Abstract
Adverse insults during intrauterine life can result in permanent changes in the physiology and metabolism of the offspring, which in turn leads to an increased risk of disease in adulthood. This is an adaptational response by the fetus to changes in the environmental signals that it receives during early life to ensure its survival and prepare itself for postnatal life. Increasing evidence suggests that the epigenetic regulation of gene expression patterns has a crucial role in the developmental programming of adult disease. This review summarizes recent studies of epigenetic mechanisms and focuses particularly on studies that explore identifiable epigenetic biomarkers in the promoters of specific disease-associated genes. Such biomarkers would enable early recognition of children who might be at risk of developing adult disease with fetal origins.
Introduction
During the late 1980s, the ‘developmental origins of adult disease hypothesis’, often called the ‘Barker hypothesis’, was originally proposed by Barker and colleagues, based on a series of retrospective epidemiological studies, which described the potential relationship of fetal growth restriction and cardiovascular and/or metabolic disease in adult life [1,2]. Since then, substantial experimental evidence from different mammalian species has reliably supported the hypothesis that an adverse in utero environment has a programming role in postnatal physiology and pathophysiology. It has been demonstrated that prenatal undernutrition [3] and hypoxia [4,5], as well as other intrauterine insults, such as exposure to toxins [6,7], result in an increased risk of developing cardiovascular and metabolic disorders in later life, including hyperphagia, obesity, endocrine and metabolic abnormalities, type 2 diabetes mellitus, insulin resistance, hypertension and ischemic heart disease.
To explain how intrauterine malnutrition influences fetal development and the susceptibility of offspring to adult disease, the thrifty phenotype hypothesis was proposed [8,9]. When there is a change in the intrauterine environment, the fetus makes adaptations to ensure its survival. Once the adverse environment persists beyond the point of reversible adaptation, the fetus is forced to make an irreversible adaptation that will result in persistent alterations to physiological and metabolic homeostatic set points. After birth, if the adjusted set point does not match the postnatal environment, adult diseases will be induced.
Although the Barker and thrifty phenotype hypotheses provide conceptual thoughts about the developmental programming of adult disease, it is still unclear how fetal developmental plasticity enables organisms to make adaptational responses to the fetal environment that can result in permanent adverse effects later in life. Recent studies have strongly suggested that epigenetic processes of gene expression patterns are influenced by the environment, and might have key roles in the developmental programming of adult disease [10]. In this review, we summarize current studies of the epigenetic mechanisms of developmental programming of adult disease and provide an explicit systematic blueprint of this field (Figure 1).
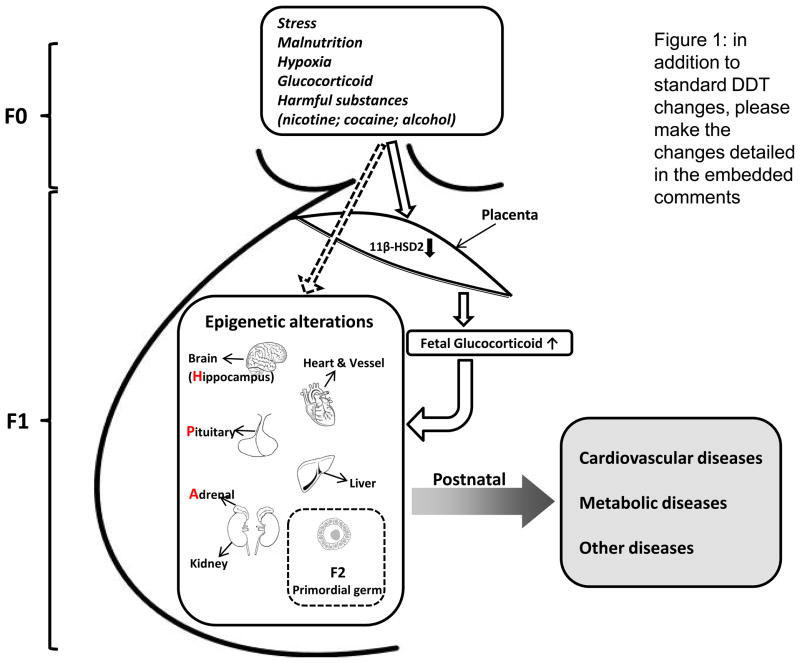
Figure 1.
Epigenetic mechanisms in the developmental programming of adult diseases. An adverse in utero environment caused by maternal stress, malnutrition, hypoxia or exposure to harmful substances results in epigentic modifications of gene expression patterns that can lead to diseases later in life. Additionally, alterations in placental 11-β-hydroxysteroid dehydrogenase-2 (11β-HSD2) might also have an important role in maternal stress-mediated epigenetic modifications in the fetal hippocampus–pituitary–adrenal (HPA) axis via its regulation of glucocorticoid levels. The epigenetic alterations that originated in utero eventually result in a postnatal phenotype with heightened susceptibility to diseases. Furthermore, the epigenetic alterations that occur in primordial germ cells can be inherited and might influence subsequent generations.
Epigenetic mechanisms of developmental programming of adult disease
Links between epigenetic process and developmental programming
Epigenetic modification relates to the stable and heritable patterns of gene expression that do not involve changes in DNA sequence [11]. Unlike genetic information, which is extremely stable, epigenetic modifications mark the effects of early environmental events and ensure sustained and reversible responses to transient stimuli, which result in modified gene expression patterns and phenotypes later in life. Several pieces of evidence support the hypothesis that epigenetic modifications are sensitive to environmental stimuli. First, monozygotic twins have an identical genotype but their phenotype is not usually identical. This manifests as discordances in the frequency or onset of disease. This could be explained by the accumulation of epigenetic changes that occur over their lifetimes; for example, in older twins, the overall content and distribution of methylated cytosines and histone acetylation are remarkably different, even though they were epigenetically indistinguishable at a younger age [12]. Studies of monozygotic twins have also revealed an association between epigenetic differences and discordance in diseases, such as bipolar disorder, Silver–Russell syndrome, Beckwith–Wiedemann syndrome and transient neonatal diabetes mellitus [13–16]. Second, studies in assisted reproduction technology (ART) directly demonstrate that the in vitro culture environment affects global patterns of DNA methylation, which can lead to the long-term alteration of the expression of genes involved in chronic metabolic disorders, such as obesity and type 2 diabetes mellitus [17]. Third, studies of metastable epialleles (i.e. alleles that are variably expressed in genetically identical individuals owing to epigenetic modifications established during early development; they are thought to be particularly vulnerable to environmental influences [18]) in mouse strains show the environmental effect on the epigenetic regulation of gene expression and function, especially the epigenetic effect of the in vitro environment on the later health of offspring. In the agouti viable yellow (avY) mouse, hypomethylation of the agouti gene promoter increased its gene expression and resulted in a yellow coat color as well as obesity (the agouti gene regulates body weight at the level of the hypothalamus). Conversely, hypermethylation decreased the expression of agouti and resulted in mice with a brown coat color and normal weight. In the same strain of mice, if the pregnant dam was fed food containing restricted methyl donors and cofactors (such as folate), the results included agouti promoter hypomethylation, increased prevalence of yellow coat color, obesity and even cancer in her offspring [19]. By contrast, feeding the dam soy isoflavone genistein led to promoter hypermethylation, decreased the prevalence of obesity and gave her offspring a brown coat [20].
These findings illustrate the fact that environmental signals early during the developmental period might produce an inherited epigenetic modification with potential long-term consequences. Epigenetic aberrations serve as a memory of exposure to inadequate or inappropriate chemical and/or nutritional or nonchemical environments during early life. Thus, identifying the specific features and functions of the epigenetic build-up at these stages and determining the mechanisms by which environmental factors might affect them in the long term will be a major milestone in the investigation of the intrauterine origin of adult disease, and could also reveal potential intervention points and means.
DNA methylation and its reprogramming process during fetal development
Epigenetic modifications to chromatin include 5′ methylation of the cytosine residue in CpG dinucleotides of DNA, covalent modifications (including methylation, acetylation, phosphorylation and ubiquitination) of histones and the gene-regulating and chromatin-organizing activities of noncoding RNAs. These epigenetic modifications change the binding of transcription activators and repressors to specific gene promoters, and/or alter the large-scale conformation and function of chromatin itself, which modulates gene expression [21].
