Summary
The emergence of in vivo cancer biomarkers is promising tool for early detection, risk stratification, and therapeutic intervention in the esophagus, where adenocarcinoma is increasing at a rate that is faster than any other in industrialized nations. Exciting advances in target identification, probe development, and optical instrumentation are creating tremendous new opportunities for advancing techniques of molecular imaging. Progress in these areas is being made with small animal models of esophageal cancer using surgical approaches to induce reflux of acid and bile, and these findings are beginning to be evaluated in the clinic. Further identification of relevant targets, characterization of specific probes, and development of endoscopic imaging technologies are needed to further this direction in the field of molecular medicine. In the future, new methods that use in vivo cancer biomarkers for the early detection of neoplastic changes in the setting of Barrett's esophagus will become available.
Keywords: esophagus, adenocarcinoma, biomarkers, peptides, rat, endoscopy
Introduction
The incidence of esophageal adenocarcinoma is currently rising faster than that of any other cancer in the western world [1–3]. Barrett's esophagus is a pathological condition of the distal esophagus that is recognized endoscopically as salmon-colored mucosa, and is confirmed histologically as intestinal metaplasia [4–6]. Barrett's mucosa is associated with acid reflux, and has an increased relative risk ~30 to 125 times higher for progression into cancer [7–9]. Moreover, clinical symptoms that motivate patients to seek medical care usually occur after this disease has advanced to a late stage [10]. Screening with white light endoscopy and biopsy is currently the primary strategy for early detection of cancer in Barrett's mucosa [11,12]. However, this approach is limited by the low yield for detection of pre-malignant (dysplastic) mucosa because biopsies are collected randomly and dysplastic changes are often occur in a spatially heterogeneous fashion [13,14]. Morever, this approach consumes time, increases cost, and decreases health-related quality of life. For example, decision analysis models have suggested that endoscopic surveillance every 2 years costs ~$30,000 per life-year saved [15,16].
In addition, the histological evaluation of excised esophageal mucosa for the presence of dysplasia is performed in a qualitative, subjective manner, and even among expert pathologists, substantial intra- and inter-observer variability in grading dysplasia occurs, limiting patient and physician confidence in the interpretation [17–19]. An incorrect evaluation of biopsy specimens may result in either an unnecessary esophagectomy or in progression to frank carcinoma. This lack of clarity in the prognostic value of conventional histopathology on the natural history of mucosa at risk for progression into adenocarcinoma demands the development of new criteria for pathological evaluation [20,21]. In addition, new therapies aimed at inhibiting specific molecular targets have created widespread enthusiasm for a new class of anti-neoplastic agents that do not have systemic toxicities, and a greater understanding of the relationship between gene amplification and protein expression and the efficacy of targeted therapies is needed [22,23]. These factors combined with poor survival rates associated with late stage adenocarcinoma provide significant motivation for further development of in vivo cancer biomarkers to improve screening of Barrett's mucosa, risk stratification of disease, and therapeutic options for cancer.
Metaplasia-Dysplasia-Adenocarcinoma Sequence
Metaplasia is a conversion of normal squamous epithelium of the esophagus into a columnar morphology, is seen endoscopically as an extension of the gastroesophageal junction, and is recognized on histology by the presence of Goblet cells [4–6]. Dysplasia is a pre-cancerous condition that results from transformation of normal mucosa and has a latency period of between 7 to 14 years before progressing to adenocarcinoma, which develops in <1% of patients with Barrett's per year. This window of opportunity provides sufficient time for most patients who are at increased risk for developing cancer to be screened by endoscopy for detection and localization of dysplasia. The early detection and subsequent resection of dysplastic mucosa can prevent progression to cancer. Wide area endoscopy can be used to visualize large surface areas in esophagus to localize lesions that need further evaluation by either physical or optical biopsy. Dysplastic glands can also be present below the mucosa surface, and the invasion of cells below the muscularis into the sub-mucosa represents a more severe stage of disease with a worse prognosis.
Molecular Biomarkers
Molecular biomarkers are measurable parameters that provide useful information about the tissue phenotype and have different levels of expression in neoplastic compared to normal mucosa. There has been great progress in unraveling the sequence of genetic changes that lead to clonal selection and growth advantages for cells that transform to cancer in the esophagus [24,25]. Moreover, the timing of molecular changes that occur early, such as alterations in p53 and p16, and late, such as loss of heterozygosity and cell cycle checkpoints, has been clarified [26,27]. These changes lead to genomic instability and cumulative genetic errors [26,27], and provide a number of molecular targets that can be used for early detection and therapy of cancer and for predicting future behavior. Furthermore, these indicators can be used to risk stratify genetic events, such as homozygous deletion, intra-gene mutation, chromosome rearrangement, loss of heterozygosity, and aberrant CpG island methylation, that facilitate progression to cancer. In addition, gene expression profiles can evaluate the occurrence of DNA amplification in transformed tissue on a global scale [28,29]. These loci of mutations are present in increased numbers and produce abnormal mRNA and proteins, allowing for dysregulation in cell growth. There are a number of important molecular biomarkers in esophagus that are promising targets for in vivo cancer detection, risk stratification, and therapeutic intervention, including cyclooxygenase-2 (COX-2), HER2/neu (erbB2), mesenchymal epithelial transition factor (MET), and matrix metalloproteinases (MMPs) which will be discussed here.
