Abstract
Objective
Test the hypothesis that cardiac microRNAs (miRs) profiling in severe heart failure patients at the time of ventricular assist device (VAD) placement would differentiate those who remained VAD-dependent from those with subsequent left ventricular (LV) recovery.
Background
The relationship of myocardial miR expression to ventricular recovery is unknown.
Methods
We studied 28 patients with nonischemic cardiomyopathy requiring VAD support consisting of Test and Validation cohorts from two institutions; 14 with subsequent LV recovery and VAD removal and 14 clinically matched VAD-dependent patients. Apical core myocardium was studied for expression of 376 miRs by PCR-based array and RT-PCR methods. Samples from seven non-failing hearts were used in confirmatory studies.
Results
By PCR-array, ten miRs were differentially expressed between LV recovery and VAD-dependent patients in the Test cohort. RT-PCR confirmed lower expression in LV recovery patients for 4 miRs (-15b, -1.5 fold; -23a, -2.2 fold; -26a, -1.4 fold; and -195, -1.8 fold; all p<0.04 vs. VAD-dependent). The Validation cohort similarly showed lower miRs expression in LV recovery patients: -23a (-1.8 fold) and -195 (-1.5 fold) both p<0.03). Furthermore, miR-23a and -195 expression in non-failing hearts was similar to LV recovery patients (both p<0.04 vs. VAD dependent). LV recovery patients also had significantly smaller cardiomyocytes by quantitative histology in both cohorts.
Conclusions
Lower cardiac expression of miRs-23a and -195 and smaller cardiomyocyte size at the time of VAD placement were associated with subsequent LV functional recovery. Differential expression of miRs at VAD placement may provide markers to assess recovery potential.
Keywords: cardiomyopathy, heart-assist device, microRNA, hypertrophy
INTRODUCTION
Despite the beneficial effects of mechanical unloading achieved after ventricular assist device (VAD) placement on molecular markers and cardiomyocyte structure and function in the failing heart (1–7), only a small number of VAD-supported patients recover sufficient function to allow permanent removal of the VAD (8–10). Current data suggest that an unappreciated percentage of VAD-supported patients have a potential for recovery but may not be adequately tested for the ability to permanently and successfully remove mechanical support (9, 10). Clinical variables may assist in the prediction of recovery (9–12) but there are few studies that identify biochemical markers associated with functional recovery sufficient to allow device removal (13, 14).
MicroRNAs (miRs) are short (21–23 nucleotides long) non-coding RNAs that may play potent and widespread roles in post-transcriptional regulation of gene expression. miRs affect diverse pathways including apoptosis, cell growth and proliferation, oncogene suppression and activation, and stem cell activation (15). Recent human and animal studies have shown significant alterations in cardiac miR expression patterns with heart failure, while others demonstrated miR involvement in key heart failure pathways (16–24).
In this study, we investigated patterns of cardiac miR expression at the time of VAD placement to identify patterns associated with functional recovery from severe heart failure in VAD-supported patients. We also investigated whether this ‘recovery potential’ expression profile was induced in failing hearts by introduction of VAD support. Finally, we assessed this miR profile in nonfailing control hearts.
METHODS
Patient Selection and Tissue Collection
All studies were performed under protocols approved by the University of Pittsburgh, Texas Heart Institute, and Cleveland Clinic Foundation Institutional Review Boards.
Test Cohort
The Test Cohort consisted of 14 heart failure patients with LV tissue samples (Cores) obtained and banked at the time of VAD placement. The Recovered group (R; n=7) consisted of patients who recovered sufficient cardiac function to allow LVAD removal. Recovered patients were compared with VAD implanted patients who remained VAD-dependent (Dependent, D, n=7), and who were retrospectively matched for clinical variables associated with recovery so as to closely approximate those variables in R group patients. Patients with ischemic heart disease were excluded from all groups reported in this study. Of the 7 explanted and discharged patients, 1 suffered unexplained death ~1.5 months after discharge and 1 died ~ 3 months post-explantation due to Staphylococcus aureus mediastinitis with preserved cardiac function. The remaining 5 have survived free from heart failure more than 6 months.
Validation Cohort
The Validation cohort tissue provided by the Texas Heart Institute arose from 7 Recovered (R), and 7 Dependent (D) patients (as defined for the Test cohort, and matched for clinical variables). All 7 explanted patients survived and were free of heart failure symptoms for more than 6 months.
Patient Selection; Pre- and Post-VAD
Paired LV samples, collected at the time of VAD implantation (Pre-VAD) and cardiac transplantation (Post-VAD), were obtained from an independent third group of 6 patients (University of Pittsburgh) with severe heart failure from nonischemic cardiomyopathy.
