Abstract
The interaction between α-actinin and palladin, two actin-crosslinking proteins, is essential for proper bidirectional targeting of these proteins. As a first step toward understanding the role of this complex in organizing cytoskeletal actin, we have characterized binding interactions between the EF hand domain of α-actinin (Act-EF34) and peptides derived from palladin, and generated a NMR-derived structural model for the Act-EF34/palladin peptide complex. The critical binding site residues are similar to an actinin binding motif previously suggested for the complex between Act-EF34 and titin Z-repeats. The structure-based model of the Act-EF34/palladin peptide complex expands our understanding of binding specificity between the scaffold protein α-actinin and various ligands, which appears to require an α-helical motif containing four hydrophobic residues, common to many α–actinin ligands. We also provide evidence that the Family-X mutation in palladin, associated with a highly penetrant form of pancreatic cancer, does not interfere with α-actinin binding.
Introduction
A dynamic network of actin-associated proteins modulates the structural and dynamic properties of the actin-based cytoskeleton. Several actin binding proteins function as scaffolds that interact with a number of proteins to regulate a wide range of cellular processes, including cell growth, differentiation, adhesion, and motility. α-Actinin is an actin-crosslinking protein that has been shown to function as a platform for the assembly of multi-protein complexes1 and is uniquely positioned as an anchor between the actin cytoskeleton and the cytoplasmic domains of several cell surface adhesion proteins.2 Due to the substantial overlap in subcellular distribution and the emerging functions of palladin in actin filament regulation, the α-actinin-palladin interaction has gained special interest.
Palladin is an actin-associated protein, cloned independently in the Otey3 and Carpen4 labs, that localizes to many actin-containing structures, including stress fibers, focal adhesions, cell-cell junctions and Z-discs. Palladin binds directly to actin regulating proteins (VASP,5 profilin,6 CLP36,7 LPP,8 and Eps89) and actin cross-linking proteins (α-actinin,10 Lasp-1,11 and ezrin4). Multiple studies in both cultured cells and knockout mice suggest that palladin’s actin-organizing activity plays a central role in promoting cell motility.3, 12 Correlative evidence also supports this hypothesis, as palladin levels are upregulated in cells that are actively migrating such as developing vertebrate embryos,13 in cells along a wound-edge,14 and in metastatic cancer cells.15, 16, 17, 18 Our recent results suggest that palladin occupies an unusual functional niche, as it is a molecular scaffold for multiple actin-binding proteins,19 an actin-crosslinking protein,20 and a regulator of transcriptional activity.21 Similar to α-actinin, palladin has emerged as a key player in organizing actin arrays within migrating cells, through both direct and indirect molecular mechanisms.
Palladin is ubiquitous in developing vertebrate tissues and is also expressed in many adult tissues.3 It exists as multiple isoforms that are expressed in tissue-specific patterns.22, 23 In addition, palladin has two close relatives that are expressed in a restricted pattern: myopalladin is found only in heart and skeletal muscle24 and myotilin is expressed mostly in skeletal muscle.25 All three family members bind directly to α-actinin, although apparently via different sites of interaction.10, 24, 25, 26 The α-actinin interaction site of both palladin and myotilin lies in a homologous region with no obvious domain structure or sequence homology to other α-actinin binding partners. Despite shared sequence homology at this region, myopalladin was previously shown to bind α-actinin via its C-terminal Ig domains.24 All three palladin family members bind to the EF-hand repeats 3–4 of α-actinin's C-terminal domain, which is also where titin's Z-repeat 7 interacts with α–actinin.10
Multiple recent studies suggest that disregulation of palladin expression may play a key role in the invasive cell motility that characterizes metastatic cancer cells as well as in the development of cardiovascular diseases.15, 27, 28, 29 Additionally, palladin was directly implicated in a rare inherited form of pancreatic cancer.16 In that study, a point mutation (P239S) in palladin that falls within the α-actinin binding site was identified in an inherited form of highly penetrant pancreatic cancer, suggesting that alteration of palladin/α-actinin interactions may have direct effects on cell behaviors such as motility.
Palladin’s binding partner α-actinin has also been implicated in the metastasis of multiple cancers. Similar to palladin, α-actinin exists in humans as multiple isoforms, including two that are expressed in muscle (actinin-2 and actinin-3) and two that are expressed in non-muscle cells (actinin-1 and actinin-4). Overexpression of one or both non-muscle isoforms of α-actinin has been detected in high-grade sarcomas and in cancers of the esophagus, lung, breast and colon.18, 30, 31, 32, 33, 34, 35, 36 Findings to date suggest that α-actinin can regulate the actin cytoskeleton and increase cell motility; however the specific role of α-actinin in pathological cell motility has not yet been determined.
The direct binding interaction between α-actinin and palladin, their high degree of co-localization in podosomes and other actin-based structures, and the fact that they are both upregulated in invasive cancers suggest that these proteins may have a shared function in motility and adhesion that may be disregulated in cancer cells. It is noteworthy that palladin binds to a region of α-actinin that was previously shown to be involved in auto-inhibitory contacts that regulate α-actinin interactions, suggesting a possible role of palladin to direct or target actinin and/or regulate the ability of α-actinin to bind and cross-link F-actin.37, 38 In fact M. Rönty et al. show that this interaction is critical for bidirectional targeting of both proteins to actin bundles using transfection-based targeting assays.10 Therefore we examined the complex between α-actinin and palladin in light of the previously observed high degree of co-localization between palladin and α-actinin in various subcellular structures.3, 4, 10 Palladin and α-actinin are both functionally and physically linked to both normal and pathological cell motility, however the precise molecular role of this complex in organizing the actin cytoskeleton is unknown. To advance our understanding of the biological significance of the interaction between palladin and α-actinin in cell motility and invasion, we have undertaken the first detailed structure-function analysis of the α-actinin-palladin complex.
Here we present a structural model of α–actinin bound to a palladin peptide. NMR spectroscopy was employed to assign backbone resonances of the Act-EF34 domain, in the presence and absence of a palladin peptide. Orientation constraints and mutagenesis data were then obtained on the complex to generate a structure-based model. We show that the conformation of α-actinin bound to palladin is very similar to that bound to titin Z-repeats, suggesting a similar binding mode, and postulate that ligands that recognize α–actinin may present a common minimal binding motif within a recognition helix, where specificity is dictated by the hydrophobic ‘1-4-5-8’ motif.39
Results
Quantification of Act-EF34 Binding to a Peptide Derived from Palladin
A yeast two-hybrid assay was previously used to identify the site of direct interaction between palladin and α–actinin, narrowing the binding site to a short sequence of palladin (residues 222–280) and to the C-terminal EF-hand domain of α–actinin (Act-EF34).10 This region of palladin shares a highly homologous 17 amino acid region with the α–actinin-binding region of myotilin.25 Little sequence or structural homology exists outside this region for palladin family members. Therefore peptides comprising these 17 residues of palladin, one wild-type (WT) sequence and the other containing the Family X mutation (P239S, a.k.a. FX), were synthesized for use in binding assays with the EF-34 domain of α-actinin. It is important to note that this peptide is found in all of the major isoforms of palladin (isoforms 1, 3 and 4).
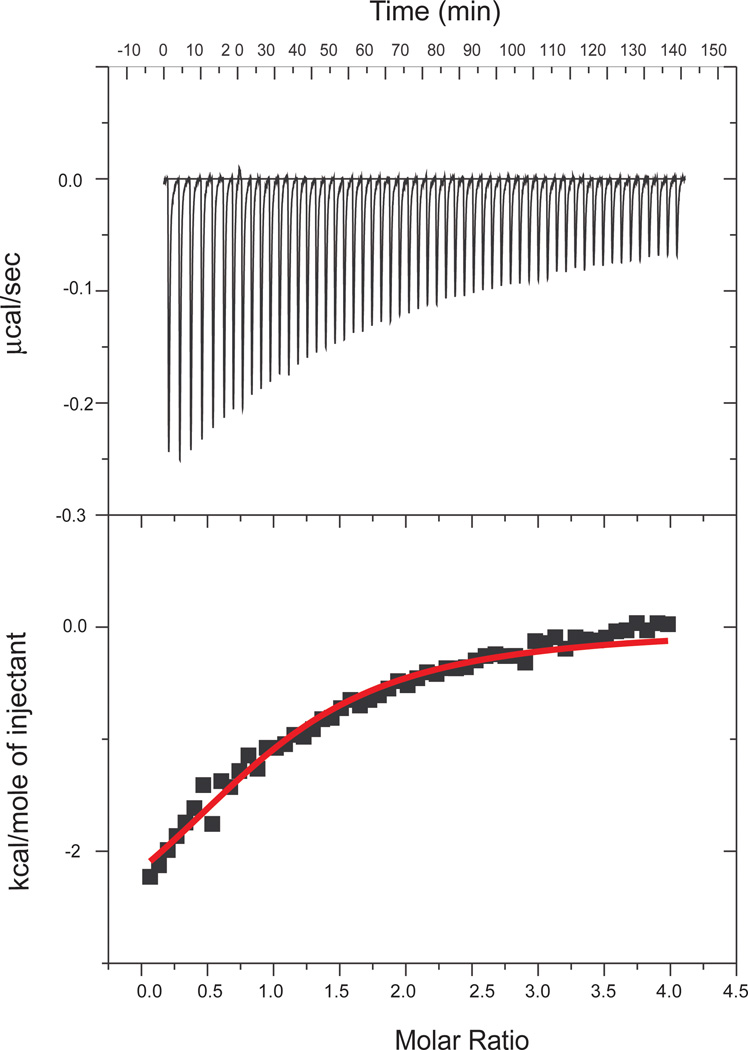
Isothermal titration calorimetry (ITC) was used to characterize the association of α–actinin (specifically, actinin-2) with both the WT and FX palladin peptides. A representative titration is shown in Figure 1, with the thermodynamic parameters of binding listed in Table 1. Both peptides have dissociation constants around 15 µM. The experimental data for Act-EF34 titrations with both WT and FX mutant palladin peptides exhibited stoichiometries close to 1, indicating 1:1 binding events with comparable affinities (within experimental error).