Cyclooxygenase-2 (COX-2)
COX-2 is a rate-limiting intra-cellular enzyme that converts arachidonic acid into prostaglandins, and regulates a broad range of cellular processes, including proliferation, angiogenesis, and apoptosis [30,31]. This biomarker is induced by injury related to acid and bile reflux in the distal esophagus, important mechanisms in Barrett's tumorigenesis [32,33]. Subsequently, COX-2 induces angiogenesis [34], which promotes tumor growth, immune surveillance, and cell adhesion through mechanisms that include stimulation of VEGF, enhancement of endothelial cell growth, induction of MMP expression, and activation of EGFR [35,36]. Furthermore, COX-2 mRNA levels have been found to be significantly higher in neoplastic esophageal mucosa than in normal. Expression of COX-2 mRNA has been found to be the highest in adenocarcinoma and to be stepwise increased with mucosal progression in the metaplasia-dysplasia-carcinoma sequence [37,38]. In addition, patients with higher levels of COX-2 are more likely to develop metastatic disease and to have cancer recurrence. Therefore, the regular use of aspirin and selective COX-2 inhibitors, such as celecoxib, has been studied as means of chemoprevetion and have been found to decrease the risk of esophageal cancer [39,40]. Furthermore, elevated COX-2 levels in squamous mucosa of patients with gastroesophageal reflux disease have been found to decrease to normal levels after anti-reflux surgery. These findings support the strategy for use of COX-2 inhibition as a means of prevention and treatment of esophageal adenocarcinoma.
HER2/neu (erbB2)
HER2/neu (erbB2) is a cell surface growth factor receptor that is normally associated with signaling involving the mitogen-activated protein kinase (MAPK) pathway and is believed to activate PI3K/Akt signaling, as well [41,42]. This proto-oncogene plays an important role in promoting neoplastic progession in the esophagus by gene amplification and is associated with inhibition of apoptosis and enhanced cell proliferation. Also, HER-2/neu signaling increases MMP activity, enhances tumorigenic and metastatic potential, and is a potent inducer of VEGF and tumor vascularity [43]. Gene amplification of HER2/neu is a common mechanism for the development of esophageal adenocarcinoma, and the associated gene is found to be amplified in 15% to 25% of esophageal cancers [44,45]. HER2/neu is also amplified in ~25 to 30% of human breast cancers, and results in the overexpression of the associated receptor that is a well-established target for Trastuzumab (Herceptin), a humanized monoclonal antibody [46,47]. In addition, small molecules, such as Lapatinib, have been developed as a therapeutic approach to inhibit the tyrosine kinase activity by blocking the ATP-binding site [48]. Clinical studies for treating patients with esophageal adenocarcinoma that are HER2/neu positive with Trastuzumab and Lapatinib are being conducted.
Mesenchymal epithelial transition factor (c-MET)
c-MET is a proto-oncogene that also encodes a tyrosine kinase receptor that is overexpressed in epithelial cells that progress to esophageal cancer [49]. Hepatocyte growth factor (HGF), also known as scatter factor, binds to the c-MET receptor and stimulates many downstream genes associated with tumor invasion, including MMPs, osteopontin, plasminogen activator, and integrins [50]. Gene expression profiles have demonstrated evidence for a significant level of c-MET activation in primary esophageal adenocarcinoma and in cells metastatic to lymph nodes [51]. This observation also occurs to a lesser extent in a subset of metaplastic and dysplastic Barrett's mucosa. Moreover, the increased expression of multiple genes in the MET pathway is associated with factors that promote invasion, including MMPs. Furthermore, oncogenic activation of the MET gene through gene amplification and overexpression represents a common mechanism in many other human cancers. These findings suggest that upregulation of the MET pathway contributes to poor outcome in patients with esophageal adenocarcinoma. Based on these results, several approaches are being developed to inhibit Met activity, including kinase and HGF inhibitors and neutralizing antibodies. Kinase inhibitors include low molecular weight molecules that prevent ATP binding and inhibit receptor activation and recruitment of the downstream effectors [52]. Moreover, HGF inhibitors bind the c-Met receptor without activation and also have anti-angiogenic effects [53]. Finally, monoclonal antibodies can neutralize HGF by impairing HGF-dependent tumor growth [54]. Thus, the c-MET receptor provides a promising target for the early detection of esophageal cancer, and represents an important in vivo cancer biomarker.
Matrix Metalloproteinases (MMPs)
MMPs are a family of proteolytic enzymes that are essential for tissue growth, development, remodeling, and resorption [55]. They can be either integrated into the plasma membrane or secreted into the mucosa, and have the ability to break down the intracellular matrix, leading to angiogenesis and cell migration, as well as the extracellular matrix, leading to tumor invasion and metastases [56]. Furthermore, MMPs can degrade the basement membrane in the initial stages of invasion into blood vessels and lymph nodes [57]. In particular, MMP-1 is an interstitial collagenase whose expression is stimulated by cytokines and regulated by tissue-inhibitor metalloproteinases (TIMPs), and imbalances in the expression of either correlates with poor clinical outcomes in esophageal cancer [58]. MMP-2 is a gelatinase that plays a role in cell migration and in the initial stages of tumor invasion by degrading type IV collagen, a major structural component of the basement membrane [59]. MMP-7 is a matrilysin that has proteolytic activity against a number of components of the extracellular matrix substrates including collagen, proteoglycan, laminin, fibronectin, and casein, and is not easily regulated by TIMPs [60]. Increased expression of MMP-7 has been found to play an important role in the spread of esophageal cancer. Furthermore, peptides have been identified techniques of phage display to bind to the fibronectin type II (FN2) module of the catalytic domain and to inhibit the activity of MMP-2 [61]. These results are promising for the future development of novel methods for in vivo detection and therapy using these molecular targets.
