cells. Like cohesin, Smc5/6 is a dynamic complex, and occupancy at a particular locus cannot be equated with functionality.
Whereas Smc5/6 is enriched at lesion-containing loci in wild-type cells, this is abolished in nse1-C216S cells treated with HU or MMS and significantly attenuated in HU-arrested cells treated with caffeine. In nse2-SA cells, an enhanced enrichment at all loci is seen with each regimen. As a generalization, nse1-C216S reduced this enhanced enrichment, but the pattern of Smc5/6 dynamics in the double mutants varies depending on the nature of the genotoxic stress. For HU, where replication forks stall, there is a general reduction, but this is most potent at the replication origins themselves. However, this observation does not necessarily mean that the effects at each locus have equal pathological significance in terms of HU resistance.
Under conditions of global alkylation damage in MMS-treated cells, lesions can impede the replisome, requiring lesion bypass or recombinational repair. In these cells, both the enhanced enrichment in nse2-SA cells and the reduction in Smc5/6 enrichment in nse2-SA nse1-C216S cells is most profound at the tRNA genes and subtelomeric repeats. A similar pattern was observed in HU-arrested cells treated with caffeine to induce global fork collapse, a much more lethal treatment regimen compared with MMS, as cell cycle checkpoints are also inhibited (Wang et al., 1999).
The effects at these loci match regions where wild-type Smc5/6 complexes enrich in ChIP-on-chip studies (Pebernard et al., 2008b). Therefore these loci likely reflect important regions of Smc5/6 function. tRNA genes are highly expressed and subject to spontaneous fork stress due to collision of RNA polymerase III and the replisome (Deshpande and Newlon, 1996; Szilard et al., 2010). Because Smc5/6 appears to be crucial in replication fork stability, it has been proposed that the complex is extremely important in regions of the genome that undergo obligate or preferred unidirectional DNA replication (Murray and Carr, 2008). The tRNA genes may represent one such region, although perhaps the most profound are the subtelomeric and telomeric repeats. On each chromosome end, these loci must be replicated from a single origin, and so if this stalls or collapses, it cannot be rescued from an adjacent origin by convergent replication. This phenomenon may also be related to Smc5/6's proposed role in the ALT pathway of telomere maintenance (Potts and Yu, 2007).
We conclude that although Smc5/6 facilitates HR, HR can proceed in a Smc5/6-independent manner so long as the complex is not recruited to lesions. What then functions in the absence of Smc5/6 at sites of recombination? Smc5/6 may be an “add-on” regulator of HR, and simply HR may proceed without any requirement for enrichment of Smc5/6 at the lesion. Given the shared loader (Lindroos et al., 2006) and related structure, cohesin may substitute for Smc5/6. Consistent with this, we have observed synthetic lethality between mutant alleles of cohesin and Smc5/6 (unpublished data). Furthermore, a yet-to-be-described regulator of HR may substitute for Smc5/6, such as components of the PRR pathway. Finally, nse1-C216S may allow otherwise dysfunctional complexes to contribute to HR. However, several observations make this unlikely. First, the suppression is neither gene nor allele specific. Moreover, the molecular defects of the suppressed mutants are diverse, and with the reduced enrichment at loci that contain lesions, such intracomplex suppression would require Smc5/6 to contribute to HR from a distance.
Why then is Smc5/6 essential for cell viability? Smc5/6 is associated with chromosomes throughout interphase and has considerable spatiotemporal overlap with cohesin (Lindroos et al., 2006; Pebernard et al., 2008b). There is interplay between the dynamics of cohesin and that of Smc5/6, including a disruption to the cohesin cycle following DNA damage in Smc5/6 mutants (Outwin et al., 2009). Such fundamental elements of chromosome structure are likely to contribute to all major events on the chromosome. In human cells, cohesin also plays a role as a transcriptional insulator (Rubio et al., 2008; Wendt et al., 2008). An insulator function for Smc5/6 may protect a damaged locus by regulating protein recruitment to enable HR to proceed without interference by transcription, replication, and perhaps even other repair pathways. Furthermore, Smc5/6 has been implicated in regulating the topology of longer chromosomes in S. cerevisiae (Kegel et al., 2011). Given that the longer chromosomes in S. cerevisiae are smaller than other eukaryotic chromosomes, such a function in chromosome topology may extend genome wide in other eukaryotes. We have observed strong interactions between Smc5/6 mutants and topoisomerase II in S. pombe, although this appears to be independent of Top2's catalytic function (Germe et al., 2009; Outwin et al., 2009; Tapia-Alveal et al., 2010).
