Lamin B1 is essential for neuronal migration and progenitor proliferation during the development of the cerebral cortex. The observation of distinct phenotypes of Lmnb1- and Lmnb2-knockout mice and the differences in the nuclear morphology of cortical neurons in vivo suggest that lamin B1 and lamin B2 play distinct functions in the developing brain.
Abstract
Neuronal migration is essential for the development of the mammalian brain. Here, we document severe defects in neuronal migration and reduced numbers of neurons in lamin B1–deficient mice. Lamin B1 deficiency resulted in striking abnormalities in the nuclear shape of cortical neurons; many neurons contained a solitary nuclear bleb and exhibited an asymmetric distribution of lamin B2. In contrast, lamin B2 deficiency led to increased numbers of neurons with elongated nuclei. We used conditional alleles for Lmnb1 and Lmnb2 to create forebrain-specific knockout mice. The forebrain-specific Lmnb1- and Lmnb2-knockout models had a small forebrain with disorganized layering of neurons and nuclear shape abnormalities, similar to abnormalities identified in the conventional knockout mice. A more severe phenotype, complete atrophy of the cortex, was observed in forebrain-specific Lmnb1/Lmnb2 double-knockout mice. This study demonstrates that both lamin B1 and lamin B2 are essential for brain development, with lamin B1 being required for the integrity of the nuclear lamina, and lamin B2 being important for resistance to nuclear elongation in neurons.
INTRODUCTION
The patterning of the cerebral cortex during embryonic development involves the rapid expansion of the pool of neuronal progenitors in the ventricular zone and their radial migration as neurons into the cortical plate (Gupta et al., 2002; Ayala et al., 2007; Wynshaw-Boris, 2007). Successive waves of neurons migrate to form distinct layers within the cortical plate, with each new layer located more superficially than earlier layers (Gupta et al., 2002). The positioning of neurons into cortical layers is crucial for their identity and their ability to form proper connections within the brain (Wynshaw-Boris, 2007), and defects in neuronal migration have catastrophic consequences for the organization of the brain (Gupta et al., 2002; Ayala et al., 2007; Wynshaw-Boris, 2007). The study of neurodevelopmental defects in humans has helped to identify cytoplasmic proteins required for neuronal migration (e.g., LIS1, NDE1, NDEL1; Gupta et al., 2002; Ayala et al., 2007; Wynshaw-Boris, 2007). These proteins regulate a network of microtubules and dynein motors and are essential for moving the cell nucleus in the direction of the leading edge of the neuron—a process called nuclear translocation (Vallee and Tsai, 2006; Tsai et al., 2007; Wynshaw-Boris, 2007). Defects that prevent nuclear translocation block the migration of neurons to the cortical plate and lead to severe neurodevelopmental abnormalities (Solecki et al., 2006; Wynshaw-Boris, 2007).
Nuclear translocation depends on effective connections between the cell nucleus and the microtubule network in the cytoplasm (Tanaka et al., 2004; Wynshaw-Boris, 2007; Pawlisz et al., 2008). Although the cytoplasmic factors regulating neuronal migration have been thoroughly investigated, it is only recently that nuclear proteins with roles in this process have been identified. First, Zhang et al. (2009) identified key roles for the SUN proteins and nesprins in neuronal migration. These proteins of the nuclear envelope are components of the LINC complex, a molecular bridge that links the nucleus to the cytoskeleton (Fridkin et al., 2009; Méjat and Misteli, 2010). Zhang et al. (2009) documented abnormalities in neuronal migration in mice lacking both SUN1 and SUN2 and in mutant mice expressing inactive forms of Syne-1/Nesprin 1 and Syne-2/Nesprin 2. Shortly thereafter, Coffinier et al. (2010a) discovered that lamin B2, a protein of the nuclear lamina, is critical for neuronal migration during embryonic development. In Lmnb2-knockout mice (Lmnb2–/–), the layering of neurons within the cerebral cortex was abnormal, and the cerebellum was reduced in size and devoid of folds (Coffinier et al., 2010a). The latter observations provided the first indication that the nuclear lamina is important for the development of the mammalian brain.
In addition to lamin B2, the nuclear lamina of most differentiated somatic cells contains lamins A, C, and B1 (Dechat et al., 2008). There is little reason to believe that LMNA, the gene encoding lamins A and C, is crucial for the development of the brain. Lmna is expressed late in mouse development (Rober et al., 1989), and Lmna-deficient mice develop normally, surviving for 5–6 wk before succumbing to cardiomyopathy and/or muscular dystrophy (Sullivan et al., 1999). Hundreds of clinically significant mutations have been identified in LMNA, but none have been linked to neurodevelopmental abnormalities. Most LMNA mutations in humans cause muscular dystrophy and/or cardiomyopathy, but some cause lipodystrophy, peripheral neuropathy, and progeria (Worman et al., 2009).
Whether lamin B1 is involved in brain development has been an open question. Vergnes et al. (2004) reported that lamin B1–deficient (Lmnb1Δ/Δ) mice were small, exhibited abnormalities in the lungs and skeleton, and died soon after birth. Lmnb1Δ/Δ fibroblasts manifest nuclear blebbing and early senescence (Vergnes et al., 2004). The possibility of CNS disease in Lmnb1Δ/Δ embryos was not investigated, but the mutant mice had an abnormally shaped cranium (Vergnes et al., 2004). This finding, together with the neurodevelopmental abnormalities in Lmnb2–/– mice, suggested that lamin B1 might be important for brain development. In the present study, we investigated that possibility, taking advantage of both the original Lmnb1Δ/Δ mouse model and newly developed conditional knockout alleles for Lmnb1 and Lmnb2 (Yang et al., 2011).
RESULTS
The abnormally shaped cranium in Lmnb1Δ/Δ embryos (Vergnes et al., 2004) led us to consider a potential role for lamin B1 in brain development. Histological analyses revealed that the brains of newborn Lmnb1Δ/Δ mice were abnormal (Figure 1). The layering of neurons in the cerebral cortex was absent, with reduced numbers of cells (Figure 1, A–C); no lamination was observed in the hippocampus; and the cerebellum was reduced in size, with no foliation (Figure 1A). The number of cortical neurons was reduced, as judged by immunostaining for the neuronal marker NeuN (Figure 1C). The neurodevelopmental abnormalities in Lmnb1Δ/Δ mice suggested that Lmnb1 is important during embryonic development. Indeed, β-galactosidase staining at E15.5 revealed Lmnb1 expression throughout the cerebral cortex (Figure 1D). At the same stage, Lmnb2 expression was prominent in the ventricular zone of the cortex (Coffinier et al., 2010a), and Lmna expression in the brain was minimal (although it was detected in the surrounding mesenchyme; Figure 1D).
