Abstract
The serotonergic (5-HT) system has been implicated in various physiological processes and neuropsychiatric disorders, but in many aspects its role in normal and pathologic brain function is still unclear. One reason for this might be the lack of appropriate animal models which can address the complexity of physiological and pathophysiological 5-HT functioning. In this respect, rats offer many advantages over mice as they have been the animal of choice for sophisticated neurophysiological and behavioral studies. However, only recently technologies for the targeted and tissue specific modification of rat genes - a prerequisite for a detailed study of the 5-HT system - have been successfully developed. Here, we describe a rat transgenic system for inducible gene manipulations in 5-HT neurons. We generated a Cre driver line consisting of a tamoxifen-inducible CreERT2 recombinase under the control of mouse Tph2 regulatory sequences. Tissue-specific serotonergic Cre recombinase expression was detected in four transgenic TPH2-CreERT2 rat founder lines. For functional analysis of Cre-mediated recombination, we used a rat Cre reporter line (CAG-loxP.EGFP), in which EGFP is expressed after Cre-mediated removal of a loxP-flanked lacZ STOP cassette. We show an in-depth characterisation of this rat Cre reporter line and demonstrate its applicability for monitoring Cre-mediated recombination in all major neuronal subpopulations of the rat brain. Upon tamoxifen induction, double transgenic TPH2-CreERT2/CAG-loxP.EGFP rats show selective and efficient EGFP expression in 5-HT neurons. Without tamoxifen administration, EGFP is only expressed in few 5-HT neurons which confirms minimal background recombination. This 5-HT neuron specific CreERT2 line allows Cre-mediated, inducible gene deletion or gene overexpression in transgenic rats which provides new opportunities to decipher the complex functions of the mammalian serotonergic system.
Introduction
5-hydroxytryptamine (5-HT, serotonin) has been implicated in a wide variety of emotional, cognitive and behavioral processes. Psychopharmacotherapeutic agents targeting molecules of the serotonergic system are often used for the treatment of a wide spectrum of psychiatric disorders. Although this clearly demonstrates the functional relevance of 5-HT for physiological as well as disease processes, there is no well-defined framework for comprehending any of its roles [1].
The understanding of the 5-HT system's function and its underlying molecular mechanisms has been strongly accelerated by using reverse genetic approaches in transgenic mouse models [2]–[11]. However, in mice the analysis of certain phenotypes reaches its limits, as complex behavioral tasks involving higher order cognitive functions are difficult to perform. Indeed, most behavioral and electrophysiological studies are traditionally conducted in rats and therefore many behavioral tests are only validated for this species. The rat behavioral repertoires and the related neural correlates have been well described and physiological interventions, microsurgery and toxicology studies as well as evaluation of higher order functions are in general more sophisticated and informative in rats than in mice [12]. As a consequence, most of the research on serotonergic functioning has been accomplished using rats despite the fact that only few rats with specific genetic manipulations of the 5-HT system are available [13].
Recently, it has become feasible to manipulate the rat's genome with conditional transgenesis [14]. In the near future, technological advances in the rat such as zinc finger nucleases [15] and the development of germline competent rat embryonic stem cells [12] will enable researchers to spatially and temporally control gene manipulation. For this purpose, it will be necessary to control gene expression or gene deletion with tissue-specific Cre-driver lines which allow the recombination of loxP-flanked target sequences in the rat genome. To specifically manipulate target genes within the serotonergic system, Cre drivers could be linked to regulatory sequences of 5-HT neuron specific genes such as Pet-1 or Tph2.
In the present study, we generated and characterized four transgenic TPH2-CreERT2 rat lines in which a 177 kb genomic sequence of the mouse Tph2 gene controls tissue-specific expression of the CreERT2 recombinase. Cre-mediated recombination of loxP flanked target genes was functionally characterized with the Cre reporter line pCaggs-loxP.lacZ.loxP-EGFP (CAG-loxP.EGFP). After tamoxifen treatment of double transgenic TPH2-CreERT2/CAG-loxP.EGFP rats, efficient EGFP expression and hence recombination occured specifically in 5-HT neurons while background recombination in the absence of tamoxifen could not be identified.
Methods
Generation of TPH2-CreERT2 transgenic rats
A PAC (L065) which contains the full-length mouse Tph2 gene (107 kb) with 51 kb upstream and 19 kb downstream DNA sequences was modified as previously described [16]. The purified, linearized TPH2-CreERT2 DNA was microinjected into the pronucleus of oocytes of Sprague-Dawley rats (Charles River Laboratories, Germany). Transgenic founder rats were identified by PCR genotyping of tail tips. The TPH2-CreERT2 transgenic rats were bred with the Cre reporter line CAG-loxP.EGFP (Schönig et al, in preparation) to generate double-transgenic TPH2-CreERT2/CAG-loxP.EGFP rats. In brief CAG-loxP.EGFP rats harbour a loxP-flanked lacZ reporter gene, controlled by the ubiquitously active CAG promoter [17], [18]. The lacZ DNA fragment precludes the transcription of a second reporter gene EGFP. Cre mediated recombination can be monitored in double-transgenic TPH2-CreERT2/CAG-loxP.EGFP by EGFP expression.