Figure 1.
ITC binding isotherm obtained for the interaction of palladin (WT) with Act-EF34. Fifty-five 5 µL aliquots of palladin (1 mM) were injected into the calorimeter cell containing Act-EF34 (600 µM) at 27 °C.
Table 1.
Thermodynamic parameters for the binding of various peptides to Act-EF34a
| Ligand | Kd (× 10−6 M) |
ΔH (kcal/mol) |
−TΔS (kcal/mol) |
ΔG (kcal/mol*K) |
|---|---|---|---|---|
| Palladin (WT)* | 15.7 ± 7.7 | −4.5 ± 0.2 | 10.1 | −6.6 |
| Palladin (FX) * | 19.5 ± 4.0 | −6.0 ± 0.2 | 12.4 | −6.5 |
| Titin Zr140 | 0.2–0.3 | n.d. | n.d. | n.d. |
| Titin Zr340 | >4.0 | n.d. | n.d. | n.d. |
| Titin Zr740 | 0.100–0.250 | −15.24 | 5.74 | −9.50 |
| Actinin N-term38 | 0.568 ± 0.27 | n.d. | n.d. | n.d. |
n.d. - not determined or reported
ITC was also used to evaluate the thermodynamic characteristics of complex formation between Act-EF34 and palladin peptides. Interestingly, a significant decrease in entropy coincides with palladin binding. A potential source of this change could be accounted for by alteration of dynamic flexibility that takes place upon binding or a general reduction in the conformational dynamics due to ordering of water. This is supported by our CD and NMR data that provide evidence of reduced conformational dynamics of Act-EF34 and support a coil to helix transition of the palladin peptide as will be discussed in following sections.
Binding of Palladin to α-Actinin Induces Helical Structure
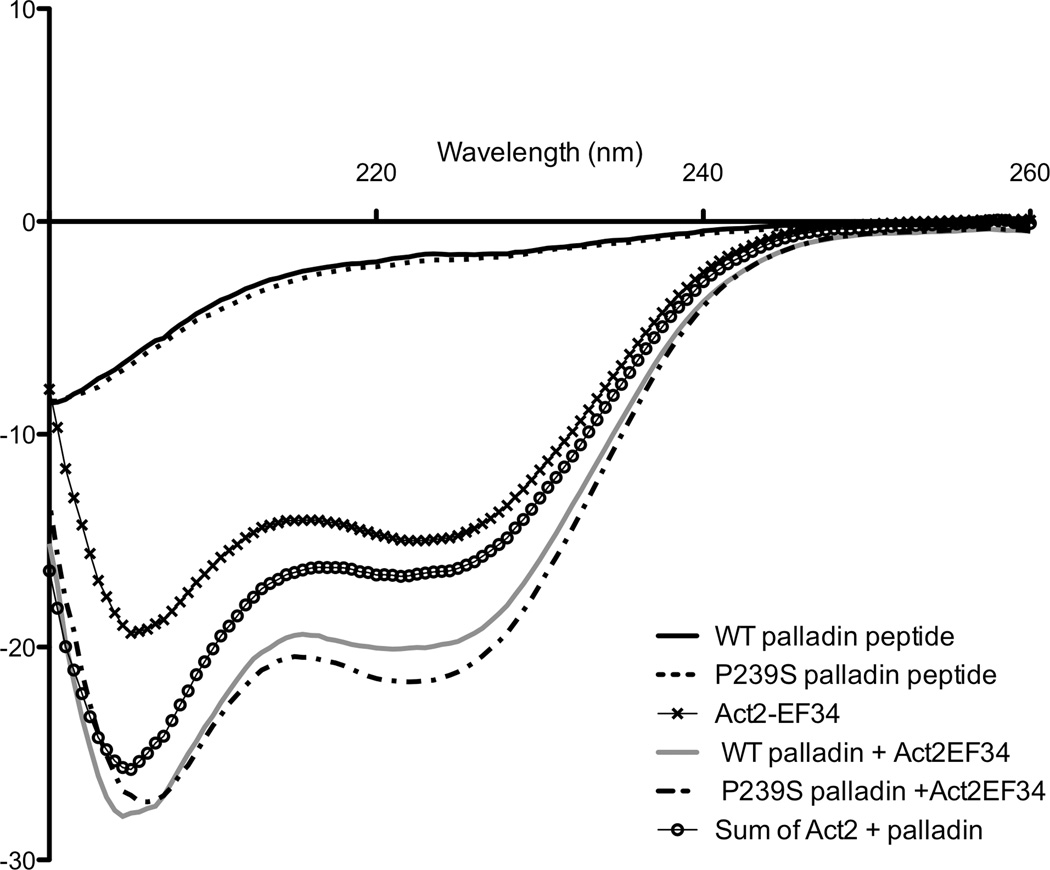
Changes in the conformation of Act-EF34 upon binding to both WT and FX palladin peptides were monitored by far-UV circular dichroism. The CD spectra of the free palladin peptides (WT and FX) in aqueous solution are typical of random coil peptides. Upon complex formation with either palladin peptide, the spectrum of the complex gains significant helical signal (Fig. 2). This gain of signal is greater than the sum of the two isolated components, indicating that complex formation induces changes in the secondary structure of either component solely or in both partners. A similar gain in α-helical signal was also observed for the complex of Act-EF34 with titin’s seventh Z-repeat.40 Several lines of evidence in the aforementioned study of the Act-EF34—Zr7 complex revealed that the Zr7 adopts a helical conformation upon association with Act-EF34;39, 40, 41 therefore it is possible that the palladin peptide also adopts a helical conformation upon binding actinin.
Figure 2.
Gain of signal for the Act-EF34/palladin complex recorded by far-UV CD spectroscopy. The concentration of each component was 20 µM, and the path length was 2 mm. The spectra of the isolated components, WT palladin (—), FX palladin (- -), Act2-EF34 (✕), WT complex (—), and FX complex (–·-) are shown. The sum of the spectra for the isolated Act-EF34 and palladin peptide is also reported for comparison (○).
Interactions between Act-EF34 and either WT or FX palladin peptides were further characterized by NMR spectroscopy. Complexes were formed by titrating 15N-labeled Act-EF34 with unlabeled peptide (upon reaching saturation at 5-fold molar excess). Two-dimensional 1H-15N HSQC spectra show that upon complex formation, the spectrum of Act-EF34 changes considerably and is in fast exchange on the NMR timescale. In addition, the chemical shift changes in backbone resonances of Act-EF34 observed upon the addition of the palladin peptide are almost identical to that obtained with FX palladin peptides. As noted previously,40 the spectrum of isolated Act-EF34 contains more peaks than what is expected from its sequence, implying that Act-EF34, which comprises the last 72 residues of α-actinin, can adopt more than one conformation. However the number of backbone resonances in the spectrum of both the WT and FX palladin peptide complexes with Act-EF34 decreases to 59 consistent with the molecular size of the EF34 domain (72 residues in total, with 6 prolines). These results indicate that Act-EF34 undergoes a change in conformational dynamics on binding to the palladin peptide.
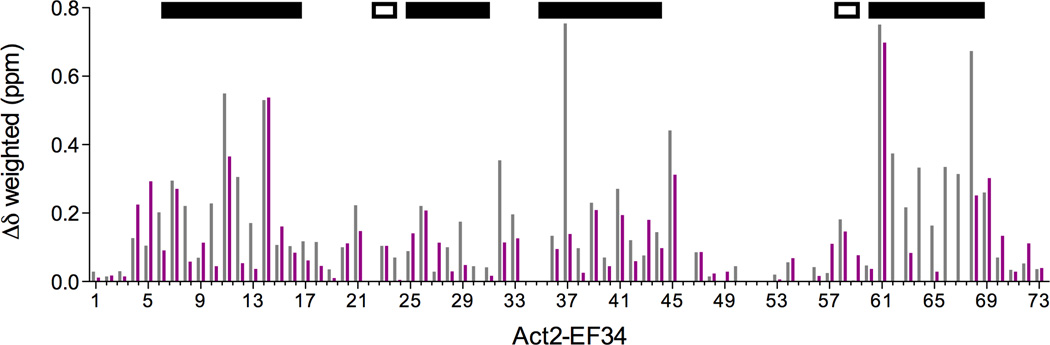
To further characterize the residues of Act-EF34 that are affected by palladin binding, chemical shift perturbation analysis was performed. Backbone chemical shift assignments were obtained for the complex of Act-EF34 with palladin peptide to compare with previously determined chemical shift assignments for unbound Act-EF34 (BMRB ID: 17626, 17627). Chemical shift perturbations (CSP) induced by both WT and FX palladin peptides are very similar to that observed for actinin in complex with the Zr7 peptide of titin, despite the lack of sequence homology between palladin and titin peptides (Fig. 3b).39, 40 The largest CSP values were associated with backbone NH resonances of residues in the C-terminal helix (residues 61–69), especially those on the hydrophobic face of helix 4. However, significant changes were also observed in region of the N-terminal helix (residues 4, 5, 7, 11, and 14 in particular), suggesting that both the N- and C-terminus of Act-EF34 interact with the peptide. Our findings showing similar chemical shift perturbations induced in Act-EF34 upon complex formation with palladin and titin peptides, suggests a similarity in their mode of binding. Furthermore, Atkinson et al. proposed a ‘1-4-5-8’ motif for conserved hydrophobic residues in the titin Z-repeats that are critical for binding Act-EF34.39 As illustrated in Table 2, alignment of several Act-EF34 ligands lends further credence to this proposed binding motif that now includes all of the palladin family members. Titrations with peptides composed of the myotilin and myopalladin sequences result in identical chemical shift perturbations, offering further support for this common motif.