Methylation appears to be the most dominate and best-studied epigenetic modification of DNA. DNA methylation in mammalian and other vertebrates generally occurs on position 5 of the pyrimidine ring of cytosine nucleotides in the symmetrical dinucleotide CpG sequence [22]. The overall frequency of CpG dinucleotides in the vertebrate genome is approximately 20% and the clustered CpG-rich areas (so-called CpG islands) usually occur in the 5′ end and the promoter regions of genes [23]. Approximately half of all transcribed genes have CpG islands, which includes all genes that are widely expressed; approximately 40% of genes are expressed in a tissue-specific manner. Of CpGs located outside of CpG islands, 80% are methylated, but CpG islands found in the promoter regions of active genes are normally not methylated [23]. DNA methylation of CpG islands is usually associated with transcriptional silencing of the associated gene [24]. In addition, DNA methylation in some non-CpG island CpG dinucleotides sometimes also has an important role in regulating the expression of certain genes, depending on the regulating role of the epigenetically modified loci in the promoter. Whereas DNA methylation that occurs in the promoter regions generally leads to gene repression by inhibiting transcription factor binding either directly or indirectly via the recruitment of methyl-CpG-binding domains (MBDs), the increased methylation in the binding sites of gene repressors results in an increase in gene activity. Additionally, DNA methylation-mediated downregulation of the repressors themselves might lead to an increase in the activities of the target genes of those repressors. Furthermore, DNA methylation can result in increased gene activities through its regulation of miRNA expression.
DNA methylation is a reversible and dynamic process [26], catalyzed by DNA methyltransferases (DNMTs). The major de novo methyltransferases, DNMT3a and DNMT3b, are principally responsible for establishing cytosine methylation at previously unmethylated CpG sites [26], particularly during embryogenesis. Genome-wide alterations in methylation by mutations in DNMTs or knockouts of the methylase genes cause a significant reduction in the level of methylated cytosines, abnormal development and embryonic lethality [27]. Other than DNMT3a and DNMT3b, DNMT1 recognizes methylation associated with the parental strand and imposes this pattern on the newly formed daughter strand of DNA; thus, methylation patterns imposed on the genome at precursor cells could be maintained by DNMT1, which is referred to as the ‘symmetrical methylation rule’ [28]. Reduction of DNMT1 expression can result in genome-wide hypomethylation in all tissues and leads to tumor development in later life [29].
During the process of DNA methylation, methylated-CpG dinucleotides are recognized by a family of MBD proteins, including MBD1–4 and methyl-CpG-binding protein 2 (MeCP2). MeCP2 binds to single methylated-CpG dinucleotides and causes chromatin remodeling and gene silencing by recruiting chromatin-remodeling corepressor complexes to the regions of DNA that bind MeCP2 [30]. Inactivating mutations in either DNMT3b or MeCP2 are associated with immune deficiency, centromeric instability and facial anomalies (ICF) syndrome and Rett syndrome [31–34].
Given that the epigenetic information can be modified and faithfully transmitted during cell development and differentiation, questions arise as to how and when the epigenetic regulation of gene expression patterns serve as a link between prenatal environmental cues, gene expression and later health. The fact that DNA methylation patterns experience a reprogramming period during gametogenesis and early embryo genesis provides a crucial window for the epigenetic-mediated developmental programming of adult disease [35]. The first phase of methylation reprogramming occurs immediately after fertilization, when the entire paternal genome (except for paternally imprinted genes, heterochromatin around centromeres and some repetitive elements) is rapidly demethylated [36]. This naturally demethylation process then slows down generally and keeps until the morula stage [36]. Simultaneously, the maternal genome undergoes a relatively slow demethylation compared with the paternal genome [37]. After five cell cycles at the first differentiate event, de novo methylation begins. After this point, the methylation level is higher in the inner cell mass that gives rise to somatic tissues than in the trophoectoderm, which forms the future placenta [37]. At a later stage of development, the asymmetry pattern of DNA methylation in somatic tissues is maintained and there are tissue-specific patterns of both DNA methylation [38] and histone modification [39,40]. After de novo remethylation, the primordial germ cells migrate through the allantois to the developing germinal ridges, where they eventually differentiate into mature gametes, thus completing this cycle of epigenetic reprogramming. During gametogenesis, primordial germ cells undergo another DNA methylation reprogramming process. Before their migration to the genital ridge, new patterns of methylation are established (before birth in the male germ line and after birth in the female line). The early primordial germ cells then erase their somatic-like epigenetic patterns [41]. This demethylation process completes around day 12–13 after fertilization. Once this occurs, the remethylation starts again and shows a sex-specific timing. In the male, de novo methylation begins at day 15.5–18.5 of gestation, continues postnatally and is completed in pachytene spermatocytes [42]. Little is known about overall DNA remethylation during oogenesis, and most studies performed to date have concentrated on imprinted genes. In females, imprints are acquired at different times before ovulation in maturing oocytes [43]. A growing body of evidence suggests that epigenetic gene regulation is influenced by the environment and is one of the molecular mechanisms with an important role in the foundation of developmental programming [10]. The differential timing of remethylation during gametogenesis might have implications for studies of sex-dependant changes caused by prenatal insults, which we review below.
Early embryogenesis is therefore a crucial period for the establishment of epigenotypes. As key nutrients are needed for the methylation process, dietary excess or deficiencies might alter the epigenetic patterns, as might as other environmental factors, such as glucocorticoid (GC) exposure, maternal cigarette smoking, maternal hypoxia or maternal drug abuse. These environmentally induced epigenetic alterations can be maintained throughout a lifetime, influencing gene expression patterns and causing changes in phenotypic traits [44].
Other epigenetic modification mechanisms and their crosstalk with DNA methylation
In eukaryotic cells, the basic unit of chromatin is the nucleosome, which comprises 146 bp of DNA wrapped around eight histone proteins; in turn, each histone protein comprises two copies each of histone (H)2A, H2B, H3 and H4 [45]. Modulation of the structure of chromatin is crucial for the regulation of gene expression because it determines the accessibility and the sequential recruitment of regulatory factors to the underlying DNA. Histones undergo a variety of reversible post-translational modifications, including acetylation and methylation of conserved lysine and arginine residues on the amino terminal tail domain, phosphorylation of serines and threonines, ubiquitinylation and ADP ribosylation [46]. The most common histone modifications are acetylation and methylation of lysine residues in the amino termini of H3 and H4. Histone acetylation, for instance, is generally associated with euchromatin, a relaxed, higher order chromatin structure, and this is thought to permit or facilitate the access of gene regulatory proteins, which include transcription and elongation factors. By contrast, hypoacetylated histone is associated with heterochromatin, a highly condensed inactive chromatin structure [46]. Histone methylation can be a marker for both active and inactive regions of chromatin. Methylation of lysine 9 on the N terminus of histone H3 (H3K9) is a hallmark of silent DNA and is globally distributed throughout heterochromatic regions, such as centromeres and telomeres. By contrast, methylation of lysine 4 of histone H3 (H3K4) denotes activity and is found predominantly at promoters of active genes [47]. Because lysine methylation can be monomeric, dimeric or trimeric, and histones might also be subject to other post-translational modifications, such as phosphorylation [48], this enormous variation leads to many possible combinations of different modifications. This might constitute a ‘histone code’ [49], which can be read and interpreted by different cellular factors.
miRNAs, a class of small noncoding RNAs, are also important players in the epigenetic control of gene expression [50]. miRNA genes are transcribed into primary miRNAs and then processed to precursor miRNAs. An endoribonuclease dicer cleaves precursor miRNAs into the final short fragments called matured miRNAs. miRNAs bind to their target mRNAs and lead to degradation of the target or translational suppression of the target genes [51].
The interactions of DNA methylation, histone modifications and miRNA might serve as a self-reinforcing network to regulate gene expression patterns in a sophisticated feedback manner (Figure 2). First, DNA methylation-induced chromatin silencing might be facilitated by the coupling action of covalent modifications of histone and vice versa [52]. For example, H3K9 methylation is a prerequisite for DNA methylation [53], and DNA methylation might also trigger H3K9 methylation [54]. Interactions among histone deacetylases (HDACs), histone methyltransferases and methylcytosine-binding proteins lead to the recruitment of DNMTs [55], although it is not yet clear what initiates the recruitment of the different epigenetic modifiers to their specific target sequences. The relation and consequences of DNA methylation and histone modifications might be more complicated than is currently thought. Individual histone modification, DNA methylation or their interactions might mediate bidirectional effects on gene transcriptional outcomes and the epigenetic marks might not be uniformly associated with ‘on’ or ‘off’ transcriptional states but instead with the potential for phenotypic plasticity [56]. Furthermore, the true causal effects of epigenetic modifications in the developmental programming of phenotypic and functional changes remain to be determined [57]. Second, miRNAs have been shown to provide post-transcriptional regulation of chromatin via targeting the enzymes involved, including histone methyltransferases, MeCPs, chromo-domain-containing proteins, HDACs and DNMT enzymes [51,58–60]. Furthermore, the miRNA-initiated transcriptional gene silencing complex can guide the histone methyltransferase to specific chromatic regions and form silenced heterochromatin [61]. Third, miRNA genes are also targeted by epigenetic modification. For example, an aberrant pattern of methylation of CpG islands near or within miRNA genes could result in misregulated expression of key miRNAs and ultimately in pathogenic alterations, including tumorigenesis [62]. Lastly, primary miRNAs are either transcribed by RNA polymerase (Pol) II as independent transcriptional units or coexpressed from the intronic regions of host gene transcripts that might serve as a feedback regulation loop of miRNA regulating the expression of host genes as well as its own.
