Summary
In a microarray analysis of the RpoS regulon in mammalian host-adapted Borrelia burgdorferi, bb0728 (cdr) was found to be dually-transcribed by the sigma factors σ70 and RpoS. The cdr gene encodes a coenzyme A disulfide reductase (CoADR) that reduces CoA-disulfides to CoA in an NADH-dependent manner. Based on the abundance of CoA in B. burgdorferi and the biochemistry of the enzyme, CoADR has been proposed to play a role in the spirochete’s response to reactive oxygen species (ROS). To better understand the physiologic function(s) of Bb CoADR, we generated a B. burgdorferi mutant in which the cdr gene was disrupted. RT-PCR and 5′-RACE analysis revealed that cdr and bb0729 are co-transcribed from a single transcriptional start site upstream of the bb0729 coding sequence; a shuttle vector containing the bb0729-cdr operon and upstream promoter element was used to complement the cdr mutant. Although the mutant was no more sensitive to hydrogen peroxide than its parent, it did exhibit increased sensitivity to high concentrations of t -butyl-hydroperoxide, an oxidizing compound that damages spirochetal membranes. Characterization of the mutant during standard (15% oxygen, 6% CO2) and anaerobic (<1% O2, 9–13% CO2) cultivation at 37°C revealed a growth defect under both conditions that was particularly striking during anaerobiosis. The mutant was avirulent by needle inoculation and showed decreased survival in feeding nymphs, but displayed no survival defect in unfed flat nymphs. Based on these results, we propose that Bb CoADR is necessary to maintain optimal redox ratios for CoA/CoA-disulfide and NAD+/NADH during periods of rapid replication throughout the enzootic cycle, to support thiol-disulfide homeostasis, and to indirectly protect the spirochete against peroxide-mediated membrane damage; one or more of these functions are essential for infection of the mammalian host by B. burgdorferi.
Keywords: RpoS, CoADR, Lyme disease
Introduction
Borrelia burgdorferi, the causative agent of Lyme disease, persists in an enzootic cycle involving a tick vector and a vertebrate host, usually a rodent(Lane et al., 1991; Steere et al., 2004; Tilly et al., 2008). As the spirochete transitions between these two distinct host milieus, it must sense and adjust to changes in ambient temperature, oxygen tension, availability of carbon sources and other nutrients, and the presence of mammalian immune factors (Seshu and Skare, 2000; Tilly et al., 2001; Anguita et al., 2003; Pal and Fikrig, 2003; Seshu et al., 2004; Boylan et al., 2006; Dunham-Ems et al., 2009; Xu et al., 2010). The sigma factor RpoS is critical to the adaptations made byB. burgdorferi within feeding nymphs and the mammalian host, controlling the reciprocal up-regulation of genes necessary for the establishment of infection and the down-regulation of genes required within the vector (Hubner et al., 2001; Caimano et al., 2004; Caimano et al., 2005; Caimano et al., 2007; Ouyang et al., 2008; Ouyang et al., 2009). In a microarray analysis of the RpoSregulonin mammalian host-adapted spirochetes, we identified a small subset of genes that can be dually-transcribed bythe housekeeping sigma factor σ70 and the alternate sigma factor RpoS(Caimano et al., 2007). Among these isbb0728 (cdr), which encodes a Coenzyme A disulfide reductase (CoADR) (Boylan et al., 2006; Caimano et al., 2007). First identified and characterized in Staphylococcus aureus (delCardayre and Davies, 1998; delCardayre et al., 1998),CoADRs subsequently have been annotated in the genomes of a number of organisms and biochemically characterized in S. aureus, Bacillus anthracis, B. burgdorferi, and Pyrococcus horikoshii (delCardayre and Davies, 1998; delCardayre et al., 1998; Harris et al., 2005; Hummel et al., 2005; Boylan et al., 2006; Mallett et al., 2006; Wallen et al., 2008). These enzymes reduce CoA-disulfides to CoA in an NAD(P)H-dependent manner in which the specificity for a particular pyridine nucleotide is dependent upon the organism(Wallen et al., 2008); the B. burgdorferi CoADR (Bb CoADR) utilizes NADH exclusively (Boylan et al., 2006). Interestingly, cdr is one of the few genes identified in the RpoS regulon encoding a protein with a predicted metabolic function(Caimano et al., 2007).
Pathogens grown in an aerobic environment must deal with the reactive oxygen species (ROS) generated both intracellularly and extracellularly as a major component of the mammalian innate immune defense against invading pathogens(Cabiscol et al., 2000; Imlay, 2003; Imlay, 2008; Sorci and Faivre, 2009).In addition to other damaging activities, ROS can cause inappropriate disulfide bond formation, inactivating protein function, and damaging or killing a cell (Imlay, 2002; Sevier and Kaiser, 2002; Kadokura et al., 2003). To combat this threat, organisms maintain abundant low-molecular weight thiols that function with specific Flavoprotein Disulfide Reductases (FDRs) and Thioredoxin Fold Proteins (TFPs) to sustain thiol-disulfide homeostasis(Holmgren, 1989; Carmel-Harel and Storz, 2000a; Holmgren et al., 2005; Imlay, 2008). Eukaryota and many Gram negative bacteria utilize glutathione and thioredoxin for this purpose (Holmgren, 1989; Carmel-Harel and Storz, 2000b; Holmgren et al., 2005). Gram positive bacteria and other more distantly related prokaryotes maintain pools of alternative low molecular weight thiols such as mycothiols(Newton et al., 1996; Newton et al., 2008), bacillithiols(Newton et al., 2009; Gaballa et al., 2010) or CoA(Newton et al., 1996; Hummel et al., 2005; Boylan et al., 2006; Nicely et al., 2007).B. burgdorferi encodes thioredoxin (Trx; bb0061) and a thioredoxin reductase (TrxB; bb0515) (Fraser et al., 1997) that is predicted to be NADPH-dependent, but the most abundant low-molecular weight thiol in the spirochete is CoA, which is found almost entirely in the reduced form (CoASH)(Boylan et al., 2006). This suggests the involvement of CoADR in the maintenance of the cell’s thiol/disulfide balance(delCardayre and Davies, 1998; delCardayre et al., 1998; Harris et al., 2005; Boylan et al., 2006; Wallen et al., 2008).Boylan, et al. (2006)also have reported that CoASH can directly reduce hydrogen peroxide(H2O2) in vitro ; the CoASH in the assay was regenerated by the addition of NADH and Bb CoADR. Direct involvement in the detoxification of peroxides has not previously been reported for CoA or CoADRs.
Given B. burgdorferi ‘ssmall genome and limited biosynthetic capabilities, one also would anticipate an important role for Bb CoADR in intermediary metabolism(Gherardini et al., 2010). Bb CoADR is one of the few enzymes identified in the spirochete capable of regenerating the NAD+ required for glycolysis, theorganism’s sole means of energy production (Fraser et al., 1997). Additionally, CoA is a ubiquitous cofactor with an essential role as one of the predominant acyl group carriers in the intermediary metabolism of all organisms (Jackowski, 1996; Wolfe, 2005). Although the spirochete does not utilize acetyl-CoA for oxidative phosphorylation, it does require this molecule for a number of important anabolic pathways, including biosynthesis of phospholipids, glycolipids, lipoproteins and peptidoglycan for the maintenance and remodeling of its cell envelope (Fraser et al., 1997; Gherardini et al., 2010; Xu et al., 2010). B. burgdorferi lacks the common pathways (e.g., the AMP-forming acetyl-CoA synthetase [AMP-ACS] pathway, β-oxidationof fatty acids, and degradation of certain amino acids) for the production of acetyl-CoA (Fraser et al., 1997; Xu et al., 2010); its sole source of this essential compound appears to be synthesis from acetate and CoA via the Ack-Pta (acetate kinase-phosphate acetyltransferase) pathway (Xu et al., 2010; Sze and Li, 2011). The reduction of CoA disulfides by Bb CoADR would ensure adequate levels of CoASH for the production of acetyl-CoA, particularly during times of stress or increased replication, with the added bonus of yielding NAD+ for energy generation.
The only previously published study of BbCoADR was a biochemical analysis using recombinant enzyme(Boylan et al., 2006). To elucidate the physiological function of Bb CoADR, we generated a B. burgdorferi mutant in which the cdr gene was disrupted. Characterization of the mutant during standard (15% O2, 5% CO2) and anaerobic (<1% O2, 9–13% CO2)cultivation revealed a growth advantage under both conditions that, surprisingly, was particularly striking during anaerobiasis. In contrast to the prediction of (Boylanet al. (2006), the mutant was no more sensitive to hydrogen peroxide (H2O2) than its parent; it was, however, more sensitive to high concentrations of t -butyl-hydroperoxide, an oxidizing compound that damages spirochetal membranes(Boylan et al., 2008; Boylan and Gherardini, 2008).The mutant was avirulent by needle inoculation and showed diminished survival in feeding nymphs but no survival defect in flat nymphs. Based on these results, we propose that Bb CoADR is required to maintain optimal redox ratios for CoA/CoA-disulfide and NAD+/NADH during periods of rapid replication, to support thiol-disulfide homeostasis, and to indirectly protect the spirochete against peroxide-mediated membrane damage; one or more of these functions are essential for infection of the mammalian host by B. burgdorferi.