Quantification of transgene copy number
Copy number quantification of the TPH2-CreERT2 transgene per cell was done via genomic quantitative real-time PCR (qPCR) for each TPH2-CreERT2 line. For amplification and data collection, we used the Rotor-Gene Q-system (Qiagen). All reactions were carried out in a total volume of 25 µL and were measured in triplicates. Each reaction mixture contained 5 ng of genomic DNA, 12.5 µl Rotor-Gene Fast SYBR Green Master Mix (Qiagen) and 300 nM forward and reverse primers. The amplification protocol consisted of an initial denaturation step at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 60°C for 10 s and 72°C for 10 s. SYBR Green fluorescence was detected at 72°C. Each amplification reaction was checked for the absence of nonspecific PCR products by melting curve analysis followed by agarose gel electrophoresis.
The absolute target copy numbers were determined using 1∶2 dilution series of genomic mouse DNA harbouring defined numbers of Cre transgenes [19] as an external standard. For each sample, the amount of Cre transgene and reference gene (ApoB) was measured in each transgenic line. The following primers were used: Cre3: 5′ TCG CTG CAT TAC CGG TCG ATG C 3′; Cre4: 5′ CCA TGA GTG AAC GAA CCT GGT CG 3′; ApoB_for: 5′ ATC TCA GCA CGT GGG CTC 3′; ApoB_rev 5′ TCA CCA GTC ATT TCT GCC TTT G 3′.
In vivo induction of Cre-mediated recombination with tamoxifen
Tamoxifen (Sigma) was dissolved in neutral oil at a final concentration of 20 mg/ml. For recombination analysis, double-transgenic TPH2-CreERT2/CAG-loxP.EGFP rats (8–12 weeks) were given a protocol of alternating daily tamoxifen injections (40 mg/kg) for a total of five consecutive days. The protocol was designed with single injections on days 1, 3 and 5 and two tamoxifen injections twelve hours apart on days 2 and 4. Control animals were injected with neutral oil (vehicle) using the same schedule. Rats were sacrificed 14 days after the last injection.
All experimental procedures were approved by the local Animal Welfare Committee (Regierungspräsidium Karlsruhe 35-918581/G-107/09) and carried out in accordance with the local Animal Welfare Act and the European Communities Council Directive of 24 November 1986 (86/609/EEC).
Immunohistochemistry
Transgenic TPH2-CreERT2 founder rats were characterized by immunohistochemistry using DAB staining (Vectastain Elite ABC kit) with a rabbit α-Cre primary antibody (Covance, 1∶2500). Founder line #15 was further characterized with dual-label fluorescent immunohistochemistry in TPH2-CreERT2 and TPH2-CreERT2/CAG-loxP.EGFP rats. The following primary antibodies were used: chicken α-βgalactosidase (Abcam, 1∶10000), rabbit α-GFP (Invitrogen, 1∶1000), rabbit α-Cre (Covance, 1∶1000), mouse α-GAD67 (Millipore, 1∶500), mouse α-TH (Millipore, 1∶500), mouse α-NeuN (Millipore, 1∶4000), mouse α-GFAP (Sigma, 1∶2000), rabbit α-TPH2 (Dianova, 1∶5000), and mouse α-TPH1 (Sigma, 1∶2000) antibodies. Tryptophane hydroxylase 2 (TPH2) is the rate-limiting enzyme of 5-HT synthesis in the brain and specific to serotonergic neurons. The anti-tryptophan hydroxylase 1 (TPH1) antibody crossreacts with TPH2 and detects both isoenzymes. Secondary antibodies were AF488 donkey α-rabbit (Invitrogen, 1∶1000 for TPH2 and 1∶5000 for GFP and Cre), Cy3 donkey α-mouse (Jackson ImmunoResearch, 1∶200 for TPH1), Cy3 donkey α-chicken (Jackson ImmunoResearch, 1∶1000 for βgalactosidase) and AF488 donkey α-mouse (Invitrogen, 1∶200 for GAD67, NeuN, TH and GFAP). Sections were examined using a Nikon C1Si-CLEM confocal laser-scanning microscope (Nikon Imaging Center, BioQuant, Heidelberg, Germany). Confocal image stacks for both channels were acquired sequentially, and projected on average using ImageJ software.
Statistical methods
Coronal slices of 3 adult TPH2-CreERT2/CAG-loxP.EGFP rats (8–10 weeks old) per group were processed with dual-label fluorescent immunohistochemistry detecting GFP and TPH. Image stacks of slices that showed TPH staining were acquired using a confocal laser-scanning microscope. The ratio of GFP+/TPH+ neurons to all TPH+ neurons was calculated separately for caudal, median and dorsal raphe nuclei. Confidence-bounds (CI) for recombination efficacy and background recombination in adult rats were calculated using the Clopper-Pearson method based on significance level 95%.