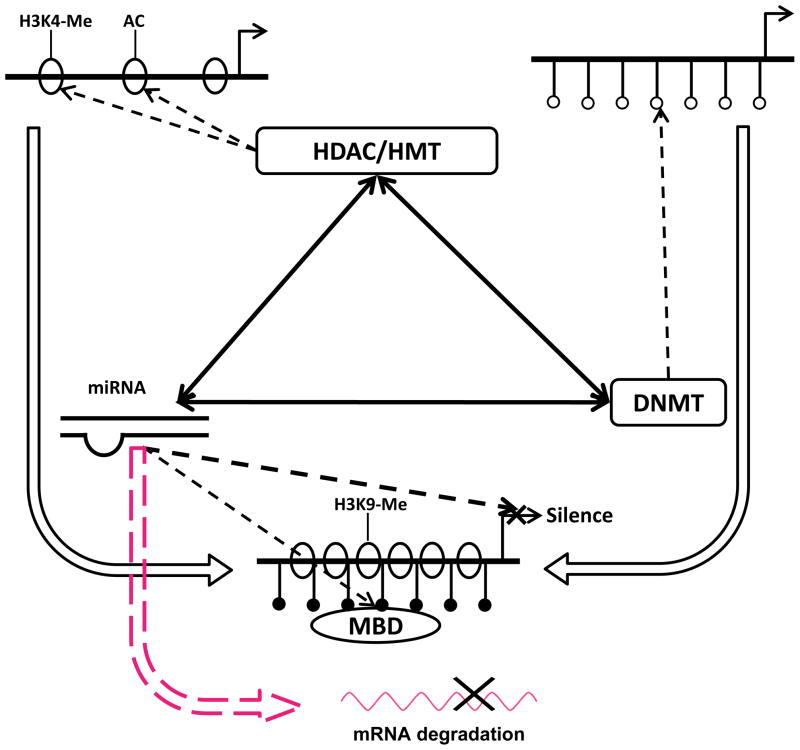
Figure 2.
The relationship between DNA methylation, histone modifications and miRNA. DNA methylation-caused chromatin silencing can be facilitated by the coupling action of covalent modifications of histone and vice versa. Interactions between histone deacetylases (HDAC), histone methyltransferases (HMT) and methyl CpG-binding proteins (MBD) lead to the recruitment of DNA methyltransferases (DNMT). miRNAs can suppress protein expression by increasing the degradation of mRNAs. Additionally, miRNAs can modulate gene transcriptional activities via their regulation of HMT, MBD, HDAC and DNMT. Furthermore, miRNA genes themselves are also targeted by epigenetic modifications. Abbreviations: AC, acetylation; H3K4, histone H3 lysine 4; H3K9, histone H3 lysine 9; Me, methylation.
Techniques for detecting epigenetic mechanisms
The most commonly used techniques for detecting DNA methylation are bisulfite genomic sequencing [63] and methylation-specific PCR [64]. Bisulfite genomic sequencing is a useful approach for a comprehensive investigation of the methylation status of CpG islands in multiple clones, whereas methylation-specific PCR is used for studying CpG sites in non-CpG island, sequence-specific transcription factor binding sites. HPLC is another reliable and reproducible method for measuring DNA methylation levels in tissues. This method has been recently improved by reducing the starting DNA requirement from 50 mg to 3 mg, which enables the method to be used with low DNA-yield samples [65]. Additionally, DNA methylation microarrays have been used to examine extensive DNA methylation in large segments of DNA [66]. Others methods used to measure DNA methylation include DNA fingerprinting [67], methylation-specific quantum dot fluorescence resonance energy transfer (MS-qFRET) [68], high performance capillary electrophoresis (HPCE) [69] and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry [70]. For detecting histone modifications, chromatin immunoprecipitation (ChIP) and ChIP on single nucleotide polymorphisms (SNP)-chip are widely used [71]. Similar to DNA methylation, techniques for measuring histone modifications are developing rapidly. New techniques, such as pathology tissue-ChIP (PAT-ChIP) [72] and the purification of histone ubiquitin ligases have been used recently [73]. Furthermore, a method using nanofluidics and multi-color fluorescence microscopy has been used to detect DNA and histone modifications simultaneously in individual chromatin fragments at approximately 10 Mbp/min [74]. This technique provides an unprecedented opportunity for genome-wide, simultaneous analyses of multiple epigenetic marks on single molecules using femtogram quantities of samples. Methods for detecting miRNA include computational approaches for miRNA target prediction and molecular biology-based analysis methods, such as cloning, northern blotting, RT-PCR and microarrays [75]. Currently, new techniques based on electronic and optical signal transduction appear to be promising candidates for use in emerging miRNA analysis applications [75].
Evidence of epigenetic mechanisms in developmental programming of adult disease
Epigenetic mechanisms in malnutrition models
It has been demonstrated that intrauterine growth restriction (IUGR) is accompanied by changes in the quantity and activity of enzymes responsible for modifying chromatin. In the IUGR rat model, global decreases in DNA methylation and increases in H3 acetylation on lysine 9 (K9) and K14 are observed in the brain at birth [76]. These changes are accompanied by a concomitant decrease in DNMT1, MeCP2 and HDAC1 [76]. Liver chromatin modifications are also induced by IUGR and present as persistent increases in acetylation of H3K9 and K14 [77], as well as reduced hepatic expression of DNMT1 [78].
Maternal diet during pregnancy can also elicit changes within the epigenome. Studies of offspring of low protein (LP)-fed dams have identified changes in DNA methylation of the genes encoding the GC receptor (GR) and the peroxisome proliferator-activated receptor-α (PPARα), leading to the increased expression of their corresponding transcripts in the liver [79]. Bogdarina et al. used a LP diet rat model to study the intrauterine programming of hypertension. They demonstrated decreased methylation of the gene promoter for angiotensin receptor, type 1b (Agtr1b) [80]. Correlating with an increase in the expression of the Agtr1b mRNA transcript and receptor protein expression, these changes are thought to augment the regulation of blood pressure and contribute to the development of hypertension later in life. A more recent study in mice reported that a maternal LP diet can differentially methylate over 200 promoter regions within the fetal liver, including the liver-X-receptor α (Lxra) gene [81]. Follow-up studies in the offspring showed a reduction in the levels of hepatic DNA promoter methylation at the gene encoding the GR, a reduction in H3 dimethylation and increases in H3 acetylation at this locus, which were associated with increased transcription [78].
Maternal calorie restriction has been associated with hypermethylation of the oncogene H-Ras in adult offspring [82]. Changes in histone modifications of offspring have also been demonstrated in maternal calorie restriction models, including losses in acetylation and increases in dimethylation of H3 at the glucose transporter type 4 (GLUT4) locus. These changes persist into adulthood, and are thought to lead to the development of type 2 diabetes mellitus in the adult offspring [83]. Pancreas/duodenum homeobox protein 1 (Pdx-1) has been shown to be epigenetically regulated in fetuses from intrauterine arterial ligation dams with deacetylation of core histones and a reduction in upstream transcription factor-1 (USF-1) binding [84]. With aging, these offspring exhibited further changes in the histone marks and increased DNA methylation of the Pdx-1 locus, which were both associated with a progressive reduction in Pdx-1 expression [84]. Global changes in DNA methylation were observed in sheep offspring of dams fed with alterations in vitamin B and methionine during the periconceptional period [85]. In addition, supplementation of the diet of pregnant mice with methyl donors altered the methylation of genes implicated in allergic airway disease [86]. It has been shown that the effects of neonatal leptin on hepatic gene expression and epigenetic status in adult offspring are directionally dependent on the nutritional status of the animal in utero [87]. Additionally, studies in sheep demonstrated that periconceptional undernutrition resulted in epigenetic changes in the hypothalamic proopiomelanocortin and GR genes in the fetus [88].
