Abstract
ADAM 3 is a sperm surface glycoprotein that has been implicated in sperm-egg adhesion. Because little is known about the adhesive activity of ADAMs, we investigated the interaction of ADAM 3 disintegrin domains, made in bacteria and in insect cells, with murine eggs. Both recombinant proteins inhibited sperm-egg binding and fusion with potencies similar to that which we recently reported for the ADAM 2 disintegrin domain. Alanine scanning mutagenesis revealed a critical importance for the glutamine at position 7 of the disintegrin loop. Fluorescent beads coated with the ADAM 3 disintegrin domain bound to the egg surface. Bead binding was inhibited by an authentic, but not by a scrambled, peptide analog of the disintegrin loop. Bead binding was also inhibited by the function-blocking anti-α6 monoclonal antibody (mAb) GoH3, but not by a nonfunction blocking anti-α6 mAb, or by mAbs against either the αv or β3 integrin subunits. We also present evidence that in addition to the tetraspanin CD9, two other β1-integrin-associated proteins, the tetraspanin CD81 as well as the single pass transmembrane protein CD98 are expressed on murine eggs. Antibodies to CD9 and CD98 inhibited in vitro fertilization and binding of the ADAM 3 disintegrin domain. Our findings are discussed in terms of the involvement of multiple sperm ADAMs and multiple egg β1 integrin-associated proteins in sperm-egg binding and fusion. We propose that an egg surface “tetraspan web” facilitates fertilization and that it may do so by fostering ADAM–integrin interactions.
INTRODUCTION
ADAM proteins contain pro-, metalloprotease-, disintegrin-, and cysteine-rich domains followed by an epidermal growth factor repeat, a spacer region, a transmembrane domain, and a cytoplasmic tail. Twenty-nine ADAMs have been identified to date (Black and White, 1998; Schlondorff and Blobel, 1999; Stone et al., 1999; Primakoff and Myles, 2000; White et al., 2001) (also see the table of ADAMs at http://www.med.virginia.edu/∼jag6n/whitelab.html). Despite the mounting information about their sequences and expression patterns, little is yet known about the functions of ADAMs, particularly of their disintegrin domains.
Thirteen ADAMs appear to be testis-specific (see the above-mentioned Web site). Of these, five (ADAMs 1, 2, 3, 5, and 18) have been shown to be proteolytically processed during spermatogenesis and epididymal transport such that their forms on mature fusion-competent sperm lack pro and metalloprotease domains and therefore begin with a disintegrin domain (Blobel et al., 1990; Phelps et al., 1990; Hunnicutt et al., 1997; Lum and Blobel, 1997; Waters and White, 1997; Yuan et al., 1997; Frayne et al., 1998).
Recombinant forms of the disintegrin domain from the testis-specific ADAM mouse (m) ADAM 2 (fertilin β) have been shown to bind to the egg and to inhibit sperm-egg binding and fusion. An aspartic acid at position 9 of the mADAM 2 disintegrin loop has been found to be important for these activities (Bigler et al., 2000; Zhu et al., 2000). We and others have suggested that the ADAM 2 disintegrin domain can interact with a β1 integrin on the egg (Almeida et al., 1995; Evans et al., 1997; Bigler et al., 2000; Chen and Sampson, 1999; Chen et al., 1999a; Takahashi et al., 2000). In keeping with the involvement of a β1 integrin, we recently showed that antibodies to CD9, a β1-integrin-associated tetraspanin protein, inhibit not only sperm-egg binding and fusion but also binding of the ADAM 2 disintegrin domain to the egg (Chen et al., 1999b). This latter observation is consistent with recent results showing that CD9 must be present on the egg for successful sperm-egg fusion (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000).
The disintegrin domains of human ADAM (hADAM) 9, hADAM 15, and hADAM 23 have been reported to bind, respectively, to the α6β1, αvβ3/α5β1, and αvβ3 integrins (Zhang et al., 1998; Nath et al., 1999, 2000; Cal et al., 2000); the disintegrin domain of hADAM15 is unique among ADAMs in that it contains the αvβ3/α5β1 integrin binding sequence Arg Gly Asp (RGD) in the middle of its disintegrin loop (Kratzschmar et al., 1996). Very recent studies have indicated that the disintegrin domains of hADAM 12, hADAM 15 (in an RGD-independent manner), and mADAM 15 can bind to the α9β1 integrin (Eto et al., 2000).
The focus of the present study is mADAM 3, a sperm surface protein that is also known as cyritestin. Like mADAM 2, mADAM 3 is found in the equatorial region of mature fusion competent sperm (Linder and Heinlein, 1997; Yuan et al., 1997; Takahashi and White, unpublished data). The equatorial region is the microdomain of the sperm plasma membrane that makes initial contact with the egg during sperm-egg binding and fusion (Wilson and Snell, 1998). Peptide analogs of the disintegrin loop of mADAM 3 inhibit sperm-egg binding and fusion (Linder and Heinlein, 1997; Yuan et al., 1997; Takahashi and White, unpublished data). Antibodies against the disintegrin loop of mADAM 3 inhibit in vitro fertilization (Yuan et al., 1997; Takahashi and White, unpublished data). Male mice lacking ADAM 3 are infertile (Shamsadin et al., 1999), as are male mice lacking mADAM 2 (Cho et al., 1998). However, whereas sperm from ADAM 2 null mice are significantly compromised (80% reduced) in their ability to bind to the egg plasma membrane, sperm from ADAM 3 null mice were reported to be competent to bind to the egg plasma membrane (Shamsadin et al., 1999).
Because little is yet known about the functions, sequence requirements, and receptors of ADAM disintegrin domains, we initiated a comparative analysis of the disintegrin domains of mADAMs 2 and 3. We were particularly interested in comparing these two ADAMs because they are both testis-specific, because they belong to the same branch of the ADAM family tree, and because neither is predicted to possess metalloprotease activity (i.e., they may be specialized for adhesive activity). Hence, we considered it possible that mADAMs 2 and 3 would interact with the egg surface in similar ways.
In the first part of this study we present evidence that the disintegrin domain of mADAM 3 inhibits sperm-egg binding and fusion with a potency similar to that of the ADAM 2 disintegrin domain. In the second part we compare the sequence requirements of the mADAM 2 and mADAM 3 disintegrin loops. In the third part we show that in addition to CD9, two other β1 integrin-associated proteins, CD81 and CD98, are present on the egg surface. We also show that antibodies to CD9 and CD98 inhibit in vitro fertilization as well as binding of the mADAM 3 disintegrin domain. Our findings have implications for the role of multiple sperm ADAMs in mammalian fertilization. They are also consistent with a model in which an egg surface “tetraspan web” (Rubinstein et al., 1996) involving integrins and integrin-associated proteins (Maecker et al., 1997; Hemler, 1998) is involved in mammalian fertilization. We propose that a tetraspan web may foster ADAM–integrin interactions.
MATERIALS AND METHODS
Cells
Drosophila melanogastor S2 cells (Invitrogen, Carlsbad, CA) were maintained in DES complete medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (Life Technologies, Gaithersburg, MD), 100 U/ml penicillin G, and 100 μg/ml streptomycin sulfate. Mock-transfected (neomycin-resistant) P388D1 mouse macrophages and P388D1 cells transfected with cDNA encoding the human α6B integrin subunit were the generous gifts of Drs. A. M. Mercurio and L. M. Shaw (Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA). They were maintained in RPMI 1640 supplemented with 15% heat-inactivated fetal bovine serum, 25 mM HEPES (pH 7.4), and 300 μg/ml G418 sulfate (Life Technologies), as described previously (Chen et al., 1999a). The P388D1 macrophage cells were periodically examined by fluorescence-activated cell sorter analysis with the anti-α6 monoclonal antibody (mAb) GoH3 to ensure high level expression of the α6 integrin.
Peptides
Peptides (14 residues) corresponding to the predicted disintegrin loop of mADAM3, as well as a scrambled sequence thereof, were synthesized on a peptide synthesizer (Symphony; Protein Technologies, Tucson, AZ) and purified by high performance liquid chromatography. The sequences were ADAM 3, CRKSKDQCDFPEFC; scrambled ADAM 3, CDRDCKFQEPFSKC. Peptides were amidated at the COOH terminus and acetylated at the NH2 terminus. The two terminal cysteine residues were protected with acetoamidomethyl groups. Peptides were dissolved in phosphate-buffered saline (PBS) at the concentration of 25 mM, diluted to 5 mM in 100 mM HEPES solution (pH 8.5) to adjust the pH (to 8), and finally diluted in egg medium immediately before use.