Figure 3.
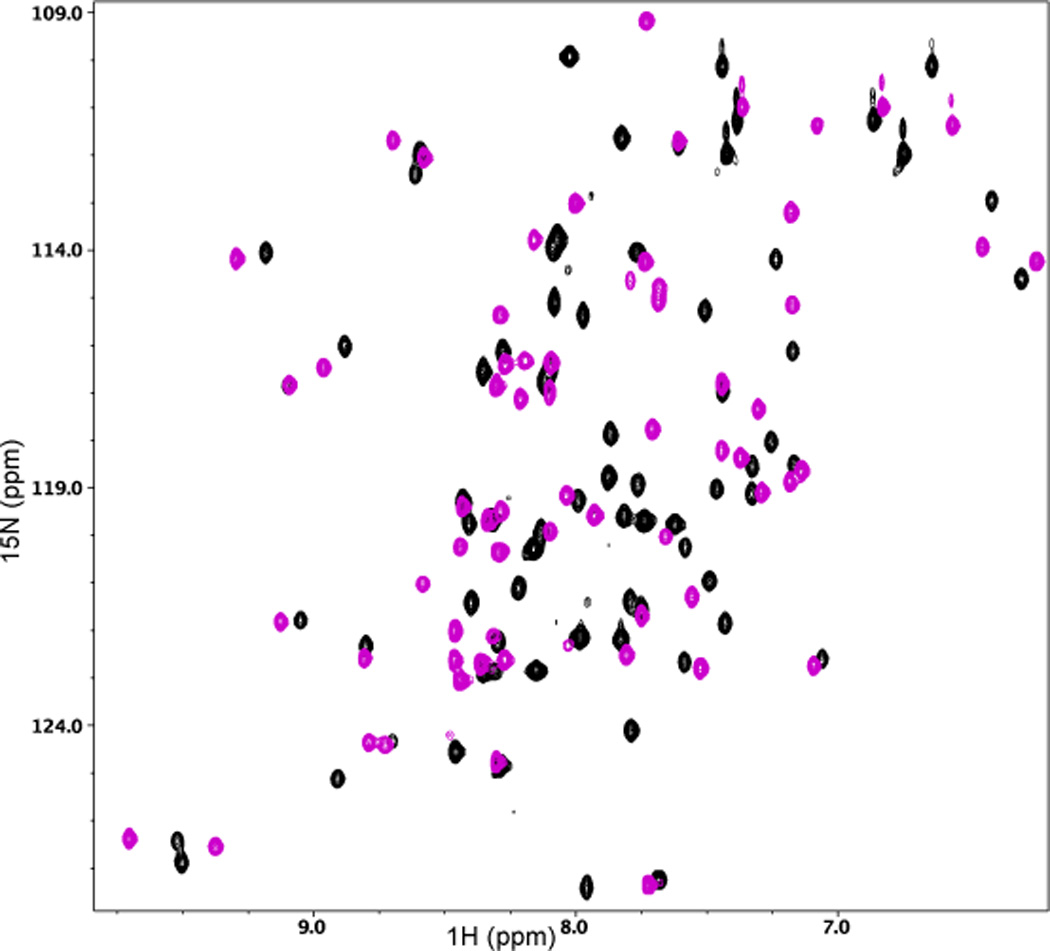
(a) 2D 1H-15N HSQC spectral overlay of 15N-labeled Act-EF34 in the absence (black) and presence of bound WT palladin peptide (magenta). (b) Chemical shift perturbation of Act-EF34 amide resonances upon complex formation with either titin Zr7 (gray) or palladin peptide (magenta). The α-helical regions or β-strands of Act-EF34 are designated by filled or open bars, respectively, above the plot. Also indicated (dotted line) is the chemical shift cut-off value used for the definition of AIRs as input for the HADDOCK calculations (0.17 ppm).
Table 2.
α-Actinin is capable of binding to a relatively diverse range of sequences. Sequence alignment of residues associated with proteins involved in binding Act-EF34, highlighting the proposed '1-4-5-8' motif. Numbers indicate positions of the hydrophobic residues involved in binding with numbering starting from the first hydrophobic residue. The residue in palladin mutated in FX is highlighted in red, while the leucines mutated in the binding motif are in blue.
| Protein | Residues | Act2-EF34 Binding Site '1--45--8' |
|---|---|---|
| Palladin | 235–251 |
|
| Myotilin | 89–105 | NQSPASFLSSILPSQPD |
| Myopalladin | 822–837 | IQNPVAFLSSVLPSLP |
| Titin ZR1 | 424–439 | AVATVVAAVDMARVRE |
| ZR3 | 519–534 | FVPKVVISAAKAKEQE |
| ZR7 | 659–674 | AVATVVAAVDQARVRE |
| Actinin N-term | 259–281 | AETAANRICKVLAVNQE |
Mapping the CSP data onto the structure of Act-EF34 is complicated by the fact that a structure of unbound Act-EF34 is not available, which may be due to the fact that Act-EF34 is conformationally dynamic in its unbound state. Evidence of multiple conformations is revealed by the presence of “doubled” backbone amides (for residues T4, T6, G56, A66, L67, Y68, and G69) in both the CBCACONH and HNCACB spectra of unbound Act-EF34, as previously observed by Y. Au and A. Pastore (unpublished). This second species is not observed once Act-EF34 is in complex with titin or palladin. The structure of Act-EF34 bound to a titin Z-repeat peptide has been solved (PDB ID: 1H8B)39, and this complex was employed for comparison purposes. To assess whether binding of the palladin peptide to Act-EF34 induced changes in the secondary structure of α-actinin, we made use of the assigned chemical shifts of Act-EF34 in both its free and bound forms, to empirically predict dihedral angles and secondary structure using the program TALOS+.42 According to this analysis, the C-terminus of Act-EF34 has a higher propensity to adopt an α-helix (residues 62–67) when bound to either palladin or titin peptides in solution (data not shown). In addition, the dihedral angles predicted for this region are more consistent with a helical secondary structure in the presence of either ligand, suggesting that peptide binding induces some additional α–helical content in the α–actinin C-terminus.
Ligand binding alters backbone dynamics of actinin
To investigate the backbone dynamics of Act-EF34, 15N longitudinal (T1) and transverse (T2) relaxation times as well as steady-state {1H}-15N nuclear Overhauser effect (NOE) data were measured for both the free and bound forms of 15N-labeled Act-EF34 (Fig. 4). The relaxation data suggest that binding of the palladin peptide to Act-EF34 results in reduced Act-EF34 NH backbone dynamics on a ps-ns time scale in regions that contact the ligand. For example, negative heteronuclear (het)NOEs were observed for the N-terminus of Act-EF34 in the unbound state, which suggests that residues 4–15 possess a higher conformational mobility relative to the rest of the Act-EF34 domain on the ps-ns timescale. However, in the bound state, these hetNOE values become positive and suggest that backbone flexibility is markedly decreased upon ligand binding.
Figure 4.
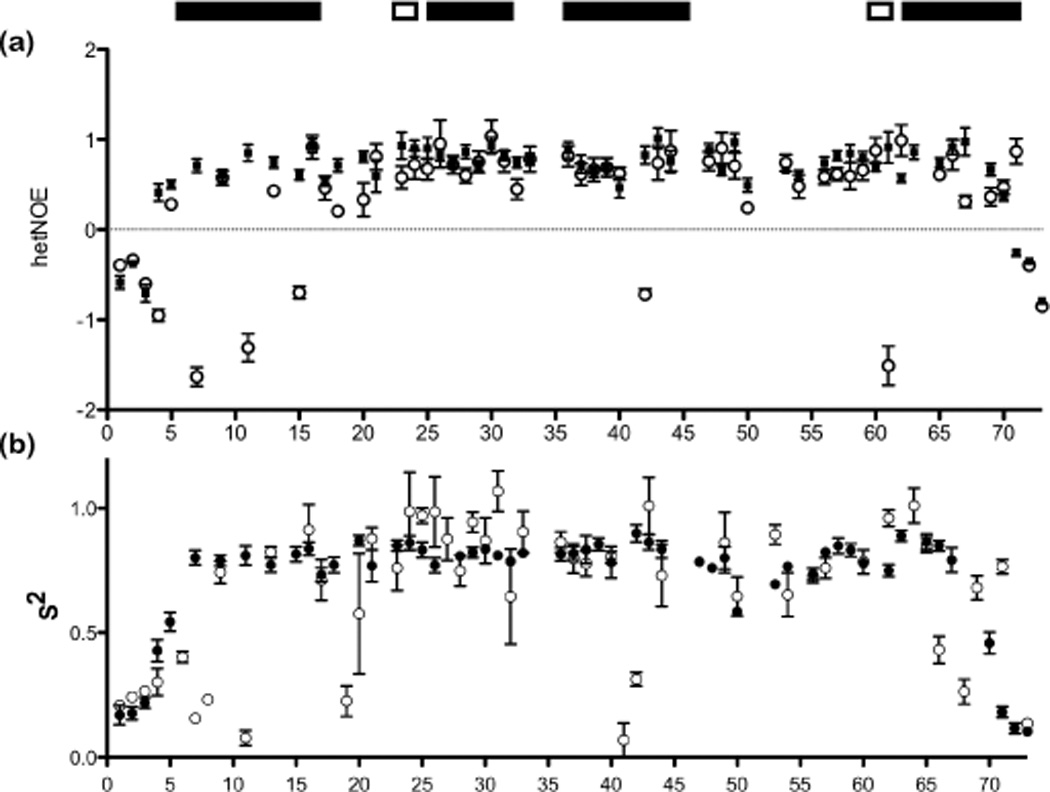
Ligand binding alters the backbone flexibility of Act2-EF34 as monitored by NMR relaxation analyses. Relaxation parameters, (a) {1H}-15N NOE values and (b) the generalized order parameter (S2) for Act-EF34 in both its palladin-bound (filled circles) and free (open circles) forms, plotted against α-actinin residue number.