Gene Expression Profiling
Recent advances in biotechnology, including high-throughput quantitative real-time PCR and cDNA microarray technology, have revolutionized our ability to analyze gene expression by providing a platform that simultaneously evaluates large numbers of genes [62,63]. These techniques have been further augmented with advances in bioinformatics to provide a powerful strategy for classifying and interpreting genomic data [64]. Gene expression profiling has been used to provide a comprehensive assessment of cellular function from cancer specimens that can be used for target identification, risk stratification, and therapeutic monitoring. This approach significantly enhances our ability to evaluate in vivo cancer biomarkers by identifying a panel of genes rather than either one or a few that may play a role in cancer progression. The use of multiple biomarkers is particularly relevant for esophageal neoplasia because of the wide variability in gene expression at spatially distinct locations within the mucosa and among different individuals. These techniques can be used to assess esophageal cancer specimens for DNA amplification to identify potential in vivo targets. For example, quantitative PCR was used to evaluate 13 distinct gene amplification events in n = 87 and n = 22 specimens of esophageal adenocarcinoma and Barrett's with high-grade dysplasia, respectively [65]. Gene amplification was then correlated with clinical variables, such as survival, stage, nodal involvement, tumor invasion, smoking history, and gender. One or more amplification events was found in 57% of the adenocarcinomas, and specific analysis of potential molecular targets, including those encoded by the amplified erbB2 (Her2), EGFR, c-Met, and MMP genes, reveals significant mRNA over-expression in adenocarcinoma compared to Barrett's metaplasia.
Small Animal Models
Small animal models are needed to understand the biology related to development of esophageal cancer because of the long natural history of this disease (10 years or more) and severe clinical consequences. Moreover, evidence from gene expression profiles of squamous epithelium, intestinal metaplasia, dysplasia and adenocarcinoma in animal models has shown significant homology to human disease [66]. With use of these and other techniques, small animal models can be studied to better understand the molecular mechanisms that affect the metaplasia-dysplasia-carcinoma sequence and to develop new treatment strategies. Moreover, Barrett's esophagus is an endoscopic diagnosis, thus imaging in small animal models can be used to establish the presence of intestinal metaplasia and is particularly useful for determining onset and to monitor progression of neoplastic transformation after a latency period. In addition, endoscopic methods can be used to guide the collection of tissue specimens and to detect mucosal changes without requiring animal sacrifice, thus minimizing the number of animals needed to perform a statistically valid study by accurately timing the onset of disease and by using each animal as its own control. Finally, endoscopic imaging in small animal models of esophageal cancer can be used to perform longitudinal studies to evaluate new options for chemoprevention and chemotherapy.
Intestinal metaplasia in the esophagus of small animal models is usually produced using surgical approaches [67,68]. In the esophagojejunostomy (EJ) model, an end-to-side anastamosis is made between the distal esophagus and the jejunum distal to the ligament of Trietz. This technique was used to show that COX-2 inhibitors (MF-Tricyclic and Sulindac) can reduce the relative risk of developing esophageal adenocarcinoma induced by acid reflux by 55% in n = 105 rats [68]. Furthermore, in the esophagoduodenostomy (ED) model, an end-to-side anastamosis is performed between the distal esophagus and the duodenum beyond to the pylorus [69]. The animals sacrificed at 30 weeks after the operation revealed that 8/11 (73%) rats supplemented with iron developed adenocarcinoma. Moreover, the total gastrectomy with esophagoduodenostomy (TGED) model involves an end-to-side anastamosis between the distal esophagus and the duodenum distal to the pylorus followed by complete resection of the stomach [70]. In this model, 34/39 (87%) developed adenocarcinoma at 28 weeks after the operation. In all of these studies, sacrifice of the animals was performed to identify the presence of neoplasia thus preventing further study to occur.
Imaging can be performed using a small animal endoscope to detect the appearance of intestinal metaplasia, dysplasia, and adenocarcinoma in a rat model for the purpose of monitoring neoplastic progression. In the esophago-gastro-jejunostomy (EGJ) model, a side-to-side anastamosis of esophago-gastric junction is made to the jejunum distal to the ligament of Trietz [71,72]. This approach retains normal stomach function and nutritional status while directing acid and bile to repeatedly flow back into the distal esophagus through the anastamosis. This esophageal reflux model has been performed in Sprague-Dawley rats at an age of 6 months. Rat chow was withheld 24 hours prior to the surgery, and anesthesia with inhaled isoflurane (3%, 0.5 ml/l volume) was used. A midline laparotomy was performed in the rat, and the gastroesophageal junction was identified and mobilized while carefully preserving the vagus nerve. A longitudinal incision ~1 cm in length was made at the esophago-gastric junction and along the anti-mesenteric border of the proximal jejunum ~3 cm distal to the ligament of Treitz. While preserving the orientation and patency of the lumen, a side-to-side esophago-gastro-jejunostomy was performed using interrupted 7-0 polypropylene sutures. In this configuration, acid and bile was directed to reflux into the distal esophagus through the weakened lower esophageal sphincter and the anastomosis, respectively.