Antibodies
The function blocking rat mAb against the integrin α6 subunit (GoH3) was purchased from either Immunotech (Westbrook, ME) or Endogen (Woburn, MA). GoH3 was reconstituted according to the manufacturers' instructions and then diluted with PBS and concentrated three times with a Centricon 30 filter to remove sodium azide. Aliquots of GoH3 (2 mg/ml in PBS) were stored at −20°C and used within 1 mo. Aliquots were thawed once. The nonfunction blocking rat mAb against integrin α6 (J1B5) was a generous gift from Dr. C. H. Damsky (University of California, San Francisco). The function-blocking hamster mAbs against the mouse integrin αv (H9.2B8) and β3 (2C9.G2) subunits were purchased from PharMingen (San Diego, CA). The rat mAb against CD9 (JF9) was a generous gift from Dr. P. W. Kincade (Oklahoma Medical Research Foundation). The hamster mAb against mouse CD81 (2F7) was purchased from Southern Biotechnology (Birmingham, AL). The rat mAb against mouse CD98 (28-19) was described previously (Tsumura et al., 1999). Cy3-conjugated goat anti-rat IgG was purchased from Zymed Laboratories (San Francisco, CA). Fluorescein-conjugated rabbit anti-hamster IgG was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
cDNA Cloning and Mutagenesis
A full-length cDNA encoding mADAM 3 was cloned from a mouse testis cDNA library (Uni-ZAP XR library, Stratagene, La Jolla, CA). As a probe, we used a portion of mADAM 3 that we previously isolated in a degenerate polymerase chain reaction (PCR) screen for mouse testis ADAMs (Wolfsberg et al., 1995). Nucleotide sequences coding amino acid residues 394–486 (the disintegrin domain of ADAM 3) were then amplified by PCR by using the cloned ADAM 3 cDNA as a template. The forward primer was 5′-TCCCCCGGGGGTGGATCGTATTGTGGTAACC-3′. The reverse primer was 5′-GCTCTAGATTCCAGGTCTGCAGCTTTTGTATC-AGG-3′. For expression in Drosophila S2 cells, the PCR fragment encoding the ADAM 3 disintegrin domain was cut with SmaI and XbaI, and subcloned into the SmaI/XbaI sites of the pMT/Bip/V5/His vector (Invitrogen). The final product encoded the disintegrin domain, a V5 tag, and a polyhistidine tag; the encoded disintegrin domain contains five extra amino acids (Arg, Ser, Pro, Trp, Pro) at its N-terminal and six extra amino acids (Ser, Arg, Gly, Pro, Phe, Glu) at its C-terminal end. For expression in Escherichia coli, the BglII/PmeI fragment from the pMT/Bip/V5/His vector containing the ADAM 3 disintegrin domain was ligated to the BamHI/SmaI sites of the vector pGEX-4T-2 (Amersham Pharmacia Biotech, Piscataway, NJ). This generated an E. coli expression vector encoding the ADAM 3 disintegrin domain (as above) followed by the V5 and polyhistidine tags.
For expression of the disintegrin domain of hADAM 15, the BamHI site in pGEX-2T/ADAM15 (Zhang et al., 1998) was changed to a BstEII site by using the QuickChange site-directed mutagenesis kit (Stratagene). The BstEII fragment from the modified pGEX-2T/ADAM15 vector was then excised and substituted with the BstEII fragment from pGEX-4T-2/ADAM2 (Bigler et al., 2000). This added hemagglutinin (HA) and polyhistidine tags to the C-terminal end of the hADAM 15 disintegrin domain. Insert DNAs from all constructs were sequenced to confirm that they encoded the predicted proteins.
The sequence of the disintegrin loop of mADAM 3 is CRKSKDQCDFPEFC. A mutant in which residues 6 and 7 of the loop were changed to alanines (KDQCD to KAACD) was generated by the QuickChange site-directed mutagenesis kit by using pMT/Bip/V5/His/ADAM 3 as the template. Additional mutations at positions 2, 5, 6, 7, 8, 9, and 12 in the ADAM 3 loop were generated in the same way. The mutations prepared in the Drosophila vector were then transferred into pGEX-4T-2 by ligation of the BglII/SmaI fragment as described above. All plasmids were sequenced to verify that the desired mutations had been introduced and that the coding region was correct. We also checked the DNA sequences of the Q7A and D9A ADAM 3 mutants from bacterial cultures used to generate the disintegrin domain proteins used in the analysis shown in Figure 5. Mutants in the ADAM 2 disintegrin domain were generated as described previously (Bigler et al., 2000).
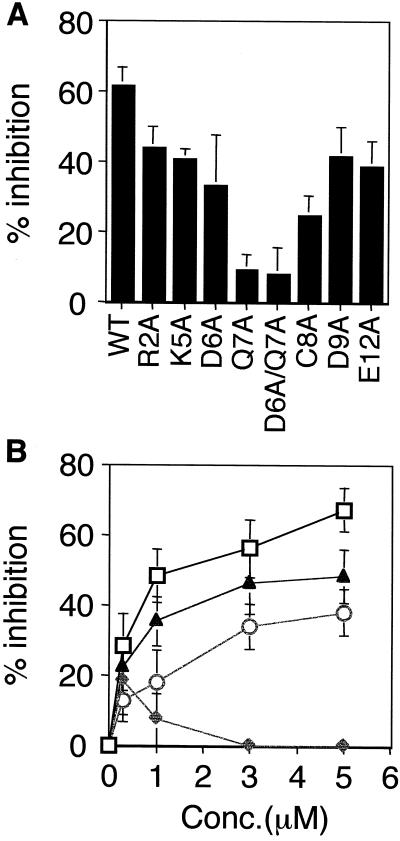
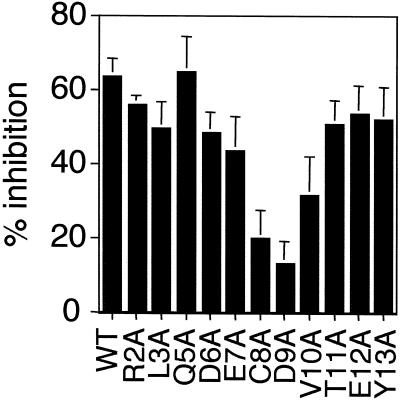
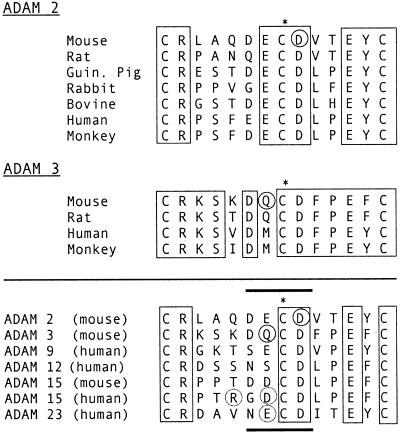
Figure 5.
Sequence-specific activity of the mADAM 3 disintegrin domain. (A) Zona-free eggs were preincubated with wt or the indicated mutant mADAM 3 disintegrin domain at a final concentration of 3 μM. Sperm-egg binding was then assayed as described in the legend to Figure 2. Data represent the average percentage of inhibitory activity of wild-type and mutant mADAM 3 disintegrin domains (compared with buffer controls). The data were compiled from three or four independent experiments. (B) Effects of the indicated concentrations of wt (□), Q7A (♦), C8A (○), and D9A (▴) mADAM 3 disintegrin domains on sperm-egg binding were determined as in A. Disintegrin domains compared within an individual experiment were prepared within 3 d of each other. Bars represent SE of the mean.
Expression and Purification of ADAM 3 Disintegrin Domain in Drosophila Cells and E. coli
We established S2 cell lines that express either the wild-type ADAM 3 disintegrin domain or a mutant ADAM 3 disintegrin domain with the substitutions D6A/Q7A in the disintegrin loop, according to the manufacturer's instructions. Cells were maintained as described above except for the addition of 300 μg/ml hygromycin B (Life Technologies). The cells were seeded at 1–2 × 106 cells/ml in DES serum-free medium in a spinner flask and incubated at room temperature (RT) until the density reached 3–5 × 106 cells/ml. Protein expression was then induced by adding 500 μM CuSO4. After 4–5 d, the supernatant from a 500-ml culture was collected by centrifugation (2000 × g, 10 min) and then filtered through an 0.2-μm filter to remove cell debris. The cleared supernatants were then applied to a 3-ml column of Talon resin (Clontech, Palo Alto, CA) by gravity flow. The resin was washed four times with 15 ml of 2 mM imidazole in 20 mM Tris/100 mM NaCl/10% glycerol (pH 8). Proteins were then eluted in five fractions by adding 3 ml of 50 mM imidazole in 20 mM Tris/100 mM NaCl (pH 8). The eluted proteins were immediately pooled and dialyzed overnight against three changes of PBS. The eluted and dialyzed proteins were then concentrated to a final volume of 200 μl with Centriprep 10 and Centricon 10 (Amicon, Beverly, MA) filters.
