Abstract
Campylobacter jejuni is the leading bacterial cause of human enteritis in many industrialized countries. There is no commercial vaccine against C. jejuni available to date. CmeC is an essential outer membrane component of CmeABC multidrug efflux pump that plays a critical role in antibiotic resistance and in vivo colonization of C. jejuni. CmeC is prevalent in C. jejuni strains and is dramatically induced and immunogenic in vivo. In this study, we analyzed CmeC sequence homology, examined in vitro immune protection of CmeC peptide antibodies, and produced full-length recombinant CmeC (rCmeC) for evaluating immunogenicity and protective efficacy of the CmeC subunit vaccine against C. jejuni using chicken model system. Amino acid sequences of CmeC from 24 diverse C. jejuni strains were determined and subjected to alignment, which revealed that CmeC is highly conserved in C. jejuni with a identity ranging from 97.3% to 100%. CmeC peptide antibodies inhibited the function of CmeABC efflux pump and enhanced susceptibility of C. jejuni to bile salts, the natural antimicrobial present in the intestine. Two full-length rCmeC proteins with N- or C-terminal His tag were produced in E. coli; the N-terminal His-tagged rCmeC with high purity and yield was obtained by single step affinity purification. The purified rCmeC was used in two vaccination trials using a chicken model of C. jejuni infection. Stimulation of CmeC-specific serum IgG responses via oral vaccination required immunization with higher doses of rCmeC (200μg) together with 70μg of mucosal adjuvant mLT (modified E. coli heat-labile enterotoxin). Subcutaneous vaccination of chickens with rCmeC remarkably stimulated both serum IgG and IgA responses. However, CmeC-specific intestinal secretory IgA response was not significantly stimulated regardless of vaccination regimen and the rCmeC vaccination did not confer protection against C. jejuni infection. Together, these findings provide further compelling evidence that CmeC is a promising subunit vaccine candidate against C. jejuni infection. However, the CmeC vaccination regimen should be optimized to enhance CmeC-specific mucosal immune response in for protection against C. jejuni.
Keywords: Campylobacter jejuni, Subunit vaccine, Multidrug efflux pump
Introduction
Campylobacter jejuni is the leading bacterial cause of human enteritis in the United States and other industrialized countries [1]. This pathogenic organism causes watery diarrhea and/or hemorrhagic colitis in humans and is also associated with Guillain-Barré syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death [2,3]. Poultry are the major reservoir of Campylobacter and thus the main source for human campylobacteriosis [1,4]. At the same time that prevalence of infection is increasing, Campylobacter has become increasingly resistant to antibiotics, including fluoroquinolones and macrolides, the major drugs of choice for treating human campylobacteriosis [5]. Despite the growing need for new antibiotics due to increasing drug resistance in Campylobacter and other bacteria, many pharmaceutical companies have been placing less emphasis on antibiotic discovery [6]. Therefore, alternative intervention strategies, such as vaccination, are needed to prevent and control Campylobacter infections. To dates, vaccines against Campylobacter infection are still not available, primarily due to the antigenic complexity of this organism and a lack of understanding of the mechanisms of pathogenesis. Information concerning protective antigens as vaccine candidates in C. jejuni is limited and vaccinations against C. jejuni using animal models including chickens have had only partial success [7–9].
It has been well established that prior infection with C. jejuni can induce protective immunity against Campylobacter infections in humans and animals, strongly supporting the feasibility of development of immunization-based approaches to control Campylobacter infections [7]. Outer membrane proteins (OMPs) of C. jejuni are considered the major mediators of pathogen-host interactions and are promising candidates for the design of protective vaccines. Recently, we characterized a unique OMP CmeC, an essential component of multidrug efflux pump CmeABC that plays a critical role in antibiotic resistance and pathogenesis of C. jejuni [10–13]. The CmeC is a promising subunit vaccine candidate against C. jejuni because of following compelling evidences. First, CmeC is essential for C. jejuni colonization in animal intestine by mediating bile resistance [10,11,13]. Compared to the wild type strain that colonized the chickens as early as day 2 post-inoculation with a density as high as 107 CFU/g feces, the isogenic CmeC mutant failed to colonize any of the inoculated chickens throughout the study [12]; the minimum infective dose for CmeABC mutant is at least 2.6×104 folds higher than that of the wild-type strain [12]. Second, PCR and immunoblotting analyses showed that CmeC is widely existed and constitutively expressed among different C. jejuni strains, suggesting that CmeC is highly conserved in terms of sequence and antigenicity [11]. Third, expression of CmeC is dramatically induced by bile salts present in the intestine, further highlighting the critical role of CmeC in Campylobacter pathogenesis [13]. This notion also is supported by a recent microarray study by Stintzi et al. [14], in which expression of cmeABC operon was found to be highly up-regulated in vivo. In addition, using multiple chicken sera for immunoblot analysis, we also demonstrated that CmeC is expressed during Campylobacter infection of chickens and elicited a specific antibody response in the host [12], supporting the feasibility of targeting CmeC for immune protection against C. jejuni colonization. Finally, we also demonstrated that inhibition of CmeABC by efflux pump inhibitors increased susceptibility of Campylobacter to various antimicrobials, prevented emergence of macrolide resistant C. jejuni, and reduced in vivo colonization of C. jejuni using a chicken model system [15,16]. Based on these observations, we hypothesize that CmeC antibodies could inhibit functions of CmeABC pump and that CmeC is a promising subunit vaccine candidate to prevent and control C. jejuni colonization in the intestine.
In this study, we determined sequence homology of CmeC in diverse Campylobacter strains as well as in vitro immune protection of CmeC peptide antibodies, which further support the feasibility of targeting CmeC for immune intervention against C. jejuni infection. We also constructed plasmids for producing full-length rCmeC and optimized conditions for purification of large quantities of rCmeC with high purity. The purified rCmeC was then used in two large chicken vaccination trials to evaluate the immunogenicity and protective efficacy of the CmeC subunit vaccine with different vaccination regimens.