FIGURE 1:
Brain abnormalities in newborn Lmnb1Δ/Δ mice. (A) Top, hematoxylin and eosin (H&E) staining of paraffin-embedded sections from newborn wild-type (WT) and Lmnb1Δ/Δ mice. ctx, cortex; hi, hippocampus; ob, olfactory bulb; st, striatum. Scale bar, 500 μm. Bottom, corresponding images at higher magnification of the hippocampus (left), and cerebellum (right); scale bar, 200 μm. (B) H&E–stained sections of the cerebral cortex from newborn WT and Lmnb1Δ/Δ mice. Roman numerals indicate the position of the cortical layers; IZ, intermediate zone; VZ, ventricular zone. (C) Immunostaining of the cerebral cortex of newborn WT, Lmnb1Δ/Δ, and Lmnb2–/– mice with an antibody against the neuronal marker NeuN (red). DNA was stained with DAPI (blue). Layers of the cortical plate are indicated on the left. (D) Lmnb1, Lmnb2, and Lmna expression patterns in the cerebral cortex at E15.5. Frozen sections of brain from Lmnb1+/Δ, Lmnb2+/–, and LmnalacZ/+ embryos were stained for β-galactosidase activity. Sections were counterstained with eosin. Brackets indicate the position of the cortical plate (CP). Scale bars, B and C, 100 μm; D, 50 μm.
At E15.5 and E17.5, the cortical plate was thinner in Lmnb1Δ/Δ embryos than in wild-type (WT) embryos (Figure 2, A–C). Immunostaining for TBR1 (a marker of cortical layer VI) revealed similar numbers of TBR1-positive (TBR1+) cells in WT and Lmnb1Δ/Δ embryos at E13.5; however, there were fewer TBR1+ cells in Lmnb1Δ/Δ embryos by E15.5, and those neurons were located more superficially than in WT brains (Figure 2D). Immunohistochemical studies of E16.5 embryos with antibodies against Otx1 (a marker of cortical layers V–VI) and TBR1 revealed that neurons expressing those markers were located more superficially in Lmnb1Δ/Δ brains (Figure 2E). The positions of the subplate and of the lateral projections, visualized by staining with the monoclonal antibodies CS56 and L1, respectively, were also more superficial than normal (Figure 2E). At E18.5, neurons expressing the layer V marker Ctip2 were also located more superficially in Lmnb1Δ/Δ embryos (Figure 2F). The abnormal positioning of those deep-layer neurons suggested a defect of the neurons born later in forming the upper layers of the cortex. To test this hypothesis, we performed neuronal birthdating experiments; pregnant mice were injected with bromodeoxyuridine (BrdU) at E13.5, and BrdU-labeled neurons were examined in E18.5 embryos (Figure 2F). In WT brains, BrdU-positive neurons (i.e., cells born at E13.5) were found in layer V, whereas in the brain of Lmnb1Δ/Δ mice, BrdU-positive neurons were scattered throughout the cortical plate (Figure 2F). Together, these studies demonstrated a defect in neuronal migration in Lmnb1Δ/Δ embryos. We used immunohistochemistry to assess Reelin expression in the marginal zone, as Reelin deficiency is known to impair neuronal migration (Rice and Curran, 2001). However, Reelin appeared to be expressed similarly in Lmnb1Δ/Δ and control embryos at E12.5–E17.5 (Supplemental Figure S1).
FIGURE 2:
Reduced size of the cortical plate and abnormal layering of cortical neurons in Lmnb1Δ/Δ embryos. (A, B) H&E staining on parasagittal sections of wild-type (WT) and Lmnb1Δ/Δ brains at E15.5 (A) and E17.5 (B). IZ, intermediate zone; MZ; marginal zone; VZ, ventricular zone. Brackets indicate the position of the cortical plate (CP). In Lmnb1Δ/Δ mice, the cortical plate was thinner and the density of neurons was reduced. (C) Ratio of cortical plate thickness to cortex thickness in Lmnb1Δ/Δ mice (shaded bars) and WT mice (white bars) at E15.5 (n = 3 WT and 4 Lmnb1Δ/Δ embryos, p = 0.0218), E16.5 (n = 2 per group), and E17.5 (n = 3 per group, p < 0.0001). Measurements were made on matching sections (≥2 sections per embryo). Errors bars indicate SD. (D) Immunostaining for the layer VI marker TBR1 on brain sections from WT and Lmnb1Δ/Δ embryos. TBR1 (green) is expressed normally in Lmnb1Δ/Δ embryos at E13.5, but at E15.5, the layer of TBR1-positive cells was thinner and more superficial than in WT embryos. DNA was stained with DAPI (blue). (E) Reduced size and abnormal positioning of cortical layers V and VI in Lmnb1Δ/Δ embryos. Parasagittal sections of brains from E16.5 WT and Lmnb1Δ/Δ embryos stained with antibodies against the layer V–VI marker Otx1 (green) and the subplate marker CS56 (red) (left) as well as TBR1 (green) and the lateral projection marker L1 (red) (right). Brackets indicate the positions of the cortical plate (left) and layer VI (right). (F) Abnormal positioning of cortical neurons in Lmnb1Δ/Δ embryos. Sections of WT and Lmnb1Δ/Δ embryos collected at E18.5 after an injection of BrdU at E13.5. Left, tissues were stained for the neuronal marker NeuN (red) and the layer V marker Ctip2 (green). Right, tissues were stained with an anti-BrdU antibody (red). Brackets mark the position of the neurons positive for Ctip2. Scale bars, A and B, 100 μm; D, 50 μm; E, F, 200 μm.
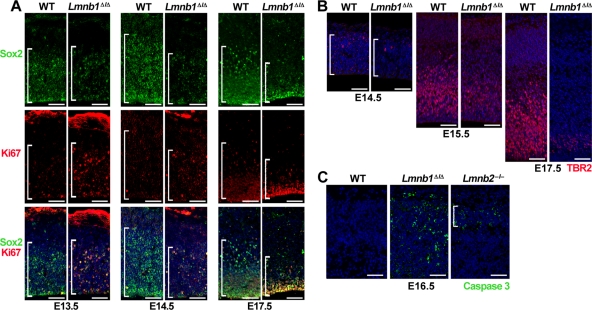
The small size of the cortical plate was due in part to reduced numbers of neuronal progenitors. At E13.5, similar numbers of Sox2+ progenitors were found in Lmnb1Δ/Δ and WT brains, but by E14.5–E15.5 their numbers were clearly reduced in Lmnb1Δ/Δ brains (167 ± 10 Sox2+ cells in E15.5 mutant embryos [n = 3] vs. 267 ± 72 in WT embryos [n = 3], per area of 430 × 470 μm; p = 0.06; Figure 3A). At the same stage, the proportion of Sox2+ cells expressing the mitotic marker Ki67 was higher in Lmnb1Δ/Δ brains, suggesting the possibility that neurons in Lmnb1Δ/Δ embryos spend more time in the S–M phase (Figure 3A). At E16.5, the numbers of Ki67+Sox2+ cells were ∼20% higher in Lmnb1Δ/Δ embryos than in WT embryos (47.8 ± 5.2% [n = 3] vs. 36.7 ± 4.6% [n = 4]; p = 0.03). In addition to producing cortical neurons, neuronal progenitors give rise to intermediate progenitors that accumulate in the subventricular zone and express TBR2 (Dehay and Kennedy, 2007). Intermediate progenitors differentiate at later stages and contribute to layers II–III of the cortical plate. At E15.5 and E17.5, we observed fewer TBR2+ cells in the subventricular zone of Lmnb1Δ/Δ embryos (Figure 3B). At E15.5, the number of intermediate progenitors was reduced by 50% in Lmnb1Δ/Δ embryos (163 ± 52 TBR2+ cells in an area of 430 × 350 μm; at least three areas evaluated per embryo; n = 3 embryos) compared with WT embryos (322 ± 49 cells; n = 3 embryos; p = 0.018). Aside from reduced proliferation, we detected apoptotic cells in the cortex of E16.5 Lmnb1Δ/Δ embryos by staining for active caspase 3 (Figure 3C). The brains of E16.5 Lmnb2–/– embryos also stained positively for active caspase 3, but the apoptotic cells were fewer in number and confined to the cortical plate; in contrast, fewer than two positive cells were observed per slice of WT cortex (Figure 3C).