Nonfailing Hearts
Non-failing human heart tissue (NF) was obtained from the Cleveland Clinic Foundation. Transmural samples of the lateral LV wall were obtained from unmatched donors whose hearts were not suitable for transplantation despite normal ventricular structure and function as measured by echocardiography. Non-failing samples were clinically matched with the Test and Validation Cohorts for age and gender (n=7, age 36±10 years, 50% female).
PCR and Histological Analysis
A screening miR PCR based array was performed on the Test cohort, followed by confirmatory individual quantitative PCR in the Test and Validation cohorts (25). miRs confirmed altered in both cohorts were tested in non-failing hearts. Measurement of cardiomyocyte size was performed in both cohorts (6, 26). Details of procedures and data analysis are provided in the Supplemental Methods section.
Statistical Analyses
miR expression data are presented as fold up or down regulation in the D group versus R (26). A negative value indicates lower expression in the R group. For confirmatory PCR, data is expressed as fold up or down expression in D versus R, or between Pre- and Post VAD samples, or normalized to the mean of NF samples for NF versus R or D. Results were compared between groups by a non-parametric one-way analysis of variance (Kruskal-Wallis test). Upon detection of overall significance, limited hypothesis driven post-hoc analyses were performed using Mann-Whitney U test. For Pre- versus Post-VAD, a paired t-test was employed. All data are reported as mean ± SD. Significance was accepted at p<0.05.
RESULTS
Patient Characteristics
Within each cohort (Table 1), parameters at the time of VAD implantation were not significantly different between R and D groups. However, there were significant differences between the cohorts: the Validation cohort had more male patients, greater LV diameters, a longer duration of heart failure prior to VAD implantation, longer time on VAD support, and more patients on rotary VAD support. Table 1 also lists clinical parameters while on VAD support, and follow up LV ejection fraction 6 months after VAD explantation in the Recovered groups of both cohorts. Test and Validation cohorts differed in the duration of support and EF at 6 months post-explantation.
Table 1. Characteristics of patients demonstrating clinical similarity between Recovered and Dependent groups prior to VAD implantation, in both Test and Validation cohorts.
Duration of congestive heart failure (CHF) was defined as the time in days from original presentation with heart failure until VAD placement. Days on VAD was defined as time to VAD explant (Recovered group) or Transplantation (Dependent group). All patients on pulsatile VAD support were on Thoratec (Pleasanton CA) extracorporeal devices. PCWP and cardiac index measured immediately prior to VAD implantation, while on inotropes.
| Clinical Characteristics At Baseline
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Test Cohort | Validation Cohort | NonFailing Controls | Test Vs Validation Cohorts | All VAD Vs NonFailing Controls | |||||
| Recovered | Dependent | p Value | Recovered | Dependent | p Value | p Value | p Value | ||
| Number of Patients | 7 | 7 | 7 | 7 | 7 | ||||
| Age (years) | 40±12 | 42±15 | NS | 27±8 | 33±9 | NS | 36±10 | NS | NS |
| Percent Ischemic | 0 | 0 | NS | 0 | 0 | NS | 0 | NS | NS |
| Inotropic Support | 100% | 100% | NS | 85% | 100% | NS | 0 | NS | <0.001 |
| Intraaortic Balloon Pump | 43% | 57% | NS | 43% | 14% | NS | 0 | NS | <0.001 |
| Other Mechanical Support (TandemHeart) | 0% | 0% | NS | 14% | 14% | NS | 0 | NS | <0.001 |
| Percent Male | 50% | 50% | NS | 71% | 71% | NS | 50% | <0.04 | NS |
| EF Prior to VAD (%) | 18±5 | 15±5 | NS | 18±5 | 17±5 | NS | 65±10 | NS | <0.001 |
| LVEDd Prior to VAD (cm) | 6.0±0.8 | 6.2±0.8 | NS | 7.0±1.0 | 7.0±1.0 | NS | Data not available | <0.02 | Data not available |
| PCWP Prior to VAD (mm Hg)* | 26±9 | 27±10 | NS | 27±5 | 27±11 | NS | Data not available | NS | Data not available |
| Cardiac Index Prior to VAD (L/min/m2)* | 1.9±0.89 | 2.0±0.73 | NS | 1.6±0.24 | 1.8±0.41 | NS | Data not available | NS | Data not available |
| Duration of CHF (days) | 62±49 | 75±58 | NS | 680±1117 | 771±802 | NS | NA | <0.001 | NA |
| Type of VAD (% Rotary) | 14% | 14% | NS | 71% | 42% | NS | NA | <0.01 | NA |
| Days on VAD | 53±31 | 61±29 | NS | 433±250 | 369±167 | NS | NA | <0.001 | NA |
| Clinical Characteristics At 6 Month Follow Up Post VAD Explantation
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Test Cohort | Validation Cohort | NonFailing Controls | Test Vs Validation Cohorts | |||||
| Recovered | Dependent | p Value | Recovered | Dependent | p Value | p Value | ||
| Number of Patients | 5 | 5 | 7 | 7 | NA | |||
| Betablocker use while on VAD | 67% | 83% | NS | 100% | 85% | NS | NA | NS |
| ACE Inhibitor use while on VAD | 83% | 83% | NS | 85% | 85% | NS | NA | NS |
| EF at 6 months post explantation (%) | 55±5 | N/A | - | 43±11 | N/A | NA | <0.04 | |
NS: not significant;
NA: Not applicable.