Materials and Methods
Bacterial strains, plasmids and culture conditions
The major bacterial strains and plasmids used in this study are listed in Table 1. Fourteen C. jejuni isolates (JL7, 10, 12, 36, 78, 81, 83, 85, 90, 91, 93, 94, 95 and 118) from various sources (human, chicken, turkey, bovine, and environment) were used for PCR amplification and sequence analysis of the complete cmeC gene. The C. jejuni strains were routinely grown in Mueller Hinton (MH) broth (Difco, Detroit, MI) or on MH agar at 42°C under microaerophilic conditions, which were generated using CampyGen Plus (Oxoid, Bashingstoke, Hampshire, England) gas pack in an enclosed jar. Escherichia coli was grown in Luria-Bertani (LB) broth (Fisher Scientific, Fairlawn, NJ) with shaking (250 rpm) or on agar at 37°C overnight. When needed, culture media were supplemented with ampicilin (100μg/ml) (Sigma, St Louis, MO).
Table 1.
Key bacterial strains and plasmids used in this project.
| Plasmids or Strains | Description | Source or Reference |
|---|---|---|
| Plasmids | ||
| pGEMT-Easy | PCR cloning vector, Ampr | Promega |
| pQE-30 | Vector for N-terminal His-tagged protein construction | Qiagen |
| pQE-70 | Vector for C- terminal His-tagged protein construction | Qiagen |
| pCmeC-NHIS | pQE-30 ligated with cmeC segment encoding mature CmeC | This study |
| pCmeC-CHIS | pQE-70 ligated with cmeC segment encoding mature CmeC | This study |
| Strains | ||
| C. jejuni | ||
| JL7 | H49024, human isolate | [29] |
| JL10 | ATCC 33291, human isolate | ATCC |
| JL12 | 15046764, bovine isolate | [29] |
| JL36 | S3B, chicken isolate | [12] |
| JL78 | W42606, human isolate | [42] |
| JL81 | F34078, human isolate | [42] |
| JL83 | M76297, human isolate | [42] |
| JL85 | F59966, human isolate | [42] |
| JL90 | M33323, human isolate | [42] |
| JL91 | W11805, human isolate | [42] |
| JL93 | M402, human isolate | [18] |
| JL94 | E46972, human isolate | [42] |
| JL95 | 19094451, ovine isolate | [29] |
| JL118 | CVM20088, chicken isolate | [43] |
| JL241 | NCTC 11168, human isolate | [44] |
| JL242 | 81-176, human isolate | [45] |
| E. coli | ||
| DH5α | F-Φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ- | Invitrogen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rk−, mk+) relA1 supE44 Δ(lac-proAB) [F′ traD36 lacIqZΔM15] | Promega |
| JL243 | JM109 containing pCmeC-NHIS | This study |
| JL348 | JM109 containing pCmeC-CHIS | This study |
PCR amplification and sequence analysis of the CmeC gene
Genomic DNA was extracted from each isolate by using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI). Primers CmeCF (5′-CAGCAAAACTTCGTTTTCGTC-3′) and CmeCR (5′ TGCCTGCTATTTACAAGGCTTA-3′) were used to amplify the entire cmeC gene. Amplified PCR products were purified by QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced in Molecular Biology Resource Facility at The University of Tennessee (Knoxville, TN). The cmeC sequences were converted to amino acid sequences and subjected to alignment together with other publicly available CmeC sequences (Accession #: ABC59229, AAT38952, CAL34515, YP_001000076, ZP_01100537, ZP_01069571, ZP_01067179, YP_178433, ZP_01071951) using DNAStar package (version 6.0). To predict the secondary structure of CmeC, the amino acid sequence of CmeC was analyzed by using the program TransMembrane protein Re-Presentation in 2 Dimensions tool (TMRPres2D) [17]. Homology Modeling was performed using an automated model on SWISS Model Workspace (http://swissmodel.expasy.org/workspace/), using NodT (1wp1.pdb) as a template.
Effect of CmeC peptide antibodies on susceptibility of C. jejuni to bile salts
We have prepared a recombinant strain of E. coli JM109 expressing a portion of CmeC (aa 41 to 248 of total 492 aa in length) and high-titer of specific rabbit polyclonal antiserum directed against this CmeC peptide was also generated [11]. This CmeC peptide antiserum was used to examine the effect of CmeC peptide antibodies on susceptibility of C. jejuni 81-176 to cholic acid, a representative bile salt. Briefly, a log-phase culture of C. jejuni 81-176 was diluted to approximately 2 × 106 colony forming unit (CFU) per milliliter in MH broth containing sublethal concentration of cholic acid (2 mg/ml). The anti-CmeC and control sera were added to cells with 1:10 dilution and cells were incubated for 6 hr at 42°C under microaerophilic conditions. Samples were diluted serially in MH and plated on MH agar plates to determine bacterial viability. Both sera were inactivated at 56°C for 30 min prior to use to abolish complement activity. Two independent experiments were performed with triplicate assays in each experiment.