The proteins purified over Talon column were subjected to a second purification by Fast Protein Liquid Chromatography (Waters 650E; Millipore, Bedford, MA) on a Superose 6 HR 10/30 column (Amersham Pharmacia Biotech). The column was run according to the manufacturer's protocol with PBS as the elution buffer. Fractions containing ADAM 3 disintegrin domains were detected by dot blot analysis with an antibody against the V5 epitope, and were then pooled and concentrated with a Centricon 10 filter. After the second purification, the yield of protein ranged from 0.5 to 2 mg/l, as determined by bicinchinonic acid protein assay. The proteins were routinely analyzed by Coomassie staining of 12% SDS-PAGE gels. The proteins purified over Talon and Fast Protein Liquid Chromatography columns were kept at 4°C for 3 wk. Some of the proteins were kept at −20°C before use.
GST-ADAM 3 and GST ADAM 3 mutant disintegrin domains were produced by induction of E. coli with 100 μM isopropyl-β-d-thiogalactoside according to the manufacturer's protocol, followed by capture of the glutathione S-transferase (GST) fusion proteins on glutathione sepharose (Amersham Pharmacia Biotech). Disintegrin domains were cleaved from GST by adding 50 U of thrombin (Sigma Chemical, St. Louis, MO)/1 ml of bed volume of glutathione sepharose. The cleaved proteins were then subjected to purification over Talon spin columns (Clontech) to remove thrombin and minor contaminants. Briefly, the protein solution (∼1 ml) was loaded onto a Talon spin column and incubated for 1 h at 4°C with rotation. The columns were then washed three times with 1 ml of wash buffer (20 mM Tris/100 mM NaCl/10% glycerol/2 mM imidazole, pH 8). The bound proteins were eluted twice with 1 ml of elution buffer (20 mM Tris/100 mM NaCl/50 mM imidazole, pH 8). The eluates were then dialyzed overnight against three changes of PBS. The dialyzed protein was concentrated with a Centricon-10 filter and stored at 4°C. GST-ADAM 2 disintegrin domains were produced and purified as described previously (Bigler et al., 2000). Protein concentrations were determined by bicinchinonic acid protein assay. Bacterially produced proteins were kept at 4°C for 2–3 wk.
Sperm-Egg Binding and Fusion Assays
Sperm-egg binding and fusion assays were performed as described previously with minor modification (Takahashi et al., 1995; Chen et al., 1999a). Briefly, zona-free oocytes were prepared by acid treatment (pH 2.7) for 15–30 s followed by a 3-h incubation in M199 medium in a 37°C/5%CO2 incubator to recover fertilizability (Takahashi et al., 1995). Recovered eggs were preloaded with 0.5 μg/ml Hoechst-33342 for 15–30 min, washed, and then treated with either disintegrin proteins or antibody for 30 min at 37°C. Cauda epididymal sperm that had been capacitated for 3 h were introduced into droplets containing eggs at a final concentration of 50,000 sperm/ml. After 30 min, eggs were washed, mounted on a coverslip, and examined by phase contrast and fluorescence microscopy (Axioplan2; Zeiss, Thornwood, NY). The number of sperm bound per egg and the percentage of eggs fused were then calculated. Within the data set for any given day the average number of sperm bound per egg and the percentage of eggs fertilized in experimentally treated samples were normalized with respect to untreated controls to calculate percentage of inhibition. Values for percentage of inhibition were then averaged between replicate experiments. Under our conditions, typical average sperm binding and fusion values for untreated controls ranged from 3 to 8 sperm bound per egg and 50 to 90% eggs fused.
Fluorescent Microbead Binding to Zona-free Eggs
Fluorescent beads were prepared as described previously (Chen et al., 1999a; Bigler et al., 2000). Briefly, 0.2 μm yellow-green fluorescent sulfated-beads (Molecular Probe, Eugene, OR) were coated with purified E. coli protein at 0.5 mg/ml or Drosophila protein at 1 mg/ml overnight at 4°C. Wild-type and mutant ADAM 3 disintegrin domains showed similar coupling efficiencies to the beads (∼42% for the constructs made in E coli). Beads were blocked with goat anti-rabbit IgG and added to droplets containing 5–15 zona-free eggs. After 30–60 min at 37°C in a CO2 incubator, the eggs were washed and observed in a fluorescence microscope. For indicated images the total integrated pixel intensity was determined using the program Scion image and divided by the total number of eggs per image. Values were then normalized with respect to the appropriate buffer control and plotted in bar graph format.
Sperm Binding to P388D1 Macrophage Cells
Sperm binding to P388D1 cells was conducted as described previously (Almeida et al., 1995; Chen et al., 1999a). Cells were plated in a 24-well culture plate for 3 h and were then treated with ADAM 3 disintegrin loop peptides, either authentic or scrambled (final concentration = 500 μM). After 30 min, sperm that had been capacitated for 3 h were introduced at a final concentration of 5 × 105 sperm/ml and incubated for 1 h. Photographs were taken with a scanning confocal laser microscope and the number of sperm bound to at least 500 macrophage cells was determined.
Immunofluorescence
Zona-free eggs prepared as described above were incubated with primary antibodies in M199 medium at 50 μg/ml for 1 h at RT. Eggs were washed three times with M199, and then incubated with either Cy3-conjugated anti-rat IgG (1:100) or fluorescein isothiocyanate-conjugated anti-hamster IgG (1:100) for 45 min at RT. After three washes in M199, eggs were mounted on a coverslip and fluorescence images were acquired with a scanning confocal laser microscope.
RESULTS
ADAM 3 Disintegrin Domains Inhibit Sperm-Egg Binding and Fusion
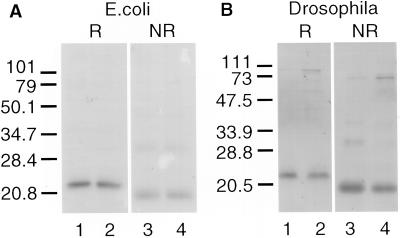
We expressed the ADAM 3 disintegrin domain in E. coli, as a GST-chimera, as well as in Drosophila cells, as a secreted protein. The proteins were purified as described in MATERIALS AND METHODS. For all experiments using ADAM 3 disintegrin domains produced in E. coli, the disintegrin domain was cleaved from the GST-chimera by treatment with thrombin, followed by repurification as described in MATERIALS AND METHODS. The released ADAM 3 disintegrin domains (wild-type and mutants; see below) ran at ∼22 kDa under reducing conditions (Figure 1A, lanes 1 and 2) and at ∼20 kDa under nonreducing conditions (Figure 1A, lanes 3 and 4). ADAM 3 disintegrin domains secreted from the Drosophila cells ran similarly, at ∼22 kDa under reducing conditions (Figure 1B, lanes 1 and 2) and at ∼20 kDa under nonreducing conditions (Figure 1B, lanes 3 and 4).
Figure 1.
Expression of recombinant mADAM 3 disintegrin domains in E. coli and Drosophila cells. mADAM 3 disintegrin domains were expressed in and purified from E. coli (A) or Drosophila cells (B) as described in MATERIALS AND METHODS. The purified proteins were analyzed on a 12% SDS gel under reducing (lanes 1 and 2) or nonreducing (lanes 3 and 4) conditions. Wild-type disintegrin domains (lanes 1 and 3) and D6A/Q7A mutant disintegrin domains (lane 2 and 4) are shown.