Screening Real Time PCR Array
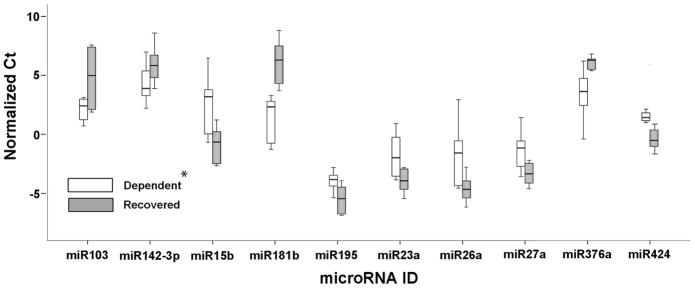
In the Test cohort, 6 of 7 patients in each group were first analyzed by Real Time PCR Array for miR expression. Of 376 miRs studied in the Test cohort, 141 were expressed at a level of ≤35 Ct in at least 1 VAD core sample; 108 were detected in every VAD core sample at a level of ≤35 Ct. Ten miRs were differentially expressed between the R and D groups (p<0.05), (Figure 1 and Table 2). miRs previously reported to play roles in heart failure relevant pathways were not differentially expressed (miRs-1, -21, -133a, -133b and -208) (15–24).
Figure 1. miRs differentially expressed between Recovered and Dependent patients as detected by RT-PCR Array in Test cohort.
Threshold detection values after normalization to small RNAs U6 and RNU44 expression. Box and whisker plot, where the range of values are represented by the whiskers, the upper and lower quartile by the box, and the median value by the bar. N=6 per group; *, p<0.05.
Table 2.
List of miRs differentially expressed (p<0.05) on screening microarray, and confirmation of expression by individual Taqman Real-Time PCR with pre-amplification.Fold change refers to Recovered (R)/Dependent (D) ratio. A negative number indicates lower expression in the Recovered group compared with Dependent. Of the 10 miRs found differentially expressed by screening array, miRs-15b, -23a, -26a, and -195 showed decreased expression by confirmatory PCR. Of these 4, miRs-23a and -195 were also differentially expressed in the Validation Cohort. In the Test Cohort N= 6 each R and D for the Screening Array and N=7 each R and D for the Confirmatory PCR.
| miR Expression
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Screening Microarray (Test Cohort) | Confirmatory PCR (Test Cohort) | Confirmatory PCR (Validation Cohort) | ||||||
| miR ID | Fold Change | p -Value | miR ID | Fold Change | p -Value | miR ID | Fold Change | p -Value |
| R/D | R/D | R/D | ||||||
| miR-181b | 11.38 | 0.002 | miR-181b* | N/A | N/A | miR-181b* | No difference | NS |
| miR-424 | −4.38 | 0.027 | miR-424 | No difference | NS | miR-424 | N/A | N/A |
| miR-376a | 5.41 | 0.028 | miR-376a | No difference | NS | miR-376a | N/A | N/A |
| miR-195 | −2.91 | 0.029 | miR-195 | −1.8 | 0.01 | miR-195 | −1.5 | 0.03 |
| miR-27a | −4.18 | 0.032 | miR-27a | No difference | NS | miR-27a | N/A | NS |
| miR-23a | −4.52 | 0.032 | miR-23a | −2.2 | 0.03 | miR-23a | −1.8 | 0.01 |
| miR-103 | 3.69 | 0.044 | miR-103 | No difference | NS | miR-103 | N/A | N/A |
| miR-26a | −14.92 | 0.044 | miR-26a | −1.4 | 0.04 | miR-26a | No difference | NS |
| miR-142-3p | 6.35 | 0.048 | miR-142-3p | No difference | NS | miR-142-3p | N/A | N/A |
| miR-15b | −12.73 | 0.048 | miR-15b | −1.5 | 0.04 | miR-15b | −1.8 | 0.09 |
miR-181b could not be validated by this method in the Test Cohort, but was not significantly altered in the Validation cohort.