Despite these hints at Smc5/6 function, we do not yet know precisely how Smc5/6 is affecting chromosome structure. The fact that most studies have focused on its role in HR-mediated DNA repair may be the reason why Smc5/6 function remains so poorly understood compared with cohesin and condensin. This is reminiscent of early studies of S. pombe Rad21, which turned out to be the kleisin subunit of cohesin but was generally believed to be a DSB repair protein in early studies (Birkenbihl and Subramani, 1992, 1995). That is, the HR defects of Smc5/6 mutants are probably a consequence of a more fundamental role for this complex in chromosome dynamics, almost certainly functioning in a coordinated manner during the cell cycle with cohesin, topoisomerase II, and possibly other determinants of chromosome structure.
MATERIALS AND METHODS
General S. pombe methods
All the strains were derived from 972h− and 975h+. Standard procedures for strain manipulation described elsewhere were used (Moreno et al., 1991; Calonge and O'Connell, 2006; Calonge et al., 2010). Strains were constructed by tetrad analysis, and multiple isolates were selected and tested. In addition, backcrosses of one or several isolates were performed to ensure the absence of suppressors. Methods to test the sensitivity to chronic drug and UV-C exposure were as described (Outwin et al., 2009; Calonge et al., 2010). Acute exposure to DNA-damaging agents was performed in liquid cultures grown to mid-logarithmic phase, where MMS (0.005%) or HU (10 mM) was added. Samples were collected after 5.5 or 4 h at 30°C, respectively. MMS was inactivated with 5% sodium thiosulfate. In addition, replication fork collapse was induced by treatment with 10 mM HU for 3 h at 30°C, followed by the addition of caffeine to 10 mM for two additional hours (Wang et al., 1999). For expression of wild-type Fml1 from the nmt1 promoter, the fml1 open reading frame was amplified by PCR and cloned into pREP1K. Residues 132–304 were deleted from the helicase domain, including the signature DEAH domain for expression of a helicase-dead derivative.
Generation of nse1-15
A genomic clone of nse1 was introduced in the leu1-based pJK148 vector (Keeney and Boeke, 1994) and was randomly mutated by propagating it in XL-1Red Escherichia coli (Stratagene). The mutated versions were integrated at leu1 in an nse1+/nse1::ura4 diploid heterozygote (Harvey et al., 2004). Selection was based initially on uracil and leucine prototrophy. To isolate haploids, cells were streaked to single colonies and tested for viable nonsporulating colonies. In addition, selection of temperature-sensitive (TS) and MMS-sensitive strains was done. The selected colonies were backcrossed to wild-type strains and were analyzed by fluorescence-activated cell sorting for DNA content. Finally, single genomic integration was assessed by Southern blot analysis. The strain nse1-15 was selected as the strongest TS phenotype at 36°C, which contains a deletion from within the nse1 ORF to 126 nucleotides 3′ to the stop codon, thus ending with the sequence QYSL(119)WRTLAF.
Generation of point mutant alleles: nse1-C216S and nse1-C199S
Site-directed mutagenesis of the selected residues was performed by preparing single-stranded template of a genomic clone of nse1 in pJK148 by the Kunkel method (Kunkel et al., 1987). The resulting mutated plasmids were sequenced, transformed, and selected for integration at leu1 in an nse1+/nse1::ura4 diploid heterozygote, based on uracil and leucine prototrophy. Haploid cells were recovered and characterized as described earlier.
Temperature shifts with temperature-sensitive mutant nse1-15
For chronic exposure to temperature stress, cells were grown at permissive temperature on yeast extract plus supplements (YES), and 10-fold serial dilutions were plated in YES medium agar plates with phloxine B. The plates were incubated at 36°C for 4 d or at 25°C for 5 d. Plates were photographed after the appropriate time. For acute exposure to temperature stress, cultures were grown at permissive temperature on liquid YES to mid-logarithmic phase. New cultures were set at OD595 of 0.2 at 25°C and then kept at 25°C or shifted to 36°C over an 8-h time course.