Production and purification of full-length rCmeC
To construct recombinant strain producing full-length N-terminal His-tagged rCmeC, a 1,444-bp fragment encoding mature 473-aa CmeC (aa 20 to 492) with the removal of the 19 aa signal peptide was PCR amplified from C. jejuni NCTC 11168 using primers CCF (5′-AAAGGATCCTGCTCTTTAAGTCCAAATTTAAATATT-3′) and CCR (5′-AAACCCGGGCTATTCTCTAAAAGACATATCTAAATT-3). The restriction sites (BamHI and SmaI, underlined in the primer sequences) were attached to the 5′ end of each primer to facilitate directional cloning of the amplified PCR product into the pQE30 vector (Qiagen, Hilden, Germany). The amplified cmeC fragment was digested with BamHI and SmaI and was cloned into the vector pQE-30, which previously had been digested with BamHI and SmaI, creating pCmeC-NHIS (Table 1). Cloning, expression, and purification of N-terminal His-tagged rCmeC were performed using the procedures described previously [10,12,18]. To construct a recombinant strain producing the full-length C-terminal His-tagged rCmeC, a 1,472-bp fragment encoding full length of 492-aa CmeC (aa 1 to 492) was PCR amplified from C. jejuni NCTC 11168 using primers CmeC_FLF (5′-ACATGCATGCATAAAATAATTTCAATTAGTG-3′, SphI site underlined) and CmeC_FLR (5′-CCTAGATCTTTCTCTAAAAGACATATCTAAA-3′, BglII site underlined). The PCR amplified cmeC fragment was digested with SphI and BglII and was cloned into the pQE-70 (Qiagen, Hilden, Germany), which previously had been digested with the same enzymes, creating pCmeC-NHIS (Table 1). The other procedures are the same as above for purifying N-terminal His-tagged rCmeC. Plasmids pCmeC-NHIS and pCmeC-CHIS were sequenced, with no frameshift and mutations in the coding sequence of cmeC detected. To optimize conditions of producing high-purity rCmeC for vaccination, 5 mM of ATP-Mg2+ was added in lysis buffer to remove contamination of molecule chaperone GroEL [19] and 0.1% Empigen BB (Sigma, St Louis, MO) was used to facilitate solubilization of rCmeC and preserve the antigenicity of rCmeC [10,12,18]. The N-terminal His-tagged rCmeC with high purity was used for vaccination trials described below.
CmeC vaccination and C. jejuni challenge: Trial 1
The experimental design is detailed in Table 2. Briefly, newly hatched broiler chickens (n = 120) were allocated randomly into 6 groups (20 per group). Each group of chickens were maintained in a sanitized wire-floored cage and provided with unlimited access to water and commercial chicken starter feed without antibiotic additives. At 1-week-old age, chickens were immunized orally with 200μl of CmeC vaccine in PBS via oral gavage. CmeC doses used were 50 and 200μg, either alone or coadministered with 10 μg of mLT (general gift from Dr. John Clements in Tulane University Medical Center). Chickens receiving PBS only or mLT only were used as control groups. Booster doses were administered 2 wk after primary immunization. Blood samples were collected from the wing vein of each chicken at 1, 3, 5, 7 wk of the experiment to monitor circulating CmeC specific IgA and IgG antibodies; intestinal lavage samples were collected and prepared at 3, and 5 wk from euthanized chickens (5 bird/group) as described previously [20] to monitor mucosal IgA antibody response. Prior to challenge, chickens were screened again with conventional culture methods to ensure that they were Campylobacter-free. At week 5, 100μl culture of C. jejuni NCTC 11168 was inoculated orally into broilers (106 CFU/bird). After challenge, cloacal swabs were collected every 2 d for 10 d. Swabs were diluted serially, and plated on MH agar containing selective supplement (SR117E; Oxoid, Bashingstoke, Hampshire, England) for enumeration of Campylobacter cells. The randomly selected colonies were tested by PCR [12] to ensure that output Campylobacter was the same as the inoculum and there were no contamination of chickens by other sources.
Table 2.
The experimental design of the first CmeC vaccination and C. jejuni challenge using broilers (Trial 1).
| Group | No. of Chickens | Day 7 Primary Immunization | Day 21 Booster Immunization | Sample Collections | C. jejuni challenge on day 35 |
|---|---|---|---|---|---|
| 1 | 20 | PBS | Blood: day 7, 21, 35, 42 Intestinal lavage: day 21, 35 |
Yes | |
| 2 | 20 | mLTa | Yes | ||
| 3 | 20 | CmeC (50 μg) | Yes | ||
| 4 | 20 | CmeC (50 μg) + mLT (10 μg) | Yes | ||
| 5 | 20 | CmeC (200 μg) | Yes | ||
| 6 | 20 | CmeC (200 μg) + mLT (10 μg) | Yes | ||
mLT: mucosal adjuvant LT-R192G, a mutant E. coli heat-labile toxin kindly provided by Dr. John D. Clements (Tulane University Medical Center).
CmeC vaccination and C. jejuni challenge: Trial 2
The second vaccination trial (Table 3) had four major modifications compared to Trial 1(Table 2). First, white leghorn chickens, which grow slower than broilers, were used in this trial. Second, to compare effects of vaccination route on immune response and protective efficacy of rCmeC, subcutaneous administration was included in this trial. Third, the dose of mucosal adjuvant mLT was increased from 10 to70μg/chicken. Finally, the dose of C. jejuni NCTC 11168 used for challenge was reduced to 105 CFU/bird.
Table 3.
Experimental design of the second CmeC vaccination and C. jejuni challenge using white leghorns (Trial 2).
| Group | No. of Chickens | Vaccination Route | Day 21 Primary Immunization | Day 35 Booster Immunization | Sample Collections | C. jejuni challenge on day 49 |
|---|---|---|---|---|---|---|
| A | 20 | Oral | mLTa (70 μg) | Blood: day 21, 35, 49, 63; Intestinal lavage: day 35, 49; Cloacal swab: day 49, 51, 53, 56, 58. | Yes | |
| B | 20 | Oral | CmeC (200 μg) | Yes | ||
| C | 20 | Oral | CmeC (50 μg)+ mLT (70 μg) | Yes | ||
| D | 20 | Oral | CmeC (200 μg) + mLT (70 μg) | Yes | ||
| E | 20 | Subcutaneous | FIAb | Yes | ||
| F | 20 | Subcutaneous | CmeC (200 μg) + FIAb | Yes | ||
mLT: mucosal adjuvant LT-R192G, a mutant E. coli heat-labile toxin kindly provided by Dr. John D. Clements (Tulane University Medical Center).