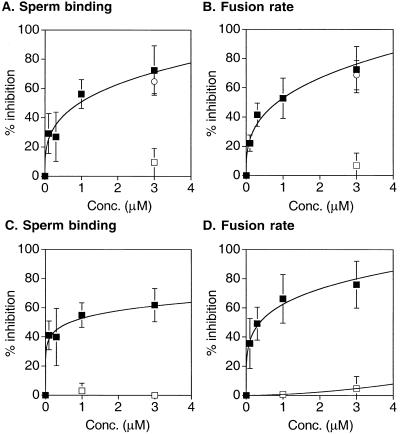
We first asked whether the ADAM 3 disintegrin domains made in E. coli and in Drosophila cells inhibit mouse sperm-egg binding and fusion. As shown in Figure 2, the ADAM 3 disintegrin domain made in E. coli (and released from its GST-chimera and repurified) inhibited sperm-egg binding (Figure 2A, solid squares) and fusion (Figure 2B, solid squares) in a dose-dependent manner. We observed ∼60 to 75% inhibition of sperm-egg binding and fusion with ∼3 μM protein. The protein prepared from Drosophila cells exhibited a similar potency in inhibiting sperm-egg binding (Figure 2C, solid squares) and fusion (Figure 2D, solid squares). The ADAM 2 disintegrin domain prepared in E. coli (and released and repurified from its GST chimera) exerted a similar effect as the ADAM 3 disintegrin domain prepared in E. coli (Figure 2, A and B, open circles).
Figure 2.
Effects of recombinant mADAM 3 disintegrin domains on sperm-egg binding and fusion. Sperm-egg binding (A and C) and fusion (B and D) were assayed with disintegrin domains expressed in E. coli (A and B) or Drosophila cells (C and D) as described in MATERIALS AND METHODS. The symbols represent solid squares, wt-mADAM 3 disintegrin domain; open circles. wt-mADAM 2 disintegrin domain; open squares, D6A/Q7A mutant mADAM 3 disintegrin domain. The data are compiled from three or four independent experiments and represent the average percentage of inhibition of the number of sperm bound per egg (A and C) and the average percentage of inhibition of fertilization rate (B and D) compared with buffer controls, as described in MATERIALS AND METHODS. Bars represent the SE of the mean.
Role of the Disintegrin Loop
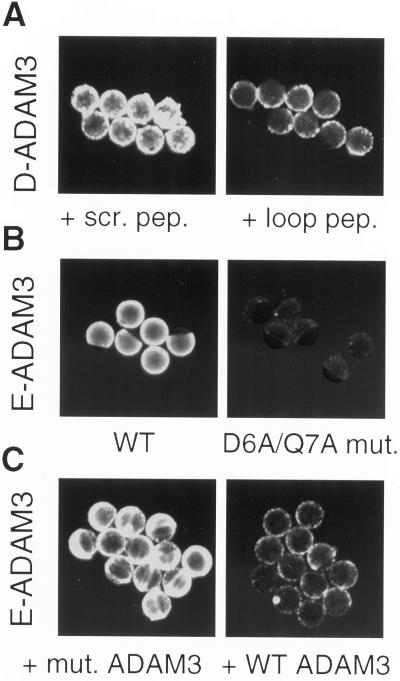
We next asked whether the disintegrin loop of mADAM 3 is important for interacting with the egg plasma membrane. To do this we coated fluorescent beads with ADAM 3 disintegrin domains. As shown in Figure 3A, left, a scrambled peptide analog of the disintegrin loop of mADAM 3 did not, whereas an authentic peptide analog of the mADAM 3 disintegrin loop did (Figure 3A, right), inhibit binding of fluorescent beads coated with the mADAM 3 disintegrin domain. As seen in Figure 3B, left, fluorescent beads coated with the wt ADAM 3 disintegrin domain bound to the egg. In contrast, an ADAM 3 disintegrin domain with mutations in its disintegrin loop that severely impair its biological activity (see below), did not (Figure 3B, right). Consistent with this observation, the mutant ADAM 3 disintegrin domain did not (Figure 3C, left), whereas the wt ADAM 3 disintegrin domain did (Figure 3C, right) inhibit binding of fluorescent beads coated with wt ADAM 3 disintegrin domain. Similar results were obtained with ADAM 3 disintegrin domains made in E. coli or in Drosophila cells.
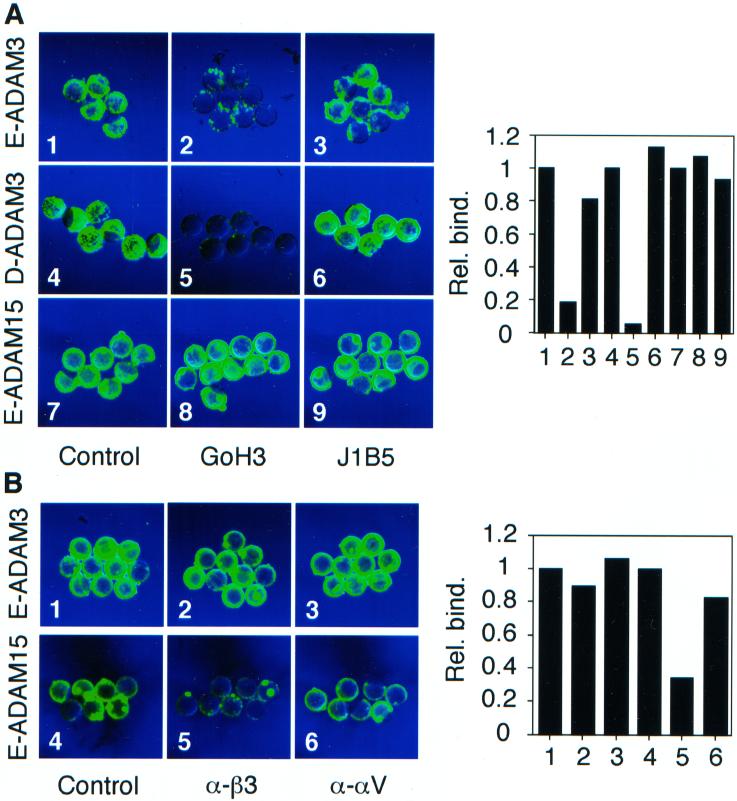
Figure 3.
Binding of beads coated with mADAM 3 disintegrin domains: role of the disintegrin loop. Fluorescent beads were coated with mADAM 3 disintegrin domains prepared, as indicated, in either Drosophila cells (D-ADAM 3) or E. coli (E-ADAM 3) and then processed for binding to zona-free murine eggs as described in MATERIALS AND METHODS. (A). Eggs were preincubated for 30 min at 37°C with either a scrambled (scr.) or an authentic (loop) mADAM 3 disintegrin loop peptide at a final concentration of 1 mM peptide. (B) Beads were coated as indicated with either wild-type or the D6A/Q7A mADAM 3 disintegrin domain and then processed for binding to eggs. (C) Eggs were preincubated for 30 min at 37°C with, as indicated, either the D6A/Q7A mutant (mut.) or the wild-type (wt) mADAM 3 disintegrin domain (20 μM), and then processed for binding of beads coated with wt-mADAM 3 disintegrin domain.
Sequence Requirements of mADAM 2 and mADAM 3 Disintegrin Loops
We recently presented data on six mutants in the disintegrin loop of the mADAM 2 disintegrin domain in which we changed each of the five charged residues to Ala and prepared one double mutant. This analysis revealed a critical importance for the aspartic acid at position 9 of the mADAM 2 disintegrin loop (Bigler et al., 2000). Here we have extended the analysis of the mADAM 2 disintegrin loop and we have conducted an analysis of the mADAM 3 disintegrin loop. As seen in Figure 4, a complete alanine scan through the disintegrin loop of mADAM 2 confirmed the critical importance of the aspartic acid at position 9. Changing this aspartic acid to glutamine also severely compromised the inhibitory activity of the mADAM 2 disintegrin domain (Bigler and White, unpublished results), indicating a critical importance for a negatively charged residue at position 9 of the mADAM 2 disintegrin loop. Changing the cysteine at position 8 of the mADAM 2 disintegrin loop also impaired the inhibitory activity of the mADAM 2 disintegrin domain.
Figure 4.
Sequence-specific activity of the mADAM 2 disintegrin domain. Zona-free eggs were preincubated for 30 min with GST-ADAM 2 or the indicated GST-ADAM 2 mutant disintegrin domain at a final concentration of 1 μM. Sperm-egg binding was then assayed as described in the legend to Figure 2. Data represent the average percentage of inhibitory activity of wild-type and mutant mADAM 2 disintegrin domains (compared with buffer controls). The data were compiled from three or four independent experiments. Bars represent the SE of the mean. Data for the R2A, D6A, E7A, D9A, and E12A mutants are from Bigler et al. (2000). Disintegrin domains compared within an individual experiment were prepared on the same day.
An alanine scan through residues near the center of the mADAM 3 disintegrin loop as well as selected residues near the ends of the mADAM 3 disintegrin loop revealed a critical importance for the glutamine at position 7 (Figure 5A). Changing the cysteine at position 8 of the loop also inhibited activity. These findings indicate that whereas an aspartic acid at position 9 is critical for the activity of the mADAM 2 disintegrin loop, a glutamine at position 7 is especially important for the activity of the mADAM 3 disintegrin loop.