Rotational diffusion analysis of the relaxation data parameters can also be used to characterize the hydrodynamic properties of proteins.43 A global fit of the backbone relaxation data produced the following values for rotational correlation times (τm): 7.5 ns for unbound Act-EF34 and 5.7 ns for Act-EF34 bound to WT palladin peptide. The smaller τm value for Act-EF34 bound to palladin is consistent with a more compact structure upon complex formation. To gain a more thorough picture of the ps-ns backbone dynamics, the relaxation data were also interpreted in terms of the Lipari-Szabo “model-free” formalism, where the generalized order parameter (S2) ranges from 0 (complete disorder in the molecular frame) to 1 (fixed bond orientation).44, 45 Model-free parameters were obtained based on the three-parameters (S2, τe, Rex,app) of the spectral density function and the τm values. Results from these analyses indicate that only a marginal increase in average order parameter is observed for unbound Act-EF34 compared to the palladin-bound Act-EF34 complex (<S2>free = 0.62 and <S2>bound = 0.69). However, a comparison of the S2 values for each residue, as shown in Fig. 4b, indicates higher backbone mobility on the ns-ps time scale in several segments of the ligand-free Act-EF34 protein with respect to the palladin-bound form. The presence of “extra” peaks in the isolated Act-EF34 domain is indicative of multiple conformations most likely resulting from conformational heterogeneity in the unbound state. Relaxation data analyses of the N-terminus and other residues involved in ligand-binding (residues 7,8,11,19,41,42,66, and 68) show significantly decreased S2 values of 0.22 (average) in the unbound form, relative to the values of S2 >0.8 obtained for the same residues in the bound form. Our data, taken together, suggest a major conformational rearrangement in Act-EF34 is observed upon complexation, resulting in a more compact and structured state.
Long-range orientation constraints suggest that the Act-EF34 complexes with palladin and titin have similar overall structures
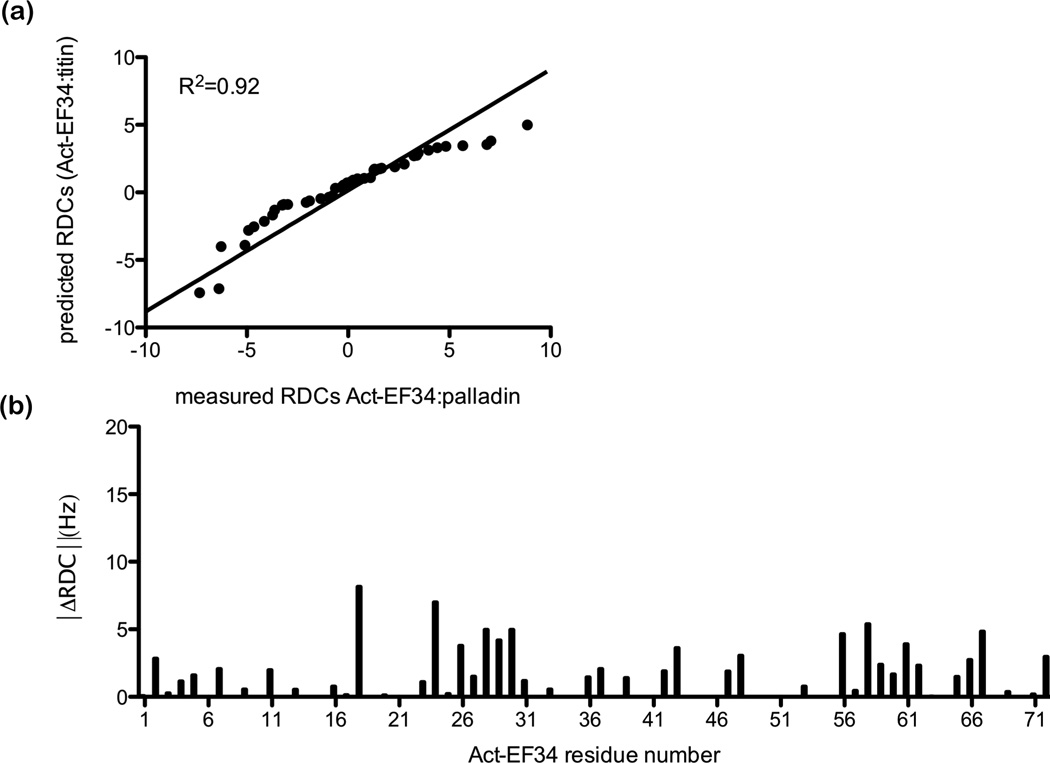
We measured one-bond backbone 1H-15N residual dipolar couplings (RDCs) for the complex of α-actinin with palladin and compared the results to RDCs back-calculated from the NMR structure of Act-EF34 bound to titin’s Z-repeat (PDB ID: 1H8B).39 RDCs are informative because they provide long-range orientation restraints for each of the amide bond vectors. Analysis of the RDC data was conducted using the program REDCAT46 using the NMR structure of the Act-EF34 bound to titin Zr7 (PDB ID: 1H8B).39 Given a molecular structure and RDC data, REDCAT uses an algorithm to solve for the elements of an order tensor that best represents the RDC data. In our case we used 45 experimentally measured RDCs for the complex of Act-EF34 with palladin peptide, excluding measurements from highly overlapped regions and regions known to be in flexible loops. An error estimate of 2 Hz was used for Act-EF34 bound to titin Zr7. REDCAT can also be used to back-calculate RDCs given elements of an order tensor and a probable structure. For Act-EF34 bound to palladin, a plot of experimental versus back-calculated RDCs is given in Fig. 5a. The correlation is reasonably good, with a R2 value of 0.92. This correlation can also be quantified in terms of a Q-factor (a goodness-of-fit measure for RDCs), where a low Q indicates better agreement.47, 48 Although a Q-factor of 0.479 was obtained for the comparison of titin-bound and palladin-bound Act-EF34, this value is not as low as that seen for structures with a high degree of structural homogeneity and corresponds to a 2.69 Å RMSD over the backbone atoms. For comparison, the calcium-insensitive EF-hands of the essential and regulatory light chains (ELC and RLC) from scallop myosin display the closest structural similarity to Act-EF34 with RMSD of 3.0 Å and 3.7 Å, respectively.39 Overall the chemical shift and RDC data indicate that the structure of Act-EF34 bound to palladin is very similar to the Act-EF34-titin complex and retains the semi-open conformation observed for other calcium-insensitive EF-hands.49, 50
Figure 5.
Comparison of titin-bound versus palladin-bound Act-EF34 revealed by residual dipolar coupling NMR (RDC) data. (a) Correlation plot of experimentally measured RDCs for Act-EF34 bound to palladin peptide versus back-calculated RDCs based on the NMR structure of Act-EF34 bound to titin Zr7 (PDB code: 1H8B) using REDCAT. (b) The absolute difference between the observed and expected dipolar couplings (ΔRDC) is plotted as a function of amino acid residue for Act-EF34.
Identification of critical palladin residues involved in actinin binding
NMR efforts failed to provide intermolecular NOEs between Act-EF34 and the palladin peptide, therefore it was not possible to calculate the structure of the complex between palladin and α-actinin based on NMR restraints alone. However, we were able to identify critical binding site residues on palladin by mutating two hydrophobic leucines in the proposed ‘1-4-5-8’ motif.39 These mutations were not predicted to alter the helicity of the peptides.51 Titration of 15N-Act-EF34 with a palladin peptide containing polar Asn residues in place of the Leu at positions 1 and 4 of the binding motif, showed no observable changes in the 1H-15N 2D HSQC spectrum in comparison with the isolated Act-EF34 domain. NMR can detect very weak interactions, due to the exquisitely sensitive dependence of the NMR chemical shift on the local environment. Therefore the lack of any changes in chemical shift upon titration of Act-EF34 with mutant palladin peptide at a molar ratio of 1:6 (Act-EF34:palladin) suggests that binding is significantly reduced or eliminated when these two residues of palladin are mutated. In addition, CD measurements revealed no change in the helicity upon addition of this palladin peptide (at same ratio) containing two binding-site mutations to Act-EF34 (data not shown), as opposed to the significant increase previously observed for both WT and FX palladin peptides bound to Act-EF34.