FIGURE 3:
Reduced numbers of neuronal progenitors in Lmnb1Δ/Δ brains. (A) Immunostaining for the neuronal progenitor marker Sox2 (top, green) and the mitotic antigen Ki67 (middle, red) at E13.5, E14.5, and E17.5. Bottom, merged images with DAPI (blue). Brackets mark the ventricular zone containing the Sox2+ cells. (B) Immunostaining for TBR2, a marker for intermediate neuronal progenitors, at E14.5, E15.5, and E17.5. Brackets mark the territory occupied by TBR2-positive cells (red). DNA was stained with DAPI (blue). (C) Immunostaining for active caspase 3 (green), a marker of apoptosis, in cerebral cortex of E16.5 WT, Lmnb1Δ/Δ, and Lmnb2–/– embryos. Bracket indicates the position of the cortical plate. Scale bars, A and B, 50 μm; C, 100 μm.
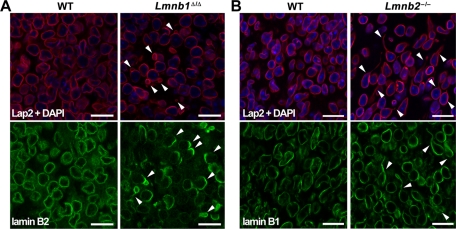
Defects in lamins A and C often lead to severe nuclear shape abnormalities in cultured fibroblasts (Muchir et al., 2004), but misshapen nuclei are seldom found in mouse tissues. For example, we observed many nuclear blebs in fibroblasts cultured from “lamin A–only mice” (LmnaLAO/LAO), but no misshapen nuclei were found in the tissues of those mice (Coffinier et al., 2010b). Vergnes et al. (2004) documented nuclear blebs in Lmnb1Δ/Δ fibroblasts, but we were skeptical that we would find misshapen nuclei in tissues of Lmnb1Δ/Δ mice. To our surprise, however, we observed severe nuclear shape abnormalities in the cerebral cortex of E16.5 Lmnb1Δ/Δ embryos. In brain sections stained for the nuclear envelope protein Lap2β or for lamin B2, 24.8 ± 6.8% of cortical neurons from Lmnb1Δ/Δ embryos (n > 350 cells evaluated per embryo, three different embryos) contained a solitary nuclear bleb versus none in WT neurons (n > 123 cells evaluated per embryo, three different embryos; p = 0.003; Figure 4A). Immunostaining for lamin B2 uncovered a second abnormality: 75 ± 6.1% of the cortical neurons in Lmnb1Δ/Δ embryos exhibited an asymmetric distribution of lamin B2 at the nuclear rim (Figure 4A; compared with none in the WT samples; same numbers of cells evaluated; n = 3 embryos/group; p < 0.0001). The nuclear bleb was invariably found in the region of the nuclear rim enriched in lamin B2 (Figure 4A).
FIGURE 4:
Lamin B1 and lamin B2 deficiencies yield distinct nuclear abnormalities in cortical neurons from E16.5 embryos. (A) Immunostaining of cortex from WT and Lmnb1Δ/Δ embryos with antibodies against Lap2β (red) or lamin B2 (green). (B) Immunostaining of cortex from WT and Lmnb2–/– embryos with antibodies against Lap2β (red) or lamin B1 (green). Arrowheads indicate nuclei with morphological abnormalities—nuclear blebs and asymmetric distribution of lamin B2 in Lmnb1Δ/Δ neurons (A) and elongated nuclei in Lmnb2–/– neurons (B). DNA was stained with DAPI (blue). Scale bar, 25 μm.
Lmnb2–/– fibroblasts do not have nuclear blebs (Coffinier et al., 2010a), but the finding of misshapen nuclei in Lmnb1Δ/Δ neurons prompted us to investigate whether Lmnb2–/– neurons might also have nuclear shape abnormalities (Figure 4B). At E16.5, cortical neurons of Lmnb2–/– mice did not have nuclear blebs, but we found cells with an elongated nucleus in lamin B2–deficient brains (Figure 4B), and there was a significant increase in the length of the nucleus in lamin B2–deficient embryonic neurons in situ compared with WT neurons (p < 0.0001; Supplemental Figure S2A). Lamin B1 was evenly distributed at the nuclear rim of Lmnb2–/– neurons—even in cells with elongated nuclei (Figure 4B).
Nuclear shape abnormalities were also observed in neurons grown from cortical explants of E13.5 embryos. Many Lmnb1Δ/Δ neurons had a single nuclear bleb, and some exhibited an asymmetric distribution of lamin B2 (Supplemental Figure S3). In the case of neurons from Lmnb2–/– embryos, we observed occasional comet-shaped nuclei with detached centrosomes (located >20 μm from the cell nucleus; Supplemental Figure S2B). Comet-shaped nuclei were never observed in neuronal progenitors from WT embryos.
Lmnb1Δ/Δ and Lmnb2–/– mice die soon after birth. To assess postnatal phenotypes in the brain, we used Lmnb1 and Lmnb2 conditional knockout alleles (Yang et al., 2011) and the Emx1-Cre transgene (Gorski et al., 2002) to generate forebrain-specific Lmnb1- and Lmnb2-knockout mice (Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl, respectively). For both conditional knockout alleles, CRE-mediated recombination excises exon 2, creating a frameshift and yielding a null allele. Specific expression of the Emx1-Cre transgene in the forebrain was documented with a CRE-activated lacZ reporter (Supplemental Figure S4A; Soriano, 1999), confirming results already in the literature (Gorski et al., 2002). Efficient forebrain-specific gene inactivation during embryogenesis was achieved with both the Lmnb1 and Lmnb2 conditional alleles. Immunohistochemical studies on the brain from an E15.5 Emx1-Cre Lmnb1fl/fl embryo revealed markedly reduced lamin B1 expression in the forebrain (Supplemental Figure S4B). Higher-magnification images of Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl brains revealed that ∼90% of E15.5 cortical cells had no lamin B1 or lamin B2, respectively (Supplemental Figure S4C).