NS, not significant;
N/A: not applicable. These measurements were not performed.
Real Time PCR Confirmation; Test Cohort
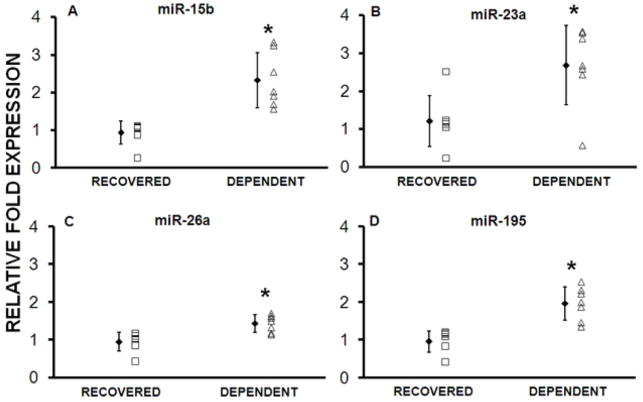
The expression of 9 differentially expressed miRs was examined using Taqman PCR in 6 patients in both groups of the Test cohort. Of the 9 miRs studied, miRs-15b, -23a, -26a and -195 were confirmed as significantly altered between R and D groups (p ≤ 0.05) (Table 2 and Figure 2). The 10th miR (miR-181b) could not be confirmed in the Test cohort due to limitation of RNA availability but was tested in the Validation cohort (see below). In the seventh patient from each group of the Test cohort, Taqman PCR was used to measure the expression of eight miRs (-1, -15b, -21, -23a, -26a, -133a, -133b, -195), and the reference small RNA RNU48.
Figure 2. Recovered patients in the Test Cohort have decreased expression of selected miRs.
Individual miRs found differentially expressed in the screening array were tested using individual PCR. miRs-15b, -23a, -26a and -195 were confirmed to be differentially expressed. *p<0.04 (n=7 per group)
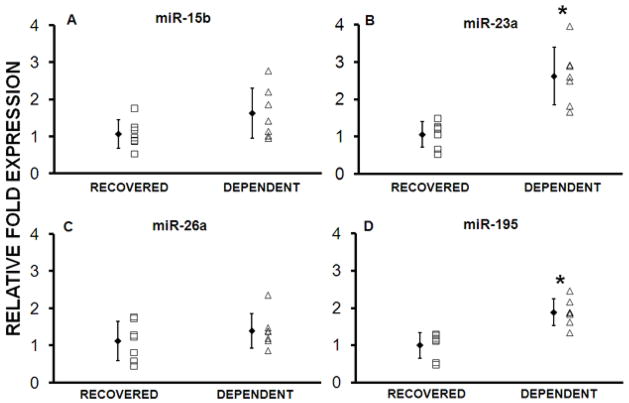
Real Time PCR Confirmation; Validation Cohort
The 4 miRs confirmed to be differentially expressed in the Test cohort, along with miR-181b, were similarly measured in the Validation cohort. miRs-23a and -195 had significant differential expression (Table 2 and Figure 3). Expression of miRs -15b, -26a, and -181b was not significantly different, although the direction of change was the same as in the Test cohort. Combining the two datasets demonstrated that miR-15b was also differentially expressed (p<0.03).
Figure 3. Recovered patients in the Validation Cohort have decreased expression of miRs-23a and -195.
miRs confirmed to be differentially expressed in the Test cohort were tested in the Validation cohort (n=7 per group). miRs-23a and -195, but not -15b and 26a were differentially expressed. *p<0.03
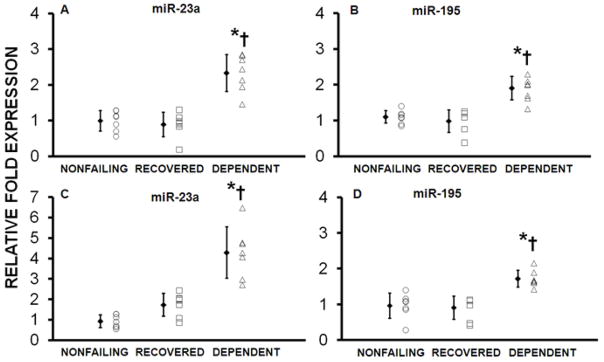
Real Time PCR Confirmation; Comparison with NF Heart Tissue
The 2 miRs (-23a and -195) found differentially expressed in both Test and Validation Cohorts were measured in NF LV tissue. Expression of both miRs-23a and -195 was significantly increased in the D groups compared with NF or R samples, but was similar between NF and R group in both cohorts (Figure 4).
Figure 4. Expression of miRs 23a and 195 was similar in Nonfailing hearts compared with Recovered, but was significantly increased in Dependent vs. Nonfailing.