Results
Predicted structure and bioinformatics of BbCoADR
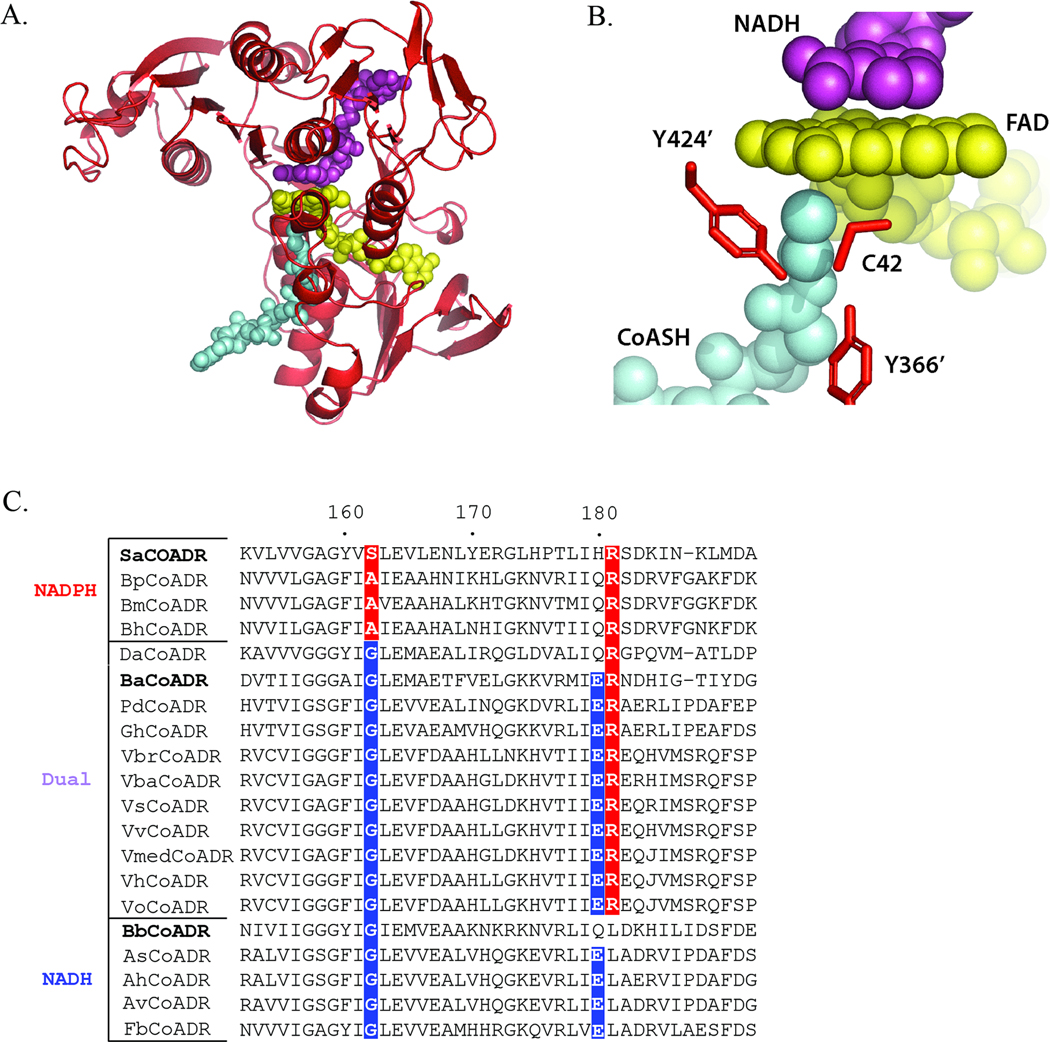
CoADR, along with NADH oxidase (Nox) and NADH peroxidase (Npx), represent the Peroxidase-Oxidase-Reductase (POR) subgroup of the Flavoprotein Disulfide Reductase (FDR) family; in addition to the POR subgroup, the FDR family includes, among others, glutathione reductase (GR; DSR subgroup) and thioredoxin reductase (TrxR; AHR subgroup) (Argyrou and Blanchard, 2004; Ojha et al., 2007). Bacterial CoADR enzymes have been well-described in S. aureus, B. burgdorferi, andB. anthracis (delCardayre and Davies, 1998; delCardayre et al., 1998; Boylan et al., 2006; Mallett et al., 2006; Wallen et al., 2008; Wallen et al., 2009). The crystal structures for oxidized Sa CoADR and Ba CoADR and for the reduced NADH and NADPH complexes of Ba CoADR have been reported previously (Mallett et al., 2006; Wallen et al., 2008; Wallen et al., 2009). Using the coordinates of the Ba CoADR-NADH complex, we generated a homology model for the Bb CoADR-NADH complex (Figure 1); Ba CoADR is 39% identical (63% similar) to Bb CoADR. Figure 1A shows the reduced Bb CoADR monomer with bound NADH, FAD and the CoASH product. The NADH, CoASH, and FAD are brought together within the predicted Bb CoADR active site formed, in part, by a cysteine residue (Cys42) from one polypeptide and two tyrosine residues (Tyr366′ and Tyr424′) from the second (Figure 1B). One functional distinction between the well-characterized bacterial CoADRs involves the pyridine nucleotide specificity: Sa CoADR has a preference for NADPH, Ba CoADR exhibits dual NAD(P)H specificity, and Bb CoADR preferentially utilizes NADH (delCardayre and Davies, 1998; delCardayre et al., 1998; Boylan et al., 2006; Wallen et al., 2008).Preference for a particular pyridine nucleotide can be indicative of an enzyme’s role in the cell; enzymes that are NAD(H)-dependent are more likely involved in the regeneration of oxidized pyridine nucleotides for glycolysis (Argyrou and Blanchard, 2004; Harris et al., 2005; Ying, 2006; Ying, 2008). Of the CoADRs thus far examined, only Bb CoADR selectively uses NADH(delCardayre and Davies, 1998; delCardayre et al., 1998; Gouet et al., 1999; Harris et al., 2005; Boylan et al., 2006; Mallett et al., 2006; Wallen et al., 2008).To determine whether Bb CoADR is unusual among CoADRs in its preference for NADH or whether NADH-selectivity is a hallmark of Gram negative bacteria and/or spirochetes, we analyzed the NAD(P)H-binding motifs of CoADR orthologs annotated in 17 distinct species, including spirochetes (from the genus Brachyspira) and both Gram-negative γ-and δ-proteobacteria (Figure 1C). NADH-selectivity of members of the FDR family depends largely upon the contribution of a Gly residue at position 162 and either a Glu or Asp residue at position 180 (positions given relative to Ba CoADR for clarity)(Karplus and Schulz, 1989; Stehle et al., 1993; Wallen et al., 2008) (Figure 1C).Based on sequence data, and in the absence of structural and functional data, we predict that, in addition to the Bb CoADR, four of the analyzed CoADRs (from three Aeromonas species and Ferrimonas balearica) also selectively utilize NADH. The rest have binding motifs consistent either with NADPH-selectivity (n = 3) or the ability to use both pyridine nucleotides (n = 10) (Figure 1C).
Figure 1.
(A) A monomer of a Bb CoADR homology model generated based on the structure of the reduced Ba CoADR-NADH complex (Wallen et al., 2008). CoASH, FAD, and NADH are colored cyan, yellow, and magenta, respectively. The orientation is the same as Figure 3 published in (Wallenet al., 2008). PDB code for the Ba CoADR structure used is 3CGD. (B) The predicted active site of Bb CoADR. Bb CoADR residues [Cys 42 (chain A) and Tyr366’ and Tyr424’ (chain B)] are indicated. CoASH, FAD, and NADH are colored the same as in (B).(C) Sequence alignment for the NAD(P)H-binding motifs of annotated CoADRs from Gram negative bacteria and spirochetes. Sequence numbering corresponds to Ba CoADR. Gly at position 162 and Asp or Glu at position 180 are favored for NADH-specificity, while Ala and Arg residues at positions 162 and 181, respectively, contribute to NADPH-specificity (Karplus and Schulz, 1989; Stehle et al., 1993; Wallen et al., 2008). A hybrid sequence of Glu180 and Arg181 is characteristic of dual NADH and NADPH specificity (Wallen et al., 2008). Enzymes are grouped according to predicted NAD(P)H substrate preference; positions important for NADPH- (red) or NADH- (blue) specificity are indicated. CoADRs with experimentally-determined pyridine nucleotide specificity are indicated in bold. Bp, Br. pilisicoli ; Bm, Br. murdochii ; Bh, Br. hyodysenteriae ; Da, D. acetoxidans ; Pd, Photobacterium damselae ; Gh, Grimontia hollisae ; Vbr, Vibrio brasiliensis ; Vba, V. bacterium ; Vs, V. splendidus ; Vv, V. vulnificus ; Vmed, V. sp. MED22; Vh, V. harveyi ; Vo, V. orientalis ; As, A. salmonicidia ; Ah, A. hydrophila ; Av, A. veronii ; Fb, F. balearica.
Construction of a cdr mutant in B. burgdorferi
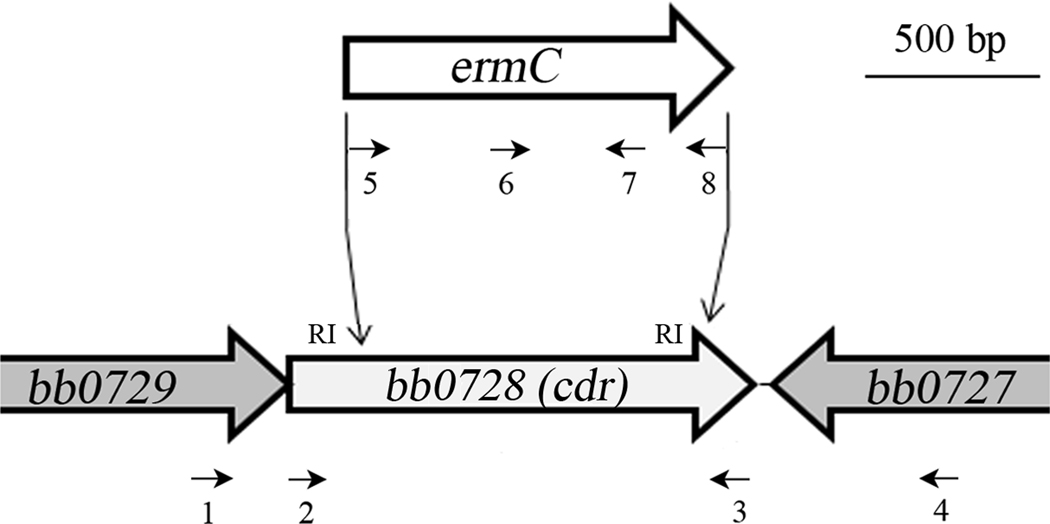
To investigate the function of Bb CoADR, we generated a cdr null mutant in c162, a virulent clone of B. burgdorferi strain 297 (Steere et al., 1984) (Table 1). For this purpose, we cloned the erythromycin-resistance gene (ermC) from pGK12 (Kok et al., 1984; Sartakova et al., 2000) into two native EcoRI sites 197 and 1203 nucleotides from the 5′ end of the 1330-nucleotide cdr open reading frame (Fraser et al., 1997) (Figure 2). Removal of this fragment leaves 44 N-terminal amino acids (10% of the 444 total) that include part of the FAD-BD1 domain containing the active site Cys42, but eliminates the NADH-binding domain, FAD-BD2, and the Interface domain including Tyr366(Mallett et al., 2006; Wallen et al., 2008; Wallen et al., 2009) (Figures 1A and 1B). Thus, this mutation is expected to completely abrogate the activity of Bb CoADR. To select the mutant, c162 cells electroporated with the cdr -knockout construct were split into two aliquots and grown in the presence of erythromycin at 37°C under standard (15% O2, 6% CO2) or anaerobic (<1% O2, 9–13% CO2) conditions. Both conditions were used in anticipation of the possibility that the mutation would render the cells sensitive to oxygen; however, erythromycin-resistant clones with the appropriate insertion (not shown) were recovered using both conditions. One clone recovered under anaerobic conditions, designated c309 (Table 1), was selected for further study.
Table 1.
B. burgdorferi clones used in this study
| Clone name |
genotype | Description | reference |
|---|---|---|---|
| c162 | clone of strain 297 | wild-type (wt) | (Caimano et al., 2007) |
| c309 | c162 cdr∷ermC | cdr mutant (mut) | This work |
| c1655 | c309 + pCE1735 (bb0729-cdr operon); missing lp28-1 |
complement (ct) | This work |
| c174 | c162 rpoS∷ermC | rpoS mutant | (Caimano et al., 2004; Eggers et al., 2004) |
Figure 2.
Generation of a cdr -mutant in a virulent clone of B. burgdorferi strain 297. An erythromycin-resistance cassette (ermC) was cloned into two endogenous EcoRI (RI) sites within bb0728 (cdr) in c162. Oligonucletoide primers used for the generation and screening of the mutant are indicated by small arrows. Numbers correspond to primers found in Table 2.
Promoter identification and generation of a complemented clone of the cdr mutant
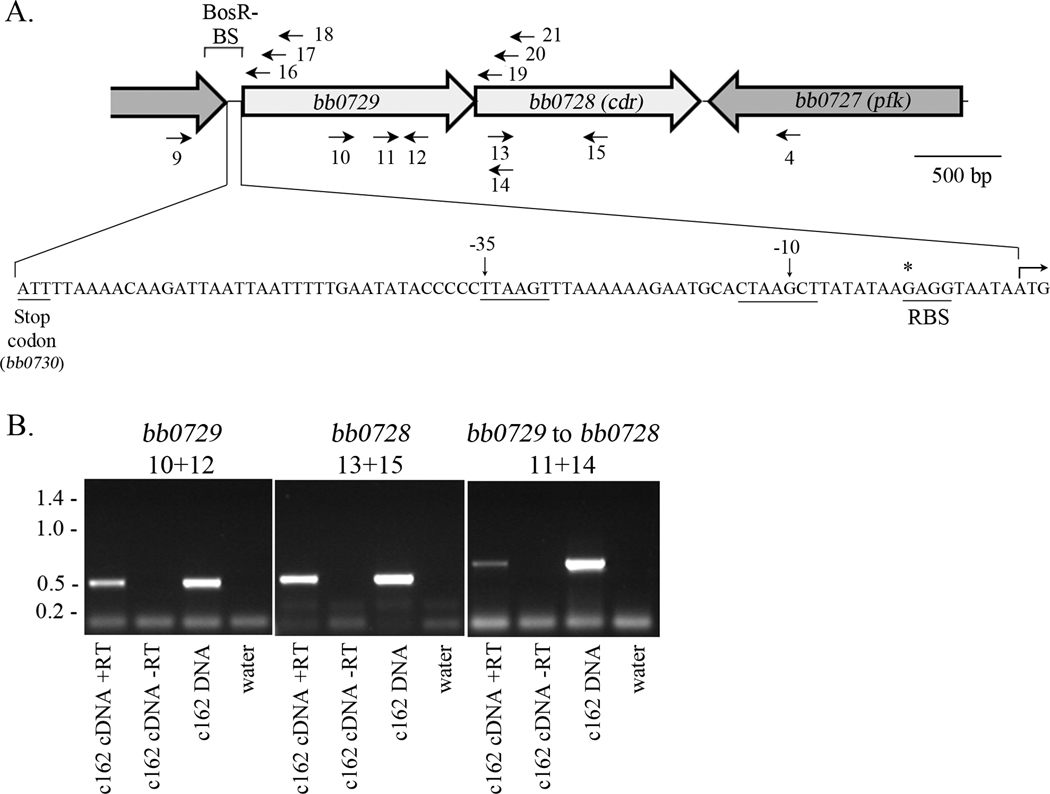
When the sequence of B. burgdorferi strain B31 was released (Fraser et al., 1997), the cdr gene was predicted to form a two-gene operon with bb0729 (Figure 3A); bb0729 has been annotated as encoding a dicarboxylate amino acid (Glu) transporter(Fraser et al., 1997), but the predicted gene product is 50% identical with TcyP, a Na+/cystine symporter of B. subtilis (Burguiere et al., 2004; Overbeek et al., 2005). As shown in Figure 3B, amplification of B. burgdorferi cDNA using primers that lie within bb0729 and bb0728 (Table 2) confirmed that these two genes are co-transcribed. To generate a complement ofc309in which the cdr gene was re-introduced under the control of its own promoter, we next had to identify the region controlling cdr expression. 5′-RACE using primers internal to either cdr (primers 16–18) or bb0729 (primers 18–21) (Figure 3A and Table 2) revealed a single strong transcriptional start at position −9 relative to the translational start of bb0729 ; this is immediately upstream of the predicted ribosomal binding site for bb0729 (illustrated in Figure 3A). In contrast, amplification upstream of the cdr open reading frame yielded no product. A comparison of the ‘extended −10’ and −35 regions of the bb0729 promoter with a consensus sequence previously identified for RpoS-dependent genes (Caimano et al., 2007) reveals that the −35 region sequence (TTAAGT) differs by two nucleotides while the ‘extended −10 region’ (TGCACTAAGCT) differs by four.