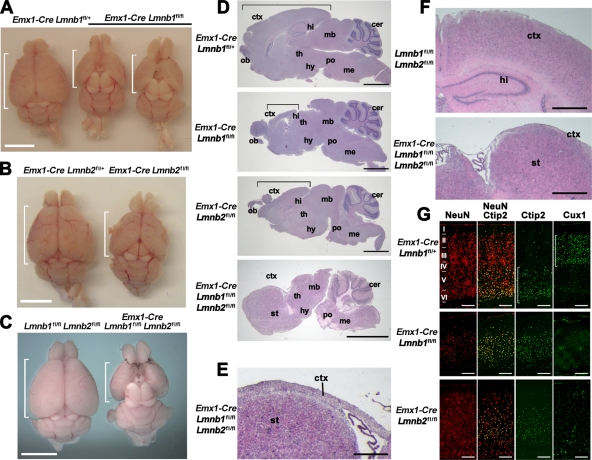
Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl mice were born at the expected Mendelian frequency, appeared to have normal longevity (mice were observed for >1 yr), and were grossly indistinguishable from WT mice. After removal of the skin, however, we observed that the cranium in both models was smaller, and the cerebral cortex was reduced in size (Figure 5, A and B). At 4 mo of age, the average length of the cortex in Emx1-Cre Lmnb1fl/fl mice was 0.65 ± 0.03 cm versus 0.90 ± 0.03 cm in WT mice (n = 3 per group; p < 0.001). The length of the cortex in Emx1-Cre Lmnb2fl/fl mice was 0.80 ± 0.0 cm versus 1.02 ± 0.02 cm in WT mice (n = 2 per group). We also bred mice lacking both lamin B1 and lamin B2 in the forebrain (Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl). The cortex of these “double-knockout” mice was significantly smaller than those of Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl mice (Figure 5, C and D). Compared to WT siblings, the double-knockout mice had very similar body weights (16.20 ± 2.96 g vs. 16.10 ± 2.39 g) at 1 mo of age, but the brain weight of the double-knockout mice was significantly smaller than that of WT mice (0.27 ± 0.01 vs. 0.47 ± 0.02 g, respectively; n = 4 and 5 females; p < 0.0001). Brain sections of 1-mo-old double-knockout mice revealed atrophy of the cortex, which was reduced to a thin layer of tissue overlaying the striatum (Figure 5, E and F). Coronal sections also showed a complete absence of the hippocampal structures (Figure 5F). Immunohistochemistry studies on E17.5 Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl embryos revealed cells that lacked both lamin B1 and lamin B2 (Supplemental Figure S4D). Immunohistochemical studies on the thin layer of tissue above the striatum in adult Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl mice failed to detect any neurons (Supplemental Figure S4E), and none of the remaining cells lacked expression of both lamin B1 and lamin B2 (Supplemental Figure S4F).
FIGURE 5:
Forebrain-specific deletion of Lmnb1 and Lmnb2 results in small forebrains and abnormal layering of cortical neurons in adult mice. (A–C) Reduced size of the forebrain in Emx1-Cre Lmnb1fl/fl (A), Emx1-Cre Lmnb2fl/fl (B), and double-knockout Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl (C) mice compared with control siblings. Brains are oriented with the olfactory lobes on top; the brackets indicate the length of the cerebral cortex. (D) H&E staining of sagittal brain sections from Emx1-Cre Lmnb1fl/fl, Emx1-Cre Lmnb2fl/fl, and Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl mice, along with a control mouse (Emx1-Cre Lmnb1fl/+). Brackets indicate the length of the cortex. (E) Higher-magnification image of the double-knockout Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl brain section to illustrate the reduced thickness of the cortex (ctx). (F) H&E staining of coronal brain sections from a control mouse and an Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl double-knockout mouse. The hippocampus is absent in the double-knockout brain, and cortical tissue is markedly reduced in size, forming just a thin layer of tissue above the striatum. (G) Immunohistochemical staining on cortical sections with the neuronal marker NeuN (red), the layer V–VI marker Ctip2, and the layer II–III marker Cux1 (both green). Roman numerals indicate cortical layers; brackets indicate layers of neurons positive for Ctip2 or Cux1. Scale bars, A–C, 5 mm; D, 2 mm; E, 250 μm; F, 500 μm; G, 200 μm. All samples are from 1-mo-old mice. cer, cerebellum; ctx, cortex; hi, hippocampus; hy, hypothalamus; mb, midbrain; me, medulla; ob, olfactory bulb; po, pons; st, striatum; th, thalamus.
We analyzed the effect of the forebrain-specific inactivation of Lmnb1 or Lmnb2 on the layering of neurons in the cortex. As expected, the layering of the cortical neurons in the adult brain was abnormal in both Emx1-Cre Lmnb1fl/fl mice and Emx1-Cre Lmnb2fl/fl mice. Immunostaining for the neuronal marker NeuN revealed reduced numbers of neurons in Emx1-Cre Lmnb1fl/fl mice, with most neurons expressing the layer V marker Ctip2 and very few neurons expressing the layer II–III marker Cux1 (Figure 5G). Emx1-Cre Lmnb2fl/fl mice had fewer cortical neurons than did WT mice (Figure 5G and Supplemental Figure S4C), and neurons failed to organize into proper layers. However, there were significantly more neurons and more Cux1-positive cells in Emx1-Cre Lmnb2fl/fl brains than in Emx1-Cre Lmnb1fl/fl brains (Figure 5G).
Misshapen cell nuclei were easily detectable in both forebrain-specific knockout models. Many neurons of Emx1-Cre Lmnb1fl/fl embryos contained a solitary nuclear bleb and exhibited an asymmetric distribution of lamin B2 (Supplemental Figure S5A). In Emx1-Cre Lmnb2fl/fl brains, we observed an increased frequency of neurons with elongated nuclei, but lamin B1 was distributed evenly at the nuclear rim (Supplemental Figure S5B). Of interest, few neurons in the cerebral cortex of adult Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl mice exhibited abnormal nuclear morphology (Figure 6A). However, in the dentate gyrus of adult Emx1-Cre Lmnb1fl/fl mice, many neurons exhibited a markedly asymmetric distribution of lamin B2 (Figure 6B). In contrast, neurons in the dentate gyrus from Emx1-Cre Lmnb2fl/fl mice exhibited normal nuclear morphology (Figure 6C).