Fig 4A and B represent data from the Test Cohort, while Figs 4C and D are from the Validation cohort. *p<0.04, Dependent vs. Recovered; †p<0.03, Dependent vs. Nonfailing control Hearts.
Cardiomyocyte Size Determination
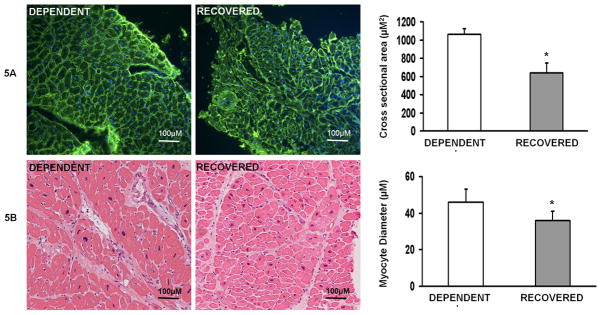
Fluorescent microscopy of wheat germ agglutinin stained cardiomyocytes from the Test cohort demonstrated that the R group had a significantly smaller cardiomyocyte cross-sectional areas when compared with the D group (Figure 5a). Independent examination by light microscopy of H+E stained samples show that the R group had significantly smaller cardiomyocyte diameters when compared with the D group (Figure 5b).
Figure 5.
Figure 5a. Recovered patients have smaller cardiomyocytes at the time of VAD implantation.
Representative images (20X) of wheat germ agglutinin (green; nuclei blue) stained VAD core samples from Test Cohort Recovered and Dependent samples, with calculated cross sectional area in μM2. (n=3 per group; *p=0.01). 5b. Representative (H+E) images and measurements of cardiomyocyte diameter (μM) in the Validation Cohort Recovered and Dependent samples (n=7 per group, *p = 0.01).
Effects of VAD Support on Selected miRs
To determine whether miRs that were differentially expressed between R and D Core samples were also responsive to mechanical unloading, the expression of select miRs was measured in a separate set of six paired cardiac samples obtained at the time of VAD implantation and subsequent cardiac transplantation. Patient characteristics are listed in Table 3.
Table 3.
Clinical characteristics of patients with paired samples obtained at the time of VAD placement (pre-VAD) and at time of transplant (Post-VAD).
| Clinical Characteristics At Baseline | |
|---|---|
| Number of Patients | 6 |
| Age (years) | 54±18 |
| Percent Ischemic | 0 |
| Inotropic Support | 100% |
| Intraaortic Balloon Pump | 50% |
| Other Mechanical Support (TandemHeart) | 0% |
| Percent Male | 50% |
| EF Prior to VAD (%) | 18±5 |
| LVEDd Prior to VAD (cm) | 6.6±1.2 |
| PCWP Prior to VAD (mm Hg) | 28±9 |
| Cardiac Index Prior to VAD (L/min/m2) | 1.8±0.69 |
|
| |
|
Clinical Characteristics While On VAD
| |
| Betablocker use while on VAD | 83% |
| ACE Inhibitor use while on VAD | 83% |
| Duration of CHF (days) | 210±108 |
| Duration of VAD support | 144±67 |
| Betablocker use while on VAD | 67% |
| ACE Inhibitor use while on VAD | 83% |
VAD support did not alter expression of any of the miRs found to be differentially expressed between the R and D groups (Table 4). To assess whether mechanical unloading alters expression of other miRs, additional miRs previously shown to play a role in heart failure (15–24) were analyzed by real-time PCR. While miRs-208 and -21 showed altered expression in response to mechanical support (Table 4), miRs-1, -133a, -133b did not. None of these miRs, however, were differentially altered between R and D groups.
Table 4.
miRs differentially expressed in Recovered hearts by screening array (*) were not altered after VAD placement in a separate group with paired samples obtained before VAD implantation and at the time of transplantation.