Aaggard-Tillery et al. [89] used a nonhuman primate model to investigate the effects of maternal diet on alternations to the epigenome that might be related to obesity. The authors identified obesity and hyperacetylation of H3K14 in the liver of offspring that were fed with a high-fat diet (35% fat compared with a control diet of 13% fat) [89]. In a separate study, they found that decreasing the high fat diet from 35% to 32% during pregnancy maintained obesity in offspring but the epigenetic changes were lost [90]. Recently, it has been shown in rats that a chronic high-fat diet in fathers causeed hypomethylation of the interleukin 13 receptor, alpha 2 (Il13ra2) gene and programming of β-cell dysfunction in F1 female offspring, demonstrating a concept of non-genetic, intergenerational transmission of metabolic sequelae of a high-fat diet from father to offspring [91].
Recent studies in humans have also shown long-term effects of maternal diet on the epigenome of the offspring. It has been demonstrated that individuals exposed to the Dutch famine of 1944 (the so-called ‘hunger winter’) have lower levels of methylation of the insulin-like growth factor 2 (IGF2) gene in adulthood [92]. Furthermore, Tobi et al. [93] identified an additional six loci that were differentially methylated after prenatal exposure to the famine; all six loci were implicated in either growth or metabolic and cardiovascular phenotypes.
Epigenetic mechanisms in other prenatal insult models
Hypoxia is a common intrauterine stress. A fetus might experience prolonged hypoxic stress under a variety of conditions, including pregnancy at high altitude, pregnancy with anemia, placental insufficiency, cord compression, preeclampsia, heart, lung and kidney disease, or hemoglobinopathy. Animal studies have suggested a possible link between fetal hypoxia and increased risk of cardiovascular disease in the resulting offspring [94–102]. Studies in rats have found that maternal hypoxia resulted in an increase in cardiac vulnerability to ischemia and reperfusion injury in male offspring [99,102,103]. In addition, it has been demonstrated that downregulation of protein kinase C epsilon [PKCε (the gene is PRKCE)] protein expression in the hearts of adult offspring is a mechanism for increased heart susceptibility to ischemia and reperfusion injury in animals exposed to hypoxia before birth [104]. A recent study showed that prenatal hypoxia caused repression of PRKCE in the heart via increased CpG methylation of specificity protein 1 (SP1) and early growth response protein 1 (Egr-1) binding sites at the PRKCE promoter [105]. This provides clear evidence of a novel mechanism of methylation in non-CpG island, sequence-specific transcription factor binding sites in the subtle epigenetic modifications of gene expression patterns in developmental programming of cardiac function in response to an adverse intrauterine environment. Although how hypoxia directly causes the increased methylation of the PKCε promoter remains unclear, recent studies suggest a link between prolonged oxidative stress and aberrant DNA methylation patterns [106]. Given the fact that cardiomyocytes are major producers of reactive oxygen species owing to their high metabolic demand, it is plausible that increased oxidative stress is a mechanism in the hypoxia-induced methylation and repression of PRKCE in the heart.
Maternal cigarette smoking is another major health concern worldwide and has been associated with adverse pregnancy outcomes for the mother, the fetus and the newborn child. The adverse consequences have been well identified in epidemiological studies, including intrauterine growth restriction, sudden infant death syndrome and cardiovascular disease in offspring [107–109]. As one of the main toxic components in cigarette smoke, nicotine is a major contributor to the development of cardiovascular disorders. It has been demonstrated that antenatal nicotine exposure caused PRKCE repression in the developing heart through an increase in methylation of the Egr-1 binding site at the PRKCE promoter [110,111]. It appears that nicotine has no direct effect but instead stimulates the release of sympathetic neurotransmitter norepinephrine in the fetal heart, resulting in PRKCE repression. Interestingly, maternal hypoxia caused an increase in methylation at both the SP1 and Egr-1 binding sites in the fetal heart, yet the direct effect of hypoxia on cardiomyocytes resulted in increased methylation only at the SP1 binding sites [105]. These findings are intriguing and suggest a lack of a direct effect of hypoxia on methylation at the Egr-1 binding site. Given the fact that fetal hypoxia also increases sympathetic activity [112], it is possible that the increased Egr-1 methylation at the PRKCE promoter in fetal hearts seen in the rat model of maternal hypoxia was mediated in part by an increase in sympathetic neurotransmitter norepinephrine content in the heart in response to hypoxic stress. Although these studies demonstrate the different patterns of promoter methylation between maternal hypoxia and nicotine treatments, they also reveal a possible common mechanism of norepinephrine-mediated increase in methylation of the Egr-1 binding site at the PRKCE promoter in the developing heart.
Another well-known insult to the developing fetus is caused by maternal cocaine administration. Antenatal cocaine exposure causes the downregulation of PKCε in the heart, resulting in heightened ischemic injury and reduced preconditioning-mediated protection in the heart [7,113]. Similar to the findings of maternal hypoxia and fetal nicotine exposure, it has been shown that cocaine increased CpG methylation of certain loci at the PRKCE promoter, leading to decreased transcription factor binding to the promoter and PRKCE repression in the heart [114,115].
Epigenetic mechanisms in the placenta
The placenta is the principal metabolic, respiratory, excretory and endocrine organ of the fetus; there is substantial molecular variation across its fetal and maternal compartments and it affects fetal growth [116]. There are approximately 80 known imprinted genes (approximately 200 predicted), a large percentage of which are expressed in trophoblasts and regulate placental growth [117]. In a study of IUGR and non-IUGR placentas, an altered expression pattern of imprinted genes, including increased pleckstrin homology-like domain, family A, member 2 (PHLDA2) and decreased mesoderm-specific transcript homolog protein (MEST), in placental response to maternal vascular underperfusion, was investigated [118]. The unbalanced expression of these two oppositely imprinted genes is one component of the adaptive response of placental tissue to chronic maternal vascular underperfusion associated with IUGR. This provides support for the hypothesis that maternally expressed genes, such as PHLDA2 and cyclin-dependent kinase inhibitor 1C (Cdkn1c) restrict fetal and placental growth to the energetic advantage of the mother, whereas paternally expressed genes, such as MEST and IGF2, augment fetal and placental development at the expense of maternal resources. In the study of IUGR, there was no evidence of altered DNA methylation in imprinting centers of PHLDA2 and MEST, which led McNinn et al. [118] to conclude that ‘the high PHLDA2/MEST mRNA ratios in this subset of IUGR may reflect altered DNA methylation in as yet uncharacterized cis acting regulatory sequences, but more likely reflects conventional transcriptional dysregulation by transacting factors in placental cytotrophoblasts’. Although the mechanisms involved in acute regulation of placental 11 β-hydroxysteroid dehydrogenase-2 (11β HSD2) are not well known, evidence suggests that epigenetic mechanisms modify expression of the gene encoding 11β-HSD2 [119].
Sex differences in the developmental programming of adult disease
The sex differences in the developmental programming of adult disease have been well observed. Epidemiological studies have shown that the association between low birth weight (LBW) and chronic kidney disease is observed in men, but not in women [120,121]. Another similar inverse association is also observed between birth weight and blood pressure in both men and women [122]. Sex-specific differences in response to fetal insults are also seen in experimental studies, with a protective status observed in the females [123]. The male offspring of LP-fed dams underwent an age-dependent loss in glucose tolerance, showing impaired glucose tolerance by 15 months of age and type 2 diabetes mellitus and insulin resistance by 17 months of age [124]; by contrast, the female offspring developed hyperinsulinemia and impaired glucose tolerance at a much later age (21 months) [125]. In response to moderate protein restriction administered during gestation in the rat, a reduction in nephron number associated with hypertension was observed only in male offspring [126]; severe protein restriction was required to induce adverse programming effects in female offspring [127]. Similarly, vascular dysfunction was enhanced only in the male offspring of nutrient-restricted dams [128]. Hypertension programmed in response to placental insufficiency in the rat resulted in hypertension in adult males, but not in females [129]. In sheep, blood pressure was higher only in male offspring after fetal exposure to GCs [130]. Only male offspring exhibited serious cardiovascular dysfunction in response to fetal hypoxia, nicotine and cocaine exposure [7,103,131].