To explore further the finding that the critical residue in the ADAM 3 disintegrin loop (in the context of single alanine mutants) appears to be the glutamine at position 7 (Figure 5A) and not, as we predicted (Figure 4), the aspartic acid at position 9, we conducted dose-response experiments with wild-type, Q7A, and D9A mADAM 3 disintegrin domains. (We also included the C8A mADAM 3 disintegrin loop mutant in this analysis.) The results, presented in Figure 5B, confirm the critical importance of the glutamine at position 7 of the ADAM 3 disintegrin loop. They also show that changing the aspartic acid at position 9 (in the mADAM 3 loop) impairs activity, albeit not as strongly as the alanine substitution at position 7. The collective results presented in Figures 4 and 5, A and B, also indicate that the central cysteines in the ADAMs 2 and 3 disintegrin loops, albeit not critical, are needed for maximal inhibitory activity.
Role of α6 Integrin Subunit
We conducted several experiments to explore whether the mADAM 3 disintegrin domain interacts with an integrin on the egg. Two integrins that are well expressed on the mouse egg surface are α6β1 and αvβ3 (Almeida et al., 1995; Evans, 1999). We first assessed the effects of the anti-α6 mAbs GoH3 (a function blocking mAb) and J1B5 (a nonfunction blocking mAb) on the ability of beads coated with the mADAM 3 disintegrin domain to bind to eggs. As seen in Figure 6A, GoH3 inhibited binding of fluorescent beads coated with the mADAM 3 disintegrin domain prepared in either E. coli (Figure 6A, panel 2) or in Drosophila cells (Figure 6A, panel 5). The mAb J1B5 had no effect on binding of beads coated with the ADAM 3 disintegrin domain prepared in either E. coli (Figure 6A, panel 3) or in Drosophila cells (Figure 6A, panel 6). Neither anti-α6 mAb inhibited binding of beads coated with the disintegrin domain of hADAM 15 (Figure 6A, panels 7–9). We next tested the effects of mAbs that target the αv and β3 integrin subunits. Neither the αv nor the β3 mAb blocked binding of beads coated with the ADAM 3 disintegrin domain (Figure 6B, panels 2 and 3). In contrast, the anti-β3 mAb inhibited binding of beads coated with the hADAM15 disintegrin domain (Figure 6B, panel 5); hADAM 15 contains the tripeptide RGD in its disintegrin loop. These results suggest that the mADAM 3 disintegrin domain can interact, either directly or indirectly, with the α6, or an α6-like, integrin on the egg surface (see DISCUSSION), whereas the hADAM 15 disintegrin domain interacts with a β3 integrin on the egg surface, most likely αvβ3 (Almeida et al., 1995).
Figure 6.
Binding of beads coated with mADAM 3 disintegrin domains: effects of anti-integrin subunit antibodies. Fluorescent beads were coated with, as indicated, mADAM 3 or hADAM 15 disintegrin domains made in either E. coli (E-ADAM) or Drosophila cells (D-ADAM) as described in the legend to Figure 3. Eggs were preincubated for 30 min at 37°C with, as indicated, either: buffer (A1, A4, and A7); function blocking anti α6 mAb, GoH3 (A2, A5, and A8); nonfunction blocking anti-α6 mAb, J1B5 (A3, A6, and A9); hamster IgG (B1 and B4); hamster anti-mouse β3 mAb (B2 and B5), or hamster anti-mouse αv mAb (B3 and B6) at 200 μg/ml, with the exception that the eggs for Drosophila ADAM3 bead binding were treated with GoH3 at 100 μg/ml. Good inhibition of binding of beads coated with Drosophila ADAM 3 was also observed with 50 μg/ml GoH3. Bead binding was then observed in a fluorescence microscope as described in the legend to Figure 3. Relative fluorescence intensities, normalized to the buffer control for each set (A1, A4, A7, B1, B4), were determined as described in MATERIALS AND METHODS and then plotted (to the right of the micrographs); numbers under each bar refer to the respective micrograph panels.
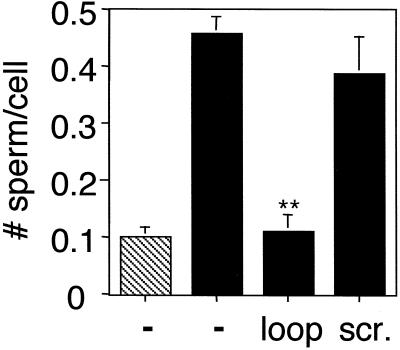
We have previously shown that mouse sperm bind more avidly to macrophage cells that have been transfected with the α6 integrin subunit than to their mock-transfected counterparts (Almeida et al., 1995; Chen et al., 1999a). We have also shown that a peptide analog of the mADAM 2 disintegrin loop inhibits sperm binding to the α6-transfected macrophages to a greater extent than a scrambled ADAM 2 disintegrin loop peptide (Almeida et al., 1995). We therefore tested the effects of an authentic and a scrambled mADAM 3 disintegrin loop peptide on sperm binding to α6-transfected macrophages. As shown in Figure 7, whereas the authentic mADAM 3 disintegrin loop peptide strongly inhibited binding, the scrambled peptide had minimal effect (Figure 7).
Figure 7.
Sperm adhesion to α6-expressing macrophage cells: effect of ADAM 3 disintegrin loop peptides. Mock-transfected (▧) and α6B-transfected P388D1 cells (▪) were, as indicated, preincubated for 30 min at 37°C with either an authentic (loop) or a scrambled mADAM 3 disintegrin loop peptide at a final concentration of 500 μM peptide. Sperm binding was then assayed as described in MATERIALS AND METHODS. The data represent the average number of sperm bound per cell. The data were averaged from three independent experiments. Bars represent the SE of the mean. Data for peptide treated groups were analyzed by Student's t test (**p < 0.01).
Role of Integrin-associated Proteins CD9, CD81, and CD98
We recently reported that JF9, a function-blocking antibody against the tetraspanin, integrin-associated protein CD9, inhibits not only in vitro fertilization but also binding of beads coated with the mADAM 2 disintegrin domain (Chen et al., 1999b). Consistent with these results, it has recently been shown that female mice that lack CD9 are infertile due to a defect in sperm-egg fusion (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000). We therefore asked whether CD9 and other integrin-associated proteins are involved in the interaction between the mADAM 3 disintegrin domain and murine eggs.
We first asked whether CD81 is present on murine eggs. CD81 is a tetraspanin that has been shown to associate with several β1 integrins, as well as with CD9 (Maecker et al., 1997; Hemler, 1998). In addition to a role in myoblast fusion (Tachibana and Hemler, 1999), CD81 binds to the glycoprotein of hepatitis C virus (Higginbottom et al., 2000; Petracca et al., 2000), an enveloped virus that fuses with host cells. Moreover, recent work has detected fertility defects in back-crossed CD81 null mice (Deng et al., 2000). We analyzed for the presence of CD98 on eggs for similar reasons. CD98 associates with the β1 integrin subunit (Fenczik et al., 1997) and it has been reported to cooperate with the α3β1 integrin (Ohta et al., 1994) during cell-cell fusion mediated by human immunodeficiency virus (Ohgimoto et al., 1995) and Newcastle disease virus as well as during cell-cell fusion of monocytes (Tsumura et al., 1999). We previously demonstrated the presence of CD9 on the egg surface by both immunoprecipitation of biotinylated cell surface proteins as well as by immunofluorescense (Chen et al., 1999b). Here we show by immunofluorscence that CD81 and CD98 are also present on the surface of murine eggs (Figure 8A).