Figure 3.
The bb0729 cdr operon. (A) Organization of the region surrounding bb0728 (cdr). Primers used to demonstrate the presence of a single transcript are indicated below the gene diagram; primers used for 5′-RACE to determine promoter location(s) are shown above the gene diagram. The region in which BosR potentially binds (Boylan et al., 2006) is indicated (BosR-BS). The transcriptional start site (*), ribosomal binding site (RBS) and −10/−35 elements, as determined by 5′-RACE, are shown within the region between bb0730 and bb0729. (B) bb0729 and the cdr gene are co-transcribed under the regulatory control of a promoter upstream of bb0729. C162 was grown to 5 × 107 cells/ml at 37°C under standard conditions. RNA was harvested, converted to cDNA, and analyzed by RT-PCR using the indicated primer pairs; the corresponding locations for the primers (Table 2) are shown in (A). Molecular weights are given in kbp.
Table 2.
Oligonucleotides used in this study.
| #a | Name | Sequence (5′-3′) | Description (reference) |
|---|---|---|---|
| 1 | bb0729-1225F | CTTTCCAGTGGGATTGGTAGGACTTG | mutant screening |
| 2 | bb0728-5′F (XhoI)b | GATCCTCGAGATGATGAAAATAATAATTATTGGGG | mutant generation |
| 3 | bb0728-3′R (BamHI) | GATCGGATCCTTCTTTCTATTTGGCAGCATTGCCAGC | mutant generation |
| 4 | bb0727-839F | CTTGATTTTGACATAGAAGGTCCTAATGG | mutant screening/ complement generataion |
| 5 | ermC-1F | CGATTCACAAAAAATAGGCACACG | mutant generation |
| 6 | ermC-464F | TCTTTGAAATCGGCTCAGGGGGGGGCC | mutant screening |
| 7 | ermC-808R | TCTGCCATTAAAAGTAATGCCAATGAGAGAGCG | mutant screening |
| 8 | ermC-1118R | AAACCGTGTGCTCTACGACCAAAAC | mutant generation/screening |
| 9 | bb0730-1159R (SphI) | GATCGCATGCAGCTTGTAAACAAATAAATAGGAAAACA | complement generation |
| 10 | bb0729-550F-RT | GGCTTAGAAAAAACTCAACCATCG | RT-PCR |
| 11 | bb0729-805F-RT | GCTTCCTACATTGCCATAGGTCTTAC | RT-PCR |
| 12 | bb0729-1002R-RT | TTGCTATTCCTTCGCTTACTCC | RT-PCR |
| 13 | bb0728-140F-RT | TGGGGGGATTCTTTGACAAC | RT-PCR |
| 14 | bb0728-140R-RT | GTTGTCAAAGAATCCCCCCA | RT-PCR |
| 15 | bb0728-680F-RT | CTACTCCTTCTGCCTTTTTTTCTCC | RT-PCR |
| 16 | bb0729-GSP3 | AAGGAGCCTTACGTATCCATCGC | 5′-RACE |
| 17 | bb0729-GSP2 | GTAGGCTCATTTTCCCAACATC | 5′-RACE |
| 18 | bb0729-GSP1 | CCTTCGGCTGTTAATCCCAATGC | 5′-RACE |
| 19 | bb0728-GSP3 | GTAAGGCAGGCCACAGGTTCC | 5′-RACE |
| 20 | bb0728-GSP2 | GATAACTTCGTGGTTAGTTTTAACAG | 5′-RACE |
| 21 | bb0728-GSP1 | GATTGGTGGAATAATAGGTTTTGCAC | 5′-RACE |
| bb0728-qRTPCR-R | GACGCTGTTATACTTGCTACCG | qRT-PCR | |
| bb0728-qRTPCR-R | GAAGCTGAGCCCAATGTGCCT | qRT-PCR | |
| bb0729-qRTPCR-F | TTGGGCGATGGATACGTAAGG | qRT-PCR | |
| bb0729-qRTPCR-F | CGCTTGTAGTCCTTCGGCTGT | qRT-PCR | |
| flaB-qRTPCR-F | CTTTTCTCTGGTGAGGGAGCTC | qRT-PCR and qPCR(Zhang et al., 2009) | |
| flaB-qRTPCR-R | GCTCCTTCCTGTTGAACACCC | qRT-PCR and qPCR(Zhang et al., 2009) | |
| flaB-probe | CTTGAACCGGTGCAGCCTGAGCA | qRT-PCR and qPCR(Zhang et al., 2009) | |
| nidogen-F | CCCCAGCCACAGAATACCAT | qPCR (Benhnia et al., 2005) | |
| nidogen-R | AAAGGCGCTACTGAGCCGA | qPCR (Benhnia et al., 2005) | |
| nidogen-Probe | CCGGAACCTTCCCACCCAGC | qPCR (Benhnia et al., 2005) | |
| Tick β-actin-F | GGTATCGTGCTCGACTC | qPCR(Zhang et al., 2009) | |
| Tick β-actin-R | ATCAGGTAGTCGGTCAGG | qPCR(Zhang et al., 2009) | |
| T7 | TAATACGACTCACTATAGG | General screening | |
| M13R | GGAAACAGCTATGACCATG | General screening |
Boylan et al., 2006; reported that the transcription factor BosR (BB0647) enhanced expression of cdr by binding somewhere within a 225-bp region upstream of bb0729. While two potential consensus sequences for BosR binding have been identified, neither element was found within the region directly upstream of bb0729 (Katona et al., 2004; Ouyang et al., 2011). Nevertheless, to ensure that any potential regulatory elements proximal to the promoter were included in the construct used for complementation of the cdr mutant, we amplified the entire bb0729-cdr operon, including the 500-bp upstream region to generate the complementing plasmid pCE1735. All transformants selected contained pCE1735 but lacked the endogenous B. burgdorferi plasmid lp28-1; after multiple unsuccessful attempts to recover a complemented clone with a complete plasmid complement, we selected one lacking lp28-1 (c1655, Table 1) for further study.
Transcriptional analysis of cdr during in vitro growth
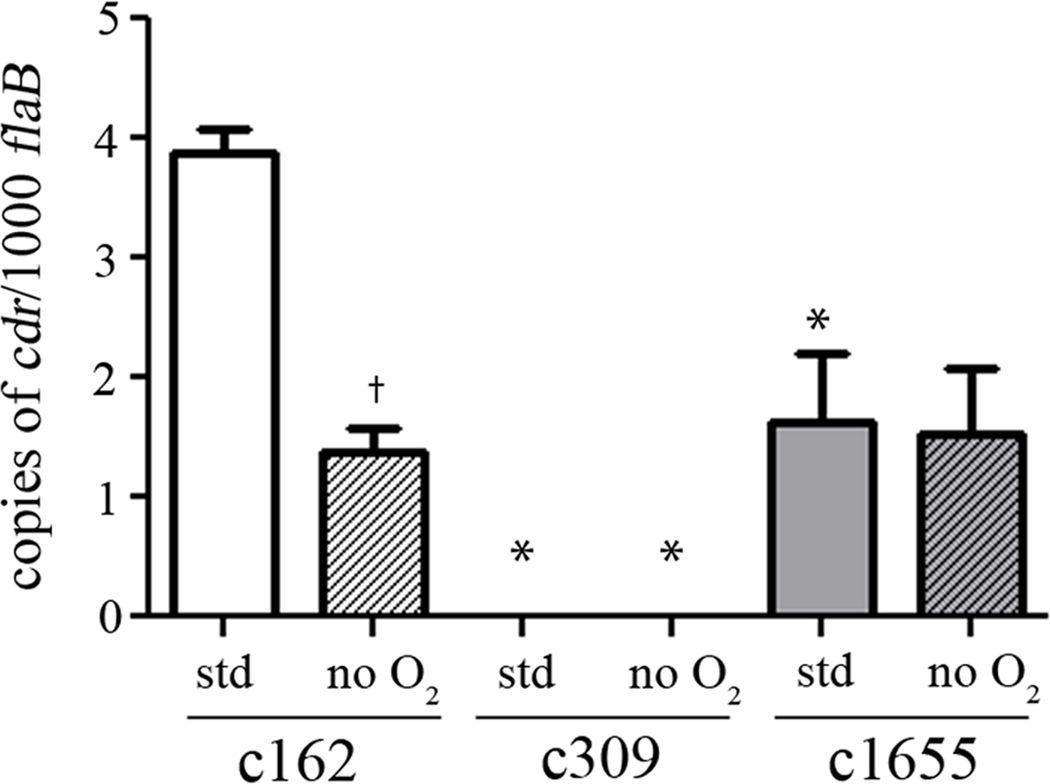
To determine whether cdr expression is sensitive to changes in oxygen tension during in vitro growth, qRT-PCR was performed on RNAs harvested from the wild-type clone, c162, cultured at 37°C under both standard (15% oxygen, 6% CO2) and anaerobic (<1% O2, 9–13% CO2) growth conditions; c309 (mut) and c1655 (ct) were included in these analyses to assess levels of cdr expression in the mutant and complement. As shown in Figure 4, while copy numbers of cdr transcripts are quite low, c162 produced significantly more transcript (P < 0.05) under standard growth conditions than when grown anaerobically (Figure 4). As expected, no transcript was detected in c309 under either condition. While transcript levels in c162and c1655were virtually identical under anaerobic conditions, transcript levels in c162 were approximately twice those in c1655 under standard conditions. This difference under standard conditions may suggest regulatory elements found within the context of the native gene that are not present in the complementing construct, however the difference did not affect complementation of the cdr mutation (see below).
Figure 4.
Transcriptional analyses of cdr expression in c162 (wt), c309 (mut), and c1655 (ct) grown in vitro under standard and anaerobic conditions. RNAs were harvested from spirochetes grown to late exponential phase at 37°C in either the presence (std) or absence (no O2) of oxygen. The levels of cdr were assayed in quadruplicate from three independently-derived samples by qRT-PCR and normalized to those of the flaB gene (84). Error bars indicate the standard error of the mean (SEM). The dagger (†) indicates significantly less cdr transcript in c162 grown under anaerobic conditions compared to the amount observed in c162 grown under standard conditions. The asterisk (*) denotes significantly less cdr in either c309 or c1655 relative to the levels observed in c162 grown under the same condition (P < 0.05).