Modeling of the Palladin-Actinin complex
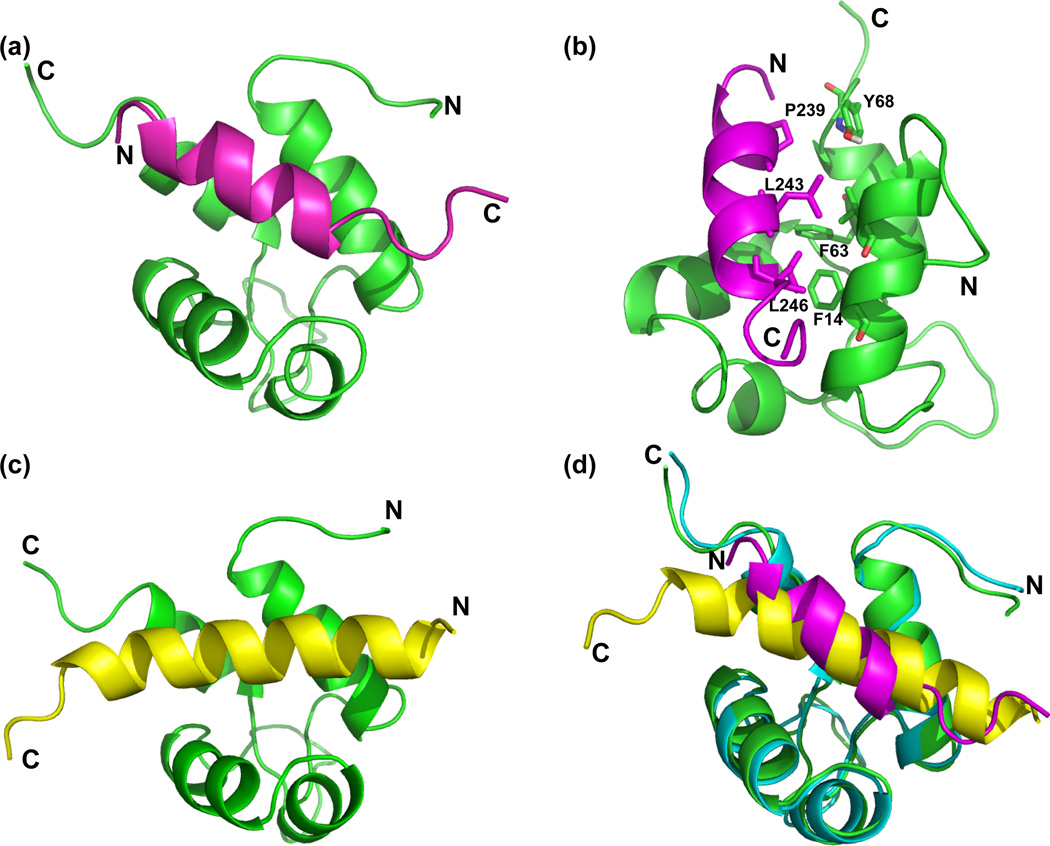
Modeling of protein-protein complexes from NMR and/or mutagenesis data has been highly successful in generating robust structures of protein complexes. To generate a model of the Act-EF34:palladin complex, the data-driven docking program, HADDOCK,52 was employed. HADDOCK offers the advantage of deriving restraints from a broad array of experimental data, while also permitting conformational rearrangements to occur during the docking process. It is straightforward to dock proteins when the conformation of the protein and/or peptide is known, and when large conformational changes do not occur upon binding. However, the experimental evidence we have obtained from RDCs, CD, and CSP indicate that the conformational change in Act-EF34 that we observe upon binding palladin is similar to that observed for titin binding. Therefore, the initial structure used in our docking protocol is that of Act-EF34 bound to titin. Based on the observed chemical shift perturbations of Act-EF34 upon complex formation with the palladin peptide, we were able to define 20 ambiguous interaction restraints (AIRs) at the interface on Act-EF34 (see Materials and Methods). As a palladin peptide containing asparagine substitutions for residues Leu 243 and 246 did not demonstrate detectable binding to Act-EF34, these palladin residues were also input as AIRs. Analysis of the final 200 water-refined HADDOCK models for the ActEF34-palladin complex resulted in 10 different clusters, with cluster 1 showing the best average HADDOCK score (Table 3). These results suggest that cluster 1 represents the best HADDOCK-derived structural ensemble for the ActEF34-palladin complex (Fig. 6a), with an average backbone RMSD value of 0.9 (± 0.5) Å.
Table 3.
Statistics of the top 3 Act-EF34 – palladin clusters obtained with HADDOCK
| Cluster | Haddock scorea | Nb | RMSD-Eminc | EvDWd | Eelecd | EAIRe | Edesolvf |
|---|---|---|---|---|---|---|---|
| 1 | −79.6 (4.6) | 12 | 0.9 (0.5) | −43 (4) | −110 (17) | 62 (29) | −21 (3) |
| 2 | −58.0 (1.4) | 7 | 7.1 (0.2) | −42 (5) | −41 (16) | 49 (19) | −12 (3) |
| 3 | −57.6 (6.5) | 5 | 7.5 (0.2) | −40 (3) | −57 (19) | 47 (28) | −11 (5) |
Final HADDOCK score calculated as the weighted sum of: 0.2*Eelec + 1.0*EvDW + 1.0*Edesolv + 0.1*EAIR.
Number of structures in a given cluster.
Overall backbone RMSD from the lowest energy structure.
The intermolecular energies (kcal mol−1) were calculated with the OPLS (Optimized Potentials for Liquid Simulations) parameters using a 8.5 Å cut-off.
HADDOCK ambiguous interaction restraint (AIR) energy (kcal mol−1)
The desolvation energy is calculated using the atomic desolvation parameter by Fernandez-Recio et al.74
Figure 6.
Comparison of the Act-EF34 complex with palladin and titin peptides. (a) Ribbon diagram showing a representative palladin complex structural model. Act-EF34 is shown in green and palladin in magenta, with termini labeled to highlight orientation. (b) Critical residues used in docking are highlighted. The backbone atoms associated with the hydrophobic Act-EF34 Phe 14, Phe 63, and Tyr 68 residues that were utilized as restraints in docking. Palladin residues, Leu 243 and 246, are hydrophobic residues in the first and fourth position of the ‘1-4-5-8’ motif. These residues were mutated, found to abolish binding to Act-EF34, and then used as restraints for docking. (c) For comparison, the ribbon diagram of the complex between Act-EF34 (green) and titin’s seventh Z-repeat in yellow is shown (Act-EF34—Zr7; PDB ID: 1H8B). (d) Structural alignment of palladin-bound (green with magenta peptide) and titin-bound (cyan with yellow) Act-EF34.
As further validation of this structural model, we compared the RDCs back calculated from this HADDOCK-derived structure of Act-EF34-palladin with those that were experimentally determined for the complex resulting in a RMSD of 0.41 Å and a Q-factor of 0.189. These values reflect a better agreement between the experimental data and model than our previous comparison between the Act-EF34 complex with titin and the experimental RDCs for Act-EF34—palladin complex. The structures of Act-EF34 bound to titin (Fig. 6b) or palladin peptides (Fig. 6a) only differ slightly, with backbone RMSD value of 0.76 Å as shown in the structural alignment (Fig. 6d). However the orientation of the peptides are reversed in these two complexes. Restraints for positioning the peptide were lacking in the preliminary stages of docking, therefore we utilized paramagnetic relaxation enhancement (PRE) measurements to verify the orientation of the palladin peptide.
Use of NMR-derived paramagnetic relaxation data to orient the palladin peptide in the palladin/Act-EF34 complex
Placement of the palladin peptide in the complex with Act-EF34 was initially determined primarily by anchoring palladin peptide residues Leu 243 and 246 in the docking procedure. 1H-15N residual dipolar coupling data and chemical shift perturbation measurements recorded on Act-EF34 in complex with the palladin peptide provide ample information for constraining actinin in the docking procedure. Site-directed mutagenesis provided important constraints for palladin; yet the relative orientation of the helical palladin peptide was not evident due to the lack of long-range distance restraints. In order to overcome this limitation, we introduced a paramagnetic center (nitroxide radical) into the palladin peptide by formation of a disulfide between MTSL and an N-terminal cysteine of the palladin peptide. Paramagnetic relaxation probes provide an efficient mechanism for relaxation of neighboring nuclei via dipolar coupling, and thus provide useful long-range distance information.53 The orientation of the peptide was rapidly determined by evaluating the intensity changes associated with backbone resonances of Act-EF34 upon addition of the MTSL-palladin peptide. Paramagnetic relaxation enhancement (PRE) was detected by comparing the peak heights in 2D 1H-15N-HSQC spectra before and after reduction of the nitroxide spin label with ascorbic acid. A decrease in the intensity of peaks was observed predominantly for residues located at both the N- and C-termini, indicating their proximity to the N-terminus of the palladin peptide. These PRE results support the HADDOCK model of the palladin-Act-EF34 complex, which orients the peptide in the same manner despite the fact that this was not used as an explicit constraint.
Discussion
In this study, we have examined the binding interaction between the minimal ligand binding module of palladin and the C-terminal EF-hand of α-actinin using NMR spectroscopy, CD and ITC. We have also evaluated whether mutation of the palladin residue 239 from proline to serine, also referred to as the Family-X mutation due to its association with highly penetrant pancreatic cancer, perturbs interactions with Act-EF34. A model representing a structure of a complex between palladin and Act-EF34 has been obtained using information derived from a variety of NMR experiments (chemical shift perturbation, residual dipolar couplings, and paramagnetic relaxation enhancement) and mutagenesis. This first structural model of a complex of palladin and α-actinin is described and further analyzed in the context of other α-actinin complexes to reveal a common recognition motif.
A general ligand recognition motif for α-actinin
Act-EF34 binds to a variety of molecules such as the Z-repeats of titin, palladin family members, and its own N-terminus in an autoinhibitory mode. Although the binding regions of the target proteins do not possess strong sequence homology, there is a conserved pattern of hydrophobic residues which comprise a ‘1-4-5-8 motif’ as shown in Table 2 and previously described.39 Residues within this motif adopt an α-helical conformation upon binding to Act-EF34 and possess a common, hydrophobic face of the helix that makes contact with the hydrophobic cleft of Act-EF34. Both titin and palladin peptides occupy a cavity in Act-EF34 formed by the first helix and the C-terminus of the Act-EF34 domain. In our NMR-derived model of the palladin/Act-EF34 complex, the orientation of the palladin peptide within the Act-EF34 cavity is opposite that of titin. This difference in bound ligand directionality also occurs in target peptides of other representative EF-hand domain complexes. Complexes of calmodulin (CaM) with different binding partners can be used to illustrate this point, as the N-termini of the MLCK (myosin light chain kinase)54, 55 and CaMKII (Ca2+/calmodulin-dependent protein kinases II)56 peptides interact primarily with the C-terminus of CaM; whereas melittin57 and CaMKK (Calmodulin-dependent protein kinase kinase)58 peptides bind in the reverse orientation with respect to CaM. Experimental observations have linked peptide orientation to a subtle balance between hydrophobic and electrostatic effects.59, 60 Like a palindromic sequence, the ‘1-4-5-8’ motif of hydrophobic residues on the ligand can bind in either orientation. These anchoring residue interactions are often augmented by charged residues that form salt bridges with Act-EF34 and direct orientation of the peptide within the cavity. Intermolecular contacts between palladin and Act-EF34 appear to be solely dependent on hydrophobic side chains on both molecules, with the leucine side chains of palladin (243 and 246) making contact with the hydrophobic core of Act-EF34 (specifically Phe 14, Tyr 48, Tyr 60, Phe 63, and Tyr 68) (Fig. 6b).