In Vivo Molecular Probes
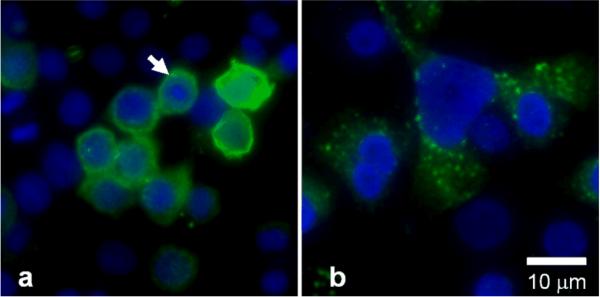
In vivo molecular probes for targeting disease in the gastrointestinal tract have been developed using techniques of phage display to identify peptides that bind selectively to cell surface antigens associated with transformed mucosa [73,74]. For the esophagus, cultured human cells, including SEG-1 (adenocarcinoma), OE-21 (squamous), and Q-hTERT (columnar) are used to provide antigenic surfaces for peptide selection. The phage library (Ph.D.-7, New England Biolabs, Beverly, MA) contains ~2.8 × 109 unique sequences with ~70 copies each. First, phage that bind non-specifically to targets expressed by non-dysplastic (normal and metaplastic) esophageal mucosa are removed from the library by biopanning against the squamous and columnar cells. The remaining phage are recovered from the supernatant and then biopanned against the esophageal adenocarcinoma cells, and those that bind are then eluted off the cells using acid. Screening of the candidates is performed by fluorescence intensity from the addition of polyclonal anti-M13 phage (primary) antibody and mouse anti-rabbit (secondary) antibody labeled with FITC to the phage that bind to cells in culture. Promising phage are selected based on the highest fluorescence target-to-background ratio using adenocarcinoma and columnar cells. The clone expressing the target sequence `ASYNYDA' with a target-to-background ratio of >5 was selected. Preferential peptide binding is shown in Fig. 1, where images of FITC-labeled target phage (green) show a) intense binding to plasma membrane (arrow) of adenocarcinoma cells, and b) poor binding to columnar cells. A DAPI counterstain (blue) is used to identify the cell nuclei, scale bar 10 μm.
Fig 1. Selective binding of target peptide to human adenocarcinoma cells.
Images of FITC-labeled target phage (green) show a) preferential binding to plasma membrane (arrow) of SEG-1 (adenocarcinoma) cells, and b) minimal binding to Q-hTERT (columnar) cells. DAPI counterstain (blue) localizes nuclei, scale bar 10 μm.
In Vivo Molecular Imaging
Small Animal Models
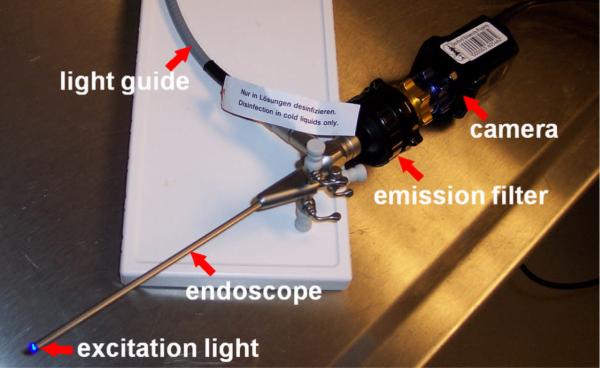
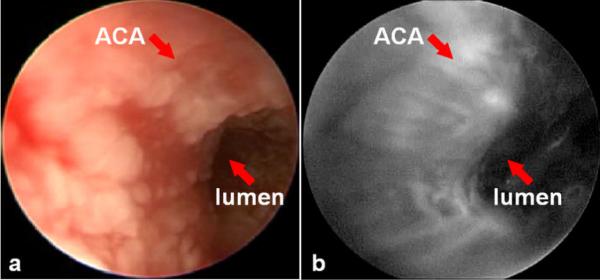
In vivo molecular imaging has been performed in small animal models of esophageal neoplasia to localize binding by targeted peptides. A rigid endoscope for imaging the rat esophagus consists of a 9.5 Fr (3 mm) diameter rigid Hopkins II 0 deg telescope with a 11.5 cm working length (Karl Storz Veterinary Endoscopy, Goleta, CA), as shown in Fig. 2. A 3 Fr (1 mm) diameter instrument channel is available to perform tissue biopsy. Fluorescence excitation light is produced with a 450 to 475 nm passband filter that can be manually placed into the optical path of a xenon (175 W) light source, and is delivered to the endoscope via a light guide. Fluorescence images are collected with 510 nm barrier filter to block the excitation light, and detected with a color CCD camera. The molecular imaging studies were performed in the surgically-altered (EGJ) rats after 36 weeks. The rats were fasted overnight, anesthetized using inhaled isoflurane, and placed in the supine position. The extremities and incisors were restrained to prevent injury to the animal. The endoscope was inserted into the distal esophagus using ~1 to 2 ml of air injected through an instrument port to dilate the esophageal lumen. The FITC-labeled target peptide was topically administered through the instrument channel onto the distal esophagus in a 1 mL solution diluted in PBS at a concentration of 100 μM. After incubation for 5 minutes, excess peptide was rinsed off with tap water, and binding was evaluated by fluorescence. A white light endoscopic image of the distal esophagus reveals the presence of an esophageal adenocarcinoma present within a patch of Barrett's esophagus, as shown in Fig. 3a. Increased fluorescence intensity was observed at the site of the lesion with an average target-to-background ratio of 5±2, as shown in Fig. 3b. A complete examination usually lasts < 2 minutes.
Fig 2. Small animal fluorescence endoscope.
Macroscopic white light and fluorescence endoscope with high spatial resolution imaging over large field of view for small animals is provided with a rigid endoscope. Light guide delivers excitation from xenon lamp, and fluorescence is detected through emission filter by a sensitive CCD camera.
Fig 3. In vivo macroscopic images of rat adenocarcinoma.
a) White light endoscopic image of the distal esophagus of a rat with an esophago-jejunal (EGJ) anastamosis reveals the presence of an esophageal adenocarcinoma (ACA) present within a patch of Barrett's esophagus. b) Fluorescence image collected after target peptide was topically administered. Unbound peptide was rinsed off after 5 minutes of incubation, and reveals increased fluorescence intensity at the site of the lesion.