Results
Generation of TPH2-CreERT2 transgenic rats
For inducible, tissue-specific expression of CreERT2 in serotonergic neurons of the rat brain, a 177 kb fragment of mouse genomic DNA containing the Tph2 gene and its regulatory elements was used [16]. The linearized CreERT2 expression cassette (Fig. 1A) was introduced into the rat genome via pronuclear microinjection of fertilized Sprague Dawley rat oocytes. Seven transgenic founder rats were identified by PCR of tail DNA. Of those, three founders did not transmit their transgene leaving four founders for characterization.
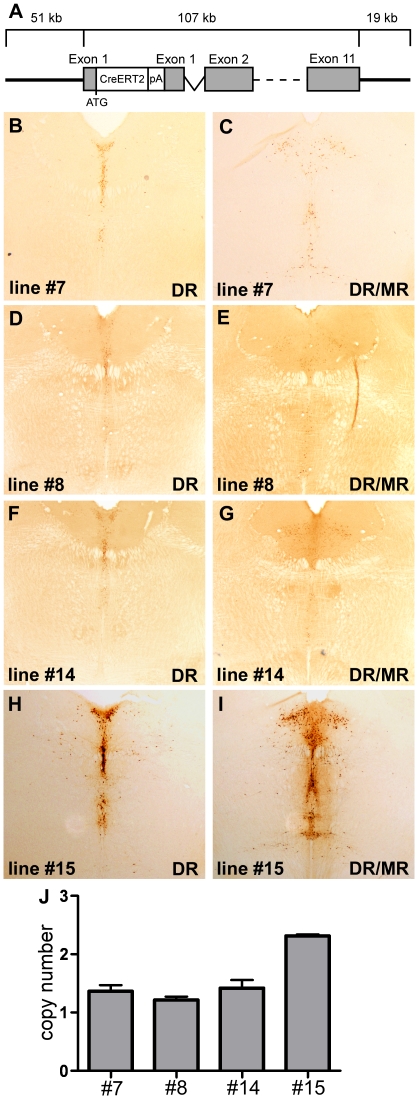
Figure 1. Copy number dependent Cre expression in TPH2-CreERT2 founder rats.
(A) Mouse TPH2-CreERT2 expression cassette for DNA microinjection. (B–I) DAB-immunohistochemistry with a Cre antibody shows weak Cre expression in the brain stem and mid-brain of TPH2-CreERT2 rat founder lines #7 (Fig. 1B,C), #8 (Fig. 1D,E), and #14 (Fig. 1F,G). Founder line #15 shows extensive Cre staining in areas where serotonergic raphe nuclei are located (Fig. 1H,I). Intensity of Cre expression correlates with the transgene copy number of TPH2-CreERT2 rat founders (Fig. 1J).
Cre expression in TPH2-CreERT2 founder rats
All four TPH2-CreERT2 founder lines showed Cre immunostaining in the raphe nuclei of the brain stem and midbrain while no Cre expression was observed outside the raphe nuclei. The efficacy of Cre expression varied notably among the founder lines. The TPH2-CreERT2 founder lines #7, #8 and #14 showed weak and mosaic Cre staining in serotonergic neurons (Fig. 1B–G) while strong Cre expression could be detected in the raphe nuclei of line #15 (Fig. 1H,I). Since large genomic DNA constructs are thought to regulate transgene expression independent of their integration site but copy number dependent [20], we determined the transgene copy number of each TPH2-CreERT2 founder by qPCR (Fig. 1J). Stronger Cre expression in founder line #15 could be correlated with increased transgene copy number while the weakly expressing founder lines #7, #8 and #14 contained only a single copy of the transgene. Cre expression in founder line #15 (Fig. 2A,C,E,G) was further investigated for their tissue-specificity with dual-label fluorescent immunohistochemistry using a 5-HT neuron specific TPH antibody and a Cre antibody (Fig. 2B,D,F,H). Colocalization of Cre and TPH demonstrated that Cre was exclusively expressed in 5-HT neurons. Hence, the transgenic rat founder line #15 showed extensive and tissue-specific Cre expression in 5-HT neurons of all raphe nuclei.
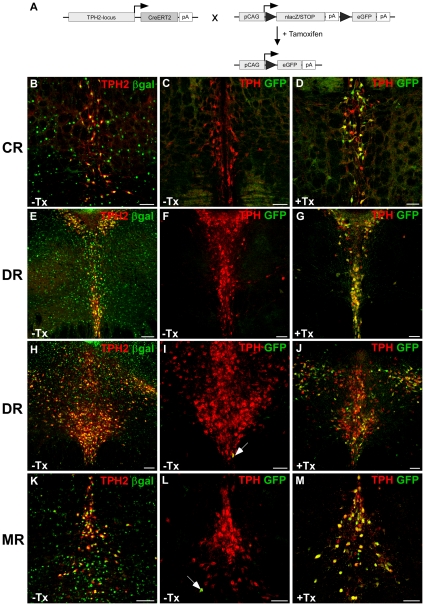
Figure 2. Cre expression is restricted to serotonergic neurons of the raphe nuclei.