Therefore, sex differences in developmental programming are observed, with female offspring exhibiting a protected status regardless of the species or specific fetal insults. Although only a few studies have begun to elucidate the mechanisms leading to sex differences in the developmental programming of adult disease, the protective phenomena observed in females in response to prenatal insults are likely to involve multiple mechanisms at different developmental stages. The first is the crucial role of sex hormones developed postnatally, which are probably associated with sex-dependent changes. Testosterone has been demonstrated to be a key factor in hypertension programmed in adult male growth-restricted offspring [132]. By contrast, estradiol is implicated to have a protective role against hypertension in adult female growth-restricted offspring [129]. For example, in the offspring of rats, increases in blood pressure induced by a maternal LP diet were influenced by estrogen levels [133]. The mechanism by which sex hormones contribute to sex differences in developmental programming might also involve modulation of some crucial regulatory systems. For example, in the renin angiotensin system (RAS), the expression of angiotensin I converting enzyme 2 (ACE2) mRNA was enhanced in normotensive adult female growth-restricted offspring to protect them from hypertension. Ovariectomy normalizes ACE2 mRNA expression to control levels and induces hypertension in female growth-restricted offspring [129]. Thus, modulation of RAS by estradiol might be one mechanism that contributes to sex differences in the developmental programming of hypertension.
Although it is plausible to suggest a primary role of sex hormones developed postnatally for the sex dichotomy seen in the developmental programming of adult disease, recent studies demonstrate that fetal insults cause sex-dependent changes in gene expression patterns in fetuses and neonates before sexual maturity [105,115]. This suggests that the sex difference in developmental programming occurs earlier during fetal development. Given the fact that male and female fetuses are probably exposed to similar concentrations of steroid hormones in utero, another mechanism is that the sex difference observed might be caused, in part, by the greater expression of estrogen receptor α (ERα) and β (ERβ) in female fetuses. This is indeed the case in the heart [105]. The greater expression levels of both ERα and ERβ in the heart of female fetuses protect the SP1 binding sites at the PRKCE promoter from being methylated by hypoxia and maintain the cardiovascular protective role of PKCε [105]. Furthermore, it has been reported that ERα can cause DNA demethylation [26], which might contribute to the protection of promoter methylation in female fetuses. The sex-specific reprogramming of DNA methylation patterns during gametogenesis and early embryo genesis [35] might also contribute to the sex differences in the fetal epigenetic response to prenatal insults. Additionally, the gene expressions in the placenta are also sex specific. It is known that some genes expressed in the placenta are normally maternally silenced and/or paternally expressed genes that promote growth, whereas others are normally maternally expressed and/or paternally silenced that limit growth, which might also be a potential mechanism that contributes to the sex-dependent development of adult disease caused by prenatal insults. In human pregnancy, the placenta of a female fetus might impart a certain degree of protection from GC excess owing to increased GC inactivation compared with males [134]. Animal studies also showed that sex-specific placental function has a role in the development of anxiety-related behaviors in male offspring resulting from stress during early gestation [135].
Inheritance of developmental programming of adult disease
An increasing number of epidemiological studies indicate that the effects of intrauterine insults can be passed on to subsequent generations, without further exposure of the F1 generation. For example, birth size is reduced in the offspring of women who themselves had a LBW [136]. Similar findings were obtained in the study of the Dutch ‘hunger winter’, in which women who were severely undernourished during the first trimester of pregnancy gave birth to babies who were on average of normal birth weight, but who themselves went on to give birth to smaller babies in the next generation [137]. Owing to the limitations of clinical studies, the evidence for true transgenerational epigenetic inheritance in humans remains to be determined. Animal studies showed that it takes three generations for fetal growth and development to return to normal after 10–12 generations of a LP diet [138]. Offspring of females administered dexamethasone during the last week of pregnancy exhibited LBW and dysregulated glucose homeostasis in adulthood, in association with alterations in hepatic gluconeogenic enzymes [139]. When these F1 animals were mated, their offspring (F2) also had LBW, disrupted glucose homeostasis and elevated liver phosphoenolpyruvate carboxykinase (PEPCK) in the absence of any gestational manipulation [140]. However, the F3 generation is unaffected.
There are several possible explanations for the intergenerational effects induced by prenatal insults. First, the environment of the uterus of undernourished mothers might affect the developing reproductive tract of the fetus. Indeed, mothers who were small at birth have reduced uterine and ovarian size [141]. It is proposed that smaller uterine size might impose a greater ‘maternal constraint’ on the fetus, thereby reducing its growth; although there is not yet any direct evidence for such an effect. Second, the transgenerational effect could be a result of alterations in primordial germ cell formation in the F1 generation. The prenatal insults are likely to affect both somatic and germ cell differentiation, leading to the exposed offspring developing the observed phenotype but also programming gametes and affecting the next generation of offspring. Third, changes in the epigenome during development might be passed on to subsequent generations [142]. For these effects to be inherited, such exposure must induce either stable chromosomal alterations or involve epigenetic modification that is maintained through germ cell maturation [143]. Evidence from rodent studies showed that nutritional and endocrinological interventions in pregnant animals (F0) resulted in phenotypic and/or epigenetic changes that persisted for at least two generations (F1 and F2) [140,144]. Altered methylation patterns of PPARα induced by prenatal nutrition were found in juvenile rats (day 34) and persisted throughout their lifespan (day 80) [145]. Moreover, these methylation patterns of the hepatic PPARα and GR were observed in the F1 and F2 generations [144]. In view of epigenetics as a possible mechanism of genetic imprinting of adult disease, future prenatal and gestational nutritional recommendations might need to be formulated on the basis of region, culture and risk assessment criteria.
The mechanisms by which epigenetic information (whether in terms of environmentally imposed methylation patterns or genomic imprinting) survives the reprogramming that occurs during embryo genesis and gametogenesis remain unresolved. A role for miRNA in spermatozoa has been proposed for the non-mendelian inheritance of one trait in the mouse, but the wider applicability of this mechanism remains uncertain [146]. Nonetheless, the transgenerational effects of prenatal insult are not passed on forever, as they are eventually diminished through several generations [138,140]. The involved mechanisms are not clear but might partially be to the result of the reversibility of epigenetic modification by the environment.
Reversibility of epigenetic changes in the developmental programming of adult disease and therapeutic opportunities
Although epigenetic DNA modifications are typically thought of as being stable once established, pharmacological manipulations of acetylation and methylation patterns have been shown in low- and high-lactation glooming (LG) offspring. Low GR expression in low-LG offspring can be reversed by the central infusion of a HDAC inhibitor, trichostatin A (TSA), which increases acetylation of H3, reduces methylation of the exon 17 region and increases NGFI-A (EGR-1) binding in the adult rat brain [147]. The higher levels of GR in high-LG offspring can be reduced by the administration of the methyl donor S-adenosyl-methionine (SAM), through increased DNA methylation of exon 17 [148]. The combined results of these studies imply that there is a degree of plasticity that remains in the adult for alterations in gene expression by epigenetic modification. Recent laboratory studies have explored the reversibility of induced phenotypic effects and whether aberrant phenotypes induced in utero or during early development can be rescued. Research exploring methods of restoring aberrant phenotypes has led to promising speculation that, ultimately, susceptible patients might be identified by means of screening for epigenetic markers during early life, and that customized interventions might then be instituted. Given that the epigenetic control of key genes is central to the developmental priming of the metabolic and cardiovascular diseases, epigenetics provides a probable target for pharmacological therapies. Several compounds have been identified that are able to regulate DNMT or HDAC activity. For example, the plant-derived isoflavone genistein can reactivate methylation-silenced genes. Similarly, administration of leptin to neonates of undernourished dams has been shown to reverse epigenetic changes and several otherwise primed metabolic sequelae, including weight gain and hyperinsulinemia, and also increased total body adiposity [149]. As a methyl donor, folate supplementation of LP-fed dams prevented the elevation of blood pressure, impaired endothelium-dependent vasodilatation and reduced nitric oxide synthase mRNA levels [150]. Fish oil administered to LP-fed dams minimized the elevation of blood pressure and the reduced microcirculation in the myocardium in the offspring [151]. Omega-3 supplementation of pregnant dams that received dexamethasone prevented intrarenal ACE activation in the offspring [152]. Vitamin D supplementation in infancy is associated with a reduced incidence of preeclampsia [153]. Insufficient vitamin D intake is also incriminated in the short height and reduced pelvic diameter in mothers of LBW babies with increased risk of hypertension [154]. Corrective effects on phenotypic changes, gene expression and associated methylation changes in PPARα have been reported after exogenous leptin administration to the neonatal offspring of undernourished rats [155]. Other studies suggest that hyperleptinemia and hypertension are reversed by dietary intervention with n-3 fatty acids and that altered behavioral responses can be reversed by pharmacological manipulation of epigenetic status [156].