Clinical Imaging
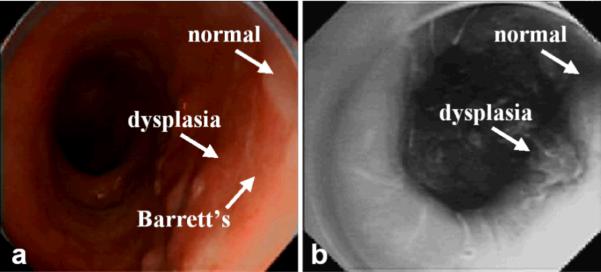
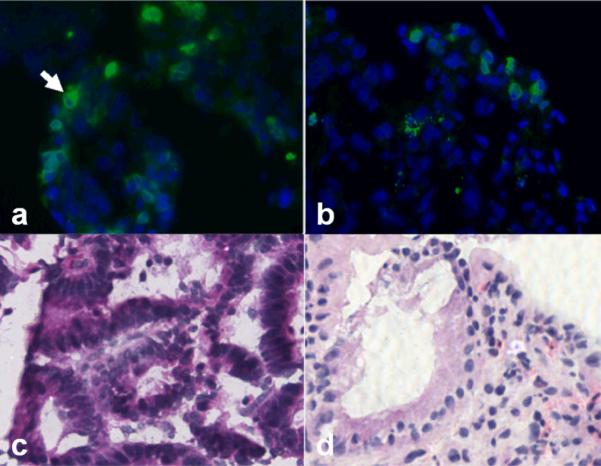
A prototype endoscope was used to collect fluorescence images of peptide binding from human subjects with Barrett's esophagus and high-grade dysplasia using the target peptide labeled with FITC. With IRB approval and informed consent, human subjects with a history of Barrett's esophagus and biopsy proven high-grade dysplasia scheduled for endoscopic mucosal resection were recruited into the study. After esophageal intubation, a 10 second video was collected in the white light mode. Then, ~3 ml of peptide at a concentration of 10 μM was administered topically to the distal esophagus using a mist spray catheter. After 5 minutes for incubation, excess peptide was gently rinsed off with water, and another 10 second video was collected of the peptide targeted fluorescence image. The mucosa of the distal esophagus was then removed by endoscopic mucosal resection. Each video stream was exported into individual frames. In 4a, a standard white light endoscopic image shows a 4 cm length of salmon pink mucosa consistent with endoscopically apparent Barrett's esophagus. No distinct architectural lesions can be appreciated. The targeted image, shown in Fig. 4b, reveals increased fluorescence intensity at a site (arrow) that was biopsied and evaluated as high-grade dysplasia on histopathology. The mean fluorescence intensities from 5 independent regions of HGD and non-dysplastic Barrett's metaplasia was found to have a target-to-background ratio of 1.52 in this set of images. Targeted biopsies were collected from sites suspicious for HGD, non-dysplastic Barrett's esophagus. The specimens were embedded in OCT media, cooled to 4°C, and cut into 10 μm horizontal sections on a cryostat. The specimens were then evaluated for fluorescence on a tabletop confocal microscope, and an adjacent section was formalin fixed and stained with H&E for evaluation by two pathologists. Fig. 5 shows a) significant binding of the FITC-labeled peptide to the surface of cells (arrow) in Barrett's mucosa with high-grade dysplasia, and b) minimal binding to Barrett's mucosa with no dysplasia. Routine histology (H&E) of the corresponding images c,d) from adjacent frozen sections are also shown for purposes of comparison, magnification 400×.
Fig 4. Macroscopic imaging of high-grade dysplasia in Barrett's metaplaisa.
Endoscopic images collected in vivo of distal esophagus in human subject with Barrett's esophagus and high grade dysplasia on a) white-light endoscopy and b) fluorescence imaging. Topically administered FITC-labeled target peptide reveals increased fluorescence intensity at the site of the lesion confirmed to be high-grade dysplasia on histopathology.
Fig 5. Binding of target peptide to esophagus.
a) Significant binding is seen to Barrett's mucosa with high-grade dysplasia, and minimal binding is seen to b) Barrett's tissue with no dysplasia on frozen section. Routine histology (H&E) of corresponding images is shown in c,d), magnification 400×.
Future Directions
In summary, a number of in vivo cancer biomarkers have been identified that are promising for use in early detection, risk stratification, and therapeutic intervention of esophageal adenocarcinoma. Moreover, molecular probes have been developed using peptide libraries that can be applied topically to the distal esophagus, and have been shown to demonstrate selective binding to neoplastic mucosa with high target-to-background ratio. Furthermore, new endoscopic instruments are being developed to localize and validate probe binding with high spatial resolution. The use of in vivo cancer biomarkers for targeted detection and therapy in patients at increased risk of developing esophageal cancer, such as those with Barrett's esophagus, is an exciting new field with great potential but remains at a very early stage of development. More progress is needed in molecular probe development, including improving target-to-background ratios, enhancing delivery mechanisms, and characterizing systemic toxicities. Also, more effort is required to better understand the prognostic value of known cancer biomarkers and new biomarkers are needed to accurately assess the risk of patients with Barrett's esophagus for progression to malignancy. In addition, there is a need for multi-spectral instruments that image over large fields of view and with deep tissue penetration to be developed to confirm binding to multiple in vivo targets. Furthermore, issues of inter-observer variability, clinical efficacy, and cost effectiveness need to be addressed, and then multi-center, randomized controlled trials are needed to validate and standardize the imaging protocols. These promising techniques have great potential to improve the detection of early neoplasia, increase efficacy of surveillance, and ultimately improve patient outcomes for cancer of the esophagus.