FIA: Freund’s incomplete adjuvant
Enzyme-linked immunoabsorbant assay (ELISA)
CmeC-specific antibodies in intestinal lavage samples and sera were measured by indirect ELISA, which was performed using a previously published protocol [21] with the following modifications. Microtiter plates (Nunc-Immuno Plate, Thermo Fisher scientific, Fairlawn, NJ) were coated with 100μl of rCmeC (30 ng/well) in coating buffer (ammonium acetate plus ammonium carbonate [pH 8.2]) overnight at room temperature. For vaccination Trial 1, to examine the levels of circulating CmeC specific IgA and IgG antibodies and mucosal IgA antibodies, chicken serum and intestinal lavage samples were routinely diluted 1:100 and 1:4, respectively. For vaccination Trial 2, chicken intestinal lavage samples were routinely diluted 1:4 for ELISA test while each serum sample was serially diluted up to 4,096-fold for ELISA analysis. Three negative serum samples from individual chickens without receiving any treatment were used in each ELISA plate as background control. End point titer was defined as the last dilution at which the optical density of sample wells exceeded the mean optical density of three control wells plus 3 × standard deviation of OD405nm. Titer was expressed as the reciprocal of the end point dilution log2.
Statistical analysis
Differences in serum and intestinal lavage OD405nm readings or antibody titers among treatment groups were analyzed by least square analysis of covariance with the data at day 0 postimmunization as the covariant; main effects were day of sample collection and treatment. Comparison of lavage OD405nm readings or antibody titers within treatment groups across time were tested by ANOVA. Two-way ANOVA followed by Least significant difference (LSD) test were used to assess significance of difference among percentages and shedding levels of the groups at each sampling point. Levels of significance for P value are 5% (0.05). All statistical analyses were performed using SAS software (v9.03, SAS Institute Inc., Cary, NC).
Results
CmeC is highly conserved in C. jejuni
The full-length CmeC genes from 14 C. jejuni strains were PCR amplified and sequenced in this study. The new sequences were translated into deduced protein sequence and aligned with the CmeC sequences from 10 C. jejuni strains deposited in the NCBI public database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). As shown in Figure 1, amino acid sequences of CmeC displayed a high level of homology (97.3% to 100% aa identity) among C. jejuni strains with diverse backgrounds, suggesting that CmeC is highly conserved in C. jejuni. The secondary structure prediction and homologous modeling of CmeC using NodT (1wp1.pdb) as a template showed that the monomer of CmeC has 4 transmembrane beta sheets (data not shown); three such CmeC monomers may interact with each other and form a functional porin with 12-stranded transmembrane beta barrel to export substrates, as what has been shown for TolC, the CmeC homolog in E. coli [22].
Figure 1.
Phylogenetic relationship of CmeC from diverse C. jejuni strains. The amino acid based dendrogram was constructed by neighbor-joining methods in MEGA 4.0. The strains with ‘JL’ prefix were used for cmeC gene amplification and sequencing in this study and the complete cmeC sequences of these strains have been deposited in GenBank. The CmeC sequences of other strains are the public database NCBI (http://www.ncbi.nlm.nih.gov/).
CmeC peptide antibodies enhanced susceptibility of C. jejuni to bile salt
Supplementation of anti-CmeC serum in MH broth containing a sublethal concentration of cholic acid resulted in moderate but unit, P significant growth reduction (~ 0.6 log10< 0.05, Figure 2) when compared to growth in the presence of control serum (pre-immune serum, negative for CmeC). This finding suggests that anti-CmeC antibodies specifically bind to surface-exposed CmeC, inhibit the function of CmeABC efflux pump, and increases the susceptibility of C. jejuni to bile salt.
Figure 2.
Growth responses of C. jejuni 81-176 to anti-CmeC serum and control serum (pre immune serum). The log-phase culture of 81-176 was diluted to approximately 2×106 cfu/ml in MH broth containing sublethal concentrations of cholic acid (2 mg/ml). Anti-CmeC and control sera were added to cells with 1:10 dilution and cells were incubated for 6 hours at 42°C under microaerophilic conditions. Samples were diluted in MH broth and plated on MH agar plates to determine bacterial viability. Each bar represents the mean value obtained from triplicate assays.
Production of high purity His-tagged rCmeC
The anti-CmeC serum used in above in vitro immune protection assay was directed against a portion of CmeC (aa 41 to 248) [11] and may not contain all protective epitopes of CmeC. To better study immune protection conferred by CmeC vaccination in this study, two E. coli constructs were constructed for producing full-length rCmeC proteins. Both N-terminal (Figure 3A) and C-terminal His-tagged rCmeC (Figure 3B) were successfully produced in E. coli and purified by Ni-NTA agarose affinity column. However, the yield of C-terminal His-tagged rCmeC was significantly lower than that of N-terminal His-tagged rCmeC; Approximately 1.5 mg of N terminal His-tagged rCmeC was consistently purified from 100 ml of induced culture using 1 ml of Ni-NTA resin. Thus, to produce large amount of rCmeC for vaccination studies, we then focused on optimization of purification for N-terminal His-tagged rCmeC. As shown in Figure 3A, a protein band with approximate MW of 60 kDa was consistently co-purified with rCmeC when following the standard protocol. Based on the molecular weight of this co-purified protein, it is likely this contaminated protein is E. coli molecule chaperone GroEL [19]. Since it has been reported that including Mg2+-ATP in washing buffer could efficiently remove the GroEL contamination [23], the purification procedure was modified to improve the purity of the extracted rCmeC. As shown in Figure 3C, addition of 5 mM of Mg2+-ATP in lysis buffer completely removed contaminated bands, resulting in eluents containing rCmeC with high purity. Large quantities of high-purity rCmeC was obtained using the modified procedure for the vaccination and ELISA in this study.