| miR ID | Selection Criteria | Fold change Pre/postVAD |
p Value |
|---|---|---|---|
| miR-181b | * | - | - |
| miR-424 | * | No difference | NS |
| miR-376a | * | No difference | NS |
| miR-195 | * | No difference | NS |
| miR-27a | * | No difference | NS |
| miR-23a | * | No difference | NS |
| miR-103 | * | No difference | NS |
| miR-26a | * | No difference | NS |
| miR-142-3p | * | No difference | NS |
| miR-15b | * | No difference | NS |
| miR-1† | Literature | No difference | NS |
| miR-21† | Literature | 1.6 | 0.04 |
| miR-133a† | Literature | No difference | NS |
| miR-133b† | Literature | No difference | NS |
| miR-208† | Literature | 1.9 | 0.02 |
DISCUSSION
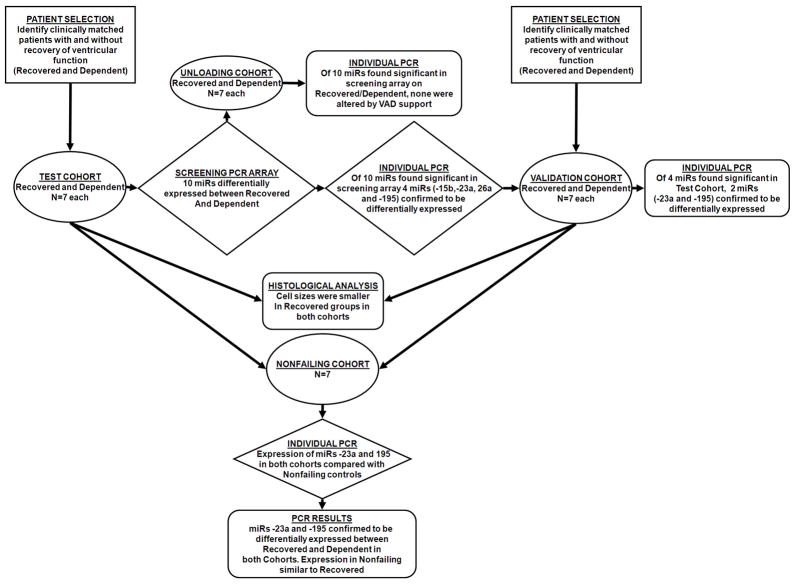
We report, in two separate patient cohorts, a pattern of myocardial miR expression at the time of VAD placement that is associated with subsequent LV functional recovery. The main findings in this study are: 1) In patients with advanced non-ischemic heart failure requiring VAD support, cardiac expression of miR-23a and -195 prior to LVAD implantation differs between hearts with eventual recovery of ventricular function and those with persistent VAD dependence. 2) This differential expression is associated with remission of heart failure independent of clinical parameters previously associated with recovery, such as LV size, duration of heart failure, and duration of VAD support. 3) Remission of heart failure is associated with a smaller cardiomyocyte size at the time of VAD placement. The experimental protocol and main findings are summarized in Figure 6.
Figure 6.
Summary of experimental protocol and major findings.
As myocardial transcriptomic biomarkers can associate with outcomes in heart failure (27), we hypothesized that biomarkers (such as miR expression patterns) exist at the time of VAD implantation that are associated with the potential for recovery. We analyzed cardiac miR expression in patients selected to match clinical variables between the Recovered and Dependent groups within each cohort (Test and Validation). However, the two cohorts differed significantly in clinical features typically associated with the development of VAD-independence. These differences between the Test and Validation cohorts make the observation of a conserved relationship between miR expression and the development of VAD-independence particularly noteworthy.
Several reports have identified specific miRs that modulate pathologic processes relevant to heart failure; hypertrophy (16, 18, 19, 23), cell death (28), fibrosis (20, 29), vascular proliferation (30), and arrhythmias (17, 31). Relevant to our observations, it is notable that a) overexpression of miR-195 occurs in pressure overload rodent hearts and that transgenic cardiac overexpression of miR-195 induces heart failure (16), and b) elevated miR-195 expression occurs in rodent and human failing hearts (21, 22, 24). Similarly, miR-23a can mediate cardiomyocyte hypertrophy (32), and is differentially expressed between failing and non-failing hearts (16, 24). Congruent with the reported function of miRs -23a and -195, our data demonstrate that hearts that eventually recover function have smaller cardiomyocyte sizes (6, 33). Intriguingly, while VAD support has been shown to decrease cardiomyocyte size (1), our data demonstrates that VAD support alone does not induce a miR expression profile associated with functional recovery. Thus a differential miR expression in failing hearts that eventually recovered VAD-independence suggests that underlying pathophysiologic processes exist at the time of VAD placement that differ between those hearts that recover function and those that do not. We did not detect differences between the Recovered and Dependent hearts in the expression of other miRs that are differentially expressed between failing and non-failing hearts (i.e. miRs-1, 21, -29, -125b, -130, and 133 (21, 24)). The fact that they did not differ with the ability to recover function suggests that cellular processes associated with a more limited set of miRs (-23a, -195, and possibly -15b) are critical in defining the potential to recover cardiac function after mechanical unloading.