(A,C,E,G) DAB-immunohistochemistry with a Cre antibody of line #15 shows Cre staining in the brain stem and mid-brain, regions which contain serotonergic somata while extraserotonergic brain regions show no staining. (B,D,F,H) Coronal sections of dual-label fluorescence immunohistochemistry with Cre and TPH1 antibodies. The TPH1 antibody crossreacts with TPH2 and detects both isoenzymes. Colocalization of TPH1 and Cre confirms exclusive Cre expression in 5-HT neurons of the raphe nuclei. Caudal raphe nuclei (CR); dorsal raphe nuclei (DR); median raphe nuclei (MR). Scale bars: 100 µm.
Inducible and efficient recombination in serotonergic neurons of double-transgenic TPH2-CreERT2/CAG-loxP.EGFP rats
Line #15 was further used to functionally characterize the temporal and spatial control of tamoxifen-induced CreERT2-mediated recombination in 5-HT neurons. We made use of a rat Cre reporter line (CAG-loxP.EGFP), which has been shown to efficiently monitor Cre-mediated recombination in forebrain principal neurons (Schönig et al, in preparation). Here, the ubiquitously active CAG-promoter [17], [18], [21] drives the expression of a double reporter. Under uninduced baseline conditions, the loxP-flanked lacZ minigene is expressed, reflecting cell-type specific CAG-promoter activity. Upon Cre-mediated recombination, lacZ is replaced with the second reporter gene enhanced green fluorescent protein (EGFP). The appearance of EGFP serves as an indicator of Cre mediated recombination in double transgenic rats.
A prerequisite for a versatile Cre reporter line is the ability to monitor recombination in a wide range of cells. We first analysed baseline expression of beta-galactosidase (βgal) in serotonergic neurons and other cell types of the brain by dual-label fluorescent immunohistochemistry to determine expression characteristics of the CAG-loxP.EGFP line. CAG-driven βgal expression was found in virtually all brain regions (Fig. 3A–E) and in many types of neurons including monoaminergic (Fig. 3F–H) and GABAergic neurons (Fig. 3I–J). In contrast to neuronal expression, βgal expression was only infrequently found in astrocytes (Fig. 3K). These results verify the utility of the CAG-loxP.EGFP reporter line for monitoring Cre-mediated recombination not only in serotonergic neurons, but also in other neuronal subtypes.
Figure 3. Baseline βgal expression in the brain of CAG-loxP.EGFP Cre reporter rats.
(A,B) X-Gal staining of sagittal sections shows ubiquitous βgal activity throughout the brain of adult CAG-loxP.EGFP rats (P90). (C–K) Dual-label fluorescence immunohistochemistry (IHC). (C–E) βgal/NeuN IHC of the cerebellum (C), cortex (D) and OB (E) shows strong colocalization of βgal with the neuronal marker NeuN. (F–H) βgal IHC with the serotonergic marker TPH2 (F), and the dopaminergic and noradrenergic marker tyrosine hydroxylase (TH) (G,H) shows abundant colocalization of βgal with 5-HT neurons in the dorsal raphe (F), with dopaminergic neurons in the ventral tegmental area and substantia nigra (G) and noradrenergic neurons in the locus coeruleus (H) confirming strong βgal expression in all monoaminergic neurons. (I,J) βgal/GAD67 IHC shows βgal expression in GABAergic neurons of the granular layer of the OB (I) and in the hippocampus (J). (K) βgal/GFAP IHC in the hippocampus shows infrequent βgal expression in glia. OB, olfactory bulb; DR, dorsal raphe nuclei; VTA, ventral tegmental area; SN, substantia nigra; LC, locus coeruleus; HC, hippocampus. Scale bars: 100 µm.
Based on these results, we generated double-transgenic TPH2-CreERT2/CAG-loxP.EGFP rats to determine recombination efficiency and tissue specificity for our rat Cre driver line TPH2-CreERT2 (Fig. 4A). Coronal brain sections from tamoxifen and vehicle treated TPH2-CreERT2/CAG-loxP.EGFP rats were analysed using dual-label fluorescent immunohistochemistry with βgal/TPH2 and GFP/TPH1 antibodies. In vehicle treated TPH2-CreERT2/CAG-loxP.EGFP rats, βgal-expression could be detected in virtually all 5-HT neurons (Fig. 4B,E,H,K). In contrast, EGFP expression could only be detected in few 5-HT neurons (Fig. 4C,F,I,L), which indicates minimal Cre-mediated background recombination in the absence of tamoxifen (Table 1). Importantly, in tamoxifen-treated TPH2-CreERT2/CAG-loxP.EGFP rats, EGFP and TPH expression colocalised in 5-HT neurons of caudal, dorsal and median raphe nuclei indicating effective Cre-mediated recombination in all raphe nuclei (Fig. 4D,G,J,M; Table 1). Extra-serotonergic brain regions showed no EGFP staining.