Figure 3.
SDS-PAGE analysis of rCmeC production and purification from E. coli. (A) Expression and purification of N- terminal His-tagged rCmeC. M, standard molecular mass markers (Bio-Rad); 0h and 3h, noninduced and 3 h-induced whole-cell lysate, respectively; E, eluted rCmeC fraction using Ni-NTA affinity chromatography (Qiagen). The putative GroEL, an E. coli molecule chaperone, was consistently co-purified with rCmeC using the standard protocol. (B) Expression and purification of C-terminal His-tagged rCmeC. FL: flow through; Ag, Ni-NTA washing fractions; E1–E3, elution fractions. (C) Efficient removal of GroEL contaminant by ATP-Mg2+ treatment. Eluted fractions (E3 to E10) during Ni- NTA purification with (right panel) or without (left panel) addition of 5 mM of ATP- Mg2+ were subjected to SDS-PAGE analysis.
Immunological responses and protective efficacy following CmeC vaccination
In Trial 1, oral vaccination of chickens with or without mLT adjuvant did not significantly (P > 0.05) enhance IgG and IgA levels in serum and in intestinal lavage at different days postimmunization (Figure 4). Despite the variations observed among individual chickens, chickens in group 6 (200μg rCmeC + mLT) at 28 d postimmunization showed consistently and slightly higher serum IgG level (P = 0.57) than PBS control group. Interestingly, all groups displayed relatively higher IgG and IgA levels immediately prior to primary CmeC immunization (Figure 4), likely due to the effect of maternal antibodies [21,24]. Consistent with the patterns of these antibody responses, challenge of chickens with NCTC 11168 at 28 days after primary CmeC vaccination did not show a significant difference of colonization among groups (P > 0.05) (Data not shown). All chickens in each group were quickly colonized by C. jejuni NCTC 11168 two days after challenge with an average shedding level of ~107 CFU/g feces, likely due to the high dose of inoculums used for challenge (106 CFU/bird).
Figure 4.
Chicken immunological responses to rCmeC vaccination (Trial 1). (A) Serum IgG response elicited by rCmeC subunit vaccination in broiler chickens. All chicken serum samples were diluted 1:100 for indirect ELISA. (B) Serum IgA and intestinal IgA (the embedded figure) levels in chickens vaccinated with rCmeC. The ELISA procedure is detailed in Materials and Methods. Each bar is the average of OD405nm readings from 10 to 20 individual serum samples or 5 individual intestinal lavage samples with standard error indicated by error bar.
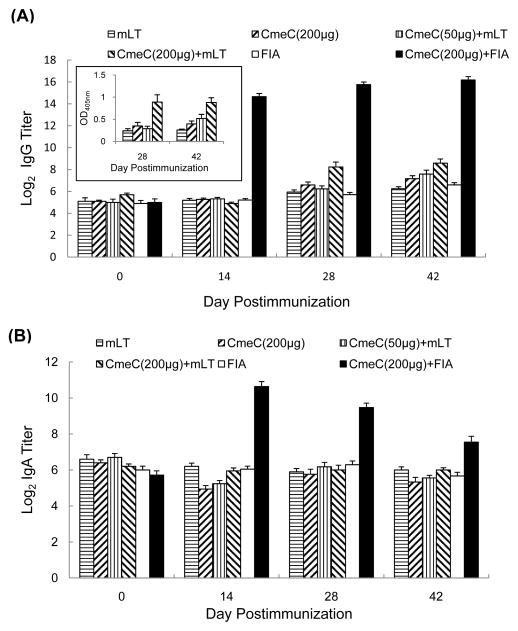
In Trial 2, oral immunization of chickens with a higher dose of CmeC (200μg) and mucosal adjuvant (70μg) significantly (P < 0.05) elevated the IgG titer than those in the control groups (receiving mLT or CmeC only) and the group administered a low dose of CmeC (50μg) plus mLT at 28 and 42 d postimmunization (Figure 5A). As shown in the embedded figure in Figure 5A, the OD405 nm values for higher dose of CmeC (200μg) together with 70μg of mLT were 0.89 ± 0.16 and 0.88 ± 0.10 at 28 d and 42 d postimmunization, respectively, which are significantly higher (P < 0.05) than the mean OD405 nm values from other groups. However, serum IgA titers were similar among the groups that received oral vaccination (Figure 5B). Subcutaneous vaccination of chickens with CmeC together with Freund’s incomplete adjuvant dramatically elevated serum IgG and IgA titers (P < 0.05) at 14, 28, and 42 d postimmunization compared with the control group vaccinated with Freund’s incomplete adjuvant only. However, CmeC vaccination did not trigger significantly higher level of intestinal secretory IgA against rCmeC, regardless of vaccination route and dosage (Figure 6). Consistent with the lack of significantly induced local IgA response to CmeC, CmeC vaccination in Trial 2 did not significantly reduce C. jejuni colonization in the intestine (Figure 7).
Figure 5.
Serum IgG and IgA titers to rCmeC (vaccination Trial 2). (A) Serum IgG titer in response to rCmeC subunit vaccination. The embedded figure shows the average of OD 405 readings of group A–F (Table 2) at 28 and 42 days postimmunization. (B) Serum IgA titer in response to CmeC subunit vaccination. Each bar is the average of log2 transformed titers or the standard error indicated by error bar.
Figure 6.
Intestinal mucosal IgA titers to CmeC (vaccination Trial 2). Each bar is the average of OD405nm readings from 5 individual intestinal lavage samples with standard error indicated by error bar.