In the context of the actinin interactions with palladin, we have identified a 17 amino acid sequence of palladin required for binding and have further identified two key residues involved in hydrophobic interaction with α-actinin via mutagenesis. Our leucine to asparagine mutations in palladin provide the first direct evidence that the hydrophobic residues at position 1 and/or 4 in the ‘1-4-5-8’ motif are necessary anchoring points for complex formation with Act-EF34. Further support for this binding motif is provided by NMR-based titration results (data not shown) with the myotilin and myopalladin peptide sequences, also containing this ‘1-4-5-8’ motif, that result in chemical shift perturbations of Act-EF34 that resemble those for palladin and titin.
Ligand-induced structural and dynamic changes in actinin
EF-hands are helix-loop-helix binding motifs found in a large number of proteins that are involved in the regulation of many cellular processes. Typically, EF-hand proteins undergo a well-established transition from a closed to an open conformation upon binding of calcium, thus exposing target peptide binding sites. Binding of target peptides to this exposed site causes little further change in the domain structure. Yet the C-terminal EF-hand of α-actinin does not bind calcium, due to amino acid substitutions that disrupt the Ca2+-binding loops, which results in ligand-binding occurring via a proposed “static” complex. As opposed to Ca2+-sensitive EF-hands, large conformational changes are not essential for binding.39 Our results provide evidence of an entropically driven transition in Act-EF34 that occurs upon ligand-binding, which results in a rigidification of the backbone that parallels an increase in secondary structure. While much attention has been focused on the calcium-dependent conformational changes in EF-hand domains, little is known about conformational dynamics for calcium-independent EF-hand proteins. Our results indicate that a major conformational rearrangement of Act-EF34 is observed upon complexation with multiple ligands associated with both the titin Z-repeats and palladin family members, and furthermore that these complexes adopt similar structures upon binding their cognate ligands.
Family X mutation does not affect binding interactions with actinin
Previous work revealed that deletion of the α–actinin binding site from palladin was critical for proper localization.10 Furthermore, expression of the FX mutant form of palladin in HeLa cells resulted in “atypical aggregates” of palladin that did not co-localize with actin.16 In this context we hypothesized that the FX mutation, which occurs in the α-actinin binding site of palladin, could disrupt this interaction and thus explain the correlation between this mutation and the disruption in cytoskeletal organization. However our results indicate that the FX mutation does not affect binding to Act-EF34 in the context of the palladin peptide. Based on our structural model, the mutated proline residue in the palladin peptide is oriented towards the actinin binding site cleft, however it does not overlap with the conserved hydrophobic binding motif. It is possible that this mutant form of palladin behaves differently in the context of full-length palladin or that the P239S mutation in FX palladin provides a novel site for phosphorylation. There may also be other proteins that interact with palladin at this site and could compete with α-actinin for binding which could be affected by this mutation. At this point it is not clear whether this mutation is loss or gain of function, so it will be of interest to further investigate the role of this mutation in palladin function.
Conclusions
We have conducted a series of NMR and biochemical studies, to generate a model of the complex between α-actinin and palladin, from which the inter-atomic interactions important for recognition are described. The NMR-derived structural model is further supported by site-directed mutagenesis. Ligand binding to Act-EF34 is accomplished via the burial of hydrophobic residues on both the exposed face of Act-EF34 and the palladin fragment, which results in a complex structure with a more compact structure and reduced backbone conformational mobility at the site of ligand binding. Our data further provide support for a common binding motif between Act-EF34 and a variety of ligands that was previously proposed by Atkinson et al. and consists of a hydrophobic ‘1-4-5-8’ motif.39 In conclusion, we have identified and evaluated the minimal residues in human palladin required for binding α-actinin and provide evidence that this ‘1-4-5-8’ motif is conserved among the other palladin family members, myotilin and myopalladin. The results presented here have set the stage for investigating the contribution of the α-actinin-palladin interaction in actin cytoskeleton architecture and regulation of cell motility in future cell-based assays.
Materials and Methods
Protein expression and purification
The 72 C-terminal amino acids of human actinin-2 (residues 823–894) was expressed and purified as described.40 Uniformly 13C,15N-labeled samples were produced using standard strategies in M-9 minimal media.
Peptides
All peptides were synthesized by K. Krajewski and B. Strahl from the UNC High-Throughput Peptide Synthesis and Arraying Facility and determined to be more than 95% pure by MALDI-MS (Matrix-assisted laser desorption/ionization mass spectrometry) and HPLC (high pressure liquid chromatography). Wildtype (WT) palladin peptide (Ac-HGQTPAAFLSALLPSQP-OH) corresponds to amino acids 235–252 of palladin isoform #4 (also referred to as the 90 kDa isoform). The Family X (FX) peptide (Ac-HGQTSAAFLSALLPSQP-OH) contains a point mutation (P239S). A third peptide containing mutations in the first and fourth positions (L{1,4}N) of the 1-4-5-8 hydrophobic binding motif was synthesized with polar asparagine residues in place of the leucines (Ac-HGQTPAAFNSANLPSQP-OH). For the PRE experiments, a single cysteine residue was added to the N-terminus of the WT palladin peptide for modification with the spin label reagent MTSL (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)methanethiosulfonate) (Toronto Research Chemicals) that attaches via a disulfide bond (Ac-CHGQTPAAFLSALLPSQP-OH). MTSL was added from a concentrated stock in acetonitrile at a molar ratio of 4:1 MTSL:peptide and incubated at room temperature for 12–16 h. Excess MTSL reagent was removed by dialysis at 4 °C for 24 h into NMR buffer with minimal amount of reducing agent (20 mM MOPS, pH 6.6, 10 mM NaCl, 0.2 mM TCEP (tris(2-carboxyethyl)phosphine), 10 % D2O, 0.01 % NaN3). The concentration of each peptide was determined using their predicted molar extinction coefficients: WT, ε214=33,763; FX, ε214=31,122 M−1cm−1; and L(1,4)N, ε214= 34,908 M−1cm−1.
CD measurements
CD spectra were recorded on an Applied Photophysics Pistar-180 CD spectropolarimeter. Spectra were recorded on samples containing 20 µM peptides and/or Act-EF34 protein in 10 mM phosphate buffer, 10 mM NaCl, containing 2 mM TCEP, at pH 6.6 and 27 °C, and were baseline corrected by subtraction of the appropriate buffer spectrum.
Isothermal Titration Calorimetry
All isothermal titration calorimetry (ITC) experiments were performed on a MicroCal VP-ITC microcalorimeter. Protein preparations were extensively dialyzed against 20 mM MOPS (pH 6.6 or 7.5), 10 or 100 mM NaCl, 2 mM TCEP and peptides were resuspended in this same buffer. The pH of the peptide and protein solutions was determined to be identical to within 0.05 pH units at room temperature. All solutions were degassed for ≈5–10 min without stirring and kept on ice before the ITC experiments. Titrations consisted of sequential injections, in which 600 µM−1 mM of the palladin peptide was added to a sample cell containing 200–600 µM Act-EF34 in the same buffer at 27 °C. The heats of dilution of palladin peptide into buffer were determined separately and subtracted from the titration prior to analysis. All data best fit to a one-binding site model using MicroCal Origin software to obtain values for the stoichiometry (N), association constant (Ka), and enthalpy change (ΔH), whereas values for the entropy change (TΔS) and free energy (ΔG) were calculated using standard thermodynamic equations.61
NMR spectroscopy
The assignments of unbound Act-EF34 have been determined previously and are available (BMRB ID: 17627). All NMR spectra were acquired on Varian Inova 700 MHz NMR spectrometer equipped with a triple resonance probes and z-axis pulsed-field gradients operating at 600- and 700*-MHz (*also equipped with cryoprobe). Sample concentrations ranged from 0.3–0.6 mM in NMR buffer containing 20 mM MOPS (pH 6.6), 10 mM NaCl, 2 mM TCEP, 0.01% NaN3, and 10% D2O. All data were collected at 27 °C and the temperature was calibrated using a 100% methanol sample (Wilmad). 1H chemical shifts were referenced to an external sample containing 0.01% DSS in NMR buffer; 15N and 13C chemical shifts were indirectly referenced.62 Sequence-specific assignments of the 1HN, 13Cα, 13Cβ, 13C, and 15N resonances for Act-EF34 in complex with WT palladin peptide were obtained manually using 1H,15N heteronuclear single quantum coherence (HSQC) spectra and a combination of triple-resonance experiments including three-dimensional HNCACB,63 CBCA(CO)NNH,64 and HNCO65 pulse sequences. NMR data were processed using the NMRPipe66 and NMRViewJ software (One Moon Scientific).67 Chemical shift perturbation analysis was performed using a weighted vector combination of shifts, calculated by applying the standard formula {ΔωH2 + (0.1*ΔωN)2}0.5 for 1H-15N shifts.