Chromatin immunoprecipitation
ChIP assays were carried out as previously described (Outwin et al., 2009). Briefly, 50 ml of cell cultures (untreated, or treated with a desired DNA-damaging agent) were cross-linked with 1% formaldehyde. Immunoprecipitation was done with 0.3 μg of anti-HA antibody (12CA5; Roche, Indianapolis, IN) for HA-tagged Nse4 or 1.5 μl of polyclonal anti–green fluorescent protein (Invitrogen, Carlsbad, CA) for yellow fluorescent protein–tagged Rad52, together with 20 μl of (1:1 slurry) protein G Dynabeads (Invitrogen) overnight. Each ChIP experiment was performed at least in triplicate with the same batch of antibody to reduce quantitative batch-to-batch variation, although qualitative differences are not observed. The primers used for quantitative PCR on the DNA samples were designed with Primer3 software and are described in Supplementary Table 1. The data were generated by quantitative PCR (Opticon 3; MJ Research, Waltham, MA) and represent means ± SEs of fold enrichment over ChIP samples from isogenic controls lacking the HA tag (n ≥ 3).
Microscopy
DNA was visualized with 1 μg/ml DAPI. Data were collected from three samples, each of 100 cells. Microscopy was performed on a Nikon E800 microscope (Melville, NY) with a 100×/1.40 Plan-Apo objective lens. Images were captured on a Spot RT/SE Camera using Spot advanced software (Spot Imaging Solutions, Sterling Heights, MI).
Mutagenicity assays
Mutation rates in the can1 gene were calculated using the method of the median from 11 cultures that were started from independent single colonies (Lee et al., 2007). Mutants were plated on EMM2 medium with 60 μg/ml canavanine. MMS-induced mutagenesis was performed on cells in liquid cultures treated with 0.05% MMS for 60 min, with the MMS then inactivated with 5% sodium thiosulfate.
Supplementary Material
Acknowledgments
We thank Nick Boddy, Greg Freyer, and Matthew Whitby for S. pombe strains; Johanne Murray, Xiaolan Zhao, Patricia Cortes, Cathie Pfleger, Karen Kuntz, Kirstin Bass, and Emily Outwin for critical discussions; and Zhen-Qiang Pan for advice on ubiqutination assays. This work was supported by National Institute of Health Grants CA100076 and GM088162.
Abbreviations used:
- BRCT
BRCA1 C-terminal domain
- ChIP
chromatin Immunoprecipitation
- CPT
camptothecin
- DAPI
4′,6-diamidino-2-phenylindole
- DSB
double-stranded DNA break
- GFP
green fluorescent protein
- HA
hemagglutinin
- HEAT
Huntington, EF3, PP2A, Tor1
- HR
homologous recombination
- HU
hydroxyurea
- MAGE
melanoma antigen
- MMS
methyl methanesulfonate
- NER
nucleotide excision repair
- 4-NQO
4-nitroquinoline-N-oxide
- OTR
outer centromere repeat
- PCNA
proliferating cell nuclear antigen
- PRR
postreplication Repair
- RING
really interesting new gene
- SMC
structural maintenance of chromosomes
- STE1
subtelomeric repeats
- SUMO
small ubiquitin-like modifier
- vRING
variant ring
- YES
yeast extract plus supplements
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0272) on October 5, 2011.
REFERENCES
- Akamatsu Y, Tsutsui Y, Morishita T, Siddique MS, Kurokawa Y, Ikeguchi M, Yamao F, Arcangioli B, Iwasaki H. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 2007;26:1352–1362. doi: 10.1038/sj.emboj.7601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- Callegari AJ, Clark E, Pneuman A, Kelly TJ. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc Natl Acad Sci USA. 2010;107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari AJ, Kelly TJ. UV irradiation induces a postreplication DNA damage checkpoint. Proc Natl Acad Sci USA. 2006;103:15877–15882. doi: 10.1073/pnas.0607343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari AJ, Kelly TJ. Shedding light on the DNA damage checkpoint. Cell Cycle. 2007;6:660–666. doi: 10.4161/cc.6.6.3984. [DOI] [PubMed] [Google Scholar]
- Calonge TM, Eshaghi M, Liu J, Ronai Z, O'Connell MJ. Transformation/transcription domain-associated protein (TRRAP)-mediated regulation of Wee1. Genetics. 2010;185:81–93. doi: 10.1534/genetics.110.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge TM, O'Connell MJ. Antagonism of Chk1 signaling in the G2 DNA damage checkpoint by dominant alleles of Cdr1. Genetics. 2006;174:113–123. doi: 10.1534/genetics.106.060970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J Biol Chem. 2011;286:5119–5125. doi: 10.1074/jbc.M110.201608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen YH, Branzei D, Zhao X. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2306–2314. doi: 10.1091/mbc.E10-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Doe CL, Osman F, Dixon J, Whitby MC. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 2004;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell. 2009a;35:657–668. doi: 10.1016/j.molcel.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H. Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J Biol Chem. 2009b;284:8507–8515. doi: 10.1074/jbc.M809139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S, Nurse P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Ann Rev Cell Biol. 1991;7:227–256. doi: 10.1146/annurev.cb.07.110191.001303. [DOI] [PubMed] [Google Scholar]