The role of cdr in defense against peroxide stress
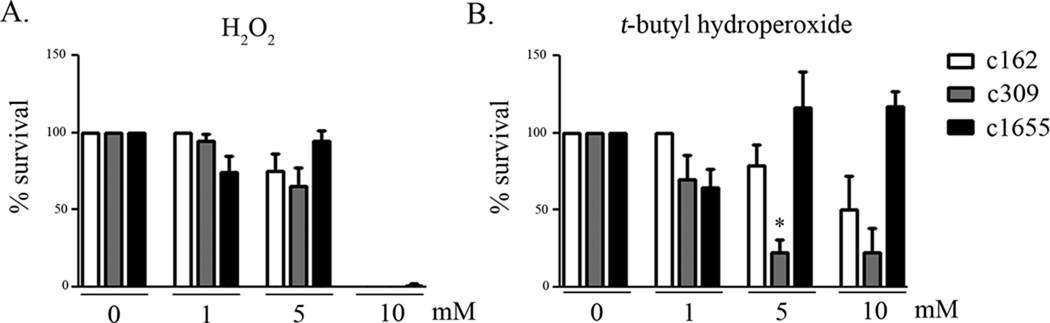
Boylan et al., 2006;previously have proposed that CoADR protects the spirochete against H2O2based on their demonstration that CoA was able to reduce this oxidant in vitro in a process that required CoADR activity to replenish the reduced CoA. To determine whether a cdr mutant is more sensitive to exogenous peroxide than the wild-type, c162, c309 (mut) and c1655 (ct) initially grown under standard conditions were exposed to concentrations of H2O2 from 0 to 10 mM (Figure 5A). Although both c162 and c309 showed modest sensitivity to exposure at 5 mM H2O2and none of the clones had measurable survival at 10 mM H2O2, we observed no significant difference in the sensitivity of c309 to H2O2-stress relative to c162 or c1655 at any concentration.
Figure 5.
Sensitivity of the cdr mutant to exogenous H2O2 or t -butyl hydroperoxide. c162 (wt), c309 (mut), and c1655 (ct) were exposed to the indicated concentration of (A) H2O2 or (B) t-butyl-hydroperoxide for 1 hr in media lacking BSA and rabbit serum. After exposure, the treated samples were diluted in fresh BSK-II and then serially-diluted 1:1 to 2.4 × 10−1 spirochetes per well. Wells were analyzed after three weeks for spirochetes. Results shown are the average of at least three replicates. Percentages are plotted relative to untreated sample of the same clone. Error bars indicate the SEM. The asterisk (*) indicates a significant difference between the mutant and wild-type clones.
The primary targets of ROS in B. burgdorferi appear to be spirochetal membranes (Boylan et al., 2008; Boylan and Gherardini, 2008). To test whether a cdr mutant is more sensitive than the wild-type to membrane damage, c162, c309, and 1655 were exposed to 0 to 10 mM of t -butyl-hydroperoxide (Figure 5B), an oxidizing agent that has been demonstrated to initiate lipid peroxidation in B. burgdorferi outer membranes in vitro (Boylan et al., 2008; Boylan and Gherardini, 2008). At every concentration of t -butyl-hydroperoxide, the mutant appeared more sensitive to exposure than the wild-type; the difference between the two was significant at 5 mM (P < 0.05). This difference was due to the absence of the cdr gene, as the complement was not significantly more sensitive than the wild-type at any concentration.
Spirochetes lacking BbCoADR exhibit a growth defect under both aerobic and anaerobic conditions
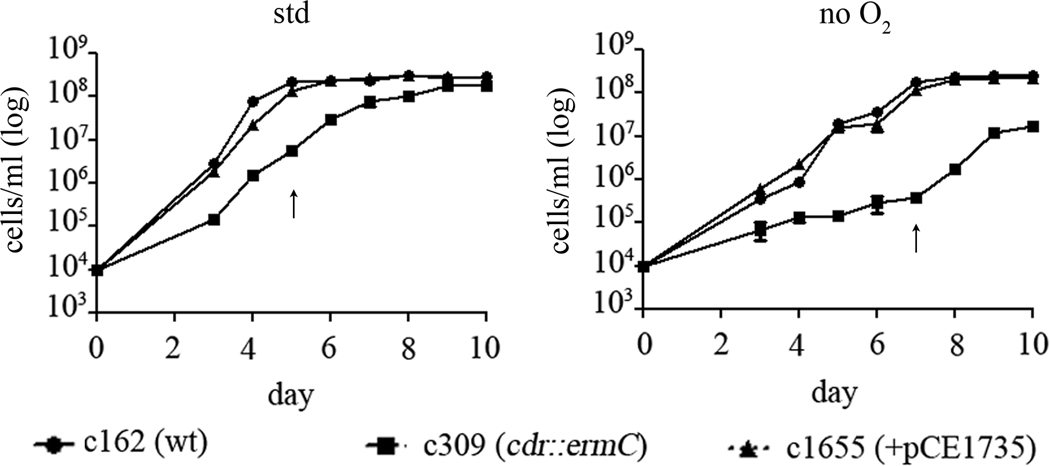
Given the potential role(s) of CoADR in both intermediary metabolism and oxidative stress response, we wanted to determine if the cdr mutant exhibited a growth defect and, if so, whether it was influenced by the presence or absence of oxygen. Accordingly, c162 (wt), c309 (mut), and 1655 (ct) were grown at 37°C under standard or anaerobic conditions (Figure 6). c162 reached equivalent densities under both conditions, although it entered stationary phase approximately 48 hours later under anaerobic conditions (arrows in Figure 6). Under standard conditions, c309 exhibited a significant growth defect (Figure 6, left panel). By day 10, the last day on which growth was measured, the density of c309 plateaued at 70% of the final density of c162. The growth defect of c309 was even more severe under anaerobic conditions (Figure 6, right panel); the density of c309 at day 10 was only 8% that of the wild-type and complemented clones and did not rise appreciably by day 14 (not shown). Under both conditions, c162 and c1655 grew indistinguishably.
Figure 6.
The cdr mutant grownin vitro exhibits a growth defect that is even more severe during anaerobiasis. c162 (wt), c309 (mut), and c1655 (ct) were grown at 37°C under standard and anaerobic conditions. Density was measured daily using a Petroff-Hauser counting chamber. Each assay was done in triplicate and error bars represent the SEM. The point at which RNA samples were taken for Figure 4 is indicated by the arrows.
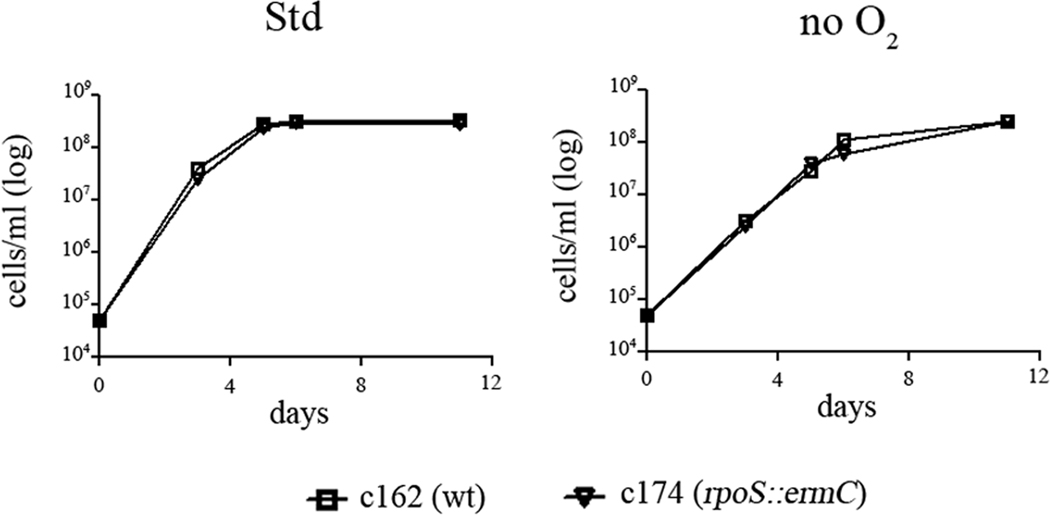
At 37°C in vitro, more than 50% of the expression of cdr is RpoS-dependent (Caimano et al., 2007). To determine if an rpoS -mutant exhibits the same in vitro growth defect as observed with the cdr mutant, we compared the growth of the rpoS -mutant c174 (Caimano et al., 2004; Eggers et al., 2004; Caimano et al., 2005)(Table 1) with that of its parent, c162, grown at 37°C under both standard and anaerobic conditions. In contrast to the cdr -mutant (Figure 6), the rpoS -mutant grew as well as the wild-type under both conditions (Figure 7). Thus, the RpoS-dependent component of cdr expression is not required for normal in vitro growth in BSK-II medium.
Figure 7.
An rpoS -mutant grown in vitro exhibits no growth defect. c162 (wt) and c174 (rpoS mutant) was grown at 37°C under standard and anaerobic conditions. Density was measured daily using a Petroff-Hauser counting chamber. Graphs shown are representative of three independent experiments.
BbCoADR is required for infectivity in the murine model
To test the infectivity of the cdr mutant, we needle-inoculated two groups of five C3H/HeJ mice with 1 × 104spirochetes of either clone c162 (wt) or c309 (mut). Eight weeks post-inoculation, infection was assessed by serology and ear tissue biopsy. All of the mice infected with c162 developed a robust immune response to multiple borrelial antigens, whereas the mice inoculated with c309 failed to seroconvert, developing antibodies to only one major 20-kDa band; this band was confirmed to be OspC by blotting against purified OspC protein (data not shown and Table 3). All mice infected with c162 also were culture-positive, while spirochetes were not recovered from mice inoculated with c309 (Table 3).
Table 3.
Results of mouse infectivity studies.
| Strain | exp | Mouse background |
Seroconversiona | Culture positive |
Visible arthritisb |
|---|---|---|---|---|---|
| C162 (wt) |
1 | C3H/HeJ | 5/5 | 5/5 | ND |
| 2 | SCID | ND | 4/4c | 4/4 | |
| 3 | SCID | ND | 5/5 | 5/5 | |
| c309 (mut) | 1 | C3H/HeJ | 0/5d | 0/5 | ND |
| (c162 cdr∷ermC) | 2 | SCID | ND | 0/5 | 0/5 |
| 3 | SCID | ND | 0/5 | 0/5 | |
| c1655 (ct) | 1 | --e | -- | -- | -- |
| (c309+pCE1735) | 2 | SCID | ND | 5/5 | 4/5 |
| 3 | SCID | ND | 5/5 | 5/5 | |
as determined by blotting Bb antigens with 8-wk mouse sera;
observed 4 weeks after infection;
one mouse died prior to evaluation;
an immune response to only one antigen, OspC, was detected;
the infection of C3H/HEJ mice with c1655 was not done (see text); ND, not determined
To confirm that the avirulence of c309 was due to the lack of CoADR, we inoculated SCID mice with 1 × 104spirochetes of clones c162, c309, or the complement, c1655. SCID mice were used because c1655 lacks 1p28-1, which is required for survival in immunocompetent mice (Labandeira-Rey and Skare, 2001; McDowell et al., 2002; Labandeira-Rey et al., 2003; Lawrenz et al., 2004). Four weeks post-inoculation, infection was assessed by culture of ear punch biopsies. As presented in Table 3, all mice inoculated with c162 or c1655 were culture-positive, whereas no spirochetes were isolated from c309. Additionally, all mice infected with c162 and nine of the ten mice inoculated with c1655 also exhibited visible swelling in the rear ankle joints, whereas none of those inoculated with c309 had visible swelling (Table 3 and data not shown).