Backbone relaxation measurements were assessed by measuring 15N T1, T2, and {1H}-15N heteronuclear NOE using standard pulse sequences.68 Data were acquired at 600 MHz for both unbound Act-EF34 (300 µM) and the complex of Act-EF34:WT-palladin (300 µM Act-EF34 and 1 mM palladin peptide). For T1 and T2 experiments, nine relaxation time points along with three duplicates were collected. In the 2D {1H}-15N NOE, a recycle delay of 4.5 s was used. Motions on the ps-ns timescale were characterized using model-free formalism.44 The Dellwo-Wand method69 was employed to fit the isotropic rotational correlation time (τm) to the relaxation rates. The global tumbling time was determined to be 7.49 ns/rad for unbound Act-EF34 and 5.69 ns/rad for palladin-bound Act-EF34, assuming spherical molecular symmetry for both bound and unbound states. Backbone relaxation rates were fitted to the five model-free spectral density functions using the relxn2.2 program developed in the lab of A. Lee.70 Effective N-H bond distance and the 15N chemical shift anisotropy values were set to 1.02 Å and −170 ppm, respectively. Akaike information criteria (AIC) was used to select the model that best describes the motion.71
1H-15N backbone residual dipolar couplings (RDCs) were measured at 300 K and 600 MHz using a sensitivity-enhanced IPAP-type 1H-15N HSQC experiment.72 Isotropic samples contained 500 mM 15N-labeled Act-EF34:WT-palladin complex in standard NMR buffer, and liquid crystalline aligned samples contained 500 mM 15N-labeled Act-EF34:WT-palladin complex in the same buffer plus 5% C12E5 PEG (polyethylene glycol) and 1-hexanol for r = 0.96 (mole ratio of C12E5:hexanol). NMR data processing and analyses were performed using NMRPipe/NMRDraw and NMRViewJ. The 1DHN value was calculated by subtracting the measured coupling in the isotropic experiment from the anisotropic experiment. Analysis of RDCs in terms of molecular structure and orientation was conducted with the program REDCAT.46 The molecular coordinates from the NMR structure of the Act-EF34 bound to titin Zr739 (PDB ID: 1H8B) were used to back-calculate the RDCs for palladin peptide-bound Act-EF34, with the best set of principal order parameters for Act-EF34 complex with Zr7 as determined by REDCAT to be Sxx (5.44 E-05), Syy (1.27 E-04), and Szz (−1.82 E-04).
The paramagnetic probe, MTSL, was attached to the palladin peptide as described above and was titrated into a sample of uniformly 15N-enriched Act-EF34. The effects of line broadening were compared as the difference in normalized height of the backbone amide HSQC peaks. 2D 1H-15N-HSQC experiments were acquired at 27 °C at 700 MHz on 0.2 mM 15N-Act2-EF34 in complex with MTSL-palladin peptide (molar ratio 1:5). An identical 1H-15N-HSQC spectrum was collected after the spin label was reduced with 3-fold excess ascorbic acid, by addition of 2 µL ascorbic acid from a concentrated stock. Line broadening was measured as the difference in the height of a backbone amide peak in the presence of the paramagnetic versus diamagnetic probe.
Docking Protocol
The program HADDOCK52 was used to dock Act-EF34 to the WT palladin peptide using ambiguous intermolecular restraints defined according to established criteria. The chemical shift perturbations observed for Act-EF34 upon complex formation with the palladin peptide were used to define ambiguous interaction restraints (AIRs) for residues at the surface. Active residues for α-actinin were defined as those having chemical shift perturbations larger than the average + 0.5 standard deviation (>0.173 ppm): Gln-9, Val-10, Ser-13, Phe-14, Ile-16, Leu-17, Glu-32, Leu-33, Pro-34, Asp-36, Gln-37, Tyr-40, Cys-41, Leu-67, and Tyr-68). Asparagine substitutions at residues leucine 9 and 12 of the palladin peptide eliminated detectable binding to Act-EF34, therefore these residues were also defined as active AIRs. Passive residues were automatically defined around the active residues by HADDOCK. AIRs were defined between the active residues of Act-EF34 and all “active” and “passive” residues of the palladin peptide. The C-terminal EF-hand domain of α-actinin-2 from the complex with the seventh Z-repeat of titin (PDB ID: 1H8B) was used as the starting structure for docking. Because no structure of the palladin peptide was available, initial peptide coordinates were generated in MacPyMOL73 using standard helical conformation. We assume that the peptide residues 238–247 adopt a helical conformation when bound to Act-EF34 given its strong propensity to form an α-helix (as predicted by PSIPRED protein structure prediction server51) and the observed shift in secondary structure upon complex formation as measured by CD. Docking was performed at the HADDOCK server website (http://haddock.chem.uu.nl/Haddock). The default HADDOCK parameters were used with 1000 initial complex structures generated for rigid body docking, and the best 200 solutions in terms of intermolecular energies were refined in water after semi-flexible simulated annealing stage.52 The clustering cut-off was set at 7.5 Å with a minimum cluster size of 4.
Supplementary Material
Acknowledgements
We thank Annalisa Pastore for providing us with the previously unpublished chemical shift assignments for the ligand-free form of Act-EF34; members of Andrew Lee’s lab for help analyzing the relaxation data; Min Qi Lu for help with initial protein purification; Greg Young, the NMR facility director at UNC-CH, for assistance in NMR data collection; and Ashutosh Tripathy, Director of the Macromolecular Interaction Facility at UNC-CH, for assistance with CD and ITC. This work was supported by grants from NIH F32 CA139926-01 and the Lineberger Comprehensive Cancer Center postdoctoral training grant (to M.R.B.), 5R01GM080568 to (S.L.C.) and RO1GM081505 (to C.A.O.).
Abbreviations
- Act-EF34
C-terminal EF hand domain of α-actinin
- FX
Family X
- ITC
isothermal titration calorimetry
- Zr7
7th Z-repeat of titin
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum correlation
- NOE
nuclear Overhauser enhancement
- CSP
chemical shift perturbation
- RDC
residual dipolar coupling
- PRE
paramagnetic relaxation enhancement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes
Chemical shift assignments for both the palladin-bound and unbound form of Act-EF34 have been deposited (BMRB ID: 17626 and 17627).
Contributor Information
Moriah R. Beck, Email: mobeck@email.unc.edu.
Carol A. Otey, Email: carol_otey@med.unc.edu.
Sharon L. Campbell, Email: campbesl@med.unc.edu.
References
- 1.Revenu C, Athman R, Rovine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5:635–646. doi: 10.1038/nrm1437. [DOI] [PubMed] [Google Scholar]
- 2.Otey CA, Carpen O. alpha-Actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 3.Parast MM, Otey CA. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mykkanen OM, Gronholm M, Ronty M, Lalowski M, Salmikangas P, Suila H, Carpen O. Characterization of human palladin, a microfilament-associated protein. Mol Biol Cell. 2001;12:3060–3073. doi: 10.1091/mbc.12.10.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boukhelifa M, Parast MM, Bear JE, Gertler FB, Otey CA. Palladin is a novel binding partner for Ena/VASP family members. Cell Motil Cytoskeleton. 2004;58:17–29. doi: 10.1002/cm.10173. [DOI] [PubMed] [Google Scholar]
- 6.Boukhelifa M, Moza M, Johansson T, Rachlin A, Parast M, Huttelmaier S, Roy P, Jockusch BM, Carpen O, Karlsson R, Otey CA. The proline-rich protein palladin is a binding partner for profilin. Febs J. 2006;273:26–33. doi: 10.1111/j.1742-4658.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 7.Maeda M, Asano E, Ito D, Hasegawa Y, Hamaguchi M, Senga T. Characterization of interaction between CLP36 and palladin. FEBS J. 2009;276:2775–2785. doi: 10.1111/j.1742-4658.2009.07001.x. [DOI] [PubMed] [Google Scholar]
- 8.Jin L, Kern MJ, Otey CA, Wamhoff BR, Somlyo AV. Angiotensin II, focal adhesion kinase, and PRX1 enhance smooth muscle expression of lipoma preferred partner and its newly identified binding partner palladin to promote cell migration. Circ. Res. 2007;100:817–825. doi: 10.1161/01.RES.0000261351.54147.de. [DOI] [PubMed] [Google Scholar]
- 9.Goicoechea S, Arneman D, Disanza A, Garcia-Mata R, Scita G, Otey CA. Palladin binds to Eps8 and enhances the formation of dorsal ruffles and podosomes in vascular smooth muscle cells. J Cell Sci. 2006;119:3316–3324. doi: 10.1242/jcs.03076. [DOI] [PubMed] [Google Scholar]
- 10.Ronty M, Taivainen A, Moza M, Otey CA, Carpen O. Molecular analysis of the interaction between palladin and alpha-actinin. FEBS Lett. 2004;566:30–34. doi: 10.1016/j.febslet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Rachlin AS, Otey CA. Identification of palladin isoforms and characterization of an isoform-specific interaction between Lasp-1 and palladin. J Cell Sci. 2006;119:995–1004. doi: 10.1242/jcs.02825. [DOI] [PubMed] [Google Scholar]
- 12.Boukhelifa M, Parast MM, Valtschanoff JG, LaMantia AS, Meeker RB, Otey CA. A role for the cytoskeleton-associated protein palladin in neurite outgrowth. Mol Biol Cell. 2001;12:2721–2729. doi: 10.1091/mbc.12.9.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo H, Liu X, Wang F, Huang Q, Shen S, Wang L, Xu G, Sun X, Kong H, Gu M, Chen S, Chen Z, Wang Z. Disruption of palladin results in neural tube closure defects in mice. Mol Cell Neurosci. 2005;29:507–515. doi: 10.1016/j.mcn.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Ronty MJ, Leivonen SK, Hinz B, Rachlin A, Otey CA, Kahari VM, Carpen OM. Isoform-Specific Regulation of the Actin-Organizing Protein Palladin during TGF-beta1-Induced Myofibroblast Differentiation. J Invest Dermatol. 2006;126:2387–2396. doi: 10.1038/sj.jid.5700427. [DOI] [PubMed] [Google Scholar]