Figure 7.
Campylobacter colonization levels after challenging CmeC-vaccinated chickens with C. jejuni NCTC 11168 (Trial 2). The letter for each type of point corresponds to the group described in Table 3. Each point represents mean log10 CFU/g feces ± the standard deviation in each group.
Discussion
Previous studies have reported that CmeC is prevalent in C. jejuni, is induced and immunogenic in vivo, and is essential for C. jejuni colonization [10–13]. These findings suggest that CmeC is a promising subunit vaccine candidate against C. jejuni colonization in the intestine. The in vitro studies in this project provided further compelling evidence that CmeC is an attractive candidate for C. jejuni vaccine development. Alignment of complete CmeC sequences from diverse strains demonstrated that CmeC is highly conserved in C. jejuni (Figure 1). This finding is consistent with a recent report in which a small portion of cmeC was PCR amplified from different Campylobacter strains for sequencing [25]. It is likely that CmeC displayed variation between different Campylobacter species, such as C. jejuni and C. coli, because alignment of partial CmeC sequence showed only 83% aa identity between C. jejuni and C. coli [25]. However, sequence analysis of full length CmeC in this study clearly indicated that the genetic variation within C. jejuni, which is responsible for ~ 90% of human campylobacteriosis [1], is extremely limited. This evidence together with in vitro inhibitory effect of CmeC peptide antiserum on the function of CmeABC efflux pump (Figure 2) provides a strong rationale to develop CmeC-based subunit vaccine.
To overcome the potential limitations of using partial CmeC peptide for vaccination studies, such as lack of enough protective epitopes, the full-length rCmeC proteins were produced and purified in this study. Expression and purification of recombinant membrane proteins in E. coli system are always coupled with problems such as low yield, insolubility, unfolding or misfolding, co-purification of contaminations, proteolytic degradation, and less biological activity. In the past decade, extensive efforts have been placed on the modification of promoter, design of fusion with tags, refolding after purification, chaperone co expression/knockout, and protease gene knockout [26–28]. In this study, the N-terminal His tagged rCmeC is located in cytoplasm due to removal of the 19-aa signal peptide. Thus, cytoplasmic rCmeC proteins which have hydrophobic transmembrane domains are likely to attract/arrest the chaperones [28]. During our purification of rCmeC, we consistently observed the co-purification of a protein with molecular weight of ~ 60 kDa together with the target rCmeC band (Figure 3A and 3C), suggesting that the GroEL (60 kDa)-GroES (10 kDa) chaperone system in E. coli [19,26] may bind newly synthesized rCmeC. Since the binding of ATP can trigger the turnover of substrates in GroEL-GroES systems by reducing the affinity of GroEL-GroES with substrates [23], 5 mM of Mg2+-ATP was added into the lysate to facilitate the disassociation of rCmeC from GroEL-GroES. This single and simple modification increased the purity of extracted rCmeC without affecting yield (Figure 3C). It is also important to mention that the detergent Empigen BB was used to facilitate solubilization of rCmeC during purification in this study because Empigen BB is a mild zwitterionic detergent and is known for its ability to preserve the antigenicity and functional activity of isolated proteins [29]. Together, the rCmeC protein with high purity has been obtained in this study for various research efforts, such as vaccination described in this study and crystallization in the future studies.
We have established a chicken model of C. jejuni infection and used this model to study the critical role of CmeC in colonization of C. jejuni [12]. We chose to use chickens for vaccination and C. jejuni challenge studies primarily for the following reasons. First, chickens are a natural host for C. jejuni. The high susceptibility of chicken to Campylobacter colonization, the ease of handling, and the low cost of chickens have provided an ideal model system to study the colonization and immunogenicity of C. jejuni in the host [8,12,15,16,30]. Second, newly hatched chickens are always Campylobacter-free, providing a clean and economical host for evaluating Campylobacter colonization [21]. Third, to date, there are no appropriate disease models for C. jejuni infection; a recently developed non-human primate model system may partly mimic symptoms caused by C. jejuni infection in humans [31]. Although C. jejuni does not cause clinical disease in chickens (an infection model, not a disease model), chickens are a sufficient and powerful colonization model to evaluate adaptation and survival of Campylobacter in response to harsh in vivo conditions (e.g. bile salts relevant to this study). Fourth, the heat labile enterotoxin (LT) has been used as a mucosal adjuvant with chicken oral vaccines [32,33], which provides a rationale for us to use mLT together with the CmeC oral vaccine in chicken model system described in this study. Finally, poultry is considered a major source for C. jejuni infection in humans [7]. Reduction of Campylobacter in poultry by vaccination of chickens will reduce the risk of exposure by humans who consume poultry products. Therefore, findings from chicken studies are also directly relevant to food safety and public health.