Figure 4. Inducible recombination is restricted to serotonergic neurons of adult TPH2-CreERT2/CAG-loxP.EGFP rats.
(A) TPH2-CreERT2 rats were bred to CAG-loxP.EGFP rats to generate double-transgenic TPH2-CreERT2/CAG-loxP.EGFP rats. Under uninduced baseline conditions, the loxP-flanked lacZ minigene is expressed reflecting cell-type specific CAG-promoter activity. Upon Cre-mediated recombination (+ Tamoxifen), lacZ is replaced with the second reporter gene enhanced green fluorescent protein (EGFP). The appearance of EGFP serves as an indicator of Cre mediated recombination in double transgenic rats. TPH2-CreERT2/CAG-loxP.EGFP rats were daily injected with tamoxifen (40 mg/kg) or vehicle for five consecutive days starting between P60–90. Coronal sections show dual-label fluorescence immunohistochemistry for TPH/βgal (B,E,H,K) and TPH/GFP (C,F,I,L) in vehicle-treated rats (-Tx) and TPH/GFP in tamoxifen-treated (+Tx) rats (D,G,J,M). Colocalization is visualized at the level of caudal raphe nuclei (CR) (B–D), dorsal raphe nuclei (DR) (E–J) and median raphe nuclei (MR) (K–M) using confocal images. In vehicle-treated rats, TPH2-CreERT2/CAG-loxP.EGFP rats display strong basal, non-recombined βgal expression in TPH2+ 5-HT neurons (B,E,H,K) making these rats ideally suited to monitor tamoxifen-induced Cre-mediated recombination in 5-HT neurons. (C,F,I,L) Without tamoxifen treatment, background recombination, i.e. EGFP expression (arrows) hardly occurs. (D,G,J,M) After tamoxifen treatment, the majority of TPH+ 5-HT neurons in all raphe nuclei now show EGFP expression indicating Cre-mediated recombination in these neurons (GFP+/TPH+). Scale bars: 100 µm.
Table 1. Recombination efficacy and background recombination for rat TPH2-CreERT2 line #15.
| Line | absolute recombination frequency (GFP+/TPH+):TPH+ | relative recombination frequency | 95% CI | |
| + Tamoxifen | #15 CR | 145∶195 | 74% | 68–80% |
| #15 DR | 1787∶2368 | 75% | 74–77% | |
| #15 MR | 751∶937 | 80% | 78–83% | |
| #15 total | 2683∶3500 | 77% | 75–78% | |
| + Vehicle | #15 CR | 2∶175 | 1.1% | 0.0–2.7% |
| #15 DR | 9∶1921 | 0.5% | 0.2–0.8% | |
| #15 MR | 8∶506 | 1.6% | 0.05–3.4% | |
| #15 total | 19∶2602 | 0.7% | 0.4–1.1% |
Caudal (CR), median (MR) and dorsal (DR) raphe nuclei were separately and jointly (total) calculated. Confidence-bounds (CI) were calculated using the Clopper-Pearson method based on significance level 95.0%.
Discussion
In this study, we describe an inducible, tissue-specific rat transgenic CreERT2 driver line for conditional gene manipulations in serotonergic neurons. We functionally demonstrate efficient, tamoxifen-inducible, 5-HT neuron specific recombination with minimal background activity in TPH2-CreERT2 rats crossed to the rat Cre reporter line pCAG-loxP.EGFP.
Application of mouse genomic regulatory sequences for the generation of tissue-specific rat Cre driver lines
TPH2 is the rate-limiting enzyme of 5-HT synthesis and strongly and exclusively expressed in serotonergic neurons of the raphe nuclei in the brain [22]. Hence, regulatory elements of Tph2 should be suitable to direct Cre expression specifically to 5-HT neurons. We previously made use of large regulatory elements of the Tph2 locus identified on a genomic mouse PAC clone to generate a TPH2-CreERT2 mouse line that shows highly efficient, tamoxifen-inducible recombination in 5-HT neurons [16]. As not only the rat and mouse Tph2 genes are almost identical [22], but also the entire region of the mouse Tph2 locus on chromosome 10 is highly homologous to the rat locus on 7q22 (NCBI Blast), we decided to use the same 177 kb TPH2-CreERT2 construct to generate transgenic rats. We demonstrate the fidelity of the mouse Tph2 locus to direct Cre expression selectively to serotonergic neurons in transgenic rats. With this TPH2-CreERT2-expression cassette, transgenic 5-HT neuron-specific Cre expression is likely not dependent on the genomic site of integration since all founder lines showed Cre expression in the raphe nuclei. More likely, the efficacy of Tph2-controlled Cre expression appears to depend on the transgenic copy number. This is in accordance with previous reports showing that large genomic DNA constructs allow copy-number dependent transgene expression independent of the genomic integration site of the construct [20], [23], [24]. Efficient serotonergic Cre expression could only be found in founder line #15 which contained 2–3 transgene copies compared to single copies in all other founder lines.