Given that nutritional factors, including deficiency or supplementation of dietary methyl donors, can have a profound impact on pregnancy outcome and offspring health through epigenetic mechanisms, it is important to consider carefully dietary intake of supplements and methyl donors during pregnancy. Adequate intake of folates throughout the life span is associated with a lower risk of cardiovascular disease and certain cancers. However, the long-term effect of folic acid supplementation on susceptibility to chronic diseases has not yet been clearly defined. Therefore, despite the promising prospect of epigenetic therapy, there are still many concerns regarding the clinical approach in humans. First, animal studies inducing global demethylation have indicated that, although the risk of colorectal cancers is reduced, the risk of lymphoma and sarcoma is simultaneously increased [157]. This indicates the potential carcinogenicity and mutagenicity of current epigenetic therapies. In addition, HDAC inhibitors and DNA methylation inhibitors are cytotoxic agents and induce cell-cycle arrest and apoptosis by upregulating the cyclin-dependent kinase inhibitors p21 and/or p53 [158]. Given that current epigenetic therapy compounds produce nonspecific modification of genes and transposable elements, their adverse effects on other gene expression are a major concern. As we have reviewed here, epigenetic modulation of gene expression is a complicated network involving DNA methylation, histone modification and miRNA regulation, and each disorder-related abnormal epigenetic pattern has its own characterization; therefore, finding the most sensitive adjustable epigenetic target is a main challenge for researchers. Although more work is needed to characterize all of these effects and the mechanisms involved, current findings emphasize that epigenetic intervention at crucial periods during development is still an area of potential pharmacological therapeutic benefit.
Concluding remarks
In summary, prenatal insults, including malnutrition, hypoxia, GC exposure, smoking and drug abuse, can not only restrict the growth of fetus, but also cause serious impacts on their health in later postnatal life, such as increased risk of metabolic and cardiovascular disease. Furthermore, owing to the possibility of transgenerational inheritance of the prenatal insult-induced detrimental effects, more than one generation could be affected. Fortunately, the phenomenon of the developmental programming of adult disease has been demonstrated by epidemiological and experimental studies and has attracted extensive attention from sociologists, obstetrician-gynecologists, pediatricians and researchers. In addition to avoiding possible prenatal insults, a timely and valid therapy approach is also important for correcting the existing consequences induced by prenatal insults to improve the later health of offspring. The developing technology of epigenetic manipulation could help to screen for epigenetic biomarkers during early life, and the reversibility of epigenetic action provides a promising window for designing therapy invention. Although current understanding of the epigenetic mechanisms in developmental programming of adult disease is in its infancy, it is one of the most promising research areas for improving reproductive health.
Acknowledgments
This work was supported in part by NIH grants HL82779 (LZ), HL83966 (LZ), HL89012 (LZ), and HD31226 (LZ). We apologize to all authors whose work could not be cited owing to space constraints.
Biographies
Lubo Zhang
Lubo Zhang is Professor of Pharmacology and Physiology at Loma Linda University School of Medicine. He received his PhD in pharmacology from Iowa State University in 1990, and was the President of the US Western Pharmacology Society in 2008. He has been a member of various study sections of the grant review boards for the US National Institutes of Health and American Heart Association for more than 15 years. Dr Zhang is the author or co-author of over 460 scientific articles, book chapters and abstracts. His research interests focus on the epigenetic mechanisms in developmental programming of adult cardiovascular disease.

Ma Chen
Ma Chen received her MD in 2002 and her PhD in pharmacology in 2008 from Wuhan University, China. She is currently a postdoctoral fellow in the Center for Perinatal Biology at Loma Linda University School of Medicine. Her research has focused on the epigenetic mechanisms of developmental programming of adult disease, and she has published several papers in this field. She attended the 15th IUPHAR World Congress of Pharmacology in 2008 and the Society for Gynecologic Investigation 57th Annual Meeting in 2010.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 2.Barker DJ, et al. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341:938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- 3.Jones AP, Friedman MI. Obesity and adipocyte abnormalities in offspring of rats undernourished during pregnancy. Science. 1982;215:1518–1519. doi: 10.1126/science.7063860. [DOI] [PubMed] [Google Scholar]
- 4.Bae S, et al. Effect of maternal chronic hypoxic exposure during gestation on apoptosis in fetal rat heart. Am J Physiol Heart Circ Physiol. 2003;285:H983–H990. doi: 10.1152/ajpheart.00005.2003. [DOI] [PubMed] [Google Scholar]
- 5.Ducsay CA, et al. Long-term hypoxia enhances ACTH response to arginine vasopressin but not corticotropin-releasing hormone in the near-term ovine fetus. Am J Physiol Regul Integr Comp Physiol. 2009;297:R892–R899. doi: 10.1152/ajpregu.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruin JE, et al. Long-term consequences of fetal and neonatal nicotine exposure: a critical review. Toxicol Sci. 2010;116:364–374. doi: 10.1093/toxsci/kfq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer KD, et al. Prenatal cocaine exposure abolished ischemic preconditioning-induced protection in adult male rat hearts: role of PKCepsilon. Am J Physiol Heart Circ Physiol. 2009;296:H1566–H1576. doi: 10.1152/ajpheart.00898.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 9.Gluckman PD, et al. Predictive adaptive responses and human evolution. Trends Ecol Evol. 2005;20:527–533. doi: 10.1016/j.tree.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Dolinoy DC. Epigenetic gene regulation: early environmental exposures. Pharmacogenomics. 2007;8:5–10. doi: 10.2217/14622416.8.1.5. [DOI] [PubMed] [Google Scholar]
- 11.Egger G, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 12.Poulsen P, et al. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- 13.Rosa A, et al. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B. 2008;147:459–462. doi: 10.1002/ajmg.b.30616. [DOI] [PubMed] [Google Scholar]
- 14.Gicquel C, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 15.Gaston V, et al. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2001;9:409–418. doi: 10.1038/sj.ejhg.5200649. [DOI] [PubMed] [Google Scholar]
- 16.Kant SG, et al. Monozygous triplets discordant for transient neonatal diabetes mellitus and for imprinting of the TNDM differentially methylated region. Hum Genet. 2005;117:398–401. doi: 10.1007/s00439-005-1304-1. [DOI] [PubMed] [Google Scholar]
- 17.Katari S, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolinoy DC, et al. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 19.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Dolinoy DC. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 2008;66 (Suppl 1):S7–S11. doi: 10.1111/j.1753-4887.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg AD, et al. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 23.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod Fertil Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 24.Jones L, et al. RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kass SU, et al. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 26.Delcuve GP, et al. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 27.Li E, et al. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 28.Reik W, et al. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 30.Swales AK, Spears N. Genomic imprinting and reproduction. Reproduction. 2005;130:389–399. doi: 10.1530/rep.1.00395. [DOI] [PubMed] [Google Scholar]
- 31.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 32.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 34.Gartler SM, et al. Normal histone modifications on the inactive X chromosome in ICF and Rett syndrome cells: implications for methyl–CpG binding proteins. BMC Biol. 2004;2:21. doi: 10.1186/1741-7007-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 36.Santos F, et al. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 37.Branco MR. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev. 2008;22:1567–1571. doi: 10.1101/gad.1690508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song F, et al. Tissue specific differentially methylated regions (TDMR): changes in DNA methylation during development. Genomics. 2009;93:130–139. doi: 10.1016/j.ygeno.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shechter D, et al. Analysis of histones in Xenopus laevisI A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J Biol Chem. 2009;284:1064–1074. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicklay JJ, et al. Analysis of histones in Xenopus laevis II Mass spectrometry reveals an index of cell type-specific modifications on H3 and H4. J Biol Chem. 2009;284:1075–1085. doi: 10.1074/jbc.M807274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nafee TM, et al. Epigenetic control of fetal gene expression. Br J Obstet Gynecol. 2008;115:158–168. doi: 10.1111/j.1471-0528.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- 42.Allegrucci C, et al. Epigenetics and the germline. Reproduction. 2005;129:137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 43.Kierszenbaum AL. Genomic imprinting and epigenetic reprogramming: unearthing the garden of forking paths. Mol Reprod Dev. 2002;63:269–272. doi: 10.1002/mrd.90011. [DOI] [PubMed] [Google Scholar]