- 15.Goicoechea SM, Bednarski B, Garcia-Mata R, Prentice-Dunn H, Kim HJ, Otey CA. Palladin contributes to invasive motility in human breast cancer cells. Oncogene. 2009;28:587–598. doi: 10.1038/onc.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, Brentnall TA. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salaria SN, Illei P, Sharma R, Walter KM, Klein AP, Eshleman JR, Maitra A, Schulick R, Winter J, Ouellette MM, Goggins M, Hruban R. Palladin is overexpressed in the non-neoplastic stroma of infiltrating ductal adenocarcinomas of the pancreas, but is only rarely overexpressed in neoplastic cells. Cancer Biol Ther. 2007;6:324–328. doi: 10.4161/cbt.6.3.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suehara Y, Kondo T, Fujii K, Hasegawa T, Kawai A, Seki K, Beppu Y, Nishimura T, Kurosawa H, Hirohashi S. Proteomic signatures corresponding to histological classification and grading of soft-tissue sarcomas. Proteomics. 2006;6:4402–4409. doi: 10.1002/pmic.200600196. [DOI] [PubMed] [Google Scholar]
- 19.Otey CA, Rachlin A, Moza M, Arneman D, Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int Rev Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- 20.Dixon RDS, Arneman DK, Rachlin AS, Sundaresan N, Joseph Costello M, Campbell SL, Otey CA. Palladin is an actin crosslinking protein that uses immunoglobulin-like domains to bind filamentous actin. J Biol Chem. 2008 doi: 10.1074/jbc.M707694200. [DOI] [PubMed] [Google Scholar]
- 21.Jin L, Gan Q, Zieba BJ, Goicoechea SM, Owens GK, Otey CA, Somlyo AV. The actin associated protein palladin is important for the early smooth muscle cell differentiation. PLoS One. 2010;5:e12823. doi: 10.1371/journal.pone.0012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HV, Moser M. Comparative expression analysis of the murine palladin isoforms. Dev Dyn. 2008;237:3342–3351. doi: 10.1002/dvdy.21755. [DOI] [PubMed] [Google Scholar]
- 23.Otey CA, Dixon RDS, Stack C, Goicoechea SM. Cytoplasmic Ig-Domain Proteins: Cytoskeletal Regulators with a Role in Human Disease. Cell Motil Cytoskeleton. 2009;66:618–634. doi: 10.1002/cm.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bang ML, Mudry RE, McElhinny AS, Trombitas K, Geach AJ, Yamasaki R, Sorimachi H, Granzier H, Gregorio CC, Labeit S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol. 2001;153:413–427. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmikangas P, Mykkanen OM, Gronholm M, Heiska L, Kere J, Carpen O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum Mol Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- 26.Ronty M, Taivainen A, Moza M, Kruh GD, Ehler E, Carpen O. Involvement of palladin and alpha-actinin in targeting of the Abl/Arg kinase adaptor ArgBP2 to the actin cytoskeleton. Exp Cell Res. 2005;310:88–98. doi: 10.1016/j.yexcr.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Jin L. The actin associated protein palladin in smooth muscle and in the development of diseases of the cardiovasculature and in cancer. J Muscle Res Cell Motil. 2011;32:7–17. doi: 10.1007/s10974-011-9246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu B, Jones J, Hollingworth MA, Hruban RH, Kern SE. Invasion-specific genes in malignancy: serial analysis of primary and passaged cancers. Cancer Res. 2001;61:1833–1838. [PubMed] [Google Scholar]
- 29.Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 30.Fu L, Qin YR, Xie D, Chow HY, Ngai SM, Kwong DL, Li Y, Guan XY. Identification of alpha-actinin 4 and 67 kDa laminin receptor as stage-specific markers in esophageal cancer via proteomic approaches. Cancer. 2007;110:2672–2681. doi: 10.1002/cncr.23110. [DOI] [PubMed] [Google Scholar]
- 31.Guvakova MA, Adams JC, Boettiger D. Functional role of alpha-actinin, PI 3-kinase and MEK1/2 in insulin-like growth factor I receptor kinase regulated motility of human breast carcinoma cells. J Cell Sci. 2002;115:4149–4165. doi: 10.1242/jcs.00104. [DOI] [PubMed] [Google Scholar]
- 32.Hatakeyama H, Kondo T, Fujii K, Nakanishi Y, Kato H, Fukuda S, Hirohashi S. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics. 2006;6:6300–6316. doi: 10.1002/pmic.200600488. [DOI] [PubMed] [Google Scholar]
- 33.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, Ino Y, Ono M, Hirohashi S. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology. 2005;128:51–62. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Honda K, Yamada T, Seike M, Hayashida Y, Idogawa M, Kondo T, Ino Y, Hirohashi S. Alternative splice variant of actinin-4 in small cell lung cancer. Oncogene. 2004;23:5257–5262. doi: 10.1038/sj.onc.1207652. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto S, Tsuda H, Honda K, Kita T, Takano M, Tamai S, Inazawa J, Yamada T, Matsubara O. Actinin-4 expression in ovarian cancer: a novel prognostic indicator independent of clinical stage and histological type. Mod. Pathol. 2007;20:1278–1285. doi: 10.1038/modpathol.3800966. [DOI] [PubMed] [Google Scholar]
- 37.Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K. The crystal structure of the actin binding domain from alpha-actinin in its closed conformation: structural insight into phospholipid regulation of alpha-actinin. J Mol Biol. 2005;348:151–165. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Young P, Gautel M. The interaction of titin and alpha-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. Embo J. 2000;19:6331–6340. doi: 10.1093/emboj/19.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atkinson RA, Joseph C, Kelly G, Muskett FW, Frenkiel TA, Nietlispach D, Pastore A. Ca2+-independent binding of an EF-hand domain to a novel motif in the alpha-actinin-titin complex. Nat Struct Biol. 2001;8:853–857. doi: 10.1038/nsb1001-853. [DOI] [PubMed] [Google Scholar]
- 40.Joseph C, Stier G, O'Brien R, Politou AS, Atkinson RA, Bianco A, Ladbury JE, Martin SR, Pastore A. A structural characterization of the interactions between titin Z-repeats and the alpha-actinin C-terminal domain. Biochemistry. 2001;40:4957–4965. doi: 10.1021/bi002739r. [DOI] [PubMed] [Google Scholar]
- 41.Atkinson RA, Joseph C, Dal Piaz F, Birolo L, Stier G, Pucci P, Pastore A. Binding of alpha-actinin to titin: implications for Z-disk assembly. Biochemistry. 2000;39:5255–5264. doi: 10.1021/bi991891u. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall JB, Walker O, Fushman D. Characterization of the overall rotational diffusion of a protein from 15N relaxation measurements and hydrodynamic calculations. In: Downing AK, editor. Methods in Molecular Biology. Totowa, NJ: Humana Press Inc.; 2004. p. 278. [DOI] [PubMed] [Google Scholar]
- 44.Lipari G, Szabo A. Model-free approach to the interpretation of nuclar mangetic relaxation in macromolecules, 1: Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 45.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 46.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Mag Res. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornilescu G, Bax A. Measurement of proton, nitrogen, and carbonyl chemical shielding anisotropy in a dilute liquid crystalline phase. J Am Chem Soc. 2000;122:10143–10154. [Google Scholar]
- 49.Houdusse A, Cohen C. Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation. Structure. 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- 50.Xie X, Harrison DH, Schlichting I, Sweet RM, Kalabokis VN, Szent-Gyorgyi AG, Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature Struct. Biol. 1994;368:306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]
- 51.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 52.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 53.Kosen PA. Spin labeling of proteins. Methods Enzymol. 1989;177:86–121. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- 54.Ikura M, Clore GM, Gronenborn AM, Zhu G, Kelee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multi-dimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 55.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 56.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 57.Scaloni A, Miraglia N, Orru S, Amodeo P, Motta A, Marino G, Pucci P. Topology of the calmodulin-melittin complex. J Mol Biol. 1998;277:945–958. doi: 10.1006/jmbi.1998.1629. [DOI] [PubMed] [Google Scholar]
- 58.Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibuanuma T. A novel taget recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nature Struct. Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 59.Amodeo P, Morelli MAC, Strazzullo G, Fucile P, Gautel M, Motta A. Kinase recognition by calmodulin: modeling the interaction with the autoinhibitory region of human cardiac titin kinase. J Mol Biol. 2001;306:81–95. doi: 10.1006/jmbi.2000.4228. [DOI] [PubMed] [Google Scholar]
- 60.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 61.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interaction. Chem. Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 62.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 63.Wittekind M, Meueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J Mag Res Ser B. 1993;101:201–205. [Google Scholar]
- 64.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance 3-dimensional NMR experimentw with improved sensitivity. J Mag Res Ser B. 1994;103:203–216. [Google Scholar]
- 65.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Mag Res. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 67.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 68.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studies by N15 NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 69.Dellwo MJ, Wand AJ. Model-independent and model-dependent analysis of the global and internal dynamics of cyclosporin A. J Am Chem Soc. 1989;111:4571–4578. [Google Scholar]
- 70.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.d'Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J Biomol NMR. 2003;25:25–39. doi: 10.1023/a:1021902006114. [DOI] [PubMed] [Google Scholar]
- 72.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Mag Res. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 73.MacPyMOL: PyMOL Enhanced for Mac OS X 1.3 edit. DeLano Scientific LLC; 2006. [Google Scholar]
- 74.Fernandez-Recio J, Totrov A, Abagyan R. Identification of of protein-protein inteaction sites from docking energy landscapes. J. Mol. Biol. 2004;335:843–865. doi: 10.1016/j.jmb.2003.10.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.