References
- 1.American Cancer Society . Cancer Facts & Figures 2008. American Cancer Society; Atlanta: 2008. [Google Scholar]
- 2.Vizcaino AP, Moreno V, Lambert R, Parkin DM. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries, 1973–1995. Int J Cancer. 2002;99:860–8. doi: 10.1002/ijc.10427. [DOI] [PubMed] [Google Scholar]
- 3.Hongo M. Barrett's oesophagus and carcinoma in Japan. Aliment Pharmacol Ther. 2004;20(S8):50–4. doi: 10.1111/j.1365-2036.2004.02230.x. [DOI] [PubMed] [Google Scholar]
- 4.Hornick JL, Odze RD. Neoplastic precursor lesions in Barrett's esophagus. Gastroenterol Clin North Am. 2007;36:775–96. doi: 10.1016/j.gtc.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Mueller J, Werner M, Stolte M. Barrett's esophagus: histopathologic definitions and diagnostic criteria. World J Surg. 2004;28:148–54. doi: 10.1007/s00268-003-7050-4. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett's metaplasia. Lancet. 2000;356:2079–85. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 7.Solaymani-Dodaran M, Logan RF, West J, Card T, Coupland C. Risk of oesophageal cancer in Barrett's oesophagus and gastro-oesophageal reflux. Gut. 2004;53:1070–4. doi: 10.1136/gut.2003.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuchert MJ, Luketich JD. Barrett's esophagus-emerging concepts and controversies. J Surg Oncol. 2007;95:185–9. doi: 10.1002/jso.20635. [DOI] [PubMed] [Google Scholar]
- 9.Solaymani-Dodaran M, Logan RF, West J, Card T. Mortality associated with Barrett's esophagus and gastroesophageal reflux disease diagnoses-a population-based cohort study. Am J Gastroenterol. 2005;100:2616–21. doi: 10.1111/j.1572-0241.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 10.Cohen S, Parkman HP. Heartburn - a serious symptom. N Engl J Med. 1999;340:878–9. doi: 10.1056/NEJM199903183401109. [DOI] [PubMed] [Google Scholar]
- 11.Spechler SJ. Barrett's Esophagus. N Engl J Med. 2002;346:836–42. doi: 10.1056/NEJMcp012118. [DOI] [PubMed] [Google Scholar]
- 12.Wang KK, Sampliner RE. Updated Guidelines 2008 for the Diagnosis, Surveillance and Therapy of Barrett's Esophagus. Am J Gastroenterol. 2008;103:788–97. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 13.Falk GW, Rice TW, Goldblum JR, Richter JE. Jumbo biopsy forceps protocol still misses unsuspected cancer in Barrett's esophagus with high-grade dysplasia. Gastrointest Endosc. 1999;49:170–6. doi: 10.1016/s0016-5107(99)70482-7. [DOI] [PubMed] [Google Scholar]
- 14.Levine DS, Haggitt RC, Blount PL, et al. An endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in Barrett's esophagus. Gastroenterology. 1993;105:40–50. doi: 10.1016/0016-5085(93)90008-z. [DOI] [PubMed] [Google Scholar]
- 15.Inadomi JM, Sampliner R, Lagergren J, Lieberman D, Fendrick AM, Vakil N. Screening and surveillance for Barrett esophagus in high-risk groups: a cost-utility analysis. Ann Intern Med. 2003;138:176–86. doi: 10.7326/0003-4819-138-3-200302040-00009. [DOI] [PubMed] [Google Scholar]
- 16.Quera R, O'Sullivan K, Quigley EM. Surveillance in Barrett's oesophagus: will a strategy focused on a high-risk group reduce mortality from oesophageal adenocarcinoma? Endoscopy. 2006;38:162–9. doi: 10.1055/s-2005-921184. [DOI] [PubMed] [Google Scholar]
- 17.Reid BJ, Haggitt RC, Rubin CE, et al. Observer variation in the diagnosis of dysplasia in Barrett's esophagus. Hum Pathol. 1988;19:166–78. doi: 10.1016/s0046-8177(88)80344-7. [DOI] [PubMed] [Google Scholar]
- 18.Ormsby AH, Petras RE, Henricks WH, et al. Observer variation in the diagnosis of superficial oesophageal adenocarcinoma. Gut. 2002;51:671–6. doi: 10.1136/gut.51.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skacel M, Petras RE, Gramlich TL, et al. The diagnosis of low-grade dysplasia in Barrett's esophagus and its implications for disease progression. Am J Gastroenterol. 2000;95:3383–7. doi: 10.1111/j.1572-0241.2000.03348.x. [DOI] [PubMed] [Google Scholar]
- 20.McKenna BJ, Appelman HD. Dysplasia of the gut: the diagnosis is harder than it seems. J Clin Gastroenterol. 2002;34:111–6. doi: 10.1097/00004836-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Appelman HD. What is dysplasia in the gastrointestinal tract? Arch Pathol Lab Med. 2005;129:170–3. doi: 10.5858/2005-129-170-WIDITG. [DOI] [PubMed] [Google Scholar]
- 22.Brabender J, Marjoram P, Salonga D, et al. A multigene expression panel for the molecular diagnosis of Barrett's esophagus and Barrett's adenocarcinoma of the esophagus. Oncogene. 2004;23:4780–8. doi: 10.1038/sj.onc.1207663. [DOI] [PubMed] [Google Scholar]
- 23.Brabender J, Marjoram P, Lord RV, et al. The molecular signature of normal squamous esophageal epithelium identifies the presence of a field effect and can discriminate between patients with Barrett's esophagus and patients with Barrett's-associated adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14:2113–7. doi: 10.1158/1055-9965.EPI-05-0014. [DOI] [PubMed] [Google Scholar]