Transcriptional profiling of cdr within ticks
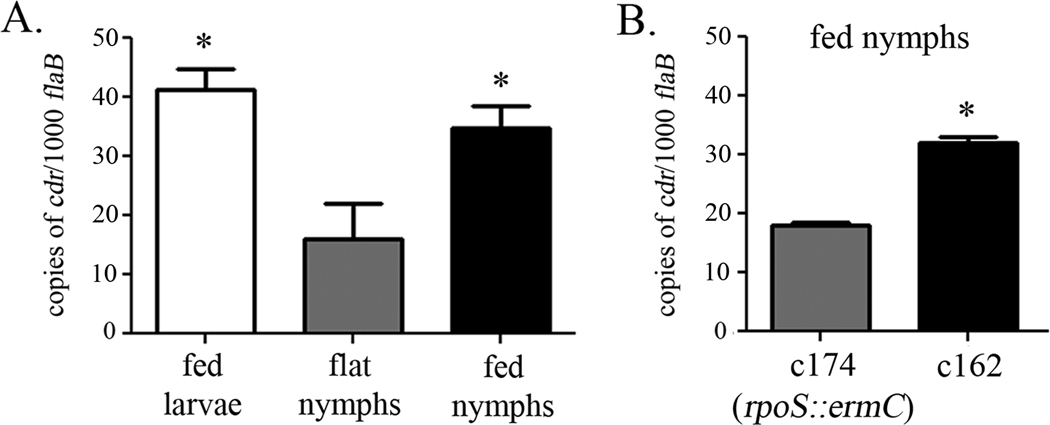
The expression profile of cdr had previously been characterized in vitro at 23°C and 37°C and in dialysis membrane chambers (DMCs) implanted into the peritoneal cavity of rats (Caimano et al., 2007). Transcriptional analyses of spirochetes within ticks were next performed to gain insight into the contribution of the gene to spirochetal physiology during the tick phase of the enzootic cycle. qRT-PCR demonstrated that cdr is expressed in all three tick phases; of note, spirochetes in fed nymphs and larvae contain on average, more than twice as much cdr transcript as spirochetes in unfed nymphs (Figure 8A). RpoS is not expressed in fed larvae or unfed nymphs, but is induced during the nymphal blood meal (Caimano et al., 2007). To determine whether the increase in cdr transcript observed within the midguts of fed nymphs was due to transcription by RpoS, we immersion-fed nymphs with both c162 (wt) and c174 (rpoS mutant)(Policastro and Schwan, 2003; Mulay et al., 2009). After a brief rest period, these ticks were allowed to feed on uninfected mice. RNA was harvested from the midguts of engorged nymphs and cdr transcript was analyzed by qRT-PCR. As shown in Figure 8B, in fed nymphs, levels of cdr in the rpoS mutant are approximately 50% of those in the wild-type, confirming the partial RpoS-dependence of cdr expression under certain conditions (Caimano et al., 2007).
Figure 8.
Expression profiling analysis of cdr expression within ticks. (A) RNA was harvested from the midguts of c162-infected fed larvae, molted flat nymphs, and fed nymphs or (B) from the midguts of either c162- or c174-infected nymphs that had been fed on a mouse. The levels of cdr transcript in a pool of five ticks from each condition were analyzed in quadruplicate by qRT-PCR and normalized to those of the constitutively produced flaB gene (Ge et al., 1997). Error bars indicate the standard error of the mean (SEM). The asterisk (*) indicates significantly more cdr transcript (P < 0.05) in fed larvae and fed nymphs as compared to unfed flat nymphs(A), or in c162 within fed nymphs as compared to c174 within fed nymphs (B).
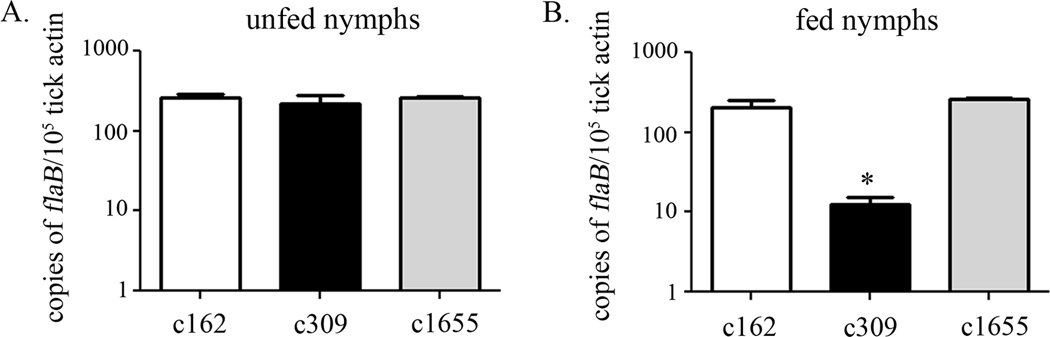
Loss of cdr affects survival of spirochete in fed, but not flat nymphs
To determine whether Bb CoADR is required within the midguts of flat and/or feeding nymphs, equivalent numbers of c162 (wt), c309 (mut), and c1655 (ct) (∼104 spirochetes/tick) were microinjected into the midguts of Ixodes scapularis nymphs (Pal et al., 2004; Kariu et al., 2011). While immersion feeding can be used to generate large numbers of infected larvae or nymphs (Policastro and Schwan, 2003), for these experiments microinjection was used instead because the number of spirochetes delivered could be standardized, 100% infectivity could be ensured, and more rapid results could be acquired (Kariu et al., 2011). After a ten day rest period, spirochete burdens in unfed nymphs were assessed by qRT-PCR analysis of B. burgdorferi flaB mRNA and normalized against the tick β-actin gene as previously described (Zhang et al., 2009). The levels of flaB mRNA in unfed nymphs were virtually identical for all three clones (Figure 9A). During feeding, however, the numbers of the cdr mutant in the midgut of fed nymphs were significantly lower (P < 0.05) than those of either the wild-type or the complemented mutant (Figure 9B). These data indicated an inability of the cdr mutant to expand appropriately within the midgut. We also observed that equivalent numbers of SCID mice became infected after being fed on by nymphs colonized with either c162 or c1655; no SCID mice fed upon by nymphs colonized with c309 became infected (data not shown).
Figure 9.
Bb CoADR promotes survival of spirochetes within fed, but not flat, nymphs. Ixodes scapularis nymphs were rectally-infused with equivalent densities of wild-type (c162), cdr mutant (c309), and complemented (c1655) clones of B. burgdorferi. (A) After a ten day ‘rest’ period, relative B. burgdorferi burdens in unfed ticks were analyzed by qRT-PCR; the number of copies of the B. burgdorferi flaB transcript were normalized to tick β-actin RNA. The remainder of the rectally-infused nymphs were then allowed to feed upon SCID mice to repletion. (B) B. burgdorferi burdens in fed nymphs were monitored by qRT-PCR. The asterisk (*) denotes a significant difference between spirochetes in the midguts of fed nymphs infused with the cdr mutant compared to those infused with either the wild-type or the complement (P < 0.05). The data represent the average of four replicates of qRT-PCR analysis on two pools of three ticks (unfed) or 1–2 ticks (fed) from a representative experiment.
Discussion
Dissecting the functional role(s) of a borrelial enzyme that potentially contributes to the spirochete’s oxidative stress response as well as intermediary metabolism is complicated by the fact that both increased oxygen levels and signals to stimulate replication are delivered simultaneously within blood and during mammalian infection (Seshu and Skare, 2000; Tilly et al., 2001; Piesman et al., 2001; Zeidner et al., 2001; Anguita et al., 2003; Pal and Fikrig, 2003; Seshu et al., 2004; Boylan et al., 2006; Boylan et al., 2008; Dunham-Ems et al., 2009; Xu et al., 2010). Using a strategy combining in vitro and in vivo analyses, we have presented evidence supporting the conclusion that CoADR functions in both capacities to promote spirochetal fitness. In particular, our observation that the cdr mutant has a much greater growth impairment when cultivated in vitro anaerobically than with oxygen strongly implies a homeostatic role for CoADR independent of defense against ROS.
The enzymatic mechanisms B. burgdorferi employs to protect itself against different types of oxidative stress are poorly understood. B. burgdorferi encodes few homologues to proteins known to participate in the oxidative stress response of other bacteria (Fraser et al., 1997; Gherardini et al., 2010; Parsonage et al., 2010) (Table 4). In addition to CoADR, B. burgdorferi encodes only a single member of the TFP class (thioredoxin, Trx; bb0061), a thioredoxin reductase (TrxB; bb0515),a Dps/Dpr homologue (NapA; bb0690) and a superoxide dismutase (SodA; bb0153)(Fraser et al., 1997; Seshu et al., 2004; Boylan et al., 2006; Li et al., 2007; Esteve-Gassent et al., 2009; Parsonage et al., 2010). Conspicuous in their absence are peroxiredoxins, glutathione peroxidases, catalases, and both glutathione biosynthetic and glutaredoxin proteins(Fraser et al., 1997; Gherardini et al., 2010; Parsonage et al., 2010) (Table 4). Based on their in vitro biochemical activities, CoADRs have been proposed to be important for maintaining the intracellular thiol-disulfide ratio and possibly in protecting the cell from ROS (delCardayre and Davies, 1998; delCardayre et al., 1998; Argyrou and Blanchard, 2004; Harris et al., 2005; Boylan et al., 2006; Wallen et al., 2008). Consistent with this notion, the B. burgdorferi cdr mutant exhibited a growth defect under in vitro and in vivo conditions in which higher levels of ROS were more likely to be present (Imlay, 2003). Furthermore, we found that in wild-type B. burgdorferi, cdr was expressed more than three times higher under standard (aerobic) conditions than under anaerobic conditions, as would be expected for a gene encoding a protein involved in the defense against noxious oxidative molecules (Stanton et al., 1999; Cabiscol et al., 2000). Thus, it appears likely that CoA and CoADR function analogously to glutathione and GR in Eukaryota and most Gram negative bacteria to maintain an appropriate intracellular thiol/disulfide ratio(Holmgren, 1989; Carmel-Harel and Storz, 2000a; Boylan et al., 2006; Imlay, 2008). It is important to note, however, that while the biochemical mechanisms whereby glutathione and Grx maintain reduced disulfide bonds in cellular proteins are well understood (Holmgren, 1989; Carmel-Harel and Storz, 2000a), it is not known whether CoASH has a role in reducing disulfide bonds in B. burgdorferi, nor what the mechanism for this activity might be.
Table 4.
Bioinformatics of genes involved in oxidative stress response and CoA biosynthesis in B. burgdorferi.
| Protein | Functional homolog/PDB entry | Comparison or reference |
|---|---|---|
| Oxidative stress responsea | ||
| BB0728 (cdr) | CoADR/3CGC (B. anthracis) | 40% identical, E value of 4 × 10−97 (Boylan et al., 2006) |
| BB0729 (tcyP) | Na+/cystine symporter (Bacillus subtilis) | 50% identical, E value of 6 × 10−111 |
| BB0515 (trxB) | thioredoxin reductase | (Fraser et al., 1997) |
| BB0061 (trxA) | thioredoxin | (Fraser et al., 1997) |
| BB0153 (sodA) | superoxide dismutase | (Esteve-Gassent et al., 2009) |
| BB0690 (dps) | Dps/NapA homolog | (Li et al., 2007) |
| Nox | NADH oxidase | Noneb |
| GshA | γ-Glu-Cys synthetase | None |
| GshB | GSH synthetase | None |
| GR | glutathione reductase | None |
| Grx | glutaredoxin | None |
| Gpx | glutathione peroxidase | None |
| AhpF | alkylhydroperoxide reductase | None |
| Prx | peroxiredoxin | None |
| CoA biosynthesisc | ||
| BB0814 (panF) | Na+/Pan symporter (E. coli) | 34% identical, E value of 2 × 10−69 |
| BB0527 (coaX; panK) |
Pan kinase/2H3G (B. anthracis) | 28% identical, E value of 2 × 10−24 |
| BB0812 (coaBC) | phosphopantothenoylcysteine synthetase/1U7U (E. coli) |
38% identical, E value of 2 × 10−27 |
| BB0702 (coaD) | phosphopantetheine adenylyltransferase/3F3M (S. aureus) |
43% identical, E value of 1 × 10−28 |
| BB0547 (coaE) | dephospho-CoA kinase/1JJV (Haemophilus influenza) |
30% identical, E value of 8 × 10−12 |
| BB0704 (acpP) | acyl carrier protein/2KWL | identical |
| PanB | ketopantoate hydroxymethyl- transferase |
None |
| PanC | Pan synthetase | None |
| PanD | aspartate decarboxylase | None |
| PanE | 2-dehydropantoate 2-reductase | None |
see also (Parsonage et al., 2010);
None indicates that there has been no homolog identified within B. burgdorferi;
see also (Boylan et al., 2006; Gherardini et al., 2010)
While CoADR may play a role in supporting intracellular thiol-disulfide homeostasis, the absence of this enzyme failed to render spirochetes more sensitive than the wild-type to high concentrations of H2O2. These results call into question the physiologic relevance of the in vitro observation that CoADR could participate in the detoxification of H2O2by reducing CoA-disulfides generated by the reaction of CoA with peroxide (Boylan et al., 2006). Membrane lipids are the primary target of ROS in B. burgdorferi (Boylan et al., 2008). The cdr mutant was more sensitive than the wild-type to high levels of t -butyl-hydroperoxide, an organic hydroperoxide known to initiate lipid peroxidation in B. burgdorferi membranes (Boylan et al., 2008; Boylan and Gherardini, 2008). The direct detoxification of organic hydroperoxides in bacteria is primarily catalyzed by peroxiredoxin TFPs distinct from the members of the FDR family that includes CoADR(Argyrou and Blanchard, 2004; Poole, 2005; Parsonage et al., 2008). In fact, B. burgdorferi contains no homologs to enzymes capable of detoxifying organic peroxides (Fraser et al., 1997; Parsonage et al., 2010). Thus, the greater sensitivity of the cdr mutant to exogenous t -butyl hydroperoxide most likely reflects the requirement for the cytoplasmic CoADR to ensure an adequate pool of acetyl-CoA or acyl-CoA (via maintenance of optimal levels of intracellular CoA) for synthesis of the lipids, glycolipids, or lipoproteins needed for membrane repair following lipid peroxidation (Gherardini et al., 2010).