FIGURE 6:
Nuclear morphology in the forebrain of adult Emx1-Cre Lmnb1fl/fl and Emx1-Cre Lmnb2fl/fl mice. (A) Immunohistochemical studies of the cortex in 1-mo-old mice with antibodies against lamin B1 (green) and lamin B2 (red); nuclear DNA is stained with DAPI (blue). Arrowheads indicate the position of nuclei of cells that lack one of the two B-type lamins. Scale bars, 50 μm. (B) Immunohistochemical studies of the dentate gyrus of 1-mo-old Emx1-Cre Lmnb1fl/+, Emx1-Cre Lmnb1fl/fl, and Emx1-Cre Lmnb2fl/fl mice with antibodies against lamin B1 (green) and lamin B2 (red). Top, low magnification; dotted lines outline the dentate gyrus; boxed areas indicate the positions of the high-magnification views on the bottom. Scale bars: top, 250 μm; bottom, 50 μm.
Recently Yang et al. (2011) showed that the absence of both lamin B1 and lamin B2 in skin keratinocytes had no effect on skin development or on keratinocyte proliferation and survival. In contrast, we found dramatic nuclear abnormalities in neurons and severe neurodevelopmental abnormalities in Lmnb1- and Lmnb2-deficient mice. In addition, no neurons were detected in the remnant of cortex found in adult Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl mice. We suspected that the different effects of lamin B deficiencies in skin and brain might relate to different levels of lamin A/C expression. Indeed, Lmna expression is almost undetectable in the embryonic brain, but Lmna expression in the skin of E15.5 and E19.5 embryos is robust (Supplemental Figure S6A). Immunostaining of embryos at E17.5 provided further evidence for robust lamin A/C expression in the skin; however, no lamin A/C expression could be detected in the cortical plate (Supplemental Figure S6B).
In contrast to the embryonic brain, which does not express significant amounts of lamin A/C (Figure 1D), all three lamin genes are expressed in the adult mouse brain (Supplemental Figure S6C). In the setting of lamin B1 deficiency, a robust signal for lamin A/C was associated with a normal distribution of lamin B2 at the nuclear rim (Supplemental Figure S6D). Of interest, some neurons of the dentate gyrus in adult mice—which retained an abnormal distribution of lamin B2 (Figure 6C and Supplemental Figure S6E)—exhibited lower levels of lamin A/C expression (Supplemental Figure S6E).
The phenotypes of the conventional knockout mice and the forebrain-specific knockout mice are summarized in Table 1.
Table 1:
Summary of the phenotypes in the setting of lamin B1 deficiency or lamin B2 deficiency.
| Lamin B1 deficiency | Lamin B2 deficiency | |
|---|---|---|
| Conventional knockout embryos | ||
| Overall phenotype | Perinatal lethal; small embryos; defects in bones and lungs | Perinatal lethal; embryos grossly normal in size; no obvious defects except in the brain |
| Brain phenotype | Reduced size, reduced populations of progenitors; disorganized layering of neurons in cortex; no foliation of cerebellum | Size close to normal; abnormal layering of neurons in cerebral cortex; no foliation of cerebellum |
| Neuronal phenotypes | Neuronal migration defects; markedly reduced number of neurons | Neuronal migration defects; number of neurons mildly reduced |
| Nuclear abnormalities in neurons | Large, solitary nuclear bleb in cortical neurons; asymmetric distribution of lamin B2 | Elongated nuclei in cortical neurons; normal distribution of lamin B1 at the nuclear rim |
| Embryonic fibroblasts | Nuclear blebs, normal distribution of lamin B2 | Elongated nuclei, normal distribution of lamin B1 along the rim |
| Forebrain-specific knockout mice | ||
| Overall phenotype | Viable; mice appear grossly normal; small cortex | Viable; mice appear grossly normal; small cortex; cortical defect more pronounced after birth |
| Cortical layers | Very small cortex with low density of neurons; upper cortical layers are missing (no Cux1+ neurons) | Small cortex with abnormal layering of cortical neurons |
| Embryonic neurons | Nuclear blebs, asymmetric distribution of lamin B2 | Elongated nuclei, normal distribution of lamin B1 at the nuclear rim |
| Adult neurons | No nuclear abnormalities, except in the dentate gyrus, where Lmna expression is low | No nuclear abnormalities detected |
DISCUSSION
In the present study, we found that proper development of the brain depends on lamin B1. Lmnb1Δ/Δ embryos have reduced numbers of neurons and abnormal layering of neurons in the cerebral cortex, as well as abnormalities in the hippocampus and cerebellum. The disorganization of cortical neurons in Lmnb1Δ/Δ embryos was caused by impaired neuronal migration, as judged by birthdating and immunohistochemical studies. In wild-type embryos, the neurons born at E13.5 (i.e., BrdU-labeled neurons) were detected 5 d later within layer V of the cortical plate, but in Lmnb1Δ/Δ embryos, many BrdU-labeled neurons were found deeper in the cortex, indicating defective migration. In addition, in Lmnb1Δ/Δ embryos, immunohistochemical markers for cortical layers V and VI were found in more superficial positions in the cortex. The neurodevelopmental abnormalities in Lmnb1Δ/Δ embryos were more severe than those described in Lmnb2–/– embryos (Coffinier et al., 2010a). Consistent with those findings, the phenotypes of forebrain-specific Lmnb1-knockout mice (Emx1-Cre Lmnb1fl/fl) were more severe than those of forebrain-specific Lmnb2-knockout mice (Emx1-Cre Lmnb2fl/fl). The presence of neurodevelopmental abnormalities in Emx1-Cre Lmnb1fl/fl mice should allay any previous concerns that the developmental defects in Lmnb1Δ/Δ brains resulted from toxic effects of the lamin B1–β-galactosidase fusion protein produced by the Lmnb1Δ allele (Vergnes et al., 2004). Emx1-Cre Lmnb1fl/fl mice have a true null allele, yet they have the same neuropathology found in Lmnb1Δ/Δ mice (e.g., disorganization of cortical layers, paucity of neurons, reduced size of the forebrain).
The reduced size of the cortex in Lmnb1Δ/Δ mice was due to reduced numbers of neuronal progenitors. This defect is likely due in part to defective cell proliferation, but this defect may not be unique to neuronal progenitors, as the entire Lmnb1Δ/Δ embryo—and not just the brain—is small (Vergnes et al., 2004). In addition to reduced cell proliferation, apoptosis contributes to the reduced cell density, as we detected widespread activation of caspase 3 in the forebrain of Lmnb1Δ/Δ embryos. Apoptosis was previously reported in association with other defects that impair neuronal migration—for example loss-of-function mutations in LIS1 (Yingling et al., 2008), NDEL1 (Sasaki et al., 2005), NDE1 (Feng and Walsh, 2004), and cyclin-dependent kinase 5 (Li et al., 2002). However, we cannot exclude a more general effect of lamin B1 deficiency on cell viability. Earlier studies with cultured cells suggested that B-type lamins could play important roles in DNA replication, mitotic spindle assembly, heterochromatin organization, and regulation of gene expression (Moir et al., 2000; Lopez-Soler et al., 2001; Hutchison, 2002; Tsai et al., 2006; Dechat et al., 2009). These functions might be crucial in the developing brain, where the expression of A-type lamins is virtually absent (Rober et al., 1989).