There are several scenarios by which lower miR-23a and-195 expression may be associated with functional recovery. Patients that recovered may have had a) less severe heart failure, b) a different disease etiology that allows greater likelihood of recovery, or c) genetic variants that underlie decreased expression of particular miRs that mediate pathophysiologic processes of heart failure, or the response to mechanical unloading. The observation that Recovered patients had miR-23a and -195 levels that resembled that of non-failing hearts and a smaller cardiomyocyte size than Dependent hearts raises the important possibility that the Recovered patients had less severe heart failure. Although we cannot exclude this possibility, we think that this is unlikely given that the standard clinical measures of heart failure (Table 1) were identical in Recovered and Dependent groups from both cohorts. Regarding the role of disease etiology, cardiac miR expression profiles differ according to heart failure etiologies (24). However we carefully matched the Recovered and Dependent groups for clinical parameters and etiology so as to prevent bias with diseases (such as myocarditis, peripartum cardiomyopathy) having a higher capacity for recovery (Supplemental Material Table s1). Finally, as the ‘Recovery’ population is only a small percentage of all VAD-supported patients, it is plausible that these patients possess infrequent miR gene variants whereby decreased miR expression is associated with recovery from heart failure, or beneficial response to mechanical unloading. Indeed, expression quantitative trait loci have been found for ~20% of miRs expressed in fibroblasts (34), and miR SNPs have been associated with congenital heart diseases susceptibility (35) and dilated cardiomyopathy incidence (36).
A major limitation of this observational study is that the list of differentially expressed miRs does not provide insight into the mechanistic processes associated with recovery. One approach to find insight into mechanistic processes would be to use a target prediction programs (i.e. Targetscan or Pictar) to develop a list of possible mRNA targets. However when analyzing miRs-23a and -195, we found little congruency in either the list of potential mRNA targets identified by the two target prediction programs, or the biologic pathways/processes identified through multiple pathway analysis programs (data not shown). The lack of convergent results may reflect either the current limitations of these bioinformatic methods, or the need to identify additional miRs that are differentially expressed between the Recovered and Dependent groups. Indeed, due to the rarity of VAD-supported patients who recover cardiac function, (8–10), the number of patients studied is small, and likely underpowered for detection of all differentially expressed miRs.
In summary, we report cardiac miR expression differences that may identify patients which have the greatest potential for recovery after VAD support. Such patients may have less severe disease, unrecognized disease etiologies that allow for recovery, or genetic variants that relate miR expression to functional recovery. This ‘recovery’ expression profile is not induced by mechanical unloading, suggesting that the patients’ potential for recovery is already established at the time of VAD placement. If confirmed in a larger cohort, such information could help guide clinical decisions regarding utilization of mechanical support devices.
Supplementary Material
Acknowledgments
FUNDING SOURCES: Research supported by funds from the Cardiovascular Institute, University of Pittsburgh (Drs Ramani and McTiernan), the Kaufman Center for Heart Failure, Cleveland Clinic Foundation (Dr. Moravec), and the National Institutes of Health (5R01HL086918-0, Dr. Gorcsan; RO1-HL/AG 61483, Dr. Taegtmeyer).
We thank Marc Simon MD, University of Pittsburgh, for providing clinical data; the clinical co-coordinators (Karen Janosko, RN, MSN; Lori DeGore, RN; Kathleen Lockard, RN MBA; Kristin Shoemaker, RN, BSN; Tracie Sabatine, RN, BSN, Pittsburgh, and Sylvia Carranza, BS, Houston), surgical staff, and the mechanical device bioengineers for their assistance.
Footnotes
No conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zafeiridis A, Jeevanandam V, Houser SR, Margulies KB. Regression of cellular hypertrophy after left ventricular assist device support. Circulation. 1998;98:656–662. doi: 10.1161/01.cir.98.7.656. [DOI] [PubMed] [Google Scholar]
- 2.Dipla K, Mattiello JA, Jeevanandam V, Houser SR, Margulies KB. Myocyte recovery after mechanical circulatory support in humans with end-stage heart failure. Circulation. 1998;97:2316–2322. doi: 10.1161/01.cir.97.23.2316. [DOI] [PubMed] [Google Scholar]
- 3.Frazier OH, Benedict CR, Radovancevic B, et al. Improved left ventricular function after chronic left ventricular unloading. Ann Thorac Surg. 1996;62:675–682. doi: 10.1016/s0003-4975(96)00437-7. [DOI] [PubMed] [Google Scholar]