- 24.Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 25.Maley CC, Galipeau PC, Li X, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 2004;64:3414–27. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 26.Koppert LB, Wijnhoven BP, van Dekken H, Tilanus HW, Dinjens WN. The molecular biology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:169–90. doi: 10.1002/jso.20359. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2006;55:1810–20. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai LA, Paulson TG, Li X, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007;46:532–42. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 29.Galipeau PC, Prevo LJ, Sanchez CA, et al. Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett's) tissue. J Natl Cancer Inst. 1999;91:2087–95. doi: 10.1093/jnci/91.24.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seibert K, Zhang Y, Leahy K, et al. Pharmacological and biochemical demonstration of the role of cyclooxygenase 2 in inflammation and pain. Proc Natl Acad Sci U S A. 1994;91:12013–7. doi: 10.1073/pnas.91.25.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirvani VN, Ouatu-Lascar R, Kaur BS, Omary MB, Triadafilopoulos G. Cyclooxygenase 2 expression in Barrett's esophagus and adenocarcinoma: Ex vivo induction by bile salts and acid exposure. Gastroenterology. 2000;118:487–96. doi: 10.1016/s0016-5085(00)70254-x. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald RC, Omary MB, Triadafilopoulos G. Altered sodium-hydrogen exchange activity is a mechanism for acid-induced hyperproliferation in Barrett's esophagus. Am J Physiol. 1998;275(1 Pt 1):G47–55. doi: 10.1152/ajpgi.1998.275.1.G47. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald RC, Omary MB, Triadafilopoulos G. Dynamic effects of acid on Barrett's esophagus. An ex vivo proliferation and differentiation model. J Clin Invest. 1996;98:2120–8. doi: 10.1172/JCI119018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Möbius C, Stein HJ, Spiess C, et al. COX2 expression, angiogenesis, proliferation and survival in Barrett's cancer. Eur J Surg Oncol. 2005;31:755–9. doi: 10.1016/j.ejso.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Buskens CJ, Sivula A, van Rees BP, et al. Comparison of cyclooxygenase 2 expression in adenocarcinomas of the gastric cardia and distal oesophagus. Gut. 2003;52:1678–83. doi: 10.1136/gut.52.12.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buskens CJ, Van Rees BP, Sivula A, et al. Prognostic significance of elevated cyclooxygenase 2 expression in patients with adenocarcinoma of the esophagus. Gastroenterology. 2002;122:1800–7. doi: 10.1053/gast.2002.33580. [DOI] [PubMed] [Google Scholar]
- 37.Vallböhmer D, Peters JH, Kuramochi H, et al. Molecular determinants in targeted therapy for esophageal adenocarcinoma. Arch Surg. 2006;141:476–81. doi: 10.1001/archsurg.141.5.476. R. [DOI] [PubMed] [Google Scholar]
- 38.Ling FC, Baldus SE, Khochfar J, et al. Association of COX-2 expression with corresponding active and chronic inflammatory reactions in Barrett's metaplasia and progression to cancer. Histopathology. 2007;50:203–9. doi: 10.1111/j.1365-2559.2007.02576.x. [DOI] [PubMed] [Google Scholar]
- 39.Heath EI, Canto MI, Wu TT, et al. Chemoprevention for Barrett's esophagus trial. Design and outcome measures. Dis Esophagus. 2003;16:177–86. doi: 10.1046/j.1442-2050.2003.00325.x. [DOI] [PubMed] [Google Scholar]
- 40.Souza RF, Spechler SJ. Barrett's esophagus: chemoprevention. Gastrointest Endosc Clin N Am. 2003;13:419–32. doi: 10.1016/s1052-5157(03)00039-4. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga E, Kimura Y, Mashino K, et al. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137–44. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 42.Davidson B, Konstantinovsky S, Kleinberg L, et al. The mitogen-activated protein kinases (MAPK) p38 and JNK are markers of tumor progression in breast carcinoma. Gynecol Oncol. 2006;102:453–61. doi: 10.1016/j.ygyno.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 43.Wang SC, Zhang L, Hortobagyi GN, Hung MC. Targeting HER2: recent developments and future directions for breast cancer patients. Semin Oncol. 2001;28:21–9. doi: 10.1053/sonc.2001.29724. [DOI] [PubMed] [Google Scholar]
- 44.Bange J, Zwick E, Ullrich A. Molecular targets for breast cancer therapy and prevention. Nat Med. 2001;7:548–52. doi: 10.1038/87872. [DOI] [PubMed] [Google Scholar]
- 45.Rauser S, Weis R, Braselmann H, et al. Significance of HER2 low-level copy gain in Barrett's cancer: implications for fluorescence in situ hybridization testing in tissues. Clin Cancer Res. 2007;13:5115–23. doi: 10.1158/1078-0432.CCR-07-0465. [DOI] [PubMed] [Google Scholar]
- 46.Ménard S, Pupa SM, Campiglio M, Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22:6570–8. doi: 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- 47.Hardwick RH, Shepherd NA, Moorghen M, Newcomb PV, Alderson D. c-erbB-2 overexpression in the dysplasia/carcinoma sequence of Barrett's oesophagus. J Clin Pathol. 1995;48:129–32. doi: 10.1136/jcp.48.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Safran H, Iannitti D, Ramanathan R, et al. Herceptin and gemcitabine for metastatic pancreatic cancers that overexpress HER-2/neu. Cancer Invest. 2004;22:706–12. doi: 10.1081/cnv-200032974. [DOI] [PubMed] [Google Scholar]