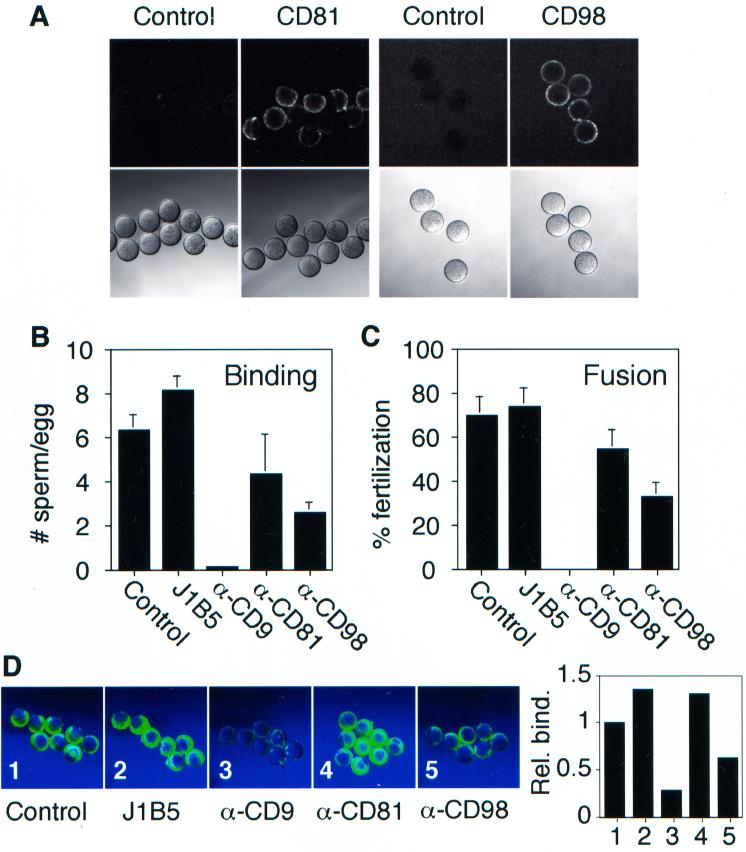
Figure 8.
Effects of antibodies to β1 integrin-associated proteins on fertilization and ADAM 3 bead binding. (A) Zona-free eggs were processed for immunofluorescence with mAbs against CD81 and CD98 as described in MATERIALS AND METHODS. The control antibody for CD81 was hamster IgG and that for CD98 was rat IgG. Sperm-egg binding (B) and fusion (C) were assayed following preincubation with 200 μg/ml the indicated antibodies as described in the legend to Figure 2. The data in B and C are from one experiment. The experiment was repeated three times with similar results. (D) Fluorescent beads coated with E. coli mADAM 3 disintegrin domain were processed for binding to zona-free eggs following a 30-min preincubation with 200 μg/ml the indicated antibodies as described in MATERIALS AND METHODS. The sample labeled control was preincubated with buffer. Scion image analysis (graph to the right in D) was performed as described in the legend to Figure 6. The anti-CD81 antibody used in the experiments shown was the mAb 2F7.
We next examined the effects of anti-CD9, anti-CD81, and anti-CD98 mAbs on sperm-egg binding (B) and fusion (C). As shown previously (Chen et al., 1999b), the anti-CD9 mAb JF9 potently inhibits (99%) sperm-egg binding (Figure 8B) and fusion (100%; Figure 8C). The anti-CD98 mAb inhibited sperm-egg binding (Figure 8B) and fusion (Figure 8D), by 63 and 60%, respectively. The anti-CD81 antibody 2F7 showed only a modest reduction in sperm-egg binding (37% inhibition) and a small reduction in fusion (17% inhibition). The anti-CD81 mAb EAT-1 (Maecker et al., 2000), at a concentration of 100 μg/ml, inhibited both sperm-egg binding and fusion by ∼40%.
We next examined the effects of mAbs against CD9, CD98, and CD81 on binding of beads coated with the mADAM3 disintegrin domain to murine eggs. As seen in Figure 8D, the mAb against CD9 strongly (Figure 8D, panel 3), and the mAb against CD98 modestly (Figure 8D, panel 5), inhibited binding of ADAM 3 disintegrin domain-coated beads. The anti-CD9 mAb and the anti-CD98 mAb also inhibited binding of beads coated with the mADAM 2 disintegrin domain (Chen et al., 1999b; Takahashi and White, unpublished data). The anti-CD9 and anti-CD98 antibodies did not inhibit binding of beads coated with the laminin E8 fragment to eggs placed in an Mn2+-containing buffer (Chen et al., 1999b; Takahashi and White, unpublished results).
DISCUSSION
The first major finding of our study is that the disintegrin domain of mADAM 3 plays an important role in sperm-egg binding and fusion. The second major finding is that the disintegrin domain of mADAM 3 interacts with the egg in much the same manner as the disintegrin domain of mADAM 2. The third major finding is that in addition to CD9 (Chen et al. 1999b, Kaji et al., 2000, Le Naour et al., 2000, Miyado et al., 2000), two other β1 integrin-associated proteins, the tetraspanin CD81 and the type II integral membrane protein CD98, are present on the egg surface. The fourth major finding is that antibodies to CD9 and CD98 significantly impair not only sperm-egg binding and fusion, but also binding of the ADAM 3 disintegrin domain. Our findings have implications for the role of multiple ADAMs in sperm-egg binding and fusion, for the role of β1 integrin-associated proteins in fertilization, and for the possible role of integrin-associated proteins in regulating ADAM–integrin interactions.
Role of Multiple Sperm ADAMs in Fertilization
ADAMs 2 and 3 are sperm surface proteins that share tissue distribution (testis only) and predicted functions. They also fall on the same branch of the ADAM family tree. Here we show that the disintegrin domain of mADAM 3 interacts with murine eggs in much the same way as we have recently reported for the disintegrin domain of mADAM 2 (Bigler et al., 2000). Both disintegrin domains inhibit sperm-egg binding and fusion with similar potencies. Both bind to the egg via their disintegrin loops. A residue near the middle of each disintegrin loop is critical for binding and biological activity. Binding of both disintegrin domains can be inhibited by function blocking antibodies against the α6 integrin subunit as well as against β1 integrin-associated proteins, notably the tetraspanin CD9 (see below). A subtle difference is the precise residue of the disintegrin loop that is critical for function (see below). Collectively, these findings suggest that ADAMs 2 and 3 may be functionally redundant during murine fertilization. This may explain, in part, why mouse sperm that lack fertilin β (ADAM 2) demonstrate residual binding (∼20%) and fusion (∼50%) activity (Cho et al., 1998) and why, reciprocally, sperm lacking ADAM 3 were reported to be able to bind to the egg plasma membrane (Shamsadin et al., 1999). Thirteen of the 29 known ADAMs (including ADAMs 2 and 3) are expressed either exclusively or predominantly in testis (http://www.people.Virginia.EDU/∼jag6n/Table_of_the_ADAMs.html). This raises the possibility that additional sperm ADAMs participate in sperm-egg binding and fusion.
Sequence Requirements of ADAM Disintegrin Domains
We discovered a subtle difference in the sequence requirements of the mADAM 2 and mADAM 3 disintegrin loops. Whereas an aspartic acid at position 9 of the mADAM 2 disintegrin loop appears to be critical, the critical residue of the mADAM 3 disintegrin loop appears to be the glutamine at position 7. These residues represent, respectively, the one immediately following and the one immediately preceding the central cysteine (asterisk, Figure 9) of the respective disintegrin loops (Figure 9). The critical aspartic acid at position 9 of the mADAM 2 disintegrin loop is conserved across all known ADAM 2 orthologues. The critical glutamine at position 7 of the mADAM 3 disintegrin loop is conserved in the rat but differs (is a methionine) in the two known primate ADAM 3 orthologues (Figure 9, top). We do not yet know whether critical residues are found in the same position in the disintegrin loops of cross species orthologues. The central cysteines in the mADAM 2 and mADAM 3 disintegrin loops (asterisk, Figure 9) appear to be required for optimal activity (Figures 4 and 5).
Figure 9.
ADAM disintegrin loops. (Top) Disintegrin loop sequences of all known ADAM 2 and ADAM 3 orthologues are shown. (Bottom) Disintegrin loop sequences of all known functional ADAM disintegrin domains are shown. Circled residues have been shown to be critical. Residues surrounded with dotted circles have been shown or implicated to contribute to activity. Identical residues within groups are boxed. The position of the central cysteine is marked with an asterisk. Accession numbers for the sequences can be obtained at http://www.people.Virginia.EDU/∼jag6n/Table_of_the_ADAMs.html.