The likely contributions of CoADR to the metabolism of B. burgdorferi can be deduced from its known biochemistry (Boylan et al., 2006). In addition to making CoASH available for formation of acetyl-CoA, a precursor for many biosynthetic reactions (Fraser et al., 1997; Wolfe, 2005; Gherardini et al., 2010; Xu et al., 2010), CoADR also supplies the NAD+ that is essential for substrate-level phosphorylation through glycolysis, the sole mechanism for ATP generation in B. burgdorferi (Fraser et al., 1997). Utilizing The SEED resource(Overbeek et al., 2005), we identified only a few B. burgdorferi proteins presumed or known to rely on NAD(H): glyceraldehyde-3-phosphate dehydrogenase (bb0057), lactate dehydrogenase (LDH) (bb0087), glycerol-3-phosphate dehydrogenase (bb0368),hydroxymethylglutaryl (HMG)-CoA reductase (bb0685), and CoADR. The other two members (Nox and Npx) of the POR subgroup of the FDR family also contribute to the obligatory role of NAD+ in the strictly fermentative energy metabolism of Streptococcus and other lactic acid bacteria (Higuchi et al., 1999; Gibson et al., 2000).While Bb CoADR is the only CoADR thus far experimentally determined to have a preference for NADH, our bioinformatics analysis indicates the presence of several additional enzymes among the γ-proteobacteria that are predicted to be NADH-specific. In general, FDR enzymes that utilize NAD(H) are more likely involved in the regeneration of oxidized pyridine nucleotides for glycolysis, whereas those that are NADPH-dependent tend to be involved in the maintenance of thiol/disulfide homeostasis(Argyrou and Blanchard, 2004; Harris et al., 2005; Ying, 2006; Ying, 2008).
In contrast to the growth defect observed during anaerobiasisin vitro, we found that the cdr mutant did not exhibit a loss of viability when maintained within the anaerobic environment of the unfed tick midgut(Seshu et al., 2004) for ten days following rectal infusion. Spirochetes cultured anaerobically in vitro replicate exponentially, whereas they replicate minimally or not at all within the unfed midgut(Zeidner et al., 2001; Piesman et al., 2003). Thus, the metabolic requirements of the spirochetes in the unfed tick differ significantly from those of organisms grown in BSK-II medium in culture, further suggesting that the defect observed in spirochetes grown anaerobically in vitro could be due to metabolic deficiencies such as inadequate NAD+ or CoASH. We also found that the cdr mutant exhibited a growth defect within the feeding nymph and within the mammalian host. Both are environments in which spirochetes encounter increased oxygen tensions and concentrations of ROS (Seshu et al., 2004; Boylan et al., 2006; Gherardini et al., 2010) but also in which they undergo marked replication (Piesman et al., 2001; Zeidner et al., 2001; Piesman et al., 2003; Hodzic et al., 2003; Dolan et al., 2004). We hypothesize that mutant spirochetes failed to thrive in both host environments at least in part because they were unable to generate sufficient energy and the precursors required to generate new cell envelopes.
Our results indicate that the metabolic importance of CoADR to B. burgdorferi is dependent upon the environment in which the spirochete is cultured. While we observed a defect with the cdr mutant grown under any condition with increased exposure to oxygen or increased replication requirements, only within the mammalian host did CoADR appear to be essential. Why is the cdr mutation not equally detrimental under all conditions? The different phenotypes observed with the cdr mutant in various environments may reflect the availability of differentially-expressed redundant metabolic pathways that enable the spirochete to overcome the lack of CoADR to varying extents. For example, although the thioredoxin-dependent reduction system has not been characterized in B. burgdorferi, it presumably functions in the maintenance of the thiol/disulfide balance as in other systems (Holmgren, 1989; Holmgren, 2000; Carmel-Harel and Storz, 2000a; Holmgren et al., 2005); perhaps under certain conditions this pathway can partially complement the function(s) of CoADR; in other organisms with two disulfide reduction systems, single mutants are often not hypersensitive to oxidants because of partial overlap in those systems (Carmel-Harel and Storz, 2000a).
The ability of B. burgdorferi to use CoADR to fulfill roles in both thiol/disulfide homeostasis and intermediary metabolism is predicated on the idea that the spirochete can synthesize and maintain a large pool of CoA. While de novo biosynthesis of pantothenate (Pan) in B. burgdorferi from α-ketoisovalerate and aspartate is absent (pan BCDE genes in E. coli), B. burgdorferi encodes a putative Pan transporter (BB0814, PanF, Na+/Pan symporter, Table 4)(Boylan et al., 2006; Gherardini et al., 2010); Pan is a precursor for CoA synthesis (Jackowski and Rock, 1981; Jackowski, 1996; Gherardini et al., 2010) and Gherardini and colleagues (Boylan et al., 2006; Gherardini et al., 2010) previously have proposed that the spirochete can synthesize CoA using Pan acquired from their environment. Based on bioinformatics, we also have identified a type III Pan kinase ortholog (PanK, BB0527; Table 4) for production of 4´-phosphopantothenate (Pan-4´-P!). The use of a type III PanK allows for uncoupling of the kinase reaction from CoASH feedback inhibition(Nicely et al., 2007; Paige et al., 2008) and is consistent with the spirochete’s dependence on CoA as the major low molecular weight thiol(Boylan et al., 2006).The spirochete also contains coaBC, coaD, and coaE loci predicted to encode proteins for conversion of Pan-4´-P! to CoASH(Boylan et al., 2006; Gherardini et al., 2010) (Table 4).Interestingly, the coaD (BB0702) locus is associated with a cluster that includes the acpP (BB0704) coding sequence; acyl carrier protein (AcpP) functions in fatty acid synthesis via the obligatory participation of the covalently bound 4´-phosphopantetheine cofactor derived from CoASH(Chan and Vogel, 2010; Gherardini et al., 2010). One other metabolite necessary for the synthesis of CoA is L-cysteine (Jackowski and Rock, 1981; Jackowski, 1996). bb0729, the upstream gene co-transcribed with cdr, has homology to the Bacillus subtilis TcyP protein (Overbeek et al., 2005), a Na+/cystine symporter(Burguiere et al., 2004) (Table 4); cystine is the oxidized form of L-cysteine. The genomic proximity of genes encoding TcyP homologs and CoADRs is found in other non-borrelial bacteria as well, including other pathogens such as Clostridium difficile. Given the potential relationship between cystine transport and CoA biosynthesis, the metabolic linkage between Bb CoADR and the product of bb0729 warrants further investigation.
Our interest in cdr stemmed in part from our earlier finding that it is one of a small number of genes within the RpoSregulon that is dually-transcribed by σ70 and RpoS following temperature shift or cultivation within DMCs (Caimano et al., 2007). Two observations made herein have afforded additional insight into the biological context and rationale for dual transcription of the gene. First, we showed that cdr also is dually-transcribed during nymphal feeding, the stage in which RpoS-dependent gene expression begins. Thus, co-transcription of this operon is the rule when both RpoS-dependent and –independent gene expression are occurring. The ability of both sigma factors to control expression of the bb0729-cdr operon is explained by our finding that cdr and bb0729 appear to be co-transcribed from a single hybrid promoter with consensus sequence elements consistent with both RpoS- and σ70-dependent promoters (Caimano et al., 2007). Second, we found that transcription of cdr in feeding larvae, when RpoS is OFF, occurs at comparable levels to that in feeding nymphs. These results clearly imply that comparable levels of the enzyme are required when spirochetes are replicating rapidly regardless of whether RpoS is ON or OFF (Piesman et al., 2001; Zeidner et al., 2001; Piesman et al., 2003; Caimano et al., 2007). We find it intriguing that σ70 is capable of achieving full transcription of the gene in the absence of RpoS during larval acquisition but is unable to compensate for its loss during nymphal feeding. This result suggests that another transcription factor either enhances σ70-dependent replication within spirochetes found in larval ticks or represses σ70-dependent replication within spirochetes found in feeding nymphs. Based (i) on data showing that BosR could bind upstream of the bb0729-cdr operon promoter and enhance transcription of cdr in an E. coli -based surrogate system (Boylan et al., 2006) and (ii) on the finding that a bosR mutant failed to regulate CoADR protein expression appropriately(Hyde et al., 2009), the Fur homolog BosR seems like a potential candidate transcription factor for in vivo regulation of cdr (Boylan et al., 2006; Hyde et al., 2009).Interestingly, however, reported consensus sequences identified for BosR-binding sites (Boylan et al., 2003; Katona et al., 2004; Ouyang et al., 2011)were not found directly upstream of the transcriptional start of the bb0729-cdr operon, and cdr was not identified as BosR-dependent in a recent microarray analysis of a bosR mutant (Ouyang et al., 2009). Thus, whether BosR directly regulatescdr transcription in vivo has yet to be determined.
If the spirochete can achieve transcript levels with σ70 sufficient for its metabolic needs in the larvae, why is cdr expression partially RpoS-dependent during the nymphal blood meal? The answer may lie in the interrelationship between the function and regulation of the RpoSregulon. RpoS-enhanced expression of cdr is not needed in the nutrient-rich BSK-II culture medium, however recent ongoing work suggests that RpoS-enhanced levels of CoADR are required in the feeding nymph or under nutrient-limiting conditions in vitro (Dunham-Ems &Radolf, unpublished data). While the link between RpoS and the CoADR is still under investigation, there is now a growing body of evidence that the physiological changes required to execute the spirochete’s infectious program are intrinsically-linked to the Ack-Pta pathway for the synthesis of acetyl-CoA from acetate (Sanjuan et al., 2009; Xu et al., 2010; Karna et al., 2011; Sze and Li, 2011). In B. burgdorferi, AckA (BB0622) converts exogenously acquired acetate into acetyl-P, which is itself converted into acetyl-CoA by the enzyme Pta (BB0589) (Xu et al., 2010; Gherardini et al., 2010). Acetyl-P also has been shown to activate the response regulator, Rrp2, by phosphorylation(Xu et al., 2010). Phosphorylated Rrp2 acts in concert with RpoN and BosR to transcribe RpoS(Hubner et al., 2001; Yang et al., 2003; Caimano et al., 2004; Fisher et al., 2005; Ouyang et al., 2008; Boardman et al., 2008; Hyde et al., 2009; Ouyang et al., 2009; Blevins et al., 2009; Mulay et al., 2009; Xu et al., 2010; Ouyang et al., 2011). Thus, an increase in acetate, such as might occur with an influx of blood, increases acetyl-P, which ultimately activates the expression of RpoS-dependent virulence factors, including CoADR. Abundant CoADR would ensure higher levels of CoASH that could be used in the formation of acetyl-CoA. Acetate, CoA, and acetyl∼P levels would work synergistically to modulate RpoS expression and, ultimately, the expression of RpoS-dependent virulence related genes.
Experimental Procedures
Computer modeling
The three-dimensional model of Bb CoADRwas generated by the SWISS-MODEL server based on the structure of the reducedBacillus anthracis CoADR (Ba CoADR)-NADH complex [PDB code: 3CGD(Wallen et al., 2008)](Arnold et al., 2006; Bordoli et al., 2009). PyMOL (Schrödinger, New York, NY) was used to generate the structural graphics.
Bacterial strains and culture conditions
All PCR primers used in this study are listed in Table 2.Routine cloning and plasmid propagation were performed using Eschericia coli strain Top10 (Invitrogen). All strains were maintained at 37°C in Luria-Bertani broth (LB) (1% Tryptone, 0.5% yeast extract, 1% NaCl) with the appropriate antibiotic. Preparation and transformation of chemically-competent E. coli were performed as described previously (Ausubel et al., 1997). Solid phase selection was performed on LB agar plates (LB with 1.5% agar) supplemented with the appropriate antibiotic.