- 49.Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006;12:3657–60. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 50.Weidner KM, Sachs M, Birchmeier W. The Met receptor tyrosine kinase transduces motility, proliferation, and morphogenic signals of scatter factor/hepatocyte growth factor in epithelial cells. J Cell Biol. 1993:121145–54. doi: 10.1083/jcb.121.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller CT, Lin L, Casper AM, et al. Genomic amplification of MET with boundaries within fragile site FRA7G and upregulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006;25:409–18. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- 52.Berthou S, Aebersold DM, Schmidt LS, et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene. 2004;23:5387–93. doi: 10.1038/sj.onc.1207691. [DOI] [PubMed] [Google Scholar]
- 53.Matsumoto K, Nakamura T. NK4 gene therapy targeting HGF-Met and angiogenesis. Front Biosci. 2008;13:1943–51. doi: 10.2741/2813. [DOI] [PubMed] [Google Scholar]
- 54.Burgess T, Coxon A, Meyer S, et al. Fully human monoclonal antibodies to hepatocyte growth factor with the rapeutic potential against hepatocyte growth factor/c-Met-dependent human tumors. Cancer Res. 2006;66:1721–9. doi: 10.1158/0008-5472.CAN-05-3329. [DOI] [PubMed] [Google Scholar]
- 55.Nagase H, Woessner JF. Matrix metalloproteinases. J Biol Chem. 1999;274:21491–4. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita K, Mori M, Kataoka A, Inoue H, Sugimachi K. The clinical significance of MMP-1 expression in oesophageal carcinoma. Br J Cancer. 2001;84:276–82. doi: 10.1054/bjoc.2000.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray GI, Duncan ME, O'Neil P, McKay JA, Melvin WT, Fothergill JE. Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer. J Pathol. 1998;185:256–61. doi: 10.1002/(SICI)1096-9896(199807)185:3<256::AID-PATH115>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 58.Tanioka Y, Yoshida T, Yagawa T, Saiki Y, Takeo S, Harada T, Okazawa T, Yanai H, Okita K. Matrix metalloproteinase-7 and matrix metalloproteinase-9 are associated with unfavourable prognosis in superficial oesophageal cancer. Br J Cancer. 2003;89:2116–21. doi: 10.1038/sj.bjc.6601372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–8. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 60.Adachi Y, Itoh F, Yamamoto H, Matsuno K, Arimura Y, Kusano M, Endoh T, Hinoda Y, Oohara M, Hosokawa M, Imai K. Matrix metalloproteinase matrilysin (MMP-7) participates in the progression of human gastric and esophageal cancers. Int J Oncol. 1998;13:1031–5. doi: 10.3892/ijo.13.5.1031. [DOI] [PubMed] [Google Scholar]
- 61.Trexler M, Briknarová K, Gehrmann M, Llinás M, Patthy L. Peptide ligands for the fibronectin type II modules of matrix metalloproteinase 2 (MMP-2) J Biol Chem. 2003;278:12241–6. doi: 10.1074/jbc.M210116200. [DOI] [PubMed] [Google Scholar]
- 62.Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 63.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 64.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 65.Miller CT, Moy JR, Lin L, Schipper M, Normolle D, Brenner DE, Iannettoni MD, Orringer MB, Beer DG. Gene amplification in esophageal adenocarcinomas and Barrett's with high-grade dysplasia. Clin Cancer Res. 2003;9:4819–25. [PubMed] [Google Scholar]
- 66.Cheng P, Gong J, Wang T, Chen J, Liu GS, Zhang R. Gene expression in rats with Barrett's esophagus and esophageal adenocarcinoma induced by gastroduodenoesophageal reflux. World J Gastroenterol. 2005;11:5117–22. doi: 10.3748/wjg.v11.i33.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X, LoCicero J, 3rd, Macri E, Loda M, Ellis FH., Jr Barrett's esophagus and associated adenocarcinoma in a mouse surgical model. J Surg Res. 2000;88:120–4. doi: 10.1006/jsre.1999.5774. [DOI] [PubMed] [Google Scholar]
- 68.Buttar NS, Wang KK, Leontovich O, et al. Chemoprevention of esophageal adenocarcinoma by COX-2 inhibitors in an animal model of Barrett's esophagus. Gastroenterology. 2002;122:1101–12. doi: 10.1053/gast.2002.32371. [DOI] [PubMed] [Google Scholar]
- 69.Goldstein SR, Yang GY, Curtis SK, et al. Development of esophageal metaplasia and adenocarcinoma in a rat surgical model without the use of a carcinogen. Carcinogenesis. 1997;18:2265–70. doi: 10.1093/carcin/18.11.2265. [DOI] [PubMed] [Google Scholar]
- 70.Ireland AP, Peters JH, Smyrk TC, et al. Gastric juice protects against the development of esophageal adenocarcinoma in the rat. Ann Surg. 1996;224:358–70. doi: 10.1097/00000658-199609000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumagai H, Mukaisho K, Sugihara H, Bamba M, Miyashita T, Miwa K, Hattori T. Cell kinetic study on histogenesis of Barrett's esophagus using rat reflux model. Scand J Gastroenterol. 2003;38:687–92. doi: 10.1080/00365520310003435. [DOI] [PubMed] [Google Scholar]
- 72.Kumagai H, Mukaisho K, Sugihara H, Miwa K, Yamamoto G, Hattori T. Thioproline inhibits development of esophageal adenocarcinoma induced by gastroduodenal reflux in rats. Carcinogenesis. 2004;25(5):723–7. doi: 10.1093/carcin/bgh067. [DOI] [PubMed] [Google Scholar]
- 73.Kelly K, Alencar H, Funovics M, Mahmood U, Weissleder R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004;64:6247–51. doi: 10.1158/0008-5472.CAN-04-0817. [DOI] [PubMed] [Google Scholar]
- 74.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–8. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]