Oral vaccination of chickens, other animals as well as humans with subunit vaccine in conjunction with adjuvant LT or mLT have been demonstrated to induce protective immunity [32–35]. Unexpectedly, oral vaccination of CmeC with mLT in two trials performed in this study did not trigger significant local immune response to CmeC and thus failed to confer protection of chickens against C. jejuni colonization. However, the findings from this study provided useful information for future development and evaluation of CmeC subunit vaccine using the chicken as a model. First, white leghorn appears a better animal host than broiler for vaccination evaluation, primarily due to its slow growth rate, which makes animal handling easier and also allows us to initiate late primary vaccination (e.g. 3 wk of age) when Campylobacter specific maternal antibodies decrease to a low level. Second, although dramatic systemic immune response to CmeC was induced using the subcutaneous vaccination route in this study, the elevated antibody titers in serum did not result in increased levels of intestinal antibodies, which suggests that vaccination via mucosal route, such as oral vaccine, should be used to induce a strong gut mucosal immune response to control Campylobacter colonization. Third, development of an oral subunit vaccine faces a common difficulty: weak immunogenicity due to the choice of adjuvant and antigen degradation in the GI tract. Previous studies have shown that Campylobacter-specific secretory and serum IgA antibodies correlate with protection against Campylobacter infection [36]. In this study, sufficient levels of CmeC-specific antibodies, particularly intestinal IgA, were not reached for protection against C. jejuni infection. Given that the mLT used in this study has been shown to be effective with oral vaccines in different animals including chickens at low dose (e.g. 5μg single dose for pig) [35], it is unlikely that the mLT fails to stimulate immune response if it reaches chicken gut together with rCmeC. We speculate that orally administered rCmeC and/or mLT may be absorbed to the upper gastrointestinal tract and significantly degraded in the intestine before it can prime the host immune system. To solve this problem, appropriate delivery systems may be explored to enhance mucosal immune response. For example, encapsulation of rCmeC using the chitosan microsphere, an effective adjuvant/carrier system [37,38], may be a promising approach to deliver rCmeC to the target site and trigger a strong local intestinal mucosal immune response. In addition, identification of protective epitopes of CmeC followed by construction of live Salmonella-vectored vaccine is also a promising approach for persistent deliver specific protective epitopes to induce local mucosal response [39]. Elucidation of the structure of CmeC by taking advantage of the high purity rCmeC obtained from this study for crystallization will facilitate construction of a Salmonella live vaccine expressing specific surface region(s) of CmeC. These hypotheses will be tested in future studies.
Successful development of CmeC-based vaccine may lead to a novel bifunctional vaccine that not only prevents Campylobacter colonization but also combats antibiotic resistance in C. jejuni. It has been demonstrated that inhibition of multidrug efflux pumps by efflux pump inhibitors (EPIs) is an effective approach to improve the clinical performance of antibiotics [40–42]. We also observed that inhibition of CmeABC pump by an efflux pump inhibitor increased susceptibilities of C. jejuni to various antibiotics and significantly reduced the frequency of emergence of macrolide resistant C. jejuni [15,16]. However, several key issues (e.g. toxicity, in vivo stability, production cost) should be well addressed before EPIs can be used clinically and accepted by medical community. Given the significant role of CmeC in multidrug resistance, we hypothesize that CmeC antibodies may function similarly as EPI by targeting an essential component of CmeABC pump and immunization of chickens with CmeC vaccine may enhance the activity of clinical antibiotics against C. jejuni. Compared to EPIs, the CmeC vaccine may be a more realistic approach to potentiate the activity of clinical antibiotics against Campylobacter. Thus, CmeC may also represent a novel vaccine that can enhance the efficacy of clinical antibiotics and even reduce the frequency of in vivo emergence of antibiotic resistant C. jejuni. This hypothesis needs to be determined after optimization of CmeC vaccination regimen in the future.
Acknowledgments
We are grateful to Dr. John D. Clements (Tulane University Medical Center) for kindly providing the mucosal adjuvant mLT-R192G as a gift for the vaccination studies in this paper. We thank Ky Van Hoang, Andree A. Hunkapiller, and Ad’lynn Martinez for technical support. This study was supported by grant 1R21AI07255101 from NIH.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Olson CK, Ethelberg S, Pelt WV, Tauxe RV. Epidemiology of Campylobacter jejuni infections in industrialized countries. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. ASM Press; Washington, DC: 2008. pp. 163–189. [Google Scholar]
- 2.Rees JH, Soudain SE, Gregson NA, Hughes RA. Campylobacter jejuni Infection and Guillain-Barre syndrome. N Engl J Med. 1995;333(21):1374–1379. doi: 10.1056/NEJM199511233332102. [DOI] [PubMed] [Google Scholar]
- 3.Nachamkin I, Allos BM, Ho T. Campylobacter species and Guillain-Barre Syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 5.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001;7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Lin J. Novel approaches for Campylobacter control in poultry. Foodborne Pathog Dis. 2009;6:755–765. doi: 10.1089/fpd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Zoete MR, van Putten JP, Wagenaar JA. Vaccination of chickens against Campylobacter. Vaccine. 2007;25:5548–557. doi: 10.1016/j.vaccine.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Jagusztyn-Krynicka EK, Laniewski P, Wyszynska A. Update on Campylobacter jejuni vaccine development for preventing human campylobacteriosis. Expert Rev Vaccines. 2009;8:625–645. doi: 10.1586/erv.09.21. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Akiba M, Sahin O, Zhang Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC. Campylobacter Antimicrob Agents Chemother. 2005;49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter0 jejuni. Antimicrob Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Sahin O, Michel LO, Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 2003;71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Cagliero C, Guo B, Barton YW, Maurel MC, et al. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Bacteriol. 2005;87:7417–7424. doi: 10.1128/JB.187.21.7417-7424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stintzi A, Marlow D, Palyada K, Naikare H, Panciera R, et al. Use of genome5 wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect Immun. 