- Frampton J, Irmisch A, Green CM, Neiss A, Trickey M, Ulrich HD, Furuya K, Watts FZ, Carr AM, Lehmann AR. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Kimata Y, Nomaguchi K, Watanabe K, Kohno K. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J Biol Chem. 2002;277:21585–21591. doi: 10.1074/jbc.M201523200. [DOI] [PubMed] [Google Scholar]
- Germe T, Miller K, Cooper JP. A non-canonical function of topoisomerase II in disentangling dysfunctional telomeres. EMBO J. 2009;28:2803–2811. doi: 10.1038/emboj.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SH, Sheedy DM, Cuddihy AR, O'Connell MJ. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol Cell Biol. 2004;24:662–674. doi: 10.1128/MCB.24.2.662-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch A, Ampatzidou E, Mizuno K, O'Connell MJ, Murray JM. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009;28:144–155. doi: 10.1038/emboj.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Strom L, Katou Y, Itoh T, Shirahige K, Sjogren C. Chromosome length influences replication-induced topological stress. Nature. 2011;471:392–396. doi: 10.1038/nature09791. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee KM, Nizza S, Hayes T, Bass KL, Irmisch A, Murray JM, O'Connell MJ. Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function. Genetics. 2007;175:1585–1595. doi: 10.1534/genetics.106.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Walicka M, Grittiths DJF, Murray JM, Watts FZ, McCready S, Carr AM. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Pavlova Y, Yates JR 3rd, Boddy MN. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–723. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Tsutsui Y, Iwasaki H, Shinagawa H. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol Cell Biol. 2002;22:3537–3548. doi: 10.1128/MCB.22.10.3537-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Carr AM. Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outwin EA, Irmisch A, Murray JM, O'Connell MJ. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol Cell Biol. 2009;29:4363–4375. doi: 10.1128/MCB.00377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR. The SMC5-6 DNA repair complex: bridging of the SMC5-6 heads by the Kleisin, NSE4, and non-Kleisin subunits. J Biol Chem. 2006;281:36952–36959. doi: 10.1074/jbc.M608004200. [DOI] [PubMed] [Google Scholar]
- Pebernard S, McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Perry JJ, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol Biol Cell. 2008a;19:4099–4109. doi: 10.1091/mbc.E08-02-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008b;27:3011–3023. doi: 10.1038/emboj.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, 3rd, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews EA, Sweeney S, Shinagawa H, Watts FZ, Lehmann AR. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol Cell Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy DM, Dimitrova D, Rankin JK, Bass KL, Lee KM, Tapia-Alveal C, Harvey SH, Murray JM, O'Connell MJ. Brc1-mediated DNA repair and damage tolerance. Genetics. 2005;171:457–468. doi: 10.1534/genetics.105.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Igawa E, Tanihigashi H, Matsubara M, Ide H, Ikeda S. Roles of base excision repair enzymes Nth1p and Apn2p from Schizosaccharomyces pombe in processing alkylation and oxidative DNA damage. DNA Repair (Amst) 2005;4:1270–1280. doi: 10.1016/j.dnarep.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuts D, Simpson LJ, Kabani S, Yamazoe M, Sale JE. Role for RAD18 in homologous recombination in DT40 Cells. Mol Cell Biol. 2006;26:8032–8041. doi: 10.1128/MCB.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C, Outwin EA, Trempolec N, Dziadkowiec D, Murray JM, O'Connell MJ. SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle. 2010;9:2065–2070. doi: 10.4161/cc.9.11.11734. [DOI] [PubMed] [Google Scholar]
- Tatebayashi K, Kato J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–57. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010;6:e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell. 1999;10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade HM, Teli T, Laursen LV, Murray JM, O'Connell MJ. A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol Genet Genomics. 2001;265:993–1003. doi: 10.1007/s004380100494. [DOI] [PubMed] [Google Scholar]
- Wang SW, Norbury C, Harris AL, Toda T. Caffeine can override the S-M checkpoint in fission yeast. J Cell Sci. 1999;112:927–937. doi: 10.1242/jcs.112.6.927. [DOI] [PubMed] [Google Scholar]
- Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 2010;29:1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, Arcangioli B, Martienssen RA. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.