In an earlier study (Coffinier et al. 2010a), we found that the size of the cerebral cortex was only slightly smaller in E16.5–19.5 Lmnb2–/– embryos than in wild-type embryos. However, the forebrain size of adult Emx1-Cre Lmnb2fl/fl mice was much smaller than the forebrain of wild-type mice, presumably because brain development proceeds for weeks after birth, and the full effects of lamin B2 deficiency on cell proliferation and survival are therefore more manifest in older mice. The impact of B-type lamins on cell viability was even more dramatic in forebrain-specific Lmnb1/Lmnb2–knockout mice. During embryonic development, it was possible to identify neurons lacking both lamin B1 and lamin B2 in the cortical plate of Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl mice. By 1 mo of age, however, the forebrain of Emx1-Cre Lmnb1fl/fl Lmnb2fl/fl mice was severely atrophic, and it was impossible to find any Lmnb1/Lmnb2–deficient neurons.
The abnormal nuclear morphology in lamin B1– and lamin B2–deficient neurons points to the reduced integrity of the nuclear envelope as a likely explanation for defective neuronal migration. The nuclei of many neurons in Lmnb1Δ/Δ embryos contained a nuclear bleb and exhibited an asymmetric distribution of lamin B2, whereas Lmnb2–/– neurons had an increased frequency of elongated nuclei. Earlier studies identified misshapen nuclei in Lmnb1Δ/Δ fibroblasts (Vergnes et al., 2004) and in HeLa cells, where Lmnb1 expression had been knocked down with small interfering RNAs (Shimi et al., 2008). In Lmnb1Δ/Δ fibroblasts, the nucleus often contains multiple blebs, whereas Lmnb1Δ/Δ neurons contain a single bleb. We suggest that the “solitary-bleb” phenotype might result from unidirectional forces applied on the nucleus during nuclear translocation. In contrast, the multiple blebs in fibroblasts might reflect multidirectional strain from the cytoskeleton as the cells spread out on the culture dish. We did not find nuclear blebs in Lmnb2–/– fibroblasts (Coffinier et al., 2010a) or in the neurons of lamin B2–deficient mice, but we did observe elongated, “comet-shaped” nuclei in the cortical neurons of Lmnb2–/– and Emx1-Cre Lmnb2fl/fl embryos. The stretched-out nuclei in cultured Lmnb2–/– neurons were often associated with a markedly displaced centrosome, suggesting that nuclear abnormalities in lamin B2–deficient neurons were linked to defective nuclear translocation. Lamin B1 was distributed evenly at the nuclear rim in lamin B2–deficient neurons.
In the present study, we demonstrated that lamin B1 deficiency is sufficient to cause severe neurodevelopmental abnormalities. In contrast, Yang et al. (2011) created mice lacking both lamin B1 and lamin B2 in skin keratinocytes and found that skin histology and keratinocyte proliferation were entirely normal. Thus, the consequences of losing the expression of the B-type lamins are significantly different in the brain and skin. Our experiments uncovered a likely mechanism for these differences—strikingly different levels of lamin A/C expression. In the skin of mouse embryos, Lmna expression is robust, as judged by β-galactosidase staining or immunohistochemistry, whereas it is minimal in the developing brain. Strong expression of Lmna in Lmnb1/Lmnb2–deficient skin is associated with protection from both skin pathology and nuclear shape abnormalities, as judged by immunohistochemical studies on frozen sections of skin. In contrast, the absence of Lmna expression in neurons of lamin B1–deficient embryos is associated with severe neuropathology and striking nuclear shape abnormalities and an asymmetric distribution of lamin B2 at the nuclear rim.
Abnormalities in nuclear shape and lamin B2 distribution were apparent in the cerebral cortex neurons of both Lmnb1Δ/Δ and Emx1-Cre Lmnb1fl/fl embryos. However, the nuclear abnormalities were absent in cortical neurons of 1-mo-old Emx1-Cre Lmnb1fl/fl mice, when Lmna expression is robust. Of interest, the asymmetric distribution of lamin B2 persisted in the dentate gyrus of adult Emx1-Cre Lmnb1fl/fl mice, where Lmna expression remains extremely low. Thus, it appears that proper lamin B2 distribution in neurons requires the expression of either lamin B1 or lamin A/C.
Because the nuclear lamina is located within the nucleus, a molecular bridge between the cytoplasmic network of microtubules and the nuclear lamina is almost certainly required to mediate nuclear translocation. Zhang et al. (2009) showed that the SUN proteins and nesprins are required for neuronal migration, and they proposed that the SUN proteins could interact with “lamin B” in the nuclear lamina. This hypothesis seems reasonable, particularly in light of recent studies showing interactions between SUN proteins and A-type lamins in nuclear movement in fibroblasts (Folker et al., 2011). Studies with Lmnb1Δ/Δ fibroblasts lend additional support for a role of B-type lamins in linking the nucleus to the cytoskeleton. Videomicroscopy studies of Lmnb1Δ/Δ fibroblasts revealed that the nucleus spins within the cell (Ji et al., 2007), suggesting that lamin B1 is crucial for tethering the nucleus to the cytoskeleton.
Finding neurodevelopmental abnormalities in lamin B1– and lamin B2–deficient mice provides new insights into the function of the nuclear lamina. Until recently, most investigators focused on A-type lamins and their involvement in muscular dystrophy, cardiomyopathy, lipodystrophy, and progeria (Worman et al., 2009). The prevailing view was that B-type lamins are essential proteins in eukaryotic cells, crucial for mitosis and many other vital functions in the cell nucleus (Dechat et al., 2008; Worman et al., 2009), but our present studies point to specific roles for the B-type lamins in brain development. It remains to be determined whether lamins B1 and B2 have unique or overlapping functions in neuronal migration and brain development. In addition, the partners for lamin B1 and lamin B2 need to be established, along with the structural features of these lamins that are crucial for their function in the brain. In particular, we wonder whether the conserved farnesyl–lipid anchors on lamin B1 and lamin B2 might play a role in keeping the nuclear lamina attached to the inner nuclear membrane during neuronal migration. Finally, our work heralds a new path for investigating the human genetics of nuclear lamins. Neuronal migration defects underlie not only lethal neurodevelopmental defects such as lissencephaly (Wynshaw-Boris and Gambello, 2001; Wynshaw-Boris, 2007), but also other neurological diseases, such as epilepsy, schizophrenia, and autism (Kahler et al., 2008; Deutsch et al., 2010; Wegiel et al., 2010). We suspect that whole-exome sequencing on patients with severe neurological diseases will eventually uncover nonsense and missense mutations in LMNB1 and LMNB2.