Interestingly, we could not identify founder lines with higher copy numbers [24]. It remains to be investigated whether this finding of low copy numbers in rat transgenesis is purely coincidental or specific for microinjected rat oocytes.
The finding that large genomic mouse sequences which have been shown to adequately control Cre expression in mouse Cre driver lines likely contain sufficient regulatory information for rat transgenesis suggest that this strategy might be applicable in a general way to generate tissue-specific rat Cre driver lines.
Functional analysis of Cre-mediated recombination in transgenic TPH2-CreERT2 rats
Novel Cre driver lines need to be functionally assessed for efficiency and tissue-specificity of Cre-mediated recombination. We previously generated a rat Cre reporter line, CAG-loxP.EGFP, which shows CAG-promoter controlled baseline, non-recombined βgal expression and EGFP reporter expression once Cre-mediated recombination of a loxP flanked lacZ STOP cassette has occurred (Schönig et al, in preparation). A major advantage of this strategy is that basal CAG-promoter activity in the tissue of interest can be readily assessed on a cellular level by monitoring βgal expression. Hence, it can be rapidly determined in advance whether CAG-loxP.EGFP rats allow for functional characterization of a new tissue-specific rat Cre driver line. We have characterized the CAG-loxP.EGFP rat Cre reporter line for its utility to monitor Cre-mediated recombination in TPH2-CreERT2 rats. We find strong βgal expression throughout the brain in all examined neuronal populations and in a portion of astrocytes. In particular, CAG-loxP.EGFP rats show abundant monoaminergic βgal expression in the absence of EGFP expression which makes CAG-loxP.EGFP rats suitable to functionally characterize tamoxifen-induced, Cre mediated recombination and background recombination in 5-HT neurons. Using TPH2-CreERT2/CAG-loxP.EGFP double transgenic rats, we functionally validate Cre-mediated recombination, i.e. EGFP expression, in 5-HT neurons of all raphe nuclei upon tamoxifen induction whereas background recombination in vehicle-treated rats was absent. Furthermore, the absence of extraserotonergic EGFP expression in TPH2-CreERT2/CAG-loxP.EGFP rats confirms tissue specificity of the TPH2-CreERT2 driver line.
Serotonergic recombination in tamoxifen-induced TPH2-CreERT2/CAG-loxP.EGFP rats was less efficient than recombination in the previously described mouse TPH2-CreERT2 line [16] (5-HT neuron specific recombination rate: mouse 90% versus rat 77%) while background recombination without tamoxifen was equally low in the rat TPH2-CreERT2 line. The lower recombination efficacy could be due to the integration site of the transgene, its copy number or missing regulatory elements in the mouse Tph2 sequence which drives CreERT2 expression in the rat brain. Alternatively, the tamoxifen dose or the induction protocol with three single daily injections and only two twice daily tamoxifen injections could potentially result in insufficient nuclear translocation of CreERT2 and thus reduced Cre-mediated recombination. The individual tamoxifen dosage of 40 mg/kg in rats is analogous to 1 mg/injection often used in mice [25]–[27]. In CreERT2 mice, it has been previously shown that the most efficient tamoxifen protocol consists of twice daily tamoxifen injections for 5 consecutive days [25], [27]. The frequency of Cre-mediated recombination in mice decreased considerably with protocols using single daily tamoxifen injections even when the protocol was extended to 10 days [25], [27]. Our initial attempts to apply the mouse protocol of twice daily tamoxifen injections to our transgenic rats failed as the rats did not well tolerate this protocol. Nonetheless, we believe that insufficient tamoxifen-mediated nuclear translocation of CreERT2 is only partially responsible for the found incomplete recombination efficacy since Cre was not expressed at all in some 5-HT neurons.
Importance of tissue specific rat Cre driver lines for rat transgenesis
Recently, a plethora of new techniques for the modification of the rat genome has been introduced including the development of germline competent embryonic rat stem cells and nuclease based methods [12], [28]–[31]. For the first time, this permits targeted integration of recombinant DNA sequences into the rat genome. In the near future, it is expected that these techniques will be applied to generate conditional loxP-flanked alleles in the rat allowing for spatial and temporal control of gene deletion with tissue-specific rat CreERT2 driver lines. This strategy is of particular importance in order to overcome lethality or induction of compensatory, homeostatic mechanisms or pleiotropy during development, inherent with traditional methods applied in rats such as ENU- or transposon mediated mutagenesis [32]–[34]. Furthermore, the CAG-loxP.EGFP line illustrates how complementary systems for tissue-specific overexpression or knock-down of target genes could be easily implemented. For inducible overexpression, the EGFP reporter cassette would be simply replaced by a candidate gene's cDNA which transcription would only be activated after Cre-mediated recombination. Alternatively, polymerase II controlled microRNAs or sponge/decoy miRNA sequences [35], [36] could be placed downstream of the loxP-flanked lacZ cassette which would allow a Cre-mediated gene knock-down. Apart from tissue specific and inducible overexpression of cDNAs, this technology enables the conditional rescue of gene knockouts, overexpression of mutated gene variants, micro RNA mediated translational repression or the study of microRNA mediated post-transcriptional gene regulation by antagonizing microRNA activity. The TPH2-CreERT2 rat line is also optimally suited for optogenetic manipulations of the serotonergic system with Cre-activated opsin genes delivered to the brain by viruses [37], [38].