The other ADAMs whose disintegrin domains have been reported to interact with integrins are hADAM 9, hADAM 12, mADAM 15, hADAM 15, and hADAM 23 (Zhang et al., 1998; Nath et al., 1999, 2000; Cal et al., 2000; Eto et al., 2000). Their respective disintegrin loop sequences are given in Figure 9 (bottom). The conserved (boxed) residues are present in all ADAM disintegrin domains except those of ADAMs 10 and 17, which are more distally related to other ADAMs (White et al., 2001). Residues so far identified as critical for function are circled. Residues R and D near the middle of the hADAM 15 disintegrin loop are circled in dots because the simultaneous change of the RGD sequence to SGA abolished its ability to support adhesion via the αvβ3 (Zhang et al., 1998), but not via the α9β1 (Eto et al., 2000), integrin. Residue E near the middle of the hADAM 23 disintegrin loop is circled in dots because substitution of this glutamic acid with an alanine, the only substitution analyzed to date, resulted in ∼50% loss of activity (Cal et al., 2000). Nothing is yet known about the sequence requirements of the hADAM 9, hADAM 12, or mADAM 15 disintegrin loops. If one analyzes the limited data available, one can tentatively generalize that residues near the central cysteine of ADAM disintegrin loops (Figure 9, thickened lines) are critical for function. With the exception of hADAM 12, one or both of the two residues preceding the central cysteine in all of the known functional ADAM disintegrin domains is a negatively charged residue. Hence, it may be that a certain number or spacing of negatively charged residues is required for optimal ADAM disintegrin domain function. Of note, although the individual mutations of D6A and E7A in the mADAM 2 disintegrin loop had only minimal effects (Figure 4), the combined mutation D6A/E7A of the loop impaired the activity of the mADAM 2 disintegrin domain by ∼50% (Bigler et al., 2000). Further work is clearly necessary to define the sequence requirements of ADAM disintegrin loops for binding of ADAM disintegrin domains to (specific) integrins. And, of course, residues outside of the disintegrin loop may be important for ADAM disintegrin domain function (Wierzbicka-Patynowski et al., 1999).
Possible Role of an Egg Surface Tetraspan Web in Sperm Binding and Fusion
Tetraspanin proteins such as CD9 and CD81 interact with each other and with β1 integrins to form tetraspan webs (Rubinstein et al., 1996). These multicomponent webs are thought to orchestrate cell surface functions such as cell signaling, perhaps in cholesterol-rich rafts or raft-like plasma membrane microdomains (Rubinstein et al., 1996; Maecker et al., 1997; Hemler, 1998). Based on the findings presented in this (Figure 8) and previous (Chen et al., 1999b; Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000) studies, it seems likely that an egg surface tetraspan web involving β1 integrins and β1 integrin-associated proteins may define or help maintain a site for sperm fusion. Even though it is not essential for fertilization (Miller et al., 2000), we consider it plausible that α6β1 is involved in, or at least is present in a tetraspan web that is intimately associated with, the process of fertilization in a normal egg (Figure 6A; Almeida et al., 1995; Chen and Sampson, 1999; Chen et al., 1999a; Bigler et al., 2000; Takahashi et al., 2000). An interesting possibility is that the newly recognized ADAM receptor, the α9β1 integrin (Eto et al., 2000), may contribute to the process of fertilization in either normal or α6 null females. With respect to the possibility of a tetraspan web involved in sperm-egg fusion, it is interesting that there is currently an active discussion as to whether some enveloped viruses fuse at raft(-like) structures in the host cell plasma membrane (Dimitrov, 2000).
Possible Role of Integrin-associated Proteins in ADAM–Integrin Interactions
Given the (above-cited) evidence that an egg surface tetraspan web involving β1 integrin(s), tetraspanins, and other β1 integrin-associated proteins appears to be involved in murine fertilization, that sperm ADAMs are clearly involved in murine fertilization, and that ADAMs can interact with integrins, a plausible model is that a tetraspan web promotes (e.g., increases the affinity or avidity of) sperm ADAM–egg integrin interactions. This model is analogous to the role that CD9 plays in increasing the affinity of the cholera toxin receptor for its ligand (Cha et al., 2000; Nakamura et al., 2000). In view of the fact that several ADAM disintegrin domains (those of ADAMs 2, 3, 9, 12, 15, and 23) have now been reported to interact with integrins, it will be interesting to see whether modulation by tetraspanins or other integrin-associated proteins (in particular cellular contexts) is a general feature of ADAM–integrin interactions.
ACKNOWLEDGMENTS
We thank Dr. Shoshana Levy for the gift of the EAT-1 and EAT-2 mAbs and for sharing information before publication, Catherine (Gibson) Rea for preparing several of the ADAM 2 mutants, and Dr. Yoshikazu Takada for the gift of the parent vector encoding the hADAM 15 disintegrin domain. We also thank Monika Tomczuk and Dr. Scott Coonrod for critical comments on the text. Y.T. especially acknowledges Dr. Naomi Yamakawa for many helpful discussions. The work was supported by a grant from the National Institutes of Health (GM-48739 to J.M.W.). Y.T. was supported by a fellowship from the Lalor Foundation.
REFERENCES
- Almeida EAC, Huovila A-PJ, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG, White JM. Mouse egg integrin α6β1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- Bigler D, Takahashi Y, Chen MS, Almeida EAC, Osbourne L, White JM. Sequence-specific interaction between the disintegrin domain of mouse ADAM 2 (fertilin β) and murine eggs: role of the α6 integrin subunit. J Biol Chem. 2000;275:11576–11584. doi: 10.1074/jbc.275.16.11576. [DOI] [PubMed] [Google Scholar]
- Black RA, White JM. ADAMs: focus on the protease domain. Curr Opin Cell Biol. 1998;10:654–659. doi: 10.1016/s0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- Blobel CP, Myles DG, Primakoff P, White JM. Proteolytic processing of a protein involved in sperm-egg fusion correlates with the acquisition of fertilization competence. J Cell Biol. 1990;111:69–78. doi: 10.1083/jcb.111.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal S, Freije JM, Lopez JM, Takada Y, Lopez-Otin C. ADAM 23/MDC3, a human disintegrin that promotes cell adhesion via interaction with the αvβ3 integrin through an RGD-independent mechanism. Mol Biol Cell. 2000;11:1457–1469. doi: 10.1091/mbc.11.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH, Brooke JS, Ivey KN, Eidels L. Cell surface monkey CD9 antigen is a coreceptor that increases diphtheria toxin sensitivity and diphtheria toxin receptor affinity. J Biol Chem. 2000;275:6901–6907. doi: 10.1074/jbc.275.10.6901. [DOI] [PubMed] [Google Scholar]
- Chen MS, Almeida EAC, Huovila A-PJ, Takahashi Y, Shaw LM, Mercurio AM, White JM. Evidence that distinct “states” of the integrin α6β1 interact with laminin and an ADAM. J Cell Biol. 1999a;144:549–561. doi: 10.1083/jcb.144.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sampson NS. Mediation of sperm-egg fusion: evidence that mouse egg α6β1 integrin is the receptor for sperm fertilin β. Chem Biol. 1999;6:1–10. doi: 10.1016/S1074-5521(99)80015-5. [DOI] [PubMed] [Google Scholar]
- Chen MS, Tung KSK, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, White JM. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin α6β1: implications for murine fertilization. Proc Natl Acad Sci USA. 1999b;96:11830–11835. doi: 10.1073/pnas.96.21.11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, O'Dell Bunch D, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG. Fertilization defects in sperm from mice lacking fertilin β. Science. 1998;281:1857–1859. doi: 10.1126/science.281.5384.1857. [DOI] [PubMed] [Google Scholar]
- Deng J, Yeung VP, Tsitoura D, Dekruyff RH, Umetsu DT, Levy S. Allergen-induced airway hyperreactivity is diminished in CD81 deficient mice. J Immunol. 2000;165:5054–5061. doi: 10.4049/jimmunol.165.9.5054. [DOI] [PubMed] [Google Scholar]
- Dimitrov DS. Cell biology of virus entry. Cell. 2000;101:697–702. doi: 10.1016/s0092-8674(00)80882-x. [DOI] [PubMed] [Google Scholar]