MATERIALS AND METHODS
Mouse experiments
The Lmnb1Δ allele (Vergnes et al., 2004) contains a βgeo gene-trap cassette in intron 5 of Lmnb1 and produces a lamin B1–βgeo fusion protein; this fusion protein lacks several major structural domains of lamin B1 and therefore is assumed to be nonfunctional. The Lmnb2-knockout allele (Lmnb2–) is a null allele created by replacing exon 1 of Lmnb2 with a lacZ reporter (Coffinier et al., 2010a). No phenotypic differences were observed between Lmnb1+/+ and Lmnb1+/Δ mice, or between Lmnb2+/+ and Lmnb2+/– mice (Vergnes et al., 2004; Coffinier et al., 2010a). Conditional knockout alleles for Lmnb1 (Lmnb1fl) and Lmnb2 (Lmnb2fl) are described elsewhere (Yang et al., 2011). In both alleles, loxP sites were introduced flanking exon 2; Cre-mediated recombination deletes exon 2 and introduces a frameshift yielding a null mutation. Emx1-Cre transgenic mice (B6.129S2-Emx1tm1(cre)Krj/J; Gorski et al., 2002) and ROSA-lacZ mice (B6.129S4-Gt(ROSA)26Sortm1Sor/J; Soriano, 1999) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice with a Lmna lacZ reporter allele were created with a mouse embryonic stem cell line (S22-2D1) that harbored a retroviral βgeo gene-trap cassette in intron 1 of Lmna (MMRRC). Lmnb1fl/fl, Emx1-Cre Lmnb1fl/+, Lmnb2fl/fl, and Emx1-Cre Lmnb2fl/+ mice were phenotypically normal. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal protocols were reviewed and approved by the Animal Research Committee at the University of California, Los Angeles.
The Lmnb1Δ allele was identified by amplifying a 400–base pair product with forward primer 5′-TCC GTG TCG TGT GGT AGG AGG-3′ and reverse primer 5′-CGG AGC GGA TCT CAA ACT CTC-3′ (located within the βgeo cassette); the wild-type (WT) allele was identified by amplifying a 600–base pair product with the same forward primer and reverse primer, 5′-GCA GGA GGG TTG GGA AAG CC-3′. The Lmnb2 knockout allele (Lmnb2–) was identified by amplifying a 550–base pair product with forward primer 5′-CGG GTT TTA CTG GAA AGC TG-3′ and reverse primer 5′-GAC AGT ATC GGC CTC AGG AA-3′ (located within the lacZ cassette); the WT allele was identified by amplifying a 350–base pair product with the same forward primer and reverse primer, 5′-CGG AGC AGC AAC CTA TCA TT-3′. The Lmnb1fl allele was identified by amplifying a 450–base pair product with forward primer 5′-AAC AAA CTT GGC CTC ACC AG-3′ (located on the distal loxP site and adjacent vector sequences) and reverse primer 5′-CTG TGG GAC AAA GAC CCA GT-3′; the WT allele was identified by amplifying a 300–base pair product with forward primer 5′-CCA CTT AGC TCG GGG AGT TT-3′ and the same reverse primer. The Lmnb2fl allele was identified by amplifying a 480–base pair product with the forward primer 5′-AAC AAA CTT GGC CTC ACC AG-3′ (located on the distal loxP site and adjacent vector sequences) and reverse primer 5′-GGT CTT GAT GCC ACT CAC CT–-3′; the WT allele was identified by amplifying a 350–base pair product with forward primer 5′-TGA GGC TTT GGA GAA AAG GA-3′ and the same reverse primer. The Cre transgene was detected by PCR with the primers 5′-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3′ and 5′-GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG-3′. Mice heterozygous for the Lmna lacZ allele were identified by amplifying the βgeo cassette with primers 5′-GAC AGT CGT TTG CCG TCT GAA TTT G-3′ and 5′-TAC CAC AGC GGA TGG TTC GGA TAA T-3′.
Histological and immunochemical staining of mouse tissues
Paraffin-embedded sections of mouse embryos (5 μm thick) were stained with hematoxylin and eosin. For immunochemical studies, mouse tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 2 h at room temperature, incubated in 30% sucrose in PBS at 4°C overnight, and frozen in O.C.T. (Tissue-Tek, Sakura Finetek). Sections 10 μm thick were fixed for 5 min in ice-cold acetone or methanol, followed by five dips in acetone and then permeabilized with 0.1% Tween-20. Background staining for mouse antibodies was minimized with the Mouse-on-Mouse Kit (Vector Laboratories, Burlingame, CA). To detect BrdU labeling, sections were pretreated with 1 N HCl for 10 min on ice, 2 N HCl for 10 min at room temperature, followed by 10 min at 37°C, and 0.1 M sodium borate (pH 8.5) for 12 min. Tissue sections were blocked with 2.5% horse serum for 1 h at room temperature and incubated overnight at 4°C with primary antibodies at the dilutions indicated in Supplemental Table S1. Alexa Fluor 488– and Alexa Fluor 568–conjugated secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, CA) were used at a 1:200 dilution, and Alexa Fluor 555–conjugated streptavidin (Molecular Probes) was diluted at 20 μg/ml. Costaining of sections with two primary antibodies from the same species was accomplished by directly labeling one of the antibodies with Alexa Fluor 555 or Alexa Fluor 647 (Molecular Probes). After counterstaining with 4′,6-diamidino-2-phenylindole (DAPI), sections were mounted with Prolong Gold antifade (Invitrogen), and images were recorded with the Axiovision software on an Axiovert 200M microscope using 5× (0.16 numerical aperture [NA], EC Plan Neofluar), 10× (0.45 NA, Plan Apochromat), 20× (0.8 NA Plan Apochromat), or 40× (0.75 NA, EC Plan Neofluar) objectives with an AxioCam MRm and an ApoTome (all from Zeiss, Thornwood, NY) or with the LCS software on a TCS-SP MP laser-scanning confocal microscope with a 63× (1.4 NA, oil) objective (all from Leica, Wetzlar, Germany).
β-Galactosidase staining
Embryos were fixed for 1 h in 4% paraformaldehyde in PBS, incubated in 30% sucrose at 4°C overnight, and embedded in O.C.T. Sections of 10 μm were cut, postfixed in paraformaldehyde, washed twice for 10 min in ice-cold PBS, and then stained for 4–16 h at 37°C in X-gal buffer (PBS, 20 mM potassium ferricyanate, 20 mM potassium ferrocyanate, 2 mM MgCl2, 0.2% NP-40, 0.1% sodium deoxycholate, and 0.8 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Nagy et al., 2003). Specificity of β-galactosidase staining was assessed by parallel experiments with Lmnb1+/+ tissues (which did not express β-galactosidase). After staining, sections were washed in PBS, postfixed in 4% paraformaldehyde in PBS, counterstained with eosin, dehydrated, and mounted in Permount (Fisher Scientific, Waltham, MA). Images were recorded on the Leica Application Suite imaging software with a MZ6 dissecting microscope and a DFC290 digital camera (all from Leica).