As we have demonstrated above, the combination of such Cre activatable “response units” with the TPH2-CreERT2 line will guide modifications specifically to serotonergic neurons.
Conditional gene manipulations in serotonergic neurons of transgenic rats
Only during the last decade, conditional transgenic mouse tools have been developed to manipulate candidate genes exclusively in 5-HT neurons using the Cre/loxP recombinatorial system [5], [16], [39]. These studies have led to important insights into the physiological role of the 5-HT system [5], [6], [8], [40]–[42]. In contrast, only few publications have addressed the involvement of the 5-HT system in impulsive behavior, cognitive flexibility, decision making, sensitivity to reward, and responsiveness to punishment and aversive signals [9], [43], all functions that have been prominently associated with 5-HT [1], [44], [45]. This comes as no surprise, since the mouse as a model organism for complex behavioral analysis of higher cognitive functions has not been the first choice for most researchers. Because of its size, ease of manipulation and breeding characteristics, the laboratory rat has been the preferred animal model for physiology, pharmacology, toxicology, nutrition, behavior, immunology and neoplasia for many decades while the mouse has emerged as the principal mammal for experimental genetics [46]. Transferring conditional genetic manipulation to the rat would greatly enhance our capabilities to dissect 5-HT functions and its implications for emotions, learning and complex behaviour. With our approach, the advantages of conditional, 5-HT neuron specific genetic manipulation – previously a mouse geneticist's province - can now be studied in the rat with all its amenities.
Acknowledgments
We thank Ariana Frömmig, Lena Wendler and Elke Hermann for excellent technical support and Sarah Gartside for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by grants from the German Ministry for Education and Research (BMBF, 01GQ1003B) National Bernstein Network for Computational Neuroscience (http://www.gesundheitsforschung-bmbf.de/en/2478.php#Heidelberg), HEALTH-F2-2007-201714 DEVANX (http://devanx.vitamib.com), and the Deutsche Forschungsgemeinschaft SFB 636 (http://www.sfb636.de). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cools R, Roberts AC, Robbins TW. Serotoninergic regulation of emotional and behavioural control processes. Trends Cogn Sci. 2008;12:31–40. doi: 10.1016/j.tics.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Murphy DL, Fox MA, Timpano KR, Moya PR, Ren-Patterson R, et al. How the serotonin story is being rewritten by new gene-based discoveries principally related to SLC6A4, the serotonin transporter gene, which functions to influence all cellular serotonin systems. Neuropharmacology. 2008;55:932–960. doi: 10.1016/j.neuropharm.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonardo ED, Hen R. Anxiety as a developmental disorder. Neuropsychopharmacology. 2008;33:134–140. doi: 10.1038/sj.npp.1301569. [DOI] [PubMed] [Google Scholar]
- 4.Alenina N, Kikic D, Todiras M, Mosienko V, Qadri F, et al. Growth retardation and altered autonomic control in mice lacking brain serotonin. Proc Natl Acad Sci U S A. 2009;106:10332–10337. doi: 10.1073/pnas.0810793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, et al. Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 2010;13:1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao ZQ, Gao YJ, Sun YG, Zhao CS, Gereau RWt, et al. Central serotonergic neurons are differentially required for opioid analgesia but not for morphine tolerance or morphine reward. Proc Natl Acad Sci U S A. 2007b;104:14519–14524. doi: 10.1073/pnas.0705740104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyasova V, Fernandez SP, Laine J, Stankovski L, Muzerelle A, et al. A Genetically Defined Morphologically and Functionally Unique Subset of 5-HT Neurons in the Mouse Raphe Nuclei. J Neurosci. 2011;31:2756–2768. doi: 10.1523/JNEUROSCI.4080-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges MR, Wehner M, Aungst J, Smith JC, Richerson GB. Transgenic mice lacking serotonin neurons have severe apnea and high mortality during development. J Neurosci. 2009;29:10341–10349. doi: 10.1523/JNEUROSCI.1963-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai JX, Han HL, Tian M, Cao J, Xiu JB, et al. Enhanced contextual fear memory in central serotonin-deficient mice. Proc Natl Acad Sci U S A. 2008;105:11981–11986. doi: 10.1073/pnas.0801329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song NN, Xiu JB, Huang Y, Chen JY, Zhang L, et al. Adult raphe-specific deletion of Lmx1b leads to central serotonin deficiency. PLoS One. 2011;6:e15998. doi: 10.1371/journal.pone.0015998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audero E, Coppi E, Mlinar B, Rossetti T, Caprioli A, et al. Sporadic autonomic dysregulation and death associated with excessive serotonin autoinhibition. Science. 2008;321:130–133. doi: 10.1126/science.1157871. [DOI] [PubMed] [Google Scholar]