- Eto K, Puzon-McLaughlin W, Sheppard D, Sehara-Fujisawa A, Zhang X-P, Takada Y. RGD-independent binding of integrin α9β1 to the ADAM 12 and -15 disintegrin domains mediates cell-cell interaction. J Biol Chem. 2000;275:34922–34930. doi: 10.1074/jbc.M001953200. [DOI] [PubMed] [Google Scholar]
- Evans JP. Sperm disintegrins, egg integrins, and other cell adhesion molecules of gamete plasma membrane interactions. Front Biosci. 1999;4:114–131. doi: 10.2741/evans. [DOI] [PubMed] [Google Scholar]
- Evans JP, Kopf GS, Schultz RM. Characterization of the binding of recombinant mouse sperm fertilin β subunit to mouse eggs: evidence for adhesive activity via an egg β1 integrin-mediated interaction. Dev Biol. 1997;187:79–93. doi: 10.1006/dbio.1997.8611. [DOI] [PubMed] [Google Scholar]
- Fenczik CA, Sethl T, Ramos JW, Hughes PE, Ginsberg MH. Complementation of dominant suppression implicates CD98 in integrin activation. Nature. 1997;390:81–85. doi: 10.1038/36349. [DOI] [PubMed] [Google Scholar]
- Frayne J, Jury JA, Barker HL, Hall L. The MDC family of proteins and their processing during epididymal transit. J Reprod Fertil Suppl. 1998;53:149–155. [PubMed] [Google Scholar]
- Hemler ME. Integrin associated proteins. Curr Opin Cell Biol. 1998;10:578–595. doi: 10.1016/s0955-0674(98)80032-x. [DOI] [PubMed] [Google Scholar]
- Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt GR, Koppel DE, Myles DG. Analysis of the process of localization of fertilin to the sperm posterior head plasma membrane during sperm maturation in the epididymis. Dev Biol. 1997;191:146–159. doi: 10.1006/dbio.1997.8700. [DOI] [PubMed] [Google Scholar]
- Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- Kratzschmar J, Lum L, Blobel CP. Metargidin, a membrane-anchored metalloprotease-disintegrin protein with an RGD integrin binding sequence. J Biol Chem. 1996;271:4593–4596. doi: 10.1074/jbc.271.9.4593. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Linder B, Heinlein UAO. Decreased in vitro fertilization efficiencies in the presence of specific cyritestin peptides. Dev Growth Differ. 1997;39:243–247. doi: 10.1046/j.1440-169x.1997.t01-1-00013.x. [DOI] [PubMed] [Google Scholar]
- Lum L, Blobel CP. Evidence for distinct serine protease activities with a potential role in processing the sperm protein fertilin. Dev Biol. 1997;191:131–145. doi: 10.1006/dbio.1997.8609. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Todd SC, Kim EC, Levy S. Differential expression of murine CD81 highlighted by new anti-mouse CD81 monoclonal antibodies. Hybridoma. 2000;19:15–22. doi: 10.1089/027245700315752. [DOI] [PubMed] [Google Scholar]
- Maecker HT, Todd SC, Levy S. The tetraspan superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG. Normal fertilization occurs with eggs lacking the integrin α6β1 and is CD9-dependent. J Cell Biol. 2000;149:1289–1296. doi: 10.1083/jcb.149.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Mitamura T, Takahashi T, Kobayashi T, Mekada E. Importance of the major extracellular domain of CD9 and the epidermal growth factor (EGF)-like domain of heparin-binding EGF-like growth factor for up-regulation of binding and activity. J Biol Chem. 2000;275:18284–18290. doi: 10.1074/jbc.M907971199. [DOI] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Stephens PE, Warn A, Hutchinson GR, Yamada KM, Docherty AJP, Murphy G. Interaction of metargidin (ADAM-15) with αvβ3 and α5β1 integrins on different hemopoietic cells. J Cell Sci. 1999;112:579–587. doi: 10.1242/jcs.112.4.579. [DOI] [PubMed] [Google Scholar]
- Nath D, Slocombe PM, Webster A, Stephens PE, Docherty AJ, Murphy G. Meltrin γ (ADAM-9) mediates cellular adhesion through α6β1 integrin, leading to a marked induction of fibroblast cell motility. J Cell Sci. 2000;113:2319–2328. doi: 10.1242/jcs.113.12.2319. [DOI] [PubMed] [Google Scholar]
- Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kawano M, Watanabe N, Ito Y. Molecular characterization of fusion regulatory protein-1 (FRP-1) that induces multinucleated giant cell formation of monocytes and HIV gp160- mediated cell fusion. FRP-1 and 4F2/CD98 are identical molecules. J Immunol. 1995;155:3585–3592. [PubMed] [Google Scholar]
- Ohta H, Tsurodome M, Matsumura H, Koga Y, Morikawa S, Kawano M, Kusugawa S, Komada H, Nishio M, Ito Y. Molecular and biological characterization of fusion regulatory proteins (FRPs); anti-FRP mAbs induced HIV-mediated cell fusion via an integrin system. EMBO J. 1994;13:2044–2055. doi: 10.1002/j.1460-2075.1994.tb06479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, Abrignani S, Grandi G. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol. 2000;74:4824–4230. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps BM, Koppel DE, Primakoff P, Myles DG. Evidence that proteolysis of the surface is an initial step in the mechanism of formation of sperm cell surface domains. J Cell Biol. 1990;111:1839–1847. doi: 10.1083/jcb.111.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- Rubinstein E, Le Naour F, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Blobel CP. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- Shamsadin R, Adham IM, Nayernia K, Heinlein UA, Oberwinkler H, Engel W. Male mice deficient for germ-cell cyritestin are infertile. Biol Reprod. 1999;61:1445–1451. doi: 10.1095/biolreprod61.6.1445. [DOI] [PubMed] [Google Scholar]
- Stone AL, Kroeger M, Sang QX. Structure-function analysis of the ADAM family of disintegrin-like and metalloproteinase-containing proteins. J Prot Chem. 1999;18:447–465. doi: 10.1023/a:1020692710029. [DOI] [PubMed] [Google Scholar]
- Tachibana I, Hemler ME. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol. 1999;146:893–904. doi: 10.1083/jcb.146.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Meno C, Sato E, Toyoda Y. Synchronous sperm penetration of zona-free mouse eggs in vitro. Biol Reprod. 1995;53:424–430. doi: 10.1095/biolreprod53.2.424. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Yamakawa N, Matsumoto K, Toyoda Y, Furukawa K, Sato E. Analysis of the role of egg integrins in sperm-egg binding and fusion. Mol Reprod Dev. 2000;56:412–423. doi: 10.1002/1098-2795(200007)56:3<412::AID-MRD12>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Tsumura H, Kawano M, Tajima M, Kusaura T, Kozuka Y, Yoshimura S, Komada H, Tsurudome M, Nishio M, Kusagawa S, Shimura K, Ito Y. Isolation and characterization of monoclonal antibodies directed against murine FRP-1/CD98/4F2 heavy chain: murine FRP-1 is an alloantigen and amino acid change at 129 (P to R) is related to the alloantigenicity. Immunol Cell Biol. 1999;77:19–27. doi: 10.1046/j.1440-1711.1999.00792.x. [DOI] [PubMed] [Google Scholar]
- Waters SI, White JM. Biochemical and molecular characterization of bovine fertilin α and β (ADAM 1 and ADAM 2): a candidate sperm-egg binding/fusion complex. Biol Reprod. 1997;56:1245–1254. doi: 10.1095/biolreprod56.5.1245. [DOI] [PubMed] [Google Scholar]
- White JM, Bigler D, Chen M, Wolfsberg TG. ADAMs. In: Beckerle M, editor. Cell Adhesion: Frontiers in Molecular Biology. Oxford: Oxford University Press; 2001. (in press). [Google Scholar]
- Wierzbicka-Patynowski I, Niewiarowski S, Marcinkiewicz C, Calvete JJ, Marcinkiewicz MM, McLane MA. Structural requirements of echistatin for the recognition of αvβ3 and α5β1 integrins. J Biol Chem. 1999;274:37809–37814. doi: 10.1074/jbc.274.53.37809. [DOI] [PubMed] [Google Scholar]
- Wilson NF, Snell WJ. Microvilli and cell-cell fusion during fertilization. Trends Cell Biol. 1998;8:93–96. doi: 10.1016/s0962-8924(98)01234-3. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Straight PD, Gerena RL, Huovila A-PJ, Primakoff P, Myles DG, White JM. Dev. Biol. 169, 378–383. 1995. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. [DOI] [PubMed] [Google Scholar]
- Yuan R, Primakoff P, Myles DG. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm-egg plasma membrane adhesion and fusion. J Cell Biol. 1997;137:105–112. doi: 10.1083/jcb.137.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-P, Kamata T, Yokoyama K, Puzon-McLaughlin W, Takada Y. Specific interaction of the recombinant disintegrin-like domain of MDC-15 (Metargidin, ADAM-15) with integrin αvβ3. J Biol Chem. 1998;273:7345–7350. doi: 10.1074/jbc.273.13.7345. [DOI] [PubMed] [Google Scholar]
- Zhu X, Bansal NP, Evans JP. Identification of key functional amino acids of the mouse fertilin beta (ADAM2) disintegrin loop for cell-cell adhesion during fertilization. J Biol Chem. 2000;275:7677–7683. doi: 10.1074/jbc.275.11.7677. [DOI] [PubMed] [Google Scholar]