- 44.Waterland RA. Does nutrition during infancy and early childhood contribute to later obesity via metabolic imprinting of epigenetic gene regulatory mechanisms? Nestle Nutr Workshop Ser Pediatr Program. 2005;56:157–171. doi: 10.1159/000086298. discussion 171–154. [DOI] [PubMed] [Google Scholar]
- 45.Quina AS, et al. Chromatin structure and epigenetics. Biochem Pharmacol. 2006;72:1563–1569. doi: 10.1016/j.bcp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Munshi A, et al. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36:75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- 47.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 48.Fischle W, et al. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 49.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 50.Szymanski M, et al. Noncoding RNA transcripts. J Appl Genet. 2003;44:1–19. [PubMed] [Google Scholar]
- 51.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 52.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 53.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 54.Tariq M, et al. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuks F, et al. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 56.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 57.Buchen L. Neuroscience: in their nurture. Nature. 2010;467:146–148. doi: 10.1038/467146a. [DOI] [PubMed] [Google Scholar]
- 58.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Benetti R, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2–dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 61.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 62.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 63.Hayatsu H. The bisulfite genomic sequencing used in the analysis of epigenetic states, a technique in the emerging environmental genotoxicology research. Mutat Res. 2008;659:77–82. doi: 10.1016/j.mrrev.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem. 2009;55:1471–1483. doi: 10.1373/clinchem.2008.121962. [DOI] [PubMed] [Google Scholar]
- 65.Armstrong KM, et al. Global DNA methylation measurement by HPLC using low amounts of DNA. Biotechnol J. 2011;6:113–117. doi: 10.1002/biot.201000267. [DOI] [PubMed] [Google Scholar]
- 66.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 67.Samuelsson JK. DNA fingerprinting techniques for the analysis of genetic and epigenetic alterations in colorectal cancer. Mutat Res. 2010;693:61–76. doi: 10.1016/j.mrfmmm.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey VJ, et al. DNA methylation detection using MS-qFRET, a quantum dot-based nanoassay. Methods. 2010;52:237–241. doi: 10.1016/j.ymeth.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 69.Anisowicz A, et al. A high-throughput and sensitive method to measure global DNA methylation: application in lung cancer. BMC Cancer. 2008;8:222. doi: 10.1186/1471-2407-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gut IG. DNA analysis by MALDI-TOF mass spectrometry. Hum Mutat. 2004;23:437–441. doi: 10.1002/humu.20023. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez BA, Huang TH. Tilling the chromatin landscape: emerging methods for the discovery and profiling of protein–DNA interactions. Biochem Cell Biol. 2005;83:525–534. doi: 10.1139/o05-055. [DOI] [PubMed] [Google Scholar]
- 72.Fanelli M, et al. Pathology tissue-chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc Natl Acad Sci U S A. 2010;107:21535–21540. doi: 10.1073/pnas.1007647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones A, et al. Purification of histone ubiquitin ligases from HeLa cells. Methods. 2011;54:315–325. doi: 10.1016/j.ymeth.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cipriany BR, et al. Single molecule epigenetic analysis in a nanofluidic channel. Anal Chem. 2010;82:2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qavi AJ, et al. Sizing up the future of microRNA analysis. Anal Bioanal Chem. 2010;398:2535–49. doi: 10.1007/s00216-010-4018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ke X, et al. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics. 2006;25:16–28. doi: 10.1152/physiolgenomics.00093.2005. [DOI] [PubMed] [Google Scholar]
- 77.Fu Q, et al. Growth retardation alters the epigenetic characteristics of hepatic dual specificity phosphatase 5. FASEB J. 2006;20:2127–2129. doi: 10.1096/fj.06-6179fje. [DOI] [PubMed] [Google Scholar]
- 78.Lillycrop KA, et al. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lillycrop KA, et al. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 80.Bogdarina I, et al. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Straten EM, et al. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Regul Integr Comp Physiol. 2010;298:R275–282. doi: 10.1152/ajpregu.00413.2009. [DOI] [PubMed] [Google Scholar]
- 82.Hass BS, et al. Effects of caloric restriction in animals on cellular function, oncogene expression, and DNA methylation in vitro. Mutat Res. 1993;295:281–289. doi: 10.1016/0921-8734(93)90026-y. [DOI] [PubMed] [Google Scholar]
- 83.Raychaudhuri N, et al. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park JH, et al. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sinclair KD, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hollingsworth JW, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Gluckman PD, et al. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci U S A. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stevens A, et al. Epigenetic changes in the hypothalamic proopiomelanocortin and glucocorticoid receptor genes in the ovine fetus after periconceptional undernutrition. Endocrinology. 2010;151:3652–3664. doi: 10.1210/en.2010-0094. [DOI] [PubMed] [Google Scholar]
- 89.Aagaard-Tillery KM, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCurdy CE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ng SF, et al. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 92.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tobi EW, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heydeck D, et al. The catecholamine sensitivity of adult rats is enhanced after prenatal hypoxia. Biol Neonate. 1994;66:106–111. doi: 10.1159/000244097. [DOI] [PubMed] [Google Scholar]
- 95.Roigas J, et al. Prenatal hypoxia alters the postnatal development of beta-adrenoceptors in the rat myocardium. Biol Neonate. 1996;69:383–388. doi: 10.1159/000244335. [DOI] [PubMed] [Google Scholar]
- 96.Butler TG, et al. Differential effects of the early and late intrauterine environment on corticotrophic cell development. J Clin Invest. 2002;110:783–791. doi: 10.1172/JCI15563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peyronnet J, et al. Long-lasting adverse effects of prenatal hypoxia on developing autonomic nervous system and cardiovascular parameters in rats. Pflugers Arch. 2002;443:858–865. doi: 10.1007/s00424-001-0766-9. [DOI] [PubMed] [Google Scholar]
- 98.Davis L, et al. Augmentation of coronary conductance in adult sheep made anaemic during fetal life. J Physiol. 2003;547:53–59. doi: 10.1113/jphysiol.2002.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li G, et al. Effect of fetal hypoxia on heart susceptibility to ischemia and reperfusion injury in the adult rat. J Soc Gynecol Investig. 2003;10:265–274. doi: 10.1016/s1071-5576(03)00074-1. [DOI] [PubMed] [Google Scholar]
- 100.Jones RD, et al. Effects of perinatal exposure to hypoxia upon the pulmonary circulation of the adult rat. Physiol Res. 2004;53:11–17. [PubMed] [Google Scholar]
- 101.Mone SM, et al. Effects of environmental exposures on the cardiovascular system: prenatal period through adolescence. Pediatrics. 2004;113:1058–1069. [PubMed] [Google Scholar]
- 102.Zhang L. Prenatal hypoxia and cardiac programming. J Soc Gynecol Investig. 2005;12:2–13. doi: 10.1016/j.jsgi.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 103.Xue Q, Zhang L. Prenatal hypoxia causes a sex-dependent increase in heart susceptibility to ischemia and reperfusion injury in adult male offspring: role of protein kinase C epsilon. J Pharmacol Exp Ther. 2009;330:624–632. doi: 10.1124/jpet.109.153239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu Y, et al. Hypoxia or nutrient restriction during pregnancy in rats leads to progressive cardiac remodeling and impairs postischemic recovery in adult male offspring. FASEB J. 2006;20:1251–1253. doi: 10.1096/fj.05-4917fje. [DOI] [PubMed] [Google Scholar]
- 105.Patterson AJ, et al. Chronic prenatal hypoxia induces epigenetic programming of PKC{varepsilon} gene repression in rat hearts. Circ Res. 2010;107:365–373. doi: 10.1161/CIRCRESAHA.110.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Franco R, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 107.Slotkin TA. Fetal nicotine or cocaine exposure: which one is worse? J Pharmacol Exp Ther. 1998;285:931–945. [PubMed] [Google Scholar]