- 4.Bartling B, Milting H, Schumann H, et al. Myocardial gene expression of regulators of myocyte apoptosis and myocyte calcium homeostasis during hemodynamic unloading by ventricular assist devices in patients with end-stage heart failure. Circulation. 1999;100:II216–223. doi: 10.1161/01.cir.100.suppl_2.ii-216. [DOI] [PubMed] [Google Scholar]
- 5.Li YY, Feng Y, McTiernan CF, et al. Downregulation of matrix metalloproteinases and reduction in collagen damage in the failing human heart after support with left ventricular assist devices. Circulation. 2001;104:1147–52. doi: 10.1161/hc3501.095215. [DOI] [PubMed] [Google Scholar]
- 6.Segura AM, Frazier OH, Demirozu Z, Buja LM. Histopathologic correlates of myocardial improvement in patients supported by a left ventricular assist device. Cardiovasc Pathol. 2010;20:139–145. doi: 10.1016/j.carpath.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Wohlschlaeger J, Levkau B, Brockhoff G, et al. Hemodynamic support by left ventricular assist devices reduces cardiomyocyte DNA content in the failing human heart. Circulation. 2010;121:989–96. doi: 10.1161/CIRCULATIONAHA.108.808071. [DOI] [PubMed] [Google Scholar]
- 8.Dandel M, Weng Y, Siniawski H, Potapov E, Lehmkuhl HB, Hetzer R. Long-term results in patients with idiopathic dilated cardiomyopathy after weaning from left ventricular assist devices. Circulation. 2005;112:I37–45. doi: 10.1161/CIRCULATIONAHA.104.525352. [DOI] [PubMed] [Google Scholar]
- 9.Simon MA, Kormos RL, Murali S, et al. Myocardial recovery using ventricular assist devices: prevalence, clinical characteristics, and outcomes. Circulation. 2005;112:I32–6. doi: 10.1161/CIRCULATIONAHA.104.524124. [DOI] [PubMed] [Google Scholar]
- 10.Dandel M, Weng Y, Siniawski H, et al. Prediction of cardiac stability after weaning from left ventricular assist devices in patients with idiopathic dilated cardiomyopathy. Circulation. 2008;118:S94–105. doi: 10.1161/CIRCULATIONAHA.107.755983. [DOI] [PubMed] [Google Scholar]
- 11.Mandarino WA, Gorcsan J, 3rd, Gasior TA, Pham S, Griffith BP, Kormos RL. Estimation of left ventricular function in patients with a left ventricular assist device. ASAIO J. 1995;41:M544–7. doi: 10.1097/00002480-199507000-00070. [DOI] [PubMed] [Google Scholar]
- 12.Gorcsan J, 3rd, Severyn D, Murali S, Kormos RL. Non-invasive assessment of myocardial recovery on chronic left ventricular assist device: results associated with successful device removal. J Heart Lung Transplant. 2003;22:1304–13. doi: 10.1016/s1053-2498(03)00056-1. [DOI] [PubMed] [Google Scholar]
- 13.Hall JL, Birks EJ, Grindle S, et al. Molecular signature of recovery following combination left ventricular assist device (LVAD) support and pharmacologic therapy. Eur Heart J. 2007;28:613–27. doi: 10.1093/eurheartj/ehl365. [DOI] [PubMed] [Google Scholar]
- 14.Razeghi P, Young ME, Ying J, et al. Downregulation of metabolic gene expression in failing human heart before and after mechanical unloading. Cardiology. 2002;97:203–209. doi: 10.1159/000063122. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Nat Acad Sci. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Ji R, Yue J, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Path. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 20.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 21.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008;45:185–92. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matkovich SJ, Van Booven DJ, Youker KA, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–71. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda S, Kong SW, Lu J, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.McGaffin KR, Sun CK, Rager JJ, et al. Leptin signalling reduces the severity of cardiac dysfunction and remodelling after chronic ischaemic injury. Cardiovasc Res. 2008;77:54–63. doi: 10.1093/cvr/cvm023. [DOI] [PubMed] [Google Scholar]
- 27.Heidecker B, Kasper EK, Wittstein IS, et al. Transcriptomic biomarkers for individual risk assessment in new-onset heart failure. Circulation. 2008;118:238–46. doi: 10.1161/CIRCULATIONAHA.107.756544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren XP, Wu J, Wang X, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terentyev D, Belevych AE, Terentyeva R, et al. miR-1 overexpression enhances Ca(2+) release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–21. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci. 2009;106:12103–8. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruckner BA, Razeghi P, Stetson S, et al. Degree of cardiac fibrosis and hypertrophy at time of implantation predicts myocardial improvement during left ventricular assist device support. J Heart Lung Transplant. 2004;23:36–42. doi: 10.1016/s1053-2498(03)00103-7. [DOI] [PubMed] [Google Scholar]
- 34.Borel C, Deutsch S, Letourneau A, et al. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011;21:68–73. doi: 10.1101/gr.109371.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Hu Z, Xu Z, et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mutat. 2009;30:1231–6. doi: 10.1002/humu.21044. [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, Rao L, Peng Y, et al. Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin Chim Acta. 2010;411:1287–90. doi: 10.1016/j.cca.2010.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.