Studies with neuronal progenitor cells
Neuronal progenitors were isolated from E13.5 sibling embryos. Cortical explants were isolated in PBS on ice and then incubated in 0.25% trypsin-EDTA (Life Technologies, Carlsbad, CA) at room temperature for 10 min; cells were further dissociated by pipetting before adding 1.5 volumes of DMEM containing 10% fetal bovine serum. Cells were plated on poly-l-lysine–coated coverslips (80,000 cells/cm2) and cultured for 4 d in neuronal differentiation medium (50% DMEM/F12-50% Neurobasal medium with B-27 and N2 supplements, all from Life Technologies). For immunocytochemistry experiments, cells were washed in PBS containing 1 mM Ca2+ and 1 mM Mg2+ (PBS/Ca/Mg) and fixed in methanol for 10 min. After permeabilizing of cells with 0.2% Triton for 5 min and blocking for 1 h in PBS/Ca/Mg containing 10% fetal bovine serum and 0.2% bovine serum albumin, the cells were incubated for 1 h with primary antibodies (Supplemental Table S1). Alexa Fluor 488–, 568–, and 647–conjugated secondary antibodies (Molecular Probes) were diluted 1:400. Nuclear DNA was stained with 2 μg/ml DAPI, and coverslips were mounted with Prolong Gold antifade reagent. Immunofluorescence images were recorded with the Axiovision software on an Axiovert 200M microscope with a 40× (0.75 NA, EC Plan Neofluar) or 63× (1.4 NA, Plan Apochromat Oil) objectives with an AxioCam MRm and ApoTome (all from Zeiss). Confocal images were recorded on the same microscope equipped with a LSM 700 laser-scanning system using the acquisition software ZEN 2010 (all from Zeiss) or on a Leica TCS-SP MP laser-scanning confocal microscope with a 63× (1.4 NA, oil) objective and the Leica Application Suite.
Cell counts and measurements
For all studies, at least three embryos per group were analyzed with sets of sections matched for the position in the brain. Brain sections stained for the markers of interest were photographed with a 20× objective using the Axiovision software, and numbers of cells positive for the different markers were recorded with the Count tool in Photoshop CS5 Extended (Adobe, San Jose, CA). Cells were counted in at least three areas of 430 × 350 μm taken from at least two sections. The thickness of the cortical plate and cortex were measured on images with the Ruler tool in Adobe Photoshop CS. Numbers of cells in brain sections of lamin B1–deficient and control embryos with nuclear blebs and an asymmetric distribution of lamin B2 were analyzed on three-dimensional (3D) reconstructions generated from stacks of confocal images with the 3D Opacity renderer in Volocity 5.4 (PerkinElmer, Waltham, MA). The 3D images were exported as TIF files, and counts were recorded with the Count tool in Adobe Photoshop CS5 Extended. Lengths of nuclei and distance of the nucleus to the centrosome in lamin B2–deficient cells and control cells were measured on 3D reconstructions from stacks of confocal images with Volocity 5.4 software.
Statistical analysis
Statistical analyses were performed in Excel 2004 (Microsoft, Redmond, WA). Results were expressed as mean ± SD or as a 95% confidence interval, and significant differences were analyzed with a two-tailed Student's t-test for independent samples at VassarStats (http://faculty.vassar.edu/lowry/VassarStats.html). The boxplot analysis of individual nuclear lengths was made in Stata 11 (StataCorp, College Station, TX).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AR050200, HL76839, HL86683, HL89781, and GM66152, a March of Dimes Grant 6-FY2007-1012, and an Ellison Medical Foundation Senior Scholar Award. C.C. is the recipient of a Scientist Development Grant from the American Heart Association (0835489N). Confocal laser scanning microscopy was performed at the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Resource Facility at the University of California, Los Angeles, supported with a National Institutes of Health/National Center for Research Resources shared resources grant (CJX1-44385-WS-29646) and a National Science Foundation Major Research Instrumentation grant (CHE-0722519). We thank the University of California, Los Angeles, Translational Pathology Core Laboratory and the Brain Research Institute Microscopic Techniques Laboratory Core for the preparation and sectioning of paraffin-embedded tissues.
Abbreviations used:
- BrdU
bromodeoxyuridine
- DAPI
4′,6-diamidino-2-phenylindole
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0504) on October 5, 2011.
REFERENCES
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci USA. 2010a;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, et al. Direct synthesis of lamin A, bypassing prelamin A processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J Biol Chem. 2010b;285:20818–20826. doi: 10.1074/jbc.M110.128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Goldman RD. Nuclear lamins and chromatin: when structure meets function. Adv Enzyme Regul. 2009;49:157–166. doi: 10.1016/j.advenzreg.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Deutsch SI, Burket JA, Katz E. Does subtle disturbance of neuronal migration contribute to schizophrenia and other neurodevelopmental disorders? Potential genetic mechanisms with possible treatment implications. Eur Neuropsychopharmacol. 2010;20:281–287. doi: 10.1016/j.euroneuro.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66:1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: a genetic look at neocortical development. Nat Rev Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- Hutchison CJ. Lamins: building blocks or regulators of gene expression? Nat Rev Mol Cell Biol. 2002;3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, Young SG, Zhang Q, Shanahan CM, Lammerding J. Cell nuclei spin in the absence of lamin B1. J Biol Chem. 2007;282:20015–20026. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- Kahler AK, et al. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- Li BS, Zhang L, Takahashi S, Ma W, Jaffe H, Kulkarni AB, Pant HC. Cyclin-dependent kinase 5 prevents neuronal apoptosis by negative regulation of c-Jun N-terminal kinase 3. EMBO J. 2002;21:324–333. doi: 10.1093/emboj/21.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjat A, Misteli T. LINC complexes in health and disease. Nucleus. 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. Review: the dynamics of the nuclear lamins during the cell cycle—relationship between structure and function. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Muchir A, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Pawlisz AS, Mutch C, Wynshaw-Boris A, Chenn A, Walsh CA, Feng Y. Lis1-Nde1-dependent neuronal fate control determines cerebral cortical size and lamination. Hum Mol Genet. 2008;17:2441–2455. doi: 10.1093/hmg/ddn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Sasaki S, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Govek EE, Tomoda T, Hatten ME. Neuronal polarity in CNS development. Genes Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, et al. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–770. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- Yang SH, Chang SY, Yin L, Tu Y, Hu Y, Yoshinaga Y, Jong PJ, Fong LG, Young SG. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of the skin and hair. Hum Mol Genet. 2011;20:3537–3544. doi: 10.1093/hmg/ddr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.