- 108.Beratis NG, et al. Increased blood pressure in neonates and infants whose mothers smoked during pregnancy. J Pediatr. 1996;128:806–812. doi: 10.1016/s0022-3476(96)70333-5. [DOI] [PubMed] [Google Scholar]
- 109.Blake KV, et al. Maternal cigarette smoking during pregnancy, low birth weight and subsequent blood pressure in early childhood. Early Hum Dev. 2000;57:137–147. doi: 10.1016/s0378-3782(99)00064-x. [DOI] [PubMed] [Google Scholar]
- 110.Lawrence J, et al. Prenatal nicotine exposure increases heart susceptibility to ischemia/reperfusion injury in adult offspring. J Pharmacol Exp Ther. 2008;324:331–341. doi: 10.1124/jpet.107.132175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lawrence J, et al. Foetal nicotine exposure causes PKCepsilon gene repression by promoter methylation in rat hearts. Cardiovasc Res. 2011;89:89–97. doi: 10.1093/cvr/cvq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Giussani DA, et al. Purinergic contribution to circulatory, metabolic, and adrenergic responses to acute hypoxemia in fetal sheep. Am J Physiol Regul Integr Comp Physiol. 2001;280:R678–R685. doi: 10.1152/ajpregu.2001.280.3.R678. [DOI] [PubMed] [Google Scholar]
- 113.Bae S, et al. Prenatal cocaine exposure increases heart susceptibility to ischaemia-reperfusion injury in adult male but not female rats. J Physiol. 2005;565:149–158. doi: 10.1113/jphysiol.2005.082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meyer K, et al. Programming of adult blood pressure by maternal protein restriction: role of nephrogenesis. Kidney Int. 2004;65:1339–1348. doi: 10.1111/j.1523-1755.2004.00511.x. [DOI] [PubMed] [Google Scholar]
- 115.Zhang H, et al. Fetal exposure to cocaine causes programming of Prkce gene repression in the left ventricle of adult rat offspring. Biol Reprod. 2009;80:440–448. doi: 10.1095/biolreprod.108.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Salafia CM, et al. Placental growth patterns affect birth weight for given placental weight. Birth Defects Res A Clin Mol Teratol. 2007;79:281–288. doi: 10.1002/bdra.20345. [DOI] [PubMed] [Google Scholar]
- 117.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 118.McMinn J, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 119.Alikhani-Koopaei R, et al. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J Clin Invest. 2004;114:1146–1157. doi: 10.1172/JCI21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hallan S, et al. Effect of intrauterine growth restriction on kidney function at young adult age: the Nord Trondelag Health (HUNT 2) Study. Am J Kidney Dis. 2008;51:10–20. doi: 10.1053/j.ajkd.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 121.Li S, et al. Low birth weight is associated with chronic kidney disease only in men. Kidney Int. 2008;73:637–642. doi: 10.1038/sj.ki.5002747. [DOI] [PubMed] [Google Scholar]
- 122.Rich-Edwards JW, et al. Longitudinal study of birth weight and adult body mass index in predicting risk of coronary heart disease and stroke in women. Br Med J. 2005;330:1115. doi: 10.1136/bmj.38434.629630.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grigore D, et al. Sex differences in the fetal programming of hypertension. Gend Med. 2008;5 (Suppl A):S121–S132. doi: 10.1016/j.genm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petry CJ, et al. Diabetes in old male offspring of rat dams fed a reduced protein diet. Int J Exp Diabetes Res. 2001;2:139–143. doi: 10.1155/EDR.2001.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fernandez-Twinn DS, et al. Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am J Physiol Regul Integr Comp Physiol. 2005;288:R368–R373. doi: 10.1152/ajpregu.00206.2004. [DOI] [PubMed] [Google Scholar]
- 126.Woods LL, et al. Modest maternal protein restriction fails to program adult hypertension in female rats. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1131–R1136. doi: 10.1152/ajpregu.00037.2003. [DOI] [PubMed] [Google Scholar]
- 127.Woods LL, et al. Programming of adult blood pressure by maternal protein restriction: role of nephrogenesis. Kidney Int. 2004;65:1339–1348. doi: 10.1111/j.1523-1755.2004.00511.x. [DOI] [PubMed] [Google Scholar]
- 128.Ozaki T, et al. Dietary restriction in pregnant rats causes gender-related hypertension and vascular dysfunction in offspring. J Physiol. 2001;530:141–152. doi: 10.1111/j.1469-7793.2001.0141m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ojeda NB, et al. Estrogen protects against increased blood pressure in postpubertal female growth restricted offspring. Hypertension. 2007;50:679–685. doi: 10.1161/HYPERTENSIONAHA.107.091785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dodic M, et al. Programming effects of short prenatal exposure to dexamethasone in sheep. Hypertension. 2002;40:729–734. doi: 10.1161/01.hyp.0000036455.62159.7e. [DOI] [PubMed] [Google Scholar]
- 131.Xiao D, et al. Prenatal gender-related nicotine exposure increases blood pressure response to angiotensin II in adult offspring. Hypertension. 2008;51:1239–1247. doi: 10.1161/HYPERTENSIONAHA.107.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ojeda NB, et al. Testosterone contributes to marked elevations in mean arterial pressure in adult male intrauterine growth restricted offspring. Am J Physiol Regul Integr Comp Physiol. 2007;292:R758–R763. doi: 10.1152/ajpregu.00311.2006. [DOI] [PubMed] [Google Scholar]
- 133.Musha Y, et al. Does estrogen affect the development of abnormal vascular function in offspring of rats fed a low-protein diet in pregnancy? Pediatr Res. 2006;59:784–789. doi: 10.1203/01.pdr.0000219126.78372.c8. [DOI] [PubMed] [Google Scholar]
- 134.Clifton VL, Murphy VE. Maternal asthma as a model for examining fetal sex-specific effects on maternal physiology and placental mechanisms that regulate human fetal growth. Placenta. 2004;25 (Suppl A):S45–S52. doi: 10.1016/j.placenta.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 135.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Klebanoff MA, et al. Low birth weight across generations. JAMA. 1984;252:2423–2427. [PubMed] [Google Scholar]
- 137.Stein AD, Lumey LH. The relationship between maternal and offspring birth weights after maternal prenatal famine exposure: the Dutch Famine Birth Cohort Study. Hum Biol. 2000;72:641–654. [PubMed] [Google Scholar]
- 138.Stewart RJ, et al. The effect of rehabilitation at different stages of development of rats marginally malnourished for ten to twelve generations. Br J Nutr. 1980;43:403–412. doi: 10.1079/bjn19800108. [DOI] [PubMed] [Google Scholar]
- 139.Nyirenda MJ, et al. Glucocorticoid exposure in late gestation permanently programs rat hepatic phosphoenolpyruvate carboxykinase and glucocorticoid receptor expression and causes glucose intolerance in adult offspring. J Clin Invest. 1998;101:2174–2181. doi: 10.1172/JCI1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Drake AJ, et al. Intergenerational consequences of fetal programming by in utero exposure to glucocorticoids in rats. Am J Physiol Regul Integr Comp Physiol. 2005;288:R34–38. doi: 10.1152/ajpregu.00106.2004. [DOI] [PubMed] [Google Scholar]
- 141.Ibanez L, et al. Reduced uterine and ovarian size in adolescent girls born small for gestational age. Pediatr Res. 2000;47:575–577. doi: 10.1203/00006450-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 142.Reik W, et al. Mammalian epigenomics: reprogramming the genome for development and therapy. Theriogenology. 2003;59:21–32. doi: 10.1016/s0093-691x(02)01269-4. [DOI] [PubMed] [Google Scholar]
- 143.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Burdge GC, et al. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2007;97:435–439. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lillycrop KA, et al. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 147.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 148.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vickers MH, et al. The effect of neonatal leptin treatment on postnatal weight gain in male rats is dependent on maternal nutritional status during pregnancy. Endocrinology. 2008;149:1906–1913. doi: 10.1210/en.2007-0981. [DOI] [PubMed] [Google Scholar]
- 150.Torrens C, et al. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- 151.Gregorio BM, et al. Maternal fish oil supplementation benefits programmed offspring from rat dams fed low-protein diet. Am J Obstet Gynecol. 2008;199:82, e81–87. doi: 10.1016/j.ajog.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 152.Wyrwoll CS, et al. Developmental programming of renal glucocorticoid sensitivity and the renin-angiotensin system. Hypertension. 2007;50:579–584. doi: 10.1161/HYPERTENSIONAHA.107.091603. [DOI] [PubMed] [Google Scholar]
- 153.Hypponen E, et al. Does vitamin D supplementation in infancy reduce the risk of pre-eclampsia? Eur J Clin Nutr. 2007;61:1136–1139. doi: 10.1038/sj.ejcn.1602625. [DOI] [PubMed] [Google Scholar]
- 154.Barker DJ, et al. Maternal and social origins of hypertension. Hypertension. 2007;50:565–571. doi: 10.1161/HYPERTENSIONAHA.107.091512. [DOI] [PubMed] [Google Scholar]
- 155.Vickers MH, et al. Neonatal leptin treatment reverses developmental programming. Endocrinology. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- 156.Weaver IC, et al. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Laird PW, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 158.Karpf AR, et al. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]