All experiments with B. burgdorferi were performed using c162 (Caimano et al., 2007), a virulent clone of B. burgdorferi strain 297 (Steere et al., 1984), or its derivatives: c174 (rpoS∷ermC) (Caimano et al., 2004; Eggers et al., 2004; Caimano et al., 2005), c309 (cdr∷ermC), or c1655 (c309+pCE1735) (Table 1). Spirochetes were routinely cultivated in modified Barbour-Stoenner-Kelly medium supplemented with 6% rabbit serum (Pel-Freez Biologicals, Rogers, AK)(BSK-II)(Samuels, 1995); when appropriate, selection and maintenance of B. burgdorferi clones was performed in media supplemented with 0.06 µg ml−1 erythromycin (c309 and c174), 400 µg ml−1 kanamycin (JH300) or 0.06 µg ml−1 erythromycin and 400 µg ml−1 kanamycin (c1655). For routine cultivation, spirochetes were maintained at 33°C in liquid culture in tightly-capped containers with a low void volume [micro aerobic(Seshu et al., 2004)]. For experiments comparing growth in the presence or absence of oxygen (see below), B. burgdorferi was grown at 37°C in containers with loose lids either in a chamber with an enhanced CO2 environment (15% O2, 6% CO2) generated by the addition of an enhanced CO2 packet (GasPak™ EZ CO2 Container System; Becton-Dickinson) (termed ‘standard’ conditions) or in a chamber with an anaerobic environment (<1% O2, 9–13% CO2) created by the inclusion of an anaerobic packet (BBL GasPak Plus Anaerobic System; Becton-Dickinson); anaerobiasis was monitored by the addition of BBL Dry Anaerobic Indicator Strips (Becton-Dickinson). For all experiments, spirochetes were passaged no more than three times before experimental manipulations were performed; plasmid contents were monitored by PCR as described previously (Eggers et al., 2002).
Generation of a bb0728 (cdr) mutant
To generate a bb0728 (cdr) mutant, the entire cdr open-reading frame was first amplified by PCR from c162 using primers 2 and 3(Table 2) and TaKaRa ExTaq high fidelity polymerase (Fisher Scientific). The amplified fragment was digested with XhoI and BamHI and ligated into digested pBSII-SK+ (Stratagene), generating pBS-cdr. The ermC gene was amplified from pGK12 (Kok et al., 1984; Sartakova et al., 2000) (provided by F.C. Cabello, New York Medical College, Valhalla, New York) using primers 5 and 8 and cloned into pCR2.1-TOPO as instructed by the manufacturer (Invitrogen, Carlsbad, CA). The cloned ermC gene then was removed from its vector by digestion with Eco RI and ligated into Eco RI-digested pBS-cdr, which cuts within the cdr gene at positions 197 and 1203. The cdr ∷ermC construct was amplified using the vector primers T7 and M13R.
Approximately 10 µg of amplified cdr∷ermC was electroporated into electrocompetent c162, as previously described (Samuels, 1995; Eggers et al., 2002). Cells were electroporated and recovered at 33°C under either microaerobic or anaerobic conditions. After overnight recovery, each batch was expanded into 40 ml BSK-II supplemented with 0.06 µg/ml erythromycin and then dispensed into two 96-well plates in 200 µl aliquots, as described previously (Caimano et al., 2004). The samples were incubated under either standard or anaerobic conditions. Erythromycin-resistant spirochetes recovered under both conditions were then screened for the anticipated mutation by PCR using primers 1, 4, 6, and 7. Multiple clones with the correct mutation were recovered; one mutant, c309, was selected for further characterization. Plasmid content was determined by PCR as described previously (Eggers et al., 2002).
Transcriptional analysis of the cdr operon
Total RNA was isolated from c162 cultivated in vitro at 37°C to approximately 5 × 107 cells/ml in BSK-II under microaerobic conditions using TRIzol reagent according to the manufacturer’s directions (Invitrogen). Contaminating DNA was removed using RQ1 RNase-free DNase (Promega) as previously described(Caimano et al., 2004). DNase-treated RNAs (1–4 µg total RNA per sample) were converted to cDNA using SuperScript First-Strand Synthesis for RT-PCR (Invitrogen) in the presence and absence of reverse transcriptase (RT) according to the manufacturer’s instructions. The primers used to detect bb0729 (10 & 12), cdr (13 & 15), and any transcript that extends between the two (11 & 14) are listed in Table 2. c162 DNA was used as a positive control and water alone was used as a negative control. Rapid amplification of the cDNA 5′-ends (5′-RACE) was performed using a 5′-RACE kit (Invitrogen) and the primers indicated in Table 2. Amplification using gene specific primers located within 5′ end of the cdr transcript yielded no product. The final product from the amplification of the region upstream of bb0729 was cloned into pCR2.1-TOPO as instructed by the manufacturer (Invitrogen). The product was sequenced using the vector primers M13R and T7.
Complementation of the cdr mutation in c309
To complement c309, the entire bb0729 and cdr open-reading frames, as well as a 500-bp segment upstream of bb0729 were PCR-amplified using primers 4 and 9. The amplified product was cloned into pCR2.1-TOPO as instructed by the manufacturer. The fragment was digested from the vector with SacI and NotI, purified from the gel using GeneClean II (Qbiogene), and ligated into SacI-NotI digested pCE323 (Eggers et al., 2005). Ten µg of the resulting vector, pCE1735, were then transformed into c309 and selected on solid-phase plates supplemented with kanamycin (Samuels, 1995). Several kanamycin-resistant colonies were selected and analyzed to ensure that they contained pCE1735 and to determine their plasmid content (Eggers et al., 2002). One clone, c1655, was selected for further characterization.
Quantitative real time reverse transcriptase (RT)-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) as previously described(Mulay et al., 2009). Contaminating genomic DNA was removed using Turbo DNAfree (Ambion, Inc. Austin, TX). DNase-treated RNAs (1–4 µg total RNA per sample) were converted to cDNA using SuperScript III (Invitrogen) in the presence and absence of reverse transcriptase (RT) according to the manufacturer’s instructions. cDNAs (+RT) were assayed in quadruplicate using iQ Supermix (Bio-Rad). The primers used to detect cdr and bb0729 transcripts are indicated in Table 2.flaB transcripts were assessed using SYBR Green or TaqMan probe-based assays. Transcript copy numbers were calculated using the iCycler post-run analysis software based on internal standard curves then normalized against copies of flaB.
Peroxide stress assays
For the oxidative stress assays, the densities of c162, c309, and c1655 were determined by darkfield microscopy using a Petroff-Hausser counting chamber. For each clone, 5 ml of BSK-II formulated without rabbit serum or BSA were inoculated to a density of 5 × 107 spirochetes per ml. Each tube was split into 4 × 1 ml aliquots and hydrogen peroxide or t -butyl hydroperoxide was added to the appropriate concentration (0–10 mM). The samples were incubated at 37°C for 1 h. After 1 h, 40 µl (2 × 106 cells) were added to 260 µl BSK-II and serially-diluted (1:1) to a final dilution of 2.4 × 10−1 spirochetes per well. Plates were placed at 37°C in a CO2-incubator and monitored for growth over a 3 week period as indicated by a color change. Wells that did not exhibit a color change were analyzed for the presence of spirochetes by darkfield microscopy. Any well that had motile spirochetes at a density above background was considered positive. Each exposure assay was done in triplicate and the assay was performed four times. Alternatively, after exposure to varying concentrations of oxidizing agent, a dilution equivalent to 100 cells of each clone was plated in solid media with the appropriate antibiotic as previously described (Samuels, 1995; Boylan et al., 2008; Boylan and Gherardini, 2008). The number of viable cells recovered at each concentration of oxidizing compound was compared to that recovered in the untreated samples. Each assay was performed in triplicate.
Growth curves
Growth kinetics of B. burgdorferi grown at 37°C in vitro under standard or anaerobic conditions were determined as follows. One hundred and fifty ml of media containing the appropriate antibiotic were inoculated with each isolate to a final density of 1 × 104 cells/ml and then distributed into ten 15 × 100 mm Petri dishes. One Petri dish each was placed in individual culture chambers under standard or anaerobic conditions. Multiple chambers were used to minimize the number of times cultures were exposed to atmospheric conditions. Starting with the third day after inoculation, one standard and one anaerobic chamber were opened and a sample of each culture removed for enumeration under darkfield using a Petroff-Hausser counting chamber (Hausser Scientific). Each B. burgdorferi clone was tested in triplicate. At intervals throughout the experiment, a 1 ml aliquot of culture was removed to confirm the consistency of the direct counts by using spectrophotometry as described previously (Samuels, 1995). For expression studies, RNA was harvested from samples in late exponential phase (Figure 6; arrows).
Mouse infectivity studies
To assess infectivity of wild-type and mutant B. burgdorferi strains, low-passage cultures were grown to mid-logarithmic density in BSK-II at 33°C. Five to eight week old C3H/HeJ or SCID (Jackson Laboratories) mice (five per group, per isolate) were inoculated intradermally with 104 spirochetes. Infection was determined four to eight weeks post-inoculation by cultivation of tissues in BSK-II medium and/or serologically, as indicated in results. For culturing, Borrelia antibiotic cocktail (0.05mg/ml sulphamethazole, 0.02 mg/ml phosphomycin, 0.05 mg/ml rifampicin, 0.01 mg/ml trimethoprin, and 0.0025 mg/ml amphotericin B) was added to the BSK-II to minimize contamination. In SCID mice, potential ankle swelling also was determined at four weeks post-inoculation. Serology was performed using whole cell lysates of wild-type B. burgdorferi grown at 37°C or purified recombinant OspC-HIS as previously described(Caimano et al., 2004; Mulay et al., 2009). All experimental procedures using mice were approved by and performed in accordance with the guidelines of the University of Connecticut Health Center Institutional Animal Care and Use Committee.
Tick-infection studies
To generate naturally-infected ticks, approximately 300–400 pathogen-free I. scapularis larvae (Oklahoma State University, Stillwater, OK) were placed on infected C3H/HeJ mice 2–3 weeks post-syringe inoculation, allowed to feed to repletion, then held in an environmental incubator until they had molted to the nymphal stage. To obtain fed nymphs, 10 to 12 infected unfedIxodes scapularis nymphs were confined to a capsule affixed to the backs of naïve C3H/HeJ mice as previously described(Mulay et al., 2009). Immersion-fed larvae and nymphs were generated as previously described (Mulay et al., 2009) according to the method described by Policastro and Schwan (Policastro and Schwan, 2003). Rectal microinjection of naïve nymphs was performed as previously described (Pal et al., 2004).
Bioinformatics
To generate the sequence alignment given in Figure 1C, BLASTP using the Bb CoADR as a query was run against all microbial genomes in the NCBI database that corresponded (6/2011) to Gram-negative organisms and spirochetes, excluding Borrelia. Candidate CoADR orthologs were identified based on conservation of key active-site residues and CLUSTALX2 (Larkin et al., 2007) was used to generate an alignment of the 17 CoADR sequences that resulted from distinct non-borrelial species. This alignment, which also includes Sa CoADR, Ba CoADR, Ph CoADR, and Bb CoADR, was then imported into ESPript (Gouet et al., 1999). Tthe SEED resource (Overbeek et al., 2005) was used for the identifaction of NAD(P)H-dependent dehydrogenases and reductases in B. burgdorferi..
Statistical analyses
To determine the statistical significance of observed differences, matching data points (n ≥ 3) were compared within GraphPad Prism v4.00 (GraphPad Software, San Diego, CA,USA) using an unpaired t -test with two-tailed P -values and a 95% confidence interval. Asterisks indicate a level of significance where P <0.05. Error bars represent the standard error of the mean (SEM).
Acknowledgments
We gratefully acknowledge the expert technical assistance of Morgan LeDoyt, Anna Allard, and Cynthia Gonzalez. We also thank Drs. Frank Gherardini (Rocky Mountain Laboratories), Julie Boylan (Rocky Mountain Laboratories), Star Dunham-Ems (University of Connecticut Health Center), Amit Luthra (University of Connecticut Health Center), and Arvind Anan (University of Connecticut Health Center) for valuable discussions and/or for sharing unpublished data.