- 12.Buehr M, Meek S, Blair K, Yang J, Ure J, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Homberg JR, Olivier JD, Smits BM, Mul JD, Mudde J, et al. Characterization of the serotonin transporter knockout rat: a selective change in the functioning of the serotonergic system. Neuroscience. 2007;146:1662–1676. doi: 10.1016/j.neuroscience.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Sato Y, Endo H, Ajiki T, Hakamata Y, Okada T, et al. Establishment of Cre/LoxP recombination system in transgenic rats. Biochem Biophys Res Commun. 2004;319:1197–1202. doi: 10.1016/j.bbrc.2004.04.204. [DOI] [PubMed] [Google Scholar]
- 15.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber T, Bohm G, Hermann E, Schutz G, Schönig K, et al. Inducible gene manipulations in serotonergic neurons. Front Mol Neurosci. 2009;2:24. doi: 10.3389/neuro.02.024.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakamata Y, Tahara K, Uchida H, Sakuma Y, Nakamura M, et al. Green fluorescent protein-transgenic rat: a tool for organ transplantation research. Biochem Biophys Res Commun. 2001;286:779–785. doi: 10.1006/bbrc.2001.5452. [DOI] [PubMed] [Google Scholar]
- 18.Michalkiewicz M, Michalkiewicz T, Geurts AM, Roman RJ, Slocum GR, et al. Efficient transgenic rat production by a lentiviral vector. Am J Physiol Heart Circ Physiol. 2007;293:H881–894. doi: 10.1152/ajpheart.00060.2007. [DOI] [PubMed] [Google Scholar]
- 19.Schönig K, Kentner D, Gossen M, Baldinger T, Miao J, et al. Development of a BAC vector for integration-independent and tight regulation of transgenes in rodents via the Tet system. Transgenic Res. 2011;20:709–720. doi: 10.1007/s11248-010-9427-0. [DOI] [PubMed] [Google Scholar]
- 20.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 21.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 22.Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 23.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 24.Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, et al. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, et al. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci. 2003;22:430–440. doi: 10.1016/s1044-7431(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 26.Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, et al. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J Neurosci. 2007;27:12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori T, Tanaka K, Buffo A, Wurst W, Kuhn R, et al. Inducible gene deletion in astroglia and radial glia-a valuable tool for functional and lineage analysis. Glia. 2006;54:21–34. doi: 10.1002/glia.20350. [DOI] [PubMed] [Google Scholar]
- 28.Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet. 2010;26:510–518. doi: 10.1016/j.tig.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li MA, Bradley A. Crafting rat genomes with zinc fingers. Nat Biotechnol. 2011;29:39–41. doi: 10.1038/nbt.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2010;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 31.Li T, Huang S, Zhao X, Wright DA, Carpenter S, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21:645–651. doi: 10.1038/nbt830. [DOI] [PubMed] [Google Scholar]
- 33.Smits BM, Mudde JB, van de Belt J, Verheul M, Olivier J, et al. Generation of gene knockouts and mutant models in the laboratory rat by ENU-driven target-selected mutagenesis. Pharmacogenet Genomics. 2006;16:159–169. doi: 10.1097/01.fpc.0000184960.82903.8f. [DOI] [PubMed] [Google Scholar]
- 34.Izsvak Z, Frohlich J, Grabundzija I, Shirley JR, Powell HM, et al. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods. 2010;7:443–445. doi: 10.1038/nmeth.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 36.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 38.Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott MM, Wylie CJ, Lerch JK, Murphy R, Lobur K, et al. A genetic approach to access serotonin neurons for in vivo and in vitro studies. Proc Natl Acad Sci U S A. 2005;102:16472–16477. doi: 10.1073/pnas.0504510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodges MR, Tattersall GJ, Harris MB, McEvoy SD, Richerson DN, et al. Defects in breathing and thermoregulation in mice with near-complete absence of central serotonin neurons. J Neurosci. 2008;28:2495–2505. doi: 10.1523/JNEUROSCI.4729-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchanan GF, Richerson GB. Central serotonin neurons are required for arousal to CO2. Proc Natl Acad Sci U S A. 2010;107:16354–16359. doi: 10.1073/pnas.1004587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao ZQ, Chiechio S, Sun YG, Zhang KH, Zhao CS, et al. Mice lacking central serotonergic neurons show enhanced inflammatory pain and an impaired analgesic response to antidepressant drugs. J Neurosci. 2007a;27:6045–6053. doi: 10.1523/JNEUROSCI.1623-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, et al. Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron. 2009;63:305–315. doi: 10.1016/j.neuron.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dayan P, Huys QJ. Serotonin in affective control. Annu Rev Neurosci. 2009;32:95–126. doi: 10.1146/annurev.neuro.051508.135607. [DOI] [PubMed] [Google Scholar]
- 45.Rogers RD. The roles of dopamine and serotonin in decision making: evidence from pharmacological experiments in humans. Neuropsychopharmacology. 2011;36:114–132. doi: 10.1038/npp.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]