2005;73:1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez A, Lin J. Effect of an effluxpump inhibitor on the function of the multidrug effluxpump CmeABC and antimicrobial resistance in Campylobacter. Foodborne Pathog Dis. 2006;3:393–402. doi: 10.1089/fpd.2006.3.393. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Martinez A. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J Antimicrob Chemother. 2006;58:966–972. doi: 10.1093/jac/dkl374. [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Zeng X, Haigh RD, Ketley JM, Lin J. Identification and Characterization of a New Ferric Enterobactin Receptor, CfrB, in Campylobacter. J Bacteriol. 2010;192:4425–4435. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiba M, Lin J, Barton YW, Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 19.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, et al. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyszynska A, Raczko A, Lis M, Jagusztyn-Krynicka EK. Oral immunization of chickens with avirulent Salmonella vaccine strain carrying C. jejuni 72Dz/92 cjaA gene lelicits specific humoral immune response associated with protection against challenge with wild-type Campylobacter. Vaccine. 2004;22:1379–1389. doi: 10.1016/j.vaccine.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Sahin O, Zhang Q, Meitzler JC, Harr BS, Morishita TY, et al. Prevalence, antigenic specificity, and bactericidal activity of poultry anti-Campylobacter maternal antibodies. Appl Environ Microbiol. 2001;67:3951–3957. doi: 10.1128/AEM.67.9.3951-3957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, et al. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci U S A. 2002;99:11103–11108. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harjes S, Scheidig A, Bayer P. Expression, purification and crystallization of human 3′-phosphoadenosine-5′-phosphosulfate synthetase 1. Acta Crystallogr D Biol Crystallogr. 2004;60:350–352. doi: 10.1107/S0907444903027628. [DOI] [PubMed] [Google Scholar]
- 24.Sahin O, Luo N, Huang S, Zhang Q. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Appl Environ Microbiol. 2003;69:5372–5379. doi: 10.1128/AEM.69.9.5372-5379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fakhr MK, Logue CM. Sequence variation in the outer membrane protein37 encoding gene cmeC, conferring multidrug resistance among Campylobacter jejuni and Campylobacter coli strains isolated from different hosts. J Clin Microbiol. 2007;45:3381–3383. doi: 10.1128/JCM.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Marco A. Protocol for preparing proteins with improved solubility by co40 expressing with molecular chaperones in Escherichia coli. Nat Protoc. 2007;2:2632–2639. doi: 10.1038/nprot.2007.400. [DOI] [PubMed] [Google Scholar]
- 27.de Marco A. Two-step metal affinity purification of double-tagged (NusA-His6) fusion proteins. Nat Protoc. 2006;1:1538–1543. doi: 10.1038/nprot.2006.289. [DOI] [PubMed] [Google Scholar]
- 28.de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;12:7–32. doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Meitzler JC, Huang S, Morishita T. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect Immun. 2000;68:5679–5689. doi: 10.1128/iai.68.10.5679-5689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conlan AJ, Coward C, Grant AJ, Maskell DJ, Gog JR. Campylobacter jejuni colonization and transmission in broiler chickens: a modelling perspective. J R Soc Interface. 2007;4:819–829. doi: 10.1098/rsif.2007.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteiro MA, Baqar S, Hall ER, Chen YH, Porter CK, et al. Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni. Infect Immun. 2009;77:1128–1136. doi: 10.1128/IAI.01056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice BE, Rollins DM, Mallinson ET, Carr L, Joseph SW. Campylobacter jejuni in broiler chickens: colonization and humoral immunity following oral vaccination and experimental infection. Vaccine. 1997;15:1922–1932. doi: 10.1016/s0264-410x(97)00126-6. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson J, Rood D, Minior D, Guillard K, Darre M, et al. Immune response to a mucosally administered aflatoxin B1 vaccine. Poult Sci. 2003;82:1565–1572. doi: 10.1093/ps/82.10.1565. [DOI] [PubMed] [Google Scholar]
- 34.Lapa JA, Sincock SA, Ananthakrishnan M, Porter CK, Cassels FJ, et al. Randomized clinical trial assessing the safety and immunogenicity of oral microencapsulated enterotoxigenic Escherichia coli surface antigen 6 with or without heat-labile enterotoxin with mutation R192G. Clin Vaccine Immunol. 2008;15:1222–1228. doi: 10.1128/CVI.00491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan L, Iosef C, Azevedo MS, Kim Y, Qian Y, et al. Protective immunity and antibody-secreting cell responses elicited by combined oral attenuated Wa human rotavirus and intranasal Wa 2/6-VLPs with mutant Escherichia coli heat-labile toxin in gnotobiotic pigs. J Virol. 2001;75:9229–6238. doi: 10.1128/JVI.75.19.9229-9238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tribble DR, Baqar S, Thompson SA. Development of a Human Vaccine. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. ASM Press; Washington, DC: 2008. pp. 429–444. [Google Scholar]
- 37.Sinha VR, Singla AK, Wadhawan S, Kaushik R, Kumria R, et al. Chitosan microspheres as a potential carrier for drugs. Int J Pharm. 2004;274:1–33. doi: 10.1016/j.ijpharm.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Arca HC, Gunbeyaz M, Senel S. Chitosan-based systems for the delivery of Vaccine antigens. Expert Rev Vaccines. 2009;8:937–953. doi: 10.1586/erv.09.47. [DOI] [PubMed] [Google Scholar]
- 39.Kwon YM, Cox MM, Calhoun LN. Salmonella-based vaccines for infectious diseases. Expert Rev Vaccines. 2007;6:147–152. doi: 10.1586/14760584.6.2.147. [DOI] [PubMed] [Google Scholar]
- 40.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas 25 aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomovskaya O, Watkins W. Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J Mol Microbiol Biotechnol. 2001;3:225–2236. [PubMed] [Google Scholar]
- 42.Ryan BM, Dougherty TJ, Beaulieu D, Chuang J, Dougherty BA, et al. Efflux in bacteria: what do we really know about it? Expert Opin Investig. 2001;10:1409–1422. doi: 10.1517/13543784.10.8.1409. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Luangtongkum T, Morishita TY, Zhang Q. Molecular typing of Campylobacter strains using the cmp gene encoding the major outer membrane protein. Foodborne Pathog Dis. 2005;2:12–23. doi: 10.1089/fpd.2005.2.12. [DOI] [PubMed] [Google Scholar]
- 44.Zeng X, Xu F, Lin J. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect Immun. 2009;77:5437–5448. doi: 10.1128/IAI.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 46.Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental pylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]