Funding for this work was provided by AI-29735 (JDR and MJC), 3R01AI029735-20S1 (JDR and MJC), AI-26756 (JDR), AI085248 (MJC), AI-10573 (KROH), AI-080615 (UP), North Carolina Biotechnology Center grant 2011-MRG-1116 (AC),and the National Research Fund for Tick-Borne Diseases (MJC). Additional funding was provided by a Quinnipiac University School of Health Sciences Faculty Research Award (C.H.E.) and a QU Summer Undergraduate Research Fellowship (R.M.).
References
- Anguita J, Hedrick MN, Fikrig E. Adaptation of Borrelia burgdorferi in the tick and the mammalian host. FEMS Microbiol Rev. 2003;27:493–504. doi: 10.1016/S0168-6445(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: advances in chemistry and function. Prog Nucleic Acid Res Mol Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Benhnia MR, Wroblewski D, Akhtar MN, Patel RA, Lavezzi W, Gangloff SC, et al. Signaling through CD14 attenuates the inflammatory response to Borrelia burgdorferi, the agent of Lyme disease. J Immunol. 2005;174:1539–1548. doi: 10.4049/jimmunol.174.3.1539. [DOI] [PubMed] [Google Scholar]
- Blevins JS, Xu H, He M, Norgard MV, Reitzer L, Yang XF. Rrp2, a sigma54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J Bacteriol. 2009;191:2902–2905. doi: 10.1128/JB.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2008;76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Gherardini FC. Determining the cellular targets of reactive oxygen species in Borrelia burgdorferi. Methods Mol Biol. 2008;431:213–221. doi: 10.1007/978-1-60327-032-8_17. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, Crane EJ, III, Gherardini FC. Borrelia burgdorferibb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol. 2006;59:475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- Boylan JA, Lawrence KA, Downey JS, Gherardini FC. Borrelia burgdorferi membranes are the primary targets of reactive oxygen species. Mol Microbiol. 2008;68:786–799. doi: 10.1111/j.1365-2958.2008.06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci U S A. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguiere P, Auger S, Hullo MF, Danchin A, Martin-Verstraete I. Three different systems participate in L-cystine uptake in Bacillus subtilis. J Bacteriol. 2004;186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Gonzalez C, Radolf JD. Alternate sigma factor, RpoS, is required for the in vivo -specific repression of the Borrelia burgdorferi lp54-borne ospA and lp6.6 genes. J Bacteriol. 2005;187:7845–7852. doi: 10.1128/JB.187.22.7845-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi, but does control expression of one or more essential virulence determinants. Infect Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000a;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000b;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- Chan DI, Vogel HJ. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J. 2010;430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- delCardayre SB, Davies JE. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J Biol Chem. 1998;273:5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- delCardayre SB, Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- Dolan MC, Piesman J, Schneider BS, Schriefer M, Brandt K, Zeidner NS. Comparison of disseminated and nondisseminated strains of Borrelia burgdorferi sensu stricto in mice naturally infected by tick bite. Infect Immun. 2004;72:5262–5266. doi: 10.1128/IAI.72.9.5262-5266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, Radolf JD. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest. 2009;119:3652–3665. doi: 10.1172/JCI39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol Microbiol. 2002;43:281–295. doi: 10.1046/j.1365-2958.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol. 2004;186:7390–7402. doi: 10.1128/JB.186.21.7390-7402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Sigma factor selectivity in Borrelia burgdorferi : RpoS recognition of the ospE /ospF /elp promoters is dependent upon the sequence of the −10 region. Mol Microbiol. 2005;59:1859–1875. doi: 10.1111/j.1365-2958.2006.05066.x. [DOI] [PubMed] [Google Scholar]
- Esteve-Gassent MD, Elliott NL, Seshu J. sodA is essential for virulence of Borrelia burgdorferi in the murine model of Lyme disease. Mol Microbiol. 2009;71:594–612. doi: 10.1111/j.1365-2958.2008.06549.x. [DOI] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, et al. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Old IG, Girons IS, Charon NW. The flgK motility operon of Borrelia burgdorferi is initiated by a σ70-like promoter. Microbiology. 1997;143 (Pt 5):1681–1690. doi: 10.1099/00221287-143-5-1681. [DOI] [PubMed] [Google Scholar]
- Gherardini FC, Boylan JA, Lawrence K, Skare JT. Metabolism and Physiology of Borrelia. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Norfolk UK: Caister Academic Press; 2010. pp. 139–166. [Google Scholar]
- Gibson CM, Mallett TC, Claiborne A, Caparon MG. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J Bacteriol. 2000;182:448–455. doi: 10.1128/jb.182.2.448-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Harris DR, Ward DE, Feasel JM, Lancaster KM, Murphy RD, Mallet TC, Crane EJ., III Discovery and characterization of a Coenzyme A disulfide reductase from Pyrococcus horikoshii. Implications for this disulfide metabolism of anaerobic hyperthermophiles. FEBS J. 2005;272:1189–1200. doi: 10.1111/j.1742-4658.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, Takahashi N, Kamio Y. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol. 1999;181:5940–5947. doi: 10.1128/jb.181.19.5940-5947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic E, Feng S, Freet KJ, Barthold SW. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect Immun. 2003;71:5042–5055. doi: 10.1128/IAI.71.9.5042-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel CS, Lancaster KM, Crane EJ., III Determination of coenzyme A levels in Pyrococcus furiosus and other Archaea: implications for a general role for coenzyme A in thermophiles. FEMS Microbiol Lett. 2005;252:229–234. doi: 10.1016/j.femsle.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith IR, Trzeciakowski JP, Skare JT. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 2009;74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol. 2002;46:111–153. doi: 10.1016/s0065-2911(02)46003-1. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S. Biosynthesis of Pantothenic Acid and Coenzyme A. In: Neidhardt FC, Curtiss R, Gross CA, Ingraham JL, Lin ECC, Low KB, editors. Esherichia coli and Salmonella typhimurium : cellular and molecular biology. Washington D.C: American Society for Microbiology; 1996. pp. 687–694. [Google Scholar]
- Jackowski S, Rock CO. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- Kariu T, Coleman AS, Anderson JF, Pal U. Methods for rapid transfer and localization of lyme disease pathogens within the tick gut. J Vis Exp. 2011 doi: 10.3791/2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karna SL, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M, Seshu J. CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect Immun. 2011;79:732–744. doi: 10.1128/IAI.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Schulz GE. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 A resolution. J Mol Biol. 1989;210:163–180. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. The fur homologue in Borrelia burgdorferi. J Bacteriol. 2004;186:6443–6456. doi: 10.1128/JB.186.19.6443-6456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J, van dV, Venema Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Seshu J, Skare JT. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun. 2003;71:4608–4613. doi: 10.1128/IAI.71.8.4608-4613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Skare JT. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RS, Piesman J, Burgdorfer W. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol. 1991;36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lawrenz MB, Wooten RM, Norris SJ. Effects of vlsE complementation on the infectivity of Borrelia burgdorferi lacking the linear plasmid lp28-1. Infect Immun. 2004;72:6577–6585. doi: 10.1128/IAI.72.11.6577-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, et al. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- Mallett TC, Wallen JR, Karplus PA, Sakai H, Tsukihara T, Claiborne A. Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Hu LT, Marconi RT. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with lyme disease spirochetes. Infect Immun. 2002;70:4196–4203. doi: 10.1128/IAI.70.8.4196-4203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, et al. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol. 2009;191:2783–2794. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Arnold K, Price MS, Sherrill C, delCardayre SB, Aharonowitz Y, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, et al. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha S, Meng EC, Babbitt PC. Evolution of function in the “two dinucleotide binding domains” flavoproteins. PLoS Comput Biol. 2007;3:e121. doi: 10.1371/journal.pcbi.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, Norgard MV. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology. 2008;154:2641–2658. doi: 10.1099/mic.0.2008/019992-0. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Deka RK, Norgard MV. BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in Borrelia burgdorferi by a Novel DNA-Binding Mechanism. PLoS Pathog. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol. 2009;74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige C, Reid SD, Hanna PC, Claiborne A. The type III pantothenate kinase encoded by coaX is essential for growth of Bacillus anthracis. J Bacteriol. 2008;190:6271–6275. doi: 10.1128/JB.00860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Fikrig E. Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes and Infection. 2003;5:659–666. doi: 10.1016/s1286-4579(03)00097-2. [DOI] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, et al. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D, Desrosiers DC, Hazlett KR, Sun Y, Nelson KJ, Cox DL, et al. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc Natl Acad Sci U S A. 2010;107:6240–6245. doi: 10.1073/pnas.0910057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D, Karplus PA, Poole LB. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J, Schneider BS, Zeidner NS. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J Clin Microbiol. 2001;39:4145–4148. doi: 10.1128/JCM.39.11.4145-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J, Zeidner NS, Schneider BS. Dynamic changes in Borrelia burgdorferi populations in Ixodes scapularis (Acari: Ixodidae) during transmission: studies at the mRNA level. Vector Borne Zoonotic Dis. 2003;3:125–132. doi: 10.1089/153036603768395825. [DOI] [PubMed] [Google Scholar]
- Policastro PF, Schwan TG. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J Med Entomol. 2003;40:364–370. doi: 10.1603/0022-2585-40.3.364. [DOI] [PubMed] [Google Scholar]
- Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Samuels DS. Electrotransformation of the spirochete Borrelia burgdorferi. Electrotransformation protocols for microorgansims. Method Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan E, Esteve-Gassent MD, Maruskova M, Seshu J. Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi. Infect Immun. 2009;77:5149–5162. doi: 10.1128/IAI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartakova M, Dobrikova E, Cabello FC. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2000;97:4850–4855. doi: 10.1073/pnas.080068797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun. 2004;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Skare JT. The many faces of Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2000;2:463–472. [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- Sorci G, Faivre B. Inflammation and oxidative stress in vertebrate host-parasite systems. Philos Trans R Soc Lond B Biol Sci. 2009;364:71–83. doi: 10.1098/rstb.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton TB, Rosey EL, Kennedy MJ, Jensen NS, Bosworth BT. Isolation, oxygen sensitivity, and virulence of NADH oxidase mutants of the anaerobic spirochete Brachyspira (Serpulina) hyodysenteriae, etiologic agent of swine dysentery. Appl Environ Microbiol. 1999;65:5028–5034. doi: 10.1128/aem.65.11.5028-5034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC, Grodzicki RL, Craft JE, Shrestha M, Kornblatt AN, Malawista SE. Recovery of Lyme disease spirochetes from patients. Yale J Biol Med. 1984;57:557–560. [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Claiborne A, Schulz GE. NADH binding site and catalysis of NADH peroxidase. Eur J Biochem. 1993;211:221–226. doi: 10.1111/j.1432-1033.1993.tb19889.x. [DOI] [PubMed] [Google Scholar]
- Sze CW, Li C. Inactivation of bb0184, Which Encodes Carbon Storage Regulator A, Represses the Infectivity of Borrelia burgdorferi. Infect Immun. 2011;79:1270–1279. doi: 10.1128/IAI.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, et al. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J Bacteriol. 2001;183:5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Rosa PA, Stewart PE. Biology of infection with Borrelia burgdorferi. Infect Dis Clin North Am. 2008;22:217–234. doi: 10.1016/j.idc.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen JR, Mallett TC, Boles W, Parsonage D, Furdui CM, Karplus PA, Claiborne A. Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking. Biochemistry. 2009;48:9650–9667. doi: 10.1021/bi900887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen JR, Paige C, Mallett TC, Karplus PA, Claiborne A. Pyridine nucleotide complexes with Bacillus anthracis coenzyme A-disulfide reductase: a structural analysis of dual NAD(P)H specificity. Biochemistry. 2008;47:5182–5193. doi: 10.1021/bi8002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, et al. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W. NAD+ and NADH in cellular functions and cell death. Front Biosci. 2006;11:3129–3148. doi: 10.2741/2038. [DOI] [PubMed] [Google Scholar]
- Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- Zeidner NS, Schneider BS, Dolan MC, Piesman J. An analysis of spirochete load, strain, and pathology in a model of tick-transmitted Lyme borreliosis. Vector Borne Zoonotic Dis. 2001;1:35–44. doi: 10.1089/153036601750137642. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang X, Kumar M, Pal U. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis. 2009;